Abstract
Although vital pulp therapy should be performed by promoting the wound-healing capacity of dental pulp, existing pulp-capping materials were not developed with a focus on the pulpal repair process. In previous investigations of wound healing in dental pulp, we found that organic dentin matrix components (DMCs) were degraded by matrix metalloproteinase-20, and DMC degradation products containing protein S100A7 (S100A7) and protein S100A8 (S100A8) promoted the pulpal wound-healing process. However, the direct use of recombinant proteins as pulp-capping materials may cause clinical problems or lead to high medical costs. Thus, we hypothesized that functional peptides derived from recombinant proteins could solve the problems associated with direct use of such proteins. In this study, we identified functional peptides derived from the protein S100 family and investigated their effects on dental pulp tissue. We first performed amino acid sequence alignments of protein S100 family members from several mammalian sources, then identified candidate peptides. Next, we used a peptide array method that involved human dental pulp stem cells (hDPSCs) to evaluate the mineralization-inducing ability of each peptide. Our results supported the selection of 4 candidate functional peptides derived from proteins S100A8 and S100A9. Direct pulp-capping experiments in a rat model demonstrated that 1 S100A8-derived peptide induced greater tertiary dentin formation compared with the other peptides. To investigate the mechanism underlying this induction effect, we performed liquid chromatography–tandem mass spectrometry analysis using hDPSCs and the S100A8-derived peptide; the results suggested that this peptide promotes tertiary dentin formation by inhibiting inflammatory responses. In addition, this peptide was located in a hairpin region on the surface of S100A8 and could function by direct interaction with other molecules. In summary, this study demonstrated that a S100A8-derived functional peptide promoted wound healing in dental pulp; our findings provide insights for the development of next-generation biological vital pulp therapies.
Keywords: dentinogenesis, computational biology, peptide, S100 proteins, wound healing, anti-inflammatory agents
Introduction
Vital pulp therapy (VPT) is receiving increasing attention because the preservation of dental pulp can prolong tooth longevity and extend healthy life expectancy in older adults (Caplan et al. 2005). Pulp capping is a VPT approach that can activate the repair process in damaged pulp tissue and preserve the vitality of such tissue. The success rate of direct pulp capping (DPC) using hydraulic calcium silicate cements (HCSCs; e.g., mineral trioxide aggregate [MTA]) is reportedly over 80% after 3 y compared with that of treatment with calcium hydroxide, which is less than 60% (Cushley et al. 2021). Other reports also indicated that HCSCs performed significantly better than calcium hydroxide (Paula et al. 2018). Thus, HCSCs are considered optimal pulp-capping materials that have been developed based on their rich calcium or phosphate contents, high biocompatibility, and excellent sealing properties. However, HCSCs were first introduced as a root perforation repairing material (Lee et al. 1993). MTA was then applied as a pulp-capping material, although it was not developed to address the problem of wound healing of the pulp. Therefore, to achieve a better success rate for the pulp capping, we considered it was essential to elucidate the pulpal wound-healing mechanism and to develop a novel material to promote this process. There have been reports of pulp-capping materials developed with the aim of achieving higher success rates while retaining the original biomaterial properties (Nakashima et al. 2003; Iohara et al. 2004; Walker et al. 2019; Machado et al. 2020). However, no pulp-capping material with novel concepts based on the pulpal wound-healing process has been commercialized since the development of HCSCs.
We previously reported that matrix metalloproteinase-20 digested organic dentin matrix components (DMCs) promoted tertiary dentin formation (Okamoto et al. 2018). Moreover, proteome analysis of the digested DMCs revealed that protein S100A7 (S100A7) and S100A8 promoted pulpal wound healing (McLachlan et al. 2004; Komichi et al. 2019). S100A7 and S100A8 are both present in dentin and pulp; they promoted tertiary dentin formation in a rat DPC model (Komichi et al. 2019). Thus, these proteins originally derived from dentin may have fundamental roles as biological pulp-capping materials that can facilitate physiological wound healing. However, the use of a recombinant protein as a clinical pulp-capping material may lead to problems in terms of administration route or high medical costs. These problems can be solved by protein-derived functional peptides in clinical applications (Sachdeva 2017; Henninot et al. 2018).
Here, we hypothesized that functional sites of proteins with the ability to promote pulpal wound healing could be effective for tertiary dentin formation in vivo, in a manner comparable to the original protein. We selected functional peptides from the protein S100 family using in silico amino acid sequence alignment and an in vitro cell-based peptide array (Kanie et al. 2016). Then, we investigated the tertiary dentin-forming abilities of the peptides using a rat DPC model; we explored the underlying mechanism by proteome analysis, which provided insights for the development of biological evidence-based materials for novel VPTs.
Materials and Methods
Amino Acid Sequence Homology Assessment in the Protein S100 Family by In Silico Sequence Alignment
Proteins S100A7, S100A8, and S100A9 have been identified in dentin–pulp complex, suggesting that they contribute to pulpal wound healing (Komichi et al. 2019; Widbiller et al. 2019). Furthermore, functional amino acid sequences might generally be conserved among proteins in the same family; thus, we used CLUSTAL-W (https://www.genome.jp/tools-bin/clustalw) to perform an amino acid sequence alignment to identify homologous regions in these proteins, based on analysis of several corresponding mammalian amino acid sequences. The amino acid sequences used in this study were registered in UniProt (https://www.uniprot.org/).
Identification of Functional Peptides by Peptide Array
After the identification of highly homologous regions via sequence alignment, we selected 58 peptides, each consisting of 9-residues (Appendix Table 1).
To investigate the selected candidate peptides’ function, we used the peptide array method. This functional peptide screening approach was described in a previous report (Kanie et al. 2016). Peptides were synthesized on cellulose membranes by Fmoc solid-phase synthesis using a peptide synthesizer (ASP222; Intavis), then placed in 96-well plates with a final concentration of each peptide of 0.25 mM/well. Human dental pulp stem cells (hDPSCs; Lonza) were seeded in the plates at 5,000 cells/well and cultured in mineralization induction medium containing α minimum essential medium (Thermo Fisher Scientific), 50 μg/mL ascorbic acid (Sigma-Aldrich), 10 mM β-glycerophosphate (Sigma-Aldrich), and 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C in 5% CO2 for 14 d. The medium was changed at 3-d intervals. After 14 d, alizarin red staining was performed and mineralized nodules were quantified using the Calcification Assessment Set (PG Research). The experiments were performed in triplicate.
Direct Pulp Capping Using Selected Peptides Derived from S100 Family Members
All animal experimental procedures were performed in accordance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines and the relevant guidelines of the Ethical Guidelines Committee for Animal Care of Osaka University Graduate School of Dentistry; the experimental procedures were approved by the committee (Approval No. 28-013-0).
This animal experiment was performed using sound teeth with the sole purpose of evaluating the effects of the peptides. Twenty-eight 8-wk-old male Wistar rats (weighing 180–220 g; Clea Japan) underwent intraperitoneal injection of sodium pentobarbital (Somnopentyl; Kyoritsu Pharmaceutical) (30 mg/kg) for general anesthesia; carprofen (Rimadyl; Pfizer) (3 mg/kg) was administered for pain relief. Rubber dam isolation was applied; the tooth and surrounding tissues were then wiped with alcohol-soaked cotton. Experimental cavities in the left and right maxillary first molars were prepared by intentional pulp exposure from the occlusal surface to the mesial pulp horn, using a round bur (#1; Dentsply Sirona). The exposed pulp was gently washed with physiological saline and hemostasis was confirmed. Next, a gelatin sponge (Spongel; Astellas Pharma) was soaked with Dulbecco’s phosphate-buffered saline (PBS; Nacalai Tesque) containing 100 µg/mL of a single candidate peptide; the soaked sponge was used for DPC. The concentrations used in the experiments were determined from in vitro cytotoxicity and cell proliferation results (Appendix Fig. 1). Each cavity was then filled with glass ionomer cement (Fuji IX; GC). Controls were PBS alone, PBS containing 1 µg/mL of the original recombinant protein, or ProRoot MTA (Dentsply Sirona). Subsequently, animals were housed in a nonstressful environment; they were allowed free intake of food and water. Four weeks after pulp capping, experimental animals were sacrificed by intraperitoneal overdose of sodium pentobarbital, followed by perfusion fixation with 4% paraformaldehyde–phosphate buffer (Nacalai Tesque). After dissection of the maxilla containing the test teeth, evaluations were performed as described in the following sections. The number of specimens was 8 in each condition.
Micro–Computed Tomography Analysis
Tertiary dentin was quantified after DPC using micro–computed tomography (CT) (R-mCT2; Rigaku). Micro-CT analysis of the test teeth was performed as described previously (Okamoto et al. 2018), with the following conditions: tube voltage of 90 kV, tube current of 160 μA, and slice width of 5 μm. Tertiary dentin formation was determined by measuring the area of tertiary dentin from binarized images; the volume of tertiary dentin was calculated by integration of each area using TRI 3D-BON (Ratok) software.
Histopathological Evaluation
After micro-CT analysis, the same specimens were immersed and fixed using the same fixative described in the DPC for 12 h; they were then subjected to demineralization in Kalkitox (Fujifilm Wako Pure Chemical Industries) for 2 d. Subsequently, specimens were dehydrated and embedded in paraffin; serial 5-μm-thick sections were prepared using a rotary microtome (RM 2155; Leica), then subjected to hematoxylin–eosin staining. Histopathological observations were performed using a light microscope (BZ-X810; Keyence).
Proteome Analysis of Pulpal Wound-Healing Induction by Protein S100–Derived Functional Peptides
To investigate the mechanism by which specific peptides promoted pulpal wound healing, liquid chromatography–tandem mass spectrometry (LC-MS/MS) was performed to quantify and compare protein expression patterns in hDPSCs. The hDPSCs (3 million cells per 100-mm dish) were cultured with 0.1 mM of functional peptide No. 1 or recombinant S100A8 (ProSpec) in mineralization induction medium for 7 d. Proteins were extracted from the cell membrane or cytoplasm using a Cell Membrane Protein Extraction Kit (Cosmo Bio). The hDPSCs cultured in the same medium supplemented with PBS were used as a control group.
Samples were processed using trypsin and lysyl endopeptidase. Ionization was performed using the UltiMate 3000 Nano LC system (Thermo Fisher Scientific) through an ESI column (0.075 × 150 mm). The mobile phase was a 5% acetonitrile solution that contained 0.1% formic acid; the flow rate was 300 nL/min. The acquired MS spectra were analyzed via Q-Exactive software (Thermo Fisher Scientific). Potential proteins were identified and quantified using peptide mass fingerprinting software (Mascot Distiller and Mascot Server v2.5; Matrix Science), based on UniProt data. Proteins identified by MS/MS were quantitatively compared using Scaffold Viewer (Proteome Software) and refined in accordance with the criteria shown in Appendix Table 2.
Statistical Analysis
Statistical analysis was performed using SPSS (IBM). The Kruskal–Wallis test was used to evaluate differences in terms of tertiary dentin formation and the amount of tertiary dentin in DPC. One-way analysis of variance and the Tukey test were used to evaluate differences in terms of cytotoxicity and cell proliferation. Student’s t test was used to evaluate differences in real-time quantitative transcription polymerase chain reaction (RT-qPCR). A P value of <0.05 was considered indicative of statistical significance.
Results
Investigation of Amino Acid Sequence Homology in the Protein S100 Family
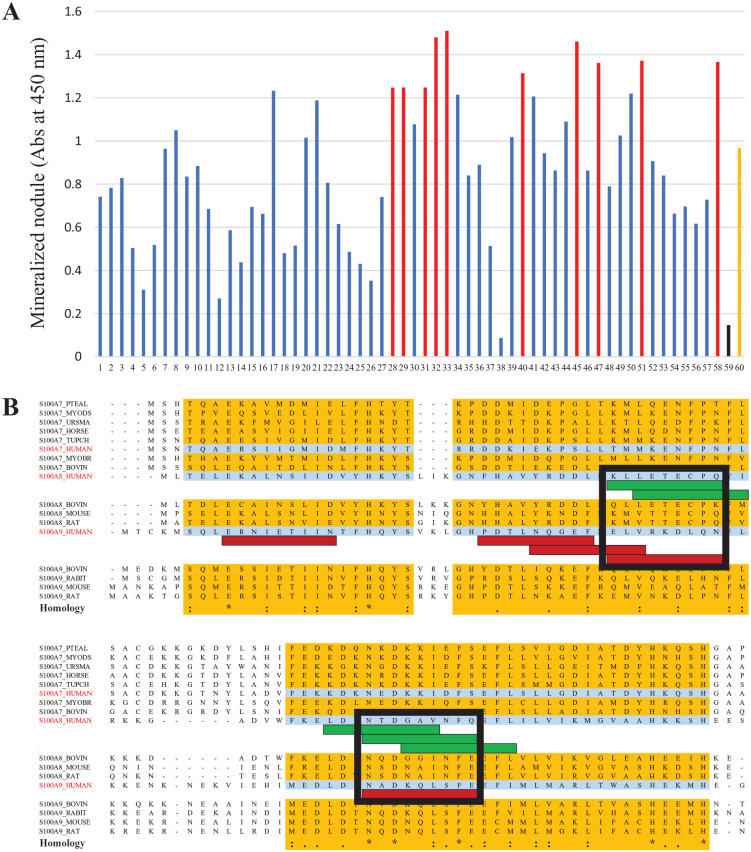
Three highly homologous regions were detected in amino acid sequences of S100A7, S100A8, and S100A9 from multiple mammalian species (Fig. 1). From these regions, 58 peptides (9 residues each) were selected. The selected peptides were subjected to peptide array analysis (Appendix Table 1).
Figure 1.
Investigation of amino acid sequence homology in the protein S100 family via sequence alignment (S100A7: polar bear, Brandt’s bat, Chinese tree shrew, black flying fox, David’s myotis, human, cow, and horse; S100A8: human, cow, mouse, and rat; S100A9: human, cow, rabbit, mouse, and rat). Three regions highlighted in yellow exhibit high homology (a, b, c). “*” indicates complete amino acid identity among all specimens. “:” indicates high amino acid homology. “.” indicates low amino acid homology. Fifty-eight peptides (9 residues each) were selected from highly homologous regions (a, b, c), using the amino acid sequences of human S100A7, S100A8, and S100A9 (Appendix Table 1).
Identification of Functional Peptides
Figure 2A shows the quantification of mineralized nodules that were induced by the 58 peptides in the peptide array analysis. Sequence alignment of 10 peptides with the highest mineralization induction potential (red bars in Fig. 2A) showed that most of them mapped to overlapping regions; green bars in the alignment indicate S100A8-derived peptides, and red bars indicate S100A9-derived peptides (Fig. 2B). On the basis of these overlapping regions, we selected the final candidate peptides from the black squares in Figure 2B. S100A8-derived peptides were No. 1 (KLLETECPQ) and No. 3 (NTDGAVNFQ); S100A9-derived peptides were No. 2 (ELVRKDLQN) and No. 4 (NTDGAVNFQ).
Figure 2.
Identification of functional peptides by the peptide array method. (A) Quantitative analysis of alizarin red staining. Red bars represent 10 peptides with the highest mineralization-inducing ability (Nos. 28, 29, 31, 32, 33, 40, 45, 47, 51, and 58). Black bar represents a random peptide (No. 59). Orange bar represents a Bone Morphogenetic Protein 2 (BMP2)-derived peptide (No. 60). (B) Selection of final candidate peptides based on additional sequence alignment information. Green bar represents an S100A8-derived peptide and red bar represents an S100A9-derived peptide. Because there were 2 overlapping sites in the sequence alignment, multiple peptides were selected from S100A8 and S100A9 (black squares); from these groups of multiple peptides, 4 final candidate peptides were selected (KLLETECPQ, ELVRKDLQN, NTDGAVNFQ, and NTDGAVNFQ).
Direct Pulp-Capping Experiments Using the Selected Peptides
No animals exhibited adverse events. Micro-CT analysis revealed that peptide No. 1 and MTA induced the highest rate of complete tertiary dentin formation in all specimens; this rate was significantly higher than the rates of tertiary dentin formation induced by peptide No. 2 or the control (Fig. 3A).
Figure 3.
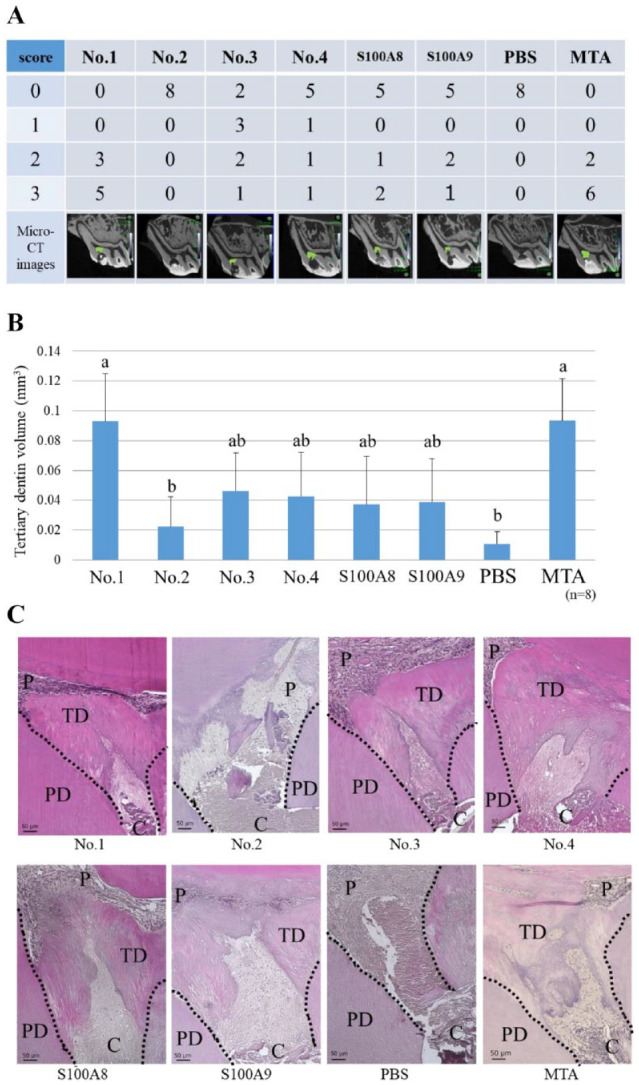
Direct pulp capping using the final candidate peptides. Each peptide (No. 1: KLLETECPQ, No. 2: ELVRKDLQN, No. 3: NTDGAVNFQ, No. 4: NADKQLSFE) (100 µg/mL) was soaked in a gelatin sponge and used as a pulp-capping material. S100A8 or S100A9 (1 µg/mL) was used as a control. (A) Micro–computed tomography (CT) evaluation of tertiary dentin bridge formation. There were significant differences between No. 1 and No. 2, between No. 1 and phosphate-buffered saline (PBS), between mineral trioxide aggregate (MTA) and No. 2, and between MTA and PBS (Kruskal–Wallis test, P < 0.05). Grade 0: no dentin bridge formation; grade 1: dentin bridge formation covers one-third of exposed pulp; grade 2: dentin bridge formation covers two-thirds of exposed pulp; grade 3: dentin bridge formation completely covers exposed pulp. Green area indicates tertiary dentin formation. (B) Quantification of tertiary dentin formation at 4 wk after direct pulp capping (DPC). Different letters indicate significant differences between groups (Kruskal–Wallis test, P < 0.05). (C) Histological images (hematoxylin–eosin staining) of tertiary dentin formed by DPC. Dotted lines indicate boundaries between tertiary dentin and primary dentin. C, cavity; P, pulp; PD, primary dentin; TD, tertiary dentin.
The volume of tertiary dentin induced by peptide No. 1 and MTA was significantly greater than by peptide No. 2 or the control, although it did not significantly differ from the volumes induced by other peptides (Fig. 3B).
Representative hematoxylin–eosin staining images showed small defects in the tertiary dentin induced by peptide Nos. 1, 3, and 4 and MTA; however, these defects did not pass through the pulp or cavity (Fig. 3C). Tertiary dentin induced by recombinant proteins also completely covered the exposed pulp, but its structure appeared sparse compared with the tertiary dentin induced by the corresponding peptides.
Proteome Analysis of Pulpal Wound Healing Induced by Functional Peptides
LC-MS/MS analysis yielded 1,833 cell membrane–derived proteins and 1,557 cytoplasm-derived proteins.
Among the cell membrane–derived proteins, 158 proteins were identified with higher expression levels in the peptide No. 1 or S100A8 condition compared with the control, whereas 250 proteins were identified with lower expression levels compared with the control. Among the cytoplasm-derived proteins, 97 with increased expression were identified, while 67 with decreased expression were identified (Excel spreadsheets at https://data.mendeley.com/datasets/h6mbkgwcbk/2).
Discussion
In current dental practice, VPT (e.g., DPC or pulpotomy) is usually performed with HCSCs, which have enabled better outcomes than treatment with calcium hydroxide (Cushley et al. 2021). However, HCSCs are limited in that they cannot reveal the biomimetic function of pulp wound healing at the cellular or molecular level.
Our previous studies regarding the mechanism of pulpal wound healing indicated that DMCs can be degraded by matrix metalloproteinase molecules; degradation products (e.g., S100A7 and S100A8) might promote wound healing (Smith et al. 2012; Yoshioka et al. 2013; Okamoto et al. 2018; Komichi et al. 2019).
In this study, sequence alignment revealed 3 regions of high amino acid sequence homology (Fig. 1). Moreover, DPC experiments in rats, using recombinant protein as pulp-capping material, showed that both proteins promoted tertiary dentin formation (Komichi et al. 2019). Therefore, the functional sequences of these proteins that affect pulpal wound healing may correspond to amino acid sequences conserved among members of the protein S100 family. In addition, the functional sequences of protein S100 family members may be conserved among mammalian species because human-derived S100 proteins were able to perform their pulp-capping functions in a rat model. Furthermore, S100A9 is expressed in dentin (Widbiller et al. 2019) and promotes epithelial wound healing and bone formation (Zreiqat et al. 2007; Basu et al. 2018). Considering the above findings, we performed sequence alignments of S100A7, S100A8, and S100A9 among multiple mammalian species.
Concerning the number of amino acid residues in test peptides, we used 9 residues because approximately 80% of existing peptide drugs contain fewer than 10 residues (Santos et al. 2016). In addition, the well-known mineralization-inducing W9 peptide (amino acid sequence: YCWSQYLCY), which has been reported to bind to receptor activator of nuclear factor (NF)–κB ligand on osteoblasts and promote osteogenic signals, is composed of 9 residues (Furuya et al. 2013). The peptide array method was originally used to identify amino acid sequences that bind to ligands in protein–protein interactions. Here, we analyzed protein–cell interactions via a peptide array–based interaction assay for solid-bound peptides and anchorage-dependent cells (PIASPAC), which can identify amino acid functional sequences that directly affect cells. Various peptides have been identified using PIASPAC (Kato et al. 2011; Kanie et al. 2016). Therefore, PIASPAC is a useful method for exploring functional regions of proteins based on current bioinformatics capabilities.
Based on alizarin red staining in the peptide array and the overlapping sequences with high homology identified in the sequence alignment analysis, we selected 4 final candidate peptides. The 3-dimensional structural sites of these overlapping sequences were examined using the molecular graphics tool PyMOL (Schrodinger); all 4 peptides were located on the surface in the hairpin region (Fig. 4). This finding indicates that peptide Nos. 1 to 4 have the potential to directly interact with other molecules.
Figure 4.
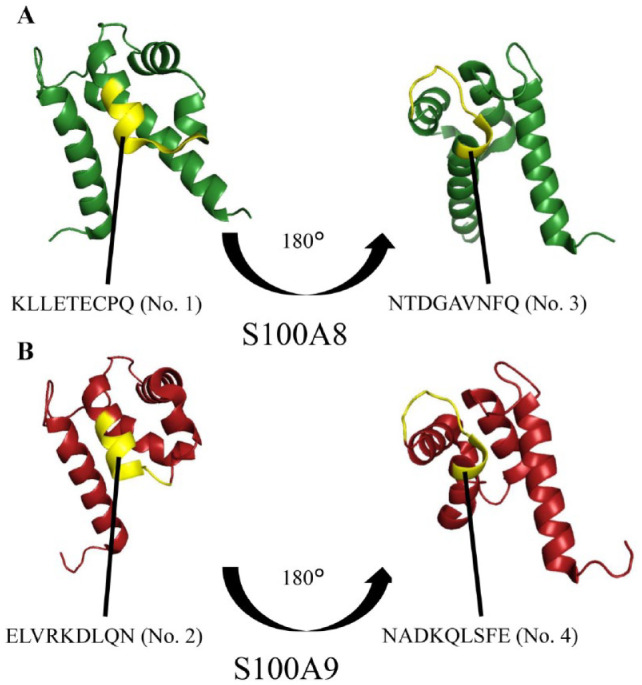
Three-dimensional locations of all peptides in the original S100A8 and S100A9 proteins. (A) Peptides No. 1 (KLLETECPQ) and No. 3 (NTDGAVNFQ) (yellow portion) were located in the hairpin area on the surface of S100A8 (green portion). (B) Peptides No. 2 (ELVRKDLQN) and No. 4 (NADKQLSFE) (yellow portion) were located in the hairpin area on the surface of S100A9 (red portion).
Because none of the peptides exhibited significant cytotoxicity or proliferation, even at the concentration of 100 µg/mL, we performed DPC experiments using this concentration. Peptide Nos. 3 and 4 induced less tertiary dentin formation than peptide No. 1; these peptides correspond to a site known as the EF-hand domain, which is a characteristic region of the protein S100 family. The EF-hand domain functions as a calcium-binding domain with a helix–loop–helix structure and contributes to protein structural stability (Chazin 2011). Therefore, this domain might not actively promote pulpal wound healing by interacting with other proteins. Compared with peptide No. 1, peptide No. 2 exhibited significantly inferior results, despite its location at a site homologous to the location of peptide No. 1 on the amino acid sequence of S100A9; the reason for this discrepancy is unclear, but chemical properties such as isoelectric point might have led to distinct functional differences. Further investigation is necessary of the mechanism by which peptide No. 1 promotes pulpal wound healing (e.g., based on its biological properties as an amino acid sequence or based on its chemical properties). In addition, because S100A8 or S100A9 was used as controls (at a molar concentration of 1/1,000 for each peptide) and exhibited inferior results compared with peptide No. 1, it was difficult to simply compare the experimental results of DPC. However, it is impractical to use recombinant proteins as pulp-capping materials at the same molar concentration as the peptides used in clinical practice because of safety and cost. Peptide No. 1 showed equivalent tertiary dentin formation to MTA. Considering MTA is a completed commercial product while the peptide is an experimental solution without any career material, a new product developed using this peptide could perform better than MTA.
In this report, DPC was performed using sound pulp/teeth. To simulate clinical situations, these experiments should be performed using a caries model or under other infectious conditions.
Results of RT-qPCR indicated that greater expression of DSPP and Nestin by peptide No. 1 was induced at an early stage of wound healing. This peptide might also affect the function of pulp cells both in vivo and in vitro (Appendix Fig. 2). To examine the mechanism by which peptide No. 1 promoted pulpal wound healing, LC-MS/MS was explored according to the Gene Ontology classification for each identified protein. In specimens incubated with peptide No. 1 or S100A8, proteins that were upregulated compared with the control were linked to Gene Ontology terms associated with promotion of the NF-κB signaling pathway. In contrast, proteins that were downregulated compared with the control were linked to Gene Ontology terms associated with promotion of the mitogen-activated protein kinase (MAPK) signaling pathway (Table). The NF-κB signaling pathway regulates immune responses, inflammation, cell proliferation, and cell differentiation (Liang et al. 2004; Oeckinghaus and Ghosh 2009; Kaileh and Sen 2010). Furthermore, this pathway regulates the odontoblast-like cell differentiation and mineralization of dental pulp cells (Lee et al. 2013; Kuramoto et al. 2019; Wu et al. 2019), promotes angiogenesis in dental pulp (Shen et al. 2021), and promotes wound healing through anti-inflammatory effects via M2 macrophages (Lawrence and Fong 2010). Therefore, we speculate that the No. 1 peptide affects the NF-κB signaling pathway by promoting pulpal wound healing through cell differentiation, mineralization, and anti-inflammatory mechanisms. The MAPK signaling pathway regulates cell proliferation, differentiation, migration, and apoptosis (Lu and Xu 2006; Krishna and Narang 2008; Keshet and Seger 2010; Sun et al. 2015). Inhibition of the MAPK signaling pathway can prevent bone destruction (Mbalaviele et al. 2006); in mouse neural tissues, this inhibition had anti-inflammatory effects (Hong et al. 2020). Thus, we suspect that inhibition of the MAPK pathway by peptide No. 1 led to the suppression of inflammation, thereby promoting pulpal wound healing. In addition, the other potent functional proteins associated with angiogenesis or the small mothers against decapentaplegic (SMAD) signaling pathway were also upregulated (Appendix Table 3). Angiogenesis can be an essential event for tissue repair, and the SMAD signaling pathway was reported to be associated with cell differentiation of the pulp tissue (Woo et al. 2016). Furthermore, another report revealed the NF-κB and SMAD signaling pathways were both upregulated by DSPP stimulation during hDPSC mineralization. This indicated that plural molecular pathways could cooperatively work for tissue repair (Lee et al. 2012; Woo et al. 2016). However, the relationship between S100A8 and the SMAD signaling pathway is still unclear. Therefore, it is necessary to elucidate the precise effect of peptide No. 1 on pulpal wound healing.
Table.
Upregulated Proteins Identified by Liquid Chromatography–Tandem Mass Spectrometry in Human Dental Pulp Stem Cells Incubated with Protein S100 Family-Derived Peptide or S100A8 for NF-κB Signaling and MAPK Signaling.
| Identified Cell Membrane–Derived Protein | Quantitative value (a.u.) | Associated Signaling Pathway | ||
|---|---|---|---|---|
| Peptide No. 1 | S100A8 | Control | ||
| Cell membrane proteins | ||||
| Flotillin 1 (FLOT1) | 10.104 | 6.3733 | 4.3079 | NF-κB signaling |
| Very long-chain (3R)–3-hydroxyacyl-CoA dehydratase (3KCR1) | 6.7361 | 5.3111 | 3.231 | |
| Protein LYRIC (LYRIC) | 5.6134 | 3.1867 | 2.154 | |
| Gap junction α1 protein (GJA1) | 3.3681 | 4.2489 | 1.077 | |
| Dual specificity mitogen-activated protein kinase kinase 1 (MAP2K1) | 0 | 2.1244 | 2.154 | MAPK signaling |
| Cadherin 2 (CDH2) | 0 | 3.1867 | 3.231 | |
| GTPase Nras (NRAS) | 0 | 7.4355 | 7.5389 | |
| Mitogen-activated protein kinase 1 | 1.1227 | 2.1244 | 3.231 | |
| Cytoplasmic proteins | ||||
| Protein S100A13 (S100A13) | 1.889 | 1.8042 | 0.93281 | NF-κB signaling |
| Nucleolar protein 3 (NOL3) | 1.889 | 1.8042 | 0.93281 | |
| Adapter molecule crk (CRK) | 2.8335 | 6.3146 | 6.5297 | MAPK signaling |
| Proteasome subunit β type 7 | 0 | 2.7062 | 3.7312 | |
MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor–κB.
In conclusion, this is the first study to demonstrate the anti-inflammatory effects of a functional peptide derived from S100A8, a protein that was previously shown to promote wound healing in dental pulp to our knowledge. Our findings may support the development of novel materials for next-generation VPT, with the potential to treat irreversible pulpitis.
Author Contributions
M. Watanabe, contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript: M. Okamoto, J. Ohshima, K. Kanie, R. Kato, K. Uto, M. Ebara, A. Yamawaki-Ogata, Y. Narita, contributed to data conception, drafted and critically revised the manuscript: S. Komichi, H. Hailing, S. Matsumoto, K. Moriyama, S. Abe, M. Morita, M. Ali, I. Kozaki, contributed to data acquisition and analysis, critically revised the manuscript: K. Takebe, contributed to conception, data acquisition and analysis, critically revised the manuscript: A, Fujimoto, contributed to data acquisition and analysis, critically revised the manuscript: Y. Takahashi, contributed to conception and design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript: M. Hayashi, contributed to conception and design, data analysis and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-docx-1-jdr-10.1177_00220345221135766 for Novel Functional Peptide for Next-Generation Vital Pulp Therapy by M. Watanabe, M. Okamoto, S. Komichi, H. Huang, S. Matsumoto, K. Moriyama, J. Ohshima, S. Abe, M. Morita, M. Ali, K. Takebe, I. Kozaki, A. Fujimoto, K. Kanie, R. Kato, K. Uto, M. Ebara, A. Yamawaki-Ogata, Y. Narita, Y. Takahashi and M. Hayashi in Journal of Dental Research
Acknowledgments
We thank Prof. Hiroyuki Honda for providing technical assistance with peptide synthesis. We thank Ryan Chastain-Gross, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. A supplemental appendix to this article is available online.
Footnotes
A supplemental appendix to this article is available online.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Grants-in-Aid for Scientific Research (22H03268 to Y. Takahashi, 21K09915 to M. Okamoto, 21K16967 to S. Komichi and 20K05227) to K. Kanie from the Japan Society for the Promotion of Science. This research was supported by AMED under grant JP22ym0126809 and JP21he0422006.
Data Sharing Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
ORCID iD: Y. Takahashi  https://orcid.org/0000-0002-7974-1252
https://orcid.org/0000-0002-7974-1252
References
- Basu A, Munir S, Mulaw MA, Singh K, Crisan D, Sindrilaru A, Treiber N, Wlaschek M, Huber-Lang M, Florian G, et al. 2018. A novel S100A8/A9 induced fingerprint of mesenchymal stem cells associated with enhanced wound healing. Sci Rep. 8(1):6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan DJ, Cai JW, Yin GS, White BA. 2005. Root canal filled versus non-root canal filled teeth: a retrospective comparison of survival times. J Public Health Dent. 65(2):90–96. [DOI] [PubMed] [Google Scholar]
- Chazin WJ. 2011. Relating form and function of EF-hand calcium binding proteins. Acc Chem Res. 44(3):171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushley S, Duncan HF, Lappin MJ, Chua P, Elamin AD, Clarke M, El-Karim IA. 2021. Efficacy of direct pulp capping for management of cariously exposed pulps in permanent teeth: a systematic review and meta-analysis. Int Endod J. 54(4):556–571. [DOI] [PubMed] [Google Scholar]
- Furuya Y, Inagaki A, Khan M, Mori K, Penninger JM, Nakamura M, Udagawa N, Aoki K, Ohya K, Uchida K, et al. 2013. Stimulation of bone formation in cortical bone of mice treated with a receptor activator of nuclear factor-kB ligand (RANKL)-binding peptide that possesses osteoclastogenesis inhibitory activity. J Biol Chem. 288(8):5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henninot A, Collins JC, Nuss JM. 2018. The current state of peptide drug discovery: back to the future? J Med Chem. 61(4):1382–1414. [DOI] [PubMed] [Google Scholar]
- Hong X, Jiang F, Li Y, Fang L, Qian Z, Chen H, Kong R. 2020. Treatment with 5-methoxytryptophan attenuates microglia-induced neuroinflammation in spinal cord trauma. Int Immunopharmacol. 88:106988. [DOI] [PubMed] [Google Scholar]
- Iohara K, Nakashima M, Ito M, Ishikawa M, Nakasima A, Akamine A. 2004. Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2. J Dent Res. 83(8):590–595. [DOI] [PubMed] [Google Scholar]
- Kaileh M, Sen R. 2010. Role of NF-kappaB in the anti-inflammatory effects of tocotrienols. J Am Coll Nutr. 29(3):S334–S339. [DOI] [PubMed] [Google Scholar]
- Kanie K, Kurimoto R, Tian J, Ebisawa K, Narita Y, Honda H, Kato R. 2016. Screening of osteogenic-enhancing short peptides from BMPs for biomimetic material applications. Materials (Basel). 9(9):730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato R, Kaga C, Kanie K, Kunimatsu M, Okochi M, Honda H. 2011. Peptide array-based peptide-cell interaction analysis. Mini Rev Org Chem. 8(2):171–177. [Google Scholar]
- Keshet Y, Seger R. 2010. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 661:3–38. [DOI] [PubMed] [Google Scholar]
- Komichi S, Takahashi Y, Okamoto M, Ali M, Watanabe M, Huang HL, Nakai T, Cooper P, Hayashi M. 2019. Protein S100-A7 derived from digested dentin is a critical molecule for dentin pulp regeneration. Cells. 8(9):1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna M, Narang H. 2008. The complexity of mitogen-activated protein kinases (MAPKs) made simple. Cell Mol Life Sci. 65(22):3525–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto H, Hirao K, Yumoto H, Hosokawa Y, Nakanishi T, Takegawa D, Washio A, Kitamura C, Matsuo T. 2019. Caffeic acid phenethyl ester (CAPE) induces VEGF expression and production in rat odontoblastic cells. Biomed Res Int. 2019:5390720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Fong C. 2010. The resolution of inflammation: anti-inflammatory roles for NF-kappaB. Int J Biochem Cell Biol. 42(4):519–523. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Monsef M, Torabinejad M. 1993. Sealing ability of a mineral trioxide aggregate for repair of lateral root perforations. J Endod. 19(11):541–544. [DOI] [PubMed] [Google Scholar]
- Lee SY, Kim SY, Park SH, Kim JJ, Jang JH, Kim EC. 2012. Effects of recombinant dentin sialoprotein in dental pulp cells. J Dent Res. 91(4):407–412. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kang YM, Heo MJ, Kim GE, Bhattarai G, Lee NH, Yu MK, Yi HK. 2013. The survival role of peroxisome proliferator-activated receptor gamma induces odontoblast differentiation against oxidative stress in human dental pulp cells. J Endod. 39(2):236–241. [DOI] [PubMed] [Google Scholar]
- Liang Y, Zhou Y, Shen PP. 2004. NF-kappaB and its regulation on the immune system. Cell Mol Immunol. 1(5):343–350. [PubMed] [Google Scholar]
- Lu ZM, Xu SC. 2006. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 58(11):621–631. [DOI] [PubMed] [Google Scholar]
- Machado M, Stafuzza TC, Vitor LLR, da Costa SA, da Costa SM, Neto NL, Oliveira TM. 2020. Pulp repair response after the use of a dentin-pulp biostimulation membrane (BBio) in primary teeth: study protocol for a randomized clinical trial. Trials. 21(1):874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbalaviele G, Anderson G, Jones A, De Ciechi P, Settle S, Mnich S, Thiede M, Abu-Amer Y, Portanova J, Monahan J. 2006. Inhibition of p38 mitogen-activated protein kinase prevents inflammatory bone destruction. J Pharmacol Exp Ther. 317(3):1044–1053. [DOI] [PubMed] [Google Scholar]
- McLachlan JL, Sloan AJ, Smith AJ, Landini G, Cooper PR. 2004. S100 and cytokine expression in caries. Infect Immun. 72(7):4102–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M, Tachibana K, Iohara K, Ito M, Ishikawa M, Akamine A. 2003. Induction of reparative dentin formation by ultrasound-mediated gene delivery of growth/differentiation factor 11. Hum Gene Ther. 14(6):591–597. [DOI] [PubMed] [Google Scholar]
- Oeckinghaus A, Ghosh S. 2009. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 1(4):a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Takahashi Y, Komichi S, Ali M, Yoneda N, Ishimoto T, Nakano T, Hayashi M. 2018. Novel evaluation method of dentin repair by direct pulp capping using high-resolution micro-computed tomography. Clin Oral Investig. 22(8):2879–2887. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Takahashi Y, Komichi S, Cooper PR, Hayashi M. 2018. Dentinogenic effects of extracted dentin matrix components digested with matrix metalloproteinases. Sci Rep. 8(1):10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paula AB, Laranjo M, Marto CM, Paulo S, Abrantes AM, Casalta-Lopes J, Marques-Ferreira M, Botelho MF, Carrilho E. 2018. Direct pulp capping: what is the most effective therapy? Systematic review and meta-analysis. J Evid Based Dent Pract. 18(4):298–314. [DOI] [PubMed] [Google Scholar]
- Sachdeva S. 2017. Peptides as ‘drugs’: the journey so far. Int J Pept Res Ther. 23(1):49–60. [Google Scholar]
- Santos GB, Ganesan A, Emery FS. 2016. Oral administration of peptide-based drugs: beyond Lipinski’s rule. ChemMedChem. 11(20):2245–2251. [DOI] [PubMed] [Google Scholar]
- Shen S, Shang LL, Liu HR, Liang QY, Liang W, Ge SH. 2021. AGGF1 inhibits the expression of inflammatory mediators and promotes angiogenesis in dental pulp cells. Clin Oral Investig. 25(2):581–592. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Scheuen BA, Takahashi Y, Ferracane JL, Shelton RM, Cooper PR. 2012. Dentine as a bioactive extracellular matrix. Arch Oral Biol. 57(2):109–121. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. 2015. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 35(6):600–604. [DOI] [PubMed] [Google Scholar]
- Walker JV, Zhuang H, Singer D, Illsley CS, Kok WL, Sivaraj KK, Gao Y, Bolton C, Liu YY, Zhao MY, et al. 2019. Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation. Nat Commun. 10(1):3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widbiller M, Schweikli H, Bruckmann A, Rosendahl A, Hochmuth E, Lindner SR, Buchalla W, Geller KM. 2019. Shotgun proteomics of human dentin with different prefractionation methods. Sci Rep. 9(1):4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo SM, Kim WJ, Lim HS, Choi NK, Kim SH, Kim SM, Jung JY. 2016. Combination of mineral trioxide aggregate and platelet-rich fibrin promotes the odontoblastic differentiation and mineralization of human dental pulp cells via BMP/Smad signaling pathway. J Endod. 42(1):82–88. [DOI] [PubMed] [Google Scholar]
- Wu Q, Li J, Song PP, Chen J, Xu YZ, Qi SC, Ma J, Pan QH. 2019. Knockdown of NRAGE induces odontogenic differentiation by activating NF-kB signaling in mouse odontoblast-like cells. Connect Tissue Res. 60(2):71–84. [DOI] [PubMed] [Google Scholar]
- Yoshioka S, Takahashi Y, Abe M, Michikami I, Imazato S, Wakisaka S, Hayashi M, Ebisu S. 2013. Activation of the Wnt/β-catenin pathway and tissue inhibitor of metalloprotease 1 during tertiary dentinogenesis. J Biochem. 153(1):43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zreiqat H, Howlett CR, Gronthos S, Hume D, Geczy CL. 2007. S100A8/S100A9 and their association with cartilage and bone. J Mol Histol. 38(5):381–391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-jdr-10.1177_00220345221135766 for Novel Functional Peptide for Next-Generation Vital Pulp Therapy by M. Watanabe, M. Okamoto, S. Komichi, H. Huang, S. Matsumoto, K. Moriyama, J. Ohshima, S. Abe, M. Morita, M. Ali, K. Takebe, I. Kozaki, A. Fujimoto, K. Kanie, R. Kato, K. Uto, M. Ebara, A. Yamawaki-Ogata, Y. Narita, Y. Takahashi and M. Hayashi in Journal of Dental Research