Abstract
Purpose
Fungal keratitis (FK) is a serious corneal infection with high morbidity. Host immune responses function as a double-edged sword by eradicating fungal pathogens while also causing corneal damage, dictating the severity, progression, and outcome of FK. However, the underlying immunopathogenesis remains elusive.
Methods
Time-course transcriptome was performed to illustrate the dynamic immune landscape in a mouse model of FK. Integrated bioinformatic analyses included identification of differentially expressed genes, time series clustering, Gene Ontology enrichment, and inference of infiltrating immune cells. Gene expression was verified by quantitative polymerase chain reaction (qPCR), Western blot, or immunohistochemistry.
Results
FK mice exhibited dynamic immune responses with concerted trends with clinical score, transcriptional alteration, and immune cell infiltration score peaking at 3 days post infection (dpi). Disrupted substrate metabolism, broad immune activation, and corneal wound healing occurred sequentially in early, middle, and late stages of FK. Meanwhile, dynamics of infiltrating innate and adaptive immune cells displayed distinct characteristics. Proportions of dendritic cells showed overall decreasing trend with fungal infection, whereas that of macrophages, monocytes, and neutrophils rose sharply in early stage and then gradually decreased as inflammation resolved. Activation of adaptive immune cells was also observed in late stage of infection. Furthermore, shared immune responses and activation of AIM2-, pyrin-, and ZBP1-mediated PANoptosis were revealed across different time points.
Conclusions
Our study profiles the dynamic immune landscape and highlights the crucial roles of PANoptosis in FK pathogenesis. These findings provide novel insights into host responses to fungi and contribute to the development of PANoptosis-targeted therapeutics for patients with FK.
Keywords: fungal keratitis (FK), time-course transcriptome, innate immunity, adaptive immunity, PANoptosis
Fungal keratitis (FK) is a severe sight-threatening corneal infection with high morbidity, which often leads to permanent blindness and eye loss if left untreated.1 It is estimated that there are 1,051,787 new FK cases globally each year, with the highest incidence in Asia and Africa, mainly in developing countries, such as India and China.1 Currently, the antifungal therapies against FK remains inefficient due to drug insensitivity and resistance, with a treatment response rate of only 7.6%.2 In order to reduce the disease burden of FK, it is essential to better understand the pathophysiology of FK for the development of novel antifungal therapies.
The innate immune system is the first line of host defense against fungi, especially macrophages and neutrophils, which are essential for recognition and clearance of fungal stimuli.3,4 Host-pathogen interactions have also been linked to innate immunity-mediated programmed cell death (PCD), which includes pyroptosis, apoptosis, and necroptosis.5 Immune cells and corneal epithelial cells can recognize pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) through multiple pattern recognition receptors (PRRs), leading to the assembly of intracellular inflammasome complexes.6 The inflammasome drives the activation of the inflammatory capspase-1 (CASP1), which further proteolytically processes pro-interleukin (IL)-1β and pro-IL-18 to their active forms (IL-1β and IL-18) and cleaves the executor molecule gasdermin D (GSDMD) into pore-forming amino-terminal domain form (GSDMD-N) triggering pyroptosis.7 Previous studies have shown that pyroptosis acts as a double-edged sword during fungal infection. On one hand, inflammasome sensors, such as NLRP3 and AIM2, play critical roles in eliminating fungal pathogens6,8; on the other hand, hyperinflammation induced by excessive cytokines may lead to tissue damage.9 Apoptosis and necroptosis are two other forms of PCD processes crucial for host defense in addition to pyroptosis. Apoptosis is triggered by activation of apical caspases caspase-8 (CASP8) or caspase-9 (CASP9), which activates executor caspases, such as caspase-3 (CASP3) and caspase-7 (CASP7).10 CASP3, an apoptotic enzyme, can also activate gasdermin E (GSDME), causing lytic cell death.11 During necroptosis, proteins with a receptor interacting protein (RIP) homotypic interaction motif (RHIM), including RIP kinase 1 (RIPK1) and RIP kinase 3 (RIPK3), play crucial roles in the phosphorylation of the mixed lineage kinase domain-like pseudokinase (MLKL), which executes cell death.12
Although the pathways leading to executor activation in these three forms of PCD were previously considered independent, there is mounting evidence that the three pathways have extensive crosstalk, resulting in a unified form of cell death, termed as PANoptosis.13 PANoptosis is defined as an inflammatory PCD pathway activated by specific triggers and regulated by the PANoptosome complex and shares key features of pyroptosis, apoptosis, and necroptosis but cannot be explained by any of these three PCD pathways alone.14 The PANoptosome serves as a molecular scaffold for concurrent engagement of key molecules from PANoptosis. Given that PANoptosis has been reported during infections with various viral, bacterial, and fungal pathogens, as well as in autoinflammatory diseases, cytokine storms, and cancers, it has significant pathophysiological relevance. However, the role of PANoptosis in FK remains elusive.
In this study, we performed time-course transcriptomic analysis of an FK mouse model to investigate the dynamic immune responses during fungal infection. We discovered that fungal infection elicited PANoptosis. Furthermore, we show that PANoptosis is linked to the inflammatory response during FK progression. Our findings reveal the dynamic immune landscape of infected corneas and show that fungal pathogens can induce PANoptosis in an FK mouse model. This study will shed light on the pathophysiology of FK and identify a novel therapeutic target for this disease.
Materials and Methods
Preparation of Aspergillus fumigatus Spore Suspension
The strain of Aspergillus fumigatus (A. fumigatus) was isolated from one patient with FK, which was obtained from the Department of Microbiology, Beijing Institute of Ophthalmology, Beijing Tongren Hospital. The typical A. fumigatus strain was cultured on potato dextrose agar (Jinzhang Technique Development Co., Ltd., Tianjin, China) at 28°C for 4 days. After scraping the conidia from the surface of PDA medium, the fresh spore suspension was harvested by washing in sterile phosphate-buffered saline (PBS). The spore solution was quantified with a cell counting chamber and diluted to a final concentration of 5 × 105 colony forming unit (CFU)/mL in PBS.
Mouse Model of Fungal Keratitis
Twelve 8-week-old female C57BL/6 mice were purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and adaptively fed for 1 week in the specific pathogen-free animal room. All the mice were randomly divided into two groups, the control group and the FK group. Only the right eyes of each mouse were selected for the experiments. Mice in the FK group were anesthetized by intraperitoneal injection of 10% chloral hydrate (2 mL/kg), and proparacaine hydrochloride (0.5%) was applied to the ocular surface 3 times for local corneal anesthesia. Then, a 33-gauge syringe was used to inject 5 µL of A. fumigatus spore solution (5 × l05 CFU/mL) into the corneal stroma until its color became uniform white. Mice in the control group were given 5 µL of PBS. The corneal tissues were collected after the mice were euthanized with an overdose of anesthesia using diethyl ether for the indicated time points. The cornea samples from the FK group at 1, 3, and 5 days post infection (dpi) were labeled as FK1, FK3, and FK5, respectively. All animal studies were performed in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Institutional Animal Care and Use Committee of Beijing Tongren Hospital Affiliated to Capital Medical University (Beijing, China; approval ID: TRLAWEC2022-S003).
Corneal Examination
FK mice were examined daily to monitor the disease progression and photographed with the slit-lamp microscope at 1, 3, 5 dpi. Corneal staining with 2% sodium fluorescein (Feiya Technique Co., Ltd., Jiangsu, China) was used to evaluate the epithelial defect. In vivo confocal microscopy (IVCM) was applied to the diagnosis of FK, which further verified that the mouse model of FK was successfully established. For clinical scores’ assessment, mice were examined at the indicated time to visually grade the disease severity on a scale ranging from 0 to 12.15 The corneal pathology scores were assigned by the following three criteria: area of opacity (0–4), density of opacity (0–4), and surface regularity (0–4). The final corneal clinical score of each mouse was calculated as the sum of the three scores described above.
Histological Analysis
The cornea tissues treated with either A. fumigatus or PBS were collected from the euthanized mice, and fixed in 4% paraformaldehyde solution, and then embedded in paraffin. Sections (5 µm thickness) were cut using a microtome (Leica RM 2165; Leica Microsystems, Nussloch, Germany) and stained with hematoxylin and eosin (H&E) for conventional histological analysis under the light microscope on 40 times objective.
RNA Extraction
Total RNA from cornea tissue samples was extracted with TRIzol reagent (catalog no. 15596018; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. RNA integrity and purity were assessed with RNA 6000 Nano LabChip Kit (catalog no. 5067-1511; Agilent Technologies, Santa Clara, CA, USA) using Agilent Bioanalyzer 2100 (Agilent Technologies) and quantified using Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA). High-quality RNA samples were used to construct sequencing library, which met the following criteria: RNA integrity number >7.0 and 28S/18S ratio >1.8.
cDNA Library Construction and RNA Sequencing
After total RNA was extracted, mRNA was purified from 5 µg total RNA using Dynabeads Oligo (dT) (Thermo Fisher Scientific) with 2 rounds of purification. Subsequently, the mRNA was fragmented into short pieces using divalent cations by the Magnesium RNA Fragmentation Module (catalog no. E6150; New England Biolabs [NEB], Ipswich, MA, USA) under elevated temperature (94°C for 5-7 minutes). Then, the cleaved RNA fragments were reverse-transcribed to create the cDNA library by SuperScript II Reverse Transcriptase (catalog no. 1896649; Invitrogen), which were next used to synthesize U-labeled second-stranded DNAs with E. coli DNA polymerase I (catalog no. M0209; NEB), RNase H (catalog no. M0297; NEB), and dUTP solution (catalog no. R0133; Thermo Fisher Scientific). After end repair of the cDNA fragments, dual-index adapters were ligated to the fragments, and size selection was performed with AMPure XP beads (catalog no. E7420; NEB). After the heat-labile UDG enzyme (catalog no. M0280; NEB), treatment of the U-labeled second-stranded DNAs, the ligated products were amplified with polymerase chain reaction (PCR) by the following conditions: initial denaturation at 95°C for 3 minutes; 8 cycles of denaturation at 98°C for 15 seconds, annealing at 60°C for 15 seconds, and extension at 72°C for 30 seconds; and then final extension at 72°C for 5 minutes. The average insert size for the final cDNA library was 300 ± 50 bp. Paired-end sequencing (PE150 module) was performed on the Illumina Novaseq 6000 platform (LC-Bio Technology CO., Ltd., Hangzhou, China) following the vendor's recommended protocol.
RNA-Seq Reads Mapping and Differentially Expressed Gene Analysis
Raw sequencing reads were filtered by Trimmomatic software to remove low-quality reads containing adapters or low-quality bases.16 Quality-controlled clean sequences were mapped to the Mus musculus reference genome (GRCm38) using the efficient splice aligner HISAT2.17 The HTSeq program was used to estimate the gene expression levels with aligned paired-end reads.18 Gene expression count matrix was deposited and publicly available in the Github website (https://github.com/xxz19900/PANoptosis-FK/). Differentially expressed genes (DEGs) were identified by the R/Bioconductor DESeq2 package with the cutoff threshold of adjusted P < 0.05 and the absolute value of log2 fold change (log2FC) >1.19 The UpSetR package was used to analyze the distribution of all DEGs in each comparison group.20
Relationship Analysis of Tissue Samples in Different Time Points
Principal component analysis (PCA) was performed with the FactoMineR and factoextra packages based on normalized count values to identify the sample clusters and distribution pattern. Correlation analysis was carried out with the cor function using the Spearman method in R to check the association of data among the biological replicates within the same group and across different groups. The correlation coefficient heatmap was visualized using the pheatmap package.
Time Series Clustering of DEGs
The time series soft cluster analysis was conducted by the fuzzy k-means algorithm in the TCseq package to assign genes to clusters according to the gene expression pattern of DEGs.21 Mean gene expression values (normalized counts) for each time point were normalized through logarithmic transformation and standardized with default settings. The optimal number of clusters was determined by evaluating the sum of squared error between clusters using the elbow method along with gap statistics.
Functional Enrichment Analyses
Gene Ontology (GO) enrichment analyses of DEGs were performed using the clusterProfiler package in R software. GO terms for biological processes (BPs) were displayed. Multiple testing corrections were applied using the Benjamini–Hochberg (BH) method. An adjusted P value threshold less than 0.05 was considered statistically significant. Dotplot was used to visualize significantly enriched GO terms.
Immune Cell Infiltration Analysis
The Immune Cell Abundance Identifier for the mouse (ImmuCellAI-mouse) is an online tool (http://bioinfo.life.hust.edu.cn/ImmuCellAI-mouse/#!/) to accurately estimate the abundance of 36 immune cell (sub)types using mouse transcriptome datasets.22 ImmuCellAI-mouse adopted a hierarchical strategy to divide 36 cell types into 3 layers. Layer 1 was composed of 7 major immune cell types: monocyte, macrophage, granulocyte, natural killer (NK) cell, dendritic cell (DC), B cell, and T cell. Second layer cells were mainly subtypes of the first layer immune cells, including macrophage subtypes (M1 and M2 macrophage), granulocyte subtypes (basophil, eosinophil, mast cell, and neutrophil), DC subtypes (conventional DC 1 [cDC1], conventional DC 2 [cDC2], monocyte-derived [MoDC], and plasmacytoid [pDC] cell), B cell subtypes (B1, follicular B, germinal center B, marginal zone B, memory B, and plasma B cell), and T cell subtypes (CD4+ T, CD8+ T, NKT, and γδ T cells). Finally, immune cells in layer 3 were subtypes of CD4+ T and CD8+ T cells, including naïve CD4+ T, CD4+ T memory (Tm), regulatory T cells (Treg), T helper, naïve CD8+ T, cytotoxic CD8+ T (Tc), CD8+ T central memory (Tcm), CD8+ T effector memory (Tem), and exhausted CD8+ T cells (Tex).
Immunohistochemistry
Slides with 5-µm thick corneal sections were deparaffinized in xylene and rehydrated in decreasing concentrations of alcohol. Antigen retrieval was performed by heating the tissues in a microwave oven using appropriate buffer (IRAK1: Tris-EDTA buffer, pH 9.0; NF-κB and IFN-γ: citrate buffer, pH 6.0). Then, 3% hydrogen peroxide was used to quench endogenous peroxidase for 20 minutes, and the samples were blocked with 5% bovine serum in PBS for 1 hour. The slides were incubated at 4°C overnight with the primary antibodies (IRAK1: catalog no. 10478-2-AP, Proteintech, 1:600; NF-κB: catalog no. ab16502, Abcam, 1:1000; IFN-γ: catalog no. bs-0480R, Bioss, Woburn, MA, USA, 1:200). The slides were washed 3 times with PBS for 5 minutes and then incubated with HRP-conjugated goat anti-rabbit IgG (catalog no. ab205718, Abcam, 1:2000) for 30 minutes at room temperature. After washing away the secondary antibodies with PBS, the sections were incubated with the 3, 3′-diaminobenzidine (DAB) kit (catalog no. P0203; Beyotime Biotech Co., Ltd.) for 10 minutes at room temperature. Sections were photographed under light microscope (400 times magnification), and five to eight fields of view per section were selected.
Immunofluorescence Staining
Corneas were enucleated, embedded in Tissue-Tek O.C.T. compound, and snap frozen in liquid nitrogen. Sections of 5 µm thickness were cut and mounted on glass slides coated with Poly-L-lysine, fixed in 4% paraformaldehyde, and blocked with PBS containing 2% BSA for 1 hour at room temperature. Then, sections were incubated with rabbit anti-mouse IFN-γ (catalog no. bs-0480R; Bioss; 1:200), followed by the Alexa Fluor 488-conjugated goat anti-rabbit IgG (catalog no. ab150077; Abcam; 1:500) secondary antibody. 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI; catalog no. 32670; Sigma-Aldrich, Darmstadt, Germany) was used to counterstain the cell nuclei. Slides were washed three times with PBS and visualized by Olympus DP72 fluorescent microscopy.
Real-Time Quantitative PCR
Total RNA was extracted from control and FK mouse corneas using TRIzol reagent (catalog no. 15596018; Thermo Fisher Scientific). Then, qPCR was performed with technical triplicates using Taq Pro Universal SYBR Master Mix (catalog no. Q712; Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer's instructions. The PCR conditions were as follows: 3 minutes at 95°C for pre-denaturation, followed by 40 cycles of 15 seconds at 95°C for annealing, and 1 minute at 60°C for extension. The expression levels of target genes were normalized for each well to the Actb mRNA levels using the 2−△△Ct method as previously described.21 The sequences for qPCR primers were listed in Supplementary Table S1.
Western Blot
Cornea tissue samples were homogenized and lysed with RIPA buffer (catalog no. P0013B; Beyotime Biotech Co., Ltd., Shanghai, China) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and further centrifugated at 14,000 × g for 15 minutes at 4°C. Then, the supernatant was taken, and the total protein concentration was determined by the BCA assay kit (catalog no. P0012; Beyotime Biotech Co., Ltd.). Protein extracts were gently mixed with loading buffer and fully denatured in a boiling water bath. Proteins were separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes (catalog no. IPVH00010; Millipore, Darmstadt, Germany). Afterward, these membranes were blocked with 5% skim milk at 37°C for 2 hours in Tris Buffered Saline with Tween (TBST), then incubated overnight at 4°C with primary antibodies against: GAPDH (catalog no. 60004-1-Ig; Proteintech, Rosemont, IL, USA; 1:200000), GSDMD (catalog no. 39754S; Cell Signaling Technology, Danvers, MA, USA; 1:1000), cleaved caspase-3 (catalog no. 9664T; Cell Signaling Technology; 1:1000), and p-MLKL (catalog no. 37333S; Cell Signaling Technology; 1:1000). Membranes were then washed three times with TBST and incubated with the appropriate horseradish peroxidase (HRP)–conjugated secondary antibodies (Abcam, Cambridge, UK; anti-rabbit [ab205718], 1:50000; Proteintech, anti-mouse [SA00001-1], 1:5000). Finally, protein bands were detected by the ECL chemiluminescent kit (catalog no. P0018S; Beyotime Biotech Co., Ltd.).
Statistical Analysis
All data processing and analyses were performed through R software. For normally distributed variables, the statistical significance was determined by Student's t-test or 1-way analysis of variance (ANOVA) followed by Tukey's HSD post hoc test when appropriate. For non-normally distributed variables, Wilcoxon rank sum test or Kruskal–Wallis was used for evaluating the significance. All figures were generated using the ggplot2 package unless indicated. All statistical P values were 2-tailed, and P < 0.05 or BH-adjusted P < 0.05 was considered statistically significant.
Results
Establishment of the Fungal Keratitis Mouse Model
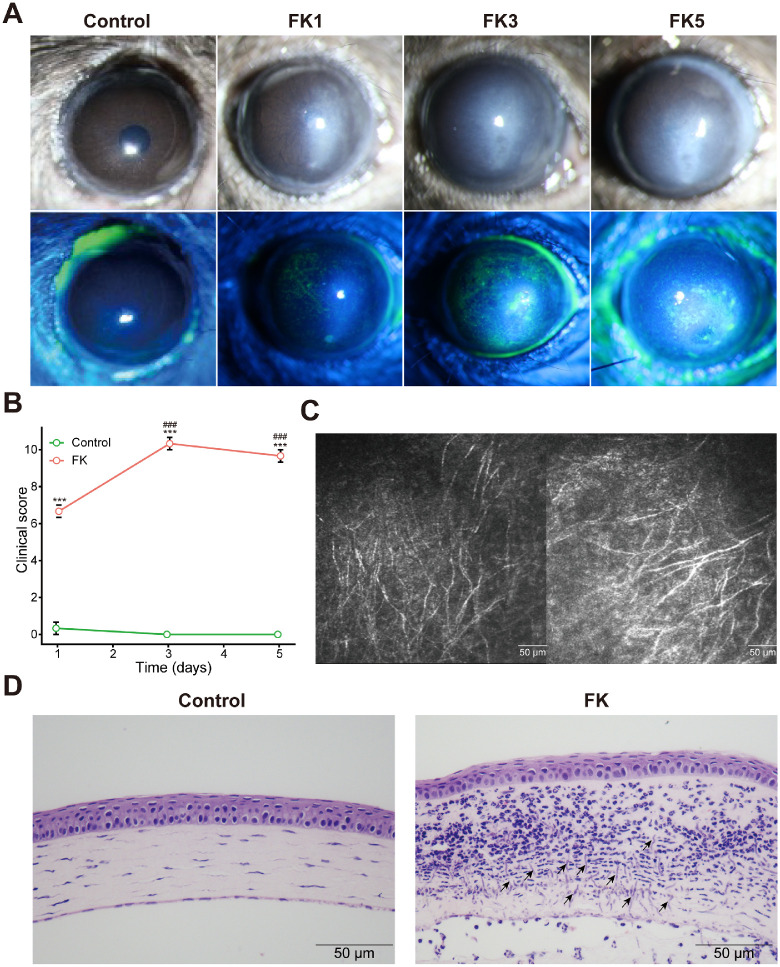
To construct the FK mouse model and observe the disease progression, A. fumigatus spore solution was injected into the corneal stroma, and clinical manifestations of FK were photographed under a slit-lamp (with or without fluorescence staining) at 1, 3, and 5 dpi. For the control group, mouse corneas remained normal in appearance with only minor changes after PBS injection, whereas all FK mice (FK1, FK3, and FK5 groups) showed symptoms of corneal opacity and stromal edema, and displayed the most severe inflammatory response at day 3, characterized by larger and denser gray-white infiltrate, diffuse spotting fluorescein staining, more severe corneal epithelial damage, and visible corneal ulcer (Fig. 1A). Consistently, clinical scores in the FK1, FK3, and FK5 groups were significantly higher than the control group, and culminated at day 3 (Fig. 1B). Furthermore, characteristic fungal hyphae were observed on the IVCM (Fig. 1C) and H&E staining (Fig. 1D), further verifying that the FK mouse model was successfully established.
Figure 1.
Establishment of the fungal keratitis mouse model. (A) Mouse corneas were observed and photographed at 1, 3, and 5 days post infection (dpi). (B) The clinical pathology score was assessed in each group. Two-way ANOVA followed by Tukey's HSD post hoc test was performed to assess differences in clinical score (***, P < 0.001 versus the control group; ###, P < 0.001 versus the FK1 group). (C) IVCM was used to diagnose the FK in mouse model. (D) Representative corneal sections from control and FK model mice were stained with hematoxylin and eosin (H&E, 400 times magnification ) for histological evaluation. Arrows showed the fungal hyphae. FK, fungal keratitis.
Time-Course Transcriptome Analysis Identified the Most Notably Altered Gene Expression Profiles of Mouse Cornea at 3 dpi During Fungal Infection
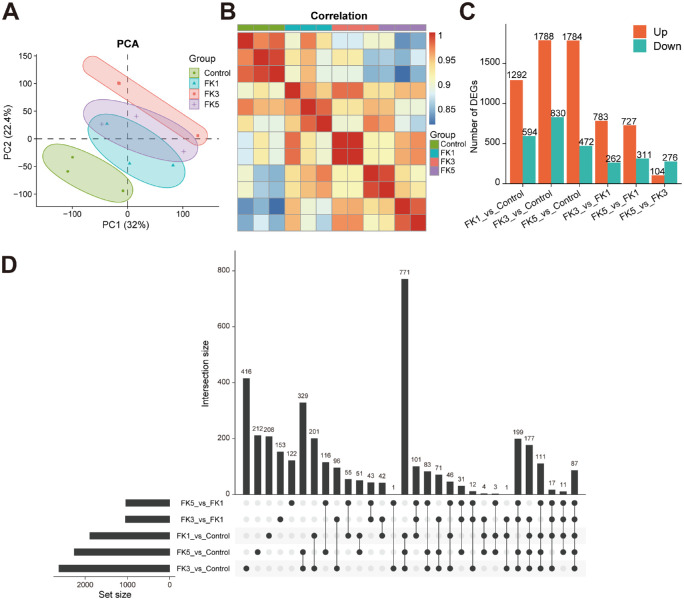
A. fumigatus infected cornea tissues were collected at 1, 3, and 5 dpi for RNA-seq analysis. To assess the host response to A. fumigatus infection, gene expression abundances were determined at each time point. To assess the quality of the transcriptome datasets, PCA was conducted to examine the distribution patterns of samples (Fig. 2A). Scatterplots were generated using the first 2 principal components (PC1 and PC2, explaining 32% and 22.4% of the total expression variation, respectively), which identified 2 well-defined groups across all samples (see Fig. 2A). The expression profiles of the control and A. fumigatus infected groups demonstrated a distinct separation along the PC1 axis, with the control samples clustering at one end of the axis, whereas the FK3 group clustered at the other end of the axis and away from the remaining time points, which was further verified in the Spearman correlation heatmap (Fig. 2B). These results suggest that clear heterogeneity exists among the 4 groups of samples and host gene expression profiles after fungal infection are most notably altered at 3 dpi.
Figure 2.
Comparative transcriptome analysis reveals the expression changes of the infected corneas in the fungal keratitis mouse model. (A) Principal component analysis of gene expression datasets in each group. (B) Spearman correlation analysis of the transcriptome sequencing samples. (C) The number of differentially expressed genes (DEGs) in each comparison group. (D) UpSet plot showing the interaction of DEGs in each comparison group. PCA, principal component analysis; DEGs, differentially expressed genes.
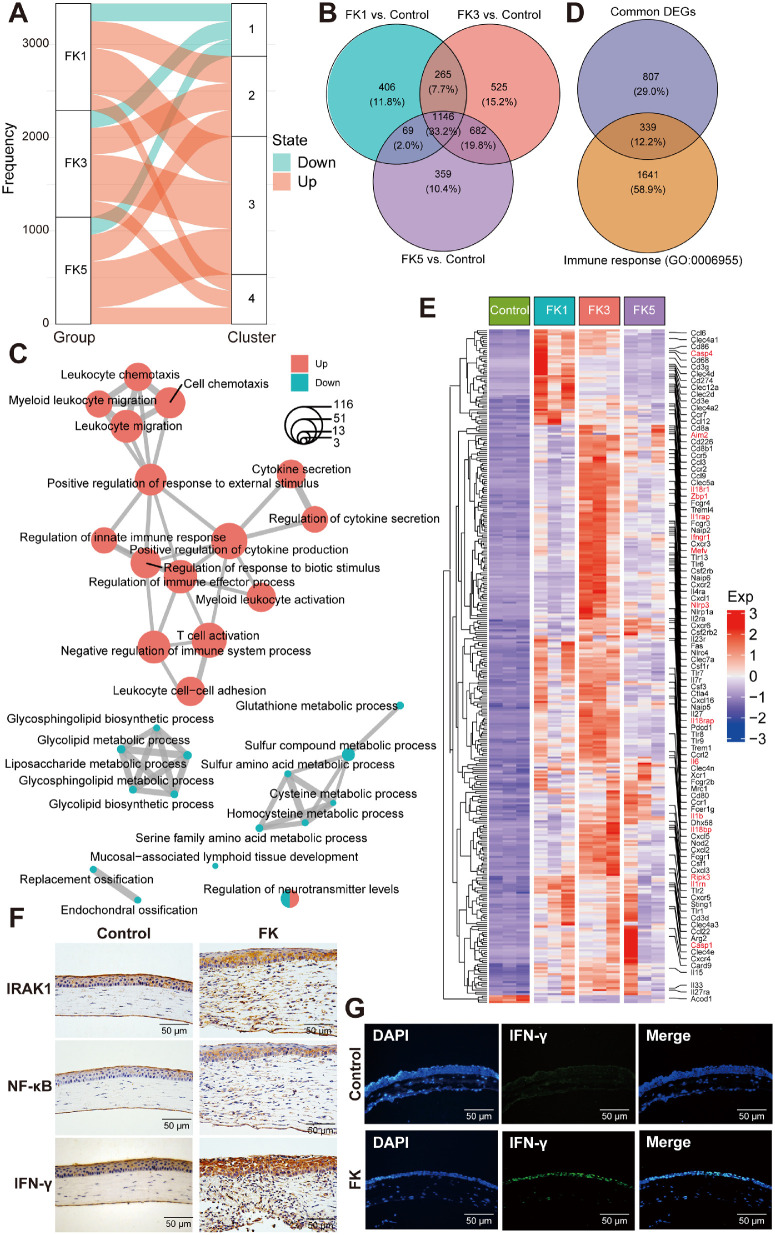
Next, DEGs were identified at different time points through pairwise comparisons among the four groups. Compared with the control group, the number of DEGs in the FK1, FK3, and FK5 groups was 1886 (1292/594; upregulated and downregulated DEGs, respectively), 2618 (1788/830), and 2256 (1784/472), respectively (Fig. 2C, Supplementary Figs. S1, S2, Supplementary Table S2). Moreover, the number of shared DEGs among all comparison groups was visualized in the UpSet plot (Fig. 2D), which displayed a high proportion of common DEGs in pairwise comparisons of the three FK groups with the control group. By contrast, pairwise comparisons within the three FK groups contributed to relatively few DEGs, with the least number of DEGs between FK3 and FK5 among all comparisons (see Fig. 2C), implying highly similar expression profiles between them. These results were consistent with the PCA, both demonstrating the most significant changes in the gene expression profiles associated with fungal infection at 3 dpi.
Gene Expression Profiles Reflected the Dynamic Host Response during Fungal Infection
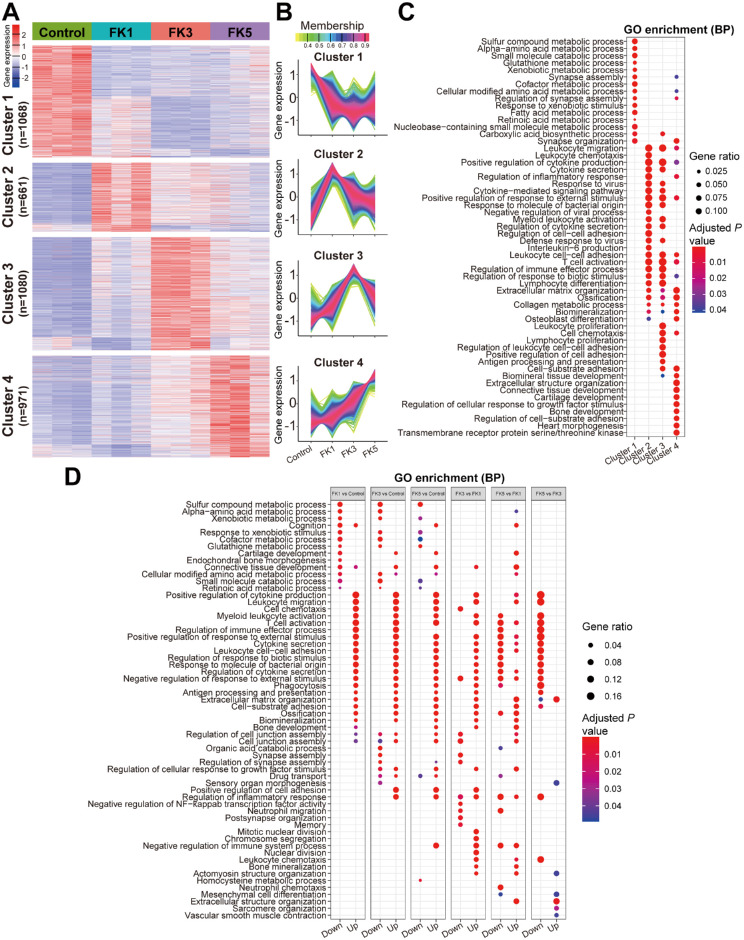
To investigate the global gene expression profiles, fuzzy k-means clustering analysis was performed among the 3780 DEGs identified by pairwise comparisons of all four groups using the TCseq tool. Results showed that they could be classified into four clusters based on the temporal gene expression patterns (Figs. 3A, 3B). The optimal cluster number (k = 4) was determined via gap statistics (Supplementary Fig. S3). Obviously, four clusters could be distinguished as transcriptional profiles with steadily changing (cluster 1 and 4) or transient (cluster 2 and 3) dynamics (see Figs. 3A, 3B). Clusters 1 and 4 (containing 1068 and 971 DEGs) displayed a steadily decreasing or increasing gene expression pattern, and their constituent genes were highly expressed in the control group and the FK5 group, respectively. Cluster 2 was composed of 661 genes, which were highly expressed at 1 dpi and corresponded primarily to the acute inflammation phase. These genes were characterized by transient upregulated gene expression patterns during this phase, whereas later (by day 5) they almost returned to the baseline. Similarly, gene expression levels of the cluster 3 (consisting of 1080 DEGs) were slightly upregulated during the early inflammatory phase and then peaked at 5 dpi.
Figure 3.
Functional enrichment analysis of DEGs in the fungal keratitis mouse model at different time points. (A) Genewise clustering heatmap showing DEGs in each comparison group. (B) Four clusters were divided according to gene expression patterns in each group. The number of DEGs in clusters 1, 2, 3, and 4 were 1068, 661,1080, and 971, respectively. (C) GO enrichment analysis of the DEGs in the four clusters. (D) GO enrichment analysis of the significant upregulated or downregulated DEGs between the indicated groups. GO, gene ontology.
Next, GO enrichment analysis was performed to explore the biological functions of the DEGs of each cluster. It was found that the gene sets in cluster 1 were mainly enriched for biological processes related to substrate metabolism, including sulfur compound, amino acid, xenobiotic, cofactor, fatty acid, retinoic acid, and small molecule metabolism. These genes were coordinately downregulated after fungal infection, suggesting that A. fumigatus infection can significantly interfere with the metabolic processes of the cornea. Cluster 2 and 3 gene sets were mainly enriched for immune-related biological processes involved in leukocyte chemotaxis, migration, and activation, T cell activation, positive regulation of response to external stimulus, response to molecule of bacterial origin, positive regulation of cytokine production, cytokine secretion, cytokine-mediated signaling pathway, and regulation of inflammatory response. In addition, genes in cluster 3 also enriched some GO terms related to adaptive immunity and cell adhesion, such as leukocyte proliferation, antigen processing, and presentation, and positive regulation of cell adhesion. Enriched GO terms in cluster 4 genes were related to tissue development-related terms, including biomineral and connective tissue development, cartilage development and bone development, regulation of cellular response to growth factor stimulus, and extracellular structure organization (Fig. 3C). Given that the gene expression levels of cluster 4 gradually increased during disease progression, this suggests that the FK mouse model is slowly undergoing the corneal wound healing.
Furthermore, GO enrichment analysis was also carried out using the significantly upregulated and downregulated DEGs in each pairwise comparison group. It was also found that significantly downregulated DEGs in the FK1, FK3, and FK5 groups compared to the control group were mainly enriched for metabolism-related GO terms, whereas significantly upregulated DEGs were mainly associated with immune responses (Fig. 3D). These findings were in agreement with the GO enrichment analysis based on the genewise clustering method (see Fig. 3C). In addition, comparative transcriptomic analysis among the three FK groups revealed that the gene expression profiles between the FK3 and FK5 groups exhibited the smallest difference and the least enriched GO terms. Significantly downregulated DEGs in the FK5 versus FK3 comparison group were also mainly enriched for the immune-related GO terms, indicating that inflammatory responses at 5 dpi were significantly reduced compared to 3 dpi, which was consistent with the above findings (see Figs. 2A, 2C).
Immune Cell Abundance Analysis Revealed Dynamic Immune Landscape of Infected Corneas During FK Progression
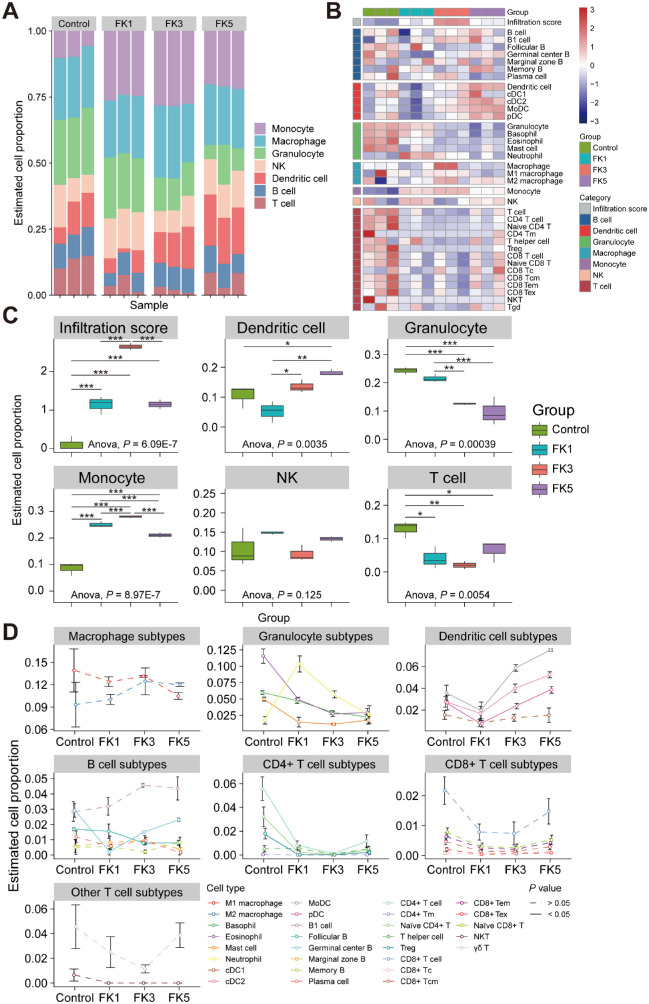
Given that the pathophysiology of FK was closely associated with immune response, we next sought to understand the role of different types of immune cells at different time points during fungal infection. By using ImmuCellAI-mouse, immune cell infiltration analysis was performed by deconvoluting the bulk corneal transcriptomic datasets, which divided the 36 immune cell types into 3 hierarchical layers. The estimated proportions of infiltrating immune cells among samples from different time points were shown in the barplot (Fig. 4A) and heatmap (Fig. 4B), indicating that the composition of immune cells had distinct alteration after fungal infection. One-way ANOVA test was further used to compare the fraction of each immune cell type in layer 1 among the 4 groups, and it was found that the proportion of DC cells showed an overall increasing trend with the development of infection, whereas an overall decreasing trend occurred for granulocytes and T cells (Fig. 4C). Notably, the change trend of monocytes was observed to first increase and then decrease, and peaked at 3 dpi, which was consistent with the overall change trend of the overall immune infiltration score (see Fig. 4C). However, there was no significant difference in the proportions of NK cells among the four groups. Dynamics of the immune cell subtypes at different time points showed that estimated proportions of granulocyte subtypes, including eosinophils, basophils, and mast cells, gradually decreased with the progression of FK, whereas the proportion of neutrophils rose sharply at 1 dpi and then gradually decreased (Fig. 4D). For DC subtypes, the proportions of MoDC, cDC2, and pDC cells gradually increased with fungal infection. Interestingly, the two cell populations in the B cell subtypes exhibited opposite trends, with a progressively increasing proportion of germinal center B cells and decreasing proportion of follicular B cells, suggesting the conversion of follicular B cells to germinal center B cells. However, the proportions of most T cells did not change significantly with the course of fungal infection, although several CD4+ T cell subtypes (naïve CD4+ T and Treg cells) were significantly reduced after infection. Overall, however, the proportions of CD4+ and CD8+ T cell subtypes were slightly elevated, although the differences were not statistically significant (see Fig. 4D). Other immune cell subtypes were not significantly altered with the progression of FK.
Figure 4.
Analysis of infiltrating immune cells in the infected corneas using ImmuCellAI-mouse software. (A) Barplot showing the infiltrating immune cell composition of the cornea tissues. (B) Heatmap showing the immune cells in each group. (C) Comparisons of the immune cell proportions among different groups. One-way ANOVA followed by Tukey's HSD post hoc test was performed to assess differences in the proportion of infiltrating immune cells (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Line charts showing dynamic changes in the abundance of different immune cell subtypes during disease progression. Dashed and solid lines indicated the absence and presence of statistically significant differences (1-way ANOVA test) among four groups, respectively. NK, natural killer; DC, dendritic cell; Treg, regulatory T cells; Tm, memory T cells; Tc, cytotoxic T lymphocytes; Tcm, central memory T cells; Tem, effector memory T cells; Tex, exhausted T cells; ANOVA, analysis of variance.
Shared Immune Responses Across Different Time Points Highlighted the Crucial Role of PANoptosis in FK Pathogenesis
To further identify the key genes involved in the immune response, DEGs at three different time points (each in comparison to the control group) were next intersected to obtain the gene set associated with mouse corneal antifungal immunity. First, the alluvial plot showed that a substantial proportion of DEGs (83.4%, 2867/3438) in the 3 FK groups were significantly upregulated and distributed in clusters 2, 3, and 4, whereas only a few DEGs were downregulated, all located in cluster 1 (Fig. 5A). Next, the Venn diagram showed that 1146 DEGs were shared by all 3 FK groups at different time points (Fig. 5B). Furthermore, GO enrichment analysis of these common DEGs revealed that these upregulated genes were enriched in terms related to immune responses, such as leukocyte migration, positive regulation of cytokine production, regulation of innate immune response, myeloid leukocyte and T cell activation, and leukocyte cell-cell adhesion, whereas the downregulated genes enriched GO terms involving glycolipid, glycosphingolipid, liposaccharide, and sulfur compound metabolic process (Fig. 5C). These findings are in good agreement with the pairwise and groupwise enrichment analyses (see Figs. 3C, 3D). Subsequently, immune response-related genes were focused by intersecting shared DEGs with gene set in GO term “immune response” (GO:0006955), resulting in a total of 339 DEGs (Fig. 5D, Supplementary Table S3). Heatmap was further used to demonstrate the expression patterns of these DEGs in each group and found that the expression levels of a large number genes of PRRs (Toll-like receptors: Tlr1, Tlr2, Tlr6, Tlr7, Tlr8, Tlr9, Tlr13, etc.; NOD-like receptors: Ciita, Naip2, Naip5, Naip6, Nod2, Nlrc4, Nlrp1a, Nlrp3, etc.; C-type lectin receptors: Clec2d, Clec4a1, Clec4a2, Clec4a3, Clec4d, Clec4e, Clec4n, Clec5a, Clec7a, Clec12a, etc.; DNA sensors: Aim2, Sting1, Zbp1, etc.; RNA sensors: Dhx58, etc.), cytokines and their receptors (Csf1:Csf1r, Il1b:Il1rap, Tnfsf8:Tnfrsf8, Tnfsf9:Tnfrsf9, etc.), chemokines and their receptors (Ccl3:Ccr1, Ccl12:Ccr2, Ccl3:Ccr5, Cxcl1:Cxcr2, Cxcl16:Cxcr6, etc.), cell adhesion molecules (Cd80/Cd86:Ctla4, Cd274:Pdcd1, Icam1:Itgal, etc.), FcγR-mediated phagocytosis (Fcgr1, Fcgr2b, Fcgr3, Fcgr4, Ptprc, etc.), and antifungal immunity genes (Card9, etc.) increased significantly after fungal infection (Fig. 5E). Additionally, several classical inflammatory markers (IRAK1, NF-κB, and IFN-γ) were selected to assess their tissue distribution using immunohistochemistry, and a large number of IRAK1-, NF-κB-, and IFN-γ-positive cells were observed in the corneal epithelium (Fig. 5F), which was also verified for the expression distribution of IFN-γ using immunofluorescence staining (Fig. 5G). Accordingly, the expression levels of many interferon-stimulated genes (ISGs) were significantly elevated, such as Ifi203, Ifi204, Ifi205, Ifi206, Ifi207, Ifi211, Ifi213, Ifi44, Ifih1, Ifit1, Ifit3, Ifitm3, Ifitm6, and Isg15. Interestingly, increased expression of several PANoptosis-related genes was also observed, including Zbp1, Aim2, Mefv, Casp1, Ripk3, Il1b, etc. (marked in red in Fig. 5E). These results highlighted the essential roles of ISGs and PANoptosis during fungal infection.
Figure 5.
Shared innate immune responses among different FK groups. (A) Alluvial plots showing the distribution of upregulated and downregulated DEGs across different clusters. (B) Venn diagram of DEGs among the three FK comparison groups. (C) Enrichmap plot showing the interaction networks between enriched biological processes analyzed by the clusterProfiler package. The color of the dots represented the state of DEGs being upregulated or downregulated, and the size of the dot indicated the number of DEGs enriched in the corresponding GO terms. (D) Venn diagram showing the total number of shared DEGs between common DEGs within three FK groups and genes in the GO term “immune response” (GO: 0006955). (E) Heatmap displaying expression patterns of the 339 shared immune-related DEGs among the four groups. (F) Immunohistochemistry staining of several representative immune markers (IRAK1, NF-κB, and IFN-γ) in mouse corneas with or without fungal infection. (G) Representative immunofluorescence images showing the increased expression levels of IFN-γ in corneal epithelial cells after A. fumigatus infection. IRAK1, interleukin-1 receptor-associated kinase 1; NF-κB, nuclear factor-κB; IFN-γ, interferon-gamma.
Experimental Validation of PANoptosis
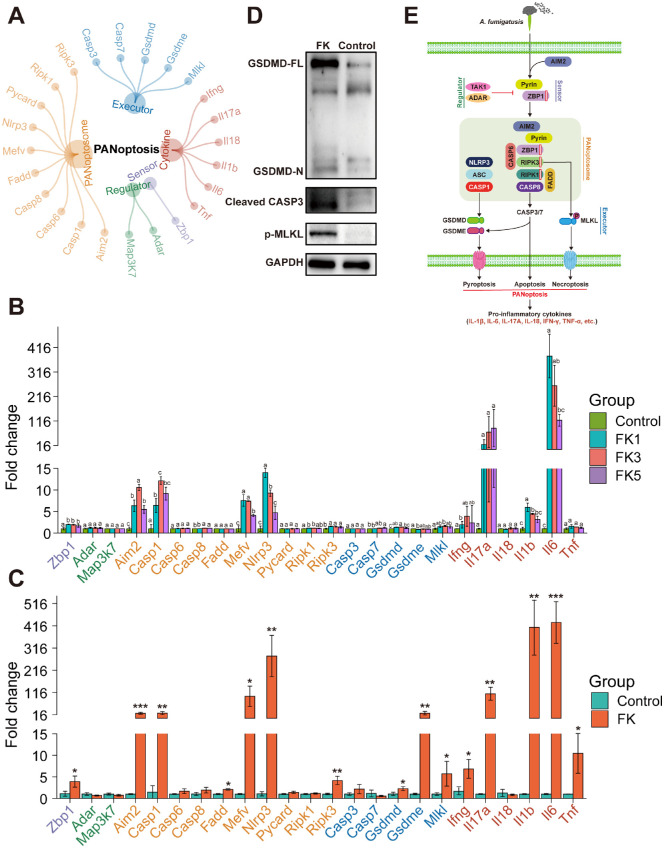
To explore whether PANoptosis was triggered in FK mice, the expression levels of several PANoptosis-related genes were further measured at both the mRNA and protein levels. First, a manually curated gene set of PANoptosis was constructed, consisting of 1 sensor, 2 regulators, 10 PANoptosome components, 5 executors, and 6 cytokines (Fig. 6A). RNA-seq analysis showed that the expression levels of Zbp1, Aim2, Casp1, Casp8, Mefv, Nlrp3, Ripk1, Ripk3, Casp7, Gsdmd, Mlkl, Ifng, Il1b, and Il6 were significantly upregulated (P < 0.05, 1-way ANOVA test), implying the activation of PANoptosis (Fig. 6B). Moreover, the expression levels of 24 PANoptosis-related genes were validated by qPCR and 15 genes were confirmed to be significantly increased (P < 0.05, Student's t-test), including Zbp1, Aim2, Casp1, Fadd, Mefv, Nlrp3, Ripk3, Gsdmd, Gsdme, Mlkl, Ifng, Il17a, Il1b, Il6, and Tnf (Fig. 6C). Both RNA-seq and qPCR detected the most prominent expression changes of Il17a (110-fold increase) and Il6 (431-fold), resulting in excessive inflammation. Western blot analysis of PANoptosis activation markers showed that pyroptosis (cleaved GSDMD), apoptosis (cleaved CASP3), and necroptosis (phosphorylated MLKL) were triggered after A. fumigatus infection in mouse corneas (Fig. 6D). These findings suggested that AIM2, pyrin, and ZBP1 can form a PANoptosome complex along with RIPK1, RIPK3, CASP6, CASP8, FADD, NLRP3, ASC, and CASP1 to sense fungal pathogens and induce PANoptosis, similar to what has been observed during bacterial and viral infections (Fig. 6E).
Figure 6.
Activation of AIM2-, pyrin-, and ZBP1-mediated PANoptosis by A. fumigatus infection. (A) The PANoptosis gene set contained 24 genes, including 1 sensor, 2 regulators, 10 PANoptosome components, 5 executors, and 6 cytokines. (B) Dynamic changes in the expression of PANoptosis-related genes across the four groups analyzed by RNA-seq datasets. Relative gene expression levels (mean fold change ± SD, n = 3) were compared to the control group (set to 1) at 1, 3, and 5 dpi. One-way ANOVA followed by Tukey's HSD post hoc test was performed to assess differences in the expression levels of PANoptosis-related genes. Different letters indicated significantly different groups (P < 0.05). (C) The qPCR validation of PANoptosis-related genes in the mouse corneas. Statistical analysis was performed using unpaired two-sided Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (D) Western blot analysis of executors involved in the PANoptosis. (E) Schematic representation of the activation of PANoptosis during A. fumigatus infection. ZBP1, Z-DNA-binding protein 1; ADAR, adenosine deaminase RNA-specific binding protein; TAK1, transforming growth factor β-activated kinase 1, encoded by the Map3k7 gene in mice; AIM2, absent in melanoma 2; CASP1/3/6/7/8, caspase 1/3/6/7/8; FADD, Fas-associated protein with death domain; pyrin, encoded by the Mefv gene in mice; ASC, apoptosis-associated speck-like protein with CARD domain, encoded by the Pycard gene in mice; NLRP3, NLR family pyrin domain containing 3; RIPK1/3, receptor-interacting protein kinase 1/3; GSDMD/E, gasdermin D/E; MLKL, mixed lineage kinase domain-like protein; IL-1β/6/17A/18, interleukin 1β/6/17A/18; TNF-α, tumor necrosis factor α.
Discussion
The host immune response against fungal pathogens not only plays an antifungal role, but excessive inflammation will cause concurrent organ and tissue damage, especially in the corneas of patients with FK, which determines the severity, progression, and outcome of the disease.4,23,24 Previous studies have reported the characteristics of innate and adaptive immune responses, which contributes to our understanding of the underlying pathogenesis of FK.25–29 However, it is difficult to obtain an integrated immune landscape at both the molecular and cellular levels upon fungal infection of the cornea. To address this issue, here, we successfully established a mouse model with typical symptoms of FK caused by A. fumigatus and used time-course transcriptome analysis to profile the immune responses in the infected mouse corneas at different time points. Our study not only illustrated the dynamics of infiltrating immune cells during disease progression, but also revealed the critical factors responsible for antifungal immunity, highlighting the crucial role of PANoptosis in the pathogenesis of FK.
In this study, a comprehensive characterization of the dynamic immunological hallmarks of FK mice across different time points was presented. First, the FK mice showed an overall acute inflammatory response, along with a concerted change trend in the clinical score, transcriptional alteration, and immune cell infiltration score, which peaked at 3 dpi. Second, the progression of fungal infection was accompanied by disrupted substrate metabolism, broad immune activation, and corneal wound healing in corneas of FK mice, evidenced by the sharply decreased expression of multiple metabolism-related genes (cluster 1), significantly increased expression of innate and adaptive immune genes (clusters 2 and 3), and gradually increased expression of development-related genes (cluster 4). Third, the mouse cornea could be infiltrated with a large number of innate immune cells (DC, granulocyte, monocyte, macrophage, and NK cells) and a relatively small number of adaptive immune cells (T and B cells). DC subtypes showed an overall increasing trend with fungal infection, such as MoDC, cDC2, and pDC, whereas granulocyte subtypes (eosinophils, basophils, and mast cells) had an overall decreasing trend. However, the proportions of neutrophil rose sharply in early stage and then gradually decreased as inflammation resolved, and the same trend was also observed in monocytes and macrophages. The conversion of B cell subsets (from follicular to germinal center B cells) and the rising proportion of total CD4+ and CD8+ T cell subsets suggested that B and T cells, despite their relatively low proportion in immune infiltrating cells, might be crucial in the removal of fungal pathogens. Finally, shared immune responses across different time points were observed, exemplified by the increased expression of various innate immune gene families, particularly the ISGs and PANoptosis-related genes.
Potent antifungal immune responses in FK were characterized by moderate and widespread activation of innate immune signaling as well as expansion of distinct immune cell subsets.4,23 After fungal infection disrupted the corneal integrity, the antifungal innate immune response is initiated through the recognition of PAMPs by PRRs,30,31 thereby inducing the production of chemokines and cytokines to recruit local innate immune cells.32 Our transcriptome datasets systematically demonstrated the elevated expression of multiple PRRs (e.g. Dectin-1, TLR9, and NLRP3), cytokines (e.g. IL-1β, and CSF1), and chemokines (e.g. CCL3, CCL12, and CXCL1; see Fig. 5E). Previous studies have demonstrated the critical roles of phagocytes (neutrophils, monocytes, macrophages, and DCs) in the early antifungal defense,3,30,33 among which neutrophils, which are recruited to the site of infection during the acute phage, may account for more than 90% of the total infiltrating immune cells,34 whereas the proportion of other phagocytes is relatively small. Subsequently, these activated phagocytes can trigger the activation of adaptive immune cells (T cells and B cells).34 Our findings demonstrated the synchronous changes in the proportions of neutrophils, monocytes, and macrophages during fungal infection, and all peaked at 3 dpi, which was consistent with disease severity, suggesting the key roles of innate immune cells in inducing inflammatory responses in FK. Interestingly, multiple DC subtypes, such as MoDC, cDC2, and pDC, were found to show an overall upward trend with fungal infection. DCs, the most potent antigen-presenting cells, are a subclass of lympho-myeloid hematopoietic cells generated from bone marrow that serve as a crucial link between the innate sensing of pathogens and the activation of adaptive immunity, which are divided into four major DC subsets: pDC, MoDC, cDC1, and cDC2.35 The pDCs are a unique type of sentinel cell that can recognize nucleic acids derived from pathogens and respond by rapidly and massively producing type I interferon.36 Jamali et al. have demonstrated the vital role for resident corneal pDCs in the control of herpes simplex keratitis.37 MoDC and cDC2 cells engage in the priming of naïve CD4+ T cells through antigen presentation on MHC class II, whereas cDC1 cells are specialized in the presentation of exogenous antigen on MHC class I to prompt naïve CD8+ T cells into cytotoxic T lymphocytes (CTLs).35 Correspondingly, the proportions of CD4+ T cells, CD8+ T cells, and B cells were all found to be elevated in late stages of fungal infection, albeit with low proportions. The conversion of follicular B cells to germinal center B cells in a subset of B cells could even be observed, suggesting that the humoral immunity was primed for antibody production,38 although further studies are warranted in the future. Collectively, our data support an important role for adaptive immune responses in antifungal immunity, although its role in ocular infections has long been overlooked.28
Recent studies have shown that PANoptosis plays an important role in detecting and eliminating various pathogens, including viruses, bacteria, and fungi.14,39–41 As a unique type of immune PCD, PANoptosis is regulated by multifaceted PANoptosome complexes that are assembled through the integration of components from different PCD pathways.39 Identification of the dynamic composition of these PANoptosome complexes under various contexts remains a challenging and active area of research.39 Our data demonstrate that after A. fumigatus infection, AIM2-, pyrin-, and ZBP1-mediated PANoptosis is activated in mouse corneas. This finding was consistent with the study by Lee and colleagues,42 that showed the previously unknown critical interactions among AIM2, pyrin, and ZBP1 that drives the assembly of AIM2-mediated multi-protein complexes during herpes simplex virus 1 and Francisella novicida infections. Our study adds to our understanding of PANoptosome components in fungal infection and further extends AIM2, pyrin as vital components of this complex.43 Furthermore, regulation of PANoptosis is essential given its critical role in activating inflammatory PCD and cytokine release. Recently, TAK1 and ADAR1 have been successively identified as negative regulators of PANoptosis.44,45 However, the gene expression levels of these two regulators were not significantly altered after fungal infection, albeit the slight elevation of ADAR1 expression in the transcriptome data, suggesting that this might be related to excessive inflammation in mouse corneas. Moreover, the activation of PANoptosis will lead to the release of a large number of proinflammatory cytokines,46 which may be related to the immunopathogenesis of FK. Gene expression levels of IFN-γ, IL-1β, IL-6, IL-17A, and TNF-α were markedly increased dramatically after fungal infection, resulting in excessive corneal inflammation. However, no significant alteration of IL-18 expression was observed. As the hallmark cytokines of pyroptosis, pro-IL-1β and pro-IL-18 will be processed into their active forms by the activated caspase-1.47 Further experiments are needed to explore the mechanisms underlying the distinct expression patterns of these two cytokines with similar functions. Interestingly, enhanced expression of ISGs family was also observed, which was often induced by type I interferons.48 However, analyses of the role of interferon and ISGs in defense against fungal infections have been relatively limited.48,49 Stevens et al. proposed that IFN-γ could be used as a broad-spectrum antifungal agent for immunotherapy,50 but clinical trials need to be carried out in the future to verify its therapeutic efficacy.51 Overall, our time-course transcriptome data show that PANoptosis activation is important in host defense against A. fumigatus infection and that balancing this activation is critical to prevent hyperinflammation in FK. It will be valuable to investigate whether small molecules targeting PANoptosis could be used to curb overt FK-associated tissue and organ damages.
There are several limitations in this study. First, this study did not adopt a time-series study design to verify the dynamic expression patterns of PANoptosis-related genes in RNA-seq data due to the limited number of experimental animals. Larger sample sizes are warranted to verify our findings in the future. Second, our study only relied on bioinformatic approaches to infer the proportions of infiltrating immune cells, although these algorithms have been proven to be accurate and reliable, it would be better to use experimental methods for biological validation. However, if traditional flow cytometry is used to quantify the proportion of immune cells, a large number of experimental animals will be consumed to obtain enough mouse corneal tissues. Novel research approaches, such as single-cell transcriptome sequencing, will provide high-resolution immune landscape in the future, although this technology currently suffers from the disadvantages of high cost, large initial cell numbers, and limited number of detected genes. Third, in order to verify the specific molecular mechanism, genetic manipulation should be performed to perturb the expression of key genes involved in PANoptosis to assess the cellular phenotypic changes.
In summary, our study profiles the dynamic immune landscape at both the molecular and cellular levels and highlights the critical role of PANoptosis in driving inflammation during A. fumigatus infection. These findings advance our understanding of host responses to fungal pathogens and provide new directions for the development of targeted therapeutics for patients with FK, emphasizing the potential for therapeutic modulation of PANoptosis in controlling infectious diseases.
Supplementary Material
Acknowledgments
Supported by grants from the National Key Research and Development Program of China (2021YFC2301000), the National Natural Science Foundation of China (81970765), and the Special Foundation for Young Scientists of Beijing Tongren Hospital (2021-YJJ-ZZL-034), and the Foundation for Fostering the National Natural Science Foundation of Capital Medical University (PYZ22107).
Disclosure: X. Xu, None; Y. Wei, None; J. Pang, None; Z. Wei, None; L. Wang, None; Q. Chen, None; Z. Wang, None; Y. Zhang, None; K. Chen, None; Y. Peng, None; Z. Zhang, None; J. Liu, None; Y. Zhang, None; Z.-B. Jin, None; Q. Liang, None
References
- 1. Brown L, Leck AK, Gichangi M, Burton MJ, Denning DW.. The global incidence and diagnosis of fungal keratitis. Lancet Infect Dis. 2021; 21: e49–e57. [DOI] [PubMed] [Google Scholar]
- 2. Xie L, Zhong W, Shi W, Sun S.. Spectrum of fungal keratitis in north China. Ophthalmology. 2006; 113: 1943–1948. [DOI] [PubMed] [Google Scholar]
- 3. Ratitong B, Pearlman E.. Pathogenic Aspergillus and Fusarium as important causes of blinding corneal infections – the role of neutrophils in fungal killing, tissue damage and cytokine production. Curr Opin Microbiol. 2021; 63: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niu L, Liu X, Ma Z, et al.. Fungal keratitis: Pathogenesis, diagnosis and prevention. Microb Pathog. 2020; 138: 103802. [DOI] [PubMed] [Google Scholar]
- 5. Jorgensen I, Rayamajhi M, Miao EA.. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017; 17: 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briard B, Karki R, Malireddi RKS, et al.. Fungal ligands released by innate immune effectors promote inflammasome activation during Aspergillus fumigatus infection. Nat Microbiol. 2019; 4: 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shi J, Zhao Y, Wang K, et al.. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015; 526: 660–665. [DOI] [PubMed] [Google Scholar]
- 8. Karki R, Man SM, Malireddi RKS, et al.. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015; 17: 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caffrey AK, Obar JJ.. Alarmin(g) the innate immune system to invasive fungal infections. Curr Opin Microbiol. 2016; 32: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Opdenbosch N, Lamkanfi M.. Caspases in cell death, inflammation, and disease. Immunity. 2019; 50: 1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rogers C, Erkes DA, Nardone A, et al.. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun. 2019; 10: 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weinlich R, Oberst A, Beere HM, Green DR.. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017; 18: 127–136. [DOI] [PubMed] [Google Scholar]
- 13. Kuriakose T, Man SM, Malireddi RK, et al.. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016; 1: aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christgen S, Zheng M, Kesavardhana S, et al.. Identification of the PANoptosome: A molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis). Front Cell Infect Microbiol. 2020; 10: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu TG, Wilhelmus KR, Mitchell BM. Experimental keratomycosis in a mouse model. Invest Ophthalmol Vis Sci. 2003; 44: 210–216. [DOI] [PubMed] [Google Scholar]
- 16. Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anders S, Pyl PT, Huber W.. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conway JR, Lex A, Gehlenborg N.. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017; 33: 2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu X, Gao Z, Yang F, et al.. Antidiabetic effects of Gegen Qinlian Decoction via the gut microbiota are attributable to its key ingredient berberine. Genomics Proteomics Bioinformatics. 2020; 18: 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miao YR, Xia M, Luo M, et al.. ImmuCellAI-mouse: A tool for comprehensive prediction of mouse immune cell abundance and immune microenvironment depiction. Bioinformatics. 2021; 38: 785–791. [DOI] [PubMed] [Google Scholar]
- 23. Mills B, Radhakrishnan N, Karthikeyan Rajapandian SG, et al.. The role of fungi in fungal keratitis. Exp Eye Res. 2021; 202: 108372. [DOI] [PubMed] [Google Scholar]
- 24. Lakhundi S, Siddiqui R, Khan NA. Pathogenesis of microbial keratitis. Microb Pathog. 2017; 104: 97–109. [DOI] [PubMed] [Google Scholar]
- 25. Chidambaram JD, Kannambath S, Srikanthi P, et al.. Persistence of innate immune pathways in late stage human bacterial and fungal keratitis: Results from a comparative transcriptome analysis. Front Cell Infect Microbiol. 2017; 7: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Q, Zhang J, Gong M, et al.. Transcriptome analysis of the gene expression profiles associated with fungal keratitis in mice based on RNA-seq. Invest Ophthalmol Vis Sci. 2020; 61: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hazlett LD. Role of innate and adaptive immunity in the pathogenesis of keratitis. Ocul Immunol Inflamm. 2005; 13: 133–138. [DOI] [PubMed] [Google Scholar]
- 28. Zhang H, Chen H, Niu J, Wang Y, Xie L.. Role of adaptive immunity in the pathogenesis of Candida albicans keratitis. Invest Ophthalmol Vis Sci. 2009; 50: 2653–2659. [DOI] [PubMed] [Google Scholar]
- 29. Hu J, Hu Y, Chen S, et al.. Role of activated macrophages in experimental Fusarium solani keratitis. Exp Eye Res. 2014; 129: 57–65. [DOI] [PubMed] [Google Scholar]
- 30. Brown GD. Innate antifungal immunity: The key role of phagocytes. Annu Rev Immunol. 2011; 29: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hardison SE, Brown GD.. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012; 13: 817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peng L, Zhong J, Xiao Y, et al.. Therapeutic effects of an anti-IL-6 antibody in fungal keratitis: Macrophage inhibition and T cell subset regulation. Int Immunopharmacol. 2020; 85: 106649. [DOI] [PubMed] [Google Scholar]
- 33. Serbina NV, Jia T, Hohl TM, Pamer EG.. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008; 26: 421–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karthikeyan RS, Leal SM Jr., Prajna NV, et al.. Expression of innate and adaptive immune mediators in human corneal tissue infected with Aspergillus or Fusarium. J Infect Dis. 2011; 204: 942–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collin M, Bigley V. Human dendritic cell subsets: An update. Immunology. 2018; 154: 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reizis B. Plasmacytoid dendritic cells: Development, regulation, and function. Immunity. 2019; 50: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jamali A, Hu K, Sendra VG, et al.. Characterization of resident corneal plasmacytoid dendritic cells and their pivotal role in herpes simplex keratitis. Cell Rep. 2020; 32: 108099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Young C, Brink R.. The unique biology of germinal center B cells. Immunity. 2021; 54: 1652–1664. [DOI] [PubMed] [Google Scholar]
- 39. Pandian N, Kanneganti TD.. PANoptosis: A unique innate immune inflammatory cell death modality. J Immunol. 2022; 209: 1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Briard B, Malireddi RKS, Kanneganti TD. Role of inflammasomes/pyroptosis and PANoptosis during fungal infection. PLoS Pathog. 2021; 17: e1009358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Place DE, Lee S, Kanneganti TD. PANoptosis in microbial infection. Curr Opin Microbiol. 2021; 59: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee S, Karki R, Wang Y, et al.. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature. 2021; 597: 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Banoth B, Tuladhar S, Karki R, et al.. ZBP1 promotes fungi-induced inflammasome activation and pyroptosis, apoptosis, and necroptosis (PANoptosis). J Biol Chem. 2020; 295: 18276–18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malireddi RKS, Gurung P, Kesavardhana S, et al.. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J Exp Med. 2020; 217: e20191644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karki R, Sundaram B, Sharma BR, et al.. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021; 37: 109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bertheloot D, Latz E, Franklin BS. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell Mol Immunol. 2021; 18: 1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi JJ, Gao WQ, Shao F.. Pyroptosis: Gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017; 42: 245–254. [DOI] [PubMed] [Google Scholar]
- 48. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015; 15: 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Niedzwiedzka-Rystwej P, Ratajczak W, Tokarz-Deptula B, Deptula W. Mechanisms of type I interferon action and its role in infections and diseases transmission in mammals. Acta Biochim Pol. 2017; 64: 199–205. [DOI] [PubMed] [Google Scholar]
- 50. Stevens DA, Brummer E, Clemons KV.. Interferon-gamma as an antifungal. J Infect Dis. 2006; 194: 33–37. [DOI] [PubMed] [Google Scholar]
- 51. Delsing CE, Gresnigt MS, Leentjens J, et al.. Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: A case series. BMC Infect Dis. 2014; 14: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.