Abstract
The promoters of Drosophila genes encoding DNA replication-related proteins contain transcription regulatory element DRE (5′-TATCGATA) in addition to E2F recognition sites. A specific DRE-binding factor, DREF, positively regulates DRE-containing genes. In addition, it has been reported that DREF can bind to a sequence in the hsp70 scs′ chromatin boundary element that is also recognized by boundary element-associated factor, and thus DREF may participate in regulating insulator activity. To examine DREF function in vivo, we established transgenic flies in which ectopic expression of DREF was targeted to the eye imaginal discs. Adult flies expressing DREF exhibited a severe rough eye phenotype. Expression of DREF induced ectopic DNA synthesis in the cells behind the morphogenetic furrow, which are normally postmitotic, and abolished photoreceptor specifications of R1, R6, and R7. Furthermore, DREF expression caused apoptosis in the imaginal disc cells in the region where commitment to R1/R6 cells takes place, suggesting that failure of differentiation of R1/R6 photoreceptor cells might cause apoptosis. The DREF-induced rough eye phenotype was suppressed by a half-dose reduction of the E2F gene, one of the genes regulated by DREF, indicating that the DREF overexpression phenotype is useful to screen for modifiers of DREF activity. Among Polycomb/trithorax group genes, we found that a half-dose reduction of some of the trithorax group genes involved in determining chromatin structure or chromatin remodeling (brahma, moira, and osa) significantly suppressed and that reduction of Distal-less enhanced the DREF-induced rough eye phenotype. The results suggest a possibility that DREF activity might be regulated by protein complexes that play a role in modulating chromatin structure. Genetic crosses of transgenic flies expressing DREF to a collection of Drosophila deficiency stocks allowed us to identify several genomic regions, deletions of which caused enhancement or suppression of the DREF-induced rough eye phenotype. These deletions should be useful to identify novel targets of DREF and its positive or negative regulators.
The promoters of Drosophila genes related to DNA replication, such as those for the 180-kDa catalytic subunit and 73-kDa subunit polypeptide of DNA polymerase α and proliferating-cell nuclear antigen (PCNA), contain a common 8-bp palindromic sequence (5′-TATCGATA), named the DNA replication-related element (DRE) (21) in addition to E2F recognition sites (13, 54). The DRE requirement for promoter activation has been confirmed in both cultured cells and transgenic flies (22, 51, 52). Studies using the latter have shown that DRE is required for the function of the PCNA promoter throughout development except in adult females. We found a specific DRE-binding factor (DREF) consisting of an 80-kDa polypeptide homodimer, and molecular cloning of its cDNA has led to confirmation that DREF is a trans activator for DRE-containing genes (22).
Recently, we found that the N-terminal fragment of the DREF polypeptide containing a region responsible for DRE binding and dimer formation acts as dominant negative effector against DREF (23). Expression of a dominant-negative form of DREF in cells of salivary glands and eye imaginal discs using the GAL4-upstream activation site (UAS) system inhibited endoreplication of larval salivary gland cells and significantly reduced DNA replication in the second mitotic wave, respectively (23). The results indicate that DREF is required for DNA replication in both the mitotic cell cycle and the endo cell cycle. However, progression of DNA replication requires a number of genes involved in growth signal transduction, cell cycle regulation, DNA replication itself, or transcription regulation. So far, we have not obtained clues to determine which of these genes is a critical target of the dominant negative form of DREF.
Screening for the DRE sequence in the Drosophila genome permitted us to estimate that more than 103 Drosophila genes are regulated by DREF (29). We classified these genes into five groups by their functional category: (i) DNA replication related, (ii) translation related, (iii) signal transduction/cell cycle regulation related, (iv) transcription factor, and (v) others. Category iv contains specific transcriptional regulators required for normal development or determination of cell differentiation, such as zeste, caudal, Antennapedia, serendipity-beta, and some RNA-biding proteins. These observations suggest a possibility that DREF may play an important role in cell differentiation other than activation of genes involved in cell proliferation.
More recently, Hart et al. have proposed a novel function of DREF as an antagonist of boundary element-associated factor (BEAF), which is involved in boundary activity of the scs′ region of the Drosophila hsp 70 gene (18, 19). Staining of polytene chromosomes with anti-DREF and anti-BEAF antibodies revealed that about half of the signals from the two proteins overlapped and that the other half were from only BEAF or only DREF. Furthermore, using a chromatin precipitation method, they demonstrated that DREF can bind to the same sequences as BEAF, establishing the chromatin boundary in the scs′ special chromatin domain present in Drosophila melanogaster 87A7 hsp 70. From the results, they assumed that competition of binding between DREF and BEAF is important for regulation of activity at the chromatin boundary.
To clarify in vivo functions of DREF, we decided to establish transgenic flies expressing DREF in eye imaginal disc cells by using the GAL4-UAS system for two reasons. The first reason is that details of cell proliferation and differentiation during Drosophila eye development have been characterized (8, 50). The cells of the eye imaginal disc, sac-like with a monolayer epithelium, proliferate and are not differentiated until the mid-third-instar larva stage. Differentiation and retinal pattern formation commence when the morphogenetic furrow (MF) sweeps across the disc from posterior to anterior. Ahead of the MF, cells proliferate asynchronously. As the cells are drawn into the MF, they are arrested in a prolonged G1 phase. A subset of these cells become committed to differentiate and begin to express neural cell markers. The first cells to be determined will form the R8 photoreceptor cells of the adult ommatidium. Additional cells are recruited sequentially, forming a precluster whose members are destined for specific fates. Cells in the precluster do not reenter the cell cycle, but all uncommitted cells divide one more time, generating a reservoir of cells for subsequent differentiation events. Thus, the Drosophila eye imaginal disc provides an ideal system to examine the effects of ectopic expression of proteins on the regulation of cell proliferation, cell cycle progression, and commitment of cell differentiation.
The second reason is that if overexpression of DREF in eye imaginal discs confers a specific phenotype in the eyes of adult flies, the flies can be utilized to screen for modifiers of DREF activity or its target genes. Such an approach has been successfully undertaken with dE2F-overexpressing flies (40).
In the present study, the consequences of ectopic expression of DREF polypeptide in cells of eye imaginal discs were investigated. Ectopic expression of DREF in the cells posterior to the MF induced ectopic DNA synthesis and apoptosis in normally postmitotic cells, inhibited differentiation of the photoreceptor cells, and resulted in a severe rough eye phenotype in the adult flies. Furthermore, we succeeded in identifying mutations that enhance or suppress the DREF-induced phenotype. Candidates for DREF interaction include cell cycle regulators and regulators of chromatin-structure, giving us a clue to resolve the molecular mechanisms of how DREF activates many genes related to cell proliferation.
MATERIALS AND METHODS
Fly strains.
Fly stocks were maintained at 25°C on standard food. The Canton S fly was used as a wild-type strain.
The dE2F91, dE2F7102, and dE2F729 alleles described previously (13) were kindly supplied by N. Dyson. The reaper, hid, and grim genes lie at locus 75C1-2. The deficiency lines, Df(3L)WR4 and Df(3L)WR10 uncovering the 75B-75C locus have been described previously (48). Flies bearing GMR-DIAP1 and GMR-DIAP2 transgenes were provided by G. Rubin. Enhancer trap lines carrying the lacZ markers D120 (scabrous) (31), BB02 (31), ro156, X63 (rhomboid) (14), P82 (7), AE127 (seven-up) (32), and H214 (klingon) (5) were obtained from Y. Hiromi. The transgenic fly carrying UAS-DREF on the second chromosome and the transgenic fly (line 16) carrying pGMR-GAL4 on the X chromosome were described previously (23, 41). Alleles of the following trithorax and Polycomb group genes were obtained from the Bloomington, Ind., stock center: trx1, pho1, sxc1, mor1, osa2, brm2, esc1, Dll9, Dll5, kto1, Pc1, Pc3, Pc7, Psc1, Pcl7, E(Pc)1, and su(z)21.b7.
pGMR constructs.
The GAL4 coding sequence was isolated using PCR with the primers 5′-ATCGAATTCGGTACCAGATGAAGCTACTGTCTTCTAT CGA and 5′-ATAAGATCTGCGGCCGCTTACTCTTTTTTTGGGTTTG GTG and the pGaTB plasmid as a template. Products were gel purified, cut with EcoRI and BglII, and cloned into EcoRI-BglII sites of the pGMR vector (20). The pGMR-DREF construct was made by carrying out PCR on pDCDREF2.2 as a template with the primers 5′-ACAGGATTCAAGATGAGCGAAGGGGTACCA and 5′-GTTCTCGAGTAATTGTTGTGATGATGCAGA. The products were gel purified, cut with BglII and XhoI, and cloned into the BglII-XhoI sites of the pGMR vector.
Scanning electron microscopy.
Adult flies were anesthetized, mounted on stages, and observed under a Hitachi S-100 scanning electron microscope in the low-vacuum mode.
Preparation for transmission electron microscopy.
Adult heads were fixed in 2% glutaraldehyde–2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at 4°C overnight. After being washed in 0.1 M sodium cacodylate buffer (pH 7.2) at 4°C overnight, they were postfixed in 1% OsO4 in the same buffer for 1.5 h at 4°C and then dehydrated in a graded ethanol series. After clearing in propylene oxide, the heads were embedded in Epon and sectioned with an ultramicrotome. These thin sections were stained with uranyl acetate and Reynolds' lead solution. They were then observed under a transmission electron microscope.
Histology of adult eyes.
Adult eyes were fixed in Bouin's fixative, embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Immunohistology.
Third-instar larvae were dissected in Drosophila Ringer's solution, and imaginal discs were fixed in 4% paraformaldehyde–phosphate-buffered saline (PBS) for 20 min at room temperature. After being washed with PBS–0.3% Triton X-100 (PBS-T), the samples were blocked for 30 min at room temperature with PBS-T containing 10% normal goat serum. The samples were incubated with culture supernatant of hybridoma-producing anti-Elav monoclonal antibody at a 1:2 dilution, mouse anti-β-galactosidase monoclonal antibody (Promega) at a 1:1,000 dilution, rabbit polyclonal antibody specific for DREF at a 1:1,000 dilution, or culture supernatant of hybridoma-producing anti-DREF monoclonal antibody at a 1:200 dilution at 4°C for 16 h. The anti-Elav monoclonal antibody was kindly donated by Y. Hiromi (36). After extensive washing with PBS-T, the imaginal discs were incubated with an alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (IgG) (Promega) or an alkaline phosphatase-conjugated goat anti-rabbit IgG (Promega) at a 1:2,000 dilution for 2 h at room temperature and washed with PBS-T. Color was developed with a solution containing 100 mM Tris-HCl (pH 9.5), 100 mM NaCl, 5 mM MgCl2, 0.34 mg of nitroblue tetrazolium salt per ml, and 0.175 mg of 5-bromo-4-chloro-3-indolylphosphate toluidinium salt (X-phosphate). The tissues were washed with PBS and mounted in 90% glycerol–PBS for microscopic observation. In the case of the enhancer trap line, β-galactosidase was detected using mouse anti-β-galactosidase antibody at a 1:1,000 dilution as the first antibody and peroxidase-conjugated goat anti-mouse IgG (E-Y laboratory) at a 1:500 dilution as the second antibody. Staining was visualized in PBS-T containing 0.5 mg of diaminobenzidine per ml, 2.5 mM CoCl2 and 0.003% H2O2.
BrdU labeling.
Cells in S phase were detected by a 5-bromo-2′-deoxyuridine (BrdU) labeling method as described previously (49) with minor modifications. Third-instar larvae cultured at 25°C were dissected in Grace's medium and then incubated in the presence of 20 mg of BrdU (Boehringer) for 30 min. The samples were fixed in Carnoy's fixative (ethanol-acetic acid-chloroform, 6:3:1) for 15 min at 25°C and further fixed in 80% ethanol–50 mM glycine buffer (pH 2.0) at −20°C for 2 h. Incorporated BrdU was visualized using an anti-BrdU antibody and an alkaline phosphatase detection kit (Boehringer). The time of color development for alkaline phosphatase was precisely regulated to be identical for all samples.
Apoptosis assay.
Third-instar larvae were dissected in Drosophila Ringer's solution, and imaginal discs were fixed in 4% paraformaldehyde–PBS for 30 min at room temperature. After they were washed with PBS, endogenous peroxidase activity was blocked by treatment at room temperature for 30 min with methanol containing 0.3% H2O2. The samples were then washed with PBS and permeabilized by being incubated in a solution containing 0.1% sodium citrate and 0.1% Triton X-100 on ice for 2 min. After extensive washing, the TUNEL reaction was carried out using an in situ cell death detection kit, (POD; Boehringer Mannheim) as specified by the manufacturer.
Modifier screen.
We observed that the DREF-expressing transgenic flies that developed at 28°C exhibited a more severe rough eye phenotype than did the transgenic flies that developed at 25°C, because transcriptional stimulation activity of GAL4 is temperature dependent. Therefore, we performed a genetic screen for modifier of DREF as following. Female flies expressing DREF (GMR-GAL4/GMR-GAL4; UAS-DREF/UAS-DREF; +/+) were crossed with individual males carrying the deficiency chromosome, mutant alleles of the E2F gene, or mutant alleles of the trithorax and Polycomb group genes, and F1 progeny were developed at either 25 or 28°C.
RESULTS
Expression pattern of DREF in the eye imaginal disc.
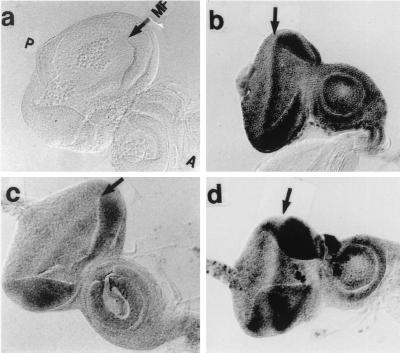
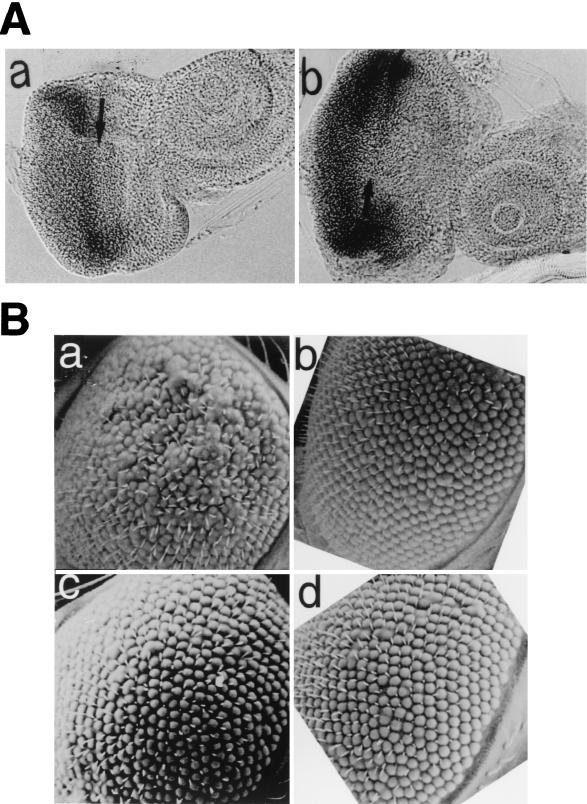
We first examined the distribution of endogenous DREF in wild-type eye imaginal discs. Anti-DREF polyclonal antibody (Fig. 1b), monoclonal antibody 2 (Fig. 1c) or monoclonal antibody 4 (Fig. 1d) all strongly stained cells in front of the MF, when the cells were entering the G1 phase of the second mitotic cycle. In addition, an elevated level of DREF was observed in the cells just posterior to the MF, corresponding to the S-phase zone in the second mitotic wave. Relatively low levels of DREF were present in the postmitotic cells undergoing differentiation. The expression pattern is very similar to that of dE2F (1, 4). The results suggest that DREF is involved in differentiation processes in addition to cell proliferation.
FIG. 1.
Immunostaining of eye imaginal discs with anti-DREF antibodies. (b to d) Eye imaginal discs from wild-type third-instar larva were stained with rabbit anti-DREF polyclonal antibody (b), mouse anti-DREF monoclonal antibody 2 (c), and mouse anti-DREF monoclonal antibody 4 (d). (a) Negative control without primary antibody. P, posterior; A, anterior; MF, morphogenetic furrow. The time of color development for alkaline phosphatase is 1 h.
Ectopic expression of DREF perturbs normal eye development.
Glass protein is a transcription factor that is expressed in all cells in and posterior to the MF (33). The pGMR vector contains a multimer of Glass-binding sites that are sufficient to drive the expression of coding regions placed downstream of these binding sites. To investigate the consequence of the ectopic expression of DREF, we established a transgenic line bearing pGMR-GAL4 crossed with a transgenic fly carrying DREF cDNAs under the control of a GAL4-binding sequence (pUAS-DREF1–709). Immunostaining using anti-DREF polyclonal antibody revealed elevated levels of DREF expressed in all cells in and posterior to the MF of the third-instar larvae of the progeny, as expected (Fig. 2b).
FIG. 2.
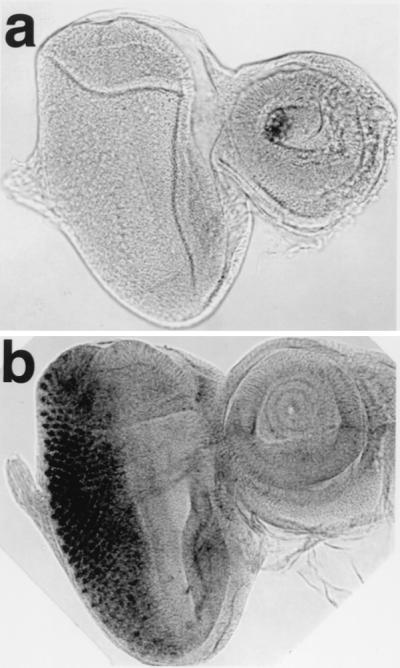
Ectopic expression of DREF protein with the GMR promoter. Immunostaining of eye imaginal discs with the anti-DREF antibody is shown. (a) GMR-GAL4/+; +/+, (b) GMR-GAL4/+; UAS-DREF/+. The time of color development for alkaline phosphatase is 5 min.
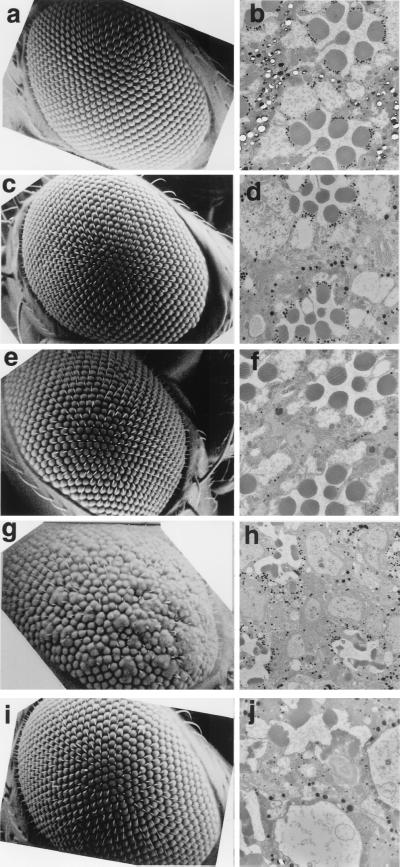
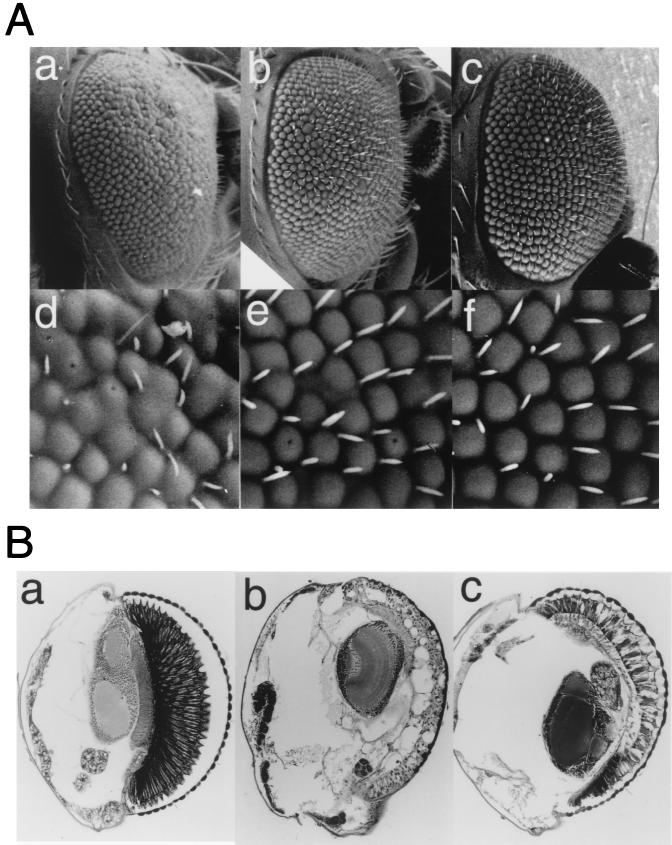
The eyes of adults carrying one copy of pGMR-GAL4 and one copy of pUAS-DREF1–709 were severely rough in appearance, and most bristles were missing (Fig. 3g), whereas the eyes of flies carrying either of one copy of pGMR-GAL4 or one copy of pUAS-DREF1–709 appeared normal (Fig. 3c and c). In addition, the organized array of each ommatidium was destroyed and some ommatidia were fused each other. Examination of retinal sections revealed the number of ommatidia to be reduced and the number and shape of the photoreceptor cells in each ommatidia to also be abnormal. Pigment cells were found to be missing in all ommatidia, and some remaining tissues were vacuolated in transgenic flies carrying one copy of pGMR-GAL4 and one copy of pUAS-DREF1–709 (Fig. 3h). To confirm that ectopic expression of DREF-directly affects eye development, we established transgenic flies carrying pGMR-DREF1–709. Flies carrying pGMR-DREF1–709 did not exhibit an apparent rough eye phenotype on inspection by scanning electron microscopy (Fig. 3i); however, the abnormal photoreceptor cells and missing pigment cells were seen in sections of eyes from transgenic flies carrying pGMR-DREF1–709 (Fig. 3j), indicating that abnormality of eye development was indeed induced by overexpression of DREF polypeptide and not by overexpression of GAL4 protein. Furthermore, the eye phenotype in these flies suggests that DREF overexpression affects eye development without causing the apparent rough eye under scanning electron microscopy.
FIG. 3.
Scanning and transmission electron micrographs of adult compound eyes. (a and b) Wild-type eye. (c and d) GMR-GAL4/+;+/+. (e and f) +/+. UAS-DREF/+. (g and h) GMR-GAL4/+; UAS-DREF/+. (i and j) +/+; GMR-DREF#22/GMR-DREF#22. These flies were developed at 28°C. The rough eye phenotype is evident in panels g and h.
Expression of DREF induces an ectopic S phase posterior to the MF.
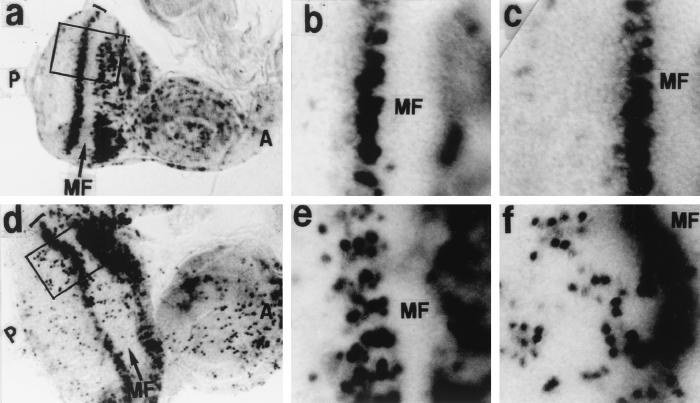
To assess whether expression of DREF in late G1 can drive cells destined to become postmitotic cells into the S phase, imaginal discs were pulse-labeled with BrdU for 30 min in vitro and stained with an anti-BrdU antibody (Fig. 4). No significant difference between control and DREF-expressing discs was observed in the region anterior to the MF, where the GMR promoter was inactive. In discs of wild-type or transgenic flies expressing solely the GAL4 protein, the S-phase cells were apparent as a dense narrow band with BrdU signals posterior to the MF. In control discs, a 30-min pulse of BrdU incorporation led to labeling of cells in one or two columns posterior to the MF (Fig. 4b), while in imaginal discs of transgenic flies expressing DREF, the equivalent of five columns were labeled (Fig. 4e). Therefore, overexpression of DREF either expanded the S phase or induced an extra S phase. In addition, some cells in the region more posterior to the MF, where cells are committed to the neuronal fate and normally differentiate into specific cells such as photoreceptors, were labeled with BrdU (Fig. 4d), and the quantitative measurement of BrdU-staining cells is summarized in Table 1. Most of the nuclei of cells with BrdU incorporated occupied a more apical position in the imaginal discs than did nuclei in the synchronous S-phase cells posterior to the MF and in uncommitted cells (Fig. 4f), suggesting that DREF overexpression caused ectopic S phase in some cells that are normally postmitotic.
FIG. 4.
Patterns of BrdU incorporation in eye imaginal discs. (a to c) GMR-GAL4/+; +/+. (d to f) GMR-GAL4/+; UAS-DREF/+. The eye disc was stained with an anti-BrdU antibody. MF and arrows indicate the position of the MF. Panels b and e show high-magnification basal views of the same focal planes as panels a and d, respectively. Panels c and f show BrdU signals at the apical focal planes of a and d, respectively.
TABLE 1.
BrdU staining cells in the region posterior to the MF
| Disc | No. of BrdU-staining cells
|
|
|---|---|---|
| Canton S | GMR-GAL4/+; UAS-DREF/+; +/+ | |
| 1 | 14 | 90 |
| 2 | 16 | 105 |
| 3 | 12 | 82 |
| 4 | 17 | 69 |
| 5 | 9 | 74 |
| Mean | 13 | 84 |
Expression of DREF interferes with photoreceptor differentiation.
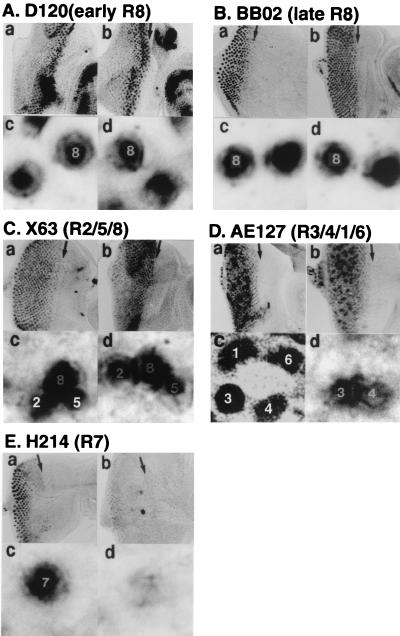
It has been reported that overexpression of dE2F in eye imaginal disc causes ectopic S phases in differentiation-uncommitted reservoir cells, although committed cells in the eye disc are relatively resistant to the effects of ectopic E2F expression (1, 11). However, overexpression of DREF appears to result in the occurrence of an extra round of S phase in the committed cells (Fig. 4f). Therefore, we analyzed the effects of DREF expression on photoreceptor specifications in detail. In wild-type discs, developmentally uncommitted cells are sequentially recruited into clusters that comprise ommatidial precursors. Cluster formation is first observed within the MF, where cells are in G1. Cells either leave the cell cycle and differentiate or undergo a final synchronous round of cell division. Overt ommatidial organization starts in the MF when cells are grouped into equally spaced concentric aggregates, which converted into preclusters. Photoreceptor cells have been found to be generated in a stereotyped order: R8 is generated first, with movement posterior from the MF, then cells are added pairwise (R2 and R5, R3 and R4, and R1 and R6), and R7 is the last photoreceptor to be added to each cluster. Several enhancer trap lines expressing a nucleus-localized form of Escherichia coli β-galactosidase depend on the specific enhancer-promoter located nearby the P-element. They were used here to determine the identities of each photoreceptor. We used seven enhancer trap lines, D120 (inserted in scabrous), BB02, ro156, X63 (inserted in rhomboid), P82, AE127 (inserted in seven-up), and H214 (inserted in klingon), specifically expressing the β-galactosidase marker in photoreceptor cells (R) of early R8, late R8, late R8, R2/R5/R8, R3/R4/R7, R3/R4/R1/R6, and R7, respectively. The imaginal discs from F1 larva from mating of enhancer trap lines and DREF-expressing transgenic flies were immunohistochemically stained with the anti-β-galactosidase antibody. In ommatidia of DREF-expressing animals, nuclei of early R8 (D120, Fig. 5A), late R8 (BB02, Fig. 5B), and R2/R5/R8 (X63, Fig. 5C) demonstrated a similar staining pattern to nuclei of control ommatidia. With AE127, the ommatidia of DREF-expressing progeny were found to contain R3 and R4 nuclei but did not contain R1 or R6 nuclei (Fig. 5D, d). In addition, signals for R7 cells were not detected in imaginal discs expressing DREF (Fig. 5E, d). The results indicate that expression of DREF inhibited the differentiation of R1, R6, and R7 photoreceptor cells.
FIG. 5.
Immunostaining of eye imaginal discs with an anti-β-galactosidase antibody. Wild-type (a and c) or GMR-GAL4; UAS-DREF transgenic (b and d) flies were crossed with enhancer trap lines carrying D120 (inserted in scabrous) (A), BB02 (B), X63 (inserted in rhomboid) (C), AE127 (inserted in seven-up) (D), or H214 (inserted in klingon) (E) specifically expressing the β-galactosidase marker in photoreceptor cells (R) of early R8, late R8, R2/R5/R8, R3/R4/R1/R6, and R7, respectively, and F1 larvae were immunohistochemically stained with the anti-β-galactosidase antibody.
Expression of DREF leads to apoptosis.
Failure of normal cell cycle progression and disturbance of differentiation processes are known to cause apoptosis (17). For example, it has been reported that overexpression of dE2F and dDP in eye imaginal discs using a GMR promoter induces apoptosis and that this counterbalances cells that enter an abnormal S phase (11). DREF expression leads to ectopic DNA synthesis and inhibition of differentiation. Therefore, we investigated whether overexpression of DREF can induce apoptosis in eye imaginal disc cells. In wild-type discs of third-instar larvae, there were very few apoptotic cells (Fig. 6a). In contrast, staining of eye imaginal discs from transgenic flies expressing DREF revealed apoptotic cells to be significantly increased in the region posterior to the MF (Fig. 6b). Apoptosis seemed to begin in the imaginal disc cells in the region where commitment to R1/R6 cells takes place, suggesting that failure of differentiation into R1/R6 might induce apoptosis.
FIG. 6.
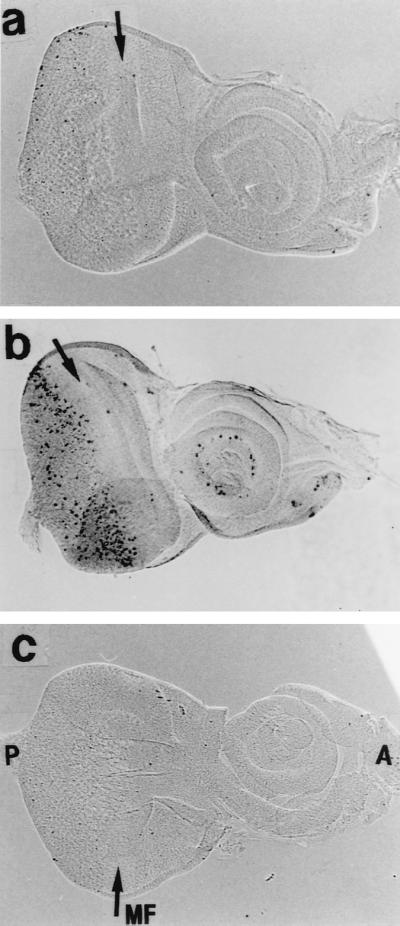
Detection of apoptotic cells in eye imaginal discs. The TUNEL method was used with (a and b) and without (c) terminal deoxynucleotidyltransferase. (a) GMR-GAL4/+;+/+. (b and c) GMR-GAL4/+; UAS-DREF/+. MF and arrows indicate the position of the MF.
The rough eye phenotype was suppressed when we crossed the transgenic line expressing DREF with those expressing DIAP1 (Drosophila homologue of the baculovirus inhibitor of apoptosis 1) (Fig. 7A, b and e) (47), DIAP2 (data not shown), or baculovirus p35 protein (Fig. 7A, c and f) (9). Horizontal sections of eyes of adult flies also showed that the DREF-induced eye degeneration was at least partially suppressed by expression of p35 proteins (Fig. 7B, b and c). The result indicates that the rough eye phenotype resulted at least partially from ectopic induction of apoptosis.
FIG. 7.
Suppression of the DREF-induced rough eye phenotype by expression of DIAP1 and p35. (A) Scanning electron micrographs of adult compound eyes. (a and d) GMR-GAL4/+; UAS-DREF/+;+/+. (b and e) GMR-GAL4/+; UAS-DREF/+; UAS-DIAP1/+. (c and f) GMR-GAL4/+; UAS-DREF/+; GMR-p35/+. (B) Horizontal sections of adult Drosophila eyes stained with hematoxylin-eosin. (a) Canton S. (b) GMR-GAL4/+; UAS-DREF/; +/+. (c) GMR-GAL4/+; UAS-DREF/+; GMR-p35/+. These flies were developed at 28°C.
dE2F mutations suppress the rough eye phenotype induced by expression of DREF.
The transgenic lines expressing DREF in the eye imaginal discs exhibit a rough eye phenotype but normal viability and fertility. Therefore, they can be used as a genetic screen to identify mutations that enhance or suppress the rough eye phenotype. We first performed a pilot experiment to test their validity for this purpose by using dE2F mutant flies. We previously demonstrated that dE2F is one of the genes under regulation by DREF. Expression of DREF in the eye imaginal discs activates the dE2F gene promoter, which can be monitored by measuring the expression of β-galactosidase in the eye imaginal disc of a dE2F mutant, dE2F729, in which the lacZ gene had been inserted near the translation initiation site of the dE2F gene in the same orientation as the dE2F gene (Fig. 8A) (4, 13, 39). We therefore predicted that a half-dose reduction of dE2F would affect the rough eye phenotype caused by DREF overexpression. Consistent with our prediction, the rough eye phenotype was suppressed when a half-dose reduction of the dE2F was achieved by crossing GMR-GAL4/UAS-DREF flies for loss-of-function mutant flies of dE2F (dE2F91, dE2F729, and dE2F7172) (Fig. 8B, b, c, and d).
FIG. 8.
A half-dose reduction of the dE2F gene suppresses the DREF rough eye phenotype. (A) Overexpression of DREF activates lacZ expression under the control of the dE2F gene promoter. Female flies expressing DREF (GMR-GAL4/GMR-GAL4; UAS-DREF/UAS-DREF;+/+) or female flies expressing GAL4 only (GMR-GAL4/GMR-GAL4; +/+; +/+) were crossed with dE2F729 male flies. F1 third-instar larvae developed at 28°C were dissected, and eye imaginal discs were used for detection of lacZ. In dE2F729 flies, the P element-carrying lacZ gene is inserted 48 nucleotides upstream of the initiator methionine of the dE2F gene in the same orientation as the dE2F gene; therefore, lacZ expression directed by the dE2F gene promoter can be detected in eye imaginal discs from dE2F729/+ larvae. (a) GMR-GAL4; +/+; dE2F729/+ eye disc stained with anti-β-galactosidase antibody. (b) GMR-GAL4; UAS-DREF/+; dE2F729/+ eye disc stained with anti-β-galactosidase antibody. (B) Scanning electron micrographs of adult compound eyes. Female flies expressing DREF (GMR-GAL4/GMR-GAL4; UAS-DREF/UAS-DREF; +/+) were crossed with dE2F729, dE2F7172, or dE2F91 male flies (w/Y; +/+; dE2F−/TM3), and F1 progeny developing at 28°C without TM3 balancer were used for analysis of eye phenotype. (a) GMR-GAL4/+; UAS-DREF/+; +/+. (b) GMR-GAL4/+; UAS-DREF/+; dE2F91/+. (c) GMR-GAL4/+; UAS-DREF/+; dE2F729/+. (d) GMR-GAL4/+; UAS-DREF/+; dE2F7172/+. In dE2F7172 flies, the P element is inserted 33 nucleotides downstream of the initiator methionine. The E2F91 allele contained a C-to-T transition at nucleotide +91 that converted Gln-31 of dE2F to a stop codon.
Effect of mutations in Polycomb and trithorax group genes on the rough eye phenotype induced by expression of DREF.
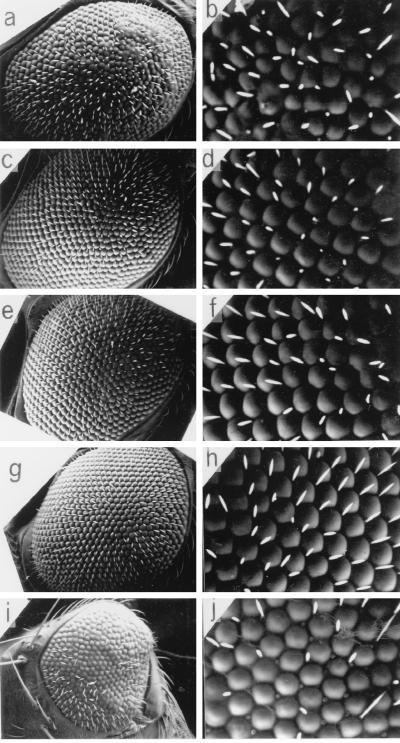
Recently, Hart et al. proposed a novel function of DREF as a modulator of boundary element activity (19). Therefore, DREF might activate the transcription of DRE-containing genes through interaction with proteins involved in modification of chromatin structure (for example, establishment, maintenance, or cancellation of the chromatin boundary). The Polycomb group proteins are required to preserve the transcriptionally silenced state, whereas the trithorax group genes are needed to perpetuate the transcriptionally active state (6, 30, 35, 42). The function of these factors is not limited to homeotic gene regulation; rather, they are involved in the control of diverse developmental processes. Several observations suggest that they change the chromatin structure, establishing a configuration that is either permissive or nonpermissive for transcription. We therefore examined the effects of mutations in Polycomb or trithorax group genes on the rough eye phenotype induced by expression of DREF. As shown in Fig. 9, a half-dose reduction of the trithorax group genes brahma (brm) (Fig. 9c and d), osa (Fig. 9e and f), and moira (mor) (Fig. 9g and h) significantly suppressed the rough eye phenotype. Mutation of enhancer of Polycomb [E(Pc)] weakly suppressed the rough eye phenotype (data not shown). Genetic crossing of the DREF-expressing strain with Dll9, a hypomorphic allele, resulted in a weakly enhanced rough eye phenotype (data not shown) and crossing with Dll5 resulted in a severe small eye phenotype (Fig. 9i and j). Mutations in other trithoax and Polycomb group genes tested, including Polycomb (Pc), Polycomb-like (Pcl), suppressor of zesta [Su(z)], Posterior sex combs (Psc), multiple wing hair (mwh), super sex comb (sxc), trithorax (trx), and kohtalo (kto), had no effect on the GMR-GAL4; UAS-DREF phenotype, suggesting that the effect is specific to only certain members of the trithorax class of genes (data not shown).
FIG. 9.
Scanning electron micrographs of adult compound eyes. Female flies expressing DREF (GMR-GAL4/GMR-GAL4; UAS-DREF/UAS-DREF; +/+) were crossed with brm2/TM6B, osa2/TM6B, mor1/TM6B, or Dll5/SM5 male flies, and F1 progeny developing at 28°C without balancer chromosome were used for analysis of eye phenotype. (a and b) GMR-GAL4/+; UAS-DREF/+; +/+. (c and d) GMR-GAL4/+; UAS-DREF/+; brm2/+. (e and f) GMR-GAL4/+; UAS-DREF/+; osa2/+. (g and h) GMR-GAL4/+; UAS-DREF/+; mor1/+. (i and j) GMR-GAL4/+; UAS-DREF/Dll5; +/+.
brm is the Drosophila homologue of the yeast SWI2/SNF2 gene, and the BRM complex containing OSA and MOR is an essential coactivator for the trithorax group protein Zesta (24). Therefore, suppression of the DREF-induced rough eye phenotype by a half-dose reduction of some members of the BRM complex genes suggests that it may contribute to the regulation of DREF activity.
Identification of genomic regions whose deletions modify the rough eye phenotype induced by expression of DREF.
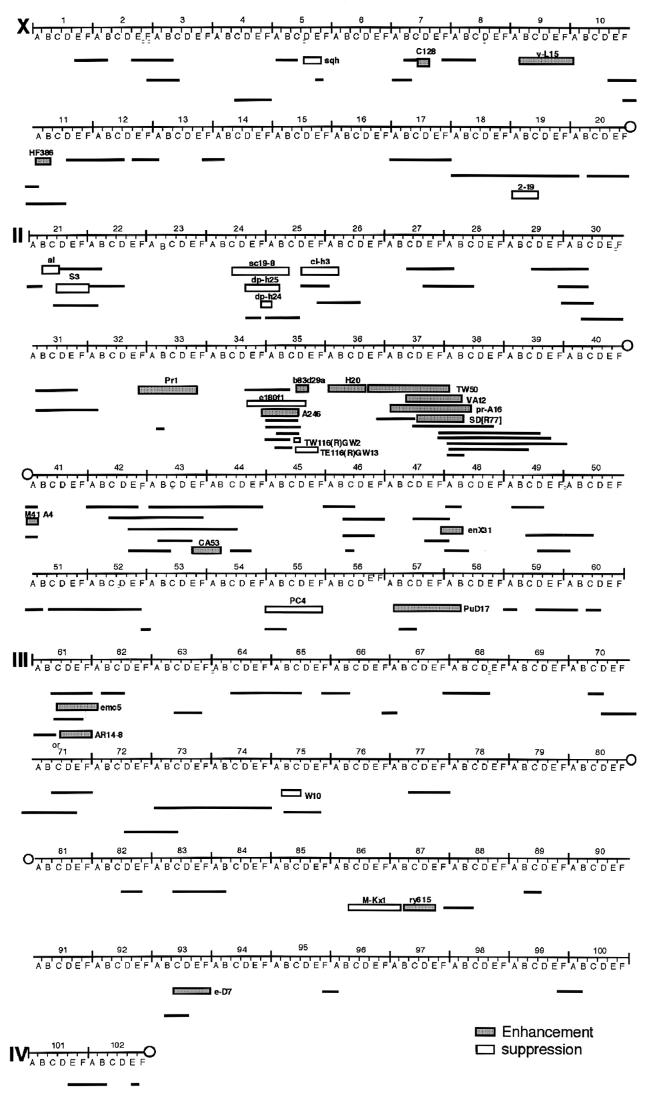
A collection of Drosophila deficiency stocks was used to cross with the transgenic flies expressing DREF, and the eye morphology of their F1 progeny was compared with that of F1 progeny between transgenic flies and Canton S. A total of 132 deficient lines were examined, permitting us to screen approximately 61% of the euchromatic region. To determine whether these deletions specifically modify the rough eye phenotype induced by DREF overexpression, crosses were made with transgenic flies having a rough eye phenotype induced by overexpression of human p53 (53). Expression of p53 inhibited cell cycle progression in the S-phase zone behind the MF and induced extensive apoptosis, but photoreceptor cell differentiation appeared to be normal. Apoptosis is a phenomenon induced in common by p53 and DREF overexpression. Therefore, if a dominant enhancer (or suppressor) of the p53-induced rough eye phenotype also enhances (or suppresses) the DREF-induced rough eye phenotype, the deletion might contain a gene involved in the apoptotic pathway. In fact, we identified nine deletions for dominant enhancers (21D2-3;21F2-22A1, 34F4;35D7, 37B2-12;38D2-5, 37C2-5;38B2-38C1, 41A, 43E6;44B6, 48A;48B, 57B4;58B, and 87B11-13;87E8-11) and one for a dominant suppressor (75B3-6;75C) of both DREF- and p53-induced rough eyes (Fig. 10). Furthermore we could determine dominant modifiers specific to the DREF function. Ten lines specifically suppressed the rough eye phenotype induced by DREF expression, and 11 lines demonstrated specific enhancement. These deletions should be useful to identify novel targets of DREF and its positive or negative regulators.
FIG. 10.
Summary of the deficiencies screened. The numbered divisions and lettered subdivisions of salivary gland polytene chromosomes are marked. The boxes indicate the influence of each deficiency on the DREF-induced rough eye phenotype: black lines, no effect; shaded box, enhancement; open box, suppression. The deletions for dominant enhancers and suppressor of both DREF- and p53-induced rough eye phenotype are indicated by italics.
DISCUSSION
Involvement of the DRE-DREF system in the regulation of a considerable variety of genes has been suggested by the results of DNA database searches. In about 3.5% of the Drosophila genome, 73 copies of 5′-TATCGATA sequences were found to be localized within 0.6 kb upstream regions of 61 genes, including those involved in transcription, translation, growth signal transduction, cell cycle regulation, and transcriptional regulation, in addition to genes related to DNA replication (29). Genes related to cell proliferation, such as those for PCNA, DNA polymerase α 180- and 73-kDa subunits, CycA, D-raf, and E2F, have been shown to be under DRE-DREF system regulation (34, 38, 39). In addition, we have reported that ectopic expression of the truncated form of DREF, possessing dominant negative activity, in salivary glands or eye imaginal discs significantly reduces DNA replication. Evidence suggests that DREF is required for DNA replication in both the endo and mitotic cell cycles. However, it has hitherto not been clear whether the accumulation of DREF is sufficient to induce DNA replication as well as ectopic expression of dE2F to drive normally quiescent cells to enter S phase (1, 12, 37).
Here we demonstrated that ectopic expression of DREF in eye imaginal discs, using the glass-responsive GMR promoter, induced ectopic DNA synthesis and apoptosis in the cells behind the morphogenetic furrow. The results appear to be very similar to phenomena observed in eye imaginal discs expressing dE2F/dDP (11). One possible molecular mechanism is direct up-regulation of genes involved in DNA replication such as those encoding PCNA and DNA polymerase α. Another possible mechanism is indirect up-regulation of genes related to DNA replication or genes required for the G1/S transition by means of activation of dE2F expression. We previously demonstrated that overexpression of DREF in eye imaginal disc cells enhanced the promoter activity of dE2F (39). Therefore, overexpression of DREF may result in E2F accumulation behind the morphogenetic furrow and consequently induce an ectopic S phase. This view is consistent with the observation that reduction of the dE2F gene dose at least partially suppressed the rough eye phenotype induced by DREF. However, it should be noted that there are some differences between the DREF- and dE2F/dDP-induced phenotypes: (i) In eye imaginal discs expressing dE2F/dDP under control of the GMR promoter, most S phases are seen in uncommitted cells, whereas in imaginal discs expressing DREF, nuclei of cells under differentiation process located apically with respect to the imaginal disc appear to incorporate BrdU; (ii) Du et al. (11) have reported that coexpression of baculovirus p35 protein, an inhibitor of cell death, strongly enhances the dE2F/dDP phenotype, indicating that the majority of cells ectopically entering S phase as a result of E2F/dDP expression are eliminated by apoptosis, while the DREF phenotype, in contrast, was significantly suppressed by coexpression of p35; and (iii) overexpression of dE2F/dDP did not grossly interfere with the initiation of neuronal cell differentiation or the stepwise recruitment of cells into preclusters, whereas expression of DREF perturbed photoreceptor specifications of R1, R6, and R7, whose differentiation normally begins after those of R8, R2, R5, R3, and R4. The molecular mechanisms underlying these differences are unclear, but we speculate that elevated amounts of DREF not only affect DNA replication-related genes but also disturb the transcription of genes required for normal processes of differentiation, consequently inducing apoptosis.
In the present study, we focused on the use of our flies for screening interaction with Polycomb and trithorax group genes. It is known that DREF and BEAF share binding sites with functions as boundary elements in Drosophila (scs′ region of hsp70, BE76, and BE28) (19, 45) and also the promoter region of DNA polymerase α 180-kDa subunit gene. Polycomb and trithorax group proteins appear to regulate chromatin boundary activity by establishing higher-order domains of chromatin organization required for the assembly of functional insulators at the nuclear matrix (15, 16, 25, 28), and the present genetic screening showed that a half-dose reduction of the trithorax group genes brm, osa, mor, and E(Pc) suppressed while a half-dose reduction of Dll enhanced the DREF-induced rough eye phenotype.
brm is the Drosophila homologue of the yeast SWI2/SNF2 gene, isolated as a dominant suppressor of Pc mutations (26, 44). osa shows strong genetic interactions with brm, suggesting that its gene product may cooperate closely with the BRM complex (43, 46). The product of mor (3, 26) is homologous to yeast Swi3p and was also recently identified as a BRM complex-associated protein, BAP (10). BRM, OSA, and MOR are part of a large multiprotein complex containing several other proteins with extensive homology to yeast SWI/SNF or the related chromatin-remodeling complex, and therefore the BRM complex has been predicted to be one of the Drosophila chromatin-remodeling complexes. Kal et al. (24) demonstrated that the BRM complex containing OSA and MOR is an essential coactivator for the trithorax group protein Zeste, a sequence-specific transcription regulator (2, 27). Several homeotic and other genes carrying the Zeste-binding sequence in their promoter regions thus appear to be candidate target genes (or chromatin regions) for the BRM complex.
Suppression of the DREF-induced rough eye phenotype by a half-dose reduction of some members of BRM complex genes suggests that DREF activity may be positively regulated by the BRM complex. The molecular basis for any interaction is not yet clear, but several possibilities exist. The first is that genetic interaction may result from a direct physical interaction between the two. To test this possibility, we are now undertaking biochemical analyses. An alternative possibility is that the BRM complex is a regulator of the expression of some genes critical for induction of the rough eye phenotype by DREF overexpression. This is difficult to test because we estimate that several hundred genes are under DREF control. Finally, it is possible that the BRM complex generally activates transcription by remodeling chromatin. However, we observed that a half-dose reduction of brm, mor, or osa did not change the severity of the rough eye phenotype induced by BEAF32 (55), and Staehling-Hampton et al. have reported that mutations in brm, mor or osa enhance the rough eye phenotype induced by overexpression of dE2F/dDP/p35 (40). Therefore, suppression of the DREF-induced rough eye phenotype by reduction of the dosage of brm, mor, or osa may reflect specific interactions between DREF and the BRM complex. Further studies are necessary to clarify the molecular basis and biological meaning of the interaction between DREF and the BRM complex.
We have also observed that the Dll mutation enhances the rough eye phenotype induced by BEAF32 expression (55). Thus, the product of Dll might participate in the regulation of chromatin boundaries such as the hsp 70 scs′ region, where DREF and BEAF bind.
In the present study, we found that genomic regions suppressed the DREF-induced rough eye phenotype when they were heterozygous for deletions: 5D1-2; 5E, 19A5; 19D3, 24C2-8; 25C8-9, 24E2-4; 25B2-5, 24F4; 25C3-5, 25D2-4; 26B2-5, 35D1; 35D4, 35D2; 35F1-2, 55A; 55F and 86C1; 87B1-5. These presumably contain genes for targets of DREF or positive regulator of DREF. We also found 11 genomic regions that enhanced the DREF-induced rough eye phenotype when they were heterozygous for deletions; 7D1; 7D5-6, 9B1-2; 10A1-2, 11A2; 11B9, 21A1; 21B7-8, 32F1-3; 33F1-2, 35D2-4; 35E2-6, 36A8-9; 36E1-2, 36E4-36F1; 38A6-7, 57B4; 58B, 61C3-4; 62A8, 61C3-4; 61A8. These may contain genes that negatively regulate DREF functions, although it is not clear which genes in these regions are responsible for modification of the DREF-induced rough eye phenotype. In some cases, we observed contradictory effects of deficiencies on the DREF-induced rough eye phenotype. For example, SD[R77] deficiency enhanced the rough eye phenotype whereas similarly sized and overlapped deficiencies exerted no effect. There also appear to be cases of overlapping and contained deficiencies (e1801f1 and A246) that have opposite effects. Because the deficiency chromosome has been balanced for the long-term, several mutations may be accumulated on these deficiency lines. Therefore, genes that genetically interact with DREF should be screened with other types of mutant collections mutagenized by P-element insertion or EMS. In any event, the transgenic flies established in the study and the information on genomic regions affecting the DREF-induced rough eye phenotype should help us to identify novel targets of DREF and its positive and negative regulators in Drosophila.
ACKNOWLEDGMENTS
We are grateful to S. Hayashi, G. Rubin, N. Dyson, and Y. Hiromi for fly stocks; N. Perrimon for pUAST; M. Moore for critical reading of the manuscript; and S. Ito and M. Nozaki for technical assistance.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Asano M, Nevins J R, Wharton R P. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 2.Biggin M D, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 3.Brizuela B J, Kennison J A. The Drosophila homeotic gene moira regulates expression of engrailed and HOM genes in imaginal tissues. Mech Dev. 1997;65:209–220. doi: 10.1016/s0925-4773(97)00081-6. [DOI] [PubMed] [Google Scholar]
- 4.Brook A, Xie J E, Du W, Dyson N. Requirements for dE2F function in proliferating cells and in post-miotic differentiating cells. EMBO J. 1996;15:3676–3683. [PMC free article] [PubMed] [Google Scholar]
- 5.Butler S J, Ray S, Hiromi Y. klingon, a novel member of the Drosophila immunoglobulin superfamily, is required for the development of the R7 photoreceptor neuron. Development. 1997;124:781–792. doi: 10.1242/dev.124.4.781. [DOI] [PubMed] [Google Scholar]
- 6.Caldas C, Aparicio S. Cell memory and cancer-the story of the trithorax and Polycomb group genes. Cancer Metastasis Rev. 1999;18:313–329. doi: 10.1023/a:1006333610078. [DOI] [PubMed] [Google Scholar]
- 7.Carthew R W, Rubin G M. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 8.Cohen S M. Imaginal disc development. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 747–841. [Google Scholar]
- 9.Crook N E, Clem R J, Miller L K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosby M A, Miller C, Alon T, Watson K L, Verrijzer C P, Goldman-levi R, Zak N B. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W, Xie J-E, Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J. 1996;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- 12.Duronio R J, Brook A, Dyson N, O'Farrell P H. E2F-induced S phase requires cyclin E. Genes Dev. 1996;10:2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- 13.Duronio R J, O'Farrell P H, Xie J E, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- 14.Freeman M, Kimmel B E, Rubin G M. Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development. 1992;116:335–346. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- 15.Gerasimova T I, Corces V G. Boundary and insulator elements in chromosomes. Curr Opin Genet Dev. 1996;6:185–192. doi: 10.1016/s0959-437x(96)80049-9. [DOI] [PubMed] [Google Scholar]
- 16.Gerasimova T I, Corces V G. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 17.Harrington E A, Fanidi A, Evan G I. Oncogenes and cell death. Curr Opin Genet Dev. 1994;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 18.Hart C M, Zhao K, Laemmli U K. The scs′ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart C M, Cuvier O, Laemmli U K. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–383. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- 20.Hay B A, Wolff T, Rubin G M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 21.Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base-pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase α and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- 22.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, Ikeda M, Nishimoto Y, Matsukage A. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J Biol Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 23.Hirose F, Yamaguchi M, Matsukage A. Targeted expression of the DNA binding domain of DRE-binding factor, a Drosophila transcription factor, attenuates DNA replication of the salivary gland and eye imaginal disc. Mol Cell Biol. 1999;19:6020–6028. doi: 10.1128/mcb.19.9.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kal A J, Mahmoudi T, Zak N B, Verrijzer C P. The Drosophila Brahma complex is an essential coactivator for the trithorax group protein Zesta. Genes Dev. 2000;14:1058–1071. [PMC free article] [PubMed] [Google Scholar]
- 25.Kennison J A. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 26.Kennison J A, Tamkun J W. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laney J D, Biggin M D. Zeste, a nonessential gene, potentially activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev. 1992;6:1531–1541. doi: 10.1101/gad.6.8.1531. [DOI] [PubMed] [Google Scholar]
- 28.Lyco F, Paro R. Chromosomal elements conferring epigenetic inheritance. Bioessays. 1999;21:824–832. doi: 10.1002/(SICI)1521-1878(199910)21:10<824::AID-BIES4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Matsukage A, Hirose F, Hayashi Y, Hamada K, Yamaguchi M. The DRE sequence TATCGATA, a putative promoter-activating element for Drosophila melanogaster cell-proliferation-related genes. Gene. 1995;166:233–236. doi: 10.1016/0378-1119(95)00586-2. [DOI] [PubMed] [Google Scholar]
- 30.Messmer S, Franke A, Paro R. Analysis of the functional role for Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;41:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 31.Mlodzik M, Baker N E, Rubin G M. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- 32.Mlodzik M, Hiromi Y, Weber U, Goodman C S, Rubin G M. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- 33.Moses K, Rubin G M. glass encodes a site-specific DNA binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 1991;5:583–593. doi: 10.1101/gad.5.4.583. [DOI] [PubMed] [Google Scholar]
- 34.Ohno K, Hirose F, Sakaguchi K, Nishida Y, Matsukage A. Transcriptional regulation of the Drosophila cycA gene by the DNA replication-related element (DRE) and DRE binding factor (DREF) Nucleic Acids Res. 1996;24:3942–3946. doi: 10.1093/nar/24.20.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- 36.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 37.Royzman I, Whittaker A J, Orr-Weaver T L. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu J-R, Choi T-Y, Kwon E-J, Lee W-H, Nishida Y, Hayashi Y, Matsukage A, Yamaguchi M, Yoo M-A. Transcriptional regulation of the Drosophila-raf protooncogene by the DNA replication-related element (DRE)/DRE-binding factor (DREF) system. Nucleic Acids Res. 1997;24:794–799. doi: 10.1093/nar/25.4.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawado T, Hirose F, Takahashi Y, Sasaki T, Shinomiya T, Sakaguchi K, Matsukage A, Yamaguchi M. The DNA replication-related element (DRE)/DRE-binding factor system is a transcriptional regulator of the Drosophila E2F gene. J Biol Chem. 1998;273:26042–26051. doi: 10.1074/jbc.273.40.26042. [DOI] [PubMed] [Google Scholar]
- 40.Staeling-Hampton K, Ciampa P J, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi Y, Hirose F, Matsukage A, Yamaguchi M. Identification of three conserved regions in the DREF transcription factors from Drosophila melanogaster and Drosophila virilis. Nucleic Acids Res. 1999;27:510–516. doi: 10.1093/nar/27.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tillib S, Petruk S, Sedkoy Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax-and Polycomb group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treisman J E, Luk A, Rubin G M, Heberlein U. Eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumkun J W, Deuring R, Scott M P, Kissinger M, Pattatutti A M. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SW12. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 45.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 46.Vazquez M, Moore L, Kennison J A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 47.Wang S L, Hawkins C J, Yoo S J, Muller H A, Hay B A. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 48.White K, Grether M, Abrams J, Young L, Farrell K, Steller H. Genetic control of cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 49.Wilder E L, Perrimon N. Dual function of wingless in the Drosophila leg imaginal disc. Development. 1995;121:477–488. doi: 10.1242/dev.121.2.477. [DOI] [PubMed] [Google Scholar]
- 50.Wolff T, Ready D F. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- 51.Yamaguchi M, Hayashi Y, Nishimoto Y, Hirose F, Matsukage A. A nucleotide sequence essential for the function of DRE, a common promoter element for Drosophila DNA replication-related genes. J Biol Chem. 1995;270:15808–15814. doi: 10.1074/jbc.270.26.15808. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi M, Hirose F, Matsukage A. Roles of multiple promoter elements of the proliferating cell nuclear antigen gene during Drosophila development. Genes Cells. 1996;1:47–58. doi: 10.1046/j.1365-2443.1996.03003.x. [DOI] [PubMed] [Google Scholar]
- 53.Yamaguchi M, Hirose F, Inoue Y H, Shiraki M, Hayashi Y, Nishi Y, Matsukage A. Ectopic expression of human p53 inhibits entry into S-phase and induces apoptosis in the Drosophila eye imaginal disc. Oncogene. 1999;18:6767–6775. doi: 10.1038/sj.onc.1203113. [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi M, Hayashi Y, Matsukage A. Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J Biol Chem. 1995;270:25159–25165. doi: 10.1074/jbc.270.42.25159. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi, M., H. Yoshida, F. Hirose, Y. I. Inoue, Y. Hayashi, M. Yamagishi, Y. Nishi, K. Tamai, K. Sakaguchi, and A. Matsukage. Ectopic expression of BEAF32A in the Drosophila eye imaginal disc inhibits differentiation of photoreceptor cells and induces apoptosis. Chromosoma, in press. [DOI] [PubMed]