Abstract
The detailed mechanisms of COVID-19 infection pathology remain poorly understood. To improve our understanding of SARS-CoV-2 pathology, we performed a multi-omics and correlative analysis of an immunologically naïve SARS-CoV-2 clinical cohort from blood plasma of uninfected controls, mild, and severe infections. Consistent with previous observations, severe patient populations showed an elevation of pulmonary surfactant levels. Intriguingly, mild patients showed a statistically significant elevation in the carnosine dipeptidase modifying enzyme (CNDP1). Mild and severe patient populations showed a strong elevation in the metabolite L-cystine (oxidized form of the amino acid cysteine) and enzymes with roles in glutathione metabolism. Neutrophil extracellular traps (NETs) were observed in both mild and severe populations, and NET formation was higher in severe vs. mild samples. Our correlative analysis suggests a potential protective role for CNDP1 in suppressing PSPB release from the pulmonary space whereas NET formation correlates with increased PSPB levels and disease severity. In our discussion we put forward a possible model where NET formation drives pulmonary occlusions and CNDP1 promotes antioxidation, pleiotropic immune responses, and vasodilation by accelerating histamine synthesis.
Keywords: CNDP1, Carnosinase, COVID-19, SARS-CoV-2, NETs, ROS, Neutrophils, Cystine, Glutathione, Carnosine
1. Introduction
To date the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic has reached 587 million cases with 6.5 million deaths. Initial reports of the SARS-CoV-2 novel coronavirus-2019 (nCOV-2019) outbreak indicated the presence of a novel beta-coronavirus from bronchoalveolar lavage fluid causing fever and atypical pneumonia among infected patients [1]. Severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) and Middle Eastern respiratory syndrome coronavirus (MERS-CoV) share similar transmission and symptom characteristics with SARS-CoV-2 but demonstrate considerably higher mortality rates [2]. Despite the potential for SARS-CoV-2 to develop into acute respiratory distress syndrome (ARDS), the rate of asymptomatic infection is estimated to be 30–40% [3,4]. The penetrance of SARS-CoV-2 in the global population, and disparity in host response between individuals, underscores the need to characterize mechanisms and markers associated with immunopathology.
The measurement of biomolecules from blood cells and plasma using mass spectrometry has accelerated our understanding of coronavirus disease −2019 (COVID-19) infections [5,6]. While the sensitivity and reliability of mass spectrometry measurements has increased substantially in the past decade, mass analyzers lack an amplification mechanism similar to nucleic acid deep sequencing approaches that employ the logarithmic polymerase chain reaction. The lower sensitivity of “-omics” approaches employing mass spectrometry can still lead to discovery of prevailing differences between experimental and control populations. This is achievable by imposing appropriate quality control measures during sample preparation and analysis [7]. We hypothesized that this might be the case for SARS-CoV-2 infections where infection severity varies greatly but is largely associated with extreme differences in inflammatory processes including neutrophil influx proximal to the alveolar-capillary barrier [8,9].
In the study described herein, we characterized plasma proteins and metabolites from healthy controls (HC), and from both mild, and severe SARS-CoV-2 infections. All samples from this study originated from individuals infected with SARS-CoV-2 during March of 2020 (prior to vaccination and recurrent infections), thus the likelihood of any established adaptive immunity was low. As a result, our study likely demonstrates the natural course of infection with the ancestral strain of SARS-CoV-2 without the influence of poised adaptive immune responses against the virus.
Prior efforts towards identification of protective mechanisms have focused on adaptive immunity, and clearly showed that antibodies directed against SARS-CoV-2 Spike protein neutralize virus in natural infection and vaccine recipients [[10], [11], [12]]. Few studies have worked to identify innate physiological mechanisms that compensate for stress imposed by the infection. Our results indicate that viral infection triggers antioxidation pathways in mild and severe patient cohorts. Patient populations with severe infection show elevation of antioxidant pathway enzymes including up-regulation of cellular superoxide dismutase, glutathione modifying enzymes, and oxidized glutathione metabolite derivatives. Importantly, carnosine dipeptidase 1 (CNDP1), an enzyme with roles regulating oxidative stress and histamine levels was elevated in the mild patient population, but not HC or those with severe clinical outcomes. These results raise the possibility that one of the features that distinguish severe from mild infections is the ability to suppress a threshold of oxidative stress, vascular occlusions, and neutrophil extracellular trap (NET) formation [13]. NETs function as antiviral and antimicrobial DNA-protein complexes released by diverse infection types including bacterial [14], parasitic [15], and viral infections [5,16,17]. Importantly, NET levels were elevated in severe infection when compared to mild infection and HC populations. However, NET formation was observed in a subpopulation of mild infections, suggesting NETs might be a candidate for post-infection sequelae mechanisms in non-hospitalized patients. Additionally, NET levels were highly correlated with surfactant protein leakage from the pulmonary microcapillaries. These results suggest that the extent of NET complex formation might drive the degree of pulmonary capillary permeability. Conversely, elevated CNDP1 levels were associated with lower pulmonary surfactant levels and reduced NET formation.
2. Results
2.1. Proteomics analysis of patient plasma
We sought to characterize the host response among patients infected with SARS-CoV-2 through plasma proteomics analysis. A total of 151 patient samples were analyzed, including 40 healthy controls (HCs), 77 mild cases (a PCR-positive COVID 19 test but did not require hospitalization), and 34 severe cases that required oxygen, hospitalization, and/or other support (see methods). Prior to tandem-mass-tag (TMT) labeling [18] and mass spectrometry analysis, abundant plasma proteins were immunodepleted to improve sensitivity. Additionally, TMT-labeled peptides were subjected to high pH, reversed-phase chromatography to enhance the depth of protein identification and quantitation (Fig. 1). A total of 2766 proteins were quantified in our study. A comparison of HC to mild infections revealed a statistically significant difference (adjusted p < 0.05) among 209 different proteins with 141 proteins elevated in mild cases and 68 proteins decreased relative to HCs. Comparing severe cases to HCs showed a statistically significant difference in 1377 different proteins (adjusted p < 0.05) with 1129 proteins elevated and 248 proteins decreased in individuals with severe infection. When comparing mild to severe patients, there were 1318 proteins statistically different (adjusted p < 0.05), with an elevation in 1097 proteins and a decrease in 221 proteins in severe relative to mild. All protein mean log2 fold changes between experimental groups and corresponding p values are located in Supplemental Table 1.
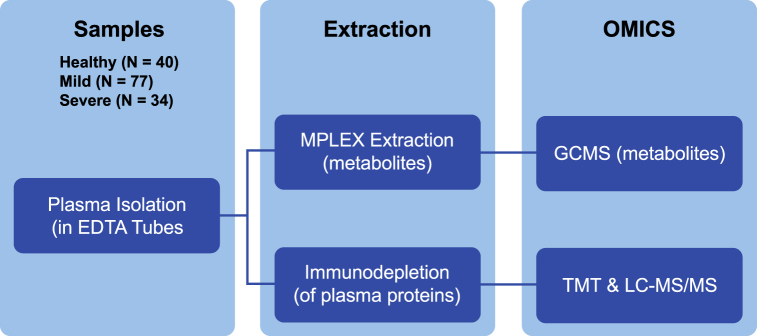
Fig. 1.
Multi-omics experimental design of COVID-19 plasma samples. (Left)- Plasma from healthy controls (N = 40), mild (N = 77), and severe (N = 34) disease patient samples were taken in EDTA-coated tubes. Each individual plasma sample was subjected to MPLEx extraction to isolate metabolites or immunodepletion (to remove abundant plasma proteins). Following extraction metabolites were subjected to GC-MS analysis (as individual, non-pooled samples). Immunodepleted plasma proteins were TMT-labeled (individually) and analyzed by LC-MS/MS.
2.2. Indicators of pulmonary distress and neutrophil activation in severe infections
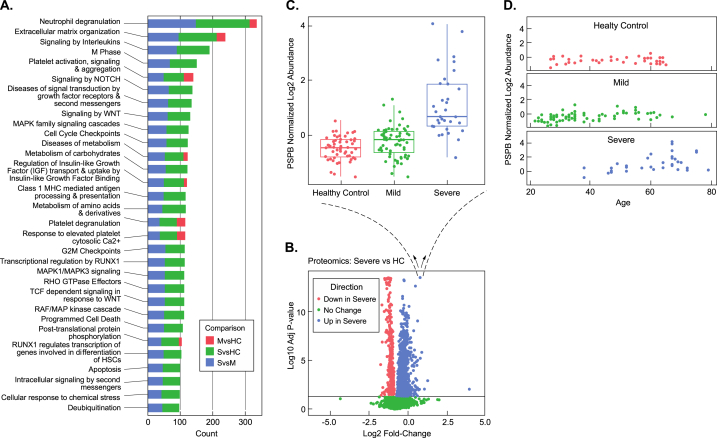
A pathway enrichment analysis of proteins from samples derived from HC, mild, and severe cases strikingly showed the differential expression of proteins that mediate activation of neutrophil degranulation in mild and severe cases relative to HC samples (adjusted p < 0.0001) (Fig. 2A). In addition to differential regulation of neutrophil degranulation pathways, proteins involved in extracellular matrix organization (adjusted p < 1.88E-09) were also elevated in COVID-19 patients relative to HCs. As expected, proteins involved in immune response pathways, platelet activation, and degranulation were also increased in COVID-19 patients, especially in severe cases. Other notable activated pathways included biological oxidation pathways that were significant in the severe cases when compared to HCs (adjusted p = 0.0008) (Supplemental Table 2).
Fig. 2.
Pulmonary surfactant, correlations with severity and age, and pathways activated in severe disease. (A) Pathway analysis of plasma proteins changing between mild (M), severe (S) and healthy controls (HC). (B) Volcano plot analysis of severe vs. healthy controls (HC). (C) Blow-up of individual, statistically-significant datapoint elevated in the severe (S) samples showing a boxplot of the log2 abundance distribution of pulmonary surfactant B protein (PSPB) levels across all patients in the study. (D) Normalized log2 abundance of PSPB vs. Age in healthy Controls (HC)-top, mild (M)-middle, and severe (S) samples-bottom.
Consistent with reported disruption of the alveolar-capillary barrier observed in ARDS patient samples and other COVID-19 plasma proteomics studies [5], we observed a statistically significant elevation in pulmonary surfactant B (PSPB) when comparing samples from severe infections to HC (adjusted p = 3.92E-14) (Fig. 2B&C). PSPB levels from mild patient samples were not statistically significant relative to HC (adjusted p = 0.53). Consistent with previous reports [19], we also observed an increase in Surfactant Protein D in the severe relative to the mild infections (adjusted p = 1.39E-05) and HC samples (adjusted p = 3.07E-05). Among the severe infection patients, PSPB levels increased with age (Fig. 2D) (p = 2E-04). We next evaluated the individual characteristics of the severe patient population mean PSPB z-scores as a function of their respective clinical treatment or outcome (extracorporeal membrane oxygenation (ECMO), intubation, oxygen, and mortality) (Supplemental Fig. 1A). Among the severe group, the mortality and ECMO group had higher levels of PSPB compared to patients receiving intubation and oxygen (Supplemental Fig. 1A).
2.3. Extracellular traps in severe and mild infections
Strong activation of both neutrophil degranulation and oxidation pathways is consistent with the reported physiological environment associated with triggering NET formation in viral infection and during free radical formation in vitro [13]. NETs have been identified in COVID-19 lung tissue, sputum, and sera of infected patients [17,[20], [21], [22], [23], [24]]. Importantly, NETs occlude pulmonary vasculature in COVID-19 patients [25].
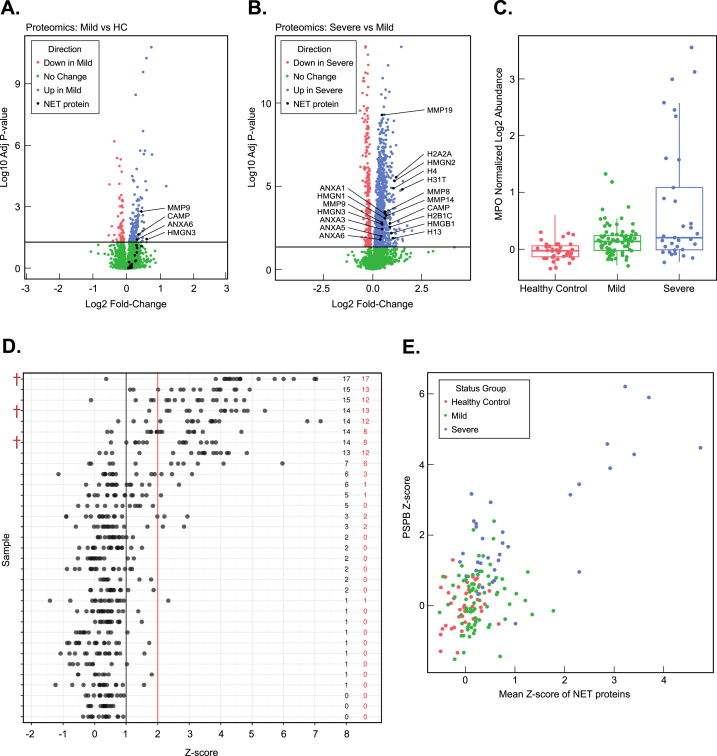
To characterize the proteome of NETs in our COVID-19 patients, we evaluated the mean relative levels and statistical significance of 18 NET protein components identified from previous proteomics screens [26,27] including: DNA-binding proteins (Histones (H13, H4, H31T, H2B1C, and H2A2) and HMGNs (HMGN1, HMGN2, HMGN3, and HMGB1) Neutrophil collagenases (MMP8, MMP19, MMP14, and MMP9)), CAMP (Cathelicidin antimicrobial peptide), and Annexins (ANXA1, ANXA3, ANXA5, and ANXA6) in samples from HC, mild, and severe samples (Supplemental Fig. 2). Only four NET proteins (MMP9 (adjusted p = 0.002), CAMP (adjusted p = 0.02), HMGN3 (adjusted p = 0.04), and ANXA6 (adjusted p = 0.04)) were elevated in the mild patient samples (Fig. 3A) relative to HC. In contrast, all of the 18 NET complex proteins assessed were enriched in the severe relative to the mild patient samples (adjusted p < 0.05) (Fig. 3B) suggesting that NET complexes are more predominant in severe cases and/or are cleared more rapidly in mild cases. Additionally, we analyzed levels of the core NET protein myeloperoxidase (MPO), which showed strong elevation in the severe population relative to HC and mild samples (p = 5.57 E−08 and 3.57 E−06) and no statistically significant difference between mild and HC samples (adjusted p = 0.187) (Fig. 3C). These results are consistent with recent reports of NETs in COVID-19 patient plasma [17,28].
Fig. 3.
Neutrophil Extracellular Traps in mild and severe disease, correlations with mortality and PSPB levels. (A) Volcano plot analysis of mild (blue) vs. HC (red) plasma samples with the positions of (18) different NET protein components labeled in black. Proteins in green have a mean difference between mild and healthy controls (HC) with (p > 0.05) NET protein components with p < 0.05 were labeled. (B) Volcano plot analysis of severe (blue) vs. mild (red) plasma samples with the positions of (18) different NET protein components labeled in black. Proteins in green have a mean difference between mild and severe with (p > 0.05) NET protein components with p < 0.05 were labeled. (C) Boxplot analysis of Myeloperoxidase (MPO) log2 abundance in healthy control, mild, and severe samples. Each data point in the Boxplot is derived from a single patient sample. (D) Analysis of NET protein z-score distributions among severe patient plasma samples. Numbers in red to the right indicate the number of NET proteins from the severe plasma sample with a z-score>2. Numbers to the right (in black) indicate the number of NET proteins from the individual severe plasma sample with a z-score >1. Individual NET protein z-score positions are shown as black dots. Severe patient mortalities are indicated with a red obelisk (†). (E) PSP Z-score plotted vs. mean NET protein z-score for each indivudal patient.
We then investigated if NET proteins were both present and showed a statistically significant difference among individual patient samples. We addressed this question by evaluating the distribution of NET proteins from individual HC, mild, and severe patient samples by calculating z-scores relative to the HC mean abundance levels. NET proteins with a z-score >2 (see methods) were visualized in a string-plot from each patient to evaluate if: 1) Individual severe patient samples contributed more to the mean NET protein levels than others, 2) Multi-protein NET complexes could be observed from individual severe patient samples, 3) Randomly distributed NET proteins are observed across all severe patient samples. Importantly, an evaluation of NET protein z-score distributions showed: 1) That not all severe patients present detectable NET proteins in plasma (Fig. 3D) and 2) A subset of the severe patients have detectable and significantly elevated levels of most of the NET proteins we monitored (Fig. 3D). Importantly, these results suggest that NET formation is isolated to a subset of severe patients and several components of the NET complex are detectable among this subset of the severe patients. Additionally, our results indicate that some mild patients have NET complexes with the number of detectable NET proteins and magnitude of NET protein z-scores considerably lower than that observed in severe cases (Supplemental Fig. 3).
We next evaluated the individual characteristics of the severe patient population mean NET protein z-scores as a function of their respective clinical treatment or outcome (extracorporeal membrane oxygenation (ECMO), intubation, oxygen, and mortality). Patients that were either provided supplemental oxygen or intubated showed a clear trend towards lower mean NET protein z-scores compared to patients that eventually died in the hospital and/or were placed on ECMO treatment, which displayed higher mean NET protein z-scores (Supplemental Fig. 1B). Only three patients died among the 151 patient cohort. Importantly, all three of the patients that died were in the severe group and were among those with the highest mean NET protein z-scores (Fig. 3D). Consistent with age as a risk factor for severe disease [29,30], nine of 33 total severe patients with the highest NET protein z-scores were 7.5 years older (mean value) than that of the 24 severe cases with lower mean NET protein z-scores (p = 0.0124). Collectively, these results strongly suggest that formation of NETs is a marker of disease severity, age, or mortality during infection.
The precise role of NET complexes in severe COVID-19 infections is currently unknown but NETs have been reported in autopsies and suspected of contributing to increased pulmonary capillary permeability and pulmonary occlusions [25]. Given the consistent observation of pulmonary surfactant B protein in COVID-19 patient plasma, we analyzed the relationship between NET formation and plasma surfactant protein levels by plotting the mean NET protein z-score for each patient against their individual PSPB protein z-scores. Mean NET protein z-scores and PSPB levels were highly correlated with r = 0.70 (Fig. 3E).
2.4. Mild infections show elevated plasma levels of carnosine dipeptidase enzyme (CDNP1)
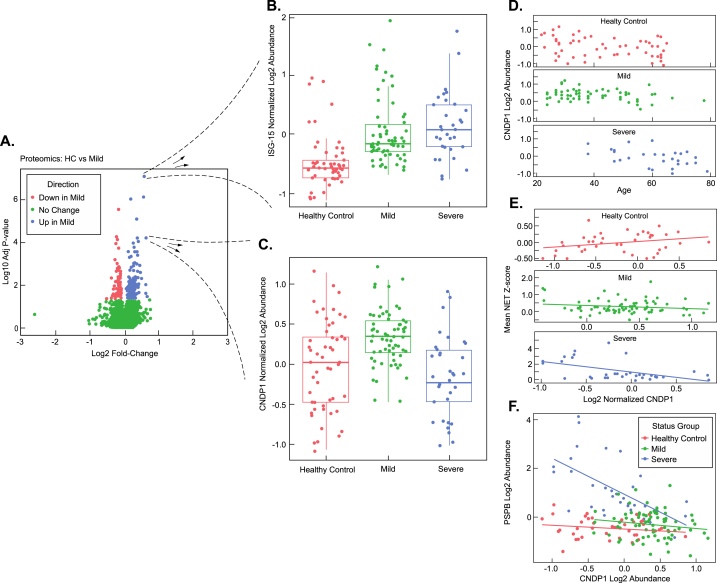
We next examined proteins specifically elevated in mild infections. We sought to determine if mild infections displayed signatures of neutrophil degranulation similar to severe infections. ISG-15, a ubiquitin-like protein, is a major component of neutrophil granules and is secreted in response to virus or IFN alpha-beta [31,32]. Consistent with the neutrophil degranulation pathway activation observed in severe COVID-19 samples (Fig. 2A), we observed that ISG-15 was elevated in mild samples relative to HC (adjusted p = 1.86E-06) (Fig. 4A&B). We then sought to identify candidate proteins among the mild infection samples that might highlight mechanistic differences between mild and severe disease. Analysis of a volcano plot between HCs and mild patient populations clearly showed multiple proteins elevated or decreased during a mild infection (Fig. 4A). The carnosine dipeptidase 1 (CNDP1) was elevated in mild patients relative to HCs (adjusted p = 6.12E-11) but was not statistically significant between HC and severe patient samples (Fig. 4C) (adjusted p = 0.621). This result was particularly interesting, as CNDP1 was among three proteins observed with increased differential expression unique to mild infections when comparing all three sample populations (HC, mild, and severe) among 2766 total quantified proteins and might represent a compensatory pathway specific to mild patient populations. The other two proteins with similar expression patterns (elevated in mild, but similar in HC and severe) were Trem-like transcript protein (TRML2) and Proto-oncogene tyrosine-protein kinase receptor (RET). CNDP1 processes the antioxidant carnosine into alanine and histidine (a substrate for histamine synthesis) and is necessary for survival in drosophila upon oxidative stress challenge [33]. CNDP1 levels were highly correlated with age in the severe group with CNDP1 levels diminished in older patients with severe infections, but not mild infections or HCs (Fig. 4D). We next evaluated the individual characteristics of the severe patient population mean CNDP1 z-scores as a function of their respective clinical treatment or outcome (extracorporeal membrane oxygenation (ECMO), intubation, oxygen, and mortality) (Supplemental Fig. 1C). Among the severe group, the mortality and ECMO group had lower levels of CNDP1 compared to patients receiving intubation and oxygen (Supplemental Fig. 1C).
Fig. 4.
Elevated Carnosine Dipeptidase Enzyme (CDNP1) in mild infections and correlation analysis of CNDP1 to age, NETs, and PSPB levels. (A) Volcano plot analysis of mild (M) vs. Healthy controls (HC) (B) Blow-up of individual, statistically-significant datapoint elevated in mild and severe samples showing a boxplot of the log2 abundance distribution of ISG-15 protein across all patients in the study. (C) Blow-up of individual, statistically-significant datapoint elevated in mild samples, but not severe samples, showing a boxplot of the log2 abundance distribution of the CNDP-1 protein across all patients in the study. (D) Analysis of CNDP1 levels vs. Age in healthy Control, mild, and severe Infections. (E) Analysis of mean NET protein z-score vs. CNDP1 log2 abundance in healthy controls, mild, and severe infections. (F) PSPB log2 abundance vs. CNDP1 log2 abundance for each indivudal patient sample.
While we observed a mean elevation in CDNP1 levels among mild infection samples, we found that some individual severe patients had CNDP1 levels matching or exceeding that of the mean CNDP1 levels in the mild infection group (Fig. 4C). Given this result, we sought to evaluate a potential relationship between NET protein complex formation and CNDP1 levels among the patient sample groups (Fig. 4E). Therefore, we plotted mean NET protein z-scores observed in individual HC, mild, and severe patients against their CNDP1 levels (assuming a linear model) (Fig. 4E). In HC samples, the slope of CNDP1 levels vs. mean NET z-scores was positive (slope = 0.463, p = 0.036) (Fig. 4E). In contrast, in mild and severe patient samples, the slope of CNDP1 levels vs. mean NET z-scores was negative (mild disease slope = −0.137, p = 0.016, severe disease slope = −0.188, p = 0.004) (Fig. 4E). These results suggest that as CNDP1 levels increase, NET complex levels decrease in both mild and severe infections.
We reasoned that if CNDP1 was protective in the patient population with mild infection, then we might observe a negative correlation between lung surfactant protein levels (indicator of more severe disease phenotype) and CNDP1 (expression is potentially protective) in plasma. A comparison between CNDP1 and PSPB levels across all patient samples (HC, mild, and severe) showed a negative correlation (r = −0.335) (Fig. 4F. When CNDP1 and PSPB levels were compared among the severe patient samples (alone) the negative correlation was much stronger with (r = −0.601) Fig. 4F). A comparison of CNDP1 and PSPB levels in HC and mild populations showed no significant correlation (HC r = −0.169, mild r = −0.151) (Fig. 4F).
Interestingly, a second, more promiscuous dipeptidase, cytosolic non-specific dipeptidase 2 (CNDP2, also known as carnosine dipeptidase 2), was also detected in our study and was elevated in the severe patient population relative to HC (adjusted p = 0.001) but was not elevated in mild infections relative to HC (adjusted p = 0.659). CNDP2 and PSPB levels showed a positive correlation (r = 0.339) (Supplemental Fig. 4A).
There was no significant correlation between CNDP1 and CNDP2 levels (r = −0.06) (Supplemental Fig. 4B). We observed a positive correlation between mean NET protein z-scores and CNDP2 levels (r = 0.59) (Supplemental Figure 4C). CNDP1 and CNDP2 share 54% amino acid sequence identity (Supplemental Fig. 5). A manual investigation of peptide fragments quantified and identified from this study showed that the two proteins (CNDP1 and CNDP2) were unambiguously identified (Supplemental Fig. 5). Both CNDP1 and CNDP2 play essential roles in modifying potent antioxidants. These results highlight the intriguing possibility that each of the CNDP dipeptidases might have a different mechanistic role in regulating NET protein complexes.
2.5. L-cystine and anti-oxidant Enzyme Elevation in COVID-19 patient plasma
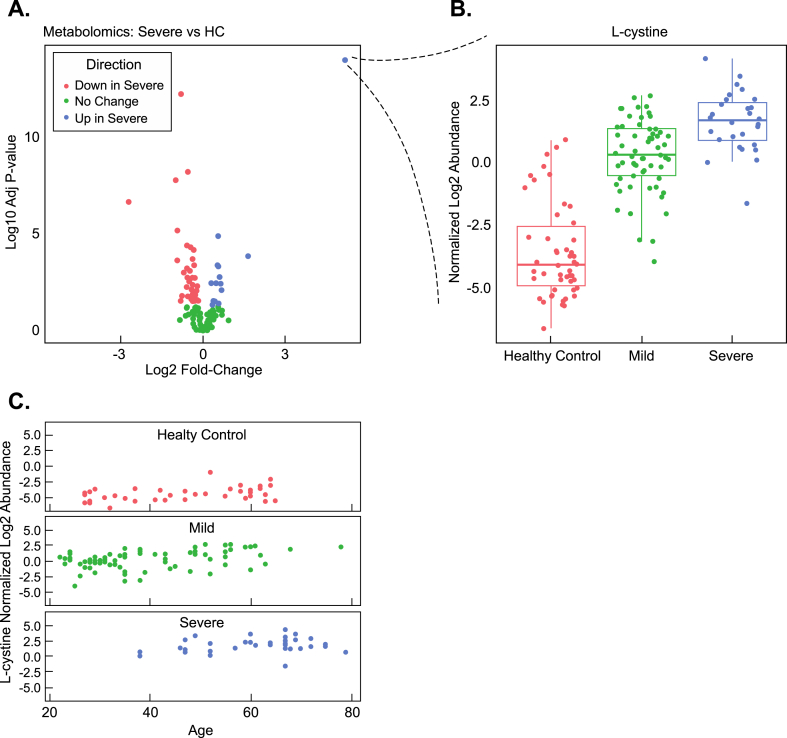
In addition to analyzing protein levels between the previously mentioned patient populations, we quantified metabolite levels using gas chromatography-mass spectrometry (GC-MS)-based metabolomics analysis. In total, we quantified 205 metabolites across all patient samples (Supplemental Table 3). Comparing mild infection to HC showed a statistically significant difference in 22 metabolites (adjusted p < 0.05) with 8 metabolites elevated in mild infection and 14 metabolites reduced. In contrast, a statistically significant difference among 71 total metabolites, with detection of higher quantities of 24 metabolites and decreased levels of 47 metabolites, was identified when severe infection plasma samples were compared to HC (adjusted p < 0.05) (Fig. 5A). Comparing severe vs. mild infections showed a difference in 56 metabolites with an elevation of 18 metabolites in severe relative to mild infection and an elevation of 38 metabolites in mild infection plasma relative to severe (adjusted p < 0.05). A common elevated metabolite observed in both the mild and severe infection patient plasma with a mean fold-change of 23.75 (adjusted p < 1E-15) between mild infection and HC and 60-fold mean increase (adjusted p < 1E-15) in severe infection plasma relative to HC was L-cystine, the disulfide-bonded form of the amino acid cysteine (Fig. 5A and B). Cystine levels were 2.8-fold higher in severe vs. mild infection patient plasma (adjusted p = 5.2E-07). L-cystine levels were associated with age in all severity groups (p = 0.03) (Fig. 5C). l-cysteine (reduced L-cystine) levels were elevated in both mild (p = 0.003) and severe (p = 0.0004) relative to HC (Supplemental Table 3). We next evaluated the individual characteristics of the severe patient population mean L-cystine z-scores as a function of their respective clinical treatment or outcome (extracorporeal membrane oxygenation (ECMO), intubation, oxygen, and mortality) (Supplemental Fig. 1D). Consistent with L-cystine functioning as a general marker of infection, L-cystine levels were elevated among all severe clinical outcome populations (Supplemental Fig. 1D).
Fig. 5.
L-cystine and Anti-oxidant Enzyme Elevation in COVID-19 Patient Plasma and correlative analysis with age. (A) Volcano plot analysis of severe vs. healthy controls (HC) metabolites (B) Blow-up of individual, statistically-significant mean datapoint from the volcano plot showing the boxplot distribution of normalized log2 abundance of L-cystine (disulfide bonded form of cysteine). (C) Normalized log2 L-cystine abundance vs. age in healthy controls (HC), mild, and severe infections.
NET formation is suppressed by antioxidants in vitro and in vivo [34,35]. Glutathione is the cell's most potent antioxidant. L-cystine forms in the extracellular space from reduced glutathione. Upon transport into cells, L-cystine is an eventual substrate for glutathione synthesis. As increased levels of NET proteins were detected in a sub-population of our severe infection patient samples, we worked to evaluate relationships between mean NET protein z-scores observed in the severe samples and L-cystine log2-fold abundance relative to HCs. Interestingly, we observed that mean NET protein z-scores were correlated to L-cystine levels (r = 0.40) (Supplemental Figure 4C).
Reduced glutathione is eventually converted to L-cystine. Hence L-cystine might serve as an indicator of glutathione production and cycling during conditions of oxidative stress. Because we observed NET proteins in the severe infection population and L-cystine elevation among all COVID-19 patient populations, we analyzed the levels of proteins with key roles in the Cystine-Glutathione metabolic cycle and other enzymes with antioxidant activities-including those with roles in superoxide to peroxide conversion. Consistent with a cellular response to oxidative stress, we observed a statistically significant increase in enzymes critical for glutathione synthesis including glutathione S-reductase (GSHR) (mild vs. HC adjusted p = 3.73E-09, severe vs. HC adjusted p = 4.53E-14) and glutamate-cysteine ligase (GSH1) (mild vs. HC adjusted p = 1E-4, severe vs. HC p = 2E-11) (Supplemental Fig. 6). Cellular SOD, which converts superoxide to peroxide, was elevated in severe infection samples relative to HC (Supplemental Fig. 6) (adjusted p = 0.0002). In contrast to cellular SOD (SODC) levels, mitochondrial SOD (SODM) and extracellular SOD (SODE) were unchanged relative to HCs (Supplemental Fig. 6) (adjusted p > 0.05).
Potential Study Limitations: In this study we worked to characterize signatures of pathology from the plasma of patients infected with SARS-CoV-2. In addition, we worked to identify candidate compensatory markers employed by individuals with mild disease. We paired our multi-omics analysis with statistics and correlative analyses. While these studies deliver convincing candidate markers for SARS-CoV-2 infection severity, we note a few potential limitations of our study. First, we have not validated these observations using independent techniques. For example, while our proteomics results are consistent with past studies of NET complexes [26,27], we did not analyze patient samples for MPO-DNA complexes or citrullinated histone levels. Second, our study captures samples from individuals in the acute phase of their infections. As a result, a sub-population of the mild infection group could have progressed to a more severe state or long-term sequelae weeks or months after the original infection. Hence some of the observations we have ascribed to mild or severe infections in this acute phase study will benefit from the clarity provided by future longitudinal studies of SARS-CoV-2 infections. NET formation is clearly associated with disease severity in this study and others. However, it should be noted that age is a strong confounding variable for both severe disease and NET formation as most severe SARS-CoV-2 infections are among older populations.
3. Discussion
Severe COVID-19 patients can develop ARDS which is characterized by hypoxemia and lung stiffness often related to increased permeability of the alveolar-capillary barrier, and an influx of neutrophils into the pulmonary space [1]. Mouse models of ARDS recapitulate similar aspects of severe Coronavirus disease [35,36]. Multiple studies using bacterial-induced ARDS display evidence of increased microcapillary permeability [35,36]. Disruption of the alveolar-capillary barrier likely leads to both infiltration of leukocytes from the capillaries to the pulmonary space and exchange of alveolar contents to the blood. Consistent with this observation, a multi-omics analysis of plasma conducted prior to the study here found pulmonary surfactant B in SARS-CoV-2 patient plasma [5]. In addition, ELISA-based analysis of SARS-CoV-2 patients has shown an increase in pulmonary surfactant D [37,38]. Taken together with results shown in our current study, these studies show that patients with severe SARS-CoV-2-induced ARDS exhibit an increase in surfactant B and surfactant D levels in severe infections. These results suggest that plasma surfactant B and D levels are logical markers of ARDS indicating increased permeability of the alveolar-capillary barrier.
Bacteria-induced mouse models of ARDS show NET formation in the pulmonary capillaries [36]. Similar to these ARDS models, general comparisons of SARS-CoV-2 patient populations vs. HCs have reported detection of NET components including DNA [17], myeloperoxidase (detected by ELISA), and histones (detected by immunofluorescence). In addition, plasma proteomics studies of SARS-CoV-2 and HC populations have reported elevated NET proteins in SARS-CoV-2 patients [5]. Our study demonstrates that the most abundant NETs are found in a sub-population of severe SARS-CoV-2 infections who have the worst clinical outcomes. Importantly, our results also show that individuals with severe ARDS display multiple, detectable NET proteins. Patients with multiple elevated NETs included all fatalities in the study and were older than the severe patients with reduced NET protein levels. Consistent with limited disease, few of the mild infection patients in our study showed elevated NET protein levels compared to HCs. However, a sub-population of the mild infection samples showed NET proteins with elevated z-scores. That mild patient populations showed a clear initiation of NET formation is important and raises critical questions about NET formation during post-infection sequelae, breakthrough infections in vaccinated populations, and infection with SARS-CoV-2 variants of concern.
NETs are found in pulmonary capillary occlusions in mouse models of ARDS [36] and autopsies of coronavirus-induced ARDS [39]. Occlusions and endothelial cell disruption within the pulmonary capillaries are hypothesized as a possible mechanism to explain the increased permeability observed in ARDS [40]. In our study, comparison of mean NET protein z-scores to pulmonary surfactant B levels showed a strong correlation consistent with a direct relationship, suggesting that as NET formation increases, pulmonary capillary permeability increases. These results support the model of NETs driving permeability, potentially through the formation of capillary occlusions. In addition to significant correlations between the NET proteins and PSPB, we identified significant correlations between mean NET z-scores and 389 other proteins (r > 0.6) (Supplemental Table 4). Among the other significant correlations were multiple established NET proteins characterized from other studies. We anticipate that within our correlative analysis to NETs there are other proteins that might stimulate NET formation.
Recent studies from SARS-CoV-2 patient neutrophils show elevated release of reactive oxygen species [41]. Free radicals and oxidative stress pathways trigger NET formation. One of the primary sources of ROS in neutrophils is the NADPH oxidase enzyme [[42], [43], [44], [45], [46]]. We observed a clear correlation between mean NET z-protein scores and NAPDH protein levels in our study (Supplemental Table 4). In addition to activation of oxidation pathways driving NET formation, we observed a substantial elevation in antioxidant factors that might dampen NET formation. Specifically, we found that the glutathione substrate L-cystine was elevated in both the mild and severe infection patient samples. Activated neutrophils import L-cystine to produce glutathione [47]. Importantly, our proteomics experiments showed a complementary elevation of enzymes critical for glutathione metabolism including GSHR (glutathione reductase), GSH1 (Glutamate-cysteine ligase), and CNDP2 (promiscuous dipeptidase) in the severe infection patients, with an elevation of GSHR and GSH1 in the mild infection patients. Taken together, the most logical interpretation of these results is that glutathione synthesis is elevated in the blood plasma of all SARS-COV-2 patient populations and this likely represents a mechanism to suppress over-activation of neutrophils (and NET formation), and other harmful processes triggered by oxidation.
In addition to observing multiple, elevated signatures of glutathione synthesis in our studies, we observed an elevation of the carnosinase CNDP1 in mild infection samples. Unlike CNDP2, CNDP1 lacks a reported activity against Cysteine-Glycine dipeptides in glutathione metabolism and instead, specifically digests the antioxidant dipeptide carnosine (alanine-histidine) [48]. Hence, CNDP1 activity is unlikely to accelerate glutathione synthesis. Instead, CNDP1's narrow metabolic activity targeting carnosine in mild infection patients is likely to drive independent pathways working separately from the multiple enzymes observed driving glutathione synthesis. The substrate of CNDP1 (carnosine) has strong antioxidant activities [[49], [50], [51]]. It is currently unclear how carnosinase activity would have a direct role in reducing oxidation, NET formation, or capillary permeability. We offer a few possibilities based on established functions for CNDP1: First, CNDP1 could accelerate vasodilation through conversion of carnosine to histidine, and subsequent conversion of histidine (by histidine decarboxylase) to histamine [52] to counteract occlusions in the microvasculature (potentially caused by NETs), Second, elevated histamine levels could have pleiotropic effects on immune responses, resulting in both pro and anti-inflammatory reactions, Third, adducted metabolites of carnosine could be cleaved and processed into histidine and alanine metabolites to re-form carnosine through carnosine synthetase for subsequent rounds of antioxidant activity, Fourth, CNDP1 could have additional, unknown biological functions unrelated to carnosinase activity.
The unique upregulation of CNDP1 in mild, but not severe infection patients raises the possibility that CNDP1 has a protective role in patients with mild infections. In support of this possibility, our results clearly show that CNDP1 levels are negatively correlated with PSPB levels and, in both mild and severe infection populations, as CNDP1 levels increase, mean NET protein z-scores decrease. To our knowledge, CNDP1 would be one of the first candidate enzymes to regulate NET formation in the context of infection. Future studies in primary human neutrophils might reveal the influence of CNDP1 in combination with carnosine on NET formation. Drosophila with targeted deletions in CNDP1 display a shortened lifespan and have increased sensitivity to oxidative stress [33]. Importantly, administration of carnosine suppresses NET formation and capillary permeability in a mouse model of ARDS induced by bacteria [35] and suppresses capillary permeability, lung pathology, and neutrophil myeloperoxidase activity in mice infected with H9N2 influenza [53]. Taken together, these studies and ours highlight the possibility of CNDP1 up-regulation as part of a protective mechanism distinct to individuals with mild infections to suppress oxidative stress, vascular occlusions, or NET formation.
4. Methods
Lead Contact: Further requests for resources and reagents should be directed to Vincent R. Gerbasi (Robertvince.gerbasi@pnnl.gov).
4.1. Materials availability
This study did not generate any unique reagents.
4.2. Data and code availability
All mass spectrometry datasets generated during this study have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) under code MSV000089330.
4.3. Experimental model and subject details
Study design and participation were approved by the University of Wisconsin Health Sciences review board under IRB protocol # 2015-0802-CR006. Consent was obtained for all participants prior to enrolment. Plasma samples were collected from (n = 40) HC patients within similar age ranges to patients with mild or severe SARS-CoV-2 infections. Study participants with infections were identified by a positive RT-PCR-based test (developed in-house) for SARS-CoV-2 collected by nasal swab. Mild infections (n = 77) were characterized as patients with a positive test but not requiring supplemental oxygen or other significant support. Severe infections were characterized as those individuals requiring supplemental oxygen, ECMO, intubation, or mortality (n = 34).
5. Methods details
5.1. Analyte extraction
Plasma samples were isolated in EDTA-coated tubes and were subjected to both proteomic analysis preparation including immunodepletion, trypsinization, TMT-labeling, and basic-RP-fractionation and metabolomic analysis preparation using the Metabolite, Protein, Lipid Extraction method, essentially described previously [54,55]. Briefly, plasma samples were prepared and batched in a random order placing National Institute of Standards and Technology (NIST) Standard Reference Material (SRM) for Human Plasma (SRM 1950) (NIST Office of Reference Materials, Gaithersburg, MD) aliquots at the beginning and end of each batch [56]. Plasma samples were thawed in a biosafety cabinet (BSC) and 110 μL of plasma was aliquoted into chloroform compatible microfuge tubes (Sorenson Biosciences, Salt Lake City, UT) along with 190 μL of a metabolite internal standard (in water) prepared as previously described [[57], [58], [59]] containing Leucine-13C6 (4 μg/mL), Creatine-D3 H2O (methyl-D3) (4 μg/mL), L-Leucine-D10 (4 μg/mL), L-Tryptophan-2,3,3-D3 (40 μg/mL), L-Tyrosine Ring-13C6 (4 μg/mL), L-Phenylalanine Ring-13C6 (4 μg/mL), N-BOC-L-tert-Leucine (4 μg/mL), N-BOC-L-Aspartic Acid (4 μg/mL), Propionic Acid 13C3 (8 μg/μL), Succinic Acid-2,2,3,3-d4 (4 μg/mL), Salicylic Acid D6 (4 μg/mL), Caffeine-d3 (1-methyl-d3) (4 μg/mL). 1190 μL of pre-made, cold chloroform/methanol mix (2:0.975 chloroform/methanol (v/v)) was added to each sample and NIST plasma along with 10 μL of SPLASH lipid standard in methanol (AVANTI polar lipids, Birmingham, AL) to make a final ratio of 3:4:8 H2O:methanol:chloroform (v/v). Viruses and other pathogens were inactivated at this point [60]. Samples were vortexed and incubated −20 °C for 10 min and centrifuged at 10 k x g for 10 min to separate the analytes. The upper polar phase containing metabolites was collected into glass vials along with 200 μL of the lower non-polar phase (by carefully breaking through the protein interlayer). The remaining non-polar phase was removed and 500 μL of cold methanol was added to the tube containing the protein. The samples were vortexed and centrifuged again to pellet the protein and the remaining supernatant was added to the metabolites and dried in vacuo and stored at −20 °C until derivatization and GC-MS analysis.
5.2. Metabolomic analysis
The extracted metabolites from MPLEx were chemically derivatized based on the method reported previously [60]. To protect carbonyl groups and reduce the number of tautomeric isomers, methoxyamine (20 μL of a 30 mg/mL stock in anhydrous pyridine) was added to each sample, followed by incubation at 37 °C with shaking for 90 min. To derivatize hydroxyl and amine groups to trimethylsilylated (TMS) forms, N-methyl-N-(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane (80 μL) was added to each vial, followed by incubation at 37 °C with shaking for 30 min. The derivatized samples were analyzed by gas chromatography-mass spectrometry (GC-MS; Agilent 7890A GC with 5975 MSD single quadrupole; Agilent Technologies, Santa Clara, CA). Separation of metabolites were done by HP-5MS GC column (30 m × 0.25 mm × 0.25 μm; Agilent Technologies) and 1 μL of samples was injected in splitless mode. The helium flowrate was adjusted by retention time locking function, and the injection port temperature was held at 250 °C throughout the analysis. The GC oven was held at 60 °C for 1 min after injection, and the temperature was then increased to 325 °C by 10 °C/min, followed by a 10 min hold at 325 °C. Data were collected over the mass range 50–600 m/z. A mixture of FAMEs (C8–C28) was analyzed together with the samples for retention index alignment purposes during subsequent data analysis. GC-MS data files were converted to CDF format and they are deconvoluted and aligned by Metabolite Detector [61]. Identification of metabolites was done by matching with PNNL metabolomics databases – augmented version of Agilent Fiehn metabolomics database [62]. The database contains mass spectra and retention index information of over 1000 authentic chemical standards and they were cross-checked with commercial GC-MS databases such as NIST20 spectral library and Wiley 11th version GC-MS databases [63,64]. Three unique fragmented ions were selected and used to integrate peak area values, and a few metabolites were curated manually, when necessary.
5.3. Plasma protein processing, TMT-labeling, and mass spectrometry
Plasma protein samples were analyzed separately (not pooled). Major plasma proteins from each individual sample were immunodepleted using a Multiple Affinity Removal System (MARS) column (Hu-14 4.6 × 100 mm, Agilent Technologies, Santa Clara, CA) column in-line with an Agilent 1200 HPLC system and analyzed as described previously [65]. Flow-through fractions were collected and concentrated from the depletion column as described previously [55]. Depleted proteins were quantified using the BCA Protein Assay and denatured in 8 M Urea, reduced in DTT, Alkylated in Iodoacetamide, and digested with trypsin. Following trypsinization, peptides were subjected to 16-plex TMT-labeling with an internal control sample and blank in each block of 14 samples analyzed. TMT-labeling reactions were quenched and desalted over C18 cartridges. Samples were fractionated into 96 fractions by basic reverse phase [66] and concatenated into 24 fractions by smart pooling to increase the depth of peptide identification. Fractions were concentrated and frozen prior to LC-MS analysis. Immunodepleted, TMT-labeled peptide sample fractions were randomized and analyzed on a NanoAcuity UPLC system interfaced with a Thermofisher Q-Exactive HFX Orbital trap mass spectrometer (Thermofisher). Separation was carried out in a C18 revered-phase column (70 cm × 75 μm i.d., Phenomenex Jupiter, 3 μm particle size, 300 Å pore size) for 152 min. For detailed gradient information and mass spectrometer settings please refer to previous work [55]. Briefly, precursor ions were analyzed between 300 and 1800 Thomson, at 60 K resolution, an automatic gain control (AGC) of 3E6, and an injection time of 20 ms. The top-12 most abundant precursor ions were fragmented by HCD (NCE setting = 30) with a quadrupole isolation width of 0.7 Thomson, employing 30 K resolution, an AGC target of 1E5 ions. Dynamic exclusion was enabled for 30 s. Xcalibur spectral RAW files were acquired in centroid mode. Protein identifications were made by searching the SwissProt version of Uniprot Human Database (Release November 2019) using MS-GF + combined to mzRefinery to recalibrate the mass spectra [67]. The identification parameters included (a) parent ion mass tolerance of ±6 ppm, (b) at least partial tryptic digestion with 2 missed cleaved sites allowed, (c) static modification of cysteine carbamidomethylation (+57.0215 Da) and N-terminal/lysine TMT labeling (+229.1629 Da), and (d) variable modifications of methionine, cysteine, tyrosine and tryptophan oxidation (+15.9949 Da); cysteine dioxidation (+31.9898 Da); and asparagine, glutamine and arginine deamidation/deamination (+0.98402 Da). Peptide-spectrum matches, peptides and proteins were filtered based on MS-GF probabilities to a false-discovery rate of <1%.
Quantitative information of the TMT reporter ion intensities was extracted using MASIC [68]. The median intensity of each peptide abundance for each sample was scaled to the combined internal standard TMT channel consisting of an equal concentration of plasma peptides from all patient samples as described previously [7]. Quantitation and statistical analysis of TMT results to generate graphs and plots utilized an in-house R-studio package (described below).
Quantification and Statistical Analysis: All data preprocessing was done in R version 4.1.0 with the pmartR package [69]. Proteomics data was normalized to the reference sample by computing the ratio of each sample's peptide abundances to the respective combined internal standard TMT channel calculated within each plex. Peptide-level data and metabolomics data were log2 transformed and filtered to remove peptides and metabolites without enough observations to conduct at least one statistical comparison of interest [70]. One sample from the mild and one sample from the severe infection group was removed from the proteomics dataset and five samples from the metabolomics dataset (three from mild, one HC, one severe) as outliers based on a robust Mahalanobis distance with a p-value threshold of 0.0001 [71] and visual inspection of principal component analysis and correlation with other samples. For the proteomics data, samples were normalized by the median of rank-invariant peptides at a p-value of 0.15, the method selected by the statistical procedure for analysis of proteomics normalization strategies (SPANS) algorithm for determining the optimal normalization method [72]. Data was then quantified to the protein-level using a median reference-based approach [73]. The metabolomics data was normalized using the median of all metabolites for each sample.
Statistical comparisons between the HC, mild, and severe infection groups were conducted for each protein and metabolite by fitting an analysis of variance (ANOVA) model with post-hoc pairwise comparisons, and a Benjamini-Hochberg multiple test correction [74] to adjust ANOVA p-values. Correlation values were calculated using Pearson's correlation. For each NET protein, the z-score was calculated as z-score = (log2 abundance – mean of log2 HC abundances)/(pooled standard deviation) for each sample. The interpretation of the z-score value is the number of standard deviations away from the mean of the HC group. Statistically, any protein or metabolite with a z-score >1 is in the top 16% of values, and a z-score >2 corresponds with approximately the top 2.5% of observations. Pathway enrichment analysis was conducted using the ReactomePA R package [75] and statistically significant proteins for each pairwise comparison (adjusted p < 0.05). Significant pathways were identified and prioritized based on adjusted p-values and associated gene counts. Statistical significance of age differences between the 9-highest severe patient NET protein z-scores and those 24 severe patients with lower NET protein z-scores were evaluated using a non-parametric kruskal-wallis test. All visualizations were generated using the ggplot2 [76] and trelliscopejs R packages.
Credit author statement
Vincent R. Gerbasi, Karen Corson: Conceived and designed the experiments, Analyzed and interpreted the data, Wrote the paper. Lisa M. Bramer, Kelly G. Stratton, Ernesto S. Nakayasu: Conceived and designed the experiments, Analyzed and interpreted the data, Contributed reagents, materials, analysis tools or data, Wrote the paper. Amie Eisfeld: designed the experiments, Performed the experiments Analyzed and interpreted the data, Contributed reagents, materials, analysis tools or data, Wrote the paperCarrie D. Nicora: Performed the experiments, Wrote the paper. Young-Mo Kim, Amy C. Sims: Conceived and designed the experiments, Performed the experiments, Analyzed and interpreted the data, Wrote the paper. Thomas O. Metz, Robert D. Hontz: Conceived and designed the experiments, Analyzed and interpreted the data, Wrote the paper. Marina A. Gritsenko: Conceived and designed the experiments, Performed the experiments, Contributed reagents, materials, analysis tools or data, Wrote the paper. Vanessa L. Paurus: Performed the experiments, Analyzed and interpreted the data. Andrew G. Letizia: Analyzed and interpreted the data, Wrote the paper. Katrina M.Waters: Conceived and designed the experiments, Wrote the paper. Athena A. Schepmoes: Performed the experiments, Contributed reagents, materials, analysis tools or data. Isaac K. Attah: Performed the experiments, Analyzed and interpreted the data. Yoshihiro Kawaoka, Osamu Akasaka, Michiko Koga, Takeya Tsutsumi, Morio Nakamura, Ichiro Nakachi, Rie Baba, Hiroki Tateno, Shoji Suzuki, Hideaki Nakajima, Hideaki Kato, Kazunari Ishida, Makoto Ishii, Yoshifumi Uwamino, Keiko Mitamura, Kiyoko Iwatsuki-Horimoto, Hiroshi Yotsuyanagi: Conceived and designed the experiments, Performed the experiments Analyzed and interpreted the data, Contributed reagents, materials, analysis tools or data, Wrote the paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Timothy J. Garrett at the University of Florida for sharing metabolite standard formulations. We thank Michael Perkins for assistance with generating figures. We thank Eisuke Adachi, Makoto Saito, and Hiroyuki Nagai at Institute of Medical Science of the University of Tokyo, Takayuki Ogura at Saiseikai Utsunomiya Hospital, Naoki Hasegawa, Tomoyasu Nishimura, and Katsunori Masaki at Keio University School of Medicine, Takahide Kikuchi, Daisuke Taniyama, and Kazuto Itoh at Saiseikai Central Hospital, and Shuichi Yoshida and Isano Hase at Saitama City Hospital for assistance with preparing clinical samples. VRG was supported by the Biomedical Resilience and Readiness in Adverse Operating Environment Initiative at PNNL. This work was supported by the National Security Mission Seed, under the Laboratory Directed Research and Development (LDRD) Program at Pacific Northwest National Laboratory (PNNL). PNNL is a multi-program national laboratory operated for the U.S. Department of Energy (DOE) by Battelle Memorial Institute under Contract No. DE-AC05-76RL01830. Sample testing and data analysis was funded by the U.S. Department of Defense's Joint Program Executive Office (JPEO) COVID-19 FY20/21 CARES Act to U.S. Naval Medical Research Unit TWO (NAMRU-2) via Naval Medical Research Center (Appropriation: 9700130, JON: SW713). These funds were transferred via two Military Interagency Purchase Requests (MIPRs) from NAMRU-2 to PNNL (MIPRN6281420MPNX005, MIPRN6281421MPNX003) under a Memorandum of Agreement between the U.S. Departments of Defense and Energy (MOA; USA000707-13-DPAP dtd May 01, 2013).The views expressed in this article reflect the results of research conducted by the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the United States Government. RDH, KC, and AGL are military Service member or employees of the United States Government. This work was prepared as part of their official duties. Title 17, U.S.C., §105 provides that ‘copyright protection under this title is not available for any work of the United States Government.’ Title 17, U.S.C., §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties. The study protocol was approved by the University of Wisconsin Health Sciences Institutional Review Board (IRB protocol # 2015-0802-CR006) in compliance with all applicable federal regulations governing the protection of human subjects.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13795.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Zhu N., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Q., et al. Global percentage of asymptomatic SARS-CoV-2 infections among the tested population and individuals with confirmed COVID-19 diagnosis: a systematic review and meta-analysis. JAMA Netw. Open. 2021;4(12):e2137257. doi: 10.1001/jamanetworkopen.2021.37257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sah P., et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc. Natl. Acad. Sci. U. S. A. 2021;118(34) doi: 10.1073/pnas.2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overmyer K.A., et al. Large-Scale multi-omic analysis of COVID-19 severity. Cell Syst. 2021;12(1):23–40 e7. doi: 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183(6):1479–1495 e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayasu E.S., et al. Tutorial: best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 2021;16(8):3737–3760. doi: 10.1038/s41596-021-00566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nathan C. Neutrophils and COVID-19: nots, NETs, and knots. J. Exp. Med. 2020;217(9) doi: 10.1084/jem.20201439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890 e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoury D.S., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 11.Polack F.P., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595(7867):426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoiber W., et al. The role of reactive oxygen species (ROS) in the formation of extracellular traps (ETs) in humans. Biomolecules. 2015;5(2):702–723. doi: 10.3390/biom5020702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinkmann V., et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 15.Guimaraes-Costa A.B., et al. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc. Natl. Acad. Sci. U. S. A. 2009;106(16):6748–6753. doi: 10.1073/pnas.0900226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisfeld A.J., et al. Multi-platform 'omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe. 2017;22(6):817–829 e8. doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuo Y., et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5(11) doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson A., et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 19.Almuntashiri S., et al. The potential of lung epithelium specific proteins as biomarkers for COVID-19-associated lung injury. Diagnostics. 2021;11(9) doi: 10.3390/diagnostics11091643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masso-Silva J.A., et al. Increased peripheral blood neutrophil activation phenotypes and NETosis in critically ill COVID-19 patients: a case series and review of the literature. Clin. Infect. Dis. 2021;74(3):479–489. doi: 10.1093/cid/ciab437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Middleton E.A., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136(10):1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strich J.R., et al. Fostamatinib inhibits neutrophils extracellular traps induced by COVID-19 patient plasma: a potential therapeutic. J. Infect. Dis. 2021;223(6):981–984. doi: 10.1093/infdis/jiaa789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teluguakula N. Neutrophils set extracellular traps to injure lungs in coronavirus disease 2019. J. Infect. Dis. 2021;223(9):1503–1505. doi: 10.1093/infdis/jiab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veras F.P., et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217(12) doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leppkes M., et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman E.A., et al. Caught in a trap? Proteomic analysis of neutrophil extracellular traps in rheumatoid arthritis and systemic lupus erythematosus. Front. Immunol. 2019;10:423. doi: 10.3389/fimmu.2019.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urban C.F., et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5(10):e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo Y., et al. Autoantibodies stabilize neutrophil extracellular traps in COVID-19. JCI Insight. 2021;6(15) doi: 10.1172/jci.insight.150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neant N., et al. Modeling SARS-CoV-2 viral kinetics and association with mortality in hospitalized patients from the French COVID cohort. Proc. Natl. Acad. Sci. U. S. A. 2021;118(8) doi: 10.1073/pnas.2017962118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Driscoll M., et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 31.Bogunovic D., Boisson-Dupuis S., Casanova J.L. ISG15: leading a double life as a secreted molecule. Exp. Mol. Med. 2013;45:e18. doi: 10.1038/emm.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogunovic D., et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337(6102):1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamakawa-Kobayashi K., et al. Loss of CNDP causes a shorter lifespan and higher sensitivity to oxidative stress in Drosophila melanogaster. Biomed. Res. 2020;41(3):131–138. doi: 10.2220/biomedres.41.131. [DOI] [PubMed] [Google Scholar]
- 34.Kirchner T., et al. vol. 2012. Mediators Inflamm; 2012. (The Impact of Various Reactive Oxygen Species on the Formation of Neutrophil Extracellular Traps). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka K.I., et al. Preventive effects of carnosine on lipopolysaccharide-induced lung injury. Sci. Rep. 2017;7 doi: 10.1038/srep42813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefrancais E., et al. Maladaptive role of neutrophil extracellular traps in pathogen-induced lung injury. JCI Insight. 2018;3(3) doi: 10.1172/jci.insight.98178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alay H., Laloglu E. The role of angiopoietin-2 and surfactant protein-D levels in SARS-CoV-2-related lung injury: a prospective, observational, cohort study. J. Med. Virol. 2021;93(10):6008–6015. doi: 10.1002/jmv.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong M., et al. Serum surfactant protein D in COVID-19 is elevated and correlated with disease severity. BMC Infect. Dis. 2021;21(1):737. doi: 10.1186/s12879-021-06447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schurink B., et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1(7):e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teuwen L.A., et al. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 2020;20(7):389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panda R., et al. A functionally distinct neutrophil landscape in severe COVID-19 reveals opportunities for adjunctive therapies. JCI Insight. 2022;7(2) doi: 10.1172/jci.insight.152291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beavers W.N., Skaar E.P. Neutrophil-generated oxidative stress and protein damage in Staphylococcus aureus. Pathog Dis. 2016;74(6) doi: 10.1093/femspd/ftw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs T.A., et al. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones O.T. The regulation of superoxide production by the NADPH oxidase of neutrophils and other mammalian cells. Bioessays. 1994;16(12):919–923. doi: 10.1002/bies.950161211. [DOI] [PubMed] [Google Scholar]
- 45.Paty P.B., et al. Superoxide production by wound neutrophils. Evidence for increased activity of the NADPH oxidase. Arch. Surg. 1990;125(1):65–69. doi: 10.1001/archsurg.1990.01410130071009. [DOI] [PubMed] [Google Scholar]
- 46.Shaked G., et al. Superoxide production by neutrophils from trauma patients: regulation of NADPH oxidase activity. J. Trauma. 1994;37(1):22–29. doi: 10.1097/00005373-199407000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Sakakura Y., et al. Expression and function of cystine/glutamate transporter in neutrophils. J. Leukoc. Biol. 2007;81(4):974–982. doi: 10.1189/jlb.0606385. [DOI] [PubMed] [Google Scholar]
- 48.Teufel M., et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J. Biol. Chem. 2003;278(8):6521–6531. doi: 10.1074/jbc.M209764200. [DOI] [PubMed] [Google Scholar]
- 49.Kohen R., et al. Antioxidant activity of carnosine, homocarnosine, and anserine present in muscle and brain. Proc. Natl. Acad. Sci. U. S. A. 1988;85(9):3175–3179. doi: 10.1073/pnas.85.9.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mozdzan M., et al. Antioxidant properties of carnosine re-evaluated with oxidizing systems involving iron and copper ions. Basic Clin. Pharmacol. Toxicol. 2005;96(5):352–360. doi: 10.1111/j.1742-7843.2005.pto_03.x. [DOI] [PubMed] [Google Scholar]
- 51.Prokopieva V.D., et al. Use of carnosine for oxidative stress reduction in different pathologies. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/2939087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flancbaum L., et al. The presence and significance of carnosine in histamine-containing tissues of several mammalian species. Agents Actions. 1990;31(3–4):190–196. doi: 10.1007/BF01997607. [DOI] [PubMed] [Google Scholar]
- 53.Xu T., et al. Carnosine markedly ameliorates H9N2 swine influenza virus-induced acute lung injury. J. Gen. Virol. 2015;96(10):2939–2950. doi: 10.1099/jgv.0.000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakayasu E.S., et al. MPLEx: a robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems. 2016;1(3) doi: 10.1128/mSystems.00043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicora C.D., et al. Metabolite, protein, and lipid extraction (MPLEx): a method that simultaneously inactivates Middle East respiratory syndrome coronavirus and allows analysis of multiple host cell components following infection. Methods Mol. Biol. 2020;2099:173–194. doi: 10.1007/978-1-0716-0211-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon-Manso Y., et al. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem. 2013;85(24):11725–11731. doi: 10.1021/ac402503m. [DOI] [PubMed] [Google Scholar]
- 57.Torrez Lamberti M.F., et al. Metabolomic profile of personalized donor human milk. Molecules. 2020;25(24) doi: 10.3390/molecules25245783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yazd H.S., et al. Metabolomic and lipidomic characterization of an X-chromosome deletion disorder in neural progenitor cells by UHPLC-HRMS. J Mass Spectrom Adv Clin Lab. 2021;20:11–24. doi: 10.1016/j.jmsacl.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chamberlain C.A., Hatch M., Garrett T.J. Metabolomic alteration in the mouse distal colonic mucosa after oral gavage with oxalobacter formigenes. Metabolites. 2020;10(10) doi: 10.3390/metabo10100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burnum-Johnson K.E., et al. MPLEx: a method for simultaneous pathogen inactivation and extraction of samples for multi-omics profiling. Analyst. 2017;142(3):442–448. doi: 10.1039/c6an02486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hiller K., et al. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal. Chem. 2009;81(9):3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 62.Kind T., et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao J.H., et al. Genetic and metabolic links between the murine microbiome and memory. Microbiome. 2020;8(1):53. doi: 10.1186/s40168-020-00817-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharon G., et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell. 2019;177(6):1600–1618 e17. doi: 10.1016/j.cell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alcazar O., et al. Parallel multi-omics in high-risk subjects for the identification of integrated biomarker signatures of type 1 diabetes. Biomolecules. 2021;11(3) doi: 10.3390/biom11030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mertins P., et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 2018;13(7):1632–1661. doi: 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gibbons B.C., et al. Correcting systematic bias and instrument measurement drift with mzRefinery. Bioinformatics. 2015;31(23):3838–3840. doi: 10.1093/bioinformatics/btv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monroe M.E., et al. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 2008;32(3):215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stratton K.G., et al. pmartR: quality control and statistics for mass spectrometry-based biological data. J. Proteome Res. 2019;18(3):1418–1425. doi: 10.1021/acs.jproteome.8b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb-Robertson B.J., et al. Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J. Proteome Res. 2010;9(11):5748–5756. doi: 10.1021/pr1005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matzke M.M., et al. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics. 2011;27(20):2866–2872. doi: 10.1093/bioinformatics/btr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Webb-Robertson B.J., et al. A statistical selection strategy for normalization procedures in LC-MS proteomics experiments through dataset-dependent ranking of normalization scaling factors. Proteomics. 2011;11(24):4736–4741. doi: 10.1002/pmic.201100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polpitiya A.D., et al. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24(13):1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Stat. Methodol. 1995;57(1):289–300. [Google Scholar]
- 75.Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12(2):477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 76.Ginestet C. ggplot2: elegant graphics for data analysis. J. Roy. Stat. Soc. Stat. Soc. 2011;174:245. 245. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry datasets generated during this study have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) under code MSV000089330.