Abstract
Age-related (AR) hearing loss (HL) is the most common sensory impairment with heritability of 55%. The aim of this study was to identify genetic variants on chromosome X associated with ARHL through the analysis of data obtained from the UK Biobank. We performed association analysis between self-reported measures of HL and genotyped and imputed variants on chromosome X from ∼460,000 white Europeans. We identified three loci associated with ARHL with a genome-wide significance level (p < 5 × 10−8), ZNF185 (rs186256023, p = 4.9 × 10−10) and MAP7D2 (rs4370706, p = 2.3 × 10−8) in combined analysis of males and females, and LOC101928437 (rs138497700, p = 8.9 × 10−9) in the sex-stratified analysis of males. In-silico mRNA expression analysis showed MAP7D2 and ZNF185 are expressed in mice and adult human inner ear tissues, particularly in the inner hair cells. We estimated that only a small amount of variation of ARHL, 0.4%, is explained by variants on the X chromosome. This study suggests that although there are likely a few genes contributing to ARHL on the X chromosome, the role that the X chromosome plays in the etiology of ARHL may be limited.
Keywords: age-related hearing loss, MAP7D2, LOC101928437, ZNF185, UK Biobank, X chromosome
1 Introduction
Age-related (AR) hearing Loss (HL), as known as presbycusis, is the most frequent sensory impairment affecting 25%–40% of individuals ≥65 years of age (Wolber et al., 2012). The prevalence of ARHL increases with age (Salvi et al., 2018), with males having a higher prevalence than females (Van Eyken et al., 2007). With lifespan increasing, especially in developed countries, the impact of ARHL will continue to increase.
ARHL is a complex disorder impacted by risk factors such as sex, noise exposure, ototoxic medications, and genetic susceptibility (Wiley et al., 2008; Bowl & Dawson, 2019). ARHL is also associated with several comorbidities, including cognitive decline, depression, diabetes, and hypertension (Yuan et al., 2018; Ponticorvo et al., 2019). Twin and family studies have shown that the heritability of ARHL ranges from 0.35 to 0.55 (Christensen et al., 2001; Duan et al., 2019). Genome-wide association studies (GWAS) have identified several genetic loci associated with ARHL [e.g., ARHGEF28 (MIM: 612790), CHMP4C (MIM: 610899), ZNF318 (MIM: 617512)] (Hoffmann et al., 2016; Wells et al., 2019).
To date, six early-onset non-syndromic HL genes and 21 syndromic HL genes have been identified on the X chromosome (Corvino et al., 2018); however, no X-linked genes or loci have been reported to be involved in etiology of ARHL. Although the X chromosome is the eighth largest chromosome and contains ∼5% of human genes (König et al., 2014) it is usually excluded from GWAS analyses (Wise et al., 2013).
Here we focused on examining the role of variants on chromosome X in ARHL through the analysis of genotype array and imputed data and self-reported measures of HL obtained from ∼460,000 white-European UK Biobank volunteers.
In this first report of the role of the X chromosome in ARHL, associations were identified between ARHL and variants near ZNF185, MAP7D2, and LOC101928437. In-silico analysis revealed inner ear expression of MAP7D2/Map7d2 and ZNF185/Zfp185 in mice and humans, particularly for the inner ear hair cells. Additionally, the estimated heritability of ARHL due to single nucleotide variants (SNVs) on the X chromosome with a minor allele frequency (MAF) > 0.01 is 0.4%.
2 Materials and methods
2.1 Data access and ethical approval
This research was conducted using the UK Biobank Resource (application numbers 32285 and 36,827). The UK Biobank study was conducted under generic approval from the National Health Services’ National Research Ethics Service. The present analyses were approved by the Institutional Review Boards at Yale University (2000026836) and Columbia University (AAAS3494).
2.2 Study population
The UK Biobank is a cohort of ∼500,000 volunteers between 40 and 69 years-of-age at the time of recruitment from 2006 to 2010 and are being followed-up for at least 20 years (Sudlow et al., 2015). Detailed information about the UK Biobank can be found elsewhere (Sudlow et al., 2015; Bycroft et al., 2018).
2.3 Genotyping, imputation, and quality control
The UK Biobank samples were genotyped on two arrays: Affymetrix UK BiLEVE Axiom array (50,000 individuals) and the Affymetrix UK Biobank Axiom array (450,000 individuals) (Bycroft et al., 2018). The two arrays share 733,322 autosomal and 20,214 X chromosome variants. Genotypes were phased using SHAPEIT3 (O’Connell et al., 2016) and imputation was performed with IMPUTE4 (Bycroft et al., 2018) using reference data from the Haplotype Reference Consortium and UK10K (Huang et al., 2015; McCarthy et al., 2016). The pseudoautosomal regions (PARs) and non-pseudoautosomal region (non-PAR) were phased and imputed independently. Genotyping, haplotype phasing, and imputation have been previously described (Bycroft et al., 2018).
Individuals with putative sex chromosome aneuploidy, inconsistent sex (reported sex did not match genetic sex) or were missing >3% of their genotype array data were removed. Analysis was restricted to individuals of white-European ancestry (N = 459,267) based upon principal components analysis. The genotype variants with call rate >95%, Hardy Weinberg Equilibrium p-value < 5.0 × 10−8 (estimated in non-PAR region in females and the variants out of HWE were removed in both females and males), and minor allele frequency (MAF) > 0.001 were used for whole-genome ridge regression (N = 442,313, Supplementary Table S1) implemented in REGENIE (Mbatchou et al., 2021). Principal components (PCs) of genetic data to include in association analysis were generated using a subset of genotyped markers (N = 144,905) that were pruned for linkage disequilibrium (LD) (r 2 > 0.1) (Supplementary Table S1).
SNVs on the X chromosome obtain from the genotype arrays that did not deviate from HWE and had a MAF>0.001 (1,207 PAR and 11,653 non-PAR) were analyzed. Imputed X chromosome SNVs and insertion/deletions (InDels) with a MAF>0.001 and INFO Score ≥0.3 (45,519 PAR and 2,050,039 non-PAR) were also analyzed. To estimate the contribution of common variants on X chromosome to heritability of ARHL we analyzed variants obtained from the genotype array data with MAF>0.01 in the non-PAR region (N = 18,773).
2.4 Phenotype definition
Using ICD10/ICD9 codes and self-reports, individuals with chronic otitis media, salpingitis, mastoiditis, otosclerosis, Meniere’s disease, deafness, labyrinthitis, conductive HL, ototoxic HL, head, ear or neck trauma, stroke, encephalitis, meningitis, or facial nerve disorders were excluded from the analysis. Additionally, individuals with unilateral/bilateral sensorineural or mixed conductive HL were excluded if they were diagnosed <55 years-of-age or did not have an age-of-diagnosis (Supplementary Table S2).
Four different case status were determined based on the response to a touchscreen questionnaire: 1) H-aid self-reported hearing aid use (f.3393: “Do you use a hearing aid most of the time?“); 2) H-diff self-reported hearing difficulty (f.2247: “Do you have any difficulty with your hearing?”); 3) H-noise self-reported hearing difficulty with background noise (f.2257: “Do you find it difficult to follow a conversation if there is background noise, e.g., TV, radio, children playing)?”; and 4) H-both individuals with both H-diff and H-noise. Individuals who provided inconsistent answers for H-aid, H-diff, or H-noise (e.g., reported H-diff on the first visit but not the second visit), or did not provide definite answers, (e.g., answered “Do not know”) were excluded (Supplementary Table S1). In the analysis age is defined for cases, when they answered “Yes” to a specific ARHL question for the first time during the assessments. We used a common set of controls without any hearing-related phenotypes. For the controls, age at last assessment was used in the analysis.
Information on the number of individuals available for analysis for each phenotype (H-aid, H-diff, H-noise, and H-both) as well as information on the age distribution among males and females can be found in Supplementary Table S3.
2.5 Association analysis
Univariate analysis was performed using a generalized linear mixed model (GLMM) as implemented in REGENIE 2.2.4 (Mbatchou et al., 2021). REGENIE uses a multi-stage approach by initially fitting a whole-genome regression model via ridge regression to estimate the polygenic background accounting for population structure and relatedness. A firth correction for binary traits is applied to calibrate the model in the presence of low frequency variants and unbalanced case-control ratios, as is the case for this analysis (i.e., H-aid 1:15 cases to controls). In step 2, score tests for association were performed for each genotyped and imputed variants with MAF >0.001 on the X chromosome. Genotypes for PAR X chromosomal variants were coded {0, 1, 2} for both males and females, whereas for non-PAR X chromosomal variants males were coded as {0, 2}. Imputed dosages were used for imputed genotypes. An additive effect model was used in the association analysis to adjust for age, sex, array (BiLEVE and Axiom), and two PCs. For the non-PAR variants, a sex-stratified analysis was also performed using the same model.
2.6 Heritability
Heritability was estimated for variants on the X chromosome using GCTA implementing a restricted maximum likelihood (REML) approach (Yang et al., 2011). The genetic relatedness matrix (GRM) was calculated using X chromosome genotyped variants. Age, sex, array type (BiLEVE and Axiom), and two PCs were included as covariates.
2.7 In-silico mRNA expression analysis
Human and mouse inner ear gene expressions were assessed in silico. To study expression during mouse inner ear development, an otic progenitor cells dataset was obtained from the Shared Harvard Inner-Ear Laboratory Database (SHIELD) (Shen et al., 2015). The Expression Analysis Resource (gEAR) was also used to visualize single cell RNA-seq data of the cochlear epithelium during four mouse developmental stages [embryonic day (E)14, E16, postnatal (P)1, and P7] (Kolla et al., 2020; Orvis et al., 2021) and of mouse otic neuronal lineages at three embryonic ages, embryonic day 9.5 (E9.5), E11.5, and E13.5 (Sun et al., 2022). Processing and normalization of expression values were performed using Seurat (Stuart et al., 2019) and single cells were grouped into cell clusters (Kolla et al., 2020; Sun et al., 2022). Finally, human inner ear expression data were obtained from RNA sequencing of inner ear tissue samples from three donor patients ages 45–60 (Schrauwen et al., 2016) and were processed and normalized using DESeq2 (Love et al., 2014).
3 Results
3.1 Case-control samples for association analysis
The number of cases and controls for each of the four ARHL traits are displayed in Supplementary Table S3. There is a higher proportion of male cases than female cases for each trait (two-sample z-test for proportions p < 2.23 × 10−308, Supplementary Table S3). Also, the mean age of males with HL was higher than females (two-sample independent t-test p < 2.81 × 10−33, Supplementary Table S3).
3.2 Detection of ARHL-associated variants
3.2 1 Non-PAR region
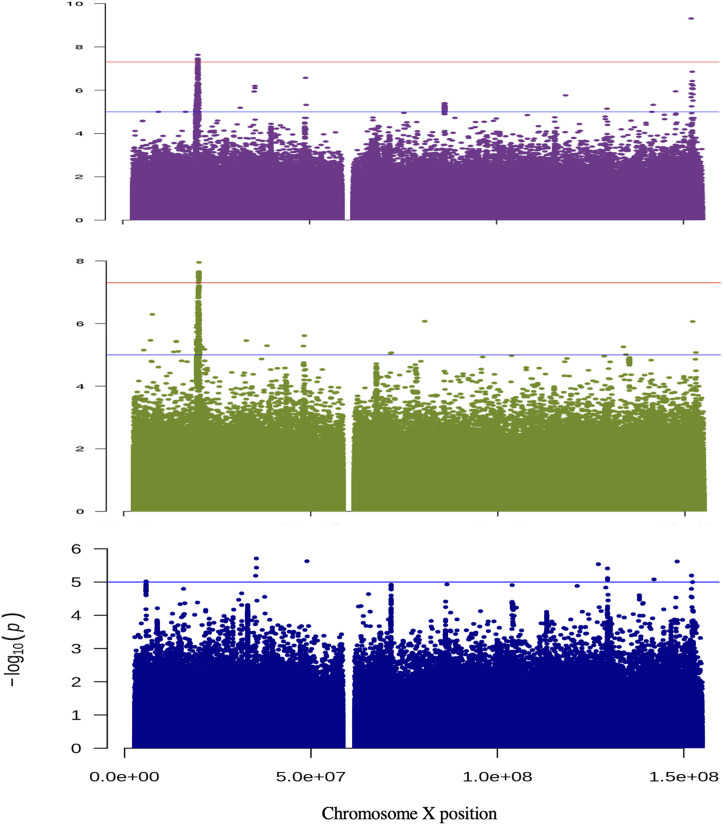
In the combined sex analysis for H-both, nine variants in two independent genomic regions reached genome-wide significance (Table 1 and Figure 1). The first region’s most significant variant is rs186256023 near ZNF185 [p = 4.9 × 10−10, odds ratio (OR) = 1.08 with confidence interval (CI) = 1.062–1.105, MAF = 0.03] and the other genomic region’s most significant variant is rs4370706 near MAP7D2 [p = 2.3 × 10−8, OR = 1.03 (CI = 1.010–1.051), MAF = 0.18]. For the other three traits, no genome-wide statistically significant variants were identified. Loci that displayed suggestive associations (5.0 × 10−8>p< 1.0 × 10−5) are shown in Supplementary Figure S1 and in Supplementary Tables S4–S7.
TABLE 1.
Variants associated with age related hearing loss.
| Combined | Females | Males | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-both | Variant | Pos | Locus | A1 | Info | Freq | Beta (OR) | SE | P | N | Beta (OR) | P | N | Beta (OR) | P | N | |
| rs186256023 | 152067381 | ZNF185 | C | 0.92 | 0.03 | 0.08 (1.083) | 0.01 | 4.9E-10 | 330,558 | 0.11 (1.116) | 8.6E-07 | 182,676 | 0.07 (1.072) | 6.4E-06 | 147,882 | ||
| rs4370706 | 20,102,906 | MAP7D2 | T | 0.99 | 0.18 | 0.03 (1.030) | 0.01 | 2.3E-08 | 330,558 | 0.05 (1.051) | 6.4E-08 | 182,676 | 0.02 (1.020) | 2.5E-03 | 147,882 | ||
| rs7882991 | 20,054,650 | MAP7D2 | A | 0.98 | 0.18 | 0.03 (1.030) | 0.01 | 3.4E-08 | 330,558 | 0.05 (1.051) | 3.3E-08 | 182,676 | 0.02 (1.020) | 4.2E-03 | 147,882 | ||
| rs10081918 | 20,053,979 | MAP7D2 | T | 0.98 | 0.18 | 0.03 (1.030) | 0.01 | 3.6E-08 | 330,558 | 0.05 (1.051) | 3.1E-08 | 182,676 | 0.02 (1.020) | 4.4E-03 | 147,882 | ||
| rs5955880 | 20,084,743 | MAP7D2 | A | 0.99 | 0.18 | 0.03 (1.030) | 0.01 | 3.9E-08 | 330,558 | 0.06 (1.062) | 2.2E-08 | 182,676 | 0.02 (1.020) | 5.2E-03 | 147,882 | ||
| rs6527961 | 20,109,072 | MAP7D2 | A | 0.99 | 0.18 | 0.03 (1.030) | 0.01 | 3.9E-08 | 330,558 | 0.06 (1.062) | 2.3E-08 | 182,676 | 0.02 (1.020) | 5.1E-03 | 147,882 | ||
| rs12009531 | 20,074,441 | MAP7D2 | A | 0.99 | 0.18 | 0.03 (1.030) | 0.01 | 4.0E-08 | 330,558 | 0.06 (1.062) | 2.4E-08 | 182,676 | 0.02 (1.020) | 5.2E-03 | 147,882 | ||
| rs7062127 | 20,061,058 | MAP7D2 | G | 0.98 | 0.18 | 0.03 (1.030) | 0.01 | 4.0E-08 | 330,558 | 0.05 (1.051) | 8.9E-08 | 182,676 | 0.02 (1.020) | 3.6E-03 | 147,882 | ||
| rs60849679 | 20,116,215 | MAP7D2 | A | 1 | 0.18 | 0.03 (1.030) | 0.01 | 4.5E-08 | 330,558 | 0.05 (1.051) | 3.7E-08 | 182,676 | 0.02 (1.020) | 4.1E-03 | 147,882 | ||
| rs5955882 | 20,110,340 | MAP7D2 | T | 0.98 | 0.18 | 0.03 (1.030) | 0.01 | 5.7E-08 | 330,558 | 0.06 (1.062) | 2.7E-08 | 182,676 | 0.02 (1.020) | 6.4E-03 | 147,882 | ||
| rs7064198 | 20,101,842 | MAP7D2 | T | 0.99 | 0.18 | 0.03 (1.030) | 0.01 | 5.3E-08 | 330,558 | 0.06 (1.062) | 2.9E-08 | 182,676 | 0.02 (1.020) | 6.1E-03 | 147,882 | ||
| H-aid | rs138497700 | 112,591,946 | LOC101928437 | G | 0.83 | 0.003 | 2.00 (7.39) | 0.58 | 4.1E-04 | 252,895 | −1.17 (0.310) | 0.228 | 148,632 | 2.6 (13.46) | 8.9E-09 | 104,263 | |
| rs141853173 | 112,449,530 | LOC101928437 | T | 0.84 | 0.003 | 1.85 (6.36) | 0.61 | 4.3E-04 | 252,895 | −0.89 (0.411) | 0.312 | 148,632 | 2.6 (13.46) | 2.3E-08 | 104,263 | ||
Abbreviations; Pos, Position in x chromosome; A1, Effect allele; Info, Imputation quality; Freq, Frequency of effect allele based on combined samples; OR, Odd ratio; SE, Standard error; P, p-Value; N, Number of included individuals.
FIGURE 1.
Manhattan plots for the analysis of non-pseudo autosomal region of the X chromosome. The results are shown for the analysis of H-Both: males and females (purple); females only (green) and males only (blue). The y-axis displays −log10 for each variant tested and the X-axis their position. The horizontal red and blue lines show the threshold for genome-wide significant (p = 5 × 10−8) and suggestive (p = 1 × 10−5) associations, respectively.
For the sex-stratified analysis of H-both, in females there are eight genome-wide significant variants within the MAP7D2 genomic region, but none the variants had a p < 10−4 in males (Table 1 and Figure 1). Of the eight variants, rs5955880 that is near MAP7D2 is the most significant [p = 2.2 × 10−8, OR = 1.06 (CI = 1.041–1.083)]. This same variant has a p = 5.2 × 10−3 in males [OR = 1.02 (CI = 1.006–1.034)].
For the sex-stratified analysis of H-aid there are two genome-wide significant variants in one genomic region in males, but none reached nominal significance in females (Table 1). The most significant SNV, rs138497700, near LOC101928437 has a p = 8.9 × 10−9 [OR = 13.46 (CI = 5.92–30.61), MAF = 0.003]. This SNV has an OR = 0.31 (CI = 0.05–2.08) and p = 0.23 in females.
We found no genome-wide significant variants in the sex-stratified analyses for H-diff and H-noise. Loci with suggestive associations (5.0 × 10−8>p< 1.0 × 10−5) are shown in Supplementary Figure S1 and Supplementary Tables S8–S15.
3.2 2 PAR regions
Among ∼45,000 genotyped and imputed variants in PAR regions, none reached genome-wide or suggestive significance in the combined or sex-specific analyses for the four ARHL traits (Supplementary Figure S3).
3.3 Heritability
The overall proportion of H-both variations explained by genotyped variants on the X chromosome was 0.4% [standard error (SE) = 0.8%, p = 3.5 × 10−6). The heritability estimates for the other ARHL traits were small and not statistically significant (p > 0.05).
3.4 In-silico mRNA expression analysis
We investigated mRNA inner ear expression for MAP7D2/Map7d2 and ZNF185/Zfp185 since they are new candidate genes for HL. Single-cell RNA (scRNA) experiments of the developing mouse cochlear epithelium show that Map7d2 and Zfp185 are both expressed in the cochlear epithelium in newborn mice (P1 developmental stage; Supplementary Figure S5). In addition, analysis of a scRNA dataset of mouse otic neuronal lineage cells (Sun et al., 2022) shows that Map7d2 is also expressed in cochleovestibular ganglion cells at E13.5 (Supplementary Figure S5). Zfp185 was not found to be expressed in otic neuronal lineage cells. Furthermore, in immortalized multipotent otic progenitor (iMOP) cells, Map7d2 expression showed a 2.5-fold upregulation when cells were cultured in epidermal growth factor (EGF) (Supplementary Table S16). Finally, MAP7D2 and ZNF185 were also found to be expressed in the adult human inner ear (Supplementary Figure S6). For LOC101928437, a non-coding RNA, there are no human or mouse inner ear data available to evaluate its expression (no known orthologue).
4 Discussion
Analyzing UK Biobank data, we studied associations between X chromosome variants and ARHL. We identified three genes (MAP7D2, LOC101928437, and ZNF185) on the X chromosome associated with ARHL.
The genomic control lambda values for all analyses show slight genomic inflation (λ = 1.02–1.09, Supplementary Figures S2, S4), consistent with other genetic analyses of datasets with large sample sizes (Yang et al., 2011). Additionally, we are only reporting results for a single chromosome, and the X chromosome has more LD than the autosomes since recombination events only occurs in women outside of the PAR (Schaffner, 2004). There may also be polygenic effects which can lead to an increased λ GC in the absence of genuine inflation (Yang et al., 2011).
We found an association between ARHL and ZNF185, which encodes an actin-cytoskeleton-associated Lin-l 1, Isl-1 and Mec-3 (LIM) domain-containing protein (Heiss et al., 1997). In keratinocytes and epidermis ZNF185 has been described as highly expressed in differentiating conditions, physically interacting with E-cadherin, a component of the adherens junctions, one of the critical cell-cell adhesive complexes crucial in the pluristratified epithelia (Smirnov et al., 2019). Although its role in the inner ear is unknown, we demonstrate it is expressed in the mouse cochlear epithelium and in adult human inner ear tissues (Supplementary Figures S5, S6). Furthermore, znf185 was found as a scRNA-seq cluster marker (gene that distinguish the different cell clusters) of neuromast support cells in zebrafish fgf3 mutants, which have increased hair cell regeneration (Lush et al., 2019).
In addition, we identified association between MAP7D2 and H-both in the combined sex analysis. The combined OR for variants in MAP7D2 was 1.03 (CI = 1.016–1.045). The MAP7 Domain Containing 2 (MAP7D2) is one of the members of the MAP7 family. It plays a vital role in regulating kinesin-1, promoting microtubule-based transport of numerous cellular cargoes entry into the axon (Hooikaas et al., 2019; Kikuchi et al., 2022). We demonstrate it is expressed in the cochlear sensory epithelium in mice, particularly in the inner and outer hair cells, in developing cochleovestibular ganglion cells and adult human inner ear tissues. In the latter, it is preferentially expressed in the cochlear duct which houses the sensory epithelium (Supplementary Figures S6). Furthermore, Map7d2 expression is upregulated when otic progenitor (iMOP) cells are cultured in EGF, a growth factor important in the growth and differentiation of hair cells (Doetzlhofer et al., 2004). This suggests Map7d2 may have a potential role in hair cell growth and differentiation.
Two variants in non-coding RNA, LOC101928437, were associated with H-aid in males. We could not evaluate inner ear expression of this non-coding RNA, since the data is unavailable. This non-coding RNA has been previously suggested to play a role in non-syndromic intellectual disability and shows a high level of brain specific expression (Zhou et al., 2015).
Although ARHL is a common disorder, little is known about the genetic susceptibility of this disorder. There is strong evidence of a difference in the prevalence of ARHL between males and females which is also observed in the UK Biobank (Wiley et al., 2008; Bowl and Dawson, 2019). Our results indicate that X chromosome SNVs are unlikely to be a factor in the observed higher prevalence in males than females. It has been previously suggested that these differences, at least partially, are due to hormonal pathways (Tadros et al., 2005; Hultcrantz et al., 2006; Souza et al., 2017).
One primary characteristic of the X chromosome in mammals is that female embryos undergo silencing of one of the two X chromosomes to equalize the dosage of X-linked genes between females and males (Lyon, 1961). This is extremely relevant to phenotypic changes, as mutations that could lead to severe complications in males are compensated by another chromosome in 50% of the female cells, which can confer protection against diseases (Cantone and Fisher, 2017). While many X-linked genes undergo X chromosome inactivation (XCI), some degree of expression heterogeneity among females has been reported: 15% of X-linked genes escape inactivation and 10% of X-linked genes exhibit variable patterns of inactivation (Carrel and Willard, 2005). Skewed XCI is present when one of the cell types is markedly in excess and may be a primary event due to chance or genetic factors (Puck and Willard, 1998) or secondary due to selection against or in favor of cells with a given genotype (Migeon and Haisley-Royster, 1998). Skewed XCI can cause phenotypic variability in both ways; it can either lead to a more severe phenotype or the expression of disease in females (Shoukat et al., 2020) or can act protective and reduce/avoid disease manifestations (Migeon, 2020). Skewed XCI is dependent on tissue and age (Zito et al., 2019), for example, the frequency of skewed XCI in peripheral blood cells increases with age, possibly due to selection (Hatakeyama et al., 2004; Kristiansen et al., 2005). Age-related X reactivation and escape XCI may also occur, and these processes could be related to neurological and oncological disease (Leppig and Disteche, 2001). It is known that skewing and escaping of XCI can occur in normal females and increases in tissues with age, therefore this process may also be involved in age-related diseases in females.
Large-scale volunteer databanks typically have limitations that include self-selection basis, no or limited numbers of children and older individuals, and study subjects mainly of European ancestry. We analyzed data from the UK Biobank for which the percent of females and average age is higher than that of the general UK population (Fry et al., 2017). HL phenotypes used in this study were self-reported. Unlike audiometric measurements, they can be less accurate and neither captured the severity nor the frequencies affected. Additionally, due to relatively small numbers of individuals of non-European ancestry, analysis was limited to white Europeans; therefore the results of this study may not be applicable to individuals from other ancestry groups.
In conclusion, we have identified significant associations between variants on chromosome X near ZNF185, MAP7D2, and LOC101928437 with ARHL in white-Europeans. The identified variants need to be replicated in an independent sample. Additionally, it would be beneficial to determine if these genes play a role in ARHL in individuals of non-European ancestry. Although common variants on the X chromosome are involved in the etiology of ARHL, based on heritability estimates their role is likely limited.
Acknowledgments
The research conducted using the United Kingdom Biobank Resource was performed under application numbers 32285 ATD and 36827 SML.
Funding Statement
This work was supported by a grant from the National Institute of Deafness and other Communication Disorders (NIDCD) DC017712 to SML and ATD.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
EN analyzed UK Biobank data and drafted the manuscript; DMC-S and GL extracted and prepared data for analysis; IS and GTW performed data interpretation and reviewed the manuscript; ATD designed experiments; revised the manuscript; and obtained funding; SML designed experiments, drafted, and finalized manuscript and obtained funding. All the authors agreed to the final version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1106328/full#supplementary-material
Abbreviations
AR, age-related; HL, hearing loss; GWAS, Genome-wide association studies; SNP, single nucleotide polymorphism; PARs, pseudoautosomal regions; non-PAR, non-pseudoautosomal region; MAF, minor allele frequency; HWE, Hardy Weinberg Equilibrium; LD, linkage disequilibrium; InDels, insertion/deletions; H-aid, hearing aid; H-diff, hearing difficulty; H-noise, hearing noise; H-both, hearing difficulty and hearing noise; OR, odds ratio; CI, confidence interval; SE, standard error; XCI, X chromosome inactivation.
References
- Bowl M. R., Dawson S. J. (2019). Age-related hearing loss. Cold Spring Harb. Perspect. Med. 9 (8), a033217. 10.1101/cshperspect.a033217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C., Freeman C., Petkova D., Band G., Elliott L. T., Sharp K., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562 (7726), 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone I., Fisher A. G. (2017). Human X chromosome inactivation and reactivation: Implications for cell reprogramming and disease. Philosophical Trans. R. Soc. Lond. Ser. B, Biol. Sci. 372 (1733), 20160358. 10.1098/rstb.2016.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L., Willard H. F. (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434 (7031), 400–404. 10.1038/nature03479 [DOI] [PubMed] [Google Scholar]
- Christensen K., Frederiksen H., Hoffman H. J. (2001). Genetic and environmental influences on self-reported reduced hearing in the old and oldest old. J. Am. Geriatrics Soc. 49 (11), 1512–1517. 10.1046/j.1532-5415.2001.4911245.x [DOI] [PubMed] [Google Scholar]
- Corvino V., Apisa P., Malesci R., Laria C., Auletta G., Franzé A. (2018). X-linked sensorineural hearing loss: A literature review. Curr. Genomics 19 (5), 327–338. 10.2174/1389202919666171218163046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer A., White P. M., Johnson J. E., Segil N., Groves A. K. (2004). In vitro growth and differentiation of mammalian sensory hair cell progenitors: A requirement for EGF and periotic mesenchyme. Dev. Biol. 272 (2), 432–447. 10.1016/j.ydbio.2004.05.013 [DOI] [PubMed] [Google Scholar]
- Duan H., Zhang D., Liang Y., Xu C., Wu Y., Tian X., et al. (2019). Heritability of age-related hearing loss in middle-aged and elderly Chinese: A population-based twin study. Ear Hear. 40 (2), 253–259. 10.1097/AUD.0000000000000610 [DOI] [PubMed] [Google Scholar]
- Fry A., Littlejohns T. J., Sudlow C., Doherty N., Adamska L., Sprosen T., et al. (2017). Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am. J. Epidemiol. 186 (9), 1026–1034. 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama C., Anderson C. L., Beever C. L., Peñaherrera M. S., Brown C. J., Robinson W. P. (2004). The dynamics of X-inactivation skewing as women age. Clin. Genet. 66 (4), 327–332. 10.1111/j.1399-0004.2004.00310.x [DOI] [PubMed] [Google Scholar]
- Heiss N. S., Gloeckner G., Bächner D., Kioschis P., Klauck S. M., Hinzmann B., et al. (1997). Genomic structure of a novel LIM domain gene (ZNF185) in Xq28 and comparisons with the orthologous murine transcript. Genomics 43 (3), 329–338. 10.1006/geno.1997.4810 [DOI] [PubMed] [Google Scholar]
- Hoffmann T. J., Keats B. J., Yoshikawa N., Schaefer C., Risch N., Lustig L. R. (2016). A large genome-wide association study of age-related hearing impairment using electronic Health records. PLoS Genet. 12 (10), e1006371. 10.1371/journal.pgen.1006371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooikaas P. J., Martin M., Mühlethaler T., Kuijntjes G.-J., Peeters C. A. E., Katrukha E. A., et al. (2019). MAP7 family proteins regulate kinesin-1 recruitment and activation. J. Cell Biol. 218 (4), 1298–1318. 10.1083/jcb.201808065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J. L., Danecek P., Malerba G., Trabetti E., Zheng H.-F., Gambaro G., Richards J. B., Durbin R., Timpson N. J., Marchini J., Soranzo N. UK10K Consortium (2015). Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 6, 8111. 10.1038/ncomms9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultcrantz M., Simonoska R., Stenberg A. E. (2006). Estrogen and hearing: A summary of recent investigations. Acta Oto-Laryngologica 126 (1), 10–14. 10.1080/00016480510038617 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Sakamoto Y., Uezu A., Yamamoto H., Ishiguro K.-I., Shimamura K., et al. (2022). Map7D2 and Map7D1 facilitate microtubule stabilization through distinct mechanisms in neuronal cells. Life Sci. Alliance 5 (8), e202201390. 10.26508/lsa.202201390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolla L., Kelly M. C., Mann Z. F., Anaya-Rocha A., Ellis K., Lemons A., et al. (2020). Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 11 (1), 2389. 10.1038/s41467-020-16113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- König I. R., Loley C., Erdmann J., Ziegler A. (2014). How to include chromosome X in your genome-wide association study. Genet. Epidemiol. 38 (2), 97–103. 10.1002/gepi.21782 [DOI] [PubMed] [Google Scholar]
- Kristiansen M., Knudsen G. P. S., Bathum L., Naumova A. K., Sørensen T. I. A., Brix T. H., et al. (2005). Twin study of genetic and aging effects on X chromosome inactivation. Eur. J. Hum. Genet. EJHG 13 (5), 599–606. 10.1038/sj.ejhg.5201398 [DOI] [PubMed] [Google Scholar]
- Leppig K. A., Disteche C. M. (2001). Ring X and other structural X chromosome abnormalities: X inactivation and phenotype. Seminars Reproductive Med. 19 (2), 147–157. 10.1055/s-2001-15395 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lush M. E., Diaz D. C., Koenecke N., Baek S., Boldt H., St Peter M. K., et al. (2019). ScRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. ELife 8, e44431. 10.7554/eLife.44431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373. 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- Mbatchou J., Barnard L., Backman J., Marcketta A., Kosmicki J. A., Ziyatdinov A., et al. (2021). Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53 (7), 1097–1103. 10.1038/s41588-021-00870-7 [DOI] [PubMed] [Google Scholar]
- McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A. R., Teumer A., et al. (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48 (10), 1279–1283. 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B. R., Haisley-Royster C. (1998). Familial skewed X inactivation and X-linked mutations: Unbalanced X inactivation is a powerful means to ascertain X-linked genes that affect cell proliferation. Am. J. Hum. Genet. 62 (6), 1555–1557. 10.1086/301858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon B. R. (2020). X-linked diseases: Susceptible females. Genet. Med. Official J. Am. Coll. Med. Genet. 22 (7), 1156–1174. 10.1038/s41436-020-0779-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J., Sharp K., Shrine N., Wain L., Hall I., Tobin M., et al. (2016). Haplotype estimation for biobank-scale data sets. Nat. Genet. 48 (7), 817–820. 10.1038/ng.3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J., Gottfried B., Kancherla J., Adkins R. S., Song Y., Dror A. A., et al. (2021). gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Methods 18 (8), 843–844. 10.1038/s41592-021-01200-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticorvo S., Manara R., Pfeuffer J., Cappiello A., Cuoco S., Pellecchia M. T., et al. (2019). Cortical pattern of reduced perfusion in hearing loss revealed by ASL-MRI. Hum. Brain Mapp. 40 (8), 2475–2487. 10.1002/hbm.24538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puck J. M., Willard H. F. (1998). X inactivation in females with X-linked disease. N. Engl. J. Med. 338 (5), 325–328. 10.1056/NEJM199801293380611 [DOI] [PubMed] [Google Scholar]
- Salvi R., Ding D., Jiang H., Chen G.-D., Greco A., Manohar S., et al. (2018). Hidden age-related hearing loss and hearing disorders: Current knowledge and future directions. Hear. Balance Commun. 16 (2), 74–82. 10.1080/21695717.2018.1442282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S. F. (2004). The X chromosome in population genetics. Nat. Rev. Genet. 5 (1), 43–51. 10.1038/nrg1247 [DOI] [PubMed] [Google Scholar]
- Schrauwen I., Hasin-Brumshtein Y., Corneveaux J. J., Ohmen J., White C., Allen A. N., et al. (2016). A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear. Res. 333, 266–274. 10.1016/j.heares.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Scheffer D. I., Kwan K. Y., Corey D. P. (2015). Shield: An integrative gene expression database for inner ear research. Database J. Biol. Databases Curation 2015, bav071. 10.1093/database/bav071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukat H. M. H., Ghous G., Tarar Z. I., Shoukat M. M., Ajmal N. (2020). Skewed inactivation of X chromosome: A cause of hemophilia manifestation in carrier females. Cureus 12 (10), e11216. 10.7759/cureus.11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A., Lena A. M., Cappello A., Panatta E., Anemona L., Bischetti S., et al. (2019). ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene 38 (10), 1625–1638. 10.1038/s41388-018-0509-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza D. da S., Luckwu B., Andrade W. T. L. de, Pessoa L. S. de F., Nascimento J. A. do, Rosa M. R. D. da. (2017). Variation in the hearing threshold in women during the menstrual cycle. Int. Archives Otorhinolaryngology 21 (4), 323–328. 10.1055/s-0037-1598601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M., et al. (2019). Comprehensive integration of single-cell data. Cell 177 (7), 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. (2015). UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12 (3), e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang L., Zhu T., Wu B., Wang G., Luo Z., et al. (2022). Single-cell transcriptomic landscapes of the otic neuronal lineage at multiple early embryonic ages. Cell Rep. 38 (12), 110542. 10.1016/j.celrep.2022.110542 [DOI] [PubMed] [Google Scholar]
- Tadros S. F., Frisina S. T., Mapes F., Frisina D. R., Frisina R. D. (2005). Higher serum aldosterone correlates with lower hearing thresholds: A possible protective hormone against presbycusis. Hear. Res. 209 (1–2), 10–18. 10.1016/j.heares.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Van Eyken E., Van Camp G., Van Laer L. (2007). The complexity of age-related hearing impairment: Contributing environmental and genetic factors. Audiology Neuro-Otology 12 (6), 345–358. 10.1159/000106478 [DOI] [PubMed] [Google Scholar]
- Wells H. R. R., Freidin M. B., Zainul Abidin F. N., Payton A., Dawes P., Munro K. J., et al. (2019). GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK biobank. Am. J. Hum. Genet. 105 (4), 788–802. 10.1016/j.ajhg.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley T. L., Chappell R., Carmichael L., Nondahl D. M., Cruickshanks K. J. (2008). Changes in hearing thresholds over 10 years in older adults. J. Am. Acad. Audiology 19 (4), 281–292. 10.3766/jaaa.19.4.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise A. L., Gyi L., Manolio T. A. (2013). eXclusion: Toward integrating the X chromosome in genome-wide association analyses. Am. J. Hum. Genet. 92 (5), 643–647. 10.1016/j.ajhg.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolber L. E., Steves C. J., Spector T. D., Williams F. M. K. (2012). Hearing ability with age in northern European women: A new web-based approach to genetic studies. PloS One 7 (4), e35500. 10.1371/journal.pone.0035500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., Visscher P. M. (2011). Gcta: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88 (1), 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Weedon M. N., Purcell S., Lettre G., Estrada K., Willer C. J., et al. (2011). GIANT ConsortiumGenomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. EJHG 19 (7), 807–812. 10.1038/ejhg.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Sun Y., Sang S., Pham J. H., Kong W.-J. (2018). The risk of cognitive impairment associated with hearing function in older adults: A pooled analysis of data from eleven studies. Sci. Rep. 8 (1), 2137. 10.1038/s41598-018-20496-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Shi Z., Cui M., Li J., Ma Z., Shi Y., et al. (2015). A new role for LOC101928437 in non-syndromic intellectual disability: Findings from a family-based association test. PloS One 10 (8), e0135669. 10.1371/journal.pone.0135669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito A., Davies M. N., Tsai P.-C., Roberts S., Andres-Ejarque R., Nardone S., et al. (2019). Heritability of skewed X-inactivation in female twins is tissue-specific and associated with age. Nat. Commun. 10 (1), 5339. 10.1038/s41467-019-13340-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.