SUMMARY
Variations in the composition of the intestinal bacterial microbiome correlate with acquisition of some sexually transmitted pathogens. To experimentally assess the contribution of intestinal dysbiosis to rectal lentiviral acquisition, we induce dysbiosis in rhesus macaques (RMs) with the antibiotic vancomycin prior to repeated low-dose intrarectal challenge with simian immunodeficiency virus (SIV) SIVmac239X. Vancomycin administration reduces T helper 17 (TH17) and TH22 frequencies, increases expression of host bacterial sensors and antibacterial peptides, and increases numbers of transmitted-founder (T/F) variants detected upon SIV acquisition. We observe that SIV acquisition does not correlate with measures of dysbiosis but rather associates with perturbations in the host antimicrobial program. These findings establish a functional association between the intestinal microbiome and susceptibility to lentiviral acquisition across the rectal epithelial barrier.
Graphical Abstract
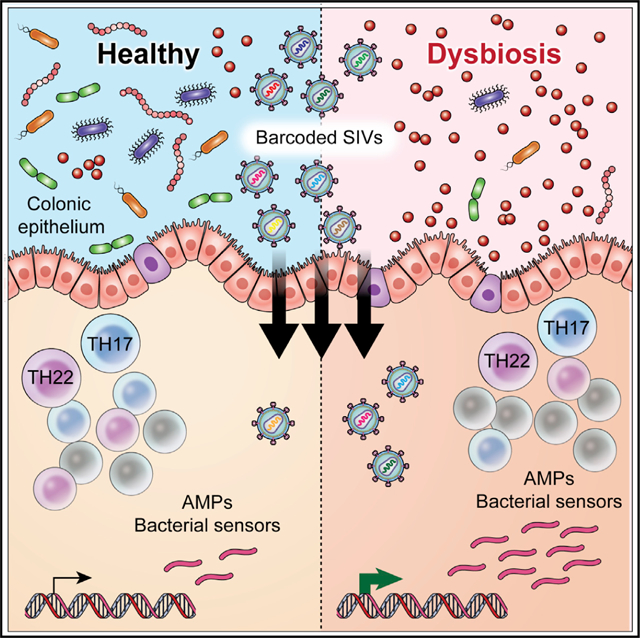
In brief
Ortiz et al. explore the contribution of intestinal dysbiosis to rectal SIV acquisition in rhesus macaques. They find that dysbiosis disrupts antimicrobial immunity and increases susceptibility to rectal SIV acquisition. These findings establish an association between the intestinal microbiome and susceptibility to lentiviral acquisition.
INTRODUCTION
Sexual transmission accounts for 90% of new global HIV-1 infections, with HIV infections among men who have sex with men (MSM) accounting for an estimated 23% of these.1 As with other sexually transmitted infections (STIs), active inflammation of vaginal, penile, or rectal tissue is associated with increased HIV-1 susceptibility and transmission potential between sexual partners.2,3 Although the presence of other STIs are a confounding risk for HIV-1 acquisition,4 only recently have variations in the commensal bacterial microbiome come forward as a potential risk factor.5–9 Commensal microbiome composition is shaped by diet, disease, sexual practices, and genetic predisposition and can contribute to both local and systemic immunity.10 Understanding whether and how microbiome composition specifically contributes to sexual HIV acquisition and transmission is essential for the rational design of pre-exposure prophylaxes.
The reasons for increased risk of HIV infection in MSM are not entirely understood. An estimated 70% of infections in MSM occur across the rectal epithelium.11 Although vulnerability of rectal columnar epithelia to physical abrasions contributes to increased susceptibility,12 there are likely other factors. In females, a Lactobacillus-dominant cervicovaginal microbiome is associated with a significantly reduced risk of HIV acquisition.8 The benefits of a Lactobacillus-dominant microbiome are multi-factorial, including low pH; competition with and antagonism of non-Lactobacilli; the production of short-chain fatty acids; and the establishment of an anti-inflammatory cervicovaginal mucosa.13 Recently, several publications have highlighted microbial signatures specific to sexually active MSM including an increased Prevotella:Bacteroides ratio and increased measures of α-diversity.5–7 These microbial signatures correlate with rectal inflammation and immune activation; however, it remains unclear whether these features increase the risk of rectal HIV acquisition.
In captive non-human primates (NHPs), sexual HIV transmission can be modeled atraumatically using simian immunodeficiency virus (SIV) variants, thus permitting a controlled experimental setting.14,15 In an HIV envelope (Env) containing the simian-human immunodeficiency virus model of transmission, Sui et al. demonstrated that accelerated infection of rhesus macaques (RMs) in a low-dose rectal challenge model was associated with significantly lower Bacteroides:Prevotella ratios and Firmicute frequencies.16 In turn, these taxonomic differences inversely correlate with markers of inflammation in the rectal mucosa. Though solidifying the correlational relationship between the microbiome and rectal lentiviral susceptibility, both this study as well as retrospective analyses in MSM5–7 have yet to establish causation.
To experimentally assess the contribution of the rectal microbiome to lentiviral acquisition, we administered vancomycin (Vanc) to RMs concurrent with repeated low-dose rectal SIVmac239X challenge. The SIVmac239X challenge virus contains 10 individual bar codes, allowing us to track the numbers of transmitted-founder (T/F) variants that establish infection.17 We observe that Vanc-induced dysbiosis is associated with perturbations in the host antimicrobial program, including low frequencies of rectal T helper 17 (TH17) and TH22 cells and increased expression of antimicrobial peptide and bacterial sensor genes. Moreover, although bacterial dysbiosis increased susceptibility to SIV acquisition, the host antimicrobial program predicted susceptibility more so than measures of dysbiosis. These results provide a mechanistic framework by which the intestinal microbiome contributes to increased risk of rectal HIV acquisition.
RESULTS
Vanc induces intestinal dysbiosis in healthy macaques
To determine whether intestinal dysbiosis contributes to enhanced HIV acquisition, we designed a low-dose intrarectal SIV challenge model incorporating repeated Vanc treatment prior to SIV challenge (Figure 1A). The design was composed of 2 parts—a pre-challenge phase and a challenge phase. During the pre-challenge phase, RMs (Table S1) were treated with (n = 8) or without (n = 8) oral 10 mg/kg Vanc for 5 days to induce intestinal bacterial dysbiosis.18 Animals were biopsied 1 week before and 2 days after treatment cessation to assess the effect of Vanc treatment on the microbiome and immunity. Animals were then rested for 1 month prior to the initiation of the challenge phase to allow surgical recovery prior to intrarectal challenge. In the Vanc group, RMs were treated with Vanc and challenged with 4 TCID50 SIVmac239X on a 28 day cycle initiated 7 days after Vanc treatment (3 days after the last dose). Plasma was assessed for viral load at days 7 and 14 post-challenge, and in the event of unsuccessful challenge, Vanc treatment was reinitiated at day 21 post-challenge. Control RMs were challenged every 14 days until infected.
Figure 1. Vancomycin induces intestinal dysbiosis in healthy macaques.

(A) Study design. Our study was composed of a pre-challenge phase, where baseline responses to vancomycin treatment were established for each animal, and a challenge phase. Samples were collected as depicted.
(B) Relative fecal bacterial abundance of control and vancomycin (Vanc) animals 7 days after Vanc treatment (3 days after the last dose). Taxa are color grouped by phylum and individually shaded by family. Significant differences in phylum abundance between groups are indicated in the figure legend.
(C) Fold change frequencies of fecal ASVs identified by MetaLonDA as longitudinally, differentially abundant over the pre-challenge period. Data show Vanc: control frequency fold change color grouped by phylum and individually shaded by family.
(D) Longitudinal observed, Chao1, and Shannon indices of fecal bacterial α-diversity. Lines represent mean per group.
(E) Principal-coordinate analysis (PCoA) of unweighted and weighted UniFrac, Bray-Curtis, and Jaccard measures of fecal bacteria β-diversity.
Data points are derived from 1 biological replicate, n = 7–8 per group. Significance methods as follows: LEfSe (B), MetaLonDA (C), unpaired two-way t tests (D), and Adonis (E).
To confirm that Vanc treatment induced bacterial dysbiosis, we assessed the stool microbiome during the pre-challenge period by 16S rDNA sequencing (Table S2). Consistent with our previous report,18 Vanc-treated animals exhibited reduced Fibrobacteres, Firmicutes, and Spirochaetes and increased Fusobacteria and Proteobacteria 7 days after Vanc treatment (3 days after the last dose) compared with controls and assessed by linear discriminant analysis effect size (LEfSe) (Figure 1B). To determine how Vanc disrupted taxa longitudinally, we compared amplicon sequence variant (ASV) frequencies in our treated animals by MetaLonDA at days 0 and 7 relative to treatment initiation. Among Firmicutes, many Clostridia taxa were significantly depleted, including losses of Ruminococcaceae and Lachnospiraceae ASVs (Figure 1C; Table S3). Though Firmicutes were significantly depleted as a phylum after treatment, we observed relative increases in Erysipelotrichaceae and Lactobacillaceae ASVs. Changes in Bacteroidetes were also disparate, driven largely by fluctuations in Prevotellaceae ASVs. Proteobacteria displayed an expansion, largely attributable to increased abundances of Gammaproteobacteria, including Enterobacteriaceae and Moraxellaceae. These results confirm that Vanc treatment induces a significant intestinal dysbiosis, including an enrichment of Prevotellaceae and Gammaproteobacteria as is commonly observed in MSM.5–7 Perturbations to the microbiome were further reflected by changes in α- and β-diversity. Observed and Chao1 measures of α-diversity were lower in Vanc animals compared with controls post-treatment (Figure 1D). Unweighted and weighted UniFrac, Bray-Curtis, and Jaccard distances too differed between control and Vanc animals post-treatment (Figure 1E). These results establish that Vanc treatment induces significant alterations to the intestinal microbiome.
Vanc treatment disrupts intestinal antimicrobial immunity in healthy macaques
We next assessed how Vanc treatment altered intestinal immunity among rectal biopsies (RBs), the anatomic site where low-dose SIVmac239X challenge occurred. Rectal CD4+ T cell frequencies did not differ either in response to treatment or between experimental and control animals (Figure 2A). We next measured SIV co-receptor expression (CCR5) and T cell activation (HLA-DR),19 proliferation (Ki-67),16 and PMA/ionomycin-induced cytokine expression (interferon γ [IFNγ], interleukin-2 [IL-2], IL-17, IL-22) and degranulation (CD107a) among rectal memory CD4+ T cells (CD4+ TM) and CD8+ T cells (CD8+ TM). As a fold change from baseline, we observed significantly increased Ki-67 expression in CD4+ TM from Vanc animals compared with controls (Figure 2B). We also observed significantly lower frequencies of IL-17-producing rectal CD4+ TM in response to stimulation in Vanc animals compared with controls (Figure S1A). No differences in phenotype nor function were observed among CD8+ TM (Figures S1B and S1C).
Figure 2. Vanc treatment disrupts intestinal antimicrobial immunity in healthy macaques.
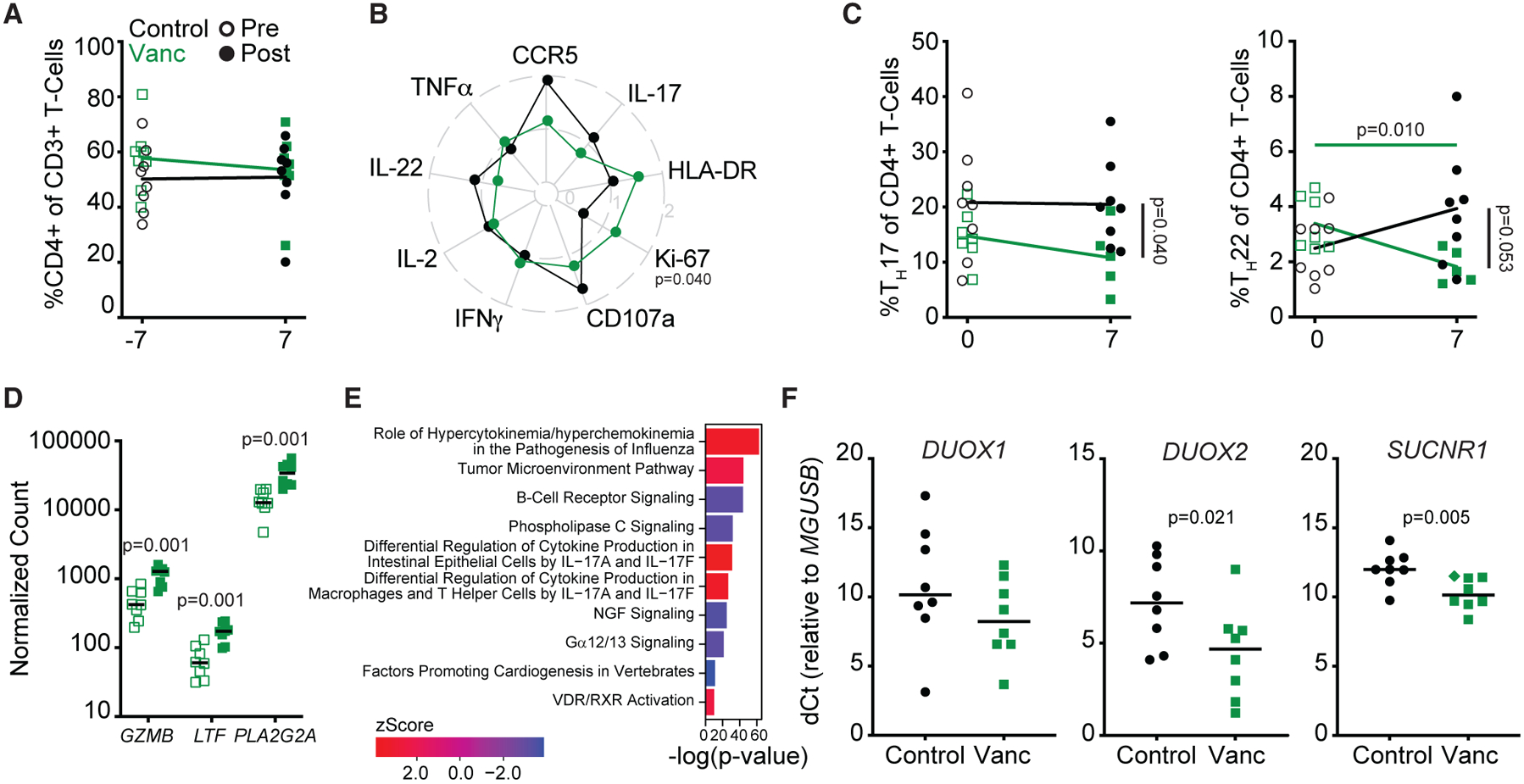
(A) Longitudinal, rectal CD4+ T cell frequencies.
(B) Radar plot depicting fold change from baseline (post- versus pre-Vanc) expression of rectal CD4+ TM activation markers and stimulation-induced cytokines.
(C) Longitudinal rectal TH17 (left) and TH22 (right) cell frequencies.
(D) NanoString-quantified counts of the top three rectal genes identified as longitudinally, differentially expressed in response to Vanc.
(E) p values and Z scores of the top 5 longitudinally increased and decreased pathways (Z score >|±2|) as identified by IPA from NanoString-quantified transcript counts in rectal homogenates.
(F) Relative expression of DUOX1, DUOX2, and SUCNR1 in rectal homogenates post-Vanc. Diamond denotes undetected transcript normalized to the limit of detection.
Lines in (A)–(D) and (F) represent mean per group. Data points are derived from 1 biological replicate, n = 5–8 per group. Significance methods as follows: unpaired or paired two-way t tests (A–D and F) and Fisher’s exact test (E). Horizontal p values color coded by group with longitudinal significance. Vertical p value denotes significance post-treatment.
TH17 and TH22 are critical in regulating intestinal antimicrobial immune responses, epithelial permeability, and physiologic hypoxia.20–23 Whereas TH17 cells can express IL-22 in addition to IL-17, TH22 cells, by definition, do not express Il-17.24,25 As a frequency of rectal CD4+ T cells, we observe that both TH17 and TH22 cells are lower in Vanc animals relative to controls post-treatment, with Vanc specifically diminishing TH22 frequencies relative to pre-treatment (Figure 2C).
To characterize the effect of Vanc more broadly, we assessed the rectal immunological transcriptome by NanoString (Table S4). Cross-sectional analysis of post-treatment samples revealed that transcriptomes largely clustered by treatment status (Figure S2A). Longitudinally, clustering was less clear. Whereas some post-treatment samples clustered with each other and away from their paired pre-treatment sample, other samples clustered by individual animal, suggesting variability in the ability of RMs to respond to Vanc (Figure S2B). Of the 770 genes included in the NanoString analysis, 90 transcripts differed between control and treated animals post-treatment and 72 differed longitudinally in treated animals, with 42 transcripts identified by both comparisons (Figures S2C–S2E). Longitudinally, the 3 genes significant after false discovery rate correction—PLA2G2A, GZMB, and LTF—all with known antimicrobial function,26–28 were uniformly upregulated (Figure 2D) and also differed by cross-sectional analysis (Figures S2D and S2E).
Ingenuity pathway analysis (IPA) of NanoString-quantified transcripts identified differentially expressed pathways for both the longitudinal and cross-sectional comparisons (Table S5). Among the pathways differentially expressed longitudinally, differential regulation of cytokine production in intestinal epithelial cells by IL-17A and IL-17F was upregulated, as was differential regulation of cytokine production in macrophages and T helper cells by IL-17A and IL-17F (Figure 2E), with the epithelial-specific pathway additionally identified in our cross-section analysis (Figure S2F). To further explore Vanc-associated effects on host antimicrobial responsiveness, we measured expression of the pro-inflammatory bacterial sensors DUOX1/229,30 and SUCNR131,32 in rectal homogenates of post-treatment animals. By qRT-PCR, homogenates from Vanc animals exhibited increased expression (lower dCt) of DUOX2 and SUCNR1 compared with controls (Figure 2F). These results demonstrate that Vanc treatment in RMs leads to disruption of the host antimicrobial response without otherwise altering canonical measures of HIV/SIV susceptibility.
Dysbiosis-responsive antimicrobial immune parameters promote SIV susceptibility
The use of a low-dose, intrarectal challenge model with barcoded SIVmac239X allowed us to monitor two outcome-based measures of susceptibility—the number of challenges required for successful infection and the number of T/F variants acquired upon infection.17 Whereas all Vanc-treated animals were infected within 6 challenges, full infection of control animals was only achieved after 11 challenges (Mantel-Cox p = 0.29, hazard ratio = 0.65; Figure 3A). Despite a statistical lack of difference in time to acquisition, Vanc animals exhibited a significantly increased number of T/F variants upon infection, with 4/8 animals acquiring 2 or more variants compared with control animals (1/8) (p = 0.039; Figure 3B). No significant differences were observed in peak viral load nor in time to peak between the two groups; however, 4/8 Vanc-treated animals showed no detectable viral load prior to 14 days post-challenge (Figure 3C).
Figure 3. Vanc-mediated intestinal dysbiosis contributes to rectal SIV acquisition through disruption of host antimicrobial immunity.
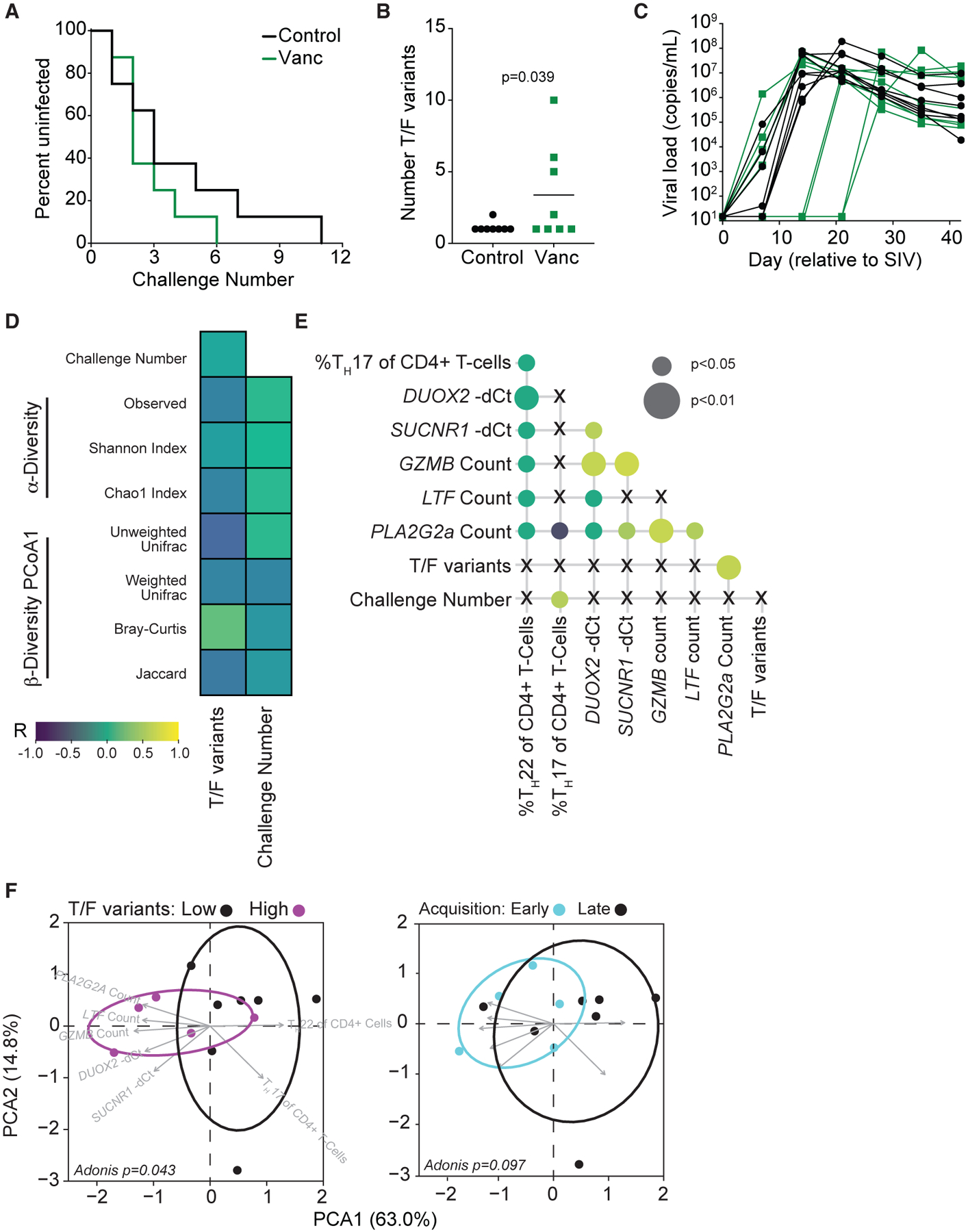
(A) Survival curve depicting the remaining percentage of uninfected animals after each challenge.
(B) Number of T/F variants detected in acute SIV infection. Lines represent mean per group.
(C) Longitudinal SIV plasma viral load in acute SIV infection.
(D) Correlation heatmap of post-treatment measures of intestinal dysbiosis versus measures of SIV susceptibility for all animals.
(E) Correlation matrix of identified Vanc-responsive measures of antimicrobial immunity versus measures of SIV susceptibility for all animals.
(F) PCA of identified Vanc-responsive measures of antimicrobial immunity, grouped by acquired T/F variant acquisition (left) or time to acquisition (right), for all animals..
Data points are derived from 1 biological replicate, n = 5–8 per group. Significance methods as follows: Mantel-Cox log-rank (A), unpaired one-way (B) or two-way (C) t test, Pearson correlation (D and E), and Adonis (F).
Although Vanc treatment induced significant intestinal dysbiosis (Figure 1), we did not observe an association between any of our measures of diversity and either challenge number or acquired T/F variant number (Figure 3D). We instead observe an association network between challenge number, acquired T/F variant number, and the measures of antimicrobial immunity found to be disrupted in Vanc animals (Figure 3E). We considered that as a network, these parameters may enhance susceptibility to SIV infection. As such, we assessed whether these limited parameters distinguish between animals that acquired low (1) versus high (>1) numbers of T/F variants and those that acquired SIV at early (1–2) versus late (>2) challenge numbers. Indeed, by principal-component analysis (PCA), we observe that this network is sufficient to cluster animals by T/F variant acquisition (Figure 3F). Whereas increased frequencies of TH17 and TH22 cells are associated with low numbers of T/F variant acquisition, increased antimicrobial gene expression is associated with increased T/F variant acquisition. When considering early- versus late-acquiring animals, we observe a trend toward clustering based on these parameters alone (Figure 3F). Our results demonstrate that perturbed intestinal antimicrobial immunity as an outcome of dysbiosis is associated with increased risk of lentiviral acquisition.
DISCUSSION
The HIV-1 epidemic continues to expand in MSM, with HIV-1 infections among MSM accounting for 23% of new global infections and 64% of new infections in western and central Europe and North America in 2020.1 Recent findings demonstrate that MSM display a distinct rectal microbiome compared with women and with men who have sex with women.5–7 As variations in the cervicovaginal microbiome contribute to female susceptibility to HIV infection,8,33 the rectal microbiome composition too may influence susceptibility to HIV infection in MSM. We experimentally demonstrate that dysbiosis of the intestinal microbiome contributes to increased susceptibility to rectal SIV acquisition in the RM SIV model of HIV-1 infection. We observe that measures of dysbiosis themselves do not predict susceptibility; rather, intestinal antimicrobial cell frequencies and gene expression patterns that occur after dysbiosis predict susceptibility.
Our approach to measure SIV susceptibility in two ways—T/F acquisition and time to acquire SIV from a low-dose inoculum—allowed us to assess a contribution of the microbiome to susceptibility across the rectal epithelium independent of sexual history or practice. The sexual act is itself neither sterile nor atraumatic, and factors within exchanged anatomical secretions may themselves influence viral transmission.34 Recent participation in condomless receptive anal intercourse is associated with an enrichment in rectal mucosa gene sets associated with mucosal injury and repair, cell proliferation, and immune activation.12 Further, it is unclear to what extent the exchange of microbiomes between partners contributes to shifts in the microbiome and localized inflammation.2,35,36 Although these confounders likely contribute to variation in susceptibility, our study provides insight into the mechanisms that govern host-intrinsic rectal SIV acquisition.
TH17 and TH22 cells were perturbed in response to Vanc and, in turn, as key components of a host antimicrobial immune network that is interwoven with rectal SIV susceptibility. That TH17 and TH22 cells are perturbed in response to antibiotic treatment is not surprising as these cells are interdependent with the commensal microbiome.20,37 While TH17 and TH22 development rely on commensal-derived metabolites,22,38 the composition of the microbiome is, in turn, susceptible to antimicrobial programs regulated by these cell types.39,40 Moreover, these interactions are essential for the development and maintenance of the intestinal barrier, with IL-17 and IL-22 signaling required for intestinal stem cell regeneration, stimulation of mucin and antimicrobial peptide secretion, and regulation of epithelial hypoxia.39,41,42 Although TH17 and TH22 cells are preferentially depleted in acute progressive HIV and SIV infections,43–45 a protective role for these cells in acquisition has not previously been demonstrated. Our results demonstrate that in contrast to activated CD4+ T cells,16,19 rectal TH17 and TH22 frequencies inversely correlate with susceptibility to infection.
While observational studies in MSM have shown an overall enrichment of Prevotellaceae and increases in α-diversity compared with population controls5–7 (indices perturbed in our Vanc animals), we do not observe a direct relationship between measures of dysbiosis and measures of SIV susceptibility. We instead observe a relationship between host antimicrobial immunity and susceptibility. These results confirm previous observational reports suggesting that overt dysbiosis is not necessary for conferring increased susceptibility to infection16,46 and further suggest that pharmaceuticals that reduce TH17 and TH22 frequencies—including, but not limited to, antibiotics47—may be counterindicated in at-risk populations. Conversely, the host antimicrobial program may be improved through pharmaceuticals or diet. For example, in murine models, a high-fiber diet improved microbiota-derived short-chain fatty acid (SCFA) production, augmenting IL-22 production, enterocyte regeneration, and antimicrobial gene expression.48
Metabolites produced by the intestinal commensal microbiome control both local and systemic immunity.49 Whereas metabolites such as SCFAs and riboflavin foster beneficial immunity,22,50 pathobiont-derived uracil29,51 and succinate31,32,52 promote intestinal inflammation through DUOX2 and SUCNR1, respectively. Unrestrained DUOX2 signaling such as in the case of bacterial dysbiosis29,51 or IL-22 deficiency30 triggers intestinal inflammation and increases microbial translocation. SUCNR1 signaling in response to luminal succinate calibrates the pro-inflammatory potential of intestinal macrophages and T cells,31,32 with IL-22 shown to regulate succinate bioavail-ability.52 Although an association between rectal HIV acquisition and availability of specific metabolites has not been formally explored, Chen et al. demonstrated that among MSM, individuals that seroconvert have a distinct microbiome profile in the months prior to conversion compared with non-converters, suggestive of differential metabolomes.53
Though designed to target specific bacterial taxa, Vanc and other antibiotics are known to have extensive off-target effects.54 In addition to perturbations among bacterial communities,55 the loss of keystone taxa can trigger ecological restructuring across microbiome domains.56 We did not assess the effect of Vanc treatment on either the virome or eukaryome, nor did we assess the contribution of these other domains on SIV susceptibility. If and how a dysbiotic virome and/or eukaryome contributes to lentiviral susceptibility requires additional research. Indeed, virus species expanded in HIV-infected humans and SIV-infected NHPs may infect gastrointestinal epithelium,57,58 and as such, their presence may influence HIV acquisition in MSM.2,35,36
Collectively, our results demonstrate that bacterial dysbiosis contributes to enhanced rectal SIV susceptibility. Although experimental dysbiosis led to enhanced SIV acquisition, we show that perturbed intestinal antimicrobial immunity more accurately predicts this increased susceptibility. Though it remains unknown how MSM sexual history and practice contribute to bacterial dysbiosis,5–7 our data suggest that interventions aimed at improving intestinal TH17 and TH22 frequencies and other host antimicrobial immune responses may reduce HIV incidence in this at-risk population.
Limitations of the study
Use of outbred RMs is associated with host variability in immunity and microbiome composition as well as with practical limits on experimental manipulation and sample size. Furthermore, we did not assess the virome or eukaryome, nor did we specifically access epithelial-adherent bacterial taxa. These features may have limited our ability to detect and attribute immunological observations to the presence or activity of specific microbes. Despite these limitations, we believe that the discovery of an association between dysbiosis-associated antimicrobial immunity and rectal SIV susceptibility provides much needed insight into the contribution of the intestinal microbiome to rectal HIV susceptibility and toward the development of therapeutic interventions to decrease said susceptibility. Lastly, we were unable to sample intestinal tissue immediately prior to SIV challenge/infection as surgical wounds would increase susceptibility to rectal infection independent of the microbiome and intestinal immunity. Although this specific limitation cannot be overcome for the use of rectal challenges, the future use of alternative experimental methods to perturb the microbiome or antimicrobial immunity will further refine the contribution of dysbiosis to rectal HIV susceptibility.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jason Brenchley (jbrenchl@niaid.nih.gov).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The 16S MiSeq data were deposited in the NCBI Sequence Read Archive: PRJNA798265. Other data available from this study are available from the lead contact upon reasonable request. This paper does not report original code - all utilized coding packages are publicly available as indicated.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Sixteen healthy male RMs (Macaca mulatta), aged 6–17, were assigned to a control (n = 8) or vancomycin treatment (n = 8) group, with sample size based on previous studies of experimental manipulations of disease progression in the RM model. Groups were stratified by age, weight and genotype (Mamu-A*001, -B*008, and -B*017) and animals sampled as mixed populations (Table S1). Vancomycin-treated animals received vancomycin hydrochloride (Hospira, Inc., 10 mg/kg, p.o.) for 5 days approximately every 28 days. All animals were challenged with 4 TCID50 SIVmac239X intrarectal (i.r.) every 28 days (vancomycin-treated RMs) or 14 days (control RMs), as outlined in Figure 1.
The NIAID Division of Intramural Research Animal Care and Use Program, as part of the NIH Intramural Research Program, approved all the experimental procedures (protocol LVD26). The Program complies with all applicable provisions of the Animal Welfare Act and other federal statutes and regulations relating to animals. Animals were housed and cared for at the NIH Animal Center, under the supervision of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited Division of Veterinary Resources and as recommended by the Office of Animal Care and Use Nonhuman Primate Management Plan. Husbandry and care met the standards set forth by the Animal Welfare Act, Animal Welfare Regulations, as well as The Guide for the Care and Use of Laboratory Animals (8th Edition). The physical conditions of the animals were monitored daily. Animals in this study were exempt from contact social housing due to scientific justification, per IACUC (Institutional Animal Care and Use Committee) protocol, and were housed in non-contact, social housing where primary enclosures consisted of stainless-steel primate caging. Animals were provided continuous access to water and offered commercial monkey biscuits twice daily as well as fresh produce, eggs, and bread products twice weekly and a foraging mix consisting of raisins, nuts, and rice thrice weekly. Enrichment to stimulate foraging and play activity was provided in the form of food puzzles, toys, cage furniture, and mirrors or television.
METHOD DETAILS
Sample collection
Blood, stool, and rectal biopsies were collected longitudinally. Sampling occurred in random order. Neither the investigators nor the animal handlers were blinded to group allocation to ensure multi-lateral supervision of design and palliative treatment. Animals were sedated with Ketamine HCl at 10 mg/kg intramuscular (i.m.) for longitudinal blood and stool sampling or with Telazol at 3–4 mg/kg i.m. for tissue timepoints. For rectal biopsies, animals were further anesthetized with isoflurane gas by intubation, to effect. Successful anesthetization was monitored by response to stimuli. No animals met endpoint criteria as defined by: (a) loss of 25% body weight from baseline weight when assigned to the protocol, (b) major organ failure or medical conditions unresponsive to treatment, (c) complete anorexia for 4 days or an inability to feed or drink sufficient nutrients to maintain body weight without assistance for 7 days, (d) distress vocalization unresponsive to treatment or intervention for 7 days, or (e) tumors arising from other than experimental means that grew in excess of 10% of body weight, impaired movement, or ulcerated.
Whole blood was collected into EDTA. For longitudinal intestinal biopsies, fecal material was removed from the rectum and biopsies obtained with biopsy forceps. 10 pinch biopsies were obtained per animal for longitudinal assessments. Biopsies were transported in RPMI and washed twice in PBS prior to processing. Approximately 1mL of RM stool was collected fresh from each animal by inserting a sterile swab 2 cm into the rectum and spinning to collect available sample. Collected feces were snap-frozen and stored at −80°C until accession.
Plasma was isolated from whole blood by centrifugation. Mononuclear cells were isolated from blood by Ficoll gradient centrifugation and from rectal biopsies by straining/grinding samples through a 0.22 μm cell strainer. Three of 10 intestinal pinch biopsies were transferred to PowerBead tubes (Qiagen) and homogenized in 1 mL TRIzol (ThermoFisher Scientific) at room temperature on a Precellys 24 homogenizer (Bertin Technologies) at 5000 revolutions per minute in 4 successive 20 sec intervals. TRIzol-preserved homogenates were stored at −80°C until accession.
Immune phenotyping and functional assessment
Polychromatic flow cytometry and cell sorting were performed on stained mononuclear cells, using a BD LSRII (FACSDiva v8.0.1). Antibodies against the following antigens were used for staining at predetermined concentrations: CCR5 (clone 3A9) PE, CD3 (SP34–2) AlexaFluor-700, CD20 (2H7) APC-H7, CD28 (CD28.2) PE-CF594, CD45 (D058–1283) BV786, and Ki-67 (B56) PE-Cy7 from BD; CD95 (DX2) PE-Cy5, CD107a (H4A3) BV711, HLA-DR (L243) BV711, IL-2 (MQ1–17H12) BV650, and TNFα (Mab11) BV605 from Biolegend; and CD4 (OKT4) eFluor-450, CD8 (SK1) PerCP-eFluor-710, IFNγ (4S.B3) eFluor-450, IL-17 (eBio64DEC17) PE-Cy7, and IL-22 (IL22JOP) APC from ThermoFisher Scientific. Cell viability was assessed using the Live/Dead Aqua Fixable Dead Cell Stain (Thermo Fisher-Scientific). Cells were permeabilized with Cytofix/Cytoperm (BD) prior to intracellular staining. CD4+ and CD8+ TM were defined as CD95+ singlet, clean, live, CD3+ lymphocytes and positive/negative gating based on clearly grouped populations, historical-determined expression, and the use of internal controls. A threshold of 100 collected events in the parent population was utilized for all subset expression analyses (FlowJo 9.9.6).
16S isolation and analysis
16S rDNA was isolated and sequenced for the V4 region (primers 515F/806R) as previously described.70 Raw Illumina FASTQ files were demultiplexed using a custom Python script. Returned paired-end FASTQ reads were filtered and processed using the DADA2 package (v1.14.1) in RStudio (v1.1.463) using R (v3.6.2) to infer ASVs at a 99% identity threshold using the Silva database (v132). Before quality trimming 5,272,884 reads were included in 48 samples – samples associated with this study irrespective of whether they were considered in this manuscript - with an average of 109,852 reads per sample. Reads were trimmed to 225bp (forward) or 200 bp (reverse) and filtered to exclude sequences with degenerate bases (N), more than 2 expected errors (maxEE), or chimerism. DADA2 quality trimming resulted in 3,065,975 reads for all the samples with an average of 63,874 reads per sample. 2 samples with less than 1000 reads were omitted from further analysis. ASVs identified as non-Bacteria, mitochondria (Rickettsiales Mitochondria), and Cyanobacteria were removed from further consideration as were resultant genera at less than 3% prevalence or with no Family diversity. 16S data are deposited in the NCBI Sequence Read Archive (SRA) under project number PRJNA798265.
The empirical identification of differentially abundant bacterial taxa between groups was determined as indicated using MetaLonDA67 for longitudinal assessments or LEfSe66 for single timepoint assessments. LEfSe was run through the Huttenhower server (http://huttenhower.sph.harvard.edu/galaxy/) whereas MetaLonDA (v1.1.8) was run in RStudio using R. ASVs assessed by MetaLonDA were fitted to a negative binomial distribution with significance adjusted by the Benjamini-Hochberg method. LEfSe was run as a one-against-all multi-class analysis, with ASVs normalized to 106 reads. α- and β-diversity measurements and coordinates for PCoA were determined utilizing phyloseq (v1.30.0) in RStudio using R.
VL and T/F assessment
Plasma viral RNA levels were determined by qRT-PCR as previously described.62 The number of detectable T/F lineages was assessed using an Illumina-based sequencing approach implemented on the MiSeq instrument, as previously described,71 with primers SIV.INT.P5 (5′-GAAGGGGAGGAATAGGGGATATG) and SIV.INT.P7 (5′-CCTCCATGT GGGAACTGCTATCC).63 Viral RNA was extracted from blood plasma obtained at ramp-up and peak viral load with up to 1 × 105 templates per reaction. All sequences were compared to the 10 lineages contained within the SIVmac239X swarm17 to determine the number of T/F per animal.
RNA extraction and quantification
For rectal transcript quantification TRIzol-preserved samples were thawed and treated with 200μL chloroform to separate nucleic acid into an aqueous phase. Following separation, Total RNA was isolated from the aqueous phase using the MagMAX-96 total RNA isolation kit (ThermoFisher Scientific) per the manufacturer’s protocol. RNA concentration and purity (A260/280 R 1.8) were assessed by spectrophotometer and normalized to 50–100 ng/μL in PCR-grade H2O. For transcript quantification by Nanostring,68 preparation, hybridization, and detection of RNA samples were carried out by following the NanoString manufacturer’s instructions (NanoString Technologies) using the nCounter NHP Immunology Panel. Subsequent analyses were performed using the nSolver analysis system (v4.0.70, NanoString Technologies). NanoString generated reads were normalized to internal positive and negative controls and housekeeping genes. NanoString quantified transcripts were further ascribed to canonical gene expression pathways by IPA (Qiagen v01-19-02).69
For individual transcript quantification, cDNA was generated using the SuperScript III reverse transcriptase kit (ThermoFisher Scientific). qRT-PCR for DUOX1, DUOX2, and SUCNR1 were preformed using transcript-specific Gene Expression Assays (ThermoFisher Scientific) with TaqMan Fast Advanced Master Mix and were normalized to housekeeping gene (monkey) beta-glucuronidase (mGusB) utilizing a custom-designed mGusB TaqMan Gene Expression Assay (ThermoFisher Scientific). Custom mGusB primers and probe as follows: forward primer 5′-CTCATTTGGAATTTTGCCGATT-3′, reverse primer 5′-CCGAGTGAAGATCCCCTTTTTA-3′, and probe 5′-TGAACAGTCACCGACGAGAGTGCTGG-3′. Relative transcript measurement was assessed in triplicate on the Applied Biosystems StepOne Plus Real Time PCR System with StepOne Software (ThermoFisher Scientific), using the recommended TaqMan Fast Advanced cycling parameters.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unpaired or paired, one and two-way t-tests were used in statistical analyses of single parameter lymphocyte phenotype and function, viral load, T/F variant number, and single transcript qRT-PCR assessments (Prism v9.0, GraphPad Software Inc.). Nano-string-quantified transcripts were analyzed by Welch’s t-test with Benjamini-Yekutieli FDR (nSolver). Radar plots were generated from non-normalized expression data using ggradar (v0.2) in R studio using R. Differentially abundant taxa were identified by MetaLonDA and LEfSe as already described. Differences in α-diversity were compared by unpaired two-way t-test and in β-diversity by adonis, in RStudio using R. Relationships between bacterial diversity, immunological parameters, and susceptibility measures were assessed by Pearson correlation using Prism or ggcorplot (v0.1.3) and ggbiplot (v0.55) in R in RStudio, and further evaluated by adonis on derived Euclidian distances. No data that met minimum threshold requirements as outlined in the Methods were excluded.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CCR5 (clone 3A9) PE | BD | Cat#560932; RRID:AB_2033947 |
| CD3 (clone SP34-2) AlexaFluor-700 | BD | Cat#557917; AB_396938 |
| CD20 (clone 2H7) APC-H7 | BD | Cat#560734; RRID:AB_1727449 |
| CD28 (clone CD28.2) PE-CF594 | BD | Cat#562296; RRID:AB_11151918 |
| CD45 (clone D058-1283) BV786 | BD | Cat#563861; RRID:AB_2738454 |
| Ki-67 (clone B56) PE-Cy7 | BD | Cat#561283; RRID:AB_10716060 |
| CD95 (clone DX2) PE-Cy5 | Biolegend | Cat#305610; RRID:AB_314548 |
| CD107a (clone H4A3) BV711 | Biolegend | Cat#328640; RRID:AB_2565840 |
| HLA-DR (clone L243) BV711 | Biolegend | Cat#307644; RRID:AB_2562913 |
| IL-2 (clone MQ1-17H12) BV650 | Biolegend | Cat#500334; RRID:AB_2563878 |
| TNFa (clone Mab11) BV605 | Biolegend | Cat#502936; RRID:AB_2563884 |
| CD4 (clone OKT4) eFluor-450 | ThermoFisher Scientific | Cat#48-0048-42; RRID:AB_2016674 |
| CD8 (clone SK1) PerCP-eFluor-710 | ThermoFisher Scientific | Cat#46-0087-42; RRID:AB_1834411 |
| IFNg (clone 4S.B3) eFluor-450 | ThermoFisher Scientific | Cat#48-7319-42; RRID:AB_2043866 |
| IL-17 (clone eBio64DEC17) PE-Cy7 | ThermoFisher Scientific | Cat#25-7179-42; RRID:AB_11063994 |
| IL-22 (clone IL22JOP) APC | ThermoFisher Scientific | Cat#17-7222-82; RRID:AB_10597583 |
| Bacterial and virus strains | ||
| SIVmac239X | Del Prete et al.17 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Vancomyin hydrochloride | Hospira, Inc. | Cat#0409-6533-11 |
| Critical commercial assays | ||
| QIAamp PowerFecal Pro DNA Kit | Qiagen | Cat#51804 |
| Qiagen DNeasy Blood and Tissue Kit | Qiagen | Cat#69506 |
| MagAttract PowerMicrobiome DNA/RNA Kit | Qiagen | Cat#27500-4-EP |
| Phusion High-Fidelity PCR Master Mix | ThermoFisher Scientific | Cat# F531L |
| Agencourt AMPure XP PCR Purification | Beckman Coulter | Cat#A63882 |
| KAPA qPCR Library Quantification kit for Illumina Platforms | KAPA Biosystems | Cat # 07960336001 |
| phiX control library | Illumina | Cat#FC-110-3001 |
| MiSeq Reagent Kit v3 | Illumina | Cat#MS-102-3001 |
| Live/Dead Aqua Fixable Dead Cell Stain | Thermo Fisher-Scientific | Cat#L34957 |
| Cytofix/Cytoperm | BD | Cat#554714 |
| TRIzol | ThermoFisher Scientific | Cat#15596018 |
| MagMAX-96 total RNA isolation kit | ThermoFisher Scientific | Cat#AM1830 |
| nCounter NHP Immunology Panel | Nanostring Technologies | Cat#XT-CSO-NHPIM1-12 |
| SuperScript III Reverse Transcriptase kit | ThermoFisher Scientific | Cat#18080044 |
| DUOX1 Gene Expression Assay (Rh01047822_m1) | ThermoFisher Scientific | Cat#4351372 |
| DUOX2 Gene Expression Assay (Rh01010975_m1) | ThermoFisher Scientific | Cat#4351372 |
| SUCNR1 Gene Expression Assay (Rh07240537_m1) | ThermoFisher Scientific | Cat#4351372 |
| TaqMan Fast Advanced Master Mix | ThermoFisher Scientific | Cat#4444557 |
| QIAsymphony DSP Virus/Pathogen Midi kit | Qiagen | Cat#937055 |
| SuperScript II Reverse Transcriptase kit | ThermoFisher Scientific | Cat#18064022 |
| EagleTaq Polymerase | Roche CustomBiotech | N/A |
| Deposited data | ||
| Silva database (v132) | N/A | https://www.arb-silva.de |
| Raw 16S FASTQ files | This paper | NCBI Sequence Read Archive: PRJNA798265 |
| Experimental models: Organisms/strains | ||
| Rhesus macaque (macaca mulatta) | NIH Animal Center | N/A |
| Oligonucleotides | ||
| 16S rDNA 515F: TCGTCGGCAGCGTCAGATGTG TATAAGAGACAGGTGCCAGCMGCCGCGGTAA |
Caporaso et al.59 | N/A |
| 16S rDNA 806R: GTCTCGTGGGCTCGGAGATGT GTATAAGAGACAGGGACTACHVGGGTWTCTAAT |
Caporaso et al.59 | N/A |
| RT-PCR Internal Amplification Control | Deer et al.60 | GenBank #FJ357008 |
| mGusB Fwd primer: CTCATTTGGAATTTT GCCGATT |
Casazza et al.61 | N/A |
| mGusB Rev primer: CCGAGTGAAGATC CCCTTTTTA |
Casazza et al.61 | N/A |
| mGusB probe: TGAACAGTCACCGACG AGAGTGCTGG |
Casazza et al.61 | N/A |
| SIVmac Fwd primer (SGAG21): GTCTGCG TCAT(dP)TGGTGCATTC |
Li et al.62 | N/A |
| SIVmac Rev primer (SGAG22): CACTAG(dK) TGTCTCTGCACTAT(dP)TGTTTTG |
Li et al.62 | N/A |
| SIVmac probe (PSGAG23): FAM-CTTC(dP) TCAGT(dK)TGTTTCACTTTCTCTTCTGCG-BHQ1 |
Li et al.62 | N/A |
| T/F sense primer (SIV.INT.P5): GAAGGG GAGGAATAGGGGATATG |
Deleage et al.63 | N/A |
| T/F antisense primer (SIV.INT.P7): CCTC CATGTGGGAACTGCTATCC |
Deleage et al.63 | N/A |
| Software and algorithms | ||
| R (v3.6.2) | N/A | https://www.r-project.org/ |
| RStudio (v1.1.463) | Posit | https://posit.co |
| phyloseq (v1.30.0) | McHurdie and Holmes64 | https://github.com/joey711/phyloseq |
| DADA2 (v1.14.1) | Callahan et al.65 | https://benjjneb.github.io/dada2/index.html |
| LEfSe | Segata et al.66 | http://huttenhower.sph.harvard.edu/galaxy/ |
| MetaLonDA (v1.1.8) | Metwally et al.67 | https://github.com/aametwally/MetaLonDA |
| FACSDiva v8.0.1 | BD | Manufacturer-supplied with BD LSRII. |
| FlowJo 9.9.6 | BD | https://www.flowjo.com |
| Prism v9.0 | Graphpad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| ggradar (v0.2) | N/A | https://github.com/ricardobion/ggradar |
| nSolver analysis system (v4.0.70) | Geiss et al.68; Nanostring Technologies | https://nanostring.com/products/analysis-solutions/nsolver-advancedanalysis-software/ |
| Ingenuity Pathway Analysis (v01-19-02) | Kramer et al.69; Qiagen | https://digitalinsights.qiagen.com/products-overview/discoveryinsights-portfolio/analysis-andvisualization/qiagen-ipa/ |
| Applied Biosystems StepOne Software (v2.0) | ThermoFisher Scientific | Manufacturer-supplied with StepOne Real-Time PCR System. |
| QIASymphonySP operating system (v5.0.4) | Qiagen | Manufacturer-supplied with the QIASymphonySP. |
| QuantStudio Real-Time PCR Software (v1.6.1) | ThermoFisher Scientific | Manufacturer-supplied with the ViiA 7 Real Time PCR System. |
| ggcorplot (v0.1.3) | N/A | https://github.com/kassambara/ggcorrplot |
| ggbiplot (v0.55) | N/A | https://github.com/vqv/ggbiplot |
Highlights.
Vancomycin treatment in rhesus macaques induces intestinal bacterial dysbiosis
Vancomycin treatment reduces TH17/TH22 frequencies and perturbs antibacterial immunity
Dysbiotic macaques acquire more transmitted-founder variants upon rectal SIV challenge
Susceptibility to rectal SIV acquisition associates with the host antimicrobial program
ACKNOWLEDGMENTS
We would like to acknowledge Heather Kendall, Richard Herbert, and all the veterinary staff at the NIH Animal Center for their excellent veterinary care. We thank the NIAID Microbiome Program, NIAID Bioinformatics and Computational Biosciences Branch, NCI Quantitative Molecular Diagnostic Core, and NCI Genomics Technology Laboratory for technical and analytical assistance. Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH and from the National Cancer Institute/NIH, under contract no. 75N91019D00024/HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of DHHS, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US government.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112020.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.UNAIDS (2020). UNAIDS Data 2020. Report. https://www.unaids.org/en/resources/documents/2020/unaids-data.
- 2.Cohen CR, Lingappa JR, Baeten JM, Ngayo MO, Spiegel CA, Hong T, Donnell D, Celum C, Kapiga S, Delany S, and Bukusi EA (2012). Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med. 9, e1001251. 10.1371/journal.pmed.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaul R, Prodger J, Joag V, Shannon B, Yegorov S, Galiwango R, and McKinnon L (2015). Inflammation and HIV transmission in sub-saharan africa. Curr. HIV AIDS Rep 12, 216–222. 10.1007/s11904-015-0269-5. [DOI] [PubMed] [Google Scholar]
- 4.Cohen MS, Council OD, and Chen JS (2019). Sexually transmitted infections and HIV in the era of antiretroviral treatment and prevention: the biologic basis for epidemiologic synergy. J. Int. AIDS Soc 22 (Suppl 6), e25355. 10.1002/jia2.25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noguera-Julian M, Rocafort M, Guillén Y, Rivera J, Casadellà M, Nowak P, Hildebrand F, Zeller G, Parera M, Bellido R, et al. (2016). Gut microbiota linked to sexual preference and HIV infection. EBio-Medicine 5, 135–146. 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vujkovic-Cvijin I, Sortino O, Verheij E, Sklar J, Wit FW, Kootstra NA, Sellers B, Brenchley JM, Ananworanich J, Loeff M.S.v.d., et al. (2020). HIV-associated gut dysbiosis is independent of sexual practice and correlates with noncommunicable diseases. Nat. Commun 11, 2448. 10.1038/s41467-020-16222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong AJS, Shaffer M, Nusbacher NM, Griesmer C, Fiorillo S, Schneider JM, Preston Neff C, Li SX, Fontenot AP, Campbell T, et al. (2018). An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 6, 198. 10.1186/s40168-018-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, et al. (2017). Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 46, 29–37. 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prodger JL, Abraham AG, Tobian AA, Park DE, Aziz M, Roach K, Gray RH, Buchanan L, Kigozi G, Galiwango RM, et al. (2021). Penile bacteria associated with HIV seroconversion, inflammation, and immune cells. JCI Insight 6, e147363. 10.1172/jci.insight.147363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan PS, Fideli U, Wall KM, Chomba E, Vwalika C, Kilembe W, Tichacek A, Luisi N, Mulenga J, Hunter E, et al. (2012). Prevalence of seroconversion symptoms and relationship to set-point viral load: findings from a subtype C epidemic, 1995–2009. AIDS 26, 175–184. 10.1097/QAD.0b013e32834ed8c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley CF, Kraft CS, de Man TJ, Duphare C, Lee HW, Yang J, Easley KA, Tharp GK, Mulligan MJ, Sullivan PS, et al. (2017). The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 10, 996–1007. 10.1038/mi.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, and Lebeer S (2013). Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol. Rev 37, 762–792. 10.1111/1574-6976.12029. [DOI] [PubMed] [Google Scholar]
- 14.McDermott AB, Mitchen J, Piaskowski S, De Souza I, Yant LJ, Stephany J, Furlott J, and Watkins DI (2004). Repeated low-dose mucosal simian immunodeficiency virus SIVmac239 challenge results in the same viral and immunological kinetics as high-dose challenge: a model for the evaluation of vaccine efficacy in nonhuman primates. J. Virol 78, 3140–3144. 10.1128/jvi.78.6.3140-3144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller CJ, Alexander NJ, Sutjipto S, Lackner AA, Gettie A, Hendrickx AG, Lowenstine LJ, Jennings M, and Marx PA (1989). Genital mucosal transmission of simian immunodeficiency virus: animal model for heterosexual transmission of human immunodeficiency virus. J. Virol 63, 4277–4284. 10.1128/JVI.63.10.4277-4284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui Y, Dzutsev A, Venzon D, Frey B, Thovarai V, Trinchieri G, and Berzofsky JA (2018). Influence of gut microbiome on mucosal immune activation and SHIV viral transmission in naive macaques. Mucosal Immunol. 11, 1219–1229. 10.1038/s41385-018-0029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Prete GQ, Park H, Fennessey CM, Reid C, Lipkey L, Newman L, Oswald K, Kahl C, Piatak M Jr., Quiñones OA, et al. (2014). Molecularly tagged simian immunodeficiency virus SIVmac239 synthetic swarm for tracking independent infection events. J. Virol 88, 8077–8090. 10.1128/JVI.01026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortiz AM, Flynn JK, DiNapoli SR, Vujkovic-Cvijin I, Starke CE, Lai SH, Long ME, Sortino O, Vinton CL, Mudd JC, et al. (2018). Experimental microbial dysbiosis does not promote disease progression in SIV-infected macaques. Nat. Med 24, 1313–1316. 10.1038/s41591-018-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carnathan DG, Wetzel KS, Yu J, Lee ST, Johnson BA, Paiardini M, Yan J, Morrow MP, Sardesai NY, Weiner DB, et al. (2015). Activated CD4+CCR5+ T cells in the rectum predict increased SIV acquisition in SIVGag/Tat-vaccinated rhesus macaques. Proc. Natl. Acad. Sci. USA 112, 518–523. 10.1073/pnas.1407466112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, Hatton RD, and Weaver CT (2012). Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075. 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, et al. (2016). Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity 44, 659–671. 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, et al. (2020). Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun 11, 4457. 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai PY, Zhang B, He WQ, Zha JM, Odenwald MA, Singh G, Tamura A, Shen L, Sailer A, Yeruva S, et al. (2017). IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe 21, 671–681.e4. 10.1016/j.chom.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trifari S, Kaplan CD, Tran EH, Crellin NK, and Spits H (2009). Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol 10, 864–871. 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 25.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, and Sallusto F (2009). Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol 10, 857–863. 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 26.Okita Y, Shiono T, Yahagi A, Hamada S, Umemura M, and Matsuzaki G (2016). Interleukin-22-Induced antimicrobial phospholipase A2 group IIA mediates protective innate immunity of nonhematopoietic cells against Listeria monocytogenes. Infect. Immun 84, 573–579. 10.1128/IAI.01000-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walch M, Dotiwala F, Mulik S, Thiery J, Kirchhausen T, Clayberger C, Krensky AM, Martinvalet D, and Lieberman J (2014). Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell 157, 1309–1323. 10.1016/j.cell.2014.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Montoya IA, Cendón TS, Arévalo-Gallegos S, and Rascón-Cruz Q (2012). Lactoferrin a multiple bioactive protein: an overview. Biochim. Biophys. Acta 1820, 226–236. 10.1016/j.bbagen.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KA, Kim SH, Kim EK, Ha EM, You H, Kim B, Kim MJ, Kwon Y, Ryu JH, and Lee WJ (2013). Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811. 10.1016/j.cell.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Grasberger H, Gao J, Nagao-Kitamoto H, Kitamoto S, Zhang M, Kamada N, Eaton KA, El-Zaatari M, Shreiner AB, Merchant JL, et al. (2015). Increased expression of DUOX2 is an epithelial response to mucosal dysbiosis required for immune homeostasis in mouse intestine. Gastroenterology 149, 1849–1859. 10.1053/j.gastro.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fremder M, Kim SW, Khamaysi A, Shimshilashvili L, Eini-Rider H, Park IS, Hadad U, Cheon JH, and Ohana E (2021). A transepithelial pathway delivers succinate to macrophages, thus perpetuating their pro-inflammatory metabolic state. Cell Rep. 36, 109521. 10.1016/j.celrep.2021.109521. [DOI] [PubMed] [Google Scholar]
- 32.Gudgeon N, Munford H, Bishop EL, Hill J, Fulton-Ward T, Bending D, Roberts J, Tennant DA, and Dimeloe S (2022). Succinate uptake by T cells suppresses their effector function via inhibition of mitochondrial glucose oxidation. Cell Rep. 40, 111193. 10.1016/j.celrep.2022.111193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klatt NR, Cheu R, Birse K, Zevin AS, Perner M, Noël-Romas L, Grobler A, Westmacott G, Xie IY, Butler J, et al. (2017). Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356, 938–945. 10.1126/science.aai9383. [DOI] [PubMed] [Google Scholar]
- 34.Sharkey DJ, Tremellen KP, Jasper MJ, Gemzell-Danielsson K, and Robertson SA (2012). Seminal fluid induces leukocyte recruitment and cytokine and chemokine mRNA expression in the human cervix after coitus. J. Immunol 188, 2445–2454. 10.4049/jimmunol.1102736. [DOI] [PubMed] [Google Scholar]
- 35.Liu CM, Prodger JL, Tobian AAR, Abraham AG, Kigozi G, Hun-gate BA, Aziz M, Nalugoda F, Sariya S, Serwadda D, et al. (2017). Penile anaerobic dysbiosis as a risk factor for HIV infection. mBio 8, e00996–17. 10.1128/mBio.00996-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopera TJ, Lujan JA, Zurek E, Zapata W, Hernandez JC, Toro MA, Alzate JF, Taborda NA, Rugeles MT, and Aguilar-Jimenez W (2021). A specific structure and high richness characterize intestinal microbiota of HIV-exposed seronegative individuals. PLoS One 16, e0260729. 10.1371/journal.pone.0260729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156. 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keir M, Yi T, Lu T, and Ghilardi N (2020). The role of IL-22 in intestinal health and disease. J. Exp. Med 217, e20192195. 10.1084/jem.20192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stockinger B, and Omenetti S (2017). The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol 17, 535–544. 10.1038/nri.2017.50. [DOI] [PubMed] [Google Scholar]
- 41.Konieczny P, Xing Y, Sidhu I, Subudhi I, Mansfield KP, Hsieh B, Biancur DE, Larsen SB, Cammer M, Li D, et al. (2022). Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 377, eabg9302. 10.1126/science.abg9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, and Val-lance BA (2018). The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 39, 677–696. 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Ryan ES, Micci L, Fromentin R, Paganini S, McGary CS, Easley K, Chomont N, and Paiardini M (2016). Loss of function of intestinal IL-17 and IL-22 producing cells contributes to inflammation and viral persistence in SIV-infected rhesus macaques. PLoS Pathog. 12, e1005412. 10.1371/journal.ppat.1005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, et al. (2012). Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol. 5, 646–657. 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim CJ, Nazli A, Rojas OL, Chege D, Alidina Z, Huibner S, Mujib S, Benko E, Kovacs C, Shin LYY, et al. (2012). A role for mucosal IL-22 production and Th22 cells in HIV-associated mucosal immunopathogenesis. Mucosal Immunol. 5, 670–680. 10.1038/mi.2012.72. [DOI] [PubMed] [Google Scholar]
- 46.Bochart RM, Busman-Sahay K, Bondoc S, Morrow DW, Ortiz AM, Fennessey CM, Fischer MB, Shiel O, Swanson T, Shriver-Munsch CM, et al. (2021). Mitigation of endemic GI-tract pathogen-mediated inflammation through development of multimodal treatment regimen and its impact on SIV acquisition in rhesus macaques. PLoS Pathog. 17, e1009565. 10.1371/journal.ppat.1009565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. (2018). Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628. 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, Kumar MV, and Gewirtz AT (2018). Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microbe 23, 41–53.e4. 10.1016/j.chom.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansaldo E, Farley TK, and Belkaid Y (2021). Control of immunity by the microbiota. Annu. Rev. Immunol 39, 449–479. 10.1146/annurev-immunol-093019-112348. [DOI] [PubMed] [Google Scholar]
- 50.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. (2012). MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723. 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 51.Kim EK, Lee KA, Hyeon DY, Kyung M, Jun KY, Seo SH, Hwang D, Kwon Y, and Lee WJ (2020). Bacterial nucleoside catabolism controls quorum sensing and commensal-to-pathogen transition in the Drosophila gut. Cell Host Microbe 27, 345–357.e6. 10.1016/j.chom.2020.01.025. [DOI] [PubMed] [Google Scholar]
- 52.Nagao-Kitamoto H, Leslie JL, Kitamoto S, Jin C, Thomsson KA, Gillilland MG 3rd, Kuffa P, Goto Y, Jenq RR, Ishii C, et al. (2020). Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med 26, 608–617. 10.1038/s41591-020-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Lin H, Cole M, Morris A, Martinson J, McKay H, Mimiaga M, Margolick J, Fitch A, Methe B, et al. (2021). Signature changes in gut microbiome are associated with increased susceptibility to HIV-1 infection in MSM. Microbiome 9, 237. 10.1186/s40168-021-01168-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maier L, Goemans CV, Wirbel J, Kuhn M, Eberl C, Pruteanu M, Müller P, Garcia-Santamarina S, Cacace E, Zhang B, et al. (2021). Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124. 10.1038/s41586-021-03986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velazquez EM, Nguyen H, Heasley KT, Saechao CH, Gil LM, Rogers AWL, Miller BM, Rolston MR, Lopez CA, Litvak Y, et al. (2019). Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol 4, 1057–1064. 10.1038/s41564-019-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tipton L, Müller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, Bonneau R, and Ghedin E (2018). Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome 6, 12. 10.1186/s40168-017-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al. (2012). Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151, 253–266. 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, et al. (2016). SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19, 323–335. 10.1016/j.chom.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deer DM, Lampel KA, and González-Escalona N (2010). A versatile internal control for use as DNA in real-time PCR and as RNA in real-time reverse transcription PCR assays. Lett. Appl. Microbiol 50, 366–372. 10.1111/j.1472-765X.2010.02804.x. [DOI] [PubMed] [Google Scholar]
- 61.Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, Roederer M, Douek DC, Betts MR, and Koup RA (2009). Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection. PLoS Pathog. 5, e1000646. 10.1371/journal.ppat.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Wang S, Kong R, Ding W, Lee FH, Parker Z, Kim E, Learn GH, Hahn P, Policicchio B, et al. (2016). Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA 113, E3413–E3422. 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deleage C, Immonen TT, Fennessey CM, Reynaldi A, Reid C, Newman L, Lipkey L, Schlub TE, Camus C, O’Brien S, et al. (2019). Defining early SIV replication and dissemination dynamics following vaginal transmission. Sci. Adv 5, eaav7116. 10.1126/sciadv.aav7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMurdie PJ, and Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Metwally AA, Yang J, Ascoli C, Dai Y, Finn PW, and Perkins DL (2018). MetaLonDA: a flexible R package for identifying time intervals of differentially abundant features in metagenomic longitudinal studies. Microbiome 6, 32. 10.1186/s40168-018-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol 26, 317–325. 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 69.Krämer A, Green J, Pollard J Jr., and Tugendreich S (2014). Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530. 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ortiz AM, Flynn JK, DiNapoli SR, Sortino O, Vujkovic-Cvijin I, Belkaid Y, Sereti I, and Brenchley JM (2019). Antiretroviral therapy administration in healthy rhesus macaques is associated with transient shifts in intestinal bacterial diversity and modest immunological perturbations. J. Virol 93, e00472–19. 10.1128/JVI.00472-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fennessey CM, Pinkevych M, Immonen TT, Reynaldi A, Venturi V, Nadella P, Reid C, Newman L, Lipkey L, Oswald K, et al. (2017). Genetically-barcoded SIV facilitates enumeration of rebound variants and estimation of reactivation rates in nonhuman primates following interruption of suppressive antiretroviral therapy. PLoS Pathog. 13, e1006359. 10.1371/journal.ppat.1006359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S MiSeq data were deposited in the NCBI Sequence Read Archive: PRJNA798265. Other data available from this study are available from the lead contact upon reasonable request. This paper does not report original code - all utilized coding packages are publicly available as indicated.


