Abstract
The Raf kinase inhibitor protein (RKIP) acts as a negative regulator of the mitogen-activated protein (MAP) kinase (MAPK) cascade initiated by Raf-1. RKIP inhibits the phosphorylation of MAP/extracellular signal-regulated kinase 1 (MEK1) by Raf-1 by disrupting the interaction between these two kinases. We show here that RKIP also antagonizes the signal transduction pathways that mediate the activation of the transcription factor nuclear factor kappa B (NF-κB) in response to stimulation with tumor necrosis factor alpha (TNF-α) or interleukin 1 beta. Modulation of RKIP expression levels affected NF-κB signaling independent of the MAPK pathway. Genetic epistasis analysis involving the ectopic expression of kinases acting in the NF-κB pathway indicated that RKIP acts upstream of the kinase complex that mediates the phosphorylation and inactivation of the inhibitor of NF-κB (IκB). In vitro kinase assays showed that RKIP antagonizes the activation of the IκB kinase (IKK) activity elicited by TNF-α. RKIP physically interacted with four kinases of the NF-κB activation pathway, NF-κB-inducing kinase, transforming growth factor beta-activated kinase 1, IKKα, and IKKβ. This mode of action bears striking similarities to the interactions of RKIP with Raf-1 and MEK1 in the MAPK pathway. Emerging data from diverse organisms suggest that RKIP and RKIP-related proteins represent a new and evolutionarily highly conserved family of protein kinase regulators. Since the MAPK and NF-κB pathways have physiologically distinct roles, the function of RKIP may be, in part, to coordinate the regulation of these pathways.
The transcriptional activator nuclear factor kappa B (NF-κB) is required for the upregulation of a large number of genes in response to inflammation, viral and bacterial infection, and other stress stimuli. Genes that respond to NF-κB encode a variety of cytokines, cell adhesion molecules, and acute-phase response proteins as well as apoptotic suppressor and effector proteins. It is believed that this reprogramming of gene expression is essential for cell survival during situations of physiological crisis (61). The activation of NF-κB in response to stimulation by the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 1 beta (IL-1β) has been extensively studied (17, 30); however, the mechanisms that modulate and eventually limit these responses are still poorly understood (61).
We report here that the recently discovered protein kinase inhibitor protein RKIP (Raf kinase inhibitor protein) acts to inhibit NF-κB activation. RKIP was first identified as an interacting partner of Raf-1 and shown to function as a negative regulator of the mitogen-activated protein (MAP) kinase (MAPK) cascade initiated by Raf-1 (75, 76). The Raf-1-initiated pathway is comprised of three sequentially acting protein kinases: a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAPK. This basic relationship has now been found to be conserved in several protein kinase pathways. In the Raf-1 pathway the MAPK is ERK1/2 (extracellular signal-regulated kinase 1 and 2), the MAPKK is MEK1 (MAP/ERK kinase 1), and the MAPKKK is Raf-1 itself. Functional studies using both gain-of-function and loss-of-function approaches demonstrated that RKIP disrupts the interaction between Raf-1 and MEK1 (75, 76). Depletion of endogenous RKIP upregulated Raf-1 kinase activity and MAPK signaling, whereas ectopic expression of RKIP suppressed Raf-1 kinase activity and MAPK signaling as well as v-Raf-mediated transformation. Biochemical studies showed that RKIP efficiently dissociated preformed Raf/MEK complexes and behaved kinetically as a competitive inhibitor of MEK phosphorylation. In vivo, the association of endogenous RKIP with Raf-1 correlated inversely with Raf-1 kinase activity during serum stimulation of quiescent cells.
Active NF-κB is a dimer that can be assembled from several members of the Rel family of transcription factors, and some form of NF-κB is expressed in most cell types (61). In unstimulated cells, NF-κB is retained in the cytoplasm in an inactive form bound to a family of inhibitory proteins known as IκB (inhibitors of κB). Activation of NF-κB requires the phosphorylation and degradation of IκB, which allows the NF-κB dimer to translocate into the nucleus. Virtually all of the many stimuli that can activate NF-κB cause the phosphorylation of IκB on two serine residues (Ser-32 and Ser-36 in IκBα). This event targets IκB for rapid polyubiquitination and degradation by the 60S proteosome (2, 31).
The kinase activity that mediates the phosphorylation of IκBα has been recently identified as part of a large (700 to 900 kDa) multisubunit cytoplasmic IKK (IκB kinase) complex (25). Three components of the IKK complex involved in the phosphorylation of IκB on the serines at positions 32 and 36 have been cloned and designated as IKKα, IKKβ, and IKKγ (also known as NEMO or IKKAP1) (14, 47, 48, 58, 62, 72, 74). The α and β subunits display kinase activity for IκB, whereas no recognizable kinase domain is found in IKKγ. It has been proposed that two molecules of IKKγ associate with a catalytic IKKα/IKKβ heterodimer, and that the C terminus of IKKγ links this core particle to upstream signaling molecules (25, 47, 78). The importance of these molecules in NF-κB signaling has been confirmed by genetic analysis utilizing knockout mice (23, 39, 41, 45, 63, 68, 69).
NF-κB can be activated by several kinase signaling cascades and is subject to multiple levels of regulation. Some of the kinases in NF-κB-activating pathways are related on the basis of sequence conservation to kinases in MAPK pathways. The kinase domains of IKKα and IKKβ contain an activation loop with the motif SXXXS, common to all MAPKKs, whose phosphorylation on both serine residues activates the kinase. Furthermore, overexpression as well as interference studies with dominant-negative mutants have suggested that some kinases in the MAPKKK family, including NIK (NF-κB-inducing kinase), MEKK1 (MEK kinase 1), TAK1 (transforming growth factor beta-activated kinase 1), mixed-lineage kinase 3, and Cot/TPL2 are involved in the phosphorylation and activation of IKKs in response to specific stimuli (20, 37, 42, 46, 50). Protein kinases other than MAPKKKs, such as protein kinase C and B, have also been reported to act upstream of IKK (27, 35, 54, 60), and a homologue of IKK designated NAK (NF-κB-activating kinase) has been implicated as an upstream activator of IKK (57, 70).
Although considerable progress has been made in the identification of kinases that activate the IKK complex, little is known about negative regulators that may impinge on these pathways (61). We show here that ectopic expression of RKIP is sufficient to downregulate NF-κB activity, whereas ablation of endogenous RKIP activity upregulates the NF-κB pathway. Specifically, RKIP can negatively modulate the activating phosphorylations of IKKα and IKKβ by upstream kinases. In mechanistic terms, RKIP physically associates with NIK and TAK1 and modulates the response of the NF-κB pathway to TNF-α- and IL-1β-mediated signaling. This mode of action bears striking similarities to the interactions of RKIP with Raf-1 and MEK in the MAPK pathway.
MATERIALS AND METHODS
Cell culture and biological reagents.
293 cells, 293/IL-1R1, BOSC 23, and COS-1 cells were grown in Dulbecco's modified Eagle's medium with high glucose and glutamine and supplemented with 10% fetal bovine serum and penicillin-streptomycin. 293/IL-1R1 (Z. Cao, Tularik) is a stably transfected 293 cell line overproducing the IL-1 receptor (9). Rat1 fibroblasts were maintained in the same medium supplemented with 10% calf serum. Recombinant human TNF-α and IL-1β were purchased from Life Technologies and Calbiochem, respectively. Anti-FLAG M2 antibody and affinity resin were purchased from Sigma. Rabbit anti-NIK polyclonal antibody was from Santa Cruz Biotechnology. Monoclonal anti-actin and anti-hemagglutinin (HA) 12CA5 antibodies were purchased from Amersham and Boehringer Mannheim, respectively. Rabbit anti-TAK1 antibody was a gift from K. Matsumoto (73). MEK inhibitor U0126 was purchased from Promega.
Plasmids and protein expression.
HA-tagged RKIP, ASK1, N-terminally truncated MEKK1, TAK1, and FLAG-tagged NIK, IKKα, IKKβ, and NAK expression plasmids have been previously described (10, 24, 46, 50, 58, 70, 72, 76). To construct FLAG-RKIP, the RKIP cDNA was PCR amplified and cloned in frame into pCMV2-FLAG (Sigma). Glutathione S-transferase (GST)–IκB was expressed in Escherichia coli and purified as described (26). To construct an HA-RKIP retrovirus expression vector, HA-RKIP (75) was cloned between the EcoRI and SalI sites of the retrovirus vector pWZL-Blast (J. Morgenstern, Millenium). The reporter plasmids E-Sel lacZ, CRE-lacZ, NF-κB-Luc, AP-1-Luc, Gal4-Luc, and Hsp-Luc and effector plasmids CMV-Gal4 (amino acids 1 to 94), CMV-Gal4(Sp1), CMV-Gal4(Sap1), and CMV-RKIP have been previously reported (34, 49, 76, 77).
Construction of a stable cell line expressing HA-RKIP.
BOSC 23 cells (55) were transfected with 20 μg of plasmid pWZL-HA-RKIP (55). At 72 h posttransfection, the virus-containing medium was filtered and used to infect Rat1 fibroblasts. Forty-eight hours after infection, the cultures were trypsinized and diluted into medium containing 5 μg of blasticidin (ICN Pharmaceuticals, Inc.)/ml. Individual blasticidin-resistant colonies were ring cloned and expanded. The expression levels of HA-RKIP in each clone were monitored by immunoblotting with 12CA5 antibody.
In vitro kinase assays.
IKK assays were performed as described (43). 293 cells were transiently transfected with the indicated expression plasmids using Lipofectamine (Life Technologies) according to the manufacturer's specifications. Thirty-six hours posttransfection, the cells were treated for 5 h with 50 ng of TNF-α/ml. After TNF-α treatment, cells were washed with cold phosphate-buffered saline and lysed in 50 mM HEPES (pH 7.6), 250 mM NaCl, 10% glycerol, 1 mM EDTA, and 0.1% Nonidet P-40 supplemented with protease and phosphatase inhibitors. Cell lysates were cleared by centrifugation and incubated for 4 h at 4°C with anti-FLAG M2 antibody conjugated to agarose beads. In vitro kinase assays were performed on the immune complexes using purified recombinant GST-IκB protein as a substrate in 20 μl of kinase buffer containing 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 0.5 mM dithiothreitol, 100 μM ATP, and 5 μCi of [γ-32P]ATP at room temperature for 30 min.
Transfection and reporter gene assays.
COS-1 cells were transfected by the DEAE dextran/chloroquine method using 5 μg of DNA per 100-mm-diameter dish. 293, 293/IL-1R1, and Rat1 cells were transfected at 70 to 80% confluence with Lipofectamine using 3 μg of DNA and 2 μg of Lipofectamine per 35-mm-diameter plate. Cells were harvested 48 h after transfection, and total cell extracts were assayed for luciferase activity. Where indicated, cultures were treated with TNF-α (50 ng/ml) or IL-1β (10 ng/ml) for 6 h immediately before harvest. A Renilla luciferase gene (Promega) driven by the constitutive thymidine kinase promoter was included in all transfection experiments as an internal control to correct for transfection efficiency. Dual luciferase assays were performed using a kit obtained from Promega. Microinjection experiments with affinity-purified RKIP antiserum and NF-κB and TRE-lacZ reporter plasmids were performed as described (75, 76).
Fractionation of Rat1 S100 cytosolic extract.
To prepare S100 cytosolic extract, 8 × 107 Rat1 cells were suspended in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol) supplemented with protease inhibitors. After 15 min on ice, the cells were disrupted by five passes through a 26-gauge needle. The lysate was centrifuged at 12,500 × g for 5 min at 4°C, and the pelleted nuclei and mitochondria were discarded. The supernatant was centrifuged again at 12,500 × g for 10 min at 4°C to remove residual mitochondria and heavy membranes. Subsequently, the supernatant was centrifuged at 100,000 × g for 60 min at 4°C. Glycerol was added to the S100 supernatant to 10%, and the KCl concentration was adjusted to 0.1 M. One milliliter of this extract (total protein, 2 mg) was applied to a 1-ml HiTrap DEAE Sepharose Fast Flow column (Amersham Pharmacia Biotech) equilibrated with buffer A with 100 mM KCl. The column was washed with 3 column volumes of buffer A with 0.1 M KCl and eluted with 10 ml of 0.1 to 1 M linear KCl gradient. Fractions of 0.5 ml were collected. Chromatography was performed at 4°C using an automatic fast protein liquid chromatography station (Amersham Pharmacia Biotech).
RESULTS
RKIP is an inhibitor of the NF-κB signaling pathway.
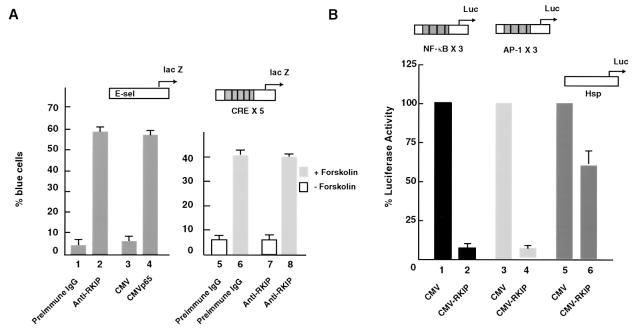
In experiments to investigate the role of RKIP in MAPK pathway signaling (75, 76), NF-κB reporters were used as controls and discrete effects on activity were noticed. We therefore performed a series of directed experiments to examine the role of RKIP on NF-κB signaling. First, we inhibited endogenous RKIP activity by antibody microinjection and monitored the effects on NF-κB activity by measuring the expression of a coinjected NF-κB lacZ reporter, E-sel lacZ (Fig. 1A). Microinjection of affinity-purified anti-RKIP antibodies strongly activated the NF-κB lacZ reporter in Rat1 fibroblasts to approximately the same extent as that elicited by the microinjection of a plasmid encoding the p65 subunit of NF-κB (Fig. 1A). This effect was specific because the injection of a control immunoglobulin G (IgG) was ineffective and the anti-RKIP IgG did not affect the expression of a cyclic AMP-dependent reporter gene, CRE × 5 lacZ (Fig. 1A, bars 1 and 7). Ectopic expression experiments corroborated these results by showing that the transfection of an RKIP expression construct diminished the basal activities of NF-κB and AP-1 reporters to approximately the same extent (Fig. 1B). The repression was specific since the effect was not observed with a reporter lacking NF-κB binding sites (Fig. 1B, bars 5 and 6).
FIG. 1.
Ablation of RKIP activates and overexpression of RKIP represses basal NF-κB activity. (A) Ablation of RKIP activity by antibody injection activates an NF-κB-dependent reporter (E-sel) but not a cyclic AMP-stimulated reporter (CRE × 5). E-sel is an E-selectin promoter DNA fragment containing one copy of the consensus NF-κB binding site (71). Quiescent Rat1 cells were microinjected with the indicated reporter plasmids and antibodies and either left unstimulated or treated with 20 μg of forskolin per ml, an activator of adenyl cyclase. (B) Overexpression of RKIP represses NF-κB- and AP-1-dependent reporters. RKIP was placed under the control of the cytomegalovirus (CMV) promoter (CMV-RKIP). RKIP or an empty vector control (CMV) was cotransfected with the indicated reporter plasmids into exponentially growing NIH 3T3 cells, and 48 h later extracts were assayed for luciferase activity. The activities of reporters in combination with the empty CMV vector were set to 100%. In all panels the means and standard deviations of at least two independent experiments are shown.
In light of the reported effects of Raf on NF-κB signaling (15, 40, 52), we considered the possibility that the observed repression by RKIP could be the result of cross talk from the Raf/MEK/ERK pathway. To address this, we examined the effect of pharmacological MEK inhibitors, such as U0126, on the repression of an NF-κB luciferase reporter. These experiments showed that significant inhibition of NF-κB signaling in unstimulated, actively growing cells can occur at concentrations of U0126 that abolish MEK activity (Fig. 2A and B, lanes 4 to 6). The effect of U0126 was specific since it had no effect on Sp1 transactivation activity (Fig. 2B, lanes 1 to 3). Cross talk between the MAPK and NF-κB pathways does occur, but its magnitude is insufficient to explain the influence of RKIP on NF-κB activity. For example, we observed that approximately 20% of the NF-κB basal activity found in exponentially cycling 293 cells could be inhibited by MEK inhibitors, and it is thus likely elicited by cross talk from the Raf/MEK/ERK pathway (Fig. 2A, lanes 1 and 2).
FIG. 2.
RKIP can inhibit NF-κB independently of MEK. (A) Pharmacological inhibition of MEK activity does not interfere with the ability of RKIP to repress basal NF-κB activity. 293 cells were cotransfected with an empty vector (cytomegalovirus [CMV]), an RKIP expression vector (CMV-RKIP), and a NF-κB reporter (NF-κB × 3Luc) as indicated. Twenty-four hours after transfection, the medium was replaced with medium containing 10 μM MEK inhibitor U0126 (lanes 2 and 4) or an equivalent volume of carrier (dimethyl sulfoxide) (lanes 1 and 3). Cells were harvested 24 h later, and luciferase activity was determined. Repression was calculated relative to the activity elicited by the reporter plus empty CMV vector (lane 1), which was assigned a value of 100%. (B) Control experiment performed in parallel to demonstrate that U0126 was active under the conditions used. A Gal4 (DNA-binding domain)-Sap1 (transactivation domain) fusion protein [CMV-Gal4(Sap1)] was used in conjunction with a Gal4 (DNA-binding site) reporter (Gal4 × 4 Luc). The Sap1 transactivation domain is a known target of ERK. Activation (lane 5) was measured relative to a Gal4-only vector [CMV-Gal4(1 to 94)] (lane 4) and was set to 1. To control for nonspecific effects of U0126, a Gal4-Sp1 (transactivation domain) fusion protein [CMV-Gal4(Sp1)] was used. This transactivation domain is known to be independent of ERK activity and was not inhibited by U0126 (lanes 1 to 3).
Interaction of RKIP with IKKs and upstream kinase activators of NF-κB.
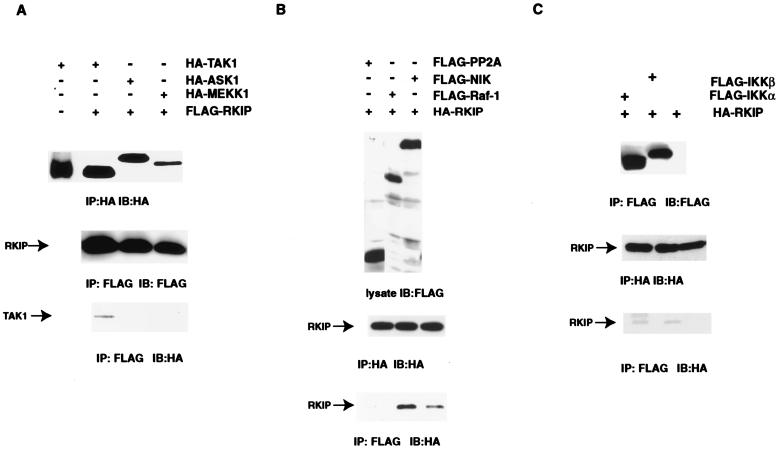
RKIP was first identified as an inhibitor of Raf-1, a kinase in the MAPKKK family. Three kinases belonging to the MAPKKK family, TAK1, MEKK1, and NIK, have recently been implicated as upstream activators of the NF-κB pathway (37, 38, 46, 50). Based on their functional relatedness to Raf-1, we considered these kinases as possible targets of RKIP, and therefore, we tested directly for evidence of physical interactions. COS-1 cells were transfected with FLAG-tagged RKIP and various HA-tagged MAPKKKs, cell lysates were immunoprecipitated with a FLAG tag-specific antibody, and coimmunoprecipitation of the MAPKKKs was visualized by immunoblotting with an HA tag-specific antibody. Clearly detectable amounts of TAK1 were coimmunoprecipitated with RKIP (Fig. 3A). The coimmunoprecipitation of TAK1 with RKIP is specific because HA-TAK1 was not found in the anti-FLAG immunoprecipitates when FLAG-RKIP was not included in the transfection (Fig. 3A). No HA tag cross-reactive material was observed with either HA-MEKK1 or HA-ASK1, a MAPKKK implicated in TNF-α-mediated activation of the JUN kinase pathway (11, 51). All the proteins being tested except for HA-MEKK1 were expressed at approximately equal levels in COS-1 cells (Fig. 3A). Using a similar approach, we also tested for interaction of RKIP with NIK and observed coimmunoprecipitation of FLAG-tagged NIK and HA-tagged RKIP. Although the binding affinity of RKIP for NIK is apparently lower than that for Raf-1, the binding is highly specific because FLAG-tagged protein phosphatase 2A (catalytic subunit) did not coimmunoprecipitate with HA-tagged RKIP in the same experiment (Fig. 3B).
FIG. 3.
Coimmunoprecipitation of NIK, TAK1, IKKα, and IKKβ with RKIP. (A) FLAG-RKIP was cotransfected into COS-1 cells with the indicated HA-tagged expression plasmids. Extracts were immunoprecipitated (IP) with anti-HA or anti-FLAG antibodies as indicated. Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to membranes, and the expression levels of HA-tagged or FLAG-tagged proteins were monitored by immunoblotting (IB) with the indicated antibodies. (B) HA-RKIP was cotransfected into COS-1 cells with the indicated FLAG-tagged expression plasmids. Extracts were analyzed as indicated above. (C) HA-RKIP was cotransfected into COS-1 cells either alone or with FLAG-tagged IKKα or IKKβ expression plasmids. Extracts were analyzed as indicated above.
The fact that both IKKα and IKKβ contain a canonical MAPKK activation loop and that MEK, a MAPKK, binds RKIP both in vivo and in vitro prompted us to extend the RKIP interaction studies to IKKs. Using the methods described above, the interaction of IKKα, IKKβ, and RKIP was investigated by coimmunoprecipitation of FLAG-tagged IKKs and HA-RKIP expressed in COS-1 cells. Detectable amounts of RKIP were coimmunoprecipitated with both IKKα and IKKβ (Fig. 3C). Although the binding is weak, the interactions that we observed between RKIP, IKKα, and IKKβ appear to be specific because we did not detect any association between RKIP and NAK, another IKK-related kinase (data not shown).
RKIP inhibits NIK- and TAK1-mediated activation of NF-κB.
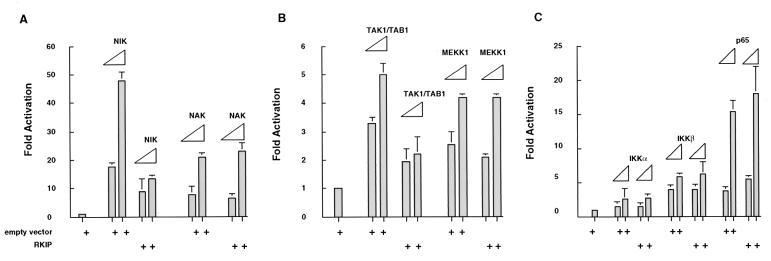
To examine the functional consequences of the above described physical associations between RKIP, NIK, TAK1, and IKKα/β, we made use of the fact that ectopic expression of these activator kinases can upregulate an NF-κB-responsive reporter. 293 cells were therefore transfected with different combinations of kinase and RKIP expression vectors in the presence of an NF-κB-luciferase reporter. For experiments examining the effects of RKIP on TAK1-mediated activation of an NF-κB reporter, TAB1, an essential TAK1 coactivator, was also included in the transfection in addition to TAK1 (50). All four upstream kinase activators of IKK, namely NIK, NAK, TAK1, and MEKK1, stimulated the NF-κB reporter (Fig. 4). As previously reported, different kinases stimulated NF-κB activity to different extents, with NIK being the most potent (50-fold), followed by NAK (20-fold), and TAK1 and MEKK1 (4- to 5-fold) (37, 38, 46, 49, 50, 70). RKIP strongly antagonized the activation of NF-κB elicited by NIK (up to fivefold) and more weakly antagonized the activation elicited by TAK1 (up to twofold). Notably, RKIP did not antagonize the activation elicited by either NAK or MEKK1 (Fig. 4A and B). The downregulation of the NIK- and TAK1-mediated activation of the NF-κB reporter by RKIP was not a result of reduced expression levels of NIK and TAK1, because we did not observe a decrease in endogenous NIK and TAK1 protein levels when RKIP was overexpressed (data not shown). Taken together, the results of these in vivo activation experiments are in complete agreement with the coimmunoprecipitation results.
FIG. 4.
RKIP inhibits NIK- and TAK1-mediated activation of NF-κB. 293 cells were cotransfected with an NF-κB luciferase reporter (NF-κB × 3), an RKIP expression vector (CMV-RKIP), or an empty vector control (cytomegalovirus [CMV]) and expression vectors for NIK, NAK, TAK1, MEKK1, IKKα, IKKβ, and NF-κB/p65 as indicated. The NIK and NAK (A), IKKα, IKKβ, and NF-κB/p65 (B), and TAK1 and MEKK1 (C) vectors were each transfected at two DNA concentrations: 0.1 and 0.5 μg per 35-mm-diameter plate. The NF-κB × 3, CMV-RKIP, and CMV vectors were held constant (50 ng for NF-κB × 3 and 2 μg for CMV-RKIP and CMV). The total DNA concentration per plate was kept constant with pUC19 plasmid DNA. Activity elicited by NF-κB × 3 cotransfected with the CMV vector control in the absence of activating kinases was set to 1. At 2 days posttransfection, cell extracts were prepared and analyzed for luciferase activity. All activities were normalized on the basis of an internal transfection control (thymidine kinase promoter-driven Renilla luciferase reporter).
In order to further delineate the position (or positions) at which RKIP acts in the NF-κB activation cascade, we examined its effect on transactivation elicited by the transfection of the p65 subunit of NF-κB. RKIP had no effect on p65-mediated activation of an NF-κB reporter (Fig. 4C). The failure to block at this downstream point indicates that RKIP does not interfere directly with the activity of NF-κB but acts upstream. Interestingly, RKIP also did not block the activation of NF-κB elicited by the overexpression of IKKα and IKKβ (Fig. 4C).
RKIP inhibits IL-1β- and TNF-α-mediated activation of NF-κB and IKK activity.
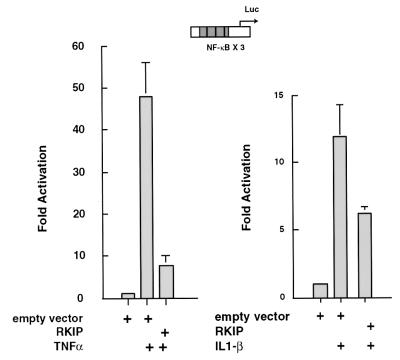
In light of previous findings that NIK and TAK1 are the physiological activators of IKK in response to the proinflammatory cytokines IL-1β and TNF-α (46, 50, 67), we also examined the effects of RKIP on TNF-α- and IL-1β-mediated stimulation of NF-κB activity. In 293 cells RKIP consistently reduced TNF-α-mediated stimulation of NF-κB activity up to fivefold (Fig. 5, left panel). Both the magnitude of TNF-α stimulation of NF-κB and the magnitude of the RKIP inhibitory effect were strikingly parallel to the results we obtained with NIK overexpression (compare Fig. 5, left panel, with Fig. 4A). Consistent with the results that TAK1 is a target of RKIP, we also observed that RKIP elicited a clear inhibition of IL-1β-mediated activation of an NF-κB reporter (Fig. 5, right panel). Importantly, the magnitude of this effect was comparable to that elicited by RKIP on ectopically expressed TAK1 (compare Fig. 5, right panel, with Fig. 4B).
FIG. 5.
RKIP inhibits TNF-α- and IL-1β-mediated activation of NF-κB. 293 (A) or 293/IL-1R1 (B) cells were transfected with an NF-κB luciferase reporter (NF-κB × 3) and an RKIP expression vector (CMV-RKIP) or empty vector control (cytomegalovirus [CMV]). Thirty hours after transfection, the cells were either left untreated or were stimulated for 6 h with TNF-α (A) or IL-1β (B) and extracts were prepared and analyzed for luciferase activity. The data presented in the figure are representative of the three experiments we performed.
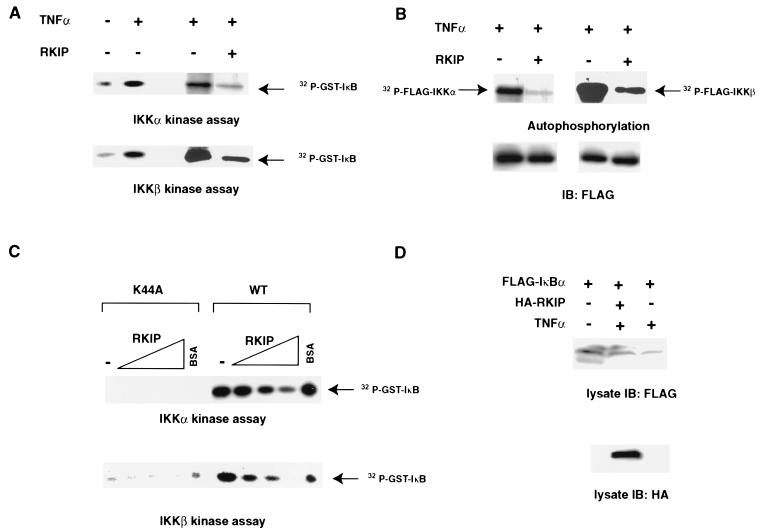
To more directly examine the effects of RKIP inhibition on the enzymatic activity of IKKs, we transfected 293 cells with FLAG-tagged IKKα or IKKβ with or without RKIP and subsequently stimulated the cells with TNF-α. IKKα and IKKβ were immunoprecipitated with an anti-FLAG antibody, and the immunoprecipitates were assayed in vitro in the presence of [γ-32P]ATP for autophosphorylation as well as phosphorylation of an exogenously added GST-IκBα substrate (Fig. 6). As reported, IKK activities toward IκBα were stimulated by TNF-α treatment (Fig. 6A). Interestingly, the inclusion of RKIP in the transfection reduced the in vitro IKK activities by four- to fivefold in both assays (Fig. 6A). In agreement with previous reports showing that the extent of IKK autophosphorylation correlated directly with kinase activities, we observed a downregulation of IKKα and IKKβ autophosphorylation in the presence of RKIP (Fig 6B). These results show that RKIP can antagonize the in vivo activation of IKK activity elicited by treatment of cells with TNF-α. To examine whether RKIP directly inhibits IKK activities, FLAG-tagged IKKs were expressed in COS-1 cells, the cells were stimulated with TNF-α, and FLAG-IKKs were purified with anti-FLAG antibody. The effects of purified bacterial recombinant RKIP on IKK activity were assayed using in vitro kinase assays with GST-IκBα as a substrate. As seen in Fig. 6C, the addition of increasing concentrations of RKIP (0 to 2.3 μM) resulted in a dose-dependent reduction of the phosphorylation of IκBα. The inhibition by RKIP was specific because addition of equivalent amounts of bovine serum albumin had no effect on the IKK activities.
FIG. 6.
Effects of RKIP on the phosphorylation of IκB proteins by IKKα and IKKβ. (A) Inhibition of IκBα phosphorylation by IKKα and IKKβ. 293 cells were transfected with the indicated FLAG-tagged IKK expression vectors with or without an RKIP expression vector. Thirty hours after transfection, cells were either left untreated or were stimulated for 10 min with TNF-α. IKK proteins were immunoprecipitated with anti-FLAG antibody and assayed for kinase activity using purified recombinant bacterially expressed full-length IκBα and [γ-32P]ATP as substrates. (B) Inhibition of IKKα and IKKβ autophosphorylation. 293 cells were transfected as described for panel A. The autophosphorylation of IKK was assayed by incubating the IKKα or IKKβ immunoprecipitates with [γ-32P]ATP. The amount of total IKK protein in each assay was determined by immunoblotting the kinase reactions with polyclonal anti-FLAG antibody. (C) Inhibition of IKK activity by RKIP in vitro. COS-1 cells were transiently cotransfected with FLAG-IKKα (1.5 μg), FLAG-IKKα K44A (1.5 μg), FLAG-IKKβ (2 μg), or FLAG-IKKβ K44A (2 μg), serum deprived for 24 h, and subsequently treated with 50 ng of TNF-α/ml for 10 min. FLAG-tagged proteins were immunoprecipitated with FLAG-M2 antibody, and aliquots of the immunoprecipitate (IP) (10 μl) were incubated on ice for 30 min with increasing concentrations of RKIP (2.5 to 22.5 μM) or 22.5 μM bovine serum albumin. The kinase reaction was initiated by adding 10 μl of a mix containing 2.5 μM GST-IκB (amino acids 5 to 55), 50 μM ATP, and 3 μCi of [32P]ATP in 1× kinase buffer. IKKα reactions were incubated for 10 min at 30°C, and IKKβ reactions were incubated for 30 min. (D) RKIP interferes with TNF-α-induced degradation of IκBα in vivo. 293 cells were transfected with the indicated expression vectors. Thirty hours after transfection, cells were either left untreated or were stimulated for 10 min with TNF-α. Cell lysates were immunoblotted with either anti-FLAG or anti-RKIP antibodies.
To examine whether overexpression of RKIP would inhibit the activation of endogenous IKKs induced by TNF-α, we made use of the fact that IκB phosphorylated by activated IKKs is rapidly targeted for degradation. 293 cells were transfected with FLAG-tagged IκBα either alone or together with HA-tagged RKIP. Cells were treated with TNF-α to activate the endogenous IKK activities, and the effects of RKIP overexpression were examined by monitoring the expression levels of FLAG-IκBα by immunoblotting with anti-FLAG antibody. As expected, the expression levels of IκBα were reduced after TNF-α treatment (Fig. 6D). Consistent with the result that RKIP inhibited IKK activities in vitro, we observed that RKIP elicited a clear inhibition of IKK-mediated degradation of IκBα in the transfected cells (Fig. 6D).
Endogenous NIK and TAK1 coimmunoprecipitate with RKIP.
To investigate whether RKIP can associate with NIK and TAK1 under physiological levels of protein expression, we immunoprecipitated RKIP from extracts of Rat1 fibroblast cells with anti-RKIP antibodies. The amounts of TAK1 and NIK proteins in the anti-RKIP immunoprecipitates were monitored with TAK1- or NIK-specific antibodies. As shown in Fig. 7A, detectable amounts of endogenous TAK1 were present in the anti-RKIP immunoprecipitate but not in the control immunoprecipitate. In light of previous findings (50) that IL-1β treatment stimulates the activity of TAK1, we decided to examine whether IL-1β could affect the association of TAK1 with RKIP. 293 cells were transfected with HA-RKIP, serum starved, and stimulated with IL-1β for 5 or 15 min. The association of endogenous TAK1 with HA-RKIP was examined by coimmunoprecipitation followed by immunoblotting with TAK1 antibodies. Although detectable amounts of TAK1 were associated with RKIP in cycling cells (Fig. 7A), no TAK1 was detected in the anti-HA immunoprecipitates from serum-deprived 293 cell lysates (Fig. 7B). Interestingly, TAK1 was found to be associated with HA-RKIP 5 min after IL-1β stimulation, and the association was drastically reduced after 15 min (Fig. 7B). Note that the association and dissociation of RKIP with TAK1 correlate well with the previously reported activation kinetics of TAK1 (67). It remains to be seen whether the RKIP could directly inhibit the kinase activity of TAK1.
FIG. 7.
Coimmunoprecipitation of endogenous TAK1 and NIK with RKIP. (A) Coimmunoprecipitation of RKIP and TAK1 from extracts of Rat1 fibroblasts. Rat1 cells (2 × 107) were lysed by sonication in phosphate-buffered saline. Protein extracts were immunoprecipitated with either anti-RKIP or control antibodies. Immunoprecipitated proteins were monitored by immunoblotting (IB) with either anti-RKIP or anti-TAK1 antibodies. The band present in the lane of the RKIP lysate is not IgG but comes from a protein that is cross-reactive with the RKIP antibody. (B) Association of TAK1 with RKIP after IL-1β treatment. 293/IL-1R1 cells were transfected with an HA-RKIP expression vector. Thirty hours after transfection, cells were serum deprived for 24 h and treated with IL-1β as indicated. Cells were harvested in hypotonic buffer and were lysed by rapid expulsion through a 25-gauge hypodermic needle. HA antibody-immunoprecipitated proteins were monitored by immunoblotting with either anti-RKIP or anti-TAK1 antibodies. (C) Fractionation of S100 cytosolic extracts. S100 extracts prepared from Rat1 cells stably transfected with HA-tagged RKIP were fractionated by DEAE Sepharose chromatography as described in Materials and Methods. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with the indicated antibodies. FT, column flowthrough. (D) Coimmunoprecipitation of RKIP, NIK, and TAK1 from DEAE Sepharose chromatography fractions. Fractions shown to contain NIK, TAK1, and HA-RKIP by immunoblotting (shown in panel C) were pooled and immunoprecipitated with either anti-HA or control anti-FLAG antibodies. The precipitated proteins were monitored by immunoblotting with the indicated antibodies.
Although we detected the association of endogenous TAK1 and RKIP in Rat1 fibroblast lysate, we failed to detect NIK in anti-RKIP immunoprecipitates or RKIP in anti-NIK immunoprecipitates despite multiple attempts. Since NIK and RKIP were coimmunoprecipitated when the tagged versions of both proteins were expressed (Fig. 3B), we considered that the NIK or RKIP antibodies may interfere with the formation of NIK/RKIP complexes. To address this possibility, we generated by retrovirus infection a series of clonal Rat1 cell lines expressing an HA-tagged RKIP cDNA. A clone expressing HA-RKIP at a level comparable to that of the endogenous RKIP protein was selected for further study (Fig. 7C). To specifically examine the interaction of HA-RKIP with NIK, we separated HA-RKIP from endogenous RKIP by ion-exchange chromatography. This was possible because the charged HA tag confers a distinct chromatographic profile on HA-RKIP to differentiate it from endogenous RKIP. While the majority of endogenous RKIP was detected in the flowthrough and wash fractions of a DEAE Sepharose column, HA-RKIP was bound to the column and, furthermore, cofractionated with TAK1 and NIK in a salt gradient elution (Fig. 7C). To further examine the association of HA-RKIP and endogenous NIK, fractions containing HA-RKIP and NIK were pooled and immunoprecipitated with an anti-HA antibody. As expected, HA-RKIP was detected in the anti-HA immunoprecipitation but not in the control immunoprecipitation (Fig. 7D). To examine whether endogenous NIK was associated with RKIP, the anti-HA immunoprecipitate was immunoblotted with anti-NIK antibodies. As shown in Fig. 7D, clearly detectable amounts of NIK were detected in the anti-HA immunoprecipitation but not the control immunoprecipitation. As a control, the same anti-HA immunoprecipitate was monitored for the presence of actin which also coeluted with HA-RKIP from the DEAE column; despite its high abundance, actin was not detected (Fig. 7D). Consistent with results obtained from coimmunoprecipitation experiments with crude Rat1 protein extracts, TAK1 was also detected in the anti-HA immunoprecipitate (Fig. 7D).
DISCUSSION
The first demonstrated function of RKIP was that of a negative regulator of the Raf/MEK/ERK signaling pathway. Since RKIP is a much more abundant protein than Raf-1, we considered the possibility that it may have additional intracellular targets (75, 76). In this study we found evidence that RKIP influences signaling by the NF-κB pathway. As was the case in the Raf/MEK/ERK pathway, RKIP activity exerts a negative effect on the NF-κB pathway.
Two distinct models could explain RKIP-mediated repression of NF-κB. First, RKIP could inhibit NF-κB activity as a result of its previously demonstrated effects on the Raf/MEK/ERK pathway. Second, RKIP could interfere directly with the pathways that lead to NF-κB activation. Several lines of evidence presented in this communication strongly suggest a direct mode of action. First, we consistently observed specific physical associations between RKIP and some, but not all, of the upstream activating kinases of the NF-κB pathway. These associations were detected both in ectopic expression systems as well as in cellular extracts under physiological levels of protein expression. Second, RKIP could inhibit NF-κB activity elicited by the activation of some, but not all, of the upstream NF-κB-activating kinases. Most importantly, the results of physical interaction and in vivo activation studies were in complete agreement. RKIP interacted with and blocked NIK and TAK1, but it neither interacted with nor blocked MEKK1 or NAK1. Importantly, the association between TAK1 and RKIP was physiologically regulated and induced upon treatment of the cells with IL-1β, a known activator of TAK1 activity. Third, RKIP could efficiently antagonize NF-κB activity in the presence of pharmacological inhibitors of MEK activity.
Our genetic epistasis analysis involving the overexpression of NIK, IKKα, IKKβ, TAK1, NAK1, MEKK1, and p65 NF-κB showed that RKIP exerted its inhibitory effects only on NIK- and TAK1-mediated activation of NF-κB. The fact that RKIP did not interfere with MEKK1-mediated activation of NF-κB indicates that RKIP does not have a general effect on NF-κB signaling through promiscuous interactions involving all MAPKKKs. The simplest interpretation of the genetic epistasis data is that RKIP acts upstream of IKK. The possibility that RKIP acts in a pathway that is parallel to those of upstream activators of IκB and that bypasses IKK is unlikely because RKIP can block activation of IKK activity by TNF-α.
Our results indicate that RKIP physically interacts with TAK1 and NIK but not with MEKK1. We also detected the physical association of RKIP with IKKα and IKKβ, both of which belong to the family of MAPKKs (43). Since the association of RKIP with TAK1, NIK, IKKα, and IKKβ has not yet been achieved in vitro using purified components, it is not known whether these interactions are direct. However, the interaction of RKIP with Raf-1 (a MAPKKK) and MEK1 (a MAPKK) has been well studied and is known to be direct (75). Thus, although the existence of a bridging partner between RKIP and any of the NF-κB pathway kinases cannot be ruled out, it would not be predicted on the basis of studies with the Raf/MEK/ERK pathway. Since TAK1, NIK, IKKα, and IKKβ have all been reported to be part of the 700-kDa TNF-α-induced IKK complex (48), it is possible that RKIP is also part of this complex.
NF-κB activation pathways involved in IL-1β and TNF-α signaling have been partially delineated and found to involve both shared and unique components (33, 50, 58, 67). Both pathways converge on NIK, which activates IKK through direct phosphorylation of IKKα. TAK1 was implicated in IL-1β signaling immediately upstream of NIK. Consistent with this view, we consistently observed a repressive effect of RKIP on TNF-α- as well as IL-1β-mediated activation of NF-κB. The inhibition may be partly due to the inhibition of IKK activation by TAK1/NIK, since we observed a decrease in IKKα and IKKβ autophosphorylation in extracts of TNF-α-treated, RKIP-overexpressing 293 cells. The inhibition may also be a result of a direct interference with IKK activity by RKIP, since we observed that RKIP inhibited the kinase activities of purified TNF-α-activated IKKα and IKKβ in vitro. A direct inhibition of IKK activity is also consistent with the result that RKIP coimmunoprecipitated with IKKα and IKKβ. Concordant with the in vitro kinase assay results, RKIP also antagonized the TNF-α-induced degradation of IκBα, presumably by interfering with the activity of endogenous IKK. Unexpectedly, RKIP did not interfere with the activation of the NF-κB reporter by ectopic expression of either IKKα or IKKβ. The discrepancy may be due to the possibility that overexpression of IKKs may result in the phosphorylation and activation of other IκBs or other unknown IKK substrates. It is thus possible that RKIP does not inhibit the phosphorylation of these other IKK substrates. Taken together, we proposed that RKIP acts as a brake on TNF-α and IL-1β signaling by antagonizing the activation of IKKs by NIK and TAK1 as well as by directly down-modulating the activity of the IKK complexes.
Prior to these studies, the IκBs and the A20 protein were the only known negative regulators of NF-κB signaling (44, 66). A20 is a cytoplasmic zinc finger protein that is induced by TNF-α in a variety of cells (53). Knockout studies in mice indicated that A20 was required for the downregulation of IKK activity after TNF-α stimulation but not IL-1β stimulation (36). Both A20 and the IKK complex were recruited to the TNF receptor after TNF-α stimulation. It was proposed that A20 antagonizes IKK activity by interacting with the IKKγ subunit of the complex (21, 79).
The data presented in this communication indicate that RKIP antagonizes NF-κB activity by a mechanism that is distinct from both IκB and A20. Unlike IκBs, which inhibit NF-κB by direct interaction with the Rel proteins, our epistasis analysis shows that RKIP acts upstream of p65. Whereas A20 was reported to act primarily on TNF-α signaling (36), RKIP can inhibit both TNF-α- and IL-1β-mediated NF-κB activation. Furthermore, epistasis analysis based on the overexpression of TRADD, TRAF2, RIP, and NIK indicated that A20 acts upstream of NIK (21), whereas our data implicate NIK as a direct target of RKIP. It remains to be determined whether RKIP is recruited to the cytokine receptors upon their stimulation. It is also not known whether RKIP can interact with IKKγ.
Proteins that are highly homologous to RKIP (85 to 95% identity) have been found in all mammalian species examined to date (18, 56). A family of RKIP-related proteins with 29 to 55% sequence identity to the mammalian proteins have also been identified in Drosophila, Caenorhabditis elegans, Saccharomyces cerevisiae, several parasites, including Onchocerca volvulus and Toxocara canis, and the flowering plants Antirrhinum and Arabidopsis (5, 6, 16, 18, 56, 59, 64). Despite limited sequence conservation between mammalian RKIP and some of these orthologues, recent studies of the crystal structures of two mammalian RKIPs and Antirrhinum CEN revealed that they displayed an almost identical novel β-fold topology (3, 4, 65). The close conservation in structure between mammalian RKIPs and plant RKIP orthologues raises the possibility that they may have similar functions. Indeed, genetic studies indicate that RKIP orthologues in plants function in signal transduction pathways that control plant architecture and development (1, 5, 6, 29, 32).
Emerging data from diverse organisms suggest that RKIP and RKIP-related proteins represent a new and evolutionarily highly conserved family of protein kinase regulators (4). The fact that RKIP impinges on both the Raf/MEK/ERK MAPK pathway and the NF-κB pathways raises the intriguing possibility that RKIP and related proteins may play roles in a number of different protein kinase signaling pathways. Since MAPK and NF-κB pathways have physiologically distinct roles, the function of RKIP may be, in part, to coordinate the regulation of these pathways.
ACKNOWLEDGMENTS
We thank Z. Cao, S. Frisch, D. Goeddel, A. H. Ichijo, A. Lin, K. Matsumoto, M. Nakanishi, M. Rothe, and D. Wallach for plasmid constructs, antibodies, and cell lines.
This work was supported by NIH grants to K.C.Y. (R01 GM64767) and J.M.S. (R01 GM55435 and GM 41690) and an AICR grant to W.K. This work was also supported in part by the COBRE award P20RR15578 from the NIH.
REFERENCES
- 1.Baccarini M, Sabatini D M, App H, Rapp U R, Stanley E R. Colony stimulating factor-1 (CSF-1) stimulates temperature-dependent phosphorylation and activation of the Raf-1 proto-oncogene product. EMBO J. 1990;9:3649–3657. doi: 10.1002/j.1460-2075.1990.tb07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin A S., Jr The NF-κ B and I κ B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Banfield M J, Barker J J, Perry A C, Brady R L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- 4.Banfield M J, Brady R L. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J Mol Biol. 2000;297:1159–1170. doi: 10.1006/jmbi.2000.3619. [DOI] [PubMed] [Google Scholar]
- 5.Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- 7.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I κ B-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 8.Bushdid P B, Brantley D M, Yull F E, Blaeuer G L, Hoffman L H, Niswander L, Kerr L D. Inhibition of NF-κ B activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature. 1998;392:615–618. doi: 10.1038/33435. [DOI] [PubMed] [Google Scholar]
- 9.Cao Z, Henzel W J, Gao X. IRAK: a kinase associated with the interleukin-1 receptor. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 10.Cardone M H, Salvesen G S, Widmann C, Johnson G, Frisch S M. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 11.Chang H Y, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets I κ B α to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 13.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive Iκ B kinase that activates the transcription factor NF-κ B. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 15.Finco T S, Baldwin A S J. NF-κ B site-dependent induction of gene expression by diverse inducers of NF-κ B requires Raf-1. J Biol Chem. 1993;268:17676–17679. [PubMed] [Google Scholar]
- 16.Gems D, Ferguson C J, Robertson B D, Nieves R, Page A P, Blaxter M L, Maizels R M. An abundant, trans-spliced mRNA from Toxocara canis infective larvae encodes a 26-kDa protein with homology to phosphatidylethanolamine-binding proteins. J Biol Chem. 1995;270:18517–18522. doi: 10.1074/jbc.270.31.18517. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh S, May M J, Kopp E B. NF-κ B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 18.Grandy D K, Hanneman E, Bunzow J, Shih M, Machida C A, Bidlack J M, Civelli O. Purification, cloning, and tissue distribution of a 23-kDa rat protein isolated by morphine affinity chromatography. Mol Endocrinol. 1990;4:1370–1376. doi: 10.1210/mend-4-9-1370. [DOI] [PubMed] [Google Scholar]
- 19.Guttridge D C, Albanese C, Reuther J Y, Pestell R G, Baldwin A S., Jr NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hehner S P, Hofmann T G, Ushmorov A, Dienz O, Leung I W-L, Lassam N, Scheidereit C, Droge W, Schmitz M L. Mixed-lineage kinase 3 delivers CD3/CD28-derived signals into the IκB kinase complex. Mol Cell Biol. 2000;20:2556–2568. doi: 10.1128/mcb.20.7.2556-2568.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. The zinc finger protein A20 inhibits TNF-induced NF-κ B-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-κ B-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinz M, Krappmann D, Eichten A, Heder A, Scheidereit C, Strauss M. NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol Cell Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of Iκ B kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 24.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 25.Israel A. The IKK complex: an integrator of all signals that activate NF-κ B? Trends Cell Biol. 2000;10:129–133. doi: 10.1016/s0962-8924(00)01729-3. [DOI] [PubMed] [Google Scholar]
- 26.Janosch P, Schellerer M, Seitz T, Reim P, Eulitz M, Brielmeier M, Kolch W, Sedivy J M, Mischak H. Characterization of IκB kinases: IκB-α is not phosphorylated by Raf-1 or PKC isozymes, but is a casein kinase II substrate. J Biol Chem. 1996;271:13868–13874. doi: 10.1074/jbc.271.23.13868. [DOI] [PubMed] [Google Scholar]
- 27.Kane L P, Shapiro V S, Stokoe D, Weiss A. Induction of NF-κ B by the Akt/PKB kinase. Curr Biol. 1999;9:601–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 28.Kanegae Y, Tavares A T, Izpisua Belmonte J C, Verma I M. Role of Rel/NF-κ B transcription factors during the outgrowth of the vertebrate limb. Nature. 1998;392:611–614. doi: 10.1038/33429. [DOI] [PubMed] [Google Scholar]
- 29.Kardailsky I, Shukla V K, Ahn J H, Dagenais N, Christensen S K, Nguyen J T, Chory J, Harrison M J, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 30.Karin M. The NF-κ B activation pathway: its regulation and role in inflammation and cell survival. Cancer J Sci Am. 1998;4(Suppl. 1):S92–S99. [PubMed] [Google Scholar]
- 31.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κ B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 33.Kopp E, Medzhitov R, Carothers J, Xiao C, Douglas I, Janeway C A, Ghosh S. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kortenjann M, Thomae O, Shaw P E. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen-activated protein kinase Erk2. Mol Cell Biol. 1994;14:4815–4824. doi: 10.1128/mcb.14.7.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lallena M-J, Diaz-Meco M T, Bren G, Paya C V, Moscat J. Activation of IκB kinase β by protein kinase C isoforms. Mol Cell Biol. 1999;19:2180–2188. doi: 10.1128/mcb.19.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee E G, Boone D L, Chai S, Libby S L, Chien M, Lodolce J P, Ma A. Failure to regulate TNF-induced NF-κ B and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the Iκ B α kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 38.Lee F S, Peters R T, Dang L C, Maniatis T. MEKK1 activates both Iκ B kinase α and Iκ B kinase β. Proc Natl Acad Sci USA. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Van Antwerp D, Mercurio F, Lee K F, Verma I M. Severe liver degeneration in mice lacking the Iκ B kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Sedivy J M. Raf-1 protein kinase activates the NF-κ B transcription factor by dissociating the cytoplasmic NF-κ B/Iκ B complex. Proc Natl Acad Sci USA. 1993;90:9247–9251. doi: 10.1073/pnas.90.20.9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z W, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. The IKKβ subunit of Iκ B kinase (IKK) is essential for NF κ B activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X, Cunningham E T, Jr, Mu Y, Geleziunas R, Greene W C. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κ B acting through the NF-κ B-inducing kinase and Iκ B kinases. Immunity. 1999;10:271–280. doi: 10.1016/s1074-7613(00)80027-8. [DOI] [PubMed] [Google Scholar]
- 43.Ling L, Cao Z, Goeddel D V. NF-kappaB-inducing kinase activates IKK-α by phosphorylation of Ser-176. Proc Natl Acad Sci USA. 1998;95:3792–3797. doi: 10.1073/pnas.95.7.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liou H C, Baltimore D. Regulation of the NF-κ B/rel transcription factor and I κ B inhibitor system. Curr Opin Cell Biol. 1993;5:477–487. doi: 10.1016/0955-0674(93)90014-h. [DOI] [PubMed] [Google Scholar]
- 45.Makris C, Godfrey V L, Krahn-Senftleben G, Takahashi T, Roberts J L, Schwarz T, Feng L, Johnson R S, Karin M. Female mice heterozygous for IKK γ/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol Cell. 2000;5:969–979. doi: 10.1016/s1097-2765(00)80262-2. [DOI] [PubMed] [Google Scholar]
- 46.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. MAP3K-related kinase involved in NF-κ B induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 47.Mercurio F, Manning A M. Multiple signals converging on NF-κ B. Curr Opin Cell Biol. 1999;11:226–232. doi: 10.1016/s0955-0674(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 48.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-κ B activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 49.Nemoto S, DiDonato J A, Lin A. Coordinate regulation of IkappaB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-κB-inducing kinase. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/mcb.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I κ B as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 51.Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 52.Norris J L, Baldwin A S., Jr Oncogenic Ras enhances NF-κ B transcriptional activity through Raf-dependent and Raf-independent mitogen-activated protein kinase signaling pathways. J Biol Chem. 1999;274:13841–13846. doi: 10.1074/jbc.274.20.13841. [DOI] [PubMed] [Google Scholar]
- 53.Opipari A W, Jr, Hu H M, Yabkowitz R, Dixit V M. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J Biol Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- 54.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. NF-κ B activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 55.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pikielny C W, Hasan G, Rouyer F, Rosbash M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron. 1994;12:35–49. doi: 10.1016/0896-6273(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 57.Pomerantz J L, Baltimore D. NF-κ B activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18:6694–6704. doi: 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an Iκ B kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 59.Robinson L C, Tatchell K. TFS1: a suppressor of cdc25 mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1991;230:241–250. doi: 10.1007/BF00290674. [DOI] [PubMed] [Google Scholar]
- 60.Romashkova J A, Makarov S S. NF-κ B is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 61.Rothwarf D M, Karin M. The NF-κ B activation pathway; a paradigm in information transfer from membrane to nucleus. Sci Signal Transduct Knowledge Environ. 1999;5:1–16. doi: 10.1126/stke.1999.5.re1. . [Online.] [DOI] [PubMed] [Google Scholar]
- 62.Rothwarf D M, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the Iκ B kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt-Supprian M, Bloch W, Courtois G, Addicks K, Israel A, Rajewsky K, Pasparakis M. NEMO/IKK γ-deficient mice model incontinentia pigmenti. Mol Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- 64.Seddiqi N, Bollengier F, Alliel P M, Perin J P, Bonnet F, Bucquoy S, Jolles P, Schoentgen F. Amino acid sequence of the Homo sapiens brain 21–23-kDa protein (neuropolypeptide h3), comparison with its counterparts from Rattus norvegicus and Bos taurus species, and expression of its mRNA in different tissues. J Mol Evol. 1994;39:655–660. doi: 10.1007/BF00160411. [DOI] [PubMed] [Google Scholar]
- 65.Serre L, Vallee B, Bureaud N, Schoentgen F, Zelwer C. Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins. Structure. 1998;6:1255–1265. doi: 10.1016/s0969-2126(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 66.Song H Y, Rothe M, Goeddel D V. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κ B activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Iries K, Ninomiya-Tsuji J, Matsumoto K. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 68.Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKKα. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka M, Fuentes M E, Yamaguchi K, Durnin M H, Dalrymple S A, Hardy K L, Goeddel D V. Embryonic lethality, liver degeneration, and impaired NF-κ B activation in IKK-β-deficient mice. Immunity. 1999;10:421–429. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 70.Tojima Y, Fujimoto A, Delhase M, Chen Y, Hatakeyama S, Nakayama K, Kaneko Y, Nimura Y, Motoyama N, Ikeda K, Karin M, Nakanishi M. NAK is an Iκ B kinase-activating kinase. Nature. 2000;404:778–782. doi: 10.1038/35008109. [DOI] [PubMed] [Google Scholar]
- 71.Whitley M Z, Thanos D, Read M A, Maniatis T, Collins T. A striking similarity in the organization of the E-selectin and beta interferon gene promoters. Mol Cell Biol. 1994;14:6464–6475. doi: 10.1128/mcb.14.10.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Iκ B kinase-β: NF-κ B activation and complex formation with Iκ B kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 73.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 74.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Complementation cloning of NEMO, a component of the Iκ B kinase complex essential for NF-κ B activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 75.Yeung K, Janosch P, McFerran B, Rose D W, Mischak H, Sedivy J M, Kolch W. Mechanism of suppression of the Raf/MEK/extracellular signal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol Cell Biol. 2000;20:3079–3085. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis K D, Rose D W, Mischak H, Sedivy J M, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 77.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 78.Zandi E, Karin M. Bridging the gap: composition, regulation, and physiological function of the IκB kinase complex. Mol Cell Biol. 1999;19:4547–4551. doi: 10.1128/mcb.19.7.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang S Q, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKγ) upon receptor stimulation. Immunity. 2000;12:301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]