Abstract
Reelin, a large extracellular protein, plays several critical roles in brain development and function. It is encoded by RELN, first identified as the gene disrupted in the reeler mouse, a classic neurological mutant exhibiting ataxia, tremors and a ‘reeling’ gait. In humans, biallelic variants in RELN have been associated with a recessive lissencephaly variant with cerebellar hypoplasia, which matches well with the homozygous mouse mutant that has abnormal cortical structure, small hippocampi and severe cerebellar hypoplasia. Despite the large size of the gene, only 11 individuals with RELN-related lissencephaly with cerebellar hypoplasia from six families have previously been reported. Heterozygous carriers in these families were briefly reported as unaffected, although putative loss-of-function variants are practically absent in the population (probability of loss of function intolerance = 1). Here we present data on seven individuals from four families with biallelic and 13 individuals from seven families with monoallelic (heterozygous) variants of RELN and frontotemporal or temporal-predominant lissencephaly variant. Some individuals with monoallelic variants have moderate frontotemporal lissencephaly, but with normal cerebellar structure and intellectual disability with severe behavioural dysfunction. However, one adult had abnormal MRI with normal intelligence and neurological profile. Thorough literature analysis supports a causal role for monoallelic RELN variants in four seemingly distinct phenotypes including frontotemporal lissencephaly, epilepsy, autism and probably schizophrenia. Notably, we observed a significantly higher proportion of loss-of-function variants in the biallelic compared to the monoallelic cohort, where the variant spectrum included missense and splice-site variants. We assessed the impact of two canonical splice-site variants observed as biallelic or monoallelic variants in individuals with moderately affected or normal cerebellum and demonstrated exon skipping causing in-frame loss of 46 or 52 amino acids in the central RELN domain. Previously reported functional studies demonstrated severe reduction in overall RELN secretion caused by heterozygous missense variants p.Cys539Arg and p.Arg3207Cys associated with lissencephaly suggesting a dominant-negative effect. We conclude that biallelic variants resulting in complete absence of RELN expression are associated with a consistent and severe phenotype that includes cerebellar hypoplasia. However, reduced expression of RELN remains sufficient to maintain nearly normal cerebellar structure. Monoallelic variants are associated with incomplete penetrance and variable expressivity even within the same family and may have dominant-negative effects. Reduced RELN secretion in heterozygous individuals affects only cortical structure whereas the cerebellum remains intact. Our data expand the spectrum of RELN-related neurodevelopmental disorders ranging from lethal brain malformations to adult phenotypes with normal brain imaging.
Keywords: autism, epilepsy, lissencephaly, RELN, Reelin
Di Donato et al. present data from seven individuals with homozygous, and 13 with heterozygous, pathogenic variants in the RELN gene. They reanalyse prior data to define a broad spectrum of RELN-associated phenotypes, from lissencephaly with or without cerebellar hypoplasia to neuropsychiatric disorders including autism and schizophrenia.
Introduction
Reelin is a large extracellular protein that plays several roles in brain development and function by regulating neuronal migration, laminar organization, dendritic morphogenesis and neurotransmission. It is encoded by RELN, a gene first identified as disrupted in the reeler mouse, a classic neurological mutant that exhibits ataxia, tremors and a characteristic ‘reeling’ gait.1–3 The neuropathological features in mouse mutants consist of a poorly laminated and ‘inverted’ cerebral cortex, small malformed hippocampi, severe cerebellar hypoplasia with an afoliar surface, and defects in dendritic and synaptic morphogenesis. Heterozygous mutants have normal brain histology but subtle behavioural defects and mild abnormalities in dendritic and synaptic morphogenesis.
In humans, biallelic pathogenic variants of RELN have been associated with a variant of lissencephaly with cerebellar hypoplasia (LIS-CBLH) that fits well with features of the reeler mutant including an uncommon frontotemporal-predominant pachygyria, moderately thick 5–10 mm cortex, small hippocampi and severe cerebellar hypoplasia with an afoliar surface. Despite the large size of the gene with 10.4 kb of coding sequence and 65 exons extending over 517.9 kb of human chromosome 7 (and hence a large target for mutations), only 11 individuals with LIS-CBLH from six families have been reported.4–9
Heterozygous or monoallelic sequence variants of uncertain significance have been associated with autism, epilepsy, schizophrenia, myoclonus-dystonia and cortical malformations including both LIS and polymicrogyria. Targeted or genome-wide association studies (GWAS) have suggested associations between RELN and autism, bipolar disorder and Alzheimer's disease. These individuals have had normal brain imaging, at least by report, as few images have been published. The relationship between RELN-related LIS-CBLH and RELN-related neurodevelopmental and neuropsychiatric disorders has remained elusive.
Here we report seven children from four families with biallelic (homozygous or compound heterozygous) and 13 individuals from seven families with monoallelic (heterozygous) variants of RELN and frontotemporal- or temporal-predominant LIS. Most individuals with monoallelic variants have moderate frontotemporal LIS that is less severe than biallelic individuals but with normal cerebellar structure and a constant association with intellectual disability (ID) and severe behavioural dysfunction. Our observations in individuals with monoallelic variants significantly expand the spectrum of phenotypes seen in humans and bridge the gap between autosomal recessive RELN-related LIS-CBLH and the diverse neurodevelopmental and neuropsychiatric disorders that have been associated with monoallelic variants in RELN.
Materials and methods
Subject recruitment
This study originated with targeted sequencing of 17 LIS-associated genes including RELN in 216 individuals with unsolved LIS ascertained over several decades,10 and continued as new subjects were referred to us. The study was approved by Institutional Review Boards at the University of Minnesota, Seattle Children’s Hospital, the University of California—San Diego, Meyer Children’s Hospital in Florence, and the Royal Children’s Hospital in Melbourne. All families provided informed consent, or we received a waiver of consent for de-identified data.
Clinical data
We obtained medical histories emphasizing development, behaviour and epilepsy, and brain imaging studies (MRI in 18, CT scan in 1) for seven individuals with biallelic variants and 13 with monoallelic variants (Table 1 and Supplementary Tables 1 and 2). Clinical reports for most subjects are included in the Supplementary material. Pedigrees for the multiplex families are shown in Supplementary Fig. 1.
Table 1.
Clinical features in individuals with monoallelic variants in RELN
| Cohort | Monoallelic (Discovery cohort) | Unaffected | |||||
|---|---|---|---|---|---|---|---|
| Patient ID | LR15-139 | LR05-375a1 | LR05-375a2 | LR02-111a1 | LR02-111a2 | LR02-111a3 | LR02-111r1 |
| Sex/relationship | F/proband | M/proband | F/sister | M/brother | M/proband | M/brother | F/aunt |
| Genotype | |||||||
| Protein variant | p.Cys539Arg | p.Leu2552Pro | same as sib | p.S3328Sfs* | same as sib | same as sib | same as a1–a3 |
| Inheritance | de novo | paternal | paternal | paternal | paternal | paternal | maternal |
| Phenotype | |||||||
| Age at last evaluation | 26 years | 1–7/12 years | 5 months | 28 years | 22 years | 20 years | 52 years |
| MRI: Gyral pattern | LIS FL-TL mod | LIS FL-TL mod | LIS FL-TL mod | LIS FL-TL mod | LIS FL-TL mod | LIS TL mod | LIS TL mod |
| Growth: OFC | 54 cm (39%) | 44 cm (−3 SD) | 41.5 cm (32%) | 47.3 cm (12%) | 43.4 cm (−2.1 SD) | ||
| Intellectual disability | moderate | severe | moderate | severe | mild | normal | |
| Seizures | no | no | no | no | no | no | no |
| Aggression unprovoked | no | yes | yes | yes | no | ||
| Autism | no | no | |||||
| Sleep disruption | no | yes severe | yes severe | no | no | ||
| Cohort | Monoallelic (Validation cohort) | ||||||
| Patient ID | LR17-364a2 | LR17-364a1 | LR17-364f | LR18-437 | LR21-446 | LR20-419 | |
| Family relationship | M/proband | F/sister | M/father | F/proband | F/proband | F/proband | |
| Genotype | |||||||
| Protein variant | p.Ile1737_Val1784delinsMet (skips exon 35) | p.Gly2536Arg | p.Gly2536Arg | p.Arg3207Cys | |||
| Inheritance | paternal | paternal | paternal | de novo | de novo | de novo | |
| Phenotype | |||||||
| Age at last evaluation | 14 years | 18 years | 35 years | 9 years | 8 years | 5 years | |
| MRI: Gyral pattern | LIS TL mod | LIS TL mod | LIS TL mod | LIS FL-TL mod | LIS FL-TL mod | LIS FL-TL mod | |
| Growth: OFC (cm) | 55.1 cm | (<25%) | 35.5 (−2.4 SD) | ||||
| Intellectual disability | moderate | moderate | mild | mild | yes | mild | |
| Seizures | yes | no | no | yes | no | yes | |
| Aggression unprovoked | yes | yes | yes | no | |||
| Autism | yes | some features | no | no | |||
| Sleep disruption Y/N | yes moderate | yes moderate | no | ||||
F = female; FL = frontal lobe; M = male; TL = temporal lobe. See Supplementary Table 2 for additional data.
Genomic data
Genomic data (Supplementary Tables 3 and 4) were obtained using a combination of whole-exome sequencing (WES),11–14 targeted sequencing panels10,15,16 and Sanger sequencing in 20 individuals from seven families. Targeted panels (17 or 82 genes) were performed in six individuals, a targeted panel (17 genes) followed by WES in five individuals, WES only in five individuals, a targeted sequencing panel (n > 100 genes) plus del-dup analysis in one individual and Sanger sequencing to detect a known familial variant in three individuals. The authors personally reviewed sequencing data for 11/12 targeted panels and 9/10 WES studies for variants in other genes involved in neurodevelopmental disorders with no other candidate variants found. A detailed description of the methods is included in the Supplementary material.
Variant annotation and analysis
The 11 sequence variants from our study and 38 from 21 previous reports were consistently annotated using the Franklin website by Genoox (https://franklin.genoox.com) for RELN transcript NM_005045.4, which includes 65 exons. We recorded the classification for each variant adapted from ACMG-AMP criteria,13 as well as the allele frequency and number of homozygous individuals in the Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org), and the scaled (PHRED) CADD (combined annotation dependent depletion) score.17,18 Some variants analysed were rare (below 1%) but recurrent: 13/46 were observed >40 times in gnomAD.
CRISPR genome editing
Canonical splice variants seen in affected individuals from the LR17-413 (c.4747+2T>G) and LR17-364 (c.5351+1G>A) families were introduced into human induced pluripotent stem cell line SC102A-1 (System Biosciences) by CRISPR-mediated homology directed repair using a donor DNA template with cDNA coordinates given in relation to transcript NM_005045.4. We used the Alt-R™ CRISPR system from Integrated DNA Technologies (IDT) with site specific gRNA and homology directed repair template sequences (Supplementary Table 5), and Cas9 enzyme for position c.4747+2 and Cpf1 enzyme for position c.5351+1. We next used IDT recommended protocols for electroporation with the Amaxa Nucleofector System for both CRISPR systems.19,20 Briefly, guides and corresponding nucleases were combined to form RNP complexes and co-transfected with single-strand homology directed repair templates into induced pluripotent stem cells by electroporation using Alt-R™ Cas9 Electroporation and Alt-R™ homology directed repair enhancers. Electroporation of induced pluripotent stem cells was performed with the Amaxa Human Stem Cell Nucleofector Kit 2 (Lonza). After 48 h of culturing, electroporated cells were seeded at low density for subsequent single clone picking a few days later. DNA was isolated from the clones and targeted loci amplified by PCR. Sanger sequencing was used to identify clones with the intended point mutations.
mRNA splicing analysis
RNA was extracted using the RNeasy Mini Kit (Qiagen) and cDNA preparation performed with the SuperScript™ VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific). Targeted cDNA amplification was done using locus specific primers (Supplementary Table 6) with detection of PCR products by gel electrophoresis and Sanger sequencing of selected products (Supplementary Figs 2 and 3 with unmodified full-length gel images shown in Supplementary Fig. 4).
Data availability
Raw sequencing data supporting the RELN variants in our lissencephaly cohorts that were generated in research laboratories are available from the corresponding author, on reasonable request. The authors confirm that remaining data supporting the findings of this study are available within the article or Supplementary material.
Results
This study grew from a previous study using a targeted sequencing panel of 17 LIS-associated genes to investigate a cohort of 216 individuals with unexplained LIS that detected biallelic pathogenic variants of RELN in four children.10 From that point, our study evolved in steps driven by new data from multiple sources that steadily built support for the pathogenicity of monoallelic variants.
Evolution of the study
As we were completing our prior study,10 we became aware of data on an ENU-generated mouse mutant that proved to have a C-terminal deletion of Reln.21,22 The cortical lamination defect was comparable to defects described in classic reeler mutants, although slightly less severe. However, size and structure of the cerebellum were normal.
The occurrence of reeler-like cortical histology with normal cerebellar structure reminded us of several children with frontal predominant LIS and normal cerebellum and prompted us to reexamine sequence data from our unexplained LIS cohort for heterozygous RELN variants filtered out of our original analysis. We found monoallelic rare variants not seen in gnomAD in six individuals from three families including one de novo variant (Table 1, discovery cohort). Brain imaging studies in all six showed frontotemporal or temporal-predominant LIS with moderately thick 5–10 mm cortex and normal cerebellum. By this time numerous reports had implicated heterozygous variants in RELN as contributing to the cause of autism, epilepsy, schizophrenia and other disorders. Our observations combined with these reports led us to hypothesize that heterozygous (hereafter, monoallelic) variants of RELN can be pathogenic in humans with a phenotype intermediate between RELN-related LIS-CBLH and other neurodevelopmental or neuropsychiatric disorders.
We then examined our large databases of individuals with brain malformations and reached out to other specialists to ascertain additional subjects with cortical malformations and RELN variants. This process identified another three siblings with biallelic variants, six individuals with monoallelic variants from four new families (Table 1, validation cohort) and a clinically unaffected carrier from our original family. Data from these individuals and families collectively demonstrate a continuous series of brain imaging and neurodevelopmental phenotypes that we describe next.
New cohort with lissencephaly variants
Genomic data—biallelic
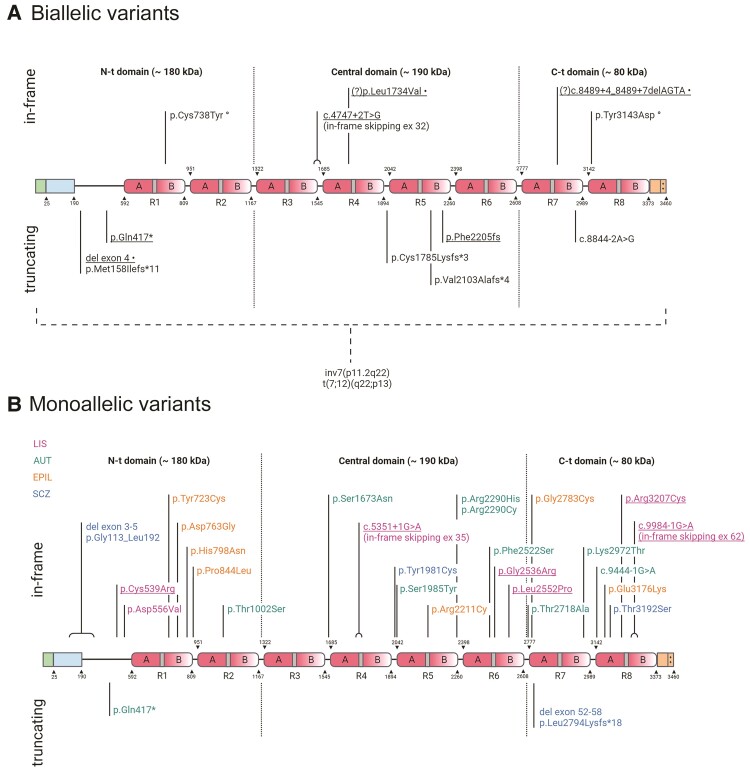
We present data on seven individuals from four unrelated families with LIS-CBLH and biallelic variants in RELN including three homozygous variants (one nonsense, one small indel leading to a frameshift and one splice site) and two compound heterozygous variants (Fig. 1A, Supplementary Tables 1, 3 and 4). We tested the effect of the homozygous canonical splice-site variant c.4747+2T>G segregating in three siblings (LR17-413a1-a3) and found mutant mRNA with skipping of exon 32 in a cell line with c.4747+2T>G that would result in an in-frame deletion of 52 amino acids in the central RELN domain (Supplementary Fig. 2). The heterozygous variants found in subject LR14-063 include a microdeletion of exon 4 inherited from her mother, and three variants inherited from her father including a probably pathogenic splice-site variant (c.8489+4_8489+7delAGTA, Fig. 1A) as well as two probably benign variants with relatively high frequencies (0.13% and 0.28% in the African American population; Supplementary Table 3, not shown in Fig. 1A).
Figure 1.
RELN protein cartoon showing pathogenic and probably pathogenic variants. Protein diagrams show the domain organization of RELN with two in vivo proteolysis sites shown as dotted lines highlighting N-terminal (N-t), Central and C-terminal (C-t) domains. The known boundaries of each domain or other functional region are indicated above or below the diagram and identified by numbers corresponding to the human RELN sequence (Uniprot no. P78509). RELN repeats are numbered (R1–R8) and their composition is marked with sub-repeats A and B separated by EGF-like domains (grey). The N-terminal region contains a signal peptide (positions 0–25 in green) and F-spondin-like domain (positions 25–190 in blue) followed by a unique sequence region (positions 190–502). The C-terminal region (3373–3460 in orange) ends with a stretch of 33 amino acids rich in basic residues (++). (A) Biallelic variants in RELN. Thirteen variants are shown, including six reported in this study (underlined). Compound heterozygous variants in one individual are marked with symbols following the description of the variant; (?) indicates two VUS identified in cis in one individual, both with potential disease relevance. Two structural chromosome rearrangements disrupting RELN are indicated below the dashed bracket. (B) Monoallelic variants in RELN. Twenty-seven variants are shown, including six reported in this study (underlined). The colour legend for associated phenotypes is also shown beneath the diagram; AUT = autism; EPIL = lateral temporal epilepsy; SCZ = schizophrenia. Created with BioRender.com.
We also re-analysed genomic data from previously published reports on 11 individuals from six families with LIS-CBLH and biallelic RELN mutations (Fig. 1A, Supplementary Tables 3 and 4) and found 2 homozygous chromosome rearrangements with breakpoints in 7q22.1 predicted to truncate RELN, three homozygous splice-site variants and two compound heterozygous missense variants in one child.4–9 Our updated analysis of the three splice-site variants predicts all to cause frameshifts with premature stop codons: skipping of exon 37 with Glu1844Profs*22 in a Saudi family, skipping of exon 43 with p.Gly2175Cysfs*11 in a British family (a genomic variant was not found and so is presumably deep intronic) and skipping of exon 55 with p.Phe2949Asnfs*8 in a Moroccan family.4,5,8 Combining our series with previous reports, 12 pathogenic variants have been reported, eight leading to protein truncation and four to in-frame mutations.
Genomic data—monoallelic
We present data on 13 individuals from seven families with frontotemporal or temporal LIS and monoallelic variants in RELN (Fig. 1B, Table 1 and Supplementary Tables 2–4), including one clinically unaffected adult relative. The six alleles include two splice-site variants (one in frame, one C-terminal deletion) and four missense variants, including one recurrent variant in two unrelated families. All CADD scores were 24.9 or higher. We studied the canonical splice-site variant c.5351+1G>A segregating in three individuals in one family (LR17-364), and found that expression of mutant mRNA caused skipping of exon 35 and in-frame loss of 46 amino acids in the fourth EGF-like domain (Supplementary Fig. 3).
While predicted to skip exon 62 of 65, the canonical splice-site mutation c.9984-1G>A may result in shortened protein partly comparable to the mouse RelnCTRdel allele that skips exon 63. Functional studies described in a preprint repository showed that the amino acid substitutions seen in two of our subjects with de novo RELN variants (p.Cys539Arg, p.Arg3207Cys) caused a severe reduction in RELN secretion (increased concentration in cell lysates, decreased in media) when transfected alone or when co-transfected with wild-type RELN.23
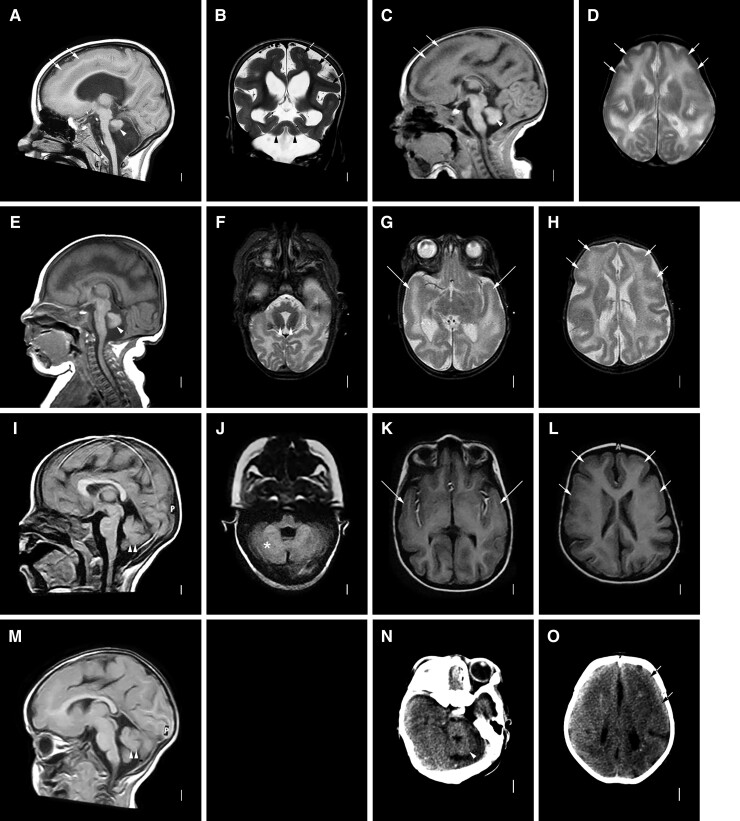
Brain imaging—biallelic
Brain MRI in six children and head CT scan in one boy demonstrated the same pattern of malformations described in previous reports including anterior predominant LIS with severe pachygyria over the frontal and anterior temporal regions that transitioned to mild pachygyria posterior to the sylvian fissure (Fig. 2). The cortex was 5–10 mm thick, always thicker frontally. The hippocampi were small with thin and open leaves. In four children, the pons was small making the ventral brainstem appear flat and the cerebellum was very small with an afoliar surface (Fig. 2A–H and N–O), as previously described. The three Egyptian siblings (LR17-413) with a splice-site variant leading to in-frame exon skipping had subtle hypoplasia of the pons and only moderate CBLH with a simplified foliar pattern, less severe than shown in any prior reports (Fig. 2I–M).
Figure 2.
Brain MRI showing LIS-CBLH with biallelic RELN variants. Brain imaging in six individuals with biallelic RELN-related LIS-CBLH. Midline sagittal MRIs in three individuals show an abnormally thin brainstem with flat pons and very small cerebellum with an afoliar surface (arrowheads in A, C and E). Midline sagittal images in two siblings with a less severe mutation show a normal brainstem and only moderately small cerebella with some foliation evident (I, M; double arrowheads point to the small cerebella). Axial (D, F–H, J–L) and coronal (B) images shows diffuse but frontal and temporal-predominant LIS with moderately thick 5–8 mm cortex. Arrows point to representative areas of pachygyria (mild LIS) only; the cortical malformation affects all brain regions. Low resolution head CT images in another child show the small cerebellum (arrowhead in N) and mild LIS (arrows in O). The imaging pattern seen in panels A–H appears similar to those shown in prior reports of individuals with biallelic mutations, while the pattern seen in panels I–M are less severe. Panels A and B are from subject LP95-137a1; C and D from LP95-137a2, E and H from LR14-063, I and L from LR17-413a1, M from LR17-413a3 and N and O from LP96-078.
Brain imaging—monoallelic
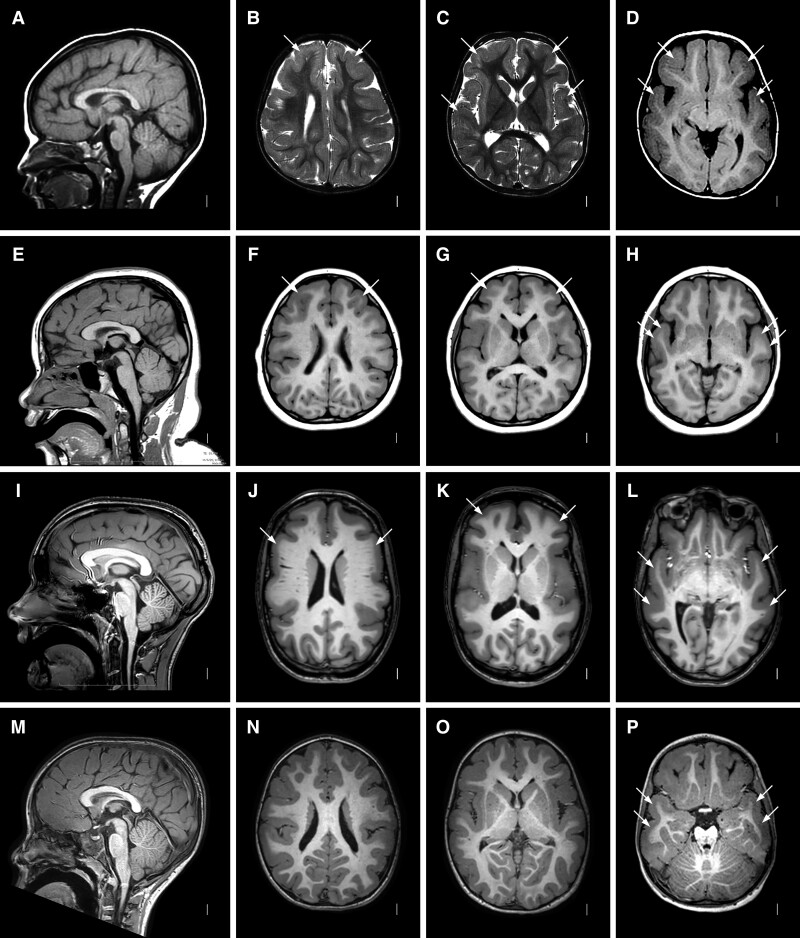
Brain MRI in 12 affected individuals and one clinically unaffected relative with monoallelic variants in RELN demonstrated the same pattern of frontotemporal LIS, although the cortical malformation was less severe and more variable than seen in the biallelic group. In 8 of 13, mild to moderate pachygyria was seen over the frontal and anterior temporal regions that transitioned to mild pachygyria or normal gyral width with mildly shallow sulci posterior to the sylvian fissures (Fig. 3 and Supplementary Fig. 5). The cortex was moderately thick (5–10 mm) over frontal and anterior temporal regions and transitioned to normal thickness posteriorly.
Figure 3.
Brain MRI showing frontotemporal LIS with monoallelic RELN variants. Images show the same pattern of LIS in four unrelated subjects including LR02-111a2 (A–D), LR21-446 (E–H), LR15-139 (I–L) and LR18-437 (M–P). Midline sagittal images (first column) show normal brainstem and cerebellum. Axial images at the level of the lateral ventricles (second column), basal ganglia (three column) and temporal lobes (fourth column) show diffuse mild LIS (pachygyria) with only moderately thick 5–8 mm cortex and consistent gradient with the malformation most severe in the frontal and temporal lobes and becoming less severe posteriorly. The posterior parietal and occipital regions appear mildly abnormal, although this is sometimes subtle. Arrows point to representative areas of pachygyria. The hippocampi also appear normal (not shown).
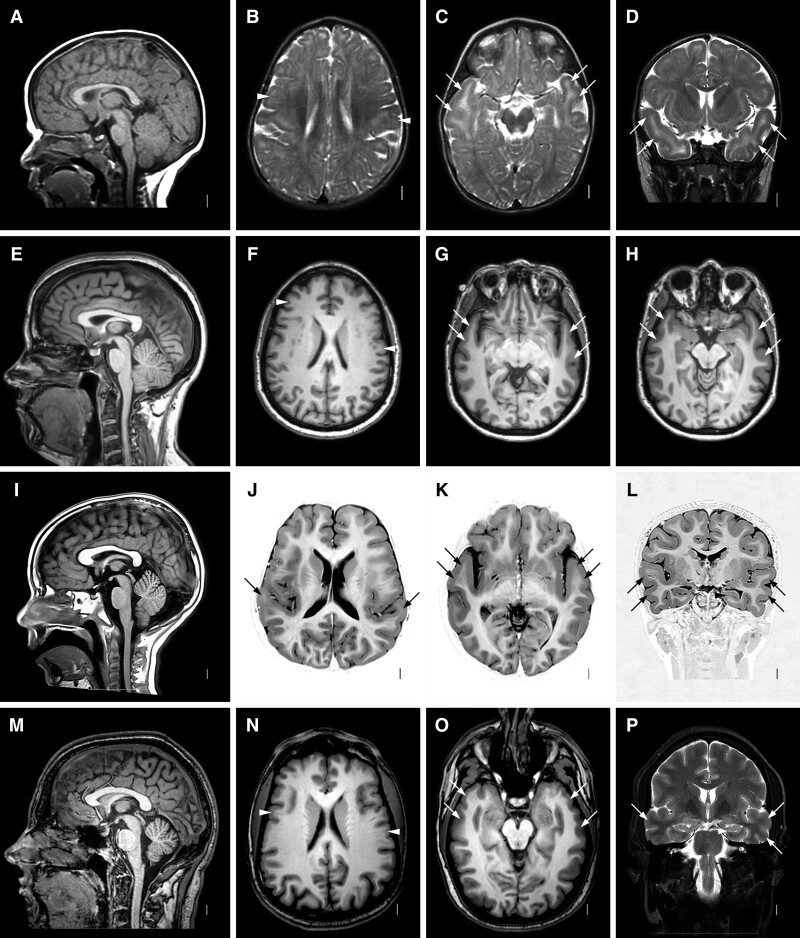
In the remaining five individuals including the asymptomatic carrier, pachygyria was seen over the anterior temporal regions only with the same moderately thick cortex (Fig. 4). The frontal lobes had mildly shallow sulci with borderline thick cortex in one boy. The hippocampi, brainstem and cerebellum appeared normal in all 13 subjects in this group.
Figure 4.
Brain MRI showing temporal LIS with monoallelic RELN variants. Images show the same pattern of LIS in four subjects from two families including LR02-111a3 (A–D), LR02-111r1 (E–H), LR17-364a2 (I–L) and LR17-364f (M–P). Midline sagittal images (first column) show normal brainstem and cerebellum. Axial images at the level of the lateral ventricles (second column), basal ganglia (third column) and temporal lobes (fourth column) show moderate but definite LIS (also known as mild pachygyria) with moderately thick 5–8 mm cortex over the temporal lobes best seen in the last column (D, H, L, P). Higher images show mild pachygyria in probands from the two families (B and J) and subtle pachygyria (or normal) in their less affected adult relatives (F and N). Arrows point to representative areas of pachygyria, while arrowheads point to areas of subtle undersulcation (F and N) or to normal hippocampi (L and P).
Clinical presentation—biallelic variants
We obtained data for five affected children plus limited data for another two children across an age range from several months to 10 years of age (Supplementary Table 1). The four children with afoliar cerebella on brain imaging all had profound ID and were non-verbal. One boy with a homozygous truncation (LP96-078) had severe spastic quadriparesis and died at home at 4 years. The two with the frameshift variant had hypotonia in infancy and were lost to follow-up.
The three siblings with small but foliated cerebella had congenital hypotonia and either severe (IQ 34) or profound ID and were effectively non-verbal although two siblings were using two words at 4–5 years of age. The oldest boy learned to walk at 7 years, but later died from complications of epilepsy at 14 years. The four children with data available all had epilepsy with seizure onset between birth and 6 months, and all had sleep disruption. Mild dysmorphic features were described in several, most probably related to facial hypotonia.
Clinical presentation—monoallelic variants
The 12 affected individuals with monoallelic variants had variable ID and a pattern of neurodevelopmental problems that included attention deficit hyperactivity disorder (ADHD), aggressive behaviour that was often unprovoked and could include self-injury, and disrupted sleep patterns (Table 1 and Supplementary Table 2). The ID varied from borderline to severe (one borderline, four mild, four moderate, two severe) and was more severe in those with frontotemporal LIS compared to those with anterior temporal LIS only. A behavioural profile emerged that comprised ADHD (6/8) that was often severe, unprovoked aggressive behaviour (6/8), anxiety disorder (3/7) and disrupted sleep patterns (5/8). Epilepsy was observed in only 3 of 12 individuals with onset at 1–4 years, and another had a single febrile seizure at 1 year of age. Several had mild hypotonia. One boy with mild ID was given a diagnosis of autism, while his older sister had some autistic features.
Incomplete penetrance and variable expressivity
The neurodevelopmental problems varied widely even among relatives carrying the same pathogenic variant. In our original family, the most severely affected boy (LR02-111a2 now 22 years) has severe ID and minimal language development. His older brother (LR02-111a1 now 27 years) has moderate ID and limited language. Their younger brother (LR02-111a3 now 20 years) has mild ID with language congruent with his developmental level and severe behaviour problems. The level of ID generally correlated with brain imaging as the two older brothers had frontotemporal LIS (Fig. 3A–D, Supplementary Fig. 5A–D), while the youngest had temporal LIS only (Fig. 5A–D). Despite the variable ID, all three brothers have severe ADHD and frequent unprovoked aggressive behaviour, while the two older brothers have had poorly regulated sleep throughout life.
Figure 5.
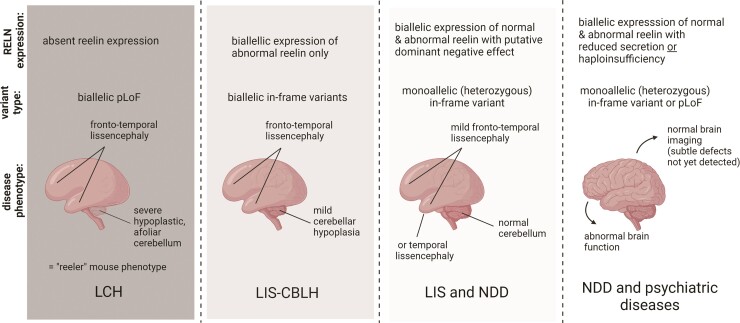
Proposed genotype-functional correlation with RELN-related phenotypes. Our data and analysis support a strong correlation between residual RELN activity and severity of the neurodevelopmental phenotype. LCH = lissencephaly with severe cerebellar hypoplasia; NDD = neurodevelopmental disorders. Created with BioRender.com.
The boys’ father (LR02-111f, not tested but monoallelic by pedigree position) was not available for evaluation but by report has had a lifelong history of mental health problems although he has worked for most of his adult life. His sister (LR02-111r1) also carries the monoallelic RELN variant. On evaluation, she has normal intellect and no history of behaviour problems (clinical report in Supplementary material). Remarkably, her brain MRI showed mild LIS limited to the anterior temporal lobes (Fig. 4E–H).
In the next family (LR05-375a1-a2, Supplementary Fig. 5A–H), the older brother had more severe ID than his younger sister, but they were lost to follow-up so further details were not available. In our third multiplex family (LR17-364), the proband had mild ID and autism, his older sister had moderate ID with autistic features but no formal diagnosis of autism, and their father had borderline ID and learning disabilities. All three had temporal only LIS on brain imaging (Fig. 4E–L).
Re-analysis of reported RELN variants
About 20 genomic studies over the past two decades have reported monoallelic or rarely putative biallelic RELN variants in diverse disorders without confirming pathogenicity. We began our re-analysis by searching published reports for variants associated with any human phenotype, excluding variants from targeted or GWAS, and found studies linking RELN to autism,24–31 epilepsy,32–39 schizophrenia,40–44 myoclonus-dystonia45 and cortical malformations including both LIS and polymicrogyria.23 Considering the incomplete penetrance and variable expressivity observed in our monoallelic cohort, we excluded inherited variants when no family history was available (mostly from autism and schizophrenia cohorts). Our search found 66 individuals from 36 families (Supplementary Table 3). The 39 alleles included two multiexon deletions (one frameshift, one in frame), one recurrent nonsense variant (p.Q417*), one splice-site variant leading to a frameshift and 35 missense variants. The CADD scores were 21 or above for 37/38 and 25 or above for 24/38 single base pair variants.
We next examined allele frequencies for the 38 single base pair variants in gnomAD v.2.1.1 and found that 18 were not seen (out of 282,912 alleles), six were observed in 1–4 alleles (variant allele frequency or VAF 0.000003977-0.00001594), 1 in 21 alleles (VAF 0.00008369) and the remaining 13 in 45–1463 alleles (VAF 0.0001595-0.005182). Five of the latter were seen in the homozygous state.
To strengthen any true associations with disease, we reasoned that those variants with higher frequencies were less likely to be pathogenic and chose to exclude variants with VAF >0.0001 in gnomAD (alleles more common than 1 in 10 000) as well as any homozygous variants. At least seven of the excluded variants have been observed in separate control or autism-inherited cohorts, while only one variant retained (p.Arg2290His) was seen in a control cohort.25 We also excluded variants with scaled CADD scores below 23, and excluded a variant reported as a second allele (p.Ile650Ser) in a child with a typical heterozygous LIS phenotype (addressed in the Supplementary material), leaving us with 24 variants for further analysis (Supplementary Table 4).
Combining these 24 alleles with six alleles from our monoallelic LIS series, we identified 30 potentially disease-causing alleles including 24 missense, three splice-site (two frameshift, one in frame) and one nonsense variant, and two multi-exon deletions. The proportion of in-frame variants compared to truncations was 4/12 (33%) in the biallelic group compared to 25/30 (83%) in the monoallelic group, a difference that is statistically significant (two-tailed P-value 0.0030 using Fisher’s exact test).
Discussion
Reelin is a large extracellular matrix glycoprotein that regulates neuronal migration and layer formation during early brain development, and subsequently axonal and dendritic outgrowth, axon guidance, dendritic spine formation and synaptogenesis, making it a strong candidate for neurodevelopmental disorders. RELN modulates synaptic function, network activity, learning and behaviour.46–49 The classic reeler phenotype in mouse and RELN-related LIS-CBLH in humans result from biallelic loss-of-function variants with loss of RELN expression. Our observations expand the human phenotype to include less severe CBLH with an in-frame mutation causing expression of a shortened mRNA.
More importantly, we report a recognizable neurodevelopmental phenotype in 12 individuals from seven families with monoallelic RELN variants, as well as an asymptomatic carrier. These data combined with re-analysis of numerous previous reports define a continuous series of brain malformations and associated neurodevelopmental and neuropsychiatric phenotypes in individuals with biallelic or monoallelic pathogenic variants of RELN.
Pathogenic RELN variants in humans—biallelic
RELN-related LIS-CBLH
In humans, biallelic variants cause RELN-related LIS-CBLH, a malformation recognizable by brain imaging associated with severe ID, hypotonia and epilepsy. Here we expand the spectrum of this disorder by reporting two children who died at 4 and 14 years of age, and three siblings with characteristic frontotemporal-predominant LIS combined with less severe CBLH associated with an in-frame variant resulting in expression of mRNA lacking one exon within the RELN central domain. These data indicate that expression of a shortened RELN protein is sufficient to partly rescue cerebellar but not cortical development, while complete absence of RELN protein results in severe CBLH with loss of cerebellar foliar structure (Fig. 2). Our analysis suggests that LIS-CBLH is the only phenotype reliably associated with biallelic RELN variants; other putative biallelic phenotypes lack sufficient evidence to support an association with RELN (see Supplementary material).
Adding our subjects to the literature (Supplementary Table 1), only 18 individuals from 10 families have been reported over >20 years.4–9. The scarcity of reports presents a conundrum as the large size of the gene (genomic region 517.9 kb), readily recognizable phenotype and lack of evidence (in mouse or human) of any prenatal lethality predict more common occurrence. The paucity of reports may be explained by reduced reproductive fitness in heterozygotes given the diverse phenotypes we report. The sparse parental data available in our cohort suggests an impact on the phenotype in monoallelic carriers of both truncating and in-frame variants.
Pathogenic RELN variants in humans—monoallelic
RELN first came to attention as a possible contributor to human neurodevelopmental disorders based on reduced expression of RELN in brain tissue from individuals with schizophrenia and on its location beneath the first autism-associated linkage peak in 7q22q36.50,51 These studies were followed by targeted and GWAS including several meta-analyses that implicated RELN variants as a cause of autism, bipolar disorder, schizophrenia and Alzheimer disease, most with marginally significant results.52–58 In addition, several targeted or genome-wide sequencing studies have suggested a role for RELN in the cause of autism, EPIL and myoclonus-dystonia.
Analysis of our new data from individuals and families with LIS combined with re-analysis of published genomic data for other phenotypes supports a causal role for monoallelic RELN variants for four seemingly distinct phenotypes including: (i) frontotemporal LIS; (ii) epilepsy especially autosomal dominant lateral temporal epilepsy; (iii) autism; and (iv) probably schizophrenia (Supplementary Tables 3 and 4). We found weak to no support for other phenotypes such as myoclonus-dystonia, bipolar disorder, Alzheimer's disease and polymicrogyria; our analysis of these data is included in the Supplementary material.
Our analysis is further supported by functional studies showing reduced secretion of RELN in vivo (serum) or in vitro for 10 subjects with one of four disorders including three subjects with LIS, four with epilepsy, two with autism and one with schizophrenia.23,29,35,43,59 We re-classified all of the variants associated with LIS or epilepsy and the de novo variants with autism as pathogenic or probably pathogenic, and the remaining variants with autism and schizophrenia as variants of unknown significance (VUS), although these may be disease-causing variants as well.
RELN-related frontotemporal-predominant lissencephaly
Our data regarding heterozygous variants in 14 individuals with frontotemporal- (adding one individual described in the preprint repository23) or temporal-predominant LIS represents a substantial and arguably dramatic expansion of the RELN-related phenotype spectrum. Our data demonstrate: (i) a recognizable and consistent phenotype observed in multiple individuals from eight unrelated families; (ii) de novo heterozygous variants in five of eight families including one recurrent variant; (iii) prediction of damaging missense variants and supportive data for several splice-site variants; and (iv) in vitro or in vivo functional studies for two de novo variants showing reduced extracellular secretion of RELN.23 However, we disagree with the interpretation of much of the data in the preprint, leading us to re-interpret some of their conclusions (Supplementary material). The Franklin algorithm classified six out of eight as VUS and the remaining two as probably pathogenic, but we now re-classify all as either pathogenic or probably pathogenic (discussed next).
All affected individuals with malformations of cortical development have a clear anterior more severe than posterior gradient with only subtle cortical malformations or normal gyral pattern in posterior regions (Figs 3 and 4). The neurodevelopmental phenotype extends from profound ID to the normal IQ range, correlating generally but not exactly with the brain imaging phenotype. The behavioural phenotype was consistent across those with moderate or mild ID and includes severe ADHD, unprovoked aggressive behaviour and disrupted sleep pattern, and one boy was given a diagnosis of autism. Only 3 of 12 (25%) had epilepsy, which was more diverse than lateral temporal epilepsy also including generalized tonic-clonic, atonic and atypical absence seizure types; one of the three had febrile seizures only, which may not be related.
RELN-related lateral temporal epilepsy
We reviewed reports of 23 individuals from nine families with epilepsy and monoallelic variants of RELN including eight familial missense variants and one splice-site variant causing a frameshift with unknown inheritance (Supplementary Tables 3 and 4).32–39 The Franklin algorithm classified all as either VUS or probably pathogenic. Functional studies found decreased RELN secretion in the four variants studied,35 and brain imaging showed subtle blurring of the cortical-white matter border in the temporal lobe in one individual.38 Our analysis re-classifies all nine variants as either pathogenic or probably pathogenic, relying on the consistent epilepsy phenotype, the shared anatomic localization between lateral temporal epilepsy and frontotemporal LIS, absence of any of the nine variants in gnomAD and functional studies showing decreased RELN secretion. The colocalization of lateral temporal epilepsy and the cortical malformation suggests that more detailed analysis of temporal lobe structure is needed in families with this epilepsy phenotype, as it is possible that subtle pachygyria or cortical thickening may be missed.
RELN-related autism
We found reports of 10 unrelated individuals with autism and monoallelic RELN variants including one recurrent nonsense (in two individuals), one splice-site (also included in the epilepsy group) and eight missense variants (Supplementary Tables 3 and 4).24–28,30,31 Two of the missense variants involved the same codon (Arg2290Cys, Arg2290His), and six were de novo. The other four were not tested for inheritance. Insufficient data was provided to allow subgrouping of the autism phenotypes. Other RELN variants observed in autism cohorts have higher frequencies in gnomAD and were excluded from our analysis, although some could be disease causing. For example, two autism-associated variants that we excluded caused decreased RELN secretion.29
The Franklin algorithm classified the nonsense variant as pathogenic, the splice-site variant as probably pathogenic and all eight missense variants as VUS, but here we re-classify the five de novo missense variants as probably pathogenic. Collectively, these data provide strong support for rare variants in RELN contributing to autism. However, more clinical data on the autism endophenotypes in individuals with autism and rare RELN variants are needed.
Several GWAS detected weakly significant association between the autism phenotype and individual single-nucleotide polymorphisms near RELN. Two meta-analyses showed either no significant association or a weak association with one single-nucleotide polymorphism (rs362691 also known as p.Leu997Val).55,57 These data add little as the RELN variants from our analysis were often de novo.
RELN-related schizophrenia
We found reports of only four individuals with schizophrenia and rare variants in RELN including two small intragenic deletions involving exons 3–5 and 52–58, and two rare missense variants with CADD scores in the top 1% of deleterious variants in the genome (Supplementary Tables 3 and 4).40–44 Two of the variants were inherited and two had no inheritance data. These data provide moderate support for RELN variants contributing to schizophrenia, and clearly more data on RELN variants in schizophrenia are needed.
Several targeted and GWAS have been performed in schizophrenia with inconsistent results for polymorphisms in RELN. However, two meta-analyses detected weak associations between schizophrenia and two single-nucleotide polymorphisms (increased or decreased risk) in Asian or white populations.53,56
Incomplete penetrance and variable expressivity
Several reports of autosomal recessive LIS-CBLH have briefly described their heterozygous parents as normal, while one report described post-traumatic stress disorder, illusions and hallucinations, panic disorder and sleep problems in several carriers. Our three multiplex families all had heterozygous parents and other family members with normal phenotypes or non-specific mental health problems, although only limited details were available.
We completed a clinical assessment and brain MRI in a woman who carries the same variant as the three affected brothers in family LR02-111 (clinical summary in Supplementary material). While she endorsed several mental health concerns, she completed high school, married and raised three sons as well as the proband (LR02-111a2) through most of his childhood and has worked her entire adult life. She has never had a seizure. However, her brain MRI showed subtle pachygyria in both anterior temporal lobes (Fig. 4M–P).
Possible dominant-negative effect
We found a significantly higher frequency of missense and other in-frame variants in the monoallelic cohort compared to the biallelic cohort (P = 0.0030), which suggests a greater impact on the phenotype as most parents of children with biallelic variants were unaffected or had less severe neuropsychiatric disorders. This difference could be explained by a dominant-negative effect. The simplest example of dominant-negative effect is given by protein homodimers in which the mutant protein A* renders A*A and A*A* dimers inactive so that heterozygotes have only ∼25% of dimer activity.60 And indeed, secreted RELN molecules form homodimers, although proteolysis at the N-t and C-t cleavage sites is likely to add complexity to the proportions of active extracellular RELN.61,62 Further support is provided by experiments in which co-transfection of wild-type and mutant RELN using LIS-related p.Cys539Arg or p.Arg3207Cys variants strongly reduced (80–90%) RELN levels in the secreted fraction.23
Interpretation of sequence variants in RELN
The ACMG-AMP consensus standards and guidelines for the interpretation of sequence variants are now widely used by molecular genomics laboratories.13 However, we find them ill-suited to analysis of monoallelic disorders with incomplete penetrance. As variants in RELN will present a common challenge for diagnostic laboratories going forward given the large size of the gene, we propose several minor modifications to existing criteria for use with monoallelic RELN variants (Supplementary Table 7). Even with these adaptations, some RELN variants that we think cause or contribute to the phenotype will still be classified as VUS and need to be interpreted using good clinical judgement, especially for autism, schizophrenia and potentially other neuropsychiatric disorders. Further studies are clearly needed to improve diagnostic accuracy.
Reln mutations in mouse models
The data we report here, including our re-analysis of previous data in humans, fit well with data from numerous mouse models with deficient Reln.
In mice, homozygous loss-of-function mutations (Relnrl/rl) cause the classic reeler phenotype of ataxia, tremors and a characteristic reeling gait first seen about 2 weeks after birth, observed in a series of spontaneous and experimentally generated mutant alleles.3,63–68 Histopathological studies in reeler mutants have shown an ‘inverted’ cortex in which successive generations of migrating neurons are unable to bypass their predecessors and accumulate in sequentially deeper positions in the cortex. In the hippocampus and dentate gyrus, cells in the pyramidal and granule layers are scattered rather than compact, and the cerebellum is small and afoliar with ectopic Purkinje cell clusters in subcortical regions and widespread loss of Purkinje and granule cells.69
Heterozygous animals with loss-of-function mutations (Reln+/rl) appear superficially normal, but detailed behavioural studies have demonstrated deficits such as decrease in prepulse inhibition of startle and signs of neophobia.70 Histopathological studies in heterozygous mice have shown normal anatomic organization and lamination, although subtle defects in neuronal migration have been found using higher-resolution methods. Histochemistry and cell counting experiments using nicotinamide-adenine dinucleotide phosphate-diaphorase (NADPH-d) have shown significantly decreased numbers of NADPH-d-positive neurons in grey matter and increased numbers in white matter of heterozygous animals compared with controls, changes that were most dramatic in the medial prefrontal cortex.70 Histochemistry and cell counting experiments using parvalbumin and glutamic acid decarboxylase 67 (GAD67) show decreased GABAergic neurons throughout the hippocampus and dentate gyrus.71 Thus, studies in heterozygous mutants support the occurrence of heterozygous neurodevelopmental and behavioural phenotypes in humans.
An alternative Reln allele in a mouse with a less severe phenotype contributed to our initial hypothesis that heterozygous RELN variants may be pathogenic in humans. Homozygous mutants for the RelnCTRdel allele had the same ‘inverted’ cerebral cortex and hippocampal dysplasia seen in classic reeler mutants (although less severe) but no cerebellar malformation or ataxia.21,22,72 These mice had an ENU-generated splice donor site mutation distal to exon 63 (Reln:c.10283+2T>C:p.L3428Lfs5*; mm39) producing a C-terminal deletion of 33 amino acids that escapes nonsense-mediated mRNA degradation. Notably, affected individuals in one of our families carry a similar splice-site variant predicted to result in loss of 65 amino acids within the C-terminal domain including 16 amino acids at the beginning of the C-terminal region (family LR02-111).
Conclusion
Combining data from our biallelic and monoallelic cohorts, we report a wide spectrum of brain malformations and associated neurodevelopmental and neuropsychiatric phenotypes in individuals with biallelic or monoallelic mutations of RELN. Our first draft genotype (plus functional studies) to phenotype correlation is shown in Fig. 5. In addition, we conclude that:
RELN-related (monoallelic) neurodevelopmental and neuropsychiatric-behavioural phenotypes are likely to be relatively common given the large size of the gene.
The spectrum of RELN-associated neurodevelopmental and neuropsychiatric phenotypes may grow even larger, adding ADHD, unprovoked aggressive behaviour and sleep disruption as these were all well documented in our monoallelic LIS cohort.
RELN-related (monoallelic) variants are associated with incomplete penetrance and variable expressivity even in families with the same mutation.
Our proposed modified sequence variant interpretation guidelines are listed in Supplementary Table 7.
Supplementary Material
Acknowledgements
The authors wish to thank the families of our subjects and in particular our original family with three affected individuals during 20 years of follow-up. We also thank Ms Kirsten Boggs and Ms Kate Pope for patient recruitment and data collection.
Abbreviations
- ADHD
attention deficit hyperactivity disorder
- CBLH
cerebellar hypoplasia
- ID
intellectual disability
- LIS
lissencephaly
- VUS
variants of unknown significance
Contributor Information
Nataliya Di Donato, Institute for Clinical Genetics, University Hospital, TU Dresden, 01307 Dresden, Germany.
Renzo Guerrini, Pediatric Neurology, Neurogenetics and Neurobiology Unit and Laboratories, Meyer Children’s Hospital, University of Florence, 50139 Florence, Italy.
Charles J Billington, Jr, Department of Pediatrics, Division of Genetics and Metabolism, University of Minnesota, Minneapolis, MN 55454, USA.
A James Barkovich, Departments of Radiology and Biomedical Imaging, Neurology, Pediatrics, and Neurosurgery, University of California, San Francisco, San Francisco, CA 94143, USA.
Philine Dinkel, Institute for Clinical Genetics, University Hospital, TU Dresden, 01307 Dresden, Germany.
Elena Freri, Department of Pediatric Neuroscience, Fondazione IRCCS Istituto Neurologico Carlo Besta, 20133 Milan, Italy.
Michael Heide, Max Planck Institute of Molecular Cell Biology and Genetics, 01307 Dresden, Germany; German Primate Center, Leibniz Institute for Primate Research, 37077 Goettingen, Germany.
Elliot S Gershon, Department of Human Genetics, The University of Chicago, Chicago, IL 60637, USA; Department of Psychiatry and Behavioral Neuroscience, The University of Chicago, Chicago, IL 60637, USA.
Tracy S Gertler, Division of Neurology, Department of Pediatrics, Ann and Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL 60611, USA.
Robert J Hopkin, Cincinnati Children’s Hospital Medical Center and University of Cincinnati College of Medicine, Department of Pediatrics, Division of Human Genetics, Cincinnati, OH 45229, USA.
Suma Jacob, Department of Psychiatry, University of Minnesota, Minneapolis, MN 55454, USA.
Sarah K Keedy, Department of Psychiatry and Behavioral Neuroscience, The University of Chicago, Chicago, IL 60637, USA.
Daniz Kooshavar, Bruce Lefory Centre, Murdoch Children’s Research Institute and University of Melbourne Department of Pediatrics, Melbourne 3052, Australia.
Paul J Lockhart, Bruce Lefory Centre, Murdoch Children’s Research Institute and University of Melbourne Department of Pediatrics, Melbourne 3052, Australia.
Dietmar R Lohmann, Institut fur Humangenetik, Universitatsklinikum Essen, 45147 Essen, Germany.
Iman G Mahmoud, Pediatric Neurology Department, Cairo University Children’s Hospital, Cairo, Egypt.
Elena Parrini, Pediatric Neurology, Neurogenetics and Neurobiology Unit and Laboratories, Meyer Children’s Hospital, University of Florence, 50139 Florence, Italy.
Evelin Schrock, Institute for Clinical Genetics, University Hospital, TU Dresden, 01307 Dresden, Germany.
Giulia Severi, Medical Genetics Unit, S. Orsola-Malpighi Hospital, 40138 Bologna, Italy.
Andrew E Timms, Center for Developmental Biology and Regenerative Medicine, Seattle Children’s Research Institute, Seattle, WA 98101, USA.
Richard I Webster, T. Y. Nelson Department of Neurology and Neurosurgery, The Children's Hospital at Westmead, Sydney 2145, Australia.
Mary J H Willis, Uniformed Services University School of Medicine and Naval Medical Center, Department of Pediatrics, San Diego, CA 92134, USA.
Maha S Zaki, Pediatric Neurology Department, Cairo University Children’s Hospital, Cairo, Egypt; Clinical Genetics Department, Human Genetics and Genome Research Division, National Research Centre, Cairo Governorate 12622, Egypt.
Joseph G Gleeson, Department of Neurosciences, Howard Hughes Medical Institute, University of California, San Diego, La Jolla, CA 92093, USA.
Richard J Leventer, Department of Neurology, Royal Children’s Hospital, Murdoch Children’s Research Institute and University of Melbourne Department of Pediatrics, Melbourne 3052, Australia.
William B Dobyns, Department of Pediatrics, Division of Genetics and Metabolism, University of Minnesota, Minneapolis, MN 55454, USA.
Funding
This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF) to N.D.D. (BMBF EGYGERF-2018-124), the Tuscany Region Call for Health 2018 to R.G. (Project DECODE-EE), the Fondazione Cassa di Risparmio di Firenze to R.G. (the BRAIN Project), the Australian Genomics Health Alliance funded by the National Health and Medical Research Council (Australia) to R.J.L. and the US National Institutes of Health-National Institute for Neurological Disorders and Stroke to J.G.G. (R01NS098004) and W.B.D. (R01NS050375). R.G. and E.P. are members of the European Reference Network for rare and complex epilepsies (EpiCARE). The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. D'Arcangelo G, Curran T. Reeler: New tales on an old mutant mouse. Bioessays. 1998;20:235–244. [DOI] [PubMed] [Google Scholar]
- 2. D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. [DOI] [PubMed] [Google Scholar]
- 3. Falconer DS. Two new mutations, trembler and reeler, with neurological actions in the house mouse (Mus musculus l). J Genet. 1951;50:192–201. [DOI] [PubMed] [Google Scholar]
- 4. Hong SE, Shugart YY, Huang DT, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26:93–96. [DOI] [PubMed] [Google Scholar]
- 5. Hong SE, Shugart YY, Huang DT, et al. Correction: Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2001;27:225–225. [DOI] [PubMed] [Google Scholar]
- 6. Chang BS, Duzcan F, Kim S, et al. The role of RELN in lissencephaly and neuropsychiatric disease. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:58–63. [DOI] [PubMed] [Google Scholar]
- 7. Zaki M, Shehab M, El-Aleem AA, et al. Identification of a novel recessive RELN mutation using a homozygous balanced reciprocal translocation. Am J Med Genet A. 2007;143A:939–944. [DOI] [PubMed] [Google Scholar]
- 8. Valence S, Garel C, Barth M, et al. RELN and VLDLR mutations underlie two distinguishable clinico-radiological phenotypes. Clin Genet. 2016;90:545–549. [DOI] [PubMed] [Google Scholar]
- 9. Zillhardt JL, Poirier K, Broix L, et al. Mosaic parental germline mutations causing recurrent forms of malformations of cortical development. Eur J Hum Genet. 2016;24:611–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Donato N, Timms AE, Aldinger KA, et al. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet Med. 2018;20:1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Donato N, Jean YY, Maga AM, et al. Mutations in CRADD result in reduced caspase-2-mediated neuronal apoptosis and cause megalencephaly with a rare lissencephaly variant. Am J Hum Genet. 2016;99:1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sadedin SP, Dashnow H, James PA, et al. Cpipe: A shared variant detection pipeline designed for diagnostic settings. Genome Med. 2015;7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ben-Salem S, Gleeson JG, Al-Shamsi AM, et al. Asparagine synthetase deficiency detected by whole exome sequencing causes congenital microcephaly, epileptic encephalopathy and psychomotor delay. Metab Brain Dis. 2015;30:687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Vita D, Mei D, Rutigliano D, et al. Familial dominant epilepsy and mild pachygyria associated with a constitutional LIS1 mutation. Am J Med Genet A. 2018;176:2808–2812. [DOI] [PubMed] [Google Scholar]
- 16. Parrini E, Marini C, Mei D, et al. Diagnostic targeted resequencing in 349 patients with drug-resistant pediatric epilepsies identifies causative mutations in 30 different genes. Hum Mutat. 2017;38:216–225. [DOI] [PubMed] [Google Scholar]
- 17. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Integrated DNA Technologies . Alt-R® CRISPR-Cas9 System: Delivery of ribonucleoprotein complexes into HEK-293 cells using the Amaxa® Nucleofector™ System. Version 3. Guides and Protocols. Integrated DNA Technologies; 2018. https://www.idtdna.com/pages/support/guides-and-protocols
- 20. Integrated DNA Technologies . Alt-R® CRISPR-Cas12a (Cpf1) System: Delivery of ribonucleoprotein complexes into HEK-293 cells using the Amaxa® Nucleofector™ System. Version 2.1. Guides and Protocols. Integrated DNA Technologies; 2018. https://www.idtdna.com/pages/support/guides-and-protocols
- 21. Ha S, Stottmann RW, Furley AJ, Beier DR. A forward genetic screen in mice identifies mutants with abnormal cortical patterning. Cereb Cortex. 2015;25:167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ha S, Tripathi PP, Mihalas AB, Hevner RF, Beier DR. C-terminal region truncation of RELN disrupts an interaction with VLDLR. Causing abnormal development of the cerebral cortex and hippocampus. J Neurosci. 2017;37:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riva M, Ferreira S, Medvedeva VP, et al. Functional characterization of RELN missense mutations involved in recessive and dominant forms of neuronal migration disorders. bioRxiv, 10.1101/2021.05.25.445586, May 25, 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- 24. Bonora E, Beyer KS, Lamb JA, et al. Analysis of reelin as a candidate gene for autism. Mol Psychiatry. 2003;8:885–892. [DOI] [PubMed] [Google Scholar]
- 25. De Rubeis S, He X, Goldberg AP, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koshimizu E, Miyatake S, Okamoto N, et al. Performance comparison of bench-top next generation sequencers using microdroplet PCR-based enrichment for targeted sequencing in patients with autism spectrum disorder. PLoS ONE. 2013;8:e74167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sanchez-Sanchez SM, Magdalon J, Griesi-Oliveira K, et al. Rare RELN variants affect Reelin-DAB1 signal transduction in autism spectrum disorder. Hum Mutat. 2018;39:1372–1383. [DOI] [PubMed] [Google Scholar]
- 30. Vanzo RJ, Prasad A, Staunch L, et al. The temple grandin genome: Comprehensive analysis in a scientist with high-functioning autism. J Pers Med. 2020;11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. [DOI] [PubMed] [Google Scholar]
- 32. Bertoli-Avella AM, Beetz C, Ameziane N, et al. Successful application of genome sequencing in a diagnostic setting: 1007 index cases from a clinically heterogeneous cohort. Eur J Hum Genet. 2021;29:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ceska K, Aulicka S, Horak O, et al. Autosomal dominant temporal lobe epilepsy associated with heterozygous reelin mutation: 3T brain MRI study with advanced neuroimaging methods. Epilepsy Behav Case Rep. 2019;11:39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dazzo E, Fanciulli M, Serioli E, et al. Heterozygous reelin mutations cause autosomal-dominant lateral temporal epilepsy. Am J Hum Genet. 2015;96:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dazzo E, Nobile C. Epilepsy-causing Reelin mutations result in impaired secretion and intracellular degradation of mutant proteins. Hum Mol Genet. 2022:665–673. [DOI] [PubMed] [Google Scholar]
- 36. Fang XQ, Zhang RR, Liu XW. Heterozygous missense mutation of the RELN gene is one of the causes of epilepsy. Neurol Res. 2022:262–267. [DOI] [PubMed] [Google Scholar]
- 37. Michelucci R, Pulitano P, Di Bonaventura C, et al. The clinical phenotype of autosomal dominant lateral temporal lobe epilepsy related to reelin mutations. Epilepsy Behav. 2017;68:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michelucci R, Dazzo E, Volpi L, et al. Autosomal dominant lateral temporal lobe epilepsy associated with a novel reelin mutation. Epileptic Disord. 2020;22:443–448. [DOI] [PubMed] [Google Scholar]
- 39. Zhang Y, Kong W, Gao Y, et al. Gene mutation analysis in 253 Chinese children with unexplained epilepsy and intellectual/developmental disabilities. PLoS ONE. 2015;10:e0141782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Costain G, Lionel AC, Merico D, et al. Pathogenic rare copy number variants in community-based schizophrenia suggest a potential role for clinical microarrays. Hum Mol Genet. 2013;22:4485–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kushima I, Aleksic B, Nakatochi M, et al. High-resolution copy number variation analysis of schizophrenia in Japan. Mol sychiatry. 2017;22:430–440. [DOI] [PubMed] [Google Scholar]
- 43. Sobue A, Kushima I, Nagai T, et al. Genetic and animal model analyses reveal the pathogenic role of a novel deletion of RELN in schizophrenia. Sci Rep. 2018;8:13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Z, Hu Z, Zhang L, et al. Identification of RELN variation p.Thr3192Ser in a Chinese family with schizophrenia. Sci Rep. 2016;6:24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Groen JL, Ritz K, Jalalzadeh H, et al. RELN rare variants in myoclonus-dystonia. Mov Disord. 2015;30:415–419. [DOI] [PubMed] [Google Scholar]
- 46. D'Arcangelo G. Reelin in the years: Controlling neuronal migration and maturation in the mammalian brain. Adv Neurosci. 2014;2014:597395. 597395. [Google Scholar]
- 47. Faini G, Del Bene F, Albadri S. Reelin functions beyond neuronal migration: from synaptogenesis to network activity modulation. Curr Opin Neurobiol. 2021;66:135–143. [DOI] [PubMed] [Google Scholar]
- 48. Lee GH, D'Arcangelo G. New insights into reelin-mediated signaling pathways. Front Cell Neurosci. 2016;10:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quattrocchi CC, Huang C, Niu S, et al. Reelin promotes peripheral synapse elimination and maturation. Science. 2003;301:649–653. [DOI] [PubMed] [Google Scholar]
- 50. Impagnatiello F, Guidotti AR, Pesold C, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. International Molecular Genetic Study of Autism Consortium (IMGSAC) . Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet. 2001;10:973–982. [DOI] [PubMed] [Google Scholar]
- 52. Bufill E, Roura-Poch P, Sala-Matavera I, et al. Reelin signaling pathway genotypes and Alzheimer disease in a Spanish population. Alzheimer Dis Assoc Disord. 2015;29:169–172. [DOI] [PubMed] [Google Scholar]
- 53. Chen N, Bao Y, Xue Y, et al. Meta-analyses of RELN variants in neuropsychiatric disorders. Behav Brain Res. 2017;332:110–119. [DOI] [PubMed] [Google Scholar]
- 54. Feher A, Juhasz A, Pakaski M, Kalman J, Janka Z. Genetic analysis of the RELN gene: Gender specific association with Alzheimer’s disease. Psychiatry Res. 2015;230:716–718. [DOI] [PubMed] [Google Scholar]
- 55. Hernandez-Garcia I, Chamorro AJ, Ternavasio-de la Vega HG, Carbonell C, Marcos M, Miron-Canelo JA. Association of allelic variants of the Reelin Gene with autistic spectrum disorder: A systematic review and meta-analysis of candidate gene association studies. Int J Environ Res Public Health. Oct 30 2020;17:8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marzan S, Aziz MA, Islam MS. Association between REELIN gene polymorphisms (rs7341475 and rs262355) and risk of schizophrenia: An updated meta-analysis. J Mol Neurosci. 2021;71:675–690. [DOI] [PubMed] [Google Scholar]
- 57. Wang Z, Hong Y, Zou L, et al. Reelin gene variants and risk of autism spectrum disorders: An integrated meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:192–200. [DOI] [PubMed] [Google Scholar]
- 58. Goes FS, Willour VL, Zandi PP, et al. Sex-specific association of the Reelin gene with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lammert DB, Middleton FA, Pan J, Olson EC, Howell BW. The de novo autism spectrum disorder RELN R2290C mutation reduces reelin secretion and increases protein disulfide isomerase expression. J Neurochem. 2017;142:89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Veitia RA, Caburet S, Birchler JA. Mechanisms of mendelian dominance. Clin Genet. 2018;93:419–428. [DOI] [PubMed] [Google Scholar]
- 61. Hattori M, Kohno T. Regulation of reelin functions by specific proteolytic processing in the brain. J Biochem. 2021;169:511–516. [DOI] [PubMed] [Google Scholar]
- 62. Kubo K, Mikoshiba K, Nakajima K. Secreted reelin molecules form homodimers. Neurosci Res. 2002;43:381–388. [DOI] [PubMed] [Google Scholar]
- 63. Andersen TE, Finsen B, Goffinet AM, Issinger OG, Boldyreff B. A reeler mutant mouse with a new, spontaneous mutation in the reelin gene. Brain Res Mol Brain Res. 2002;105:153–156. [DOI] [PubMed] [Google Scholar]
- 64. Takahara T, Ohsumi T, Kuromitsu J, et al. Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an active L1 sequence into the skipped exon. Hum Mol Genet. 1996;5:989–993. [DOI] [PubMed] [Google Scholar]
- 65. Royaux I, Bernier B, Montgomery JC, Flaherty L, Goffinet AM. Reln(rl-Alb2), an allele of reeler isolated from a chlorambucil screen, is due to an IAP insertion with exon skipping. Genomics. 1997;42:479–482. [DOI] [PubMed] [Google Scholar]
- 66. Flaherty L, Messer A, Russell LB, Rinchik EM. Chlorambucil-induced mutations in mice recovered in homozygotes. Proc Natl Acad Sci USA. 1992;89:2859–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miao GG, Smeyne RJ, D'Arcangelo G, et al. Isolation of an allele of reeler by insertional mutagenesis. Proc Natl Acad Sci USA. 1994;91:11050–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hirotsune S, Takahara T, Sasaki N, et al. The reeler gene encodes a protein with an EGF-like motif expressed by pioneer neurons. Nat Genet. 1995;10:77–83. [DOI] [PubMed] [Google Scholar]
- 69. Caviness VS. Neocortical histogenesis in normal and reeler mice: A developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. [DOI] [PubMed] [Google Scholar]
- 70. Tueting P, Costa E, Dwivedi Y, et al. The phenotypic characteristics of heterozygous reeler mouse. Neuroreport. 1999;10:1329–1334. [DOI] [PubMed] [Google Scholar]
- 71. Nullmeier S, Panther P, Dobrowolny H, et al. Region-specific alteration of GABAergic markers in the brain of heterozygous reeler mice. Eur J Neurosci. 2011;33:689–698. [DOI] [PubMed] [Google Scholar]
- 72. Ha S, Tripathi PP, Daza RA, Hevner RF, Beier DR. Reelin mediates hippocampal Cajal-Retzius cell positioning and infrapyramidal blade morphogenesis. J Dev Biol. 2020;8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data supporting the RELN variants in our lissencephaly cohorts that were generated in research laboratories are available from the corresponding author, on reasonable request. The authors confirm that remaining data supporting the findings of this study are available within the article or Supplementary material.