Abstract
The calcific aortic valve disease (CAVD) develops as an aortic valve sclerosis and progresses to an advanced form of stenosis. In many biological fields, bioinformatics becomes a fundamental component. The key mechanisms involved in CAVD are discovered with the use of bioinformatics to investigate gene function and pathways. We downloaded the original data (GSE51472) from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). After standardization, 2978 differentially expressed genes (DEGs) were identified from the data sets GSE51472 containing samples from normal, calcified, and sclerotic aortic valves. Analysis of DEGs based on the series test of clusters (STCs) revealed the two most significant patterns. Based on the result of the STC, the functional enrichment analysis of gene ontology (GO) was conducted to investigate the molecular function (MF), biological process (BP), and cell compound (CC) of the DEGs. With a p value of 0.01, DEGs associated with “chronic inflammation,” “T-cell receptor complexes,” and “antigen binding” had the highest significance within BP, CC, and MF. DEG enrichment in signaling pathways was analyzed using KEGG pathway enrichment. Using a p < 0.05 level of significance, the most enriched biological pathways related to CAVD were “Chemokine signaling pathway,” “Cytokine-cytokine receptor interaction,” “Tuberculosis,” “PI3K-Akt signaling pathway,” and “Transcriptional misregulation in cancer.” Finally, the construction of gene co-expression networks and pathway networks illustrated the pathogensis of CAVD. TLR2, CD86, and TYROBP were identified as hub genes for the development of CAVD. Moreover, “MAPK signaling pathway,” “Apoptosis,” and “Pathways in cancer” were regarded as the core pathways among the samples of normal, sclerotic and calcified aortic valve samples.
Keywords: Calcific aortic valve disease, different expressing genes, pathogenesis, hub genes, pathway-network
Impact Statement
The most common type of valvular heart disease is calcific aortic valve disease (CAVD). However, the pathogenesis of the disease is still little to known. The aortic valve samples chosen in our study represented different stages of the disease, and the differentially expressed genes (DEGs) of our interest were gradually increased or decreased with the aggravation of the disease. Therefore, all bioinformatics analyses were performed to shed light on the key genes and core pathways in the development of the disease. Further understanding of the underlying pathogenesis of CAVD will therefore be helpful for finding diagnostic and therapeutic targets.
Introduction
Calcific aortic valve disease (CAVD) constitutes the most common form of valvular heart disease. 1 CAVD includes the process from initial changes in the leaflet cellular biology to the calcification leading to left ventricular outflow obstruction in the end stage. As the disease begins, the valves thicken mildly, causing a condition called aortic valve sclerosis. As it advances, the leaflets become impaired, which becomes a condition called aortic valve stenosis. 2
In older individuals over the age of 65, CAVD prevalence is around 20–30% and in those aged 85 and over, it is 48%. Furthermore, 2–3% of people older than 65 and up to 8% of those older than 85 have significant stenosis.3,4 CAVD increases mortality risk. Surgery or valve replacement is currently the sole effective treatments for symptomatic CAVD.5 –7 There are increasing demands for a pharmacologic intervention for those senile and inoperable patients. However, it is notable that despite valve replacement treatment, the underlying mechanism of the CAVD is yet to be fully understood.
CAVD used to be thought of as an age-related degenerative disease. Nevertheless, it has become clear that CAVD is an active and complicated process, which probably involves the interaction of biochemical, immunological, and metabolic factors.6 –9 Although CAVD was reported to shares some processes with atherosclerosis, these two diseases are not synonymous, and the formation of CAVD has specific underlying mechanisms. 10 Various core pathways and activating factors have been underlined, and complex crosstalk among lipid metabolism, inflammation, neovascularization, mineralization, and osteogenesis are involved in CAVD development.11,12 Studies of these biological mechanisms could open new therapeutic avenues for CAVD.
In many areas of biology, bioinformatics is emerging as an increasingly essential tool. It aids in data mining of the biological literature, analyzing of gene expression and transcription regulation, and discovering and cataloging biological pathways and networks.13,14
Although there is not yet an efficient pharmacological treatment available to treat CAVD, trying to identify novel molecular targets would provide our patients with the possibility of novel forms of noninvasive therapies in the future. Therefore, in this study, we obtained the original data sets (GSE51472) from the publicly available Gene Expression Omnibus (GEO) database to identify DEGs among normal, sclerotic, and stenosis aortic valve samples. Increased/decreased DEGs accompanied by the valve deterioration with disease progression of CAVD were selected by using series test of cluster (STC) analyses. We then performed pathway and functional enrichment analyses and built a protein–protein interaction (PPI) network to identity core genes. The involved genes and pathway analysis may provide further insights into CAVD and open the door to understand its potential molecular mechanisms to better facilitate the targeted treatments of the disease.
Materials and methods
Data resource
Data set GSE51472 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51472). 15 The data set contained 15 samples in total, including five normal aortic valves, five sclerotic aortic valves, and five calcified aortic valves.
Identification of DEGs
The original data sets were downloaded for the subsequent analysis. All of the analysis was conducted on an online bioinformatics analysis platform called Gene-Cloud of Biotechnology Information (GCBI) (http://www.gcbi.com.cn/). GCBI allows for the comparison across two or more groups of samples in a GEO series based on R language. Statistically significant DEGs were identified by a classical analysis of variance (ANOVA) with p < 0.01 and q < 0.01. The threshold was set at the p value < 0.05.
Analysis of the STCs
Groups were arranged according to the severity of the disease. In order to identify the best characterized gene groups in the calcification process of cardiac valves, DEGs with the same variables features were organized in a trend cluster. The ordinate indicated the different grades of expression change. P value represented the significance level of the actual number of genes compared with the theoretical number of genes randomly replaced. DEG profiles with p < 0.05 were considered as STC profiles.
Analysis of DEG enrichment
The gene ontology (GO) analysis database (http://geneontology.org/) could be utilized to annotate genes and gene products and determine the characteristics of high-throughput sequencing of genomes and transcriptomes. Kyoto Encyclopedia of Genes and Genomes (KEGG) is a capable database focusing on the analysis of gene functions and pathways. In this study, the enrichment of DEGs identified through STC analysis were analyzed on the GCBI platform using the above two databases. As a cutoff criterion for functional enrichment, a p value of 0.05 was used.
Gene co-expression network and pathway-network construction
All the networks were generated using the GCBI database. The GCBI laboratory builds gene co-expression networks which illustrate the key regulatory genes and show the inter-relationship of genes, using gene expression profiles.
We could better understand the hierarchy of all pathways using the pathway-network. The pathway interaction network is constructed using the KEGG database.
Results
Identification of DEGs
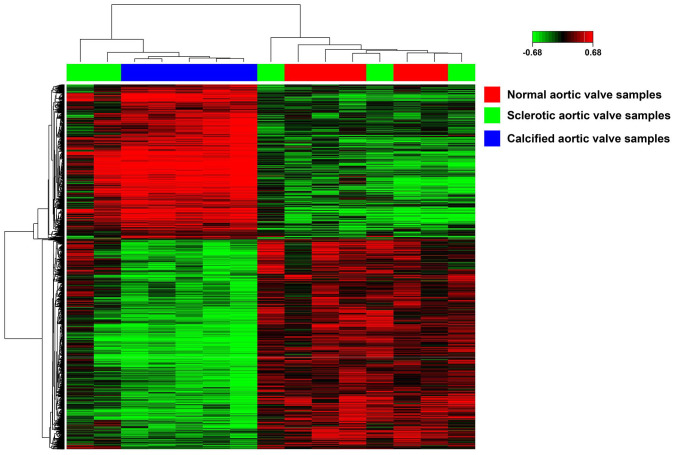
First, the original gene data file was standardized to guarantee all the following analysis reliable on GCBI online platform. Then, these data were screened to extract DEGs among normal, sclerotic, and calcified valve samples. Using p < 0.01 and q < 0.01 as cut-off criteria, 2978 DEGs have been screened from the expression profile data sets GSE51472 and shown as the heatmap in Figure 1. The top-10 DEGs are shown in Table 1.
Figure 1.
The heatmap of DEGs. Sample names are displayed on the horizontal axis. Red represents normal, green represents calcified and green represents sclerotic aortic valve samples. Clusters of DEGs are presented on the left vertical axis, and the clusters of samples are shown on the above horizontal axis. Red represents up-regulated genes and green indicated downregulated genes.
Table 1.
The top-10 DEGs among sclerotic aortic valve samples, calcified aortic valve samples, and normal aortic valve samples.
| Gene | Gene description | d score | p value | q value |
|---|---|---|---|---|
| LAMP3 | Lysosomal-associated membrane protein 3 | 5.500035 | 1.90E–05 | 6.33E–06 |
| GZMB | Granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1 | 4.486004 | 1.90E–05 | 6.33E–06 |
| XXR4 | XK, Kell blood group complex subunit-related family, member 4 | 4.051281 | 2.00E–05 | 6.67E–06 |
| PILRA | Paired immunoglobin-like type 2 receptor alpha | 4.012526 | 2.00E–05 | 6.67E–06 |
| TPBG | Trophoblast glycoprotein | 3.883444 | 2.10E–05 | 7.00E–06 |
| SH3RF2 | SH3 domain containing ring finger 2 | 3.704318 | 2.20E–05 | 7.33E–06 |
| RADIL | Ras association and DIL domains | 3.462278 | 2.40E–05 | 8.00E–06 |
| CD247 | CD247 molecule | 3.421485 | 2.50E–05 | 8.33E–06 |
| TMEM108 | Transmembrane protein 108 | 3.408931 | 2.50E–05 | 8.3E–06 |
| SLC2A5 | Solute carrier family 2 (facilitated glucose/fructose transporter), member 5 | 3.349204 | 2.60E–05 | 8.67E–06 |
STC analyses of DEGs
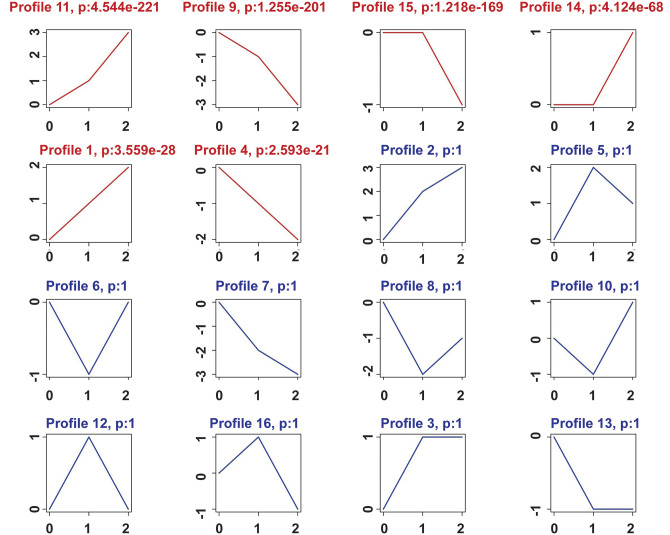
In order to refine the significantly target genes among the extracted 2978 genes, we used the 16 model patterns to show an overview of differential gene expression. As shown in Figure 2, we found six patterns of genes that showed p values (p < 0.01) with significant differences among the 16 patterns (Figure 2).
Figure 2.
The STC analysis in three groups. “Zero” represents normal aortic valve samples, “one” represents sclerotic aortic valve samples, and “two” represents calcified aortic valve samples. Patterns with red curve represent profiles with a significant p value. Patterns with blue curve represent profiles with no statistical difference. Profile number and p value were shown on the top of each pattern.
According to ascending p values, the two most significant patterns were profiles No. 9 and No. 11. The profile No. 11 contained 576 genes whose expression increased steadily, while profile No. 9 contained 555 genes whose expression reduced steadily (Figure 2).
Enrichment analyses of DEGs
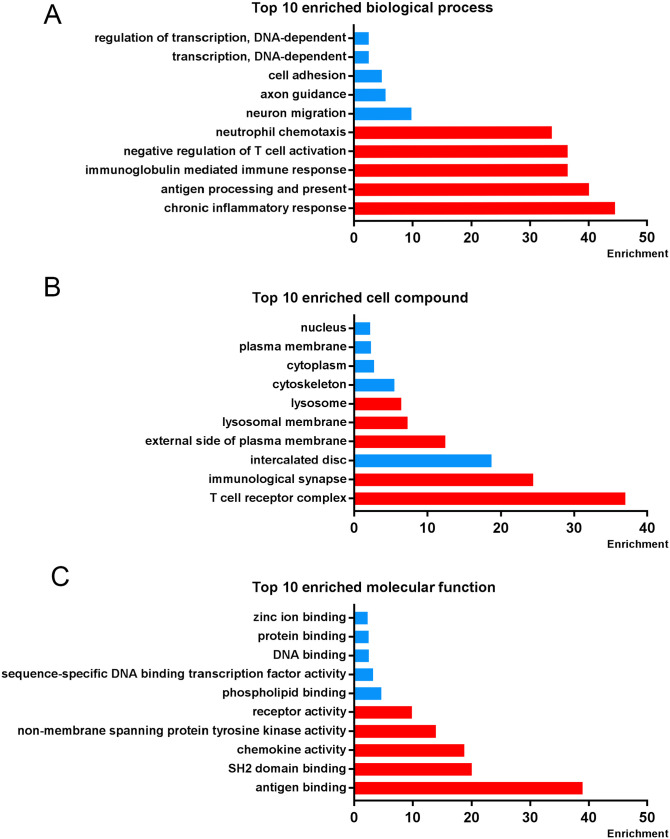
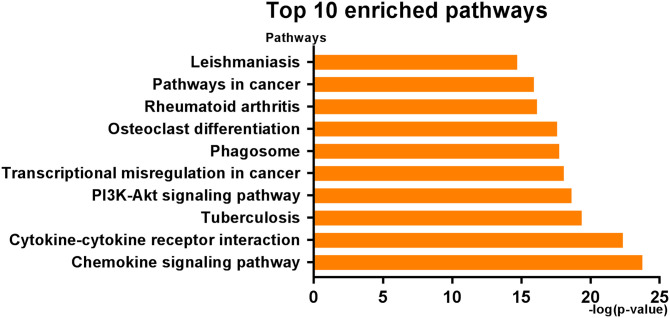
Next, No. 9 and No. 11 profiles were analyzed, respectively. GO functional enrichment analysis was conducted to investigate the BP, CC, and MF of the refined DEGs from the STC analysis. With p < 0.01 as the criterion, “chronic inflammatory response,” “antigen processing and presentation of peptide or polysaccharide antigen via MHC class II,” and “immunoglobulin mediated immune response” showed remarkable enrichment within the BP category (Figure 3(A)). For the CC category, the DEGs were significantly clustered in “T-cell receptor complex,” “immunological synapse,” and “intercalated disc” (Figure 3(B)). Furthermore, DEGs significantly grouped in “antigen binding,” “SH2 domain binding,” and “chemokine activity” for the MF category (Figure 3(C)). In order to understand the signaling pathway enrichment of DEGs, KEGG pathway enrichment was then analyzed. With the criterion of p < 0.05, the top enriched biological pathways associated with CAVD included “Chemokine signaling pathway,” “Cytokine-cytokine receptor interaction,” “Tuberculosis,” “PI3K-Akt signaling pathway,” and “Transcriptional misregulation in cancer” (Figure 4).
Figure 3.
Gene ontology (GO) enrichment analysis in selected STC profiles. The top-10 (A) biological processes, (B) cell compounds, and (C) molecular functions enrichment in three groups. Red strips represent the enrichment in profile 11 and blue strips represent the enrichment in profile 9.
Figure 4.
The top-10 KEGG pathways enrichment in three groups. (A color version of this figure is available in the online journal.)
Gene co-expression network establishment and hub genes identification
Gene co-expression network of DEGs was constructed following the enrichment analysis on the GCBI platform. As shown in Figure 5, constructing gene co-expression network explicated the hub genes in CAVD development. We found that TLR2, CD86, TYROBP, HCK, LCP2, TDO2, DYNC1LI2, PLAUR, and COL6A6 were hub genes with the highest degree (Table 2).
Figure 5.
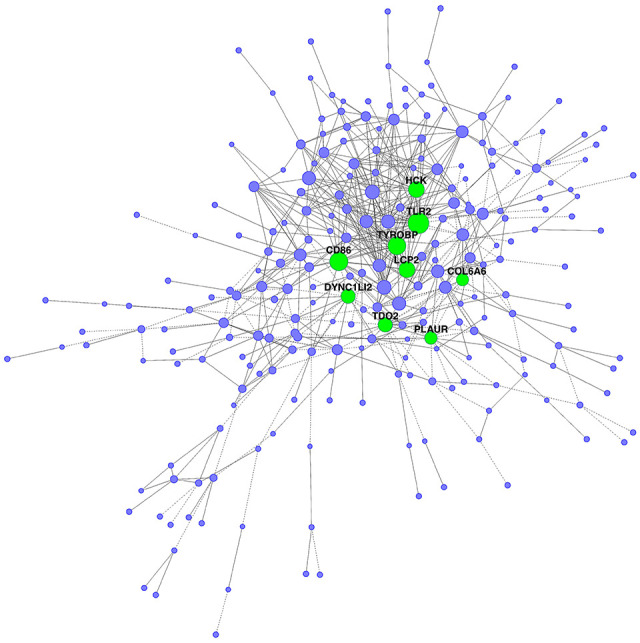
Co-expression network of DEGs in three groups. The dots represent gene and the size of the node represents degree. Green dots represent hub genes.
Table 2.
The hub genes identified by gene co-expression network in three groups.
| Gene symbol | Degree | Description |
|---|---|---|
| TLR2 | 25 | Toll-like receptor 2 |
| CD86 | 24 | CD86 molecule |
| TYROBP | 23 | TYRO protein tyrosine kinase binding protein |
| HCK | 20 | Hemopoietic cell kinase |
| LCP2 | 20 | Lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76 kDa) |
| TDO2 | 18 | Tryptophan 2,3-dioxygenase |
| DYNC1LI2 | 17 | Dynein, cytoplasmic 1, light intermediate chain 2 |
| PLAUR | 15 | Plasminogen activator, urokinase receptor |
| COL6A6 | 14 | Collagen, type VI, alpha 6 |
Construction of pathway-network
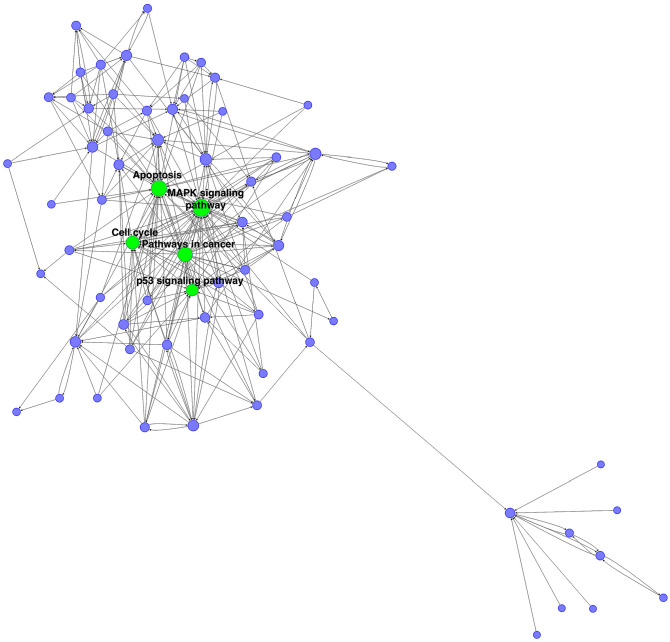
To figure out the core pathway of CAVD, we constructed the pathway-network. The pathway with the highest degree was MAPK signaling pathway. The other main pathways including “apoptosis,” “pathways in cancer,” “cell cycle,” and “p53 signaling pathway” were also with a higher degree (Figure 6).
Figure 6.
The pathway-network of DEGs in three groups. Each node represents a pathway and the size of the node represents degree. Green dots represent core pathways in the development of CAVD.
Discussion
CAVD, the typical valvular disease, puts a heavy strain on the healthcare system and causes a major challenge to public health. But now there is no reliable way to hamper or halt the development of CAVD. It is critical to understand the pathogenesis of CAVD for diagnosis, predict, and finding drug targets. The expression of thousands of genes in humans can be provided by the methodology of high-throughput sequencing and microarrays, which has been widely employed to predict the targeted diagnostic and therapeutic approaches for various diseases.
In our study, we performed STC analyses to better understand the pattern of the declared DEGs. Two respective profiles of genes whose expression increased and reduced consistently were found among normal, sclerotic, and calcified aortic valve samples. The GO analysis based on STC revealed that biological process of chronic inflammatory response was the most influenced with CAVD progression. Monocyte infiltration which participates in chronic inflammation is characteristic pathology of progressive CAVD and accelerate calcification of aortic valves. 16 The most significant cell compound is T-cell receptor complex. T-cell activation and infiltration in diseased valve has been reported as a hallmark of CAVD. 17 Concerning the molecular function, genes in profile 9 and 11 were found to be involved in antigen binding, SH2 domain binding and chemokine activity. Chemokines recruit antigen-presenting cells and immune cells together and activate immune response. The pathway with the highest level of enrichment in STC analyses was cytokine–cytokine receptor interaction, which controls the binding of cytokines. Released inflammatory cytokines mediate a variety of cellular responses that are involved in CAVD.
Following the enrichment analyses, according to gene co-expression networks, TLR2, CD86, TYROBP, HCK, LCP2, TDO2, DYNC1LI2, PLAUR, and COL6A6 were identified as the hub genes of CAVD with the highest degrees. Moreover, “MAPK signaling pathway,” “Apoptosis,” “Pathways in cancer,” “Cell cycle,” and “p53 signaling pathway” are the core pathways related to CAVD progresses.
The protein encoded by TLR2 is a receptor in the Toll-like receptor family which plays an essential role in pathogen identification, immune signal activation, and transmission. TLR2 was identified as an up-regulated gene in aortic valve stenosis when compared to the normal valve in our study. Previous studies have reported that TLR2 regulates the cellular inflammatory response and endogenous agents such as heat shock proteins as well.18 –20 In Meng’s study, the human aortic valve interstitial cells (HAVICs) responded to the agonist of TLR2 to activate nuclear factor kappa B (NF-κB) pathway and mediate the production of multiple pro-inflammatory cytokines in vitro. Bone morphogenetic protein-2 (BMP-2), a crucial molecule in bone formation, is up-regulated and causes osteogenic changes21 –23 after stimulation of the cultured HAVICs by the TLR2 agonists PGN and LPS.24 –27 Previous researches have found that biglycan induces the release of inflammatory mediators in cultured macrophages via TLR2.28,29 Cultivation of TLR2 upregulates ICAM-1 expression and increases MCP-1 production in HAVICs which express functional TLR2. 24 Furthermore, Knockdown of TLR2 in HAVICs significantly reduced the expression of ICAM-1 and MCP-1 induced by soluble biglycan. These findings indicated that TLR2 has a pivotal role in the complex pathogenesis of aortic valve stenosis.
CD86 was also referred as an up-regulated gene in aortic valve stenosis when compared to normal valve in our study. CD86 encodes a type-I transmembrane protein that belongs to the immunoglobulin superfamily. CD86 on antigen-presenting cells binds to CD28 on T-cells to activate T-cell as a costimulation signal. Moreover, CD28-CD80/CD86 signaling is indispensable for the full activation, proliferation, cytokine secretion, and survival in T-cells.30,31 CD80/CD86 co-stimulation improves the differentiation of helper T-cell and the expression of CD40 ligand. 32 Cytokines such as interferon (IFN)-γ secreted by T helper 1 cells (Th1), 33 and CD40 ligand 34 deeply influence the atherosclerotic process in murine models. Both CD86 and CD28 have been shown in macrophages of human atherosclerotic lesions. 35 Dopheide et al. 36 showed that the expression of CD86 is more elevated in dendritic cells (DCs) derived from monocytes in patients with coronary artery disease than in DCs from healthy controls. Moreover, it was reported that an increasing concentrations of the T-cell coactivators CD80 and CD86 respond to IFN-γ treatment, which inhibits osteoclast function and may lead to calcification in CAVD. 37
TYROBP contains an immunoreceptor-tyrosine-based activation motif, which is also known as DAP12. It plays a role in augmenting the inflammation by increasing circulating cytokine levels and cellular inflammatory cytokine production. 38 TYROBP is broadly distributed in dendritic cells, macrophages, and osteoclasts. It gets involved in the immune response and has a wide variety of interactions with hematopoietic cells. 39 Triggering receptors expressed on myeloid cells-1 (TREM-1), a critical DAP12-interacting protein that is expressed on hematopoietic cells, mediates inflammation amplification. TREM-1, which is recruited by DAP12, phosphorylates downstream receptor enzymes, causes intracellular calcium mobilization, and encourages the generation and release of inflammatory cytokines. 38 Blocking TREM-1/DAP12 can significantly decrease aherosclerotic plaque size, lipid content, and macrophage numbers, which indicated that TREM-1/DAP12 plays critical roles in plaque formation and inflammation in atherosclerosis. 40
In our study, mitogen-activated protein kinases (MAPKs) were the most significantly enriched pathway in CAVD. It was reported that MAPKs transmit extracellular signals to intracellular responses to regulate diverse cellular functions. Conventional MAPKs are the most common kind, and incorporate the extracellular signal-regulated kinases 1 and 2 (ERK1/2), p38 (α, β, γ, and δ), c-Jun amino-terminal kinases 1 to 3 (JNK1 to 3), and ERK5 families. These MAPK enzymes coordinate cell proliferation, differentiation, survival, and motility. 41 Previous studies have revealed that HMGB1, an agonist of TLR2, can activate multiple intracellular pro-inflammatory pathways, including p38 MAPK, JNK MAPK, and NF-κB.42 –44 In addition, the HMGB1-inducing upregulation of BMP2 and osteocalcin and calcium deposits formation were significantly dependent on NF-κB and JNK MAPK signaling pathways. 45 JNK MAPK was also found to mediate osteoblastic differentiation and mature induced by oxidized low-density lipoprotein (LDL) in VICs. 46 Moreover, Song et al. 47 found the TLR2-mediated inflammatory response in human AVICs. ERK1/2 enhanced the expression of ICAM-1 and the release of MCP-1, suggesting that ERK1/2 respond to TLR2 and mediates the secretion of the pro-inflammatory factors in human AVICs.
In conclusion, a number of genes might take part in the CAVD development, including TLR2, CD86, and TYROBP. Also, multiple pathways, such as “MAPK signaling pathway,” “Apoptosis,” and “Pathways in cancer,” are identified the core pathways among the samples of normal, sclerotic, and calcified aortic valves. Consequently, our study sheds further light on the underlying pathogenesis of CAVD, which may aid in diagnosing and treating CAVD.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Natural Science Foundation of China (81801616, 82100381).
ORCID iD: Meng-Lei Ding  https://orcid.org/0000-0002-8680-9112
https://orcid.org/0000-0002-8680-9112
References
- 1. Bach DS, Radeva JI, Birnbaum HG, Fournier AA, Tuttle EG. Prevalence, referral patterns, testing, and surgery in aortic valve disease: leaving women and elderly patients behind. J Heart Valve Dis 2007;16:362–9 [PubMed] [Google Scholar]
- 2. Freeman RV, Otto CM. Spectrum of calcific aortic valve disease: pathogenesis, disease progression, and treatment strategies. Circulation 2005; 111:3316–26 [DOI] [PubMed] [Google Scholar]
- 3. Lindroos M, Kupari M, Heikkila J, Tilvis R. Prevalence of aortic valve abnormalities in the elderly: an echocardiographic study of a random population sample. J Am Coll Cardiol 1993;21:1220–5 [DOI] [PubMed] [Google Scholar]
- 4. Stewart BF, Siscovick D, Lind BK, Gardin JM, Gottdiener JS, Smith VE, Kitzman DW, Otto CM. Clinical factors associated with calcific aortic valve disease: cardiovascular health study. J Am Coll Cardiol 1997;29: 630–4 [DOI] [PubMed] [Google Scholar]
- 5. Rodes-Cabau J. Transcatheter aortic valve implantation: current and future approaches. Nat Rev Cardiol 2011;9:15–29 [DOI] [PubMed] [Google Scholar]
- 6. Cowell SJ, Newby DE, Boon NA, Elder AT. Calcific aortic stenosis: same old story. Age Ageing 2004;33:538–44 [DOI] [PubMed] [Google Scholar]
- 7. Perin MA, Brito FS, Jr, Almeida BO, Pereira MA, Abizaid A, Tarasoutchi F, Grube E. Percutaneous aortic valve replacement for the treatment of aortic stenosis: early experience in Brazil. Arq Bras Cardiol 2009;93:299–306 [DOI] [PubMed] [Google Scholar]
- 8. O’Brien KD, Reichenbach DD, Marcovina SM, Kuusisto J, Alpers CE, Otto CM. Apolipoproteins B, (a), and E accumulate in the morphologically early lesion of “degenerative” valvular aortic stenosis. Arterioscler Thromb Vasc Biol 1996;16:523–32 [DOI] [PubMed] [Google Scholar]
- 9. Olsson M, Thyberg J, Nilsson J. Presence of oxidized low density lipoprotein in nonrheumatic stenotic aortic valves. Arterioscler Thromb Vasc Biol 1999;19:1218–22 [DOI] [PubMed] [Google Scholar]
- 10. Otto CM, O’Brien KD. Why is there discordance between calcific aortic stenosis and coronary artery disease. Heart 2001;85:601–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathieu P, Boulanger MC. Basic mechanisms of calcific aortic valve disease. Can J Cardiol 2014;30:982–93 [DOI] [PubMed] [Google Scholar]
- 12. Rajamannan NM, Evans FJ, Aikawa E, Grande-Allen KJ, Demer LL, Heistad DD, Simmons CA, Masters KS, Mathieu P, O’Brien KD, Schoen FJ, Towler DA, Yoganathan AP, Otto CM. Calcific aortic valve disease—not simply a degenerative process: a review and agenda for research from the National Heart and Lung and Blood Institute Aortic Stenosis Working Group—executive summary: calcific aortic valve disease-2011 update. Circulation 2011;124:1783–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joyce AP, Zhang C, Bradley P, Havranek JJ. Structure-based modeling of protein: DNA specificity. Brief Funct Genomics 2015;14:39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spiga E, Degiacomi MT, Dal Peraro M. New strategies for integrative dynamic modeling of macromolecular assembly. Adv Protein Chem Struct Biol 2014;96:77–111 [DOI] [PubMed] [Google Scholar]
- 15. Ohukainen P, Syvaranta S, Napankangas J, Rajamaki K, Taskinen P, Peltonen T, Helske-Suihko S, Kovanen PT, Ruskoaho H, Rysa J. MicroRNA-125b and chemokine CCL4 expression are associated with calcific aortic valve disease. Ann Med 2015;47:423–9 [DOI] [PubMed] [Google Scholar]
- 16. Zhang P, The E, Nedumaran B, Ao L, Jarrett MJ, Xu D, Fullerton DA, Meng X. Monocytes enhance the inflammatory response to TLR2 stimulation in aortic valve interstitial cells through paracrine up-regulation of TLR2 level. Int J Biol Sci 2020;16:3062–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raddatz MA, Madhur MS, Merryman WD. Adaptive immune cells in calcific aortic valve disease. Am J Physiol Heart Circ Physiol 2019;317: H141–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 1999;11:443–51 [DOI] [PubMed] [Google Scholar]
- 19. Mitchell JA, Paul-Clark MJ, Clarke GW, McMaster SK, Cartwright N. Critical role of toll-like receptors and nucleotide oligomerisation domain in the regulation of health and disease. J Endocrinol 2007;193:323–30 [DOI] [PubMed] [Google Scholar]
- 20. Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol 2007;19:3–10 [DOI] [PubMed] [Google Scholar]
- 21. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors 2004;22:233–41 [DOI] [PubMed] [Google Scholar]
- 22. Jensen ED, Nair AK, Westendorf JJ. Histone deacetylase co-repressor complex control of Runx2 and bone formation. Crit Rev Eukaryot Gene Expr 2007;17:187–96 [DOI] [PubMed] [Google Scholar]
- 23. Osman L, Yacoub MH, Latif N, Amrani M, Chester AH. Role of human valve interstitial cells in valve calcification and their response to atorvastatin. Circulation 2006;114:I547–152 [DOI] [PubMed] [Google Scholar]
- 24. Meng X, Ao L, Song Y, Babu A, Yang X, Wang M, Weyant MJ, Dinarello CA, Cleveland JC, Jr, Fullerton DA. Expression of functional Toll-like receptors 2 and 4 in human aortic valve interstitial cells: potential roles in aortic valve inflammation and stenosis. Am J Physiol Cell Physiol 2008;294:C29–35 [DOI] [PubMed] [Google Scholar]
- 25. Aikawa E, Nahrendorf M, Sosnovik D, Lok VM, Jaffer FA, Aikawa M, Weissleder R. Multimodality molecular imaging identifies proteolytic and osteogenic activities in early aortic valve disease. Circulation 2007;115:377–86 [DOI] [PubMed] [Google Scholar]
- 26. Kaden JJ, Bickelhaupt S, Grobholz R, Vahl CF, Hagl S, Brueckmann M, Haase KK, Dempfle CE, Borggrefe M. Expression of bone sialoprotein and bone morphogenetic protein-2 in calcific aortic stenosis. J Heart Valve Dis 2004;13:560–6 [PubMed] [Google Scholar]
- 27. Shao JS, Cai J, Towler DA. Molecular mechanisms of vascular calcification: lessons learned from the aorta. Arterioscler Thromb Vasc Biol 2006; 26:1423–30 [DOI] [PubMed] [Google Scholar]
- 28. Schaefer L, Babelova A, Kiss E, Hausser HJ, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Gotte M, Malle E, Schaefer RM, Grone HJ. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Invest 2005;115:2223–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Babelova A, Moreth K, Tsalastra-Greul W, Zeng-Brouwers J, Eickelberg O, Young MF, Bruckner P, Pfeilschifter J, Schaefer RM, Grone HJ, Schaefer L. Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors. J Biol Chem 2009; 284:24035–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McAdam AJ, Schweitzer AN, Sharpe AH. The role of B7 co-stimulation in activation and differentiation of CD4+ and CD8+ T cells. Immunol Rev 1998;165:231–47 [DOI] [PubMed] [Google Scholar]
- 31. Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu Rev Immunol 1996;14:233–58 [DOI] [PubMed] [Google Scholar]
- 32. Ding L, Green JM, Thompson CB, Shevach EM. B7/CD28-dependent and -independent induction of CD40 ligand expression. J Immunol 1995;155:5124–32 [PubMed] [Google Scholar]
- 33. Gupta S, Pablo AM, Jiang X, Wang N, Tall AR, Schindler C. IFN-gamma potentiates atherosclerosis in ApoE knock-out mice. J Clin Invest 1997; 99:2752–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mach F, Schonbeck U, Sukhova GK, Atkinson E, Libby P. Reduction of atherosclerosis in mice by inhibition of CD40 signalling. Nature 1998; 394:200–3 [DOI] [PubMed] [Google Scholar]
- 35. De Boer OJ, Hirsch F, van der Wal AC, van der Loos CM, Das PK, Becker AE. Costimulatory molecules in human atherosclerotic plaques: an indication of antigen specific T lymphocyte activation. Atherosclerosis 1997;133:227–34 [DOI] [PubMed] [Google Scholar]
- 36. Dopheide JF, Sester U, Schlitt A, Horstick G, Rupprecht HJ, Munzel T, Blankenberg S. Monocyte-derived dendritic cells of patients with coronary artery disease show an increased expression of costimulatory molecules CD40, CD80 and CD86 in vitro. Coron Artery Dis 2007;18: 523–31 [DOI] [PubMed] [Google Scholar]
- 37. Nagy E, Lei Y, Martinez-Martinez E, Body SC, Schlotter F, Creager M, Assmann A, Khabbaz K, Libby P, Hansson GK, Aikawa E. Interferon-gamma released by activated CD8(+) T lymphocytes impairs the calcium resorption potential of osteoclasts in calcified human aortic valves. Am J Pathol 2017;187:1413–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turnbull IR, McDunn JE, Takai T, Townsend RR, Cobb JP, Colonna M. DAP12 (KARAP) amplifies inflammation and increases mortality from endotoxemia and septic peritonitis. J Exp Med 2005;202:363–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev 2009;227:150–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang HM, Gao JH, Lu JL. Pravastatin improves atherosclerosis in mice with hyperlipidemia by inhibiting TREM-1/DAP12. Eur Rev Med Pharmacol Sci 2018;22:4995–5003 [DOI] [PubMed] [Google Scholar]
- 41. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 2011;75:50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Glushkova OV, Khrenov MO, Novoselova TV, Lunin SM, Parfenyuk SB, Alekseev SI, Fesenko EE, Novoselova EG. The role of the NF-kappaB, SAPK/JNK, and TLR4 signalling pathways in the responses of RAW 264.7 cells to extremely low-intensity microwaves. Int J Radiat Biol 2015;91:321–8 [DOI] [PubMed] [Google Scholar]
- 43. Nguyen CT, Kim EH, Luong TT, Pyo S, Rhee DK. TLR4 mediates pneumolysin-induced ATF3 expression through the JNK/p38 pathway in Streptococcus pneumoniae-infected RAW 264.7 cells. Mol Cells 2015;38:58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Byun EB, Sung NY, Park JN, Yang MS, Park SH, Byun EH. Gamma-irradiated resveratrol negatively regulates LPS-induced MAPK and NF-kappaB signaling through TLR4 in macrophages. Int Immunopharmacol 2015;25:249–59 [DOI] [PubMed] [Google Scholar]
- 45. Wang B, Li F, Zhang C, Wei G, Liao P, Dong N. High-mobility group box-1 protein induces osteogenic phenotype changes in aortic valve interstitial cells. J Thorac Cardiovasc Surg 2016;151:255–62 [DOI] [PubMed] [Google Scholar]
- 46. Li F, Zhao Z, Cai Z, Dong N, Liu Y. Oxidized low-density lipoprotein promotes osteoblastic differentiation of valvular interstitial cells through RAGE/MAPK. Cardiology 2015;130:55–61 [DOI] [PubMed] [Google Scholar]
- 47. Song R, Ao L, Zhao KS, Zheng D, Venardos N, Fullerton DA, Meng X. Soluble biglycan induces the production of ICAM-1 and MCP-1 in human aortic valve interstitial cells through TLR2/4 and the ERK1/2 pathway. Inflamm Res 2014;63:703–10 [DOI] [PMC free article] [PubMed] [Google Scholar]