Abstract
Therapeutic intervention in cyclin-dependent kinase-like 5 (CDKL5) deficiency disorders (CDDs) has remained a concern over the years. Recent advances into the mechanistic interplay of signalling pathways has revealed the role of deficient tropomyosin receptor kinase B (TrkB)/phospholipase C γ1 signalling cascade in CDD. Novel findings showed that in vivo administration of a TrkB agonist, 7,8-dihydroxyflavone (7,8-DHF), resulted in a remarkable reversal in the molecular pathologic mechanisms underlying CDD. Owing to this discovery, this study aimed to identify more potent TrkB agonists than 7,8-DHF that could serve as alternatives or combinatorial drugs towards effective management of CDD. Using pharmacophore modelling and multiple database screening, we identified 691 compounds with identical pharmacophore features with 7,8-DHF. Virtual screening of these ligands resulted in identification of at least 6 compounds with better binding affinities than 7,8-DHF. The in silico pharmacokinetic and ADMET studies of the compounds also indicated better drug-like qualities than those of 7,8-DHF. Postdocking analyses and molecular dynamics simulations of the best hits, 6-hydroxy-10-(2-oxo-1-azatricyclo[7.3.1.05,13]trideca-3,5(13),6,8-tetraen-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem: 91637738) and 6-hydroxy-10-(8-methyl-2-oxo-1H-quinolin-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem ID: 91641310), revealed unique ligand interactions, validating the docking findings. We hereby recommend experimental validation of the best hits in CDKL5 knock out models before consideration as drugs in CDD management.
Keywords: CDKL5 deficiency disorders, seizure, pharmacophore, virtual screening, molecular dynamics simulation
Introduction
Cyclin-dependent kinase-like 5 (CDKL5) deficiency disorder (CDD) is an X-linked neurodevelopmental disorder characterised by neonatal or infantile onset of epilepsy and poor neurodevelopmental features, including motor deficits.1 The CDD occurs due to missense and null CDKL5 mutations affecting the gene’s catalytic activity in brain morphogenesis.2 Therapeutic interventions in CDD have largely yielded futile results in many patients, and this is due in part to the fact that the knowledge of molecular mechanisms underlying CDD pathology is still advancing. Previous studies using CDKL5 knock-out (KO) mice revealed some insights into the molecular changes in the hippocampal and cerebral regions, affecting dendritic growth and neuronal plasticity.3 Ren et al4 investigated the perirhinal cortex of the CDKL5 KO mice and observed a dysfunction in long-term potentiation and the mechanistic role of the tropomyosin receptor kinase B (TrkB)/phospholipase C γ1 signalling cascade (TrkB/PLCγ1). The deficient TrkB PLCγ1 pathway results in poor neuronal plasticity, low levels of GluA2 subunit of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (GluA2-AMPA) receptors, insufficient dendritic morphogenesis, spine growth, and spine density. This study further evaluated the impact of the TrkB agonist, 7,8-dihydroxyflavone (7,8-DHF) on enhancing TrkB function and reverting the pathologic molecular findings. Strikingly, administration of 7,8-DHF restored long-term potentiation and visual memory, enhanced dendrite morphogenesis, increased expression levels of GluA2-AMPA and postsynaptic density 95 (PSD-95). After these findings, enhancement of TrkB signalling has remained a crucial pharmacologic consideration in the management of CDD.
The application of computational approaches in structure-based drug design has rapidly accelerated the field of synthetic chemistry, and many researchers employ these approaches as starting points for identification of lead compounds which may eventually end up as effective therapeutic regimens.5 In this study, we aimed to identify compounds with better agonistic functions to TrkB than 7,8-DHF through the incorporation of structure-based drug design technique, pharmacophore-aided virtual screening, and molecular dynamics (MD) simulation to improve the therapeutic options for managing CDD. We discovered 6 best hit compounds, with 2 ranking highest. These include 6-hydroxy-10-(2-oxo-1-azatricyclo[7.3.1.05,13]trideca-3,5(13),6,8-tetraen-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem: 91637738) and 6-hydroxy-10-(8-methyl-2-oxo-1H-quinolin-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem ID: 91641310), which outperformed 7,8-DHF in terms of binding energy and equally possessed drug-like properties, good bioavailability, and low biotoxicity qualities. These compounds can be considered as alternatives to 7,8-DHF in the development of drugs to manage CDD.
Materials and Methods
Retrieval of x-ray crystallographic structure of TrkB ligand-binding domain
The RCSB protein data bank (PDB) serves as a global repository of biologic macromolecules, archiving three-dimensional (3D) structures of diverse biomolecules for easy retrieval for research and academic purposes. These structures are experimentally validated using nuclear magnetic resonance (NMR), x-ray crystallography, electron microscopy, or micro-electron diffraction.6 The experimentally validated structure of the human TrkB ligand binding domain (PDB ID: 1wwb) was discovered and reported by Ultsch et al7 (Figure 1). This was retrieved from the PDB (https://www.rcsb.org/) for docking and simulation studies.
Figure 1.

X-ray crystallographic structure of the ligand binding domain of TrkB receptor.7,8 TrkB indicates tropomyosin receptor kinase B.
Single ligand blind docking of 7,8-DHF to TrkB
We used Discovery Studio 2021 and Chimera (version 33.02.1242) to dock the experimentally validated ligand and agonist of the human TrkB protein, 7,8-DHF (PubChem ID: 1880) (Figure 2), to the ligand binding domain of TrkB to identify the interacting residues and best conformations of the ligand within the binding pocket of 1wwb.
Figure 2.

Structure of 7,8-DHF. 7,8-DHF indicates 7,8-dihydroxyflavone.
Pharmacophore modelling of 7,8-DHF for virtual screening
Pharmacophores are collections of functionally active chemical moieties with unique spatial characteristics and identity to a chemically active compound.9 They are crucial for biologic reactions to occur, specifically, in ligand-receptor interactions. Technically, pharmacophore models are developed from a group of chemically identical compounds sharing unique and distinctive activities. In modelling pharmacophores, a range of functional groups are considered, including aromatic rings, proton donors or acceptors, electron acceptors or donors, hydrophobic clusters, and so on.9
Pharmacophore models can be either designed from ligands in isolation or from protein-ligand complexes. In this study, we modelled the pharmacophores based on interaction between the ligand binding domain of TrkB and its agonist, 7,8-DHF, using the Pharmit server (http://pharmit.csb.pitt.edu/). This server enables the modelling of pharmacophores and their screening against many large repositories of chemical compounds, including PubChem (https://pubchem.ncbi.nlm.nih.gov/), ChemBl (https://www.ebi.ac.uk/chembl/), Zinc Database (https://zinc.docking.org/), MolPort database (https://www.molport.com/), and so on, at the same time, including energy minimisation of identified compounds in preparation for virtual screening.10 The filtering and prioritisation of ligands from several chemical databases was based on similarity with pharmacophore model developed on the Pharmit server, based on the structure of 7,8-DHF and adherence to the mandatory features for determining the bioavailability and drug-likeness of a lead or potential drug compound.11,12
Creation and preparation of ligand library
The Pharmit web server was configured to include best hits from the following databases: PubChem, MCULE, ZINC database, MolPort, CHEMBL30, ChemDiv, ChemSpace, MCULE-ULTIMATE, NCI Open Chemical Repository, and LabNetwork.13 Specifically, the following features were used as thresholds to filter databases in accordance with Lipinski rule of 5, Veber, Ghose, Egan, and Muegge rules.14-17 These include molecular weight ⩽ 500 g, number of rotable bonds ⩽ 10, logP (octanol-water partition coefficient) value ⩽ 5, topological polar surface area (TPSA) ⩽ 140 Å, hydrogen bonds acceptor ⩽ 10, hydrogen bonds donor ⩽ 5, and number of aromatics ⩽ 2.17
A library of 691 unique ligands was created following pharmacophore-based screening against major databases. The 2-dimensional SDF files of these ligands were obtained on Pharmit and converted into 3D formats using the OpenBabel module of the python-based open-source tool, PyRx.18 For docking purposes, the tool was used to establish a magnetic force field around each of the 691 compounds and the reference, 7,8-DHF, and to minimise the total energy while preserving the intact molecular structure for ligand-receptor interaction. Subsequently, all ligands were converted into their corresponding pdbqt formats.
Virtual screening and postscreening analyses
We conducted the molecular docking and virtual screening analysis to identify compounds with better binding affinities than 7,8-DHF, using Autodock vina (version 1.2), while postdocking analysis was done to identify the interactions involved, interacting residues, bond types, and lengths, using Discovery Studio 2021 Client.
Computational evaluation of ADMET properties of best compounds
To evaluate the adsorption, distribution, metabolism, excretion, and toxicity of 7,8-DHF and the other compounds with best hits to the target, we conducted an ADMET study using the SWISS ADME web tool (http://www.swissadme.ch/). This examined the pharmacokinetic and toxicologic profiles of each compound.19
MD simulation
The MD simulation was carried out by using NAMD (Nanoscale Molecular Dynamics)20 and VMD (Visual Molecular Dynamics).21 This helped to determine the stability of the best poses of the 2 top hits from the docking studies in the binding site of 1wwb. Energy minimisation of 1000 steps was performed to fix the backbone atoms, while the production simulation run was carried out using 1 ns, equivalent to 500 000 steps. The simulation was performed at a constant pressure of 1 atm and temperature of 310 K using Periodic Boundary conditions. The protein structure file (PSF) of the target was generated separately from that of the ligand using VMD, while those of ligands were generated using the Charmm36 forcefield of the Charmm-GUI webserver. These were generated to define the bond types, bond angles, atom types, and the number of molecules in the simulation system. The topologies (PDB and PSF) of the protein and ligands were merged, and the complexes were solvated using VMD to generate the cubic water box. The other necessary parameters (time and Periodic Boundary conditions) for the simulation were defined in a script and run using NAMD. The RMSD (root mean square deviation), RMSF (root mean square fluctuation), and PCA (principal components analysis) of the simulation results were carried out using Bio3D on the Galaxy Europe platform,22 while the hydrogen bond (h-bond) analysis was carried out using VMD software.
Results and Discussion
The CDD is a rare disorder with X-linked inheritance pattern.23 The CDD is characterised by poor cognition, epilepsy of infancy or newborn, and suboptimal locomotive efficiency.24 Most therapeutic interventions fail to effectively manage the seizure or developmental outcome. The molecular pathomechanism of action of CDD involves aberrant synaptic plasticity and neuron morphogenesis, resulting in erratic network currents and hyperexcitability in neurons.25 The involvement of TrkB signalling in CDD pathogenesis was recently discovered using mouse KO models of CDKL5.26 Ren et al4 investigated the molecular mechanism of action of CDKL5 in the perirhinal cortex of KO mice and unravelled the intricate role of TrkB/PLCy1 signalling pathway in CDD pathology, where treatment with the TrkB agonist prodrug, 7,8-DHF resulted in restoring long-term potentiation in neurons, and improved dendritic formation and synaptogenesis. Such studies are of more relevance AS the rise of debates over the null impacts of formerly rated drugs such as ataluren (a drug which promotes read-through in nonsense-mediated genetic disorders), for management of CDDs.27,28 Till date, there has been continuous search for more chemotherapeutic agents that may be used combinatorially or as stand-alone therapies to facilitate CDKL5 functions in deficient patients. The discovery of the impact of the TrkB agonist, 7,8-dihydoflavone, in upregulating the TrkB/PLCγ signalling pathway has resulted in 1 breakthrough, with an increasing need to provide alternative compounds with more potent functions than 7,8-dihydoroflavone. Studies focusing on accentuating TrkB function may potentially generate viable therapeutic regimens as stand-alone drugs or combinatorial therapies for management of CDD. This study identified more potent agonists to TrkB than 7,8-DHF using pharmacophore-based virtual screening of 691 compounds against the ligand binding domain of the TrkB receptor.
Blind docking of 7,8-DHF to 1wwb
Blind docking is an approach used to determine ligand binding sites and best ligand conformation in ligand-receptor interactions.29 This technique is well suited for small protein targets and single ligand docking. It permits screening of the total area of the query protein to identify best residues with strongest interaction and best ligand conformation. The technique results in an unbiased mapping of ligands to peptide surfaces by screening across the entire protein surface to identify best binding kinetics.30 Following blind docking, the best conformations of the reference ligand and interacting residues within the binding pocket of 1wwb are shown in Figure 3. These residues include Aspartate 370, Alanine 363, Valine 307, Lysine 372, and Leucine 361.
Figure 3.
Interactions of 7,8-DHF with 5 specific residues on TrkB ligand binding domain: (A) 2D interactions and (B) 3D interactions. 2D indicates two dimension; 7,8-DHF 7,8-dihydroxyflavone; 3D, three dimension; TrkB, tropomyosin receptor kinase B.
Pharmacophore modelling and ligand library results
We investigated the pharmacophore characteristics of 7,8-DHF and used this to generate a model which was used to create a library of ligands on the Pharmit web server. The unique features of the pharmacophore are presented in Table 1. These include the following major properties; namely, 2 hydrogen bond donors, 2 hydrogen bond receptors, and 3 aromatic features. In total, 691 unique ligands were mined from a total of 5 databases, namely, PubChem, ChemBl, MCULE, MolPort, and ZINC.
Table 1.
Unique pharmacophore features of 7,8-DHF used in Library generation.
| Pharmacophore features | x | y | z | Radius |
|---|---|---|---|---|
| Hydrogen donor | 33.36 | 10.27 | 23.03 | 1.0 |
| Hydrogen acceptor | 33.76 | 8.7 | 20.78 | 1.0 |
| Hydrogen acceptor | 28.27 | 7.3 | 23.51 | 1.0 |
| Hydrogen acceptor | 33.76 | 8.7 | 20.78 | 1.0 |
| Aromatic | 30.11 | 9.04 | 24.16 | 1.0 |
| Aromatic | 31.48 | 8.39 | 22.29 | 1.0 |
| Aromatic | 29.66 | 11.6 | 27.56 | 1.0 |
Abbreviation: 7,8-DHF, 7,8-dihydroxyflavone.
Virtual-screening analyses
We docked a total of 691 compounds including the 7,8-DHF (reference compound), against the binding regions of 1wwb. Six compounds having the best binding affinities ranging from –9.6 to –8.8 kcal/mol, including the reference compound having –7.1 kcal/mol, are presented in Table 2. All reported compounds had better binding affinities than the reference compound. Specifically, the 2 best compounds discovered include compound 1 with PubChem ID: 91637738 and compound 2 with PubChem ID: 91641310, having –9.6 and –9.3 kcal/mol, respectively. Computationally, these 2 compounds are predicted to possess better agonistic functions to the TrkB receptor than 7,8-DHF.
Table 2.
Chemical and structural features of 6 best compounds having better binding affinities to 1wwb than 7,8-DHF.
| Compound ID | PubChem ID | Molecular formula | H-bond acceptors | H-bond donors | Binding energy | MW (g/mol) | |
|---|---|---|---|---|---|---|---|
| Compound 1 | 91637738 |

|
C24H16N4O4 | 5 | 3 | –9.6 | 424.41 |
| Compound 2 | 91641310 |

|
C22H14N4O4 | 5 | 4 | –9.3 | 398.37 |
| Compound 3 | 91639858 |

|
C23H14N4O4 | 5 | 3 | –9.3 | 410.38 |
| Compound 4 | 91639334 |

|
C22H14N4O4 | 5 | 3 | –9.2 | 398.37 |
| Compound 5 | 91640550 |

|
C21H12N4O4 | 5 | 4 | –9.0 | 384.34 |
| Compound 6 | 91639725 |

|
C21H12N4O3 | 5 | 3 | –8.8 | 368.34 |
| 7,8-DHF | 1880 |

|
C15H10O4 | 4 | 2 | –7.1 | 254.24 |
Abbreviations: 7,8-DHF, 7,8-dihydroxyflavone; MW, molecular weight.
Post virtual-screening studies of ligand-receptor interactions
We used Discovery Studio 2021 Client to analyse the atomic interactions between the best hit ligands (ie, compound 1, compound 2) including the Reference, and the amino acid residues located within the binding site of TrkB. These interactions work concertedly to generate stronger binding preference between atoms of the best hit compounds than those of the reference compound to the receptor. Multiple types of interactions were observed, including pi-pi stacked, pi-sigma, pi-cation, conventional hydrogen bond, pi-anion, alkyl, pi-alkyl, and carbon-hydrogen bond interactions (Table 3). Compounds 91639334 and 91639725 formed unfavourable donor-donor bonds with Lys X:308, which indicates the repulsion between 2 atoms, and this affects the stability of the compounds in the active site of the protein. However, this did not affect the overall binding energies of these compounds. A summary of all atomic linkages with their unique bond lengths are shown as follows:
Table 3.
Interacting residues, interaction types, and bond lengths of the 6 ligands and reference ligand, within the binding pocket of 1wwb.
| S. No. | Ligands | H-bond acceptors | H-bond donors | Interacting residues and bond lengths |
|---|---|---|---|---|
| 1 | 91637738 | 5 | 3 | Conventional hydrogen bond: Asp X:370 (2.86 Å), Leu X:361(2.25 Å) Pi-Lone pair bond: Thr X:306(2.98 Å) Pi cation bonds: Lys 372 (4.73 Å) Pi anion: Asp 370 (3.43 Å) Pi-alkyl bonds: Ile X:374(5.07 Å), Lys X:308(4.59 Å), Val X:307(4.58 Å), Ala X:363(5.11 Å), Lys X:372(5.4 Å,3.9 Å,4.87 Å,5.08 Å) Alkyl: Ile X:374(5.44 Å) Carbon-hydrogen bond: Lys X:372(2.8 Å) |
| 2 | 91641310 | 5 | 4 | Conventional hydrogen bond: Leu 361(2.2 Å), Asp X:370(2.86 Å) Pi anion bonds: Asp 370(3.43 Å) Pi cation: Lys X:372(4.77 Å) Pi-alkyl bonds: Ile X:374(5.07 Å), Lys X:308(4.52 Å), Val X:307(4.53 Å), Ala X:363(5.05 Å), Lys X:372(3.92 Å,4.92 Å,5.38 Å) Carbon-hydrogen bond: Lys X:372(2.82 Å) |
| 3 | 91639858 | 5 | 3 | Conventional hydrogen bond: Leu 361(2.18 Å), Asp X:370(2.86 Å) Pi cation: Lys 372(4.76 Å) Pi anion bonds: Asp 370(3.44 Å) Pi-alkyl bonds: Ile X:374(5.04 Å), Lys X:308(4.54 Å), Val X:307(4.53 Å), Ala X:363(5.06 Å), Lys X:372(4.9 Å,3.93 Å,4.85 Å,5.37 Å) Alkyl: Ile X:374(5.43 Å) Carbon-hydrogen bond: Lys X:372(2.84 Å) Pi-Lone pair bond: Thr X:306(2.98 Å) |
| 4 | 91639334 | 5 | 3 | Conventional hydrogen bond: Leu 361(2.18 Å), Asp X:370(2.87 Å) Pi cation: Lys 372(4.76 Å) Pi anion bonds: Asp 370(3.42 Å) Pi-alkyl bonds: Ile X:374(5.08 Å), Lys X:308(4.51 Å), Val X:307(4.53 Å), Ala X:363(5.04 Å), Lys X:372(3.91 Å,5.36 Å,4.92 Å) Carbon-hydrogen bond: Lys X:372(2.78 Å) Unfavourable donor-donor bond: Lys X:308(1.78 Å) |
| 5 | 91640550 | 5 | 4 | Conventional hydrogen bond: Leu 361(2.18 Å), Asp X:370(2.89 Å) Pi anion bonds: Asp 370(3.43 Å) Pi cation: Lys 372(4.77 Å) Pi-alkyl bonds: Ile X:374(5.05 Å), Lys X:308(4.51 Å), Val X:307(4.53 Å), Ala X:363(5.06 Å), Lys X:372(5.37 Å,3.93 Å,4.91 Å) Carbon-hydrogen bond: Lys X:372(2.83 Å) |
| 6 | 91639725 | 5 | 3 | Conventional hydrogen bond: Leu 361(2.24 Å), Asp X:370(2.9 Å) Pi anion bonds: Asp 370(3.40 Å) Pi cation: Lys 372(4.73 Å) Pi-alkyl bonds: Ile X:374(4.63 Å), Lys X:308(4.57 Å), Val X:307(4.57 Å), Ala X:363(5.08 Å), Lys X:372(4.88 Å,3.96 Å) Unfavourable Donor-donor bond: Lys X:308(1.88 Å) |
| 7 | 1880 (7,8-DHF) | 4 | 2 | Conventional hydrogen bond: Ala X:363(2.68 Å), Thr X:306(2.19 Å) Pi anion bonds: Asp X:370(4.05 Å,3.37 Å) Pi sigma bond: Val X:307(2.82 Å) Pi-alkyl bonds: Lys X:372(4.27 Å,4.72 Å), Leu X:361(5.08 Å), Ala X:363(4.43 Å,4.83 Å), Val X:307(3.93 Å) |
Abbreviation: 7,8-DHF, 7,8-dihydroxyflavone.
Computational prediction of ADMET properties of best compounds
The Swiss ADME web tool was used to analyse the physicochemical, pharmacokinetic, lipophilicity, hydrophilicity, and drug-like qualities of all best hit compounds. These parameters are indicators of absorption, distribution, metabolism, excretion, and toxicity of the compounds, and the results are shown in Table 4.
Table 4.
Absorption, distribution, metabolism, excretion, and toxicity studies of 6 ligands and reference ligand.
| PubChem ID | MW | TPSA | Consensus Log P | ESOL class | Ali class | Silicos-IT class | GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Synthetic accessibility |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 91637738 | 424.41 | 116.91 | 2.41 | Soluble | Soluble | Poorly soluble | High | No | Yes | No | No | No | No | No | 4.12 |
| 91641310 | 398.37 | 127.77 | 2.3 | Soluble | Soluble | Poorly soluble | High | No | No | Yes | No | No | No | No | 4.04 |
| 91639858 | 410.38 | 116.91 | 2.15 | Soluble | Soluble | Poorly soluble | High | No | Yes | Yes | No | No | No | No | 3.98 |
| 91639334 | 398.37 | 116.91 | 2.01 | Soluble | Soluble | Poorly soluble | High | No | No | No | No | No | No | No | 3.98 |
| 91640550 | 384.34 | 127.77 | 1.95 | Soluble | Soluble | Poorly soluble | High | No | No | Yes | No | No | No | No | 3.93 |
| 91639725 | 368.34 | 107.8 | 2.32 | Soluble | Soluble | Poorly soluble | High | No | Yes | Yes | No | No | No | No | 3.88 |
| 1880 (7,8-DHF) | 254.24 | 70.67 | 2.35 | Moderately soluble | Moderately soluble | Moderately soluble | High | Yes | No | Yes | No | No | Yes | Yes | 3.02 |
Abbreviations: BBB, blood brain barrier; MW, molecular weight; TPSA, total polar surface area; GI, gastrointestinal.
All the compounds presented in Table 4 have good drug-like properties and bioavailability, owing to nonviolation of Lipinski rule of five17 (molecular weight [MW] ⩽ 500 g/mol; MlogP ⩽ 4.15; hydrogen bond acceptors N or O less ⩽ 10; hydrogen bond donors NH or OH ⩽ 5), Ghose rules (160 g/mol ⩽ MW ⩽ 480 g/mol; –0.4 ⩽ WlogP ⩽ 5.6; 40 ⩽ MR ⩽ 130; 20 ⩽ Atoms ⩽ 70), Veber rules (Number of rotable bonds ⩽ 10; TPSA ⩽ 140 Å), Egan (WlogP ⩽ 5.88; TPSA ⩽ 131.6 Å), and Muegge (Bayer) rules (MW: 200 g/mol ⩽ MW ⩽ 600 g/mol; –2 ⩽ XlogP ⩽ 5; TPSA ⩽ 150 Å; number of rings ⩽ 7; number of carbon > 4; number of heteroatoms > 1; number of rotable bonds ⩽ 15; H-bond acceptor ⩽ 10; H-bond donor ⩽ 5).14-17,31
The MW of a compound describes the totality of atomic masses of all atoms contained in a molecule. In pharmacology, compounds with large molecular weight > 500 g/mol have poorer cellular distribution than those less than this threshold.32 All the 6 best hits possessed molecular weights within the drug-tolerable range of 368.34-424.41 g/mol. The reference ligand also possessed a tolerable value of 254.24 g/mol. The TPSA and number of rotable bonds determine the bioavailability of a drug compound, with a threshold of 140 Å according to the Veber rule.33 The potential bioavailability of the compounds was assessed using their molecular weight and TPSA values. All the compounds possessed molecular weights less than 500 g/mol and TPSA values less than 140 Å. The measure of lipophilicity of a compound is given by its partition coefficient between n-octanol and water (log Po/w) and is a classical feature in pharmacokinetics. On SwissADME server, the consensus value of log Po/w is computed as the arithmetic mean of 5 different predictive models of logP. These include XlogP, WlogP, MlogP, SILICOS-IT, and ilogP.33 This value is classically expected to be less than 5. Hydrophilicity is the measure of water solubility and is an essential parameter to be considered in absorbability of oral and parenteral drugs.34 Swiss ADME uses 3 water solubility models (ESOL, Ali, and SILICOS-IT models) to predict the general log S value, which is converted on a solubility scale into different classes thus: insoluble < –10 < poorly soluble < –6 < moderately soluble < –4 < soluble < –2 < very soluble < 0 < highly soluble.33,35,36 All the 6 best hits fell within the ‘Soluble’ ESOL and Ali solubility classes, while the reference compound, 7,8-DHF, only had moderate solubility.
Important metrics of pharmacokinetics such as gastrointestinal absorption, permeation of the blood brain barrier (BBB), substrate or nonsubstrate to permeability glycoprotein (P-gp) (an adenosine triphosphate [ATP]-binding cassette family member which protects the brain from xenobiotics influx), inhibitors of the 5 main cytochrome P-450 (CYP3A4, CYP2C9, CYP1A2, CYP2C19, and CYP2D6), and index of skin permeation are also assessed on SwissADME. All the best hits showed high level of gastrointestinal absorption, including the reference compound. Regarding their ability to permeate the BBB, all compounds 1 to 6 were nonpermeants of BBB, indicating the need for special delivery approach into the central nervous system (CNS). Three out of the 6 best hits, that is, compounds 2, 4, and 5, are potential substrates for P-gp, while the remaining 3, that is, compounds 1, 3, and 6, including the reference ligand, 7,8-DHF, are potential substrates for P-gp. Compounds that act as cytochrome P450 inhibitors are responsible for drug-interactions and inhibit metabolism and excretion of xenobiotics.37,38 Among the top 6 compounds with best binding affinity, compounds 1 and 4 were not found as inhibitors against any of the 5 major CYP450s involved in xenobiotic biotransformations, while compounds 2, 3, 5, and 6 have the potential to inhibit only CYP1A2. On the contrary, the reference compound, 7,8-DHF, was shown to inhibit CYP1A2, CYP2D6, and CYP3A4.
MD simulation
RMSD protein and ligand analyses
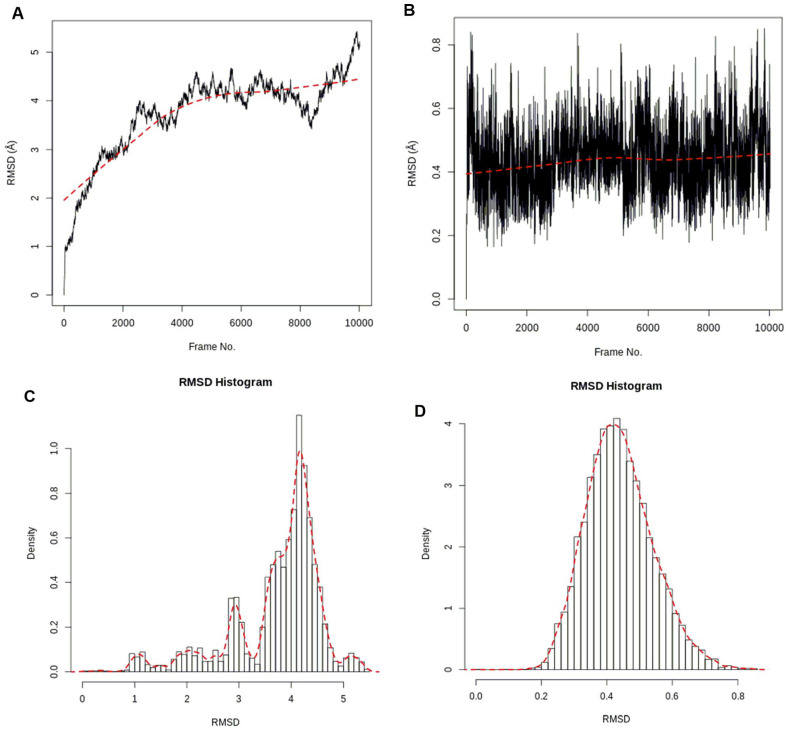
The stability of the atomic positions of the protein (1wwb) and ligands (compounds 1 and 2) in the simulation systems was captured by the RMSD. The RMSD plots and histogram of the c-alpha of the protein backbone and compound 1 are shown in Figure 4, while those of compound 2 are shown in Figure 5. For the docked complex of compound 1, the RMSD of the protein backbone rose to ~3.0 Å within the first 2500 frames (~0.25 ns) and showed stable fluctuations between 3.3 and 4.5 Å before increasing to 5.3 Å at the end of the simulation (Figure 4A). The RMSD for compound 1 in the protein’s active site showed some stability fluctuating between 0.2 and 0.8 Å after the initial rise from 0.0 Å (Figure 4B). The stabilities of the protein and ligand in the simulation system were also confirmed from the RMSD histograms (Figure 4C and D). The fluctuations can be attributed to possible imperfections in the simulation forcefield.39
Figure 4.
RMSD time series: (A) 1wwb protein and (B) compound 1. RMSD histogram: (C) 1wwb protein and (D) compound 1. RMSD indicates root mean square deviation.
Figure 5.
RMSD time series: (A) 1wwb protein and (B) compound 2. RMSD histogram: (C) 1wwb protein and (D) compound 2. RMSD indicates root mean square deviation.
For the docked complex of compound 2, the RMSD of the protein backbone remained fluctuated between 0.9 and 2.1 Å for the first 2000 frames (~0.2 ns) after the initial rise from 0.0 Å. The RMSD also fluctuated between 2.9 and 4.1 Å from 2500 to 6600 frames and remained stable at the end at ~5.3 Å (Figure 5A). The RMSD for compound 2 in the active site of the protein showed steady fluctuation between 0.6 and 1.8 Å after the initial rise from 0.0 Å (Figure 5B). The stabilities of the protein and ligand in the simulation system were also confirmed and seen in the RMSD histograms (Figure 5C and D).40
RMSF analyses
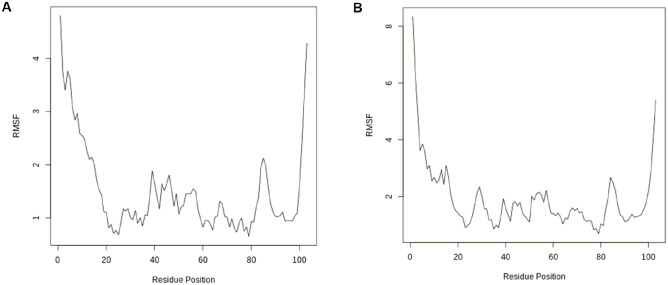
The positions of the protein structure that fluctuated more from their mean structure the least (or most) are captured in the RMSF plots for both the docked complexes of compounds 1 and 2, as shown in Figure 6. The significant fluctuations (especially those above 1 Å) observed in the plots at different residue positions correspond to the protein surface’s flexible loop sections.41 The RMSF measures the flexibility of the atoms of the protein, in this case, the c-alpha of the protein backbone. The flexibility or fluctuation is greater at both ends of the residue positions because they correspond to the protein surface’s flexible loop sections
Figure 6.
RMSF versus residue position: (A) compound 1 and (B) compound 2. RMSF indicates root mean square fluctuation.
Principal components analyses
The important dynamics of the complex system were modelled using PCA to examine the relationship between statistically meaningful conformations identified in the course of the trajectory.22,41 The essential variations in the complex were captured by grouping the principal components as eigenvectors based on the variance. The proportion of the variance found in each component is shown as eigenvalue rank plots. The PCA plots for the c-alpha backbone of the protein in the complex of compound 1 and that of compound 2 are shown in Figure 7. The first 3 PCAs (PC1, PC2, and PC3) of the complex simulation for compound 1 accounted for 69.5% of the cumulative variance annotated for each data point (Figure 7A), while that of compound 2 accounted for 79.3% (Figure 7B), as shown in their eigenvalue rank plots. Also, the observed colour variation of blue, white, and red in the PCA plots showed the conformational changes in the protein backbone captured at different data points.
Figure 7.
PCA results for the protein backbone in the 1wwb complex, comprising graphs of PC2 versus PC1, PC2 versus PC3, and PC3 versus PC1, and an eigenvalue rank plot with the cumulative variance annotated for each data point: (A) compound 1 and (B) compound 2. PCA indicates principal components analysis.
H-bond analyses
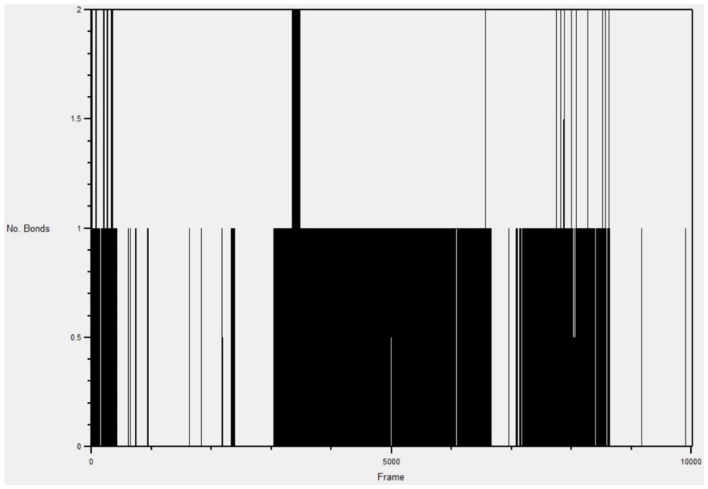
H-bond formations are important in the stabilities of the complexes during MD simulation. The number of H-bonds observed between compound 1 and the active site of 1wwb during the simulation are shown in Figure 8, while those of compound 2 is shown in Figure 9. A total of 6 h-bonds was found during the MD simulation of the compound 1-1wwb complex. The highest occupancy rate (2.62%) was observed for the interaction between compound 1 as the donor and ASP370 as the acceptor. All the h-bonds were observed as interactions with 3 amino acid residues: THR 306, ASP 370, and LEU 361. This corresponded with the observed H-bond interactions from the docking studies. The highest number of hydrogen bonds observed at any frame (time) was 3 (Figure 9).
Figure 8.
Hydrogen bond graph of compound 1 in the active site of 1wwb during simulation.
Figure 9.
Hydrogen bond graph of compound 2 in the active site of 1wwb during simulation.
The MD simulation for the compound 2-1wwb complex showed that there were 10 h-bonds within the 10 020 frames (1 ns). The highest occupancy rate (18.49%) was observed between the compound and THR306, which contributed significantly to the stability of the complex. The highest number of hydrogen bonds observed at any frame (time) was 2 (Figure 9).
Conclusion
Recent understanding of the crucial importance of tropomyosin/PLCγ signalling pathway in CDD patients has implicated it as a potential target for pharmacologic interventions in managing CDD. This was proven by the improvement pathologic molecular features in CDD on administration of a potent TrkB agonist, 7,8-DHF, in using CDKL5 KO models. Using pharmacophore-aided virtual screening of 691 ligands, this study identified 6 ligands with better binding affinity and potentially stronger agonistic properties to TrkB than 7,8-DHF, which could serve as leads for synthesis of drugs in managing CDD. Compound 1, 6-hydroxy-10-(2-oxo-1-azatricyclo[7.3.1.05,13]trideca-3,5(13),6,8-tetraen-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem ID: 91637738) had an affinity of –9.6 kcal/mol and compound 2, 6-hydroxy-10-(8-methyl-2-oxo-1H-quinolin-3-yl)-8-oxa-13,14,16-triazatetracyclo[7.7.0.02,7.011,15]hexadeca-1,3,6,9,11,15-hexaen-5-one (PubChem ID: 91641310) had an affinity of –9.3 kcal/mol, while the reference ligand, that is, 7,8-DHF (PubChem ID: 1880) had an affinity of –7.1 kcal/mol. Molecular dynamics simulation was done to confirm findings from postdocking analyses and to identify other unique chemical interactions between the 2 best ligands and the receptor under varied biomolecular environmental conditions. Computational evaluation of ADMET properties of 6 best compounds confirmed their drug-like properties, optimal bioavailability, and low toxicity potentials. In vivo experimental validation of the best hits in KO rodent models of CDD are required to validate these in silico findings.
Recommendation
One main challenge in developing drugs against CNS disorders is the potential of drugs to cross the BBB. In fact, up to 98% of all therapeutics do not cross this barrier due to less lipophilicity and relatively large molecular weights, which hamper their ability to access the brain parenchyma.42 This occurs in spite of passing the established parameters for drug-likeness. Due to this challenge, many other drug delivery processes are being exploited. These approaches include the use of nanocarriers like liposomes, magnetic and nonmagnetic nanoparticles, polymeric nanoparticles, and micelles.43 Nanodelivery has extra benefits such as reduced drug toxicity and higher safety profile, enhanced biodistribution, and the ability of drugs to safely persist within circulation for prolonged action.44 Conveyance through the BBB is also being achieved through drug modification by addition of specific ligands to target brain endothelial cells, endogenous transporters, and receptors.42 The use of venom-derived peptides to transverse the BBB transiently is also in consideration.45,46 Alternatively, these drugs may require cerebrospinal fluid (CSF) delivery. On the contrary, pro-drugs like 7,8-DHF may exhibit altered target binding and distribution causing lesser effect in humans or organ toxicity.44
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health (NIH) Fogarty Common Fund (grant number 1U2RTW010679).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: IAA developed the concept of the manuscript. GOO analysed the data. GOO and IAA wrote the first draft. SOR and EA reviewed and modified the draft. IAA, GOO, SOR and EA approved the final version of the work.
References
- 1. Olson HE, Demarest ST, Pestana-Knight EM, et al. Cyclin-dependent kinase-like 5 deficiency disorder: clinical review. Pediatr Neurol. 2019;97:18-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katayama S, Sueyoshi N, Inazu T, Kameshita I. Cyclin-Dependent Kinase-Like 5 (CDKL5): possible cellular signalling targets and involvement in CDKL5 deficiency disorder. Neural Plast. 2020;2020:6970190. doi: 10.1155/2020/6970190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gennaccaro L, Fuchs C, Loi M, et al. A GABAB receptor antagonist rescues functional and structural impairments in the perirhinal cortex of a mouse model of CDKL5 deficiency disorder. Neurobiol Dis. 2021;153:105304. [DOI] [PubMed] [Google Scholar]
- 4. Ren E, Roncacé V, Trazzi S, et al. Functional and structural impairments in the perirhinal cortex of a mouse model of CDKL5 deficiency disorder are rescued by a TrkB agonist. Front Cell Neurosci. 2019;13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Batool M, Ahmad B, Choi S. A structure-based drug discovery paradigm. Int J Mol Sci. 2019;20:2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: celebrating 50 years of the PDB with new tools for understanding and visualizing biological macromolecules in 3D. Protein Sci. 2022;31:187-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ultsch MH, Wiesmann C, Simmons LC, et al. Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC. J Mol Biol. 1999;290:149-159. [DOI] [PubMed] [Google Scholar]
- 8. Sehnal D, Bittrich S, Deshpande M, et al. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431-W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Binyamin H, Senderowitz H. Photovoltaphores: pharmacophore models for identifying metal-free dyes for dye-sensitized solar cells. npj Comput Mater. 2022;8:1-17. [Google Scholar]
- 10. Kumar BK, Faheem, Sekhar KVGC, et al. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J Biomol Struct Dyn. 2022;40:1363-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Padmi H, Kharisma V, Ansori A. Macroalgae bioactive compounds for the potential antiviral of SARS-CoV-2: an in silico study. J Pure Appl Microbiol. 2022;16:1018-1027. [Google Scholar]
- 12. Yusufkhan PS, Deshmukh SR, Farooqui M. Design, synthesis, and biological evaluation of some methyl 2-(1h-pyrazol-4-ylthio)-1, 2, 3, 4-tetrahydro-6-methylpyrimidine-5-carboxylate derivatives as potential DHFR inhibitors. Int J Health Sci. 2022;6:1018-1040. [Google Scholar]
- 13. Oladejo D, Oduselu G, Dokunmu T, et al. In silico evaluation of inhibitors of plasmodium falciparum AP2-I transcription factor and plasmodium falciparum bromodomain protein 1. 2022. https://www.researchsquare.com/article/rs-1318124/v1
- 14. Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Medi Chem. 2000;43:3867-3877. [DOI] [PubMed] [Google Scholar]
- 15. Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem. 2001;44:1841-1846. [DOI] [PubMed] [Google Scholar]
- 16. Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1:55-68. [DOI] [PubMed] [Google Scholar]
- 17. Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337-341. [DOI] [PubMed] [Google Scholar]
- 18. Saini G, Dalal V, Gupta DN, Sharma N, Kumar P, Sharma AK. A molecular docking and dynamic approach to screen inhibitors against ZnuA1 of Candidatus Liberibacter asiaticus. Mol Simulat. 2021;47:510-525. [DOI] [PubMed] [Google Scholar]
- 19. Oduselu GO, Ajani OO, Ajamma YU, Brors B, Adebiyi E. Homology modelling and molecular docking studies of selected substituted Benzo [d] imidazol-1-yl) methyl) benzimidamide scaffolds on Plasmodium falciparum adenylosuccinate lyase receptor. Bioinform Biol Insights. 2019;13:1177932219865533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33-827. [DOI] [PubMed] [Google Scholar]
- 22. Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695-2696. [DOI] [PubMed] [Google Scholar]
- 23. Simoes de Oliveira L, O’Leary H, Nawaz S, et al. Enhanced hippocampal LTP but typical NMDA receptor and AMPA receptor function in a novel rat model of CDKL5 deficiency disorder. 2022. https://www.biorxiv.org/content/10.1101/2022.06.29.497927v1 [DOI] [PMC free article] [PubMed]
- 24. Hong W, Haviland I, Pestana-Knight E, et al. CDKL5 deficiency disorder-related epilepsy: a review of current and emerging treatment. CNS Drugs. 2022;36:591-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Negraes PD, Trujillo CA, Yu NK, et al. Altered network and rescue of human neurons derived from individuals with early-onset genetic epilepsy. Mol Psychiatry. 2021;26:7047-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baltussen LL, Negraes PD. Chemical genetic identification of CDKL5 substrates reveals its role in neuronal microtubule dynamics. EMBO J. 2018;37:e99763. doi: 10.15252/embj.201899763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Devinsky O, King L, Bluvstein J, Friedman D. Ataluren for drug-resistant epilepsy in nonsense variant-mediated Dravet syndrome and CDKL5 deficiency disorder. Ann Clin Transl Neurol. 2021;8:639-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Knight EMP, Amin S, Bahi-Buisson N, et al. Safety and efficacy of ganaxolone in patients with CDKL5 deficiency disorder: results from the double-blind phase of a randomised, placebo-controlled, phase 3 trial. Lancet Neurol. 2022;21:417-427. [DOI] [PubMed] [Google Scholar]
- 29. Jofily P, Pascutti PG, Torres PH. Improving blind docking in DOCK6 through an automated preliminary fragment probing strategy. Molecules. 2021;26:1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Grimm M, Dai WT, Hou MC, Xiao ZX, Cao Y. CB-Dock: a web server for cavity detection-guided protein–ligand blind docking. Acta Pharmacol Sin. 2020;41:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615-2623. [DOI] [PubMed] [Google Scholar]
- 32. Syaban MFR, Muhammad RF, Adnani B, et al. Molecular docking studies of interaction curcumin against beta-secretase 1, amyloid A4 protein, gamma-secretase and glycogen synthase kinase-3β as target therapy for Alzheimer disease. Res J Pharm Technol. 2022;15:3069-3074. [Google Scholar]
- 33. Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ottaviani G, Gosling DJ, Patissier C, Rodde S, Zhou L, Faller B. What is modulating solubility in simulated intestinal fluids? Eur J Pharm Sci. 2010;41:452-457. [DOI] [PubMed] [Google Scholar]
- 35. Ali J, Camilleri P, Brown MB, Hutt AJ, Kirton SB. Revisiting the general solubility equation: in silico prediction of aqueous solubility incorporating the effect of topographical polar surface area. J Chem Inf Model. 2012;52:420-428. [DOI] [PubMed] [Google Scholar]
- 36. Delaney JS. ESOL: estimating aqueous solubility directly from molecular structure. J Chem Inf Comput Sci. 2004;44:1000-1005. [DOI] [PubMed] [Google Scholar]
- 37. Di L. The role of drug metabolizing enzymes in clearance. Expert Opin Drug Met. 2014;10:379-393. [DOI] [PubMed] [Google Scholar]
- 38. Huang SM, Strong JM, Zhang L, et al. New era in drug interaction evaluation: US Food and Drug Administration update on CYP enzymes, transporters, and the guidance process. J Clin Pharmacol. 2008;48:662-670. [DOI] [PubMed] [Google Scholar]
- 39. Aksimentiev A, Arkhipov A, Birnbaum R, et al. Using VMD. http://www.ks.uiuc.edu/Training/Tutorials/
- 40. Bray SA, Senapathi T, Barnett CB, Grüning BA. Intuitive, reproducible high-throughput molecular dynamics in Galaxy: a tutorial. J Cheminformatics. 2020;12:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson TO, Adegboyega AE, Ojo OA, et al. A computational approach to elucidate the interactions of chemicals from Artemisia annua targeted toward SARS-CoV-2 main protease inhibition for COVID-19 treatment. Front Med (Lausanne). 2022;9:907583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khawli LA, Prabhu S. Drug delivery across the blood–brain barrier. Mol Pharm. 2013;10:1471-1472. doi: 10.1021/mp400170b [DOI] [PubMed] [Google Scholar]
- 43. Kumar V, Kumar V, McGuire T, Coulter DW, Sharp JG, Mahato RI. Challenges and recent advances in medulloblastoma therapy. Trends Pharmacol Sci. 2017;38:1061-1084. [DOI] [PubMed] [Google Scholar]
- 44. Sethi B, Kumar V, Mahato K, Coulter DW, Mahato RI. Recent advances in drug delivery and targeting to the brain. J Control Release. 2022;350:668-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sánchez-Navarro M, Giralt E. Peptide shuttles for blood–brain barrier drug delivery. Pharmaceutics. 2022;14:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carrera-Aubesart A, Defaus S, Pérez-Peinado C, et al. Examining topoisomers of a snake-venom-derived peptide for improved antimicrobial and antitumoral properties. Biomedicines. 2022;10:2110. [DOI] [PMC free article] [PubMed] [Google Scholar]