Abstract
Background
Community-acquired carbapenem-resistant Enterobacterales (CA-CRE) are an important threat.
Methods
In CRACKLE-2, we defined patients with CA-CRE as admitted from home, without pre-existing conditions, and a positive culture within 48 h of admission. Healthcare-associated CRE (HA-CRE) were those with the lowest likelihood of community acquisition, not admitted from home and cultured >48 h after admission. Specific genetic markers in carbapenemase-producing Klebsiella pneumoniae were evaluated through random forest modelling.
Results
CA-CRE and HA-CRE were detected in 83 (10%) and 208 (26%) of 807 patients. No significant differences were observed in bacterial species or strain type distribution. K. pneumoniae (204/291, 70%) was the most common CRE species, of these 184/204 (90%) were carbapenemase producers (CPKP). The top three genetic markers in random forest models were kpi_SA15, fimE, and kpfC. Of these, kpi_SA15 (which encodes a chaperone/usher system) was positively associated (OR 3.14, 95% CI 1.13–8.87, P = 0.026), and kpfC negatively associated (OR 0.21, 95% CI 0.05–0.72, P = 0.015) with CA-CPKP.
Conclusions
Ten percent of CDC-defined CRE were CA. The true proportion of CA-CRE in hospitalized patients is likely lower as patients may have had unrecorded prior healthcare exposure. The kpi_SA15 operon was associated with the CA phenotype.
Introduction
Enterobacterales are commonly encountered pathogens causing community-associated infections, such as urinary tract infections. More recently, ESBL-producing Enterobacterales have been increasingly associated with community-acquired (CA) infections in the US and worldwide.1 Carbapenems are the preferred treatment option for invasive ESBL-producing Enterobacterales infections.2,3 Concerningly, carbapenem resistance is also increasingly observed in Enterobacterales.4,5 Several novel treatment options have been developed recently to better treat infections caused by carbapenem-resistant Enterobacterales (CRE).6–8 However, the costs, need for intravenous therapy, and concerns over further resistance development, limit the use of these agents, especially in empirical antibiotic regimens. There are several reports of CA-CRE causing bacteraemia, gastroenteritis, urinary tract infections, neonatal meningitis, and peritonitis.1,9,10 In addition, in the Consortium on Resistance against Carbapenems in Klebsiella and other Enterobacterales (CRACKLE) studies, the proportion of patients hospitalized with CRE who were admitted from home increased over time.11,12
We have previously described 1040 unique patients hospitalized in 49 US hospitals with CRE isolated from clinical cultures.11 Here, we explored possible community origins of CRE in a subset of these patients. We compared characteristics between CRE that were CA-CRE and those that were healthcare-associated (HA) CRE.
Patients and methods
Patients
CRACKLE-2 was a prospective, observational, multicentre cohort study.11 For this analysis, patients were included from CRACKLE-2 who were hospitalized in participating US hospitals between 30 April 2016 and 31 August 2017 with CRE isolated from any anatomical site in a clinical culture and who met criteria for CA-CRE or HA-CRE. Patients with CRE that were susceptible to carbapenems upon centralized susceptibility testing (‘unconfirmed CRE’) were excluded.11 The characteristics of the cohort from which these patients have been selected have been previously described.11 Patients from 24 study sites were included.
Ethics
The Institutional Review Board (IRB) of each participating healthcare system approved this study. Need for informed consent was waived by each participating IRB. The IRB approval reference number at the University of North Carolina at Chapel Hill is 19-3032.
Clinical definitions
Infections were defined using standardized criteria, as previously described.11 As the collected data did not allow application of the criteria outlined by the US Centers for Disease Control and Prevention, CA-CRE were defined here by the presence of all of the following: admission from home, index CRE culture date within 48 h of hospitalization, and absence of five documented conditions that would increase the likelihood of recent healthcare exposure (kidney disease, malignancy, liver disease, immunocompromise, or pregnancy). For the purpose of these analyses, HA-CRE were defined as those isolates with the lowest likelihood of community acquisition: admission from a long-term care or acute healthcare facility AND timing of index CRE culture more than 48 h after admission. Patients who fell into neither the CA-CRE nor the HA-CRE category were excluded from these analyses. Clinical data were obtained from electronic medical records, and outcomes were defined as previously described.11
Microbiology
Susceptibility to carbapenems was determined at local clinical laboratories following the manufacturer’s instructions for MicroScan (Beckman Coulter, Atlanta, GA, US), Vitek®2 and Etest® (both bioMérieux, Durham, NC, US), BD Phoenix™ and BBL™ discs (both Becton Dickinson, Durham, NC, US), Sensititre (Thermo Fisher, Waltham, MA, US), and using disc diffusion or in-house agar dilution tests. Carbapenem susceptibility results were confirmed in two independent central research laboratories using the Etest® and MicroScan. CRE isolates that tested susceptible to carbapenems upon central testing (‘unconfirmed CRE’) were excluded from this study.11
Whole-genome sequencing and genomic analysis
Genomic analysis was limited to carbapenemase-producing K. pneumoniae (CPKP). WGS methods and genomic analyses were described previously.11 Briefly, Snippy was used to identify core SNPs from whole genome sequences for each genome compared with the reference genome NGST258_2 (GCA_000597905.1). DNA sequences identified as being prophages, repeat regions, or regions of recombination were masked from downstream analysis using PHASTER, MUMmer, and Gubbins, respectively.13–15 A maximum-likelihood phylogenetic tree was constructed from the recombination-free alignment using RAxML (v8.2.12) using a general time-reversible model of nucleotide substitution and four discrete gamma categories of rate heterogeneity (GTRGAMMA). Node support was estimated by bootstrapping with 500 replicates. Clusters were defined as strains sharing <22 SNPs and a most-recent common ancestor based on the phylogenetic tree.16 R package ‘circlize’ was used to visualize the phylogenetic tree.17
Statistical analysis
Distributions across two groups were analysed using the Wilcoxon Rank Sum test for continuous variables and Fisher’s exact or Pearson-χ2 tests for categorical variables. P values <0.05 were considered statistically significant. The CRACKLE-2 desirability of outcome ranking (DOOR) was compared across groups, as previously described.11 Briefly, this outcome assessed three deleterious events: lack of clinical response, prolonged hospitalization (hospitalization ≥30 days after first positive culture or readmission within 30 days), and adverse events (new renal failure and/or Clostridioides difficile infection), in addition to survival at 30 days after the index culture. The best outcome was defined as being alive without deleterious events. The worst outcome was death. Two levels in-between these two extremes were: alive with 1 event, and alive with 2 or 3 deleterious events. Within the subset of patients with CPKP (n = 184), random forest analysis for the purpose of variable selection was performed on bacterial genetic markers with CA-CRE versus HA-CRE as the outcome. Seventy-five markers were a priori hypothesized to be potentially associated with colonization based on literature review (Table S1, available as Supplementary data at JAC Online). Of these 75 markers, 40 were found in >95% or <5% of strains and were excluded from analyses. Relative importance of variables in random forest models was determined using mean decrease in Gini coefficient and mean decrease in accuracy over 10 models. Logistic regression was used to determine OR and 95% confidence intervals (CI) of the top three selected genes. SAS 9.4 (SAS Institute, Cary, NC) and R 4.1.1 (R Core Team, 2020) were used for analyses.
Results
Clinical characteristics and outcomes
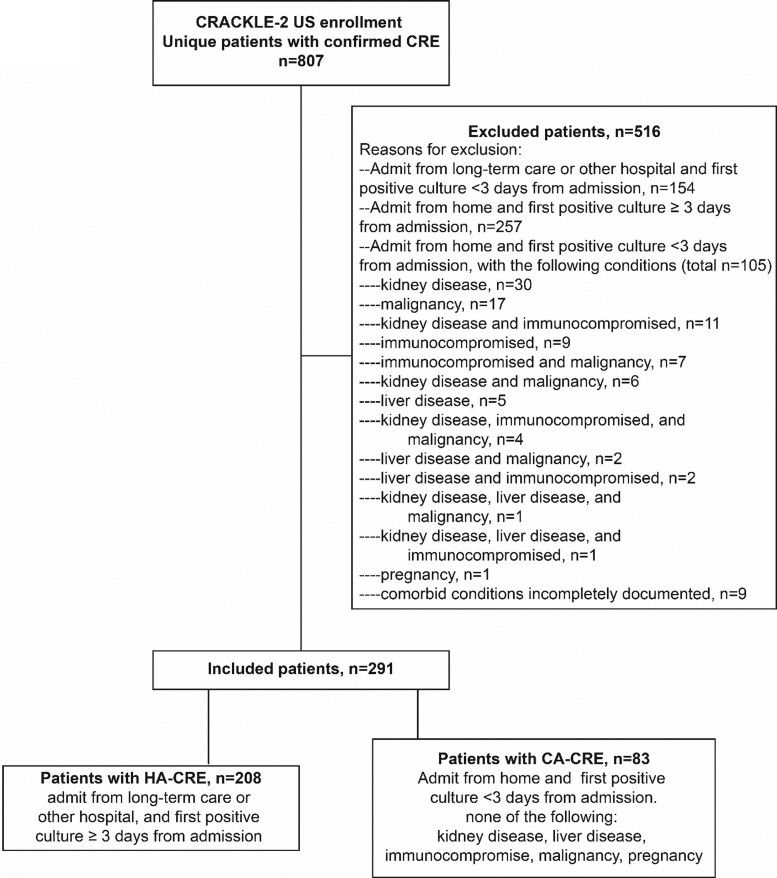
In CRACKLE-2, 807 unique patients with confirmed CRE were enrolled during the study period. Of these, 83 (10%) and 208 (26%) met the criteria for either CA-CRE or HA-CRE, respectively (Table 1 and Figure 1). Distinct variability in the proportions of HA-CRE and CA-CRE of total CRE cases was observed between study sites (Figure S1, available as Supplementary data at JAC Online). Patients with CA-CRE were more likely to be admitted at a community hospital; 28% of patients with CA-CRE were admitted at a community hospital, as compared with 13% of patients with HA-CRE (P < 0.01). The distribution of sources between CA-CRE and HA-CRE was significantly different, P < 0.001; the urine was a more common source for CA-CRE (65% versus 25% in HA-CRE), whereas HA-CRE were more frequently isolated from the respiratory tract (40% versus 7% in CA-CRE). As expected, since absence of several comorbid conditions is part of the CA-CRE definition, the Charlson comorbidity index was lower in patients with CA-CRE (median 1, IQR 0–3) versus HA-CRE (median 3, IQR 1–5), P < 0.001. Patients with CA-CRE had shorter post-culture hospital stays (median 7 days, IQR 4–12 days versus 15 days, IQR 7–29 days, P < 0.001) and lower acuity of illness on the day of positive culture (median Pitt bacteraemia score 2, IQR 0–3 versus 4, IQR 2–6, P < 0.001) than those with HA-CRE. Overall, mortality of patients with CA-CRE was significantly lower as compared with patients with HA-CRE at 30 days (6% versus 26%, P < 0.001) and at 90 days (10% versus 34%, P < 0.001). Inverse probability weighting (IPW)-adjusted DOOR analysis showed that a randomly selected patient with CA-CRE had a 66% (95% CI 56%–78%) chance of a better overall outcome as compared with a patient with HA-CRE (Table 1). Similar differences were observed in the subset of patients with infections for unadjusted DOOR probabilities (69%, 95% CI 59%–78%), and for mortality at 30 days (10% versus 35%, P < 0.001) and 90 days (10% versus 42%, P < 0.001). Of note, 90 day readmission rates in patients discharged alive were similarly high in patients with CA-CRE (34/78, 44%) and patients with HA-CRE (59/145, 41%), P = 0.69.
Table 1.
Patient characteristics with possibly community-associated CRE (CA-CRE) versus confirmed healthcare-associated CRE (HA-CRE)
| Characteristic | CA-CRE (n = 83) | HA-CRE (n = 208) | Total (n = 291) | P value |
|---|---|---|---|---|
| Region | 0.24 | |||
| Northeast | 34 (41) | 105 (50) | 139 (48) | |
| South | 25 (30) | 45 (22) | 70 (24) | |
| Midwest | 18 (22) | 36 (17) | 54 (19) | |
| West | 6 (7) | 22 (11) | 28 (10) | |
| Age, years, median (IQR) | 64 (47–77) | 65 (55–77) | 65 (54–77) | 0.59 |
| Gender | 0.78 | |||
| Male | 44 (53) | 114 (55) | 158 (54) | |
| Female | 39 (47) | 94 (45) | 133 (46) | |
| Hispanic or Latino ethnicity | 16 (19) | 19 (9) | 35 (12) | 0.02 |
| Race | 0.76 | |||
| White | 33 (40) | 97 (47) | 130 (45) | |
| Black | 27 (33) | 65 (31) | 92 (32) | |
| Othera | 11 (13) | 26 (12) | 37 (13) | |
| Unknown | 12 (14) | 20 (10) | 32 (11) | |
| Post-culture length of hospital stay, days, median (IQR) | 7 (4–12) | 15 (7–29) | 12 (6–23) | <0.001 |
| Charlson comorbidity score, median (IQR) | 1 (0–3) | 3 (1–5) | 3 (1–4) | <0.001 |
| Pitt bacteraemia score, median (IQR) | 2 (0–3) | 4 (2–6) | 3 (2–6) | <0.001 |
| Culture source | <0.001 | |||
| Urine | 54 (65) | 52 (25) | 106 (36) | |
| Wound | 10 (12) | 35 (17) | 45 (15) | |
| Blood | 6 (7) | 18 (9) | 24 (8) | |
| Respiratory | 6 (7) | 84 (40) | 90 (31) | |
| Other | 7 (8) | 19 (9) | 26 (9) | |
| Infection | 30 (36) | 86 (41) | 116 (40) | 0.41 |
| Community hospital | 23 (28) | 28 (13) | 51 (18) | <0.01 |
| Disposition after discharge | <0.001 | |||
| Death | 3 (4) | 53 (25) | 56 (19) | |
| Hospice | 2 (2) | 10 (5) | 12 (4) | |
| Home | 55 (66) | 25 (12) | 80 (27) | |
| Long-term care | 18 (22) | 69 (33) | 87 (30) | |
| Long-term acute care | 4 (5) | 38 (18) | 42 (14) | |
| Transfer to other hospital | 1 (1) | 13 (6) | 14 (5) | |
| 30 day mortality | 5 (6) | 55 (26) | 60 (21) | <0.001 |
| 90 day mortality | 8 (10) | 71 (34) | 79 (27) | <0.001 |
| 90 day readmission | 34/78 (44) | 59/145 (41) | 93/223 (42) | 0.69 |
| DOOR at 30 days | ||||
| Alive without events | 58 (70) | 66 (32) | 124 (43) | |
| Alive with one event | 19 (23) | 45 (22) | 64 (22) | |
| Alive with two or three events | 1 (1) | 42 (20) | 43 (15) | |
| Dead | 5 (6) | 55 (26) | 60 (21) |
Values shown are n (%) unless indicated otherwise.
Other races include Native Hawaiian or Pacific islander (n = 2), Asian (n = 29) and multiracial (n = 69).
Figure 1.
Flow diagram.
Microbiology
The most common carbapenemase genes in both groups were blaKPC-2 and blaKPC-3, which were found in 94% (214/227) of carbapenemase-producing Enterobacterales (Table 2). No difference in carbapenemase gene distribution was noted between CA-CRE and HA-CRE. Species distribution was similar between CA-CRE and HA-CRE (Table 2). Seventy percent of CRE isolates were K. pneumoniae (204/291), of which 66% (135/204) belonged to ST258. ST258 was similarly common in CA-carbapenem-resistant K. pneumoniae (CRKP) (36/60, 60%) and HA-CRKP (98/144, 68%). Carbapenemase genes were present in 90% of CRKP (184/204). In non-K. pneumoniae CRE, carbapenemase genes were present in 43/87 (49%) isolates; blaKPC-2 (n = 15) and blaKPC-3 (n = 22) were most commonly encountered. ST131 (n = 15, 68%) was the most common genetic lineage in E. coli, and four ST131 E. coli were CA-CRE. Five of 41 (12%) Enterobacter isolates were ST171 E. cloacae, of which four were CA-CRE.
Table 2.
Bacterial characteristics of possibly community-associated CRE (CA-CRE) and confirmed healthcare-associated CRE (HA-CRE) isolates
| Characteristic | CA-CRE (n = 83) | HA-CRE (n = 208) | Total (n = 291) | P value |
|---|---|---|---|---|
| Species | 0.28 | |||
| Klebsiella pneumoniae | 60 (72) | 144 (69) | 204 (70) | |
| ST258 | 36 (43) | 98 (48) | 134 (46) | |
| ST307 | 4 (5) | 6 (3) | 10 (3) | |
| ST15 | 3 (4) | 3 (1) | 6 (2) | |
| Enterobacter spp. | 12 (14) | 29 (14) | 41 (14) | |
| ST171 E. cloacae | 4 (5) | 1 (0) | 5 (2) | |
| Escherichia coli | 8 (10) | 14 (7) | 22 (8) | |
| ST131 | 4 (5) | 11 (5) | 15 (5) | |
| Other Klebsiella spp.a | 0 (0) | 10 (5) | 10 (3) | |
| Otherb | 3 (4) | 11 (5) | 14 (5) | |
| Carbapenemase presentc | 67 (81) | 160 (77) | 227 (78) | 0.48 |
| bla KPC-2 | 37 (45) | 82 (39) | 119 (41) | |
| bla KPC-3 | 22 (27) | 75 (36) | 97 (33) | |
| bla NDM-1 | 5 (6) | 0 | 5 (2) | |
| Otherd | 4 (5) | 5 (2) | 9 (3) |
Data shown are n (%) unless otherwise indicated.
Klebsiella aerogenes (n = 6), Klebsiella michiganensis (n = 3), Klebsiella oxytoca (n = 1).
Serratia marcescens (n = 5), Citrobacter freundii (n = 4), Raoultella ornithinolytica (n = 2), Citrobacter freundii (n = 1), Hafnia paralvei (n = 1), Providencia stuartii (n = 1).
Counts exceed 100%, as three isolates carried two different carbapenemase genes.
bla KPC-4 (n = 2), blaOXA-232 (n = 4), blaNDM-7 (n = 1), blaVIM-2 (n = 1), blaSME-2 (n = 1), blaOXA-181 (n = 1).
Molecular epidemiology of carbapenemase-producing K. pneumoniae
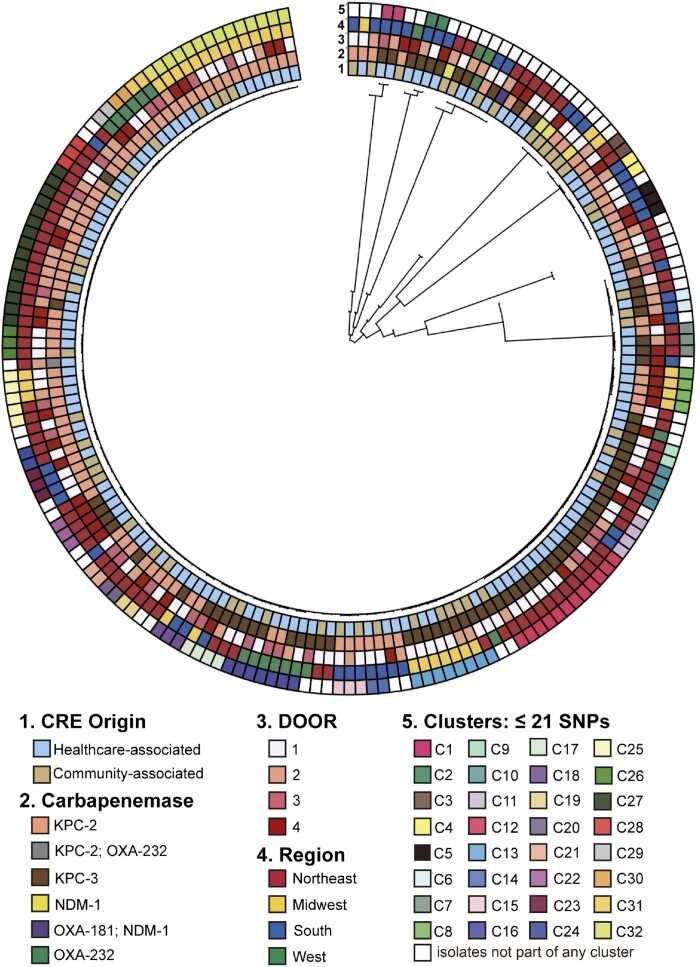
The population structure of 184 CPKP [53 (29%) CA-CPKP and 131 (71%) HA-CPKP] is shown in Figure 2. In CPKP, 131 (71%) strains belonged to ST258, and a total of 27 K-loci types were present. Most ST258 CPKP isolates had either KL107 (64/128, 50%), KL106 (38/128, 30%), or KL51 (14/128, 11%). All ST307 isolates had KL102 (10/10, 100%). Six ST15 CPKP strain were found, three of these were CA-CPKP. The O-locus O2v2 was present in 86/184 (47%) CPKP isolates; 74/131 (56%) ST258 carried O2v2 versus 12/53 (23%) of non-ST258 isolates (P < 0.001). CA-CPKP and HA-CPKP had a similar distribution of K-loci and O-loci. In total, 113 CPKP belonged to 31 clusters; 26/53 (49%) of CA-CPKP and 87/131 (66%) of HA-CPKP. 16/31 (52%) of clusters contained only HA-CPKP strains (n = 47), 14 clusters contained both CA-CPKP (n = 26) and HA-CPKP (n = 40), and a single cluster contained two CA-CPKP isolates.
Figure 2.
Maximum likelihood phylogenetic tree of carbapenemase-producing Klebsiella pneumoniae (n=184). DOOR outcomes (1) alive without events, (2) alive with 1 event, (3) alive with 2 or 3 events, or (4) death.
Bacterial genetic analysis
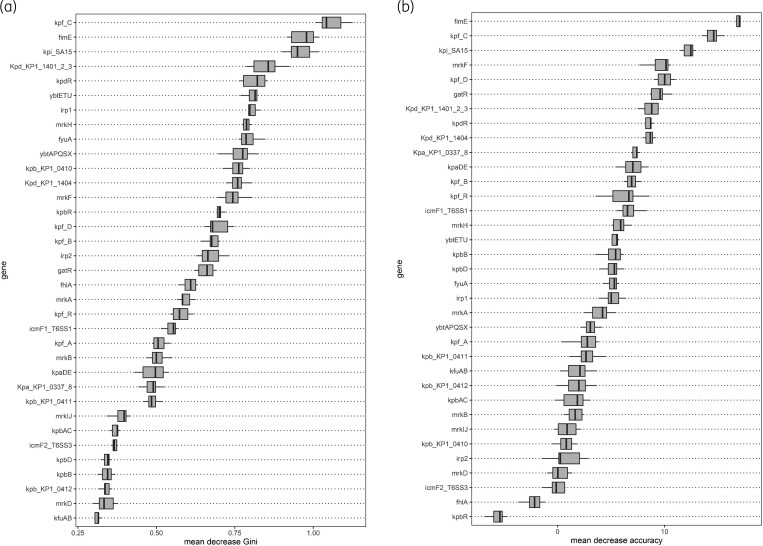
Bacterial genetic markers previously reported to be potentially associated with colonization were selected based on literature review (Table S1). Results of random forest modelling are shown in Figure 3. The top three genetic markers associated with highest mean decreases in Gini coefficient and accuracy over 10 separate models were kpi_SA15, kpfC, and fimE. The kpi_SA15 operon was more commonly present in CA-CPKP isolates (9/53, 17%) as compared with HA-CPKP (8/131, 6%), P = 0.026. kpfC was less commonly present in CA-CPKP isolates (46/53, 87%) as compared with HA-CPKP (127/131, 97%), P = 0.015. No difference in distribution of fimE between CA-CPKP (48/53, 91%) and HA-CPKP (121/131, 92%) was observed.
Figure 3.
Results of random forest modelling. Boxes indicate the first and third quartile, line indicates the median, and whiskers indicate the minimum and maximum. Results shown for each genetic marker from 10 random forest models. (a) Decrease in mean Gini coefficient. (b) Decrease in mean accuracy.
Discussion
Overall, we found that 10% of patients hospitalized with CRE represented community-associated cases. Our estimates of CA-CRE varied geographically and between participating hospitals in the same region. A rise in CA-CRE infections has been observed in other regions of the world.1,18,19 An international scoping review of CRE in the community reported estimates of CA-CRE proportion ranging from 0%–30%.20 In the four US-based studies included in this review, estimates ranged between 5.6% and 10.8%, consistent with our findings.21–24 These studies were conducted between 2008–13 and used various methods and definitions of CA-CRE to arrive at their estimates.21–24 Between 2012 and 2017, rates of CRE cases in the US have been stable and there has been no indication of widespread community transmission of CRE in the US.25 However, early recognition of CRE strains that may contribute to future community spread is of the utmost importance. A shift from predominantly healthcare-associated to community-acquired infections in the epidemiology of CRE would have a dramatic impact on our society through lives lost, increased healthcare costs, and increased antibiotic usage.
We observed several clinical differences between patients with CA-CRE and HA-CRE. This was expected, as our definition for CA-CRE not only selects for higher likelihood of community acquisition, but also for a healthier population with fewer comorbid conditions. The differences in outcomes including all-cause mortality, post-culture length-of-stay, and DOOR outcomes are most likely primarily related to host factors, rather than to reduced pathogenicity of CA-CRE. Surprisingly, rates of readmission were similarly high in patients with CA-CRE when compared with patients with HA-CRE. This may be a consequence of the impact of the detection of CRE in clinical cultures, which is known to predispose to subsequent CRE infections.26
In E. coli, we specifically looked for and found several CA-CRE isolates belonging to ST131. However, we did not find evidence that ST131 was overrepresented in the CA-CRE cohort. ST131 E. coli is a worldwide-distributed high-risk strain that is frequently associated with urinary tract infections and bacteraemia.27 Multiple reports of household and pet-to-human transmission of ST131 E. coli have been reported.27 Our cohort is limited as an evaluation of the impact of ST131 E. coli in community spread of CRE in the US, as only 22 patients with E. coli were included. While also limited by small numbers, we observed a numerical overrepresentation of ST171 E. cloacae in CA-CRE strains. ST171 has been previously identified as a high-risk CRE clone associated primarily with KPC-3 that is emerging in the US.28 Both ST131 E. coli and ST171 E. cloacae have the potential to cause increasing numbers of CA-CRE infections in the future. Monitoring these strain types going forward is warranted.
As most patients in our cohort had carbapenemase-producing K. pneumoniae, we focused our in-depth genetic analyses on this subgroup. CA-CRE and HA-CRE were mixed throughout the population structure of CRE in this cohort, and we found no evidence for a single clone accounting for most of the community transmission. Several clusters were identified, including clusters that included both patients with CA-CRE and HA-CRE implying possible intra-hospital transmission of CA-CRE isolates to hospitalized patients. We hypothesized that the ability of bacteria to successfully colonize hosts with a relatively normal gut microbiome is one of the requirements for successful community spread. Therefore, we selected bacterial genetic markers a priori that had previously been reported to be potentially associated with colonization by K. pneumoniae. These included genes encoding type 1 (fim) and type 3 (mrk) pili, as well as other fimbriae operons. fim and mrk are the best described chaperone/usher systems encoding genes resulting in assembly of type 1 and type 3 fimbriae.29,30 fimE and fimB control phase-switching of type 1 fimbriae, and absence of fimE results in overexpression of type 1 fimbriae.31 It is unclear why fimE was identified as an important variable in random forest modelling as no numeric differences were found in positivity rates for fimE between CA-CPKP and HA-CPKP strains. In random forest analysis, a high relative importance may be a consequence of interactions with other variables. Therefore, absence of fimE may be important for the CA phenotype only in the setting of other specific genetic markers. The presence of the kpi_SA15 operon was associated with CA-CRE phenotype in our cohort. This should be considered a hypothesis-generating finding. The kpi_SA15 operon contains four genes that encode components of a different chaperone/usher system.32,33 However, little is known about the functional importance of this system. kpfC encodes the usher component of the kpf chaperone–usher system. kpfR is a negative regulator of the kpf system.34 A kpfR knock-out K. pneumoniae strain exhibited enhanced biofilm formation, but decreased capsule production.34 The associations between the CA phenotype and the kpi_SA15 operon and kpfC should be further evaluated in future studies. Of these, the kpi_SA15 operon is of higher interest given the positive association with community spread and prior studies.
Limitations
This study has several limitations. First, as a secondary analysis of the CRACKLE-2 cohort, defining CA cases was limited to the available clinical variables that may have misclassified some isolates. Documentation on prior hospitalizations, emergency room visits, and long-term care stays outside the study period was unavailable. For instance, it is possible that a patient admitted from home had a recent hospitalization or long-term care stay during which CRE was acquired. It is therefore likely that we overestimated true community acquisition of CRE. Second, our definition for CA-CRE excludes patients with a history of several clinical comorbidities indicative of healthcare exposures. Thus, it is not surprising that improved clinical outcomes were observed in patients with CA-CRE, including DOOR outcomes and survival. Third, associations with bacterial genetic markers should be considered hypothesis-generating. Future studies are required to determine whether any of the bacterial characteristics we identified are truly associated with CA-CRE.
Conclusions
In summary, community spread of CRE was limited during the study period with approximately 10% of patients meeting criteria for community acquisition. True community acquisition likely occurred only in a subset of these 10% of patients with CRE. Several high-risk strain types were identified within the CA-CRE cohort. Understanding the epidemiology of CA-CRE and at-risk people for carriage of CA-CRE strains will assist with measures to control AMR spread and early detection of community spread of CRE.
Supplementary Material
Acknowledgements
The investigators would like to thank all the patients and their families.
Contributor Information
Rima Shrestha, Division of Infectious Diseases, University of North Carolina, Chapel Hill, North Carolina, USA.
Courtney L Luterbach, Division of Infectious Diseases, University of North Carolina, Chapel Hill, North Carolina, USA; Division of Pharmacotherapy and Experimental Therapeutics, University of North Carolina, Chapel Hill, North Carolina, USA.
Weixiao Dai, The Biostatistics Center, The George Washington University, Rockville, Maryland, USA.
Lauren Komarow, The Biostatistics Center, The George Washington University, Rockville, Maryland, USA.
Michelle Earley, The Biostatistics Center, The George Washington University, Rockville, Maryland, USA.
Gregory Weston, Division of Infectious Diseases, Department of Medicine, Montefiore Medical Center, Albert Einstein College of Medicine, Bronx, New York, USA.
Erica Herc, Division of Infectious Diseases, Department of Medicine, Henry Ford Hospital, Detroit, Michigan, USA.
Jesse T Jacob, Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, Georgia, USA; Emory Antibiotic Resistance Center, Atlanta, Georgia, USA.
Robert Salata, Department of Medicine, Case Western Reserve University School of Medicine, Cleveland, Ohio, USA.
Darren Wong, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Deverick Anderson, Division of Infectious Diseases, Duke University, School of Medicine, Durham, North Carolina, USA; Duke Center for Antimicrobial Stewardship and Infection Prevention, Durham, North Carolina, USA.
Kirsten B Rydell, Division of Infectious Diseases, Houston Methodist Hospital, Houston, Texas, USA.
Cesar A Arias, Division of Infectious Diseases, Houston Methodist Hospital, Houston, Texas, USA; Center for Infectious Diseases Research at Houston Methodist Research Institute and Weill Cornell Medical College, Houston, Texas, USA.
Liang Chen, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, New Jersey, USA; Department of Medical Sciences, Hackensack Meridian School of Medicine, Nutley, New Jersey, USA.
David van Duin, Division of Infectious Diseases, University of North Carolina, Chapel Hill, North Carolina, USA.
MDRO Investigators:
Gregory Weston, Belinda Ostrowsky, Judith J Lok, Robert A Bonomo, T Nicholas Domitrovic, Kristine M Hujer, Andrea M Hujer, Susan D Rudin, Steven H Marshall, Robert A Salata, Federico Perez, Eric Cober, Sandra S Richter, Rebekka Arias, Carol Hill, Vance G Fowler, Jr., Deverick J Anderson, Jesse T Jacob, Minggui Wang, Liang Chen, Samit Desai, Barry N Kreiswirth, Claudia Manca, Jose R Mediavilla, Gopi Patel, W Charles Huskins, Robin Patel, Sara Revolinski, Glenn Wortmann, Robert C Kalayjian, Angela Kim, Julia Garcia-Diaz, Bettina C Fries, Brandon Eilertson, Jason C Gallagher, Michelle Earley, Scott Evans, Lauren Komarow, Omai B Garner, Henry F Chambers, John J Farrell, Lilian M Abbo, Keith S Kaye, Courtney Luterbach, David van Duin, Jennifer H Han, Yohei Doi, David L Paterson, Darren Wong, Cesar A Arias, Blake Hanson, An Dinh, Diana Panesso, William Shropshire, Truc T Tran, Ritu Banerjee, Sorabh Dhar, Michael J Satlin, and Matthew Grant
Members of the MDRO Network Investigators
Investigators are listed by centre alphabetically. Albert Einstein College of Medicine, Bronx, New York, USA: Gregory Weston, Belinda Ostrowsky; Boston University, Boston, Massachusetts, USA: Judith J. Lok; Case Western Reserve University, Cleveland, Ohio, USA: Robert A. Bonomo, T. Nicholas Domitrovic, Kristine M. Hujer, Andrea M. Hujer, Susan D. Rudin, Steven H. Marshall, Robert A. Salata, Federico Perez; Cleveland Clinic, Cleveland, Ohio, USA: Eric Cober, Sandra S. Richter; Duke Clinical Research Institute: Rebekka Arias, Carol Hill; Duke University, Durham, North Carolina, USA: Vance G. Fowler Jr., Deverick J. Anderson; Emory University, Atlanta, Georgia, USA: Jesse T. Jacob; Fudan University, Shanghai, China: Minggui Wang; Hackensack Meridian Health, Nutley, New Jersey, USA: Liang Chen, Samit Desai, Barry N. Kreiswirth, Claudia Manca, Jose R. Mediavilla; Icahn School of Medicine at Mount Sinai, New York, New York, USA: Gopi Patel; Mayo Clinic, Rochester, Minnesota, USA: W. Charles Huskins, Robin Patel; Medical College of Wisconsin and Froedtert Memorial Lutheran Hospital, Milwaukee, Wisconsin, USA: Sara Revolinski; MedStar Washington Hospital Center, Washington, District of Columbia, USA: Glenn Wortmann; MetroHealth Medical Center, Cleveland, Ohio, USA: Robert C. Kalayjian; North Shore University Hospital, Manhasset, New York, USA: Angela Kim; Ochsner Clinic Foundation, New Orleans, Louisiana, USA: Julia Garcia-Diaz; Stony Brook University, Stony Brook, New York, USA: Bettina C. Fries; SUNY Downstate Medical Center, New York, New York, USA: Brandon Eilertson; Temple University School of Pharmacy, Philadelphia, Pennsylvania, USA: Jason C. Gallagher; The George Washington University, Rockville, Maryland, USA: Michelle Earley, Scott Evans, Lauren Komarow; University of California, Los Angeles, California, USA: Omai B. Garner; University of California San Francisco, San Francisco, California, USA: Henry F. Chambers; University of Illinois College of Medicine at Peoria, Peoria, Illinois, USA: John J. Farrell; University of Miami Miller School of Medicine and Jackson Health System, Miami, Florida, USA: Lilian M. Abbo; University of Michigan, Ann Arbor, Michigan, USA: Keith S. Kaye; University of North Carolina, Chapel Hill, North Carolina, USA: Courtney Luterbach, David van Duin; University of Pennsylvania, Philadelphia, Pennsylvania, USA: Jennifer H. Han; University of Pittsburgh, Pittsburgh, Pennsylvania, USA: Yohei Doi; University of Queensland, Queensland, Australia: David L. Paterson; University of Southern California, Los Angeles, California, USA: Darren Wong; University of Texas Health Science Center at Houston, Houston, Texas, USA: Cesar A. Arias, Blake Hanson, An Dinh, Diana Panesso, William Shropshire, Truc T. Tran; Vanderbilt University, Nashville, Tennessee, USA: Ritu Banerjee; Wayne State University, Detroit, Michigan, USA: Sorabh Dhar; Weill Cornell Medicine, New York, New York, USA: Michael J. Satlin; Yale School of Medicine, New Haven, Connecticut, USA: Matthew Grant.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI104681, R01AI143910 and by the National Institute of General Medical Sciences T32GM086330.
Transparency declarations
D.v.D. declares Advisory Board fees from Allergan, Achaogen, Qpex, Shionogi, Tetraphase, Sanofi-Pasteur, T2 Biosystems, Pfizer, NeuMedicine, Roche, MedImmune, Astellas, Merck and Melinta, and research funding from Shionogi and Merck. All remaining authors have none to declare.
Disclaimer
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Supplementary data
Figure S1 and Table S1 are available as Supplementary data at JAC Online.
References
- 1. van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: An update. Infect Dis Clin North Am 2020; 34: 709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harris PNA, Tambyah PA, Lye DC et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: A randomized clinical trial. JAMA 2018; 320: 984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tamma PD, Aitken SL, Bonomo RA et al. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72: 1109–16. [DOI] [PubMed] [Google Scholar]
- 4. Shugart A, Mahon G, Huang JY et al. Carbapenemase production among less-common Enterobacterales genera: 10 US sites, 2018. JAC Antimicrob Resist 2021; 3: dlab137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sabour S, Huang JY, Bhatnagar A et al. Detection and characterization of targeted carbapenem-resistant health care-associated threats: Findings from the antibiotic resistance laboratory network, 2017 to 2019. Antimicrob Agents Chemother 2021; 65: e0110521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Duin D, Lok JJ, Earley M et al. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis 2018; 66: 163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKinnell JA, Dwyer JP, Talbot GH et al. Plazomicin for infections caused by carbapenem-resistant Enterobacteriaceae. N Engl J Med 2019; 380: 791–3. [DOI] [PubMed] [Google Scholar]
- 8. Kaye KS, Vazquez J, Mathers A et al. Clinical outcomes of serious infections due to carbapenem-resistant Enterobacteriaceae (CRE) in TANGO II, a phase 3, randomized, multi-national, open-label trial of meropenem-vaborbactam (M-V) Vs. best available therapy (BAT). OFID 2017; 4: S534–5. [Google Scholar]
- 9. Khatri A, Naeger Murphy N, Wiest P et al. Community-acquired pyelonephritis in pregnancy caused by KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2015; 59: 4375–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang HJ, Hsieh CF, Chang PC et al. Clinical significance of community- and healthcare-acquired carbapenem-resistant Enterobacteriaceae isolates. PLoS One 2016; 11: e0151897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Duin D, Arias CA, Komarow L et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020; 20: 731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Duin D, Perez F, Rudin SD et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 2014; 58: 4035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arndt D, Grant JR, Marcu A et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 2016; 44: W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 2003; Chapter 10: Unit 10 3. [DOI] [PubMed] [Google Scholar]
- 15. Croucher NJ, Page AJ, Connor TR et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015; 43: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seemann T. Snippy: Rapid haploid variant calling and core SNP phylogeny. 2020.
- 17. Gu Z, Gu L, Eils R et al. circlize Implements and enhances circular visualization in R. Bioinformatics 2014; 30: 2811–2. [DOI] [PubMed] [Google Scholar]
- 18. Le T, Wang L, Zeng C et al. Clinical and microbiological characteristics of nosocomial, healthcare-associated, and community-acquired Klebsiella pneumoniae infections in Guangzhou, China. Antimicrob Resist Infect Control 2021; 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Devi LS, Broor S, Rautela RS et al. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum β-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: A 3-year study. Int J Appl Basic Med Res 2020; 10: 156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents 2017; 50: 127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thaden JT, Lewis SS, Hazen KC et al. Rising rates of carbapenem-resistant enterobacteriaceae in community hospitals: a mixed-methods review of epidemiology and microbiology practices in a network of community hospitals in the southeastern United States. Infect Control Hosp Epidemiol 2014; 35: 978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guh AY, Bulens SN, Mu Y et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 2015; 314: 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller BM, Johnson SW. Demographic and infection characteristics of patients with carbapenem-resistant Enterobacteriaceae in a community hospital: Development of a bedside clinical score for risk assessment. Am J Infect Control 2016; 44: 134–7. [DOI] [PubMed] [Google Scholar]
- 24. Brennan BM, Coyle JR, Marchaim D et al. Statewide surveillance of carbapenem-resistant enterobacteriaceae in Michigan. Infect Control Hosp Epidemiol 2014; 35: 342–9. [DOI] [PubMed] [Google Scholar]
- 25. Centers for Disease Control and Prevention . Antibiotic Resistance Threats in the United States. 2019.
- 26. McConville TH, Sullivan SB, Gomez-Simmonds A et al. Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One 2017; 12: e0186195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 2014; 27: 543–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant Enterobacter cloacae complex emerging as a global, diversifying threat. Front Microbiol 2019; 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: nosocomial infection, regulation and biofilm formation. Future Microbiol 2012; 7: 991–1002. [DOI] [PubMed] [Google Scholar]
- 30. Struve C, Bojer M, Krogfelt KA. Characterization of Klebsiella pneumoniae type 1 fimbriae by detection of phase variation during colonization and infection and impact on virulence. Infect Immun 2008; 76: 4055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J 1986; 5: 1389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bialek-Davenet S, Criscuolo A, Ailloud F et al. Development of a multiplex PCR assay for identification of Klebsiella pneumoniae hypervirulent clones of capsular serotype K2. J Med Microbiol 2014; 63: 1608–14. [DOI] [PubMed] [Google Scholar]
- 33. Khater F, Balestrino D, Charbonnel N et al. In silico analysis of usher encoding genes in Klebsiella pneumoniae and characterization of their role in adhesion and colonization. PLoS One 2015; 10: e0116215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gomes AEI, Pacheco T, Dos Santos CDS et al. Functional insights from KpfR, a new transcriptional regulator of fimbrial expression that is crucial for Klebsiella pneumoniae pathogenicity. Front Microbiol 2020; 11: 601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.