Abstract
Introduction
A framework that extracts oncological outcomes from large-scale databases using artificial intelligence (AI) is not well established. Thus, we aimed to develop AI models to extract outcomes in patients with lung cancer using unstructured text data from electronic health records of multiple hospitals.
Methods
We constructed AI models (Bidirectional Encoder Representations from Transformers [BERT], Naïve Bayes, and Longformer) for tumor evaluation using the University of Miyazaki Hospital (UMH) database. This data included both structured and unstructured data from progress notes, radiology reports, and discharge summaries. The BERT model was applied to the Life Data Initiative (LDI) data set of six hospitals. Study outcomes included the performance of AI models and time to progression of disease (TTP) for each line of treatment based on the treatment response extracted by AI models.
Results
For the UMH data set, the BERT model exhibited higher precision accuracy compared to the Naïve Bayes or the Longformer models, respectively (precision [0.42 vs. 0.47 or 0.22], recall [0.63 vs. 0.46 or 0.33] and F1 scores [0.50 vs. 0.46 or 0.27]). When this BERT model was applied to LDI data, prediction accuracy remained quite similar. The Kaplan–Meier plots of TTP (months) showed similar trends for the first (median 14.9 [95% confidence interval 11.5, 21.1] and 16.8 [12.6, 21.8]), the second (7.8 [6.7, 10.7] and 7.8 [6.7, 10.7]), and the later lines of treatment for the predicted data by the BERT model and the manually curated data.
Conclusion
We developed AI models to extract treatment responses in patients with lung cancer using a large EHR database; however, the model requires further improvement.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12325-022-02397-7.
Keywords: Artificial intelligence, BERT, Electronic health records database, Lung cancer, Real-world data, Retrospective study
Plain Language Summary
The use of artificial intelligence (AI) to derive health outcomes from large electronic health records is not well established. Thus, we built three different AI models: Bidirectional Encoder Representations from Transformers (BERT), Naïve Bayes, and Longformer to serve this purpose. Initially, we developed these models based on data from the University of Miyazaki Hospital (UMH) and later improved them using the Life Data Initiative (LDI) data set of six hospitals. The performance of the BERT model was better than the other two, and it showed similar results when it was applied to the LDI data set. The Kaplan–Meier plots of time to progression of disease for the predicted data by the BERT model showed similar trends to those for the manually curated data. In summary, we developed an AI model to extract health outcomes using a large electronic health database in this study; however, the performance of the AI model could be improved using more training data.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12325-022-02397-7.
Key Summary Points
| Why carry out this study? |
| The structure for extracting oncology clinical outcomes from large-scale electronic health records databases using artificial intelligence (AI) is not well established. |
| Thus, adapting AI models for various countries or regions is required. Our research planned to develop AI models (Bidirectional Encoder Representations from Transformers [BERT], Naïve Bayes, and Longformer) to extract clinical outcomes in patients with lung cancer by utilizing the unstructured/structured text data from Japanese EHR of multiple hospitals. |
| These models were developed to evaluate tumors using the University of Miyazaki Hospital (UMH) database, and this was then applied to the Life Data Initiative (LDI) data set of six hospitals. |
| What was learned from the study? |
| The BERT model exhibited higher performance compared to Naïve Bayes and Longformer models, respectively (precision [0.42 vs. 0.47 or 0.22], recall [0.63 vs. 0.46 or 0.33], and F1 scores [0.50 vs. 0.46 or 0.27]). |
| When the BERT model was applied to LDI data, prediction accuracy remained quite similar. |
| The Kaplan–Meier plots of TTP for the predicted data by the BERT model showed similar trends to those for the manually curated data. |
| Although AI models could extract treatment responses in patients with lung cancer using a large EHR database, they require further improvement by using more training data. |
Introduction
Research utilizing real-world data (RWD) obtained from various sources such as claims data, electronic health records (EHR), and disease and product registries are growing significantly [1]. Randomized clinical trials (RCTs) are usually conducted under controlled conditions and may limit the generalizability to real-world clinical practice. On the contrary, RWD more specifically exhibits real clinical practice environments such as patient demographics, treatment adherence, and concurrent treatments [2]. Compared to administrative claims databases that have been used in medical research for decades, EHR databases provide access to a wider range of variables recorded during medical examinations. However, EHR databases present inherent challenges such as unstructured data [3]. Unstructured data includes narrative data present in clinical notes, surgical records, discharge summaries, radiology reports, medical images, and pathology reports stored in EHRs. Though adequate valuable information can be extracted from unstructured data, it can often be difficult to process and analyze them owing to their association with different contexts, ambiguities, grammatical and spelling errors, and the usage of abbreviations [3].
Manual review of unstructured EHR data has been a conventional method for extracting clinical outcomes but it is a laborious and cost-intensive process [3, 4]. With the increasing number of clinical texts, methods for analyzing this type of EHR data using natural language processing (NLP) are emerging rapidly [5, 6]. Several studies have reported using the NLP-based methods to extract clinical outcomes in patients with cancer using the EHR database [7–9]. Conventional methods of NLP can extract key terms but gaining an understanding of the context of key terms is equally important to better assess the outcomes and accuracy of information; thus, advanced NLP must be combined with artificial intelligence (AI). Transformers are one of the most advanced deep learning-based architectures in AI; and Generative Pre-trained Transformer 3 (GPT-3), Bidirectional Encoder Representations from Transformers (BERT), and Longformer are some advanced transformer-based models for clinical utility. BERT was developed and openly sourced by Google [10–12]. Although studies have applied AI to extract treatment responses from EHR texts for patients with cancer [7, 13], the AI methods have not been rigorously validated for reproducibility and generalizability to evaluate treatment responses using oncology imaging data [14, 15].
Research that aims to assess outcomes from large-scale EHR databases using AI models has the potential to generate real-world evidence at a fast pace. However, a framework that can extract outcomes using AI models such as dictionaries for pre-training, preparing training data sets for a correct and false response, structure and type of AI model, validation of AI model, and application of data extracted by AI models in clinical research is not well established. The aggregation of unstructured text data in EHRs from multiple institutions is also a challenge. In particular, most studies in this field have used US EHRs. In a systematic review that examined literature reporting NLP on clinical notes for chronic disease, only 24 out of 106 studies were outside of the USA [16]. However, text data of medical records (progress notes, etc.) vary in terms of language, clinical practices, the structure of the medical record system, etc. for different countries. Therefore, adapting the AI model for various countries/regions is required, and no study has reported clinical outcomes from the Japanese EHR using AI models. The current research was planned to develop AI models (in particular, a transformer of the BERT model) for extracting clinical outcomes in patients with lung cancer by utilizing unstructured text data from the Japanese EHR of multiple hospitals. We assessed the performance of our BERT model and demonstrated its practical use in estimating the time to progression (TTP) for each line of treatment of lung cancer based on the treatment responses extracted by the BERT model.
Methods
Study Design and Population
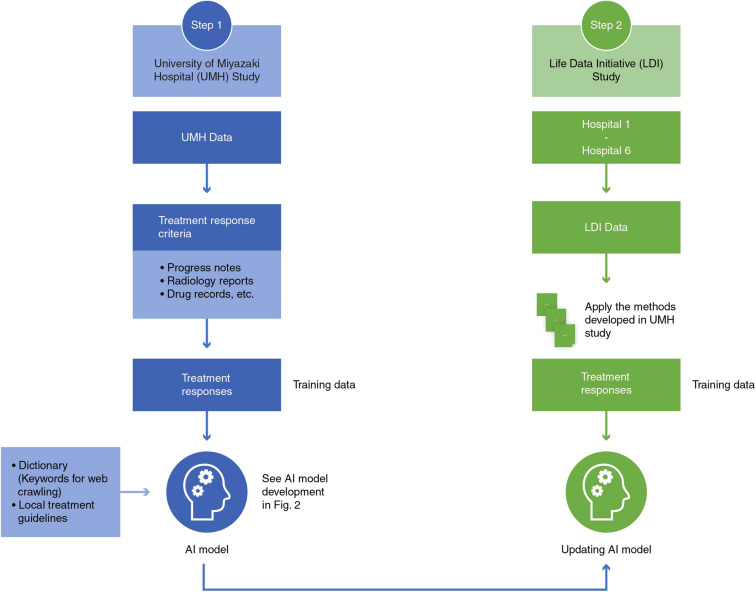
We conducted two retrospective studies. One study used the University of Miyazaki Hospital (UMH) data, and the other used the EHR database of the General Incorporated Association Life Data Initiative (LDI) which consisted of data from six hospitals. The LDI has a centralized data center for regional medical networks with an interface to receive data from each medical facility though different standards designed for exchange, integration, sharing, and for retrieving electronic health information such as medical markup language (MML), health level 7 (HL7), etc. [17, 18]. LDI was the first certified organization by the Japanese government under the Japanese Next Generation Medical Infrastructure law that enables certified organizations to collect and analyze non-anonymized medical data [19]. We developed a BERT model for assessing treatment responses in adults (at least 18 years old) with lung cancer who received anticancer drug treatment. No exclusion criteria such as type/stage of lung cancer were considered as this study was performed to develop a BERT model that interpreted relationships between words related to treatment responses and we did not aim to evaluate treatment efficacy. The BERT model was first developed using the UMH data and pre-training data, and was then applied to the LDI data and further improved using the EHR data at six hospitals (Fig. 1).
Fig. 1.
Data sources for model development. AI artificial intelligence
Data Sources
UMH: Data of eligible patients were captured from the EHR of UMH. It included structured data (patient background information, prescription, and injection information) and unstructured data (progress notes, radiology reports, and clinical summaries) between April 2018 and September 2020 (Fig. 1).
LDI: Data of eligible patients were captured from the LDI EHR of six hospitals with varying sizes (100–400 beds, n = 1; 400–800 beds, n = 2; and 800–1200 beds, n = 3) from October 2017 to January 2021. Of the six hospitals, only two were university hospitals (400–800 beds, n = 1; 800–1200 beds, n = 1) which provided designated advanced oncology care to patients and were in West Japan. Of the other three designated cancer hospitals, two (400–800 beds, n = 1; 800–1200 beds, n = 1) were in East Japan, and one with 800–1200 beds was in West Japan. However, only one hospital was not a designated cancer hospital and only housed 100–400 beds in West Japan. The variables were similar to those from the UMH data set (Fig. 1). The data used for this study was extracted from the LDI EHR system, which was connected to regional medical facilities and consisted as electronic medical records and claims data. The extracted data was analyzed in a secure system for secondary use by the NTT DATA corporation following the implementation of the Next Generation Medical Infrastructure law as certified by the Japanese government.
Model Development
UMH Study
The training data set was created by abstractors who manually evaluated treatment responses from the UMH data. Data were extracted from discharge summaries, progress notes, radiology reports, radiological test records, and drug administration records, and were tabulated electronically. The abstractors reviewed the extracted data during the study period, and recorded treatment responses for individual documents on each date. If a document was not related to treatment response, it was marked as “not evaluable”. These responses were categorized as either objective response (OR), stable disease (SD), or progressive disease (PD). The OR was defined as any shrinkage in tumor size seen in images compared to baseline. The PD was defined as any tumor progression from baseline or discontinuation of cancer treatment due to lack of efficacy or intolerance. The outcome was considered SD when neither OR nor PD was observed. The first 15 patients were evaluated by two physicians and the remaining patients were evaluated by two pharmacists who had sufficient knowledge of lung cancer treatment and the RECIST criteria [20]. Any discrepancy in tumor evaluation that was identified by the pharmacist was then addressed by the physician.
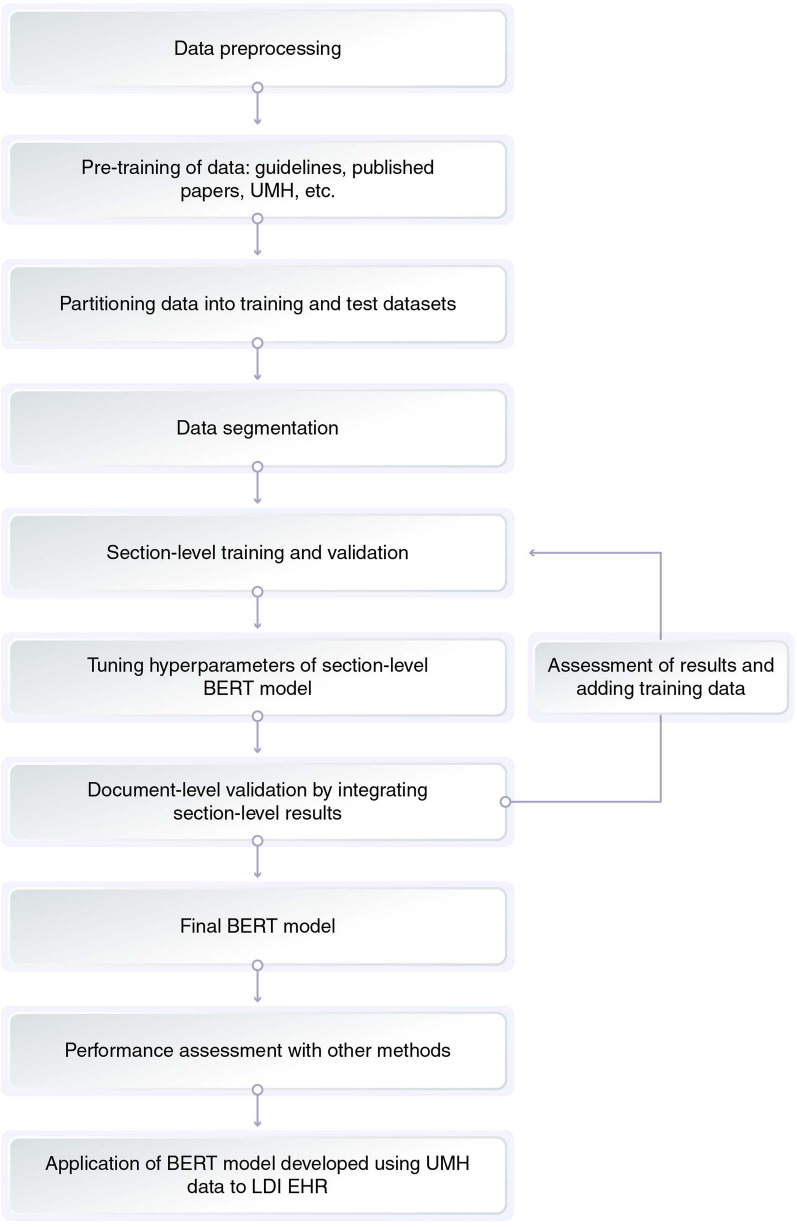
The development of the BERT model consisted of four parts: pre-training, training, validation, and tuning of the hyperparameters. Details of model development are provided in Fig. 2 and Table 1. Pre-training of the model was performed on the basis of the current guidelines, concerning papers from journals, electronic medical records of UMH, web crawling, etc. (Fig. 2 and Table 1). Each record (document) was sectioned as the BERT model could handle up to 512 tokens, and each section had a meaningful relation between sentences. One document had several topics and was divided into segments based on different topics. This helped the AI model to learn the relationships between words in a meaningful group of sentences. In other words, we prevented the AI model from learning the wrong relationship between words. The BERT model was applied to classify texts into four labels, namely OR, SD, PD, or not evaluable, and was developed on the basis of a validation approach that included section-level and document-level validation by integrating section-level results. During this process, cross-validation was performed using training and test data sets prepared by partitioning data sets into three sets of training and test data, and the same patient data was not included in both data sets. Model performance was assessed and improved by analyzing the error patterns. Hyperparameters were tuned during section-level validation. The performance of the final BERT model was assessed by comparing it with the Longfomer and Naïve Bayes model of machine learning. The Longfomer used long records without separating them into sections with shorter records whereas Naïve Bayes is a typical machine learning method and is extremely popular for document classification in various fields including medicine [10, 21, 22].
Fig. 2.
Model development. AI artificial intelligence, BERT Bidirectional Encoder Representations from Transformers, EHR electronic health records, LDI Life Data Initiative, UMH University of Miyazaki Hospital
Table 1.
Step-by-step description of model development
| Step | Description |
|---|---|
| Data preprocessing | UMH text data were processed such as deleted html tags and extra spaces, and the double-byte characters (of English letters) and numbers were converted to single byte characters |
| Pre-training of data |
The BERT model was pre-trained on the Masked Language Model Task and NExt Sentence Prediction Task via prepared corpus [20] Data used for pre-training included: - 138 documents (51,688 sentences) of clinical guidelines, published case reports, etc. - Websites of 50 URLs (6501 sentences) such as the National Cancer Center, Japan - Progress notes, radiology reports, discharge summary of 84 patients in electronic medical data of UMH (33,978 sentences) - Web crawling text (907,833 sentences) of 1156 domain words from 84 patients - General information such as news articles, blogs, and wikipedia (30 GB) |
| Partitioning of data into training and test data sets | Data was partitioned into 3 sets of training and test data for cross-validation |
| Data segmentation |
A long document was divided into several sections as follows: - The document was divided when there was a specific symbol (e.g., #, •, ★), consecutive new lines, and date at the beginning of a sentence - Past descriptions (patient history) that were completely consistent with the past records |
| Section-level training and validation |
The BERT model was trained using the section-level training data, and classified the sections into treatment responses of CR, SD, PD, or NE Owing to the high proportion of NE data, down-sampling of NE data was applied at 3 sampling proportions of 25% 50%, and 75% each. The highest F1 score was shown by 50% sampling and was selected |
| Tuning hyperparameters of section-level BERT model |
Hyperparameters of the BERT model trained at the section level were tuned with the following conditions: Learning rate, [2e−5, 5e−05, 8e−05]; warm-up proportion, 0.1; maximum number of the epoch, 20 |
| Document-level validation by integrating section-level results |
Section-level prediction results were integrated into document-level results. In case there were multiple responses within a document, 3 rules were examined: (1) The priority order of PD, OR, SD was applied considering the original definition of treatment response (2) The F1 scores of sections were totaled for each response and the response with the highest total score was selected (3) Selected the response in the section with the highest F1 score Then (3) was selected because of the highest accuracy |
| Assessment of results and adding training data | Accuracy (precision, recall, F1 score) and error patterns were analyzed. Additional training data was created by combining the expressions related to treatment responses and those with tumors or examinations to supplement the weak area based on error patterns. Then the step returned to “Section-level training and validation” until underfitting was found |
| Final BERT model | The model was fixed when model fitting was saturated |
| Performance assessment with other methods | The accuracy (precision, recall, F1 score) of the final BERT model was compared with that of the Longformer and Naïve Bayes models to evaluate which model was the best |
| Application of BERT model developed using UMH data to LDI EHR | The final BERT model using the UMH data was applied to the LDI EHR data from 6 hospitals. Afterwards, the error patterns of the BERT model were applied to LDI EHR data and evaluated. Depending on the error patterns, additional training data was created combining the expressions similar to the UMH study. The BERT model was redeveloped on the basis of this training data |
UMH University of Miyazaki Hospital, LDI Life Data Initiative, OR objective response, SD stable disease, PD progression disease, NE not evaluable, BERT Bidirectional Encoder Representations from Transformers
LDI study
The BERT model developed on the basis of the UMH data was applied to the LDI EHR of multiple hospitals. This model was improved using the same methods applied to the UMH data, and the model performance was assessed. Based on the treatment responses of each record obtained from the BERT model, the TTP for each line of treatment was estimated for each patient.
Statistical Analysis
The accuracy of AI models was calculated using accuracy, precision (positive predictive value), recall (sensitivity), and F1 scores (Fig. S1 in the supplementary material). Continuous data were summarized using descriptive statistics of mean, standard deviation, median, first quartile (Q1), and third quartile (Q3). For categorical data, frequencies (n, %) were presented. Missing values for each variable were summarized but they were not counted while calculating the summary statistics.
The TTP for each line of treatment was defined as the time from the start date of a given treatment until the date when PD was confirmed. For patients who did not have PD, TTP was censored at the date of the last record of no tumor progression or continuation of treatment. The TTP was summarized using descriptive statistics and the 95% confidence interval based on the Kaplan–Meier method.
Ethics and Approval
The study was approved by the ethics committee of the UMH (application no. 0-0845), and the opt-out consent process was granted under the ethical guidelines for the Medical and Health Research Involving Human Subjects by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Ministry of Health, Labor and Welfare (MHLW). The anonymized data were analyzed. The study was conducted following the Helsinki Declaration of 1964 and its later amendments.
The LDI data was collected by the opt-out consent process that was per the Next Generation Medical Infrastructure law, and the use of LDI data for this study was approved by the review board of LDI (application no. 2021-MIL0011).
Results
Patient Disposition and Demographics
The LDI study included EHRs of 713 patients, and the UMH study included EHRs of 85 patients. Demographic and clinical characteristics of patients are summarized in Table 2. Most patients had stage III/IV lung cancer in the UMH (56/85, 65.9%) and LDI (260/713, 36.4%) data sets, and more than 60% of patients were hospitalized at the time of analysis because of primary cancer. Recurrence of primary cancer was seen in 1.2% (1/85) and 1.1% (8/713) of patients in the UMH and LDI studies, respectively.
Table 2.
Demographic details
| Patient background | UMH study (N = 85) | LDI study (N = 713) |
|---|---|---|
| Age (years), mean ± SD | 67.3 ± 10.4 | 68.2 ± 10.2 |
| Gender | ||
| Female | 48 (56.5) | 231 (32.4) |
| Male | 37 (43.5) | 482 (67.6) |
| Body weight | ||
| N | 64 | 352 |
| Mean ± SD | 57.7 ± 10.9 | 58.7 ± 12.8 |
| Eat/smoke tobacco | ||
| Could be present | 35 (41.2) | 317 (44.5) |
| Never | 41 (48.2) | 293 (41.1) |
| Not mentioned | 9 (10.6) | 103 (14.4) |
| Primary disease stage | ||
| Stage 1 | 4 (4.7) | 62 (8.7) |
| Stage 2 | 3 (3.5) | 79 (11.1) |
| Stage 3 | 9 (10.6) | 93 (13.0) |
| Stage 4 | 47 (55.3) | 167 (23.4) |
| Not mentioned | 22 (25.9) | 312 (43.8) |
| History of hospitalization due to primary disease | ||
| Present | 52 (61.2) | 433 (60.7) |
| None | 9 (10.6) | 166 (23.3) |
| Not mentioned | 24 (28.2) | 114 (16.0) |
| Surgery for underline disease | ||
| Present | 6 (7.1) | 31 (4.3) |
| None | 0 | 559 (78.4) |
| Not mentioned | 79 (92.9) | 0 |
| No linkage data | – | 123 (17.3) |
| History of radiation therapy for the original disease | ||
| At present | 8 (9.4) | 200 (28.1) |
| None | 5 (5.9) | 513 (71.9) |
| Not mentioned | 72 (84.7) | 0 |
| Recurrence of primary disease | ||
| At present | 1 (1.2) | 8 (1.1) |
| None | 84 (98.8) | 705 (98.9) |
| Metastasis of primary disease | ||
| At present | 26 (30.6) | 116 (16.3) |
| None | 59 (69.4) | 597 (83.7) |
| Multiple-primary cancer | ||
| At present | 4 (4.7) | 158 (22.2) |
| None | 81 (95.3) | 555 (77.8) |
UMH University of Miyazaki Hospital, LDI Life Data Initiative, SD standard deviation
Training Data
The training and test data used to build and validate the BERT model in the UMH study comprised 1029 documents (Table 3). The LDI data set included 824 records in progress reports, radiation reports, and discharge summaries. In the UMH study, the treatment responses of OR, SD, and PD were recorded in 27, 22, and 17 patients, respectively. In the LDI study, OR, SD, and PD were recorded in 109, 60, and 79 patients, respectively (Table 3).
Table 3.
Response to treatment classified by the volume of source data (progress notes, radiology reports, and discharge summaries)
| UMH study | LDI study | |||
|---|---|---|---|---|
| Documents | Patients | Documents | Patients | |
| OR | 153 | 27 | 191 | 109 |
| SD | 98 | 22 | 81 | 60 |
| PD | 75 | 17 | 140 | 79 |
| Not evaluable | 703 | 31 | 412 | 215 |
| Total | 1029* | 31# | 824* | 322# |
OR objective response, SD stable disease, PD progression disease, UMH University of Miyazaki Hospital, LDI Life Data Initiative
*Represents the sum of all categories of response mentioned in all evaluated documents
#Represents the total number of patients evaluated for all categories of response; as one patient could be counted in more than one category based on extracted data from document, total patients might not equal the sum of all categories of a response
Model Performance
In the UMH study, compared to the tumor evaluation model constructed using Naïve Bayes or the Longformer model, the model constructed using BERT showed significant improvement in accuracy for the average of response, stability, and progression as indicated by higher precision (0.42 vs. 0.47 or 0.22), recall (0.63 vs. 0.46 or 0.33), and F1 scores (0.50 vs. 0.46 or 0.27). Similar trends were observed in the LDI study, i.e., higher precision (0.40 vs. 0.36 or 0.43), recall (0.54 vs. 0.26 or 0.28), and F1 (0.45 vs. 0.40 or 0.27) score in the BERT model in comparison with Naïve Bayes or the Longformer model. The accuracy showed the same relative relationship among the models as F1 scores (Table 4).
Table 4.
Performance of the final BERT model compared to other methods in the UMH study and the LDI study
| Treatment response | BERT model | Longformer model | Naïve Bayes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 | Accuracy | Precision | Recall | F1 | Accuracy | Precision | Recall | F1 | |
| UMH study | ||||||||||||
| OR | NA | 0.49 | 0.83 | 0.61 | NA | 0.33 | 0.58 | 0.42 | NA | 0.60 | 0.65 | 0.62 |
| SD | NA | 0.45 | 0.62 | 0.51 | NA | 0.32 | 0.40 | 0.39 | NA | 0.38 | 0.33 | 0.35 |
| PD | NA | 0.33 | 0.45 | 0.37 | NA | 0 | 0 | 0 | NA | 0.42 | 0.40 | 0.40 |
| NE | NA | 0.94 | 0.37 | 0.51 | NA | 0.74 | 0.56 | 0.64 | NA | 0.71 | 0.69 | 0.70 |
| Average | 0.67 | 0.42 | 0.63 | 0.50 | 0.40 | 0.22 | 0.33 | 0.27 | 0.54 | 0.47 | 0.46 | 0.46 |
| LDI study | ||||||||||||
| OR | NA | 0.54 | 0.63 | 0.58 | NA | 0.43 | 0.51 | 0.47 | NA | 0.58 | 0.55 | 0.57 |
| SD | NA | 0.18 | 0.32 | 0.23 | NA | 0.18 | 0.17 | 0.18 | NA | 0.14 | 0.10 | 0.11 |
| PD | NA | 0.47 | 0.67 | 0.55 | NA | 0.46 | 0.09 | 0.15 | NA | 0.57 | 0.49 | 0.53 |
| NE | NA | 0.83 | 0.51 | 0.64 | NA | 0.68 | 0.82 | 0.74 | NA | 0.71 | 0.80 | 0.75 |
| Average | 0.58 | 0.40 | 0.54 | 0.45 | 0.30 | 0.36 | 0.26 | 0.27 | 0.44 | 0.43 | 0.28 | 0.40 |
OR objective response, SD stable disease, PD progression disease, NA not applicable, NE not evaluable, UMH University of Miyazaki Hospital, LDI Life Data Initiative, BERT Bidirectional Encoder Representations from Transformers
Average: average among OR, SD, and PD; Accuracy: accuracy among OR, SD, and PD
When the BERT model (developed on the basis of the UMH data) was applied to LDI data, prediction accuracy decreased by 0.03 points for OR, and by 0.28 points for SD as shown by the F1 values (Table 4). This could be due to the difference in using the expressions for OR and SD by UMH and LDI institutes. On the other hand, the prediction accuracy for disease progression improved by 0.18 points, which could be attributed to the frequent use of the expression for PD, e.g., “enlargement/grow” and “aggravation” at UMH that is used commonly at LDI institutions. In patients with multiple tumors, treatment response is estimated individually for each tumor, which poses a challenge for estimating response by AI models. However, the patients with multiple lesions were fewer in LDI than in the UMH database, which might have contributed to the higher accuracy of AI models for LDI data. Overall, when the final BERT model was applied to the LDI data set, no remarkable decrease was found in precision, recall, and F1 scores. The accuracy showed similar relative relationships among the models as F1 scores.
Time to Progression
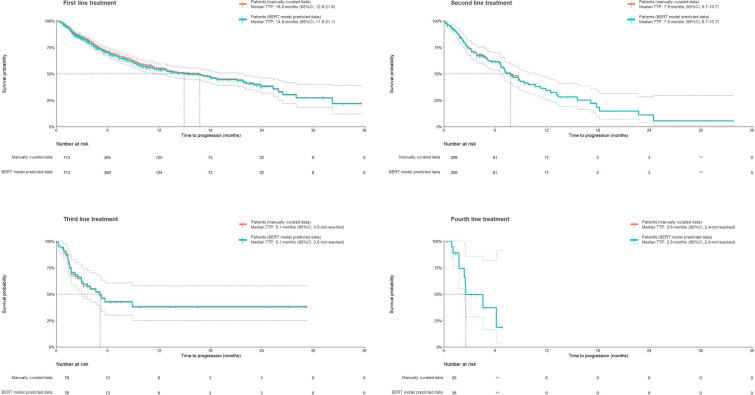
The Kaplan–Meier plots of TTP showed similar trends for the first (median 14.9 months [95% confidence interval 11.5, 21.1] and 16.8 months [12.6, 21.8]), the second (7.8 months [6.7, 10.7] and 7.8 months [6.7, 10.7]), the third (5.1 months [3.0, not reached] and 5.1 months [3.0, not reached]), and the fourth ( 2.6 months [2.4, not reached] and 2.6 months [2.4, not reached]) lines of treatment for the predicted data by the BERT model and the manually curated data (Fig. 3, Fig. S2 in the supplementary material). Table 5 demonstrates the number of patients experiencing disease progression, number of patients who discontinued treatments, and the number of censored patients who were stratified on the basis of the line of treatment.
Fig. 3.
Time to progression using treatment response estimated by the BERT model and curated manually. TTP time to progression, CI confidence interval
Table 5.
Summary of events for time to progression
| Line of treatment | Patients with event | Censored patient | Median (days) (Q1, Q3) | ||
|---|---|---|---|---|---|
| Tumor progression from baseline | Discontinuation of treatment | Total | |||
| BERT model predicted event | |||||
| 1st line (N = 713) | 62 (9%) | 179 (25%) | 241 (34%) | 472 (66%) | 454 (140, 981) |
| 2nd line (N = 209) | 15 (7%) | 71 (34%) | 86 (41%) | 123 (59%) | 237 (84, 523) |
| 3rd line (N = 78) | 7 (9%) | 23 (29%) | 30 (38%) | 48 (62%) | 155 (52, NR) |
| 4th line (N = 26) | NA | NA | 9 (35%) | 17 (65%) | 78 (54, 187) |
| Manually curated event | |||||
| 1st line (N = 713) | 57 (8%) | 177 (25%) | 234 (33%) | 479 (67%) | 510 (150, 981) |
| 2nd line (N = 209) | 14 (7%) | 72 (34%) | 86 (41%) | 123 (59%) | 237 (84, 523) |
| 3rd line (N = 78) | 8 (10%) | 22 (28%) | 30 (38%) | 48 (62%) | 155 (48, NR) |
| 4th line (N = 26) | NA | NA | 9 (35%) | 17 (65%) | 78 (54, 187) |
Q1 1st quartile, Q3 3rd quartile, NR not reached, NA data was not available for anonymization in the cell with a small number of patients
Discussion
In this study, we developed BERT models to extract treatment responses in real-world clinical practice in patients with lung cancer from a large EHR database of multiple medical institutes. The performance of the BERT model was superior to the Longformer model, and was similar or slightly better than the Naïve Bayes model. The Kaplan–Meier plots of TTP for the predicted data by the BERT model showed similar trends to those for the manually curated data.
The performance of our BERT model could be improved with some adaptations. Firstly, since there was a shortage of training data, there may be some expressions that the model has not learned yet. However, this can be improved by incorporating large training data sets. Secondly, a document may sometimes include descriptions of treatment responses for other diseases or non-pharmacological treatment. We handled this by segmenting the document into related sentences in our study as meaningful sections, but the scope for adding rules for segmentation still exists. Thirdly, text records of the last visit are often copied for future records to document any progress from previous visits, which can lead to prediction errors and can be improved by eliminating duplicated texts. We found this trend of text duplications in both UMH and LDI EHRs of multiple hospitals in Japan, but these errors have not been reported for other countries [23].
To set outcomes that bring feasibility and serve the research objective remains critical for any research that uses EHR. Achieving effective outcomes in a real-world setting should differ from those in clinical trials. Some studies have examined the RECIST response using radiology reports; however, it is important to consider that RECIST criteria are a standardized tool for evaluating tumor responses in clinical trial settings. Our study employed simplified treatment responses (OR, SD, and PD) in real-world settings. A study that used the EHR database of multiple medical institutions in the USA reported that as a result of incomplete data and insufficient clarity of radiology reports for the strict RECIST criteria, RECIST could not effectively assess PD for non-small cell lung cancer (NSCLC) [24]. On the other hand, in another study that utilized EHR of a single medical institution in the USA, a deep learning model was successfully developed to estimate the RECIST response assessments using the text of clinical radiology reports in patients with advanced NSCLC treated with programmed death 1/programmed death ligand 1 (PD-1/PD-L1) blockade [13]. This difference in the feasibility of RECIST response from EHR could be due to variations in the information recorded in the EHR, and how strictly the RECIST criteria were followed at any given institute. In Japan, recordings of the RECIST response in clinical practice are unlikely at medical institutes [20]. Rather, a real-world treatment response evaluated by physicians in clinical practice may be extracted using EHRs from multiple medical institutes that could aid in clinical decision-making. With this objective, we developed an AI model to extract treatment responses using large EHR in real-world clinical practice. We could estimate the TTP based on the treatment response extracted by our AI model.
Human curation can extract clinical outcomes from large-scale EHR data and generate RWE for efficacy and safety of the anticancer treatment. In a study by Kehl et al., machine learning (deep learning model) and human curation reported similar measurements for disease-free survival, progression-free survival, and time to improvement/response. This study used EHR data from a single institution and suggested that this model could reduce both the time and expense required to review medical records and could help accelerate efforts to generate RWE from patients with cancer [7]. In our study, the BERT model was developed on the basis of one hospital’s data with a relatively smaller set of training data applied to the EHR database of multiple hospitals with little loss in the model performance. This could be attributed largely to pre-training using the dictionary, guidelines, etc., and the additional training was based on error patterns. Recently, Rasmy et al. proposed a “Med-BERT” model, which is a BERT model adapted with pre-training data of a large EHR data set of 28,490,650 patients [25]. This model was built to benefit disease prediction studies with a small training data set. AI, including machine learning, is also used to develop various prediction models [26–28]. However, continuous improvement of the existing AI models by utilizing more extensive EHR data is important to improve the accuracy of outcomes.
A large database originating from multiple institutions offers the advantage of immediate availability of information without needing primary data collection which shortens the overall timeline of the research. However, constructing an extensively large EHR database to enable AI-based research is a tremendous challenge. In addition, the secondary use of EHR data is limited because of the sensitive nature of personal information in medical records in many countries [29]. However, in Japan, the “Act on Anonymized Medical Data that Are Meant to Contribute to Research and Development in the Medical Field” (Next Generation Medical Infrastructure law) can address this issue of data accessibility. The LDI database used in this study consists of medical records from multiple hospitals and the hospital pool is growing rapidly, allowing the application of this model in a larger and more diverse patient population of the newly included hospitals across Japan. This has the potential to provide timely RWE for decision-making.
This study has some inherent limitations such as the lack of connection among hospitals and clinics in Japan, which resulted in the inability to analyze survival time/death outcomes using Japanese EHR. In addition, imaging tests and treatments that were conducted outside the hospital were also not included. Information about death and death date (confirmed date) is available in the city government database but could not be accessed because of the personal information protection act. Our study aimed to develop AI models to extract outcomes using unstructured text data, and analyzing larger data sets was a priority over enrolling a homogenous patient cohort. The present study included patients with both small cell lung cancer (SLCLC) and non-small cell lung cancer (NSCLC) and those with early (stage I/II) and advanced stages (III/IV) of cancer. It is likely that patients with early stage of diseases have localized disease and are thereby managed surgically (with/without perioperative systemic therapy), whereas advanced diseases require multiple treatment regimens. Thus, comparing different treatment regimens in the heterogeneous population of our study is inappropriate. In addition, our study aimed to develop AI models to extract outcomes using unstructured text data, analyzing larger data sets was a priority over enrolling a homogenous population. However, future studies could enrol a homogeneous population and add another dimension by comparing different treatment regimens.
Conclusion
In the current study, we developed BERT models to extract treatment responses in real-world clinical practice in patients with lung cancer from a large EHR database of multiple medical institutes. The performance of the BERT model was superior compared to the Longformer model, and similar or slightly better than the Naïve Bayes model. The Kaplan–Meier plots of TTP for the predicted data by the BERT model showed similar trends to those for the manually curated data. However, continuous improvement of the models by using more learning data is required to improve the accuracy of outcomes.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (JPG 232 KB) Figure S1: Calculation of precision, recall, and F1 score to assess the model performance
Supplementary file2 (TIF 1595 KB) Figure S2: Time to progression for each treatment line. TTP, time to progression; CI, confidence interval
Acknowledgements
Funding
Sponsorship for this study, and the journal’s Rapid Service and Open Access fees, were all funded by Pfizer Japan Inc.
Medical Writing and Editorial Assistance
Editorial and medical writing support was provided by MedPro Clinical Research (Haruyoshi Ogata), which was supported by CBCC Global Research (Leena Patel), and was funded by Pfizer Japan Inc.
Author Contributions
Kenji Araki, Nobuhiro Matsumoto, Kanae Togo, Naohiro Yonemoto, Emiko Ohki, and Yoshiyuki Hasegawa contributed to the conception and design of the study. All authors contributed to the acquisition, analysis, or interpretation of data for the work, and participated in drafting the work or revising it critically for important intellectual content and they approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosures
The study was sponsored by Pfizer Japan Inc. Kanae Togo, Naohiro Yonemoto, Emiko Ohki, and Linghua Xu are employees of Pfizer Japan Inc. Emiko Ohki has stock ownership in Pfizer Japan Inc. Kenji Araki and Nobuhiro Matsumoto have received research funding from Pfizer. Taiga Miyazaki is consultant and advisor in Pfizer, Kyorin, and Asahi Kasei Pharma. Taiga Miyazaki serves on speakers’ bureaus for Pfizer, MSD, Astellas, Sumitomo Pharma, Kyorin, GSK, Lilly, Meiji Seika Pharma, Sanofi, Takeda Pharmaceutical, Janssen Pharmaceutical, Novartis, Boehringer Ingelheim, and AstraZeneca. Taiga Miyazaki has also received research funding from Pfizer, Astellas, Toyama Chemical, Asahi Kasei Pharma, and MSD.
Compliance with Ethics Guidelines
The study was approved by the ethics committee of UMH, and the opt-out consent process was granted under the ethical guidelines for the Medical and Health Research Involving Human Subjects by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Ministry of Health, Labor and Welfare (MHLW). The study was conducted following the Helsinki Declaration of 1964 and its later amendments.
The LDI data was collected by the opt-out consent process that was per the Next Generation Medical Infrastructure law, and the use of LDI data for this study was approved by the review board of LDI.
Data Availability
The data from the LDI data set supporting the findings of this study may be made available upon request and are subject to approval by the review board of LDI and a license agreement with the General Incorporated Association Life Data Initiative. Data from the UMH data set supporting the findings of this study may be made available upon request and are subject to approval by the ethics committee of the University of Miyazaki.
Contributor Information
Kenji Araki, Email: taichan@med.miyazaki-u.ac.jp.
Nobuhiro Matsumoto, Email: nobuhiro@med.miyazaki-u.ac.jp.
Kanae Togo, Email: kanae.togo@pfizer.com.
Naohiro Yonemoto, Email: Naohiro.Yonemoto@pfizer.com.
Emiko Ohki, Email: Emiko.Ohki@pfizer.com.
Linghua Xu, Email: Linghua.Xu@pfizer.com.
Yoshiyuki Hasegawa, Email: Yoshiyuki.Hasegawa@nttdata.com.
Daisuke Satoh, Email: Daisuke.Satoh@nttdata.com.
Ryota Takemoto, Email: Ryota.Takemoto@nttdata.com.
Taiga Miyazaki, Email: taiga_miyazaki@med.miyazaki-u.ac.jp.
References
- 1.Naidoo P, Bouharati C, Rambiritch V, et al. Real-world evidence and product development: opportunities, challenges and risk mitigation. Wien Klin Wochenschr. 2021;133(15–16):840–846. doi: 10.1007/s00508-021-01851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartlett VL, Dhruva SS, Shah ND, Ryan P, Ross JS. Feasibility of using real-world data to replicate clinical trial evidence. JAMA Netw Open. 2019;2(10):e1912869. doi: 10.1001/jamanetworkopen.2019.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tayefi M, Ngo P, Chomutare T, et al. Challenges and opportunities beyond structured data in analysis of electronic health records. Wiley Interdiscip Rev Comput Stat. 2021;13(6):e1549. doi: 10.1002/wics.1549. [DOI] [Google Scholar]
- 4.Mayer DA, Rasmussen LV, Roark CD, Kahn MG, Schilling LM, Wiley LK. ReviewR: A light-weight and extensible tool for manual review of clinical records. JAMIA Open. 2022;5(3):ooac071. [DOI] [PMC free article] [PubMed]
- 5.Dalianis H. Clinical text mining: secondary use of electronic patient records. Cham: Springer Nature; 2018. 10.1007/978-3-319-78503-5.
- 6.Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review. JMIR Med Inform. 2019;7(2):e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehl KL, Elmarakeby H, Nishino M, et al. Assessment of deep natural language processing in ascertaining oncologic outcomes from radiology reports. JAMA Oncol. 2019;5(10):1421–1429. doi: 10.1001/jamaoncol.2019.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Luo Y-H, Wampfler JA, et al. Efficient and accurate extracting of unstructured EHRs on cancer therapy responses for the development of RECIST natural language processing tools: Part I, the corpus. JCO Clin Cancer Inform. 2020;4:383–391. doi: 10.1200/CCI.19.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Luo L, Wang Y, Wampfler J, Yang P, Liu H. Natural language processing for populating lung cancer clinical research data. BMC Med Inform Decis Mak. 2019;19(5):1–10. doi: 10.1186/s12911-018-0723-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltagy I, Peters ME, Cohan A. Longformer: The long-document transformer. arXiv preprint. arXiv:2004.05150. 2020.
- 11.Devlin J, Chang M-W. Open sourcing BERT: State-of-the-art pre-training for natural language processing. Google AI Blog. https://ai.googleblog.com/2018/11/open-sourcing-bert-state-of-art-pre.html. 2020. Accessed 28 Jul 2022.
- 12.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint. arXiv:1810.04805. 2018.
- 13.Arbour KC, Luu AT, Luo J, et al. Deep learning to estimate RECIST in patients with NSCLC treated with PD-1 blockade. Cancer Discov. 2021;11(1):59–67. doi: 10.1158/2159-8290.CD-20-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rown T, Mann B, Ryder N, et al. Language models are few-shot learners. Adv Neural Inf Process Syst. 2020;33:1877–901. [Google Scholar]
- 15.Jin P, Ji X, Kang W, et al. Artificial intelligence in gastric cancer: a systematic review. J Cancer Res Clin Oncol. 2020;146(9):2339–2350. doi: 10.1007/s00432-020-03304-9. [DOI] [PubMed] [Google Scholar]
- 16.Sheikhalishahi S, Miotto R, Dudley JT, Lavelli A, Rinaldi F, Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7(2):e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshihara H. Millennial medical record project toward establishment of authentic Japanese version EHR and secondary use of medical data. J Inform Process Manag. 2018;60(11):767–778. [Google Scholar]
- 18.Yoshihara H. Millennial medical record project: secondary use of medical data for research and development based on the next generation medical infrastructure law. Jpn J Pharmacoepidemiol. 2022;27(1):3–10. doi: 10.3820/jjpe.27.3. [DOI] [Google Scholar]
- 19.Personal Information Protection Commission Japan. Act on the Protection of Personal Information. 2020. https://www.ppc.go.jp/en/legal/. Accessed 20 Dec 2022.
- 20.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Google. Pre-train procedure. https://github.com/google. Accessed 20 Dec 2022.
- 22.Chase HS, Mitrani LR, Lu GG, Fulgieri DJ. Early recognition of multiple sclerosis using natural language processing of the electronic health record. BMC Med Inform Decis Mak. 2017;17(1):24. doi: 10.1186/s12911-017-0418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metsis V, Androutsopoulos I, Paliouras G. Spam filtering with naive Bayes-which naive Bayes? In CEAS. 2006;17:28–69. [Google Scholar]
- 24.Griffith SD, Tucker M, Bowser B, et al. Generating real-world tumor burden endpoints from electronic health record data: comparison of RECIST, radiology-anchored, and clinician-anchored approaches for abstracting real-world progression in non-small cell lung cancer. Adv Ther. 2019;36(8):2122–2136. doi: 10.1007/s12325-019-00970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmy L, Xiang Y, Xie Z, Tao C, Zhi D. Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. NPJ Digit Med. 2021;4(1):1–13. doi: 10.1038/s41746-021-00455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfiky AA, Pany MJ, Parikh RB, Obermeyer Z. Development and application of a machine learning approach to assess short-term mortality risk among patients with cancer starting chemotherapy. JAMA Netw Open. 2018;1(3):e180926–e180926. doi: 10.1001/jamanetworkopen.2018.0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Q, Cai T, Hong C, et al. Performance of a machine learning algorithm using electronic health record data to identify and estimate survival in a longitudinal cohort of patients with lung cancer. JAMA Netw Open. 2021;4(7):e2114723. doi: 10.1001/jamanetworkopen.2021.14723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meropol NJ, Donegan J, Rich AS. Progress in the application of machine learning algorithms to cancer research and care. JAMA Netw Open. 2021;4(7):e2116063. doi: 10.1001/jamanetworkopen.2021.16063. [DOI] [PubMed] [Google Scholar]
- 29.Xiang D, Cai W. Privacy protection and secondary use of health data: strategies and methods. Biomed Res Int. 2021;2021:6967166. doi: 10.1155/2021/6967166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (JPG 232 KB) Figure S1: Calculation of precision, recall, and F1 score to assess the model performance
Supplementary file2 (TIF 1595 KB) Figure S2: Time to progression for each treatment line. TTP, time to progression; CI, confidence interval
Data Availability Statement
The data from the LDI data set supporting the findings of this study may be made available upon request and are subject to approval by the review board of LDI and a license agreement with the General Incorporated Association Life Data Initiative. Data from the UMH data set supporting the findings of this study may be made available upon request and are subject to approval by the ethics committee of the University of Miyazaki.