SUMMARY
Pfs230 domain 1 (Pfs230D1) is an advanced malaria transmission-blocking vaccine antigen demonstrating high functional activity in clinical trials. However, the structural and functional correlates of transmission-blocking activity are not defined. Here, we characterized a panel of human monoclonal antibodies (hmAbs) elicited in vaccinees immunized with Pfs230D1. These hmAbs exhibited diverse transmission-reducing activity, yet all bound to Pfs230D1 with nanomolar affinity. We compiled epitope binning data for seventeen hmAbs and structures of nine hmAbs complexes to construct a high-resolution epitope map and revealed that potent transmission-reducing hmAbs bound to one face of Pfs230D1 while non-potent hmAbs bound to the opposing side. The structure of Pfs230D1D2 revealed that non-potent transmission-reducing epitopes were occluded by the second domain. The hmAb epitope map delineated binary hmAb combinations that synergized for extremely high-potency transmission-reducing activity. This work provides a high-resolution guide for structure-based design of enhanced immunogens and informs diagnostics that measure the transmission-reducing response.
Graphical Abstract
In Brief
There is a limited understanding of the epitopes targeted by antibodies upon vaccination with malaria transmission-blocking vaccines in humans. Tang and Coehlo et al. characterized a panel of monoclonal antibodies elicited in vaccinees and generated a human monoclonal antibody epitope map that defines the area of functional transmission-reducing activity on the surface of the leading malaria transmission-blocking vaccine antigen Pfs230D1.
INTRODUCTION
Malaria transmission-blocking vaccines (TBV) work by eliciting antibodies that target antigens expressed during sexual stage of Plasmodium parasites or mosquitoes, interrupting development within mosquitoes1,2. Instead of providing protection to an individual vaccinee, TBV offer benefits to all members in the community by reducing the number of infectious mosquitoes lowering transmission and the infection rate in the population.
The development of TBV has primarily focused on antigens expressed on the surface of gametes, zygotes and ookinetes. The three current leading TBV candidates, Pfs48/45, Pfs230 and Pfs25, are first identified from animals vaccinated with gametes3,4 or ookinetes5. Pfs25 is expressed on the surface of zygotes/ookinetes and exclusively found on parasites in the mosquito vector5. Monoclonal antibodies to Pfs25 isolated from both mouse and human samples have been characterized and epitopes have been mapped, with varying degrees of transmission-blocking activity6,7. In clinical studies, Pfs25 vaccines have induced antibodies with marginal to moderate transmission-blocking activity in immune sera8,9. Multimerization of Pfs25 on a nanoparticle demonstrates that four doses are required to achieve statistically significant serum activity, and the antibody titers drop rapidly after the fourth dose2,10,11.
Pfs48/45 is a GPI-anchored protein expressed by parasites in both the human host and mosquito vector and is displayed on the surface of midgut gametes and zygotes12–14. Pfs48/45 belongs to the 6-Cys family with three 6-Cys domains15. Pfs48/45 domain 3 is a target of potent transmission-blocking antibodies derived from rodents16,17. The most potent antibody against Pfs48/45 murine monoclonal antibody 85RF45.1 binds to a conserved conformational epitope located away from the GPI-anchor of Pfs48/4518–20.
Pfs230 is also found on the surface of the gametes and zygotes yet lacks a transmembrane domain or signal sequence13,14. Pfs230 is proposed to form a stable complex with Pfs48/45 for surface localization13,14. Disruption of Pfs230 hinders formation of the exflagellation center by affecting emerging male gamete binding to erythrocytes, suggesting a role for Pfs230 in sexual stage progression21. Pfs230 comprises fourteen 6-Cys domains and is another member of the 6-Cys family21–23. The large number of cysteine-rich domains and size of Pfs230 complicate production of the full-length protein. Studies have focused on fragments of Pfs230 encompassing the N-terminal region domains as these regions can induce transmission blocking antibodies22–24. Rabbits immunized with recombinant Pfs230D1M that includes part of the pro-domain and first 6-Cys domain of Pfs230 with no heterologous amino acids using a scalable manufacturing platform, produced transmission-blocking antibodies24 and the immune response could be enhanced by chemical conjugation to carrier protein Exoprotein A (EPA)25. A murine monoclonal antibody, 4F12, isolated upon the immunization with P. falciparum gametes enables the identification of a potent transmission-blocking epitope in Pfs230D120,24. Pfs230D1-EPA formulated in Alhydrogel (Clinicaltrials.gov NCT02334462) induces potent transmission-blocking activity in human sera and enables the isolation of a potent and non-potent transmission-blocking human monoclonal antibodies (hmAbs). LMIV230–01 is a potent hmAb with ~85 % transmission-reducing activity (TRA) at 120 μg/ml and the co-crystal structure reveals a large, broadly-conserved epitope in Pfs230D126. In contrast, a non-potent transmission-reducing hmAb LMIV230–02 exhibits ~10 % TRA at 120 μg/ml. Competitive ELISA and epitope binning both demonstrates these two hmAb recognize non-overlapping epitopes on Pfs230D126.
Recently, Pfs230D1 has emerged as a potent transmission-blocking vaccine. Pfs230D1-EPA formulated in Alhydrogel or AS01 adjuvants has been assessed in Phase 1 and Phase 2 clinical trials27 (Clinicaltrials.gov NCT02334462, NCT02942277, NCT03917654). Here, we report the functional activities and binding epitopes of eight human antibodies isolated from the Phase 1 trial. These structures enable the construction of a human antibody epitope map that is associated with transmission-reducing activity. Furthermore, we report the crystal structure of Pfs230D1D2, containing the first two 6-cys domains, that inform the architecture of Pfs230 and uncover non-functional epitopes on Pfs230D1 that are occluded by the second 6-cys domain. The human antibody epitope map enabled identification of synergistic human monoclonal antibody combinations that enhance transmission-blocking activity. These findings provide a high-resolution guide for structural vaccinology to design immunogens that can improve the potency and durability of transmission blocking vaccines.
RESULTS
Eight Pfs230D1-specific hmAbs possess diverse functional activity with similar affinity.
Peripheral blood mononuclear cells (PBMC) were collected from Malian subjects immunized with either Pfs230D1-EPA/Alhydrogel® (Clinicaltrials.gov NCT02334462) or Pfs230D1-EPA/AS01 (Clinicaltrial.gov NCT02942277). Pfs230D1-specific single-memory B cells were isolated and 131 unique paired of B cell receptor sequences were obtained as previously described26 (Table S1). We reduced the complexity of this large panel of hmAbs by reducing sequences that arose by clonal expansion. Variable regions of heavy and light chain were cloned into a human IgG1 backbone for expression and purification of monoclonal antibodies. 63 of the 131 hmAbs showed clear interaction with Pfs230D1 by ELISA. These 63 hmAbs were screened by the standard membrane feeding assay (SMFA) to identify the transmission-reducing potential of this large panel of hmAbs (Table S2).
Four hmAbs from each adjuvant were selected for further characterization based on multiple criteria including germline usage, high expression, ability to crystallize and functional activity from initial screening: 230AL-18, 230AL-20, 230AL-26 and 230AL-37 from Alhydrogel®; 230AS-18, 230AS-26, 230AS-73 and 230AS-88 from AS01. Variable gene usage of these paired hmAb sequences showed diverse usage of both heavy and light chains (Figure 1A), with only 230AS-18 and 230AL-37 using the same gene family for heavy and light chains. 230AS-18 and 230AL-37 arose from different individuals receiving different formulations: 230AS-18 from the Pfs230D1-EPA/AS01 trial and 230AL-37 from the Pfs230D1-EPA/Alhydrogel trial. Sequence alignment of both heavy and light chains of these two hmAbs showed their complementarity-determining regions (CDRs) are different (Figure S1).
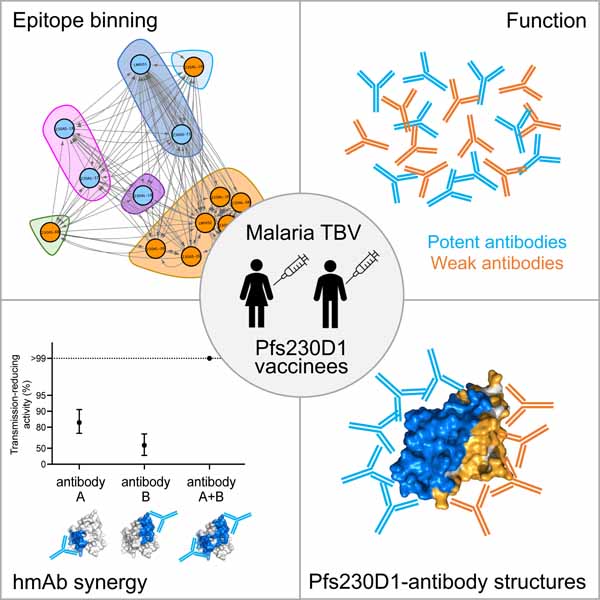
Figure 1. Human monoclonal antibodies isolated from two clinical studies on Pfs230D1-EPA vaccines possess diverse variable gene usage, functional activity and epitope bins.
A) Variable gene usage of 8 hmAbs isolated in the present study. B) Transmission-reducing activities (TRA) of individual hmAb. Data are the results of four separate feeds in the presence of human complement. The best estimated percentage TRA for each hmAb is depicted in bars. C) Epitope binning of human anti-Pfs230D1 scFvs. Biotinylated Pfs230D1 was immobilized on biosensors. Saturating scFvs are listed on the left and the competing scFvs are listed on the top. Reported scores are a percentage of total binding response of that scFv in the absence of competitor scFv. Values <50% are shaded in black. Any experiments with >100% and <0% were given a score of 100 and 0, respectively. Potential epitope bins are grouped and highlighted. D) Network analysis of epitope binning for seventeen hmAbs isolated from two immunization schemes of Pfs230D1-EPA. Data for eight hmAbs are reported in the current study and data for an additional nine reported previously 26. The blue and orange nodes represent potent and non-potent transmission-reducing hmAbs, respectively, and arrows depict epitope binning orientation with the length of the connection weighted by percent binding.
The hmAbs were evaluated for transmission-reducing activity (TRA) by standard membrane feed assay (SMFA) in the presence of human complement. Four hmAbs (230AS-18, 230AS-73, 230AL-18 & 230AL-37) exhibited >80% TRA at a concentration of 100 μg/ml (Figure 1B), called “potent” hmAb herein, while the other four hmAbs (230AS-26, 230AS-88, 230AL-20 & 230AL-26) showed no to weak TRA (<60%). In contrast, all eight hmAbs bound tightly to Pfs230D1 at nanomolar range with KD values 0.3–2.7 nM (Table 1 and S3; Figure S2). This indicates affinity alone is insufficient to explain the diverse transmission-reducing activity of these hmAbs.
Table 1. Binding affinities of IgGs towards Pfs230D1a.
| mAbs | Pfs230D1 | KD (x 10−9 ± SEM M) | ka (x 104 ± SEM 1/Ms) | kdis (x 10−4 ± SEM 1/s) |
|---|---|---|---|---|
| 230AS-18 | Wild-type | 0.85 ± 0.07 | 16.94 ± 0.09 | 1.43 ± 0.13 |
| G605S | 3.92 ± 0.17 | 10.39 ± 0.12 | 4.09 ± 0.22 | |
| G605R | 12.18 ± 0.62 | 6.54 ± 0.84 | 8.89 ± 0.39 | |
| D714N | 30.88 ± 1.30 | 7.81 ± 0.14 | 23.22 ± 0.78 | |
| 230AL-37 | Wild-type | 0.34 ± 0.01 | 53.85 ± 0.82 | 1.81 ± 0.11 |
| G605S | 0.65 ± 0.03 | 21.42 ± 0.14 | 1.47 ± 0.08 | |
| G605R | 5.01 ± 0.28 | 8.51 ± 0.09 | 4.26 ± 0.13 | |
| D714N | 14.96 ± 0.23 | 29.12 ± 0.26 | 42.18 ± 0.50 | |
| 230AL-18 | Wild-type | 2.50 ± 0.16 | 25.40 ± 0.56 | 6.19 ± 0.24 |
| G605S | 2.92 ± 0.04 | 27.31 ± 0.28 | 7.16 ± 0.80 | |
| G605R | 3.34 ± 0.08 | 25.91 ± 0.25 | 8.51 ± 0.05 | |
| 230AS-73 | Wild-type | 1.61 ± 0.08 | 31.18 ± 0.39 | 4.30 ± 0.87 |
| G605S | 1.35 ± 0.04 | 35.84 ± 0.76 | 7.77 ± 0.10 | |
| G605R | 2.03 ± 0.21 | 33.51 ± 0.93 | 6.91 ± 0.90 | |
| A699T | 3.83 ± 0.13 | 25.07 ± 0.65 | 9.51 ± 0.15 | |
| 230AS-88 | Wild-type | 1.06 ± 0.05 | 9.87 ± 0.10 | 1.02 ± 0.02 |
| 230AL-26 | Wild-type | 2.68 ± 0.23 | 8.39 ± 0.07 | 2.53 ± 0.16 |
| 230AS-26 | Wild-type | 0.58 ± 0.01 | 43.81 ± 0.38 | 2.51 ± 0.06 |
| 230AL-20 | Wild-type | 0.31 ± 0.06 | 42.87 ± 0.56 | 1.31 ± 0.17 |
The kinetics were determined using biolayer interferometry (BLI). The data were fitted using a 1:1 binding model. Three technical replicates were performed in each biological replicate. The averages for three biological replicates are shown.
The hmAbs bind to distinct regions of Pfs230D1
We used biolayer interferometry (BLI) to characterize the binding epitopes of the hmAbs on Pfs230D1 by conducting an in-tandem competitive binding assay among all eight hmAbs from the current study as well as two previously reported hmAbs (LMIV230-01 and LMIV230-02)26. Biotinylated Pfs230D1 was immobilized on the biosensors and binding of single-chain fragment variables (scFv) of all hmAbs was recorded. 230AL-18 scFv possessed a fast dissociation rate from Pfs230D1 and therefore was only used as competing antibody. As reported previously, LMIV230-01 is a potent transmission-reducing hmAb (~85 % TRA at 120 μg/ml) of which the binding epitope has been structurally defined26. In contrast, LMIV230-02 is a weak transmission-reducing hmAb (~10 % TRA at 120 μg/ml) and does not compete with binding of LMIV230-0126, which is consistent with our epitope binning results (Figure 1C).
Among the ten hmAbs, we identified six bins of competing epitopes based on the complete interaction pattern from all binary comparisons (Figure 1C and S3). Both 230AS-73 and LMIV230–01 were potent hmAb that showed similar interaction pattern suggesting they share overlapping epitopes which form Bin 1. 230AL-20 competed with LMIV230–01 and 230AS-73 in one orientation but exhibited lower competition in the alternate orientation. These properties suggest 230AL-20 segregates alone into Bin 1a. Two hmAbs, 230AS-18 and 230AL-37 that shared the same gene family for pairing exhibited a similar binding pattern of non-competing with majority of hmAb forms Bin 2. 230AL-18 alone showed a very different binding pattern and was assigned to Bin 3. Bin 4 was comprised of hmAbs 230AS-26, LMIV230–02 and 230AL-26, which strongly competed among themselves, while Bin 5 was comprised of hmAb 230AS-88 that competed with a different set of hmAbs. Bins 1, 2 and 3 comprised all the potent transmission-reducing hmAbs suggesting there is a distinct surface on Pfs230D1 that can effectively block transmission.
We extended the epitope binning analysis to incorporate data for additional hmAbs previously reported26 that have a TRA <20 % at 375 μg/ml using a network analysis. These previous reported hmAbs were isolated from the Pfs230D1-EPA/Alhydrogel clinical study and were previously referred to as GKV hmAbs. We have relabeled them here to 230AL- hmAbs to reflect their consistent source with the other hmAbs in this study. This analysis revealed all the additional no or low-potency hmAbs segregated into Bin 4, the largest bin for non-potent hmAbs (Figure 1D). These results suggest the epitopes for most non-potent transmission-reducing hmAbs clustered on the same surface of Pfs230D1.
A high-resolution human antibody epitope map reveals the potent transmission-reducing epitopes are located on one face of Pfs230D1.
To delineate the binding epitopes of each hmAb in this study, we solved eight crystal structures of Pfs230D1-scFv complexes (Figure 2; Table S4). These structures showed all the hmAbs have distinct epitopes on Pfs230D1 with partial overlap between epitopes, and 230AL-37 and 230AS-18 shared the most similar epitope (Figure 2A and S4G). The discontinuous conformational binding epitopes on Pfs230D1 for both 230AL-18 and 230AS-73 comprised contact sites of beta strands and loops (Figure 2B and 2C). On the other hand, the epitopes on Pfs230D1 for 230AL-37 and 230AS-18 only consisted of loops (Figure 2D and 2E). Altogether, the binding epitopes of these potent transmission-reducing hmAbs (230AS-18, 230AS-73, 230AL-18, 230AL-37 and LMIV230–0126 revealed a large and contiguous surface on Pfs230D1 (Figure 2A). The 230AL-18 epitope overlapped with 230AS-18, 230AS-73, 230AL-37 and LMIV230–01 (Figure S4A–D). The two hmAbs 230AS-18 and 230AL-37, which used the same variable gene families, exhibited very similar binding epitopes that highly overlapped with each other (Figure S4G). The hmAb 230AL-18 epitope comprised almost all the residues recognized by the mouse mAb 4F12 (Figure S5)20. Together, these data showed that a single large surface of Pfs230D1 contains all the potent transmission-reducing epitopes.
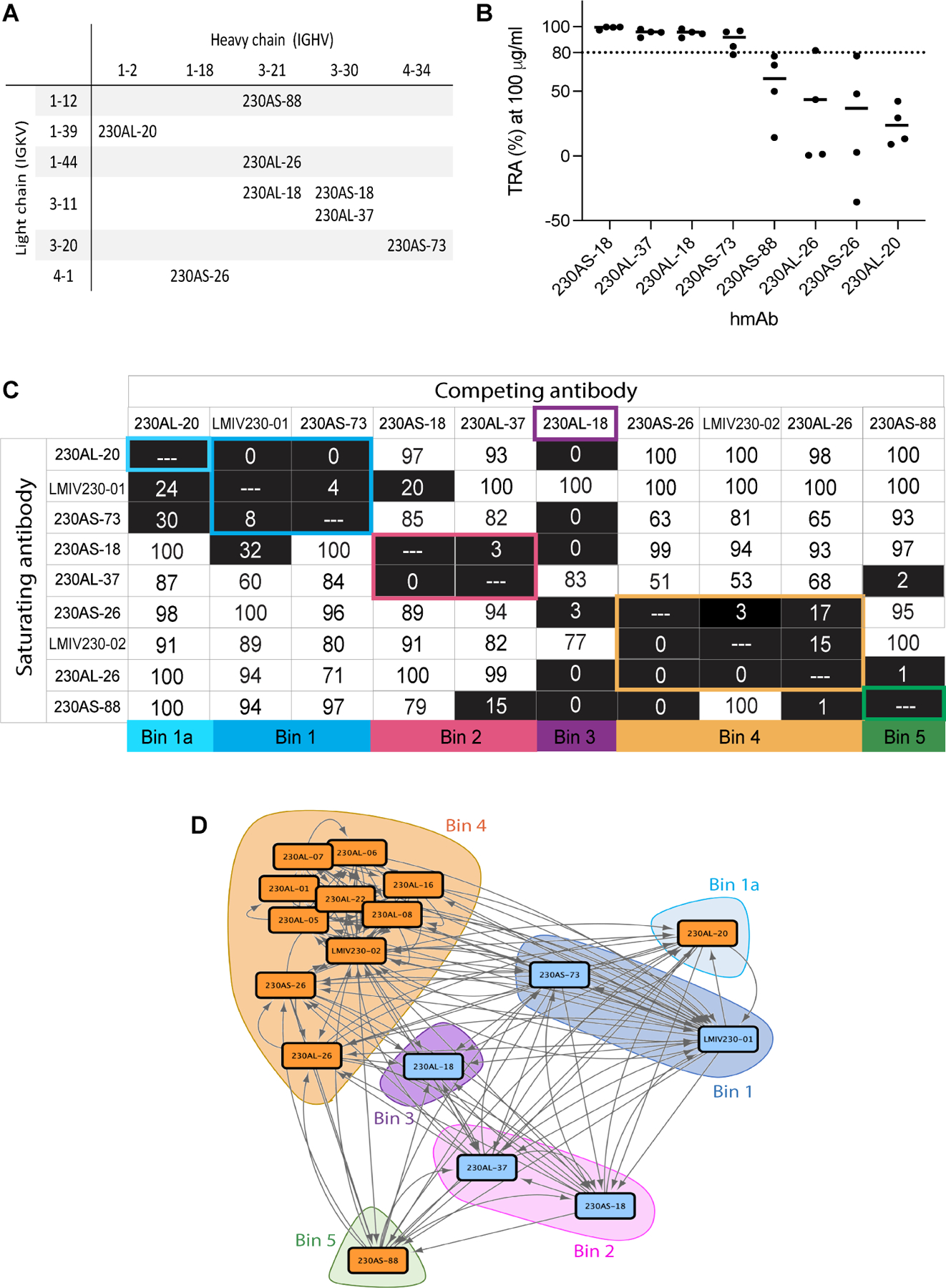
Figure 2. Structures of Pfs230D1 in complex with hmAbs revealed five potent and four weak transmission blocking antibody epitopes.
(A) Five crystal structures of Pfs230D1 in complex with potent transmission-reducing antibodies (exhibit >80% TRA at 100 μg/ml) in scFv formats. The Pfs230D1 are shown in surface representation. The scFvs are showed in ribbon representations in different color, and their binding interface on Pfs230D1 are highlighted with the corresponding color as the scFv. The structure of LMIV230–01 was previously reported (PDB:7JUM) 26. The surface representation of Pfs230D1 in the middle summarized the binding epitopes of the antibodies. (B-E) The binding epitopes of each potent transmission-reducing hmAbs. Pfs230D1 in the same orientation as (A) are shown in gray ribbons with N-terminal residue L557 labelled as N. Residues at binding interface for (B) 230AL-18, (C) 230AS-73, (D) 230AL-37 and (E) 230AS-18 are highlighted in color and selected residues are labeled. (F) Four crystal structures of Pfs230D1 in complex with weak transmission-reducing antibodies (exhibit <60 % TRA at 100 μg/ml) in scFv formats. The Pfs230D1 represented in surfaces are rotated 120° about the y-axis. The scFvs are shown in ribbon representations in different color, and their binding interface on Pfs230D1 are highlighted with the corresponding color as the scFv. The surface representation of Pfs230D1 in the middle summarized the binding epitopes of the antibodies. (G-J) The binding epitopes of each weak transmission-reducing hmAbs. Pfs230D1 in the same orientation as (F) are shown in gray ribbons with N-terminal residue L557 labelled as N. The residues at binding interface of (G) 230AL-26, (H) 230AS-26, (I) 230AS-88 and (J) 230AL-20 are highlighted in color and selected residues are labeled.
In contrast, the other four hmAbs (230AS-26, 230AS-88, 230AL-20 & 230AL-26) with no or weak TRA (<60% at 100 μg/ml) showed a large contiguous surface located on the other side of Pfs230D1 from the potent transmission-reducing face (Figure 2F). The 230AL-26 epitope overlapped with 230AS-88 and with 230AS-26 (Figure S4H–I), and while the 230AL-20 epitope did not overlap with any of the other three hmAbs, it is adjacent to 230AS-26 which extended the map. The discontinuous conformational binding epitopes on Pfs230D1 for 230AL-26 and 230AS-88 comprised contact sites of both beta strands and loops (Figure 2G and 2I). On the other hand, the epitopes on Pfs230D1 for 230AS-26 and 230AL-20 only consisted of loops (Figure 2H and 2J).
Two non-potent transmission-reducing hmAbs did not bind to full-length Pfs230 on the surface of live gametes.
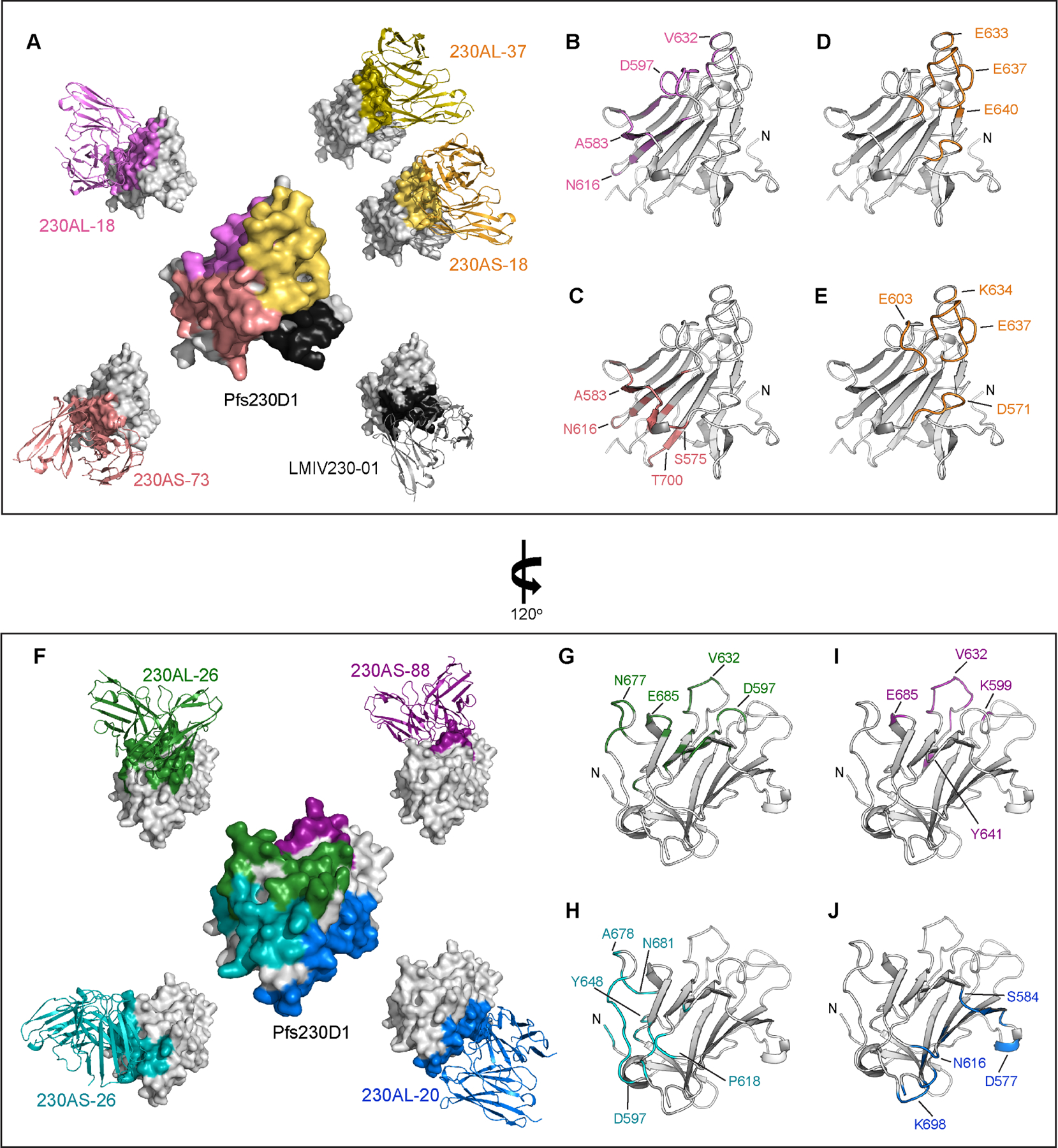
Pfs230D1 contains the first of fourteen 6-cys domains of full-length Pfs230. Presumably, the isolation of Pfs230D1 might expose surfaces which are buried by other 6-Cys domains in the full-length protein. We examined whether the hmAbs can interact with full-length Pfs230 expressed on the surface of gametes. All potent transmission-reducing hmAbs (230AS-18, 230AS-73, 230AL-18 & 230AL-37) recognized full-length Pfs230 on the surface of gametes (Figure 3A, top panel). However, two (230AS-26 & 230AL-20) out of four non-potent transmission-reducing hmAbs failed to recognize the gametes (Figure 3A, bottom panel). This suggests the binding epitopes of both 230AS-26 and 230AL-20 are not accessible in full-length Pfs230, which explains their no or low functional activity.
Figure 3. The D2 domain of Pfs230 overlapped with non-functional epitopes.
(A) Immunofluorescence of live gametes. Female NF54 P. falciparum gametes were incubated with various hmAb reported in green. The nucleus of the gametes were stained with Hoechst in blue. Scale bar is 20μm. The experiment was performed in duplicate. (B) Structure of Pfs230D1D2. D1 domain is colored in gray and D2 domain in orange. Disulfide bonds are shown in blue sticks. (C) Structure of Pfs230D1D2 and overlay with the epitope of 230AS-26. D1 domain is shown in gray surface and D2 domain is shown in orange ribbon. Binding epitopes of 230AS-26 (cyan), 230AS-88 (purple), 230AL-20 (blue) and 230AL-26 (green) are highlighted using the same color as used in the labels. (D) Binding of Pfs230D1D2 to various non-potent transmission-reducing mAbs. hmAbs were immobilized on BLI biosensors and allowed to bind Pfs230D1D2 protein. BLI responses from association and dissociation were recorded in nm.
The D2 domain of Pfs230 overlaps with non-potent epitopes.
The binding epitopes of the two hmAbs (230AS-26 & 230AL-20) that could not bind to gametes are close to the C-terminus of Pfs230D1. We therefore speculated that these epitopes are buried by other domains in full-length Pfs230 especially by the D2 domain, the second 6-Cys domain. We solved the crystal structure of Pfs230D1D2 in complex with LMIV230–01 scFv (Figure 3B; Table S4). The D2 domain adopted a typical 6-Cys fold consisting of a mix of beta strands organized in a five-on-four beta sandwich. The overall structure of D2 highly resembled the D1 domain and was stabilized by two disulfide bridges (C737-C781, C804-C862). The D2 domain was orientated ~90o to D1 as indicated by the direction of the beta sandwich. Similar spatial position between two 6-Cys domains can also be observed in dual 6-Cys proteins Pf1228 and Pf4129, regardless of the length of the linker that connects the two domains (Figure S6).
Examination of the hmAb epitopes in the context of Pfs230D1D2 structure revealed the D2 domain occluded access to the epitope of hmAb 230AS-26 (Figure 3C). Consistent with this observation, 230AS-26 did not interact with Pfs230D1D2 in a protein-protein interaction study (Figure 3D) nor with full-length Pfs230 as expressed on the parasite surface (Figure 3A) as the epitope is buried. Furthermore, LMIV230–02 shared very similar binding patterns to 230AS-26, suggesting the binding epitope of LMIV230–02 highly overlaps with 230AS-26 and is also occluded by Pfs230D2. Although the binding epitope of 230AL-20 did not overlap with the D2 domain, it was close to the C-terminus of the D2 domain. We speculate the remaining domains of full-length Pfs230 might block the epitope preventing binding of 230AL-20 to full-length Pfs230.
Polymorphisms reveal broadly conserved epitopes and potential escape variants
Analysis of 2,512 Pfs230D1 sequences (from MalariaGen30, representing diversity across Africa and Asia where P. falciparum malaria is endemic) revealed ten polymorphic residues on Pfs230D1 (Figure 4A). Some of these polymorphic residues are found within the epitopes of potent transmission-reducing hmAb (Figure 4B–E). Both 230AS-73 and 230AL-18 were unaffected by polymorphic variation in their epitopes as their binding affinities remained similar when compared across variants (Table 1 and S5). These, along with LMIV230–01 that has been previously reported26, recognize broadly conserved epitopes and are likely to be broadly transmission-reducing. The high- frequency polymorphic variant G605S/G605R (91.49%) and the low-frequency variant D714N (0.04%) were found within the epitopes of 230AS-18 and 230AL-37 (Figure 4C and 4D). Alteration of G605 substantially reduced the binding affinity of 230AS-18 and 230AL-37 by up to ~15-fold (Table 1). In addition, the low-frequency variant D714N resulted in a ~40-fold reduction in binding affinity to both hmAbs compared to the wild-type (Table 1).
Figure 4. A limited number of polymorphisms in Pfs230D1 are found within potent transmission blocking epitopes.
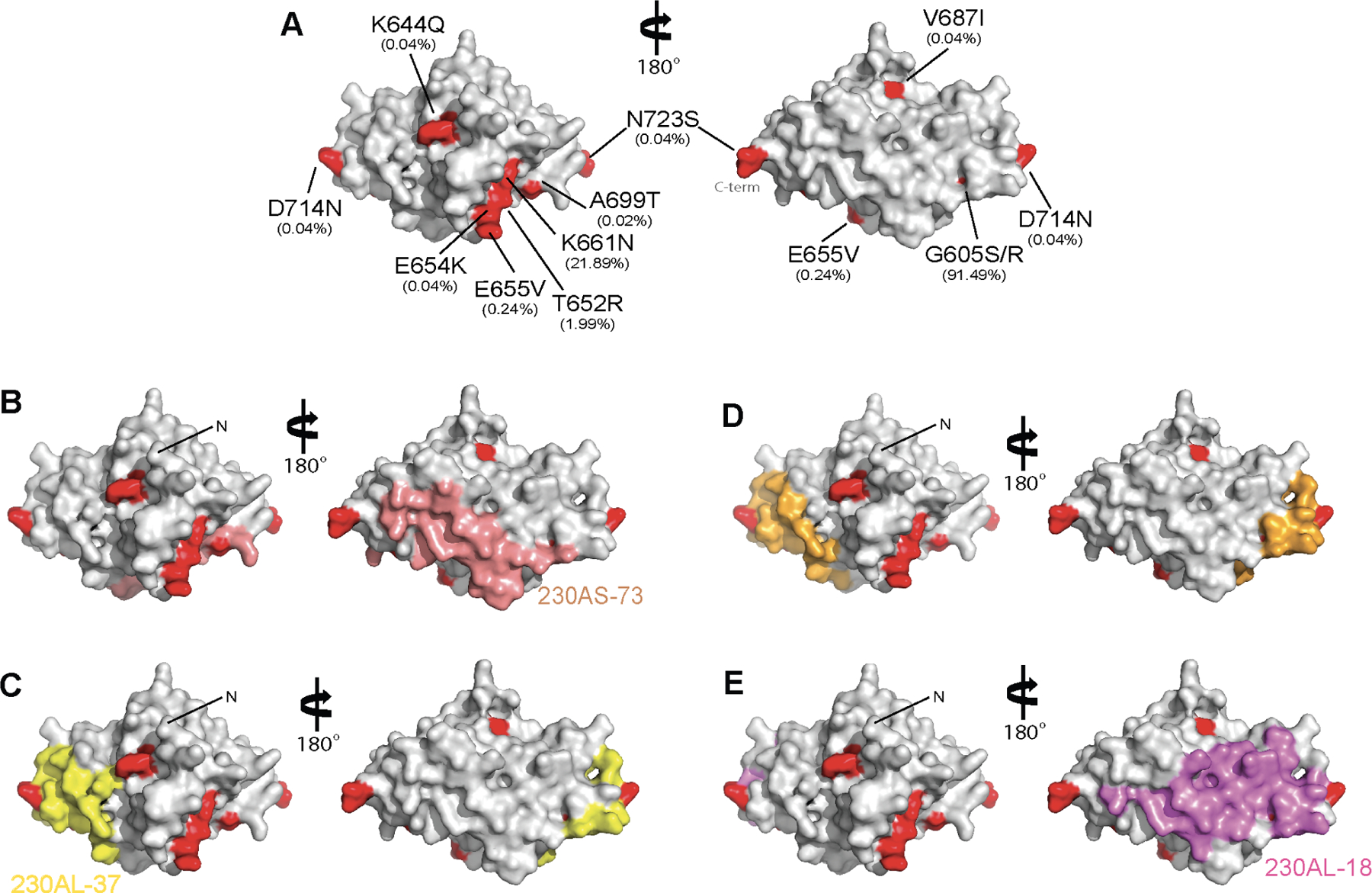
(A) Ten polymorphic residues were observed in 2,512 Pfs230D1 sequences from MalariaGen.net. Pfs230D1 is in gray surface and polymorphic residues are highlighted in red. The observed frequency is in parenthesis. (B) Surface representation of Pfs230D1 showing the binding epitope of 230AS-73 in salmon; (C) 230AL-37 in yellow; (D) 230AS-18 in gold; and (E) 230AL-18 in violet. N-terminal residue L557 are labelled as “N”. The orientation of Pfs230D1 in all panels is identical.
Residue G605 is in a loop region that is in the vicinity of the epitope for light chain CDRs of 230AS-18 and 230AL-37. Furthermore, G605 is followed by a conformationally rigid amino acid P606. Mutation of the small glycine residue into amino acids with larger side chains will likely alter the conformation of the loop and change the epitope of Pfs230D1 which becomes less favorable for the binding of 230AS-18 and 230AL-37.
Furthermore, D714 is in the loop of Pfs230D1 that protrudes to interact with the heavy chain CDR2 of 230AS-18 and 230AL-37. In wild-type Pfs230, D714 forms hydrogen bond with the highly positively charged pocket in 230AS-18 and 230AL-37. Mutation from a negatively charged aspartic acid to a neutral asparagine residue may not be compatible with the binding pocket of these two hmAbs and therefore reduce their binding affinities.
The hmAb epitope map delineates potent synergistic antibody combinations
We have identified the binding epitopes of a total of five potent transmission-reducing hmAbs, including LMIV230–01 from a previous study26, They all showed a distinct binding epitope on Pfs230D1, except for 230AS-18 and 230AL-37 that have largely overlapping epitopes (Figure S4G). We therefore selected four hmAbs (230AL-18, 230AL-37, 230AS-73 and LMIV230–01) to evaluate for additive or synergistic inhibition activity in SMFA. Four combinations allow for simultaneous binding of both hmAbs as their epitopes did not overlap: 230AL-18+230AL-37, 230AL-18+LMIV230–01, 230AL-37+230AS-73 and 230AL-37+LMIV230–01 (Figure 5A). In contrast, two combinations contain hmAbs with partially overlapping epitopes: 230AL-18+230AS-73 and 230AS-73+LIMV230–01 (Figure 5A). We first tested four hmAbs individually from 2–50 μg/ml (Figure S7); the IC80 (the concentration which gives 80% of TRA in SMFA) ranged from 2.3 to 15.0 μg/ml. Therefore, we used a fixed concentration of 5 μg/ml for each antibody when either alone or in combination (a total of 10 μg/ml of hmAbs in a feeder) in three subsequent independent SMFAs. Each individual hmAb at 5 μg/ml showed TRA from 56 to 84 % (Figure 5B, left panel). The two hmAb combinations with overlapping epitopes, 230AL-18+230AS-73 and 230AS-73+LIMV230–01, did not show a significant difference from the corresponding theoretical Bliss additive values. The other combinations of 230AL-18+230AL-37, 230AL-18+LMIV230–01, 230AL-37+230AS-73 and 230AL-37+LMIV230–01, where hmAbs could simultaneously bind, all showed significantly higher inhibition than the theoretical additive values with p<0.001 (Figure 5B, right panel). Our results indicate that these four combinations work synergistically and demonstrate the human antibody epitope map can delineate potent synergistic antibody combinations that may be suitable for use as prophylactic or therapeutic combinations.
Figure 5. Pfs230D1 hmAbs display a synergistic effect in SMFA.
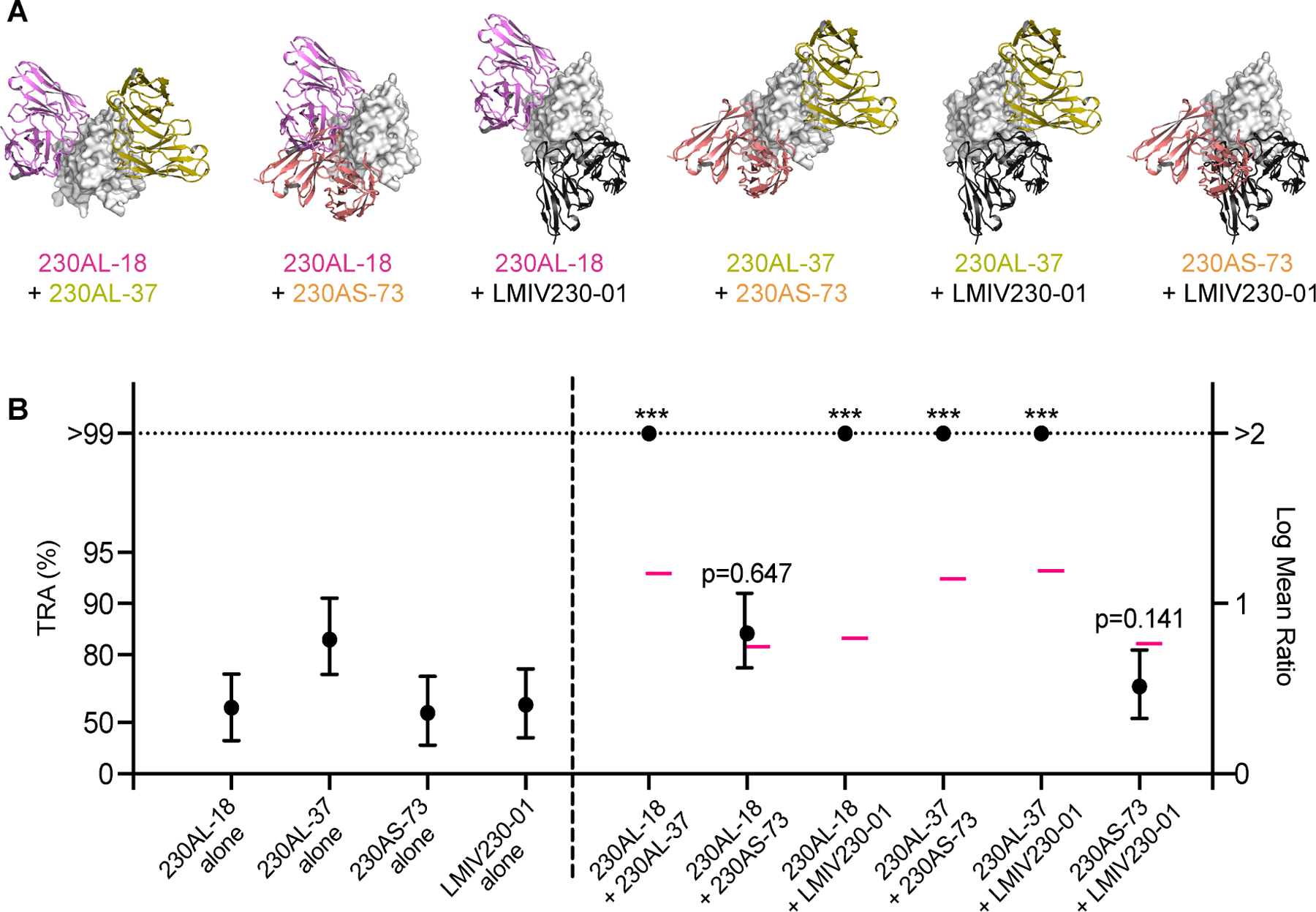
(A) Cartoons depicted the binding epitopes of six paired combinations among four hmAbs (ribbons) on Pfs230D1 (gray surface). (B) Four hmAbs were tested at 5 μg/ml either alone or in combinations of two hmAbs. The best estimated % TRA (black filled circles) and the 95% confidence interval (error bars) from three independent assays are shown. The theoretical additive values calculated using the Bliss independence model are shown in pink bars (–). Asterisks indicate that the mixture group showed significantly higher %TRA than the theoretical additive inhibition (*** p< 0.001). The ratio of mean oocyst numbers is plotted on a log scale (Log Mean Ratio, right side of y-axis), and the associated TRA (%) values are shown on the left side of the y-axis.
DISCUSSION
In 2011, the Malaria Eradication Research Agenda (MalERA) expanded the concept of transmission-blocking vaccines to the broader array of Vaccines that Interrupt Malaria Parasite Transmission (VIMT)31. TBVs have been identified as a potential tool for malaria eradication and a priority for development31. Pfs230 is found on the surface of gametocytes and zygotes and is a leading TBV candidate5,32. Children and adults in endemic areas may develop naturally acquired immune response to Pfs23033. Vaccine responses to Pfs230 may therefore be boosted during infections and this could prolong durability of vaccine activity.
In clinical studies, we demonstrated Pfs230D1 can elicit functional antibodies that inhibit oocyst formation and thus interfere with transmission of malaria parasites to mosquitoes27. Here, a total of nine epitopes were defined at atomic resolution for hmAbs that exhibited transmission-reducing activities ranging from 25% to ~100% at 100 μg/ml. We constructed a high-resolution human antibody epitope map enabled by structural, biophysical, and functional analysis of the human antibody response from two clinical studies. Five potent transmission-reducing hmAbs (TRA > 80% at 100 μg/ml) bound to one side of Pfs230D1, forming a large contiguous surface (Figure 6). In contrast, four non-potent transmission-reducing hmAbs bound to the other side of Pfs230D1, opposite to the functional surface (Figure 6). This human antibody epitope map informs fundamental host-parasite biology, vaccinology, diagnostics, parasite immune evasion and escape, and a path to the development of potent and durable TBVs. As proof-of-concept, we established that the human antibody epitope map can enable the design of potent synergistic hmAb combinations that disrupt transmission. Another approach to improve vaccine design is to leverage the epitope map to preserve the functional epitopes and eliminate the poor transmission-reducing epitopes on Pfs230D1 to direct B-cell responses.
Figure 6. Human antibody epitope map of Pfs230D1.
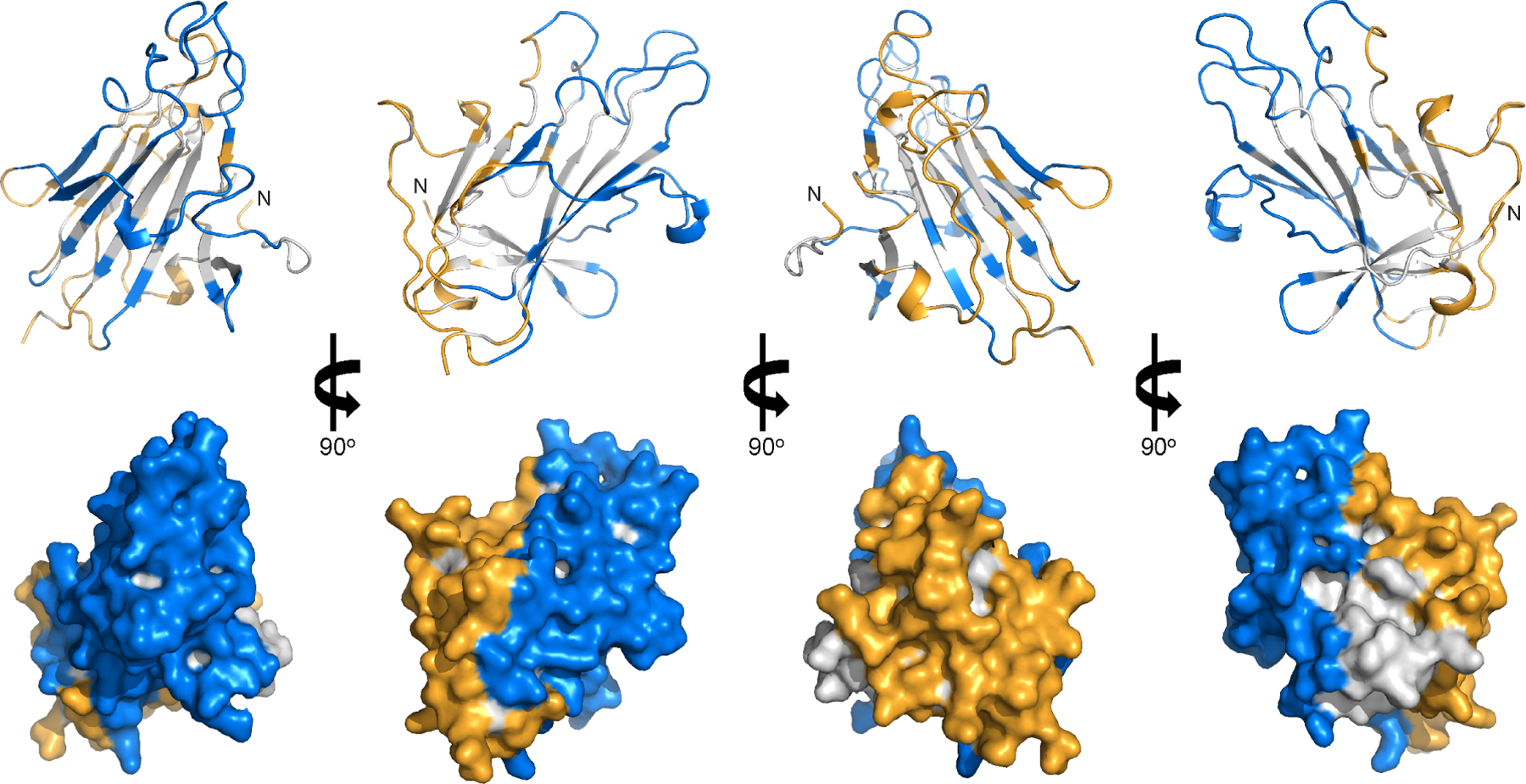
Pfs230D1 is shown as gray ribbons (top panel) and surface (bottom panel) representations. Residues in potent transmission-reducing epitopes are in blue; non-potent transmission-reducing epitopes and D2 binding epitope are in orange. The N-terminal residue L557 are labelled as “N”.
Furthermore, we determined the structure of the dual 6-cys domain Pfs230D1D2. Combining the structure of Pfs230D1D2 with the human antibody epitope map revealed an exposed surface on Pfs230D1 which has poor transmission-reducing activity and is buried in Pfs230D1D2. The position of the D2 domain largely overlapped with the non-potent antibody 230AS-26 (Figure 3C) and occluded this epitope in Pfs230D1D2. This is consistent with the results of protein-protein interaction and direct parasite binding studies where 230AS-26 interacted with neither Pfs230D1D2 protein (Figure 3D) nor full-length Pfs230 on the surface of gametes (Figure 3A). Similarly, the lack of interaction of 230AL-20 with gametes (Figure 3A) also suggests inaccessibility of the epitope in the full-length Pfs230. Finally, 230AS-88 and 230AL-26, which showed 59 and 44 %TRA at 100 μg/ml, could still bind to parasite-expressed Pfs230. It is plausible that these hmAbs may be ineffective as the orientation of the hmAb Fc is not conducive to complement activation/deposition upon binding to parasites. Nevertheless, the human antibody epitope map provides a high-resolution definition of both potent and weak transmission-reducing surfaces of Pfs230D1 that can be exploited for further development.
The antibody epitope map covers the majority of the surface area of Pfs230D1. Inevitably, polymorphic residues will be found within these epitopes. Besides being the most frequently observed (91.49%) variant, G605S/G605R was found at the edge of the epitopes of all four potent transmission-reducing hmAbs isolated in this study (Table 1, Figure 4). Although hmAbs 230AS-73 and 230AL-18 were unaffected by this polymorphic variation, 230AS-18 and 230AL-37 showed reduced binding affinities to polymorphic variants. Moreover, variants containing a low frequency D714N mutation also reduced the binding affinity of Pfs230D1 with 230AS-18 and 230AL-37. These polymorphic variants may represent escape mutants that could arise naturally upon selection driven by immune pressure. The data suggests that broadly conserved epitopes exist in Pfs230D1 and that the variants G605S/R and D714N should be monitored for vaccine escape and considered during vaccine design.
The two individuals enrolled in different immunization schemes produced highly functional antibodies with the same germline usage, very similar transmission-reducing activities, and largely similar epitopes. 230AS-18 was isolated from the Pfs230D1-EPA/AS01 trial and 230AL-37 was isolated from the Pfs230D1-EPA/Alhydrogel trial. Both antibodies arose from germline sequences IGHV3–30 and IGKV3–11, and bound to similar epitopes in Pfs230D1. 230AS-18 and 230AL-37 were two of the most potent transmission-reducing antibodies found in this study and the conserved generation of similar antibodies in different individuals adds strong supporting evidence for the ability of Pfs230D1 to raise potent transmission-reducing antibodies across diverse individuals. These results form the basis for further studies at the population level to identify the proportion of IGHV3–30/IGKV3–11 hmAbs. Lastly, the polymorphisms and potential escape mutants that affect hmAb binding all fall within the epitopes of 230AS-18 and 230AL-37. This is additional supporting evidence that the 230AS-18 and 230AL-37 epitope may be a dominant protective epitope, and the polymorphism arises in response to immune pressure leading to escape variants.
The human antibody epitope map delineated binary combinations of hmAbs that could simultaneously bind to Pfs230D1 with four combinations 230AL-18+230AL-37, 230AL-18+LMIV230–01, 230AL-37+230AS-73 and 230AL-37+LMIV230–01 demonstrating synergistic enhancement of transmission-reducing activity (Figure 5), which is in line with earlier observations that combination of the two mouse antibodies 4F12 and 5H1 enhanced transmission reduction activity20. Previous studies show Pfs230 antibody activity is enhanced by complement23,34. This complement-dependent cytotoxicity will trigger a cascade of reactions causing lysis of the target cells. As more non-competitive protective hmAbs bind to Pfs230D1 simultaneously, the cytotoxicity of the cells is expected to be increased. This is one mechanism for the synergistic enhancement of transmission-reducing activity observed. Alternately, the highly-potent antibodies may function by preventing binding of Pfs230 to interacting partners required for Pfs230 function, and the complete block of the interaction surface by multiple simultaneous binding antibodies may contribute to the synergistic effect observed. This is consistent with a complement-independent transmission-reducing activity observed for murine mAb 4F1220,24 that has an overlapping epitope with hmAb 230AL-18 (Figure S5).
This study has defined a high-resolution human antibody epitope map for the most advanced transmission-reducing vaccine antigen Pfs230D1. The epitope map provides an accurate guide for structural vaccinology to design enhanced immunogens and will inform diagnostics to measure the transmission-reducing response in vaccinees. The human antibody epitope map allows for the generation of antigens that alter the antibody response to distinct faces of Pfs230D1. An ideal antigen would focus the immune response to the potent transmission blocking epitopes and eliminate or reduce immunogenicity to the weak transmission blocking epitopes. Such immunofocusing has been possible by structure-guided or computational approaches that alter the surface of antigens35–37 or by shielding of undesirable surfaces36,38. The paired hmAb combinations that demonstrate extremely high-potency synergistic transmission-reducing activity may be evaluated for use as prophylactic transmission- reducing hmAb therapies. Finally, this work provides fundamental insight into the transmission-reducing biology of Pfs230, relevant to accelerate malaria vaccine development.
Limitations of the study
There is a technical limitation in SMFA to evaluate the synergistic effect of hmAb combinations in a dose-response manner. Previously, we reported lower %TRA estimates have low confidence and require large number of replicates to get statistically meaningful results39. In addition, performing the combination SMFA at higher concentrations is problematic as 100% inhibition is the maximum activity that can be observed. Considered the scope and the significance of the dose response results to the current study, a single dose of 5 mg/ml was reported here.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Niraj H. Tolia (niraj.tolia@nih.gov)
Material availability
All unique and stable reagents generated in this study are available via the lead contacts upon request.
Data and code availability
The crystal structures have been deposited to the Protein Data Bank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The nucleotide sequences from BCR sequencing have been deposited to the GenBank and are publicly available as of the date of publication. Accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contacts upon request.
Experimental model and subject details
Human Ethics Statement
This study was approved by the Ethics review boards from the Faculté de Médecine de Pharmacie et d’OdontoStomatologie (FMPOS), Bamako, Mali, and the US National Institute of Allergy and Infectious Diseases (NIH, Bethesda, MD, USA), as well as the Mali national regulatory authority, and U.S. Food and Drug Administration under IND 16251 (NCT02334462) and IND 17130 (NCT02942277).
Human subjects
Clinical studies NCT02334462 and NCT02942277 enrolled healthy adults (men or non-pregnant, non-breastfeeding women) from Bamako and Bancoumana Mali after informed consent in their native language. Mean age of participants was 37.2 years (range 18–55), and distribution by sex was 70% male / 30% female. Of the eight participants contributing PMBCs for this work, mean age was 37.0 (range 23–50), 6 were male and 2 were female.
Mammalian cell Iines and culture conditions
For recombinant protein expression, human Expi293F™ cells (Thermo Fisher Scientific, sex unknown, not authenticated) were cultured in suspension in GIBCO Expi293™ Expression Medium (Thermo Fisher Scientific) for 5–6 days at 37°C with 80% humidity and 8% CO2 and rotating at 120 RPM.
Method details
Immunization and samples collection
Malian adults were vaccinated with four doses of 40 µg of Pfs230D1-EPA/Alhydrogel® or Pfs230D1-EPA/AS01 at planned study days 0, 28, 168, and 540. From the Pfs230D1-EPA/Alhydrogel® trial, sera and peripheral blood mononuclear cells (PBMCs) were obtained from five participants, 14 days after the 4th dose (day 554). From the Pfs230D1-EPA/AS01 trial, collections occurred 7 days after the 3rd dose (day 175) were obtained from three participants. PBMCs (5 million cells per sample on average) were prepared for isolation of Pfs230D1-specific single memory B cells.
BCR sequencing and data processing
Amplification of BCR heavy and light chains and sequencing of Pfs230D1-specific single memory B cells was performed by iRepertoire Inc. (Huntsville, AL, USA) as previously reported 26. Briefly, nested PCR targeting heavy, and light (kappa and lambda) chains genes were performed in Pfs230D-1 single memory B cells sorted in 96 well PCR plates. Sequencing was performed using the Illumina MiSeq v2 500-cycle kit with 250 paired-end reads. Raw data were demultiplexed by Illumina dual indices. Reads were trimmed according to their base qualities with a 2-base sliding window. If the quality value in this window was lower than 20, the sequence stretch from the window to the 3′ end was trimmed from the original read. Trimmed pair-end reads were joined together through overlapping alignment with a modified Needleman-Wunsch algorithm. If paired forward and reverse reads in the overlapping region were not perfectly matched, both forward and reverse reads were thrown out without further consideration. The merged reads were mapped using a Smith-Waterman algorithm to germline V, D, J, and C reference sequences downloaded from IMGT. To define the CDR3 region, the position of CDR3 boundaries of reference sequences from the IMGT database was migrated onto reads through mapping results, and the resulting CDR3 regions were extracted and translated into amino acids. The data for each chain of the receptor pair begins from within the beginning of framework 1 and extends to the beginning of the C-region (including the isotype).
Expression and purification of human IgGs
Variable regions of Pfs230 hmAb obtained as described previously 26 were inserted into human IgG1 backbone cloned into pHLsec vector and expressed in Expi293 cells to generate IgG as secreted protein. Culture medium were diluted with equal volume of Pierce™ Protein A IgG binding buffer (Thermo Fisher Scientific, Waltham, USA) then mixed with Protein A agarose resin (Gold Biotechnology, St. Louis, USA) at room temperature for 30 min. The resin was washed with 10 bed volumes of binding buffer. IgG was then eluted with 10 bed volumes of Pierce™ Protein A IgG elution buffer (Thermo Fisher Scientific, Waltham, USA). The eluate was neutralized immediately with 0.1-fold of 1 M Tris, pH 8. IgG was further purified with buffer exchange to phosphate-buffered saline (PBS) using Superdex 200 Increase (Cytiva, Marlborough, USA) chromatography. Purified IgGs were pooled, concentrated and stored at −80°C for later use.
Expression and purification of Pfs230D1 variants
For binding study, Pfs230D1 (amino acid 542–676) variants were expressed in Expi293 cells as secreted proteins with 6xHis-tag at C-terminus. After 5 days of expression, the culture medium was mixed with Ni Sepharose Excel resin (Cytiva, Marlborough, USA). The resin was washed with Buffer A (25 mM Tris pH 7.4, 0.3 M NaCl) supplemented with 30 mM imidazole. Proteins were eluted with Buffer A supplemented with 150 mM imidazole. Eluate was concentrated and injected onto Superdex 75 Increase column (Cytiva, Marlborough, USA) equilibrated with 20 mM Tris. pH 8, 100 mM NaCl. Fractions were pooled and stored at −80°C. For crystallization, recombinant Pfs230D1 protein was expressed in Pichia pastoris as previously described 24. Pfs230D1D2 (amino acid 542–886) was expressed in Expi293 cells as secreted proteins with 6xHis-tag at C-terminus.
Purification and formation of protein complexes
The variable segment of the heavy chain sequence of hmAbs were fused to the light chain variable segment by (GGGGS)4 linker and cloned into the vector pHLsec. The plasmids were transfected into Expi293 cells with ExpiFectamine (Thermo Fisher Scientific, Waltham, USA) according to manufacturer’s protocol. All recombinant scFvs were expressed as secreted proteins. The culture medium was loaded into Ni Sepharose Excel (Cytiva, Marlborough, USA) column and the resin was washed with Buffer A (25 mM Tris pH 7.4, 0.3 M NaCl) supplemented with 30 mM imidazole. The scFvs were eluted with Buffer A supplemented with 150 mM imidazole. Eluate was concentrated and injected onto Superdex 75 Increase column (Cytiva, Marlborough, USA) equilibrated with 20 mM Tris pH 8, 100 mM NaCl. Fractions were pooled and stored at −80°C.
Standard membrane feeding assay (SMFA)
SMFA was performed to assess the ability of hmAbs to block the development of P. falciparum strain NF54 oocysts in the mosquito midgut as described previously 39. In brief, 0.15–0.2 % stage V gametocytes were mixed with indicated concentrations of hmAbs, and fed to 3- to 6-day-old female Anopheles stephensi via a parafilm membrane. SMFAs were conducted in the presence of human complement. On day 8 after the feed, mosquito midguts were dissected and the number of oocysts per midgut in 20 mosquitoes were then counted. Percent reduction in mean oocyst intensity (%TRA) of a test sample was calculated relative to the normal mouse IgGs tested at 375 μg/mL in the same assay.
Binding affinities of hmAbs to Pfs230D1
All protein samples were buffer exchanged into 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA. Purified IgG were immobilized onto Anti-hIgG Fc Capture (AHC) biosensors to capture immunogens diluted in HBS-EP+ buffer (Cytiva, Marlborough, USA) to various concentrations. Interactions were detected in OctetRed96e system (Sartorius, Göttingen, Germany) to conduct kinetic studies. Both the association and dissociation steps were 300 seconds long. Kinetic analysis of the data was conducted using Octet Data Analysis HT 12 software (Sartorius, Göttingen, Germany) and used all concentrations of antigen for the kinetic analysis. Three technical replicates were performed per biological replicate.
Epitope binning of anti-Pfs230D1 hmAbs using BLI
Streptavidin (SA) biosensors (Sartorius, Göttingen, Germany) were used to capture biotinylated Pfs230D1. Biotinylated Pfs230D1 and scFvs were diluted in HBS-EP+ buffer (Cytiva, Marlborough, USA) to a concentration of 20 nM and 150 nM, respectively. OctetRed96e system was used to conduct epitope binning experiments in in-tandem format. Briefly, immobilized Pfs230D1 on the biosensor was presented and saturated with the first antibody. The sensors were then presented to the second antibody at half of the concentration used as first antibody. Binding to non-competing epitopes is indicated if saturation with the first antibody does not block binding of the second antibody. The binning data were combined and analyzed using Octet Data Analysis HT 12 software (Sartorius, Göttingen, Germany). A prefuse force directed network weighted by 1-normalized(precent binding) was created in Cytoscape 40.
Co-crystallization and structure determination
Protein complexes were prepared by mixing antigen:scFv in 1:1 ratio. The admixture was purified by size exclusion chromatography. All crystals were grown using sitting-drop vapor diffusion at 18°C. Crystals used for diffraction data were grown in the following conditions: Pfs230D1–230AL-20: 0.16 M ammonium citrate, 18% PEG 3350; Pfs230D1-LMIV230–01-230AL-26: 0.16 M MES, pH 6.5, 1.6 M ammonium sulfate, 10 % dioxane; Pfs230D1–230AS-26: 0.2 M ammonium sulfate, 20% PEG 3350; Pfs230D1–230AS-88: 0.1 M MES, pH 6.0, 1.0 M lithium chloride, 10% PEG 6000; Pfs230D1-LMIV230–01-230AL-18: 0.2 M magnesium formate, 20% PEG 3350; Pfs230D1D2-LMIV230–01: 0.1 M Hepes, pH 6.5, 1.0 M lithium chloride, 20% PEG 6000; Pfs230D1D2–230AS-73: 0.2M sodium dihydrogen phosphate, 20% PEG 3350; Pfs230D1D2–230AL-37: 0.2 M ammonium sulfate, 20% PEG 3350; Pfs230D1–230AS-18: 0.1 M sodium acetate, pH 4.6, 20% PEG 10000. Crystals were cryoprotected in well solution supplemented with 30% PEG 400 or glycerol prior to flash freezing in liquid nitrogen.
X-ray diffraction experiments were carried out on the SER-CAT and GM/CA beamlines, Advanced Photon Source (APS), Argonne National Laboratory at 100 K. Diffraction data were processed with XDS 41. The structure of P230D1-scFv complexes were determined by molecular replacement in PHASER 42 using a model of scFvs created by the SAbPred server 43 and Pfs230D1 (PDB:7JUM) 26. Iterative model building using COOT 44, and refinement using PHENIX 45.
Immunofluorescence of live parasites
Female gametes were prepared as described previously 26. P. falciparum female gametes were incubated with 10 µg/mL of IgGs for 1 h at 37°C. Parasites were then washed with PBS and spun down at 300 × g for 5 min. Cells were then stained with Hoechst 33342 Solution (1:2,500 dilution) for 10 min, washed with PBS and spun down at 300 × g for 5 min. The pellet was diluted in RPMI medium and transferred to a 15-well uncoated Angiogenesis µ-Slide (Ibidi Gmbh, Gräfelfing, Germany). Cells were kept at 37 °C and analyzed using Leica SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany).
Quantification and statistical analysis
For SMFA, the best-estimate of %TRA, the 95% confidence interval (95%CI) and p-value from multiple feeds were calculated using a zero-inflated negative binomial (ZINB) model as before 46. Using the same model, the Bliss additivity phi term, 95%CI and p value (i.e., synergy, additive or antagonism) of two hmAb combination was calculated using the bootstrap samples 47. IC80, IgG concentration which gives 80 %TRA, was calculated from a plot where IgG concentration was transformed in a square-root scale and %TRA was transformed in a Log of mean oocyst ratio (LMR) scale, as described previously 48.
All statistical tests were performed in Prism 8 (GraphPad Software, La Jolla, CA, USA) or R (version 3.5.3, The R Foundation for Statistical Computing) and p-values <0.05 are considered significant.
Additional resources
The trials were registered in clinicaltrials.gov NCT02334462 and NCT02942277.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 230AS-18 | This paper | N/A |
| 230AL-37 | This paper | N/A |
| 230AL-18 | This paper | N/A |
| 230AS-73 | This paper | N/A |
| 230AS-88 | This paper | N/A |
| 230AL-26 | This paper | N/A |
| 230AS-26 | This paper | N/A |
| 230AL-20 | This paper | N/A |
| Bacterial and virus strains | ||
| Biological samples | ||
| Human PBMCs from subjects immunized (co-administration): 40 µg Pfs230D1M-EPA/Alhydrogel® and normal saline on D0, D28, D168, D530 | National Institute of Allergy and Infectious Diseases (NIAID) & Université des Sciences, des Techniques et des Technologies de Bamako (Mali) |
NCT02334462 |
| Human PBMCs from subjects immunized: 40 μg Pfs230D1M-EPA/AS01 on D0, D28, D168 | National Institute of Allergy and Infectious Diseases (NIAID) & Université des Sciences, des Techniques et des Technologies de Bamako (Mali) |
NCT02942277 |
| Chemicals, peptides, and recombinant proteins | ||
| 230AS-18 scFv | This paper | N/A |
| 230AL-37 scFv | This paper | N/A |
| 230AL-18 scFv | This paper | N/A |
| 230AS-73 scFv | This paper | N/A |
| 230AS-88 scFv | This paper | N/A |
| 230AL-26 scFv | This paper | N/A |
| 230AS-26 scFv | This paper | N/A |
| 230AL-20 scFv | This paper | N/A |
| LMIV230–01 scFv | This paper | N/A |
| LMIV230–02 scFv | This paper | N/A |
| Pfs230D1D2 | This paper | N/A |
| Pfs230D1 | Coelho et al. (26) | N/A |
| Protein A agarose resin | Gold Biotechnology | Cat#P-400–100 |
| Superdex 200 Increase | Cytiva | Cat#28990944 |
| Superdex 75 Increase | Cytiva | Cat#29148721 |
| Ni Sepharose Excel | Cytiva | 17371202 |
| Expi293™ Expression medium | Thermo Fisher Scientific | Cat#A1435104 |
| Opti-MEM™ I Reduced serum medium | Thermo Fisher Scientific | Cat#31985062 |
| Critical commercial assays | ||
| Anti-human Fc Capture (AHC) biosensors | Sartorius | Cat#18–5064 |
| Streptavidin (SA) biosensors | Sartorius | Cat#18–5020 |
| Pierce Protein A IgG binding buffer | Thermo Fisher Scientific | Cat#54200 |
| Pierce Protein A IgG Elution buffer | Thermo Fisher Scientific | Cat#21009 |
| ExpiFectamine™ 293 Transfection kit | Thermo Fisher Scientific | Cat#14526 |
| HBS-EP+ buffer | Cytiva | Cat#BR100826 |
| Deposited data | ||
| Crystal structure of Pfs230D1-LMIV230–01-230AL-26 scFv complex | This paper | PDB:7U9E |
| Crystal structure of Pfs230D1–230AS-26 scFv complex | This paper | PDB:7UBS |
| Crystal structure of Pfs230D1–230AS-88 scFv complex | This paper | PDB:7U9W |
| Crystal structure of Pfs230D1-LMIV230–01-230AL-18 scFv complex | This paper | PDB:7UA2 |
| Crystal structure of Pfs230D1–230AL-20 scFv complex | This paper | PDB:7UA8 |
| Crystal structure of Pfs230D1D2–230AL-37 scFv complex | This paper | PDB:7UI1 |
| Crystal structure of Pfs230D1D2–230AS-73 scFv complex | This paper | PDB:7UC8 |
| Crystal structure of Pfs230D1–230AS-18 scFv complex | This paper | PDB:7UCQ |
| Crystal structure of Pfs230D1D2-LMIV230–01 scFv complex | This paper | PDB:7UFW |
| BCR sequences | This paper | GenBank: OP962792 - OP963051; OP971844 - OP971845 |
| Experimental models: Cell lines | ||
| Expi293F™ Cells | Thermo Fisher Scientific | Cat#A14527 |
| Experimental models: Organisms/strains | ||
| Yeast: Pichia pastoris | MacDonald et al. (24) | N/A |
| Parasite: P. falciparum; NF54 strain | Miura et al. (39) | N/A |
| Parasite: P. falciparum female gametes | Coelho et al. (26) | N/A |
| Mosquito: Anopheles stephensi | Miura et al. (39) | N/A |
| Oligonucleotides | ||
| Recombinant DNA | ||
| pHLsec_230AS-18 IgG | This paper | N/A |
| pHLsec_230AL-37 IgG | This paper | N/A |
| pHLsec_230AL-18 IgG | This paper | N/A |
| pHLsec_230AS-73 IgG | This paper | N/A |
| pHLsec_230AS-88 IgG | This paper | N/A |
| pHLsec_230AL-26 IgG | This paper | N/A |
| pHLsec_30AS-26 IgG | This paper | N/A |
| pHLsec_230AL-20 IgG | This paper | N/A |
| pHLsec_LMIV230–01 IgG | This paper | N/A |
| pHLsec_LMIV230–02 IgG | This paper | N/A |
| pHLsec_230AS-18 scFv | This paper | N/A |
| pHLsec_230AL-37 scFv | This paper | N/A |
| pHLsec_230AL-18 scFv | This paper | N/A |
| pHLsec_230AS-73 scFv | This paper | N/A |
| pHLsec_230AS-88 scFv | This paper | N/A |
| pHLsec_230AL-26 scFv | This paper | N/A |
| pHLsec_30AS-26 scFv | This paper | N/A |
| pHLsec_230AL-20 scFv | This paper | N/A |
| pHLsec_LMIV230–01 scFv | This paper | N/A |
| pHLsec_LMIV230–02 scFv | This paper | N/A |
| pHLsec_Pfs230D1D2 | This paper | N/A |
| Software and algorithms | ||
| XDS | Kabsch (41) | https://xds.mr.mpg.de |
| Phenix | Adams et al. (45) | http://www.phenix-online.org |
| Coot | Emsley and Cowtan (44) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Pymol | Schrodinger | https://pymol.org/2/ |
| SAbPred server | Dunbar et al. (43) | http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/ |
| PRISM GraphPad | GraphPad Software, LLC | https://www.graphpad.com/scientific-software/prism/ |
| Octet Data Analysis HT | Sartorius | https://www.sartorius.com/en/products/protein-analysis/octet-bli-detection/octet-systems-software |
| Cytoscape | Cytoscape | https://cytoscape.org |
| Other | ||
Highlights.
Derived a human antibody epitope map of Pfs230D1 by analyzing vaccinated individuals
Potent and non-potent transmission-reducing hmAbs bind opposing faces of Pfs230D1
Protective hmAbs display synergistic enhancement of transmission-reducing activity
Pfs230D2 overlaps with non-potent transmission-reducing hmAb epitopes of Pfs230D1
ACKNOWLEDGEMENTS
This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health and by grant 01665623-GEN from PATH’s Malaria Vaccine Initiative to PED for BCR analysis and hmAb production. We thank PATH’s Malaria Vaccine Initiative for funding and collaborative support by Virginia Price, C. Richter King, Randall MacGill, and Yimin Wu for BCR analysis and hmAb production and characterization. We thank J. Patrick Gorres for proofreading and editing this manuscript. This study used the Office of Cyber Infrastructure and Computational Biology (OCICB) High Performance Computing (HPC) cluster at the National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, MD. We thank members of the Process Development Unit: Raul Herrera, Vu Nguyen and Karine Reiter for assistance in the production of Pfs230D1M. We thank the clinical trial volunteers in Mali for their participation and the protocol study team for their coordination and execution of the vaccine studies. We thank the staff members of SER-CAT and GM/CA beamlines at the Advanced Photon Source, Argonne National Laboratory for beamline supports. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions (see www.ser-cat.org/members.html), and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH1135. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
NHT, WKT, PED, CHC, IS, SAH, CAL and KM are listed as inventors on a provisional patent application related to this work.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Supplemental Table S1 Amino acid sequences of 131 paired B cell receptors. (related to Fig 1)
Supplemental Table S2 SMFA data from initial screening of 63 ELISA positive anti-Pfs230D1 hmAbs (related to Fig 1)
REFERENCES
- 1.Patel PN, and Tolia N (2021). Structural vaccinology of malaria transmission-blocking vaccines. Expert Rev Vaccines 20, 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy PE (2021). Transmission-Blocking Vaccines: Harnessing Herd Immunity for Malaria Elimination. Expert Rev Vaccines 20, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rener J, Carter R, Rosenberg Y, and Miller LH (1980). Anti-gamete monoclonal antibodies synergistically block transmission of malaria by preventing fertilization in the mosquito. Proc Natl Acad Sci U S A 77, 6797–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaushal DC, Carter R, Rener J, Grotendorst CA, Miller LH, and Howard RJ (1983). Monoclonal antibodies against surface determinants on gametes of Plasmodium gallinaceum block transmission of malaria parasites to mosquitoes. J Immunol 131, 2557–2562. [PubMed] [Google Scholar]
- 5.Carter R, Miller LH, Rener J, Kaushal DC, Kumar N, Graves PM, Grotendorst CA, Gwadz RW, French C, and Wirth D (1984). Target antigens in malaria transmission blocking immunity. Philos Trans R Soc Lond B Biol Sci 307, 201–213. [DOI] [PubMed] [Google Scholar]
- 6.Scally SW, McLeod B, Bosch A, Miura K, Liang Q, Carroll S, Reponen S, Nguyen N, Giladi E, Ramisch S, et al. (2017). Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25. Nat Commun 8, 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLeod B, Miura K, Scally SW, Bosch A, Nguyen N, Shin H, Kim D, Volkmuth W, Ramisch S, Chichester JA, et al. (2019). Potent antibody lineage against malaria transmission elicited by human vaccination with Pfs25. Nat Commun 10, 4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ockenhouse CF, Sun PF, Lanar DE, Wellde BT, Hall BT, Kester K, Stoute JA, Magill A, Krzych U, Farley L, et al. (1998). Phase I/IIa safety, immunogenicity, and efficacy trial of NYVAC-Pf7, a pox-vectored, multiantigen, multistage vaccine candidate for Plasmodium falciparum malaria. J Infect Dis 177, 1664–1673. [DOI] [PubMed] [Google Scholar]
- 9.Chichester JA, Green BJ, Jones RM, Shoji Y, Miura K, Long CA, Lee CK, Ockenhouse CF, Morin MJ, Streatfield SJ, and Yusibov V (2018). Safety and immunogenicity of a plant-produced Pfs25 virus-like particle as a transmission blocking vaccine against malaria: A Phase 1 dose-escalation study in healthy adults. Vaccine 36, 5865–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talaat KR, Ellis RD, Hurd J, Hentrich A, Gabriel E, Hynes NA, Rausch KM, Zhu D, Muratova O, Herrera R, et al. (2016). Safety and Immunogenicity of Pfs25-EPA/Alhydrogel(R), a Transmission Blocking Vaccine against Plasmodium falciparum: An Open Label Study in Malaria Naive Adults. PLoS One 11, e0163144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagara I, Healy SA, Assadou MH, Gabriel EE, Kone M, Sissoko K, Tembine I, Guindo MA, Doucoure M, Niare K, et al. (2018). Safety and immunogenicity of Pfs25H-EPA/Alhydrogel, a transmission-blocking vaccine against Plasmodium falciparum: a randomised, double-blind, comparator-controlled, dose-escalation study in healthy Malian adults. Lancet Infect Dis 18, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dijk MR, Janse CJ, Thompson J, Waters AP, Braks JA, Dodemont HJ, Stunnenberg HG, van Gemert GJ, Sauerwein RW, and Eling W (2001). A central role for P48/45 in malaria parasite male gamete fertility. Cell 104, 153–164. [DOI] [PubMed] [Google Scholar]
- 13.Kumar N (1987). Target antigens of malaria transmission blocking immunity exist as a stable membrane bound complex. Parasite Immunol 9, 321–335. [DOI] [PubMed] [Google Scholar]
- 14.Carter R, and Kaushal DC (1984). Characterization of antigens on mosquito midgut stages of Plasmodium gallinaceum. III. Changes in zygote surface proteins during transformation to mature ookinete. Mol Biochem Parasitol 13, 235–241. [DOI] [PubMed] [Google Scholar]
- 15.Carter R, Coulson A, Bhatti S, Taylor BJ, and Elliott JF (1995). Predicted disulfide-bonded structures for three uniquely related proteins of Plasmodium falciparum, Pfs230, Pfs48/45 and Pf12. Mol Biochem Parasitol 71, 203–210. [DOI] [PubMed] [Google Scholar]
- 16.Roeffen W, Teelen K, van As J, vd Vegte-Bolmer M, Eling W, and Sauerwein R (2001). Plasmodium falciparum: production and characterization of rat monoclonal antibodies specific for the sexual-stage Pfs48/45 antigen. Exp Parasitol 97, 45–49. [DOI] [PubMed] [Google Scholar]
- 17.Singh SK, Roeffen W, Mistarz UH, Chourasia BK, Yang F, Rand KD, Sauerwein RW, and Theisen M (2017). Construct design, production, and characterization of Plasmodium falciparum 48/45 R0.6C subunit protein produced in Lactococcus lactis as candidate vaccine. Microb Cell Fact 16, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lennartz F, Brod F, Dabbs R, Miura K, Mekhaiel D, Marini A, Jore MM, Sogaard MM, Jorgensen T, de Jongh WA, et al. (2018). Structural basis for recognition of the malaria vaccine candidate Pfs48/45 by a transmission blocking antibody. Nat Commun 9, 3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X, Zhang C, Zhang S, Dai S, Zhang G, Ge M, Pan Y, Sharkey SM, Graham GW, Hunt A, et al. (2018). Deconvolution of octahedral Pt3Ni nanoparticle growth pathway from in situ characterizations. Nat Commun 9, 4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh K, Burkhardt M, Nakuchima S, Herrera R, Muratova O, Gittis AG, Kelnhofer E, Reiter K, Smelkinson M, Veltri D, et al. (2020). Structure and function of a malaria transmission blocking vaccine targeting Pfs230 and Pfs230-Pfs48/45 proteins. Commun Biol 3, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eksi S, Czesny B, van Gemert GJ, Sauerwein RW, Eling W, and Williamson KC (2006). Malaria transmission-blocking antigen, Pfs230, mediates human red blood cell binding to exflagellating male parasites and oocyst production. Mol Microbiol 61, 991–998. [DOI] [PubMed] [Google Scholar]
- 22.Williamson KC, Keister DB, Muratova O, and Kaslow DC (1995). Recombinant Pfs230, a Plasmodium falciparum gametocyte protein, induces antisera that reduce the infectivity of Plasmodium falciparum to mosquitoes. Mol Biochem Parasitol 75, 33–42. [DOI] [PubMed] [Google Scholar]
- 23.Tachibana M, Wu Y, Iriko H, Muratova O, MacDonald NJ, Sattabongkot J, Takeo S, Otsuki H, Torii M, and Tsuboi T (2011). N-terminal prodomain of Pfs230 synthesized using a cell-free system is sufficient to induce complement-dependent malaria transmission-blocking activity. Clin Vaccine Immunol 18, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald NJ, Nguyen V, Shimp R, Reiter K, Herrera R, Burkhardt M, Muratova O, Kumar K, Aebig J, Rausch K, et al. (2016). Structural and Immunological Characterization of Recombinant 6-Cysteine Domains of the Plasmodium falciparum Sexual Stage Protein Pfs230. J Biol Chem 291, 19913–19922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scaria PV, Chen B, Rowe CG, Jones DS, Barnafo E, Fischer ER, Anderson C, MacDonald NJ, Lambert L, Rausch KM, et al. (2017). Protein-protein conjugate nanoparticles for malaria antigen delivery and enhanced immunogenicity. PLoS One 12, e0190312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho CH, Tang WK, Burkhardt M, Galson JD, Muratova O, Salinas ND, Alves ESTL, Reiter K, MacDonald NJ, Nguyen V, et al. (2021). A human monoclonal antibody blocks malaria transmission and defines a highly conserved neutralizing epitope on gametes. Nat Commun 12, 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Healy SA, Anderson C, Swihart BJ, Mwakingwe A, Gabriel EE, Decederfelt H, Hobbs CV, Rausch KM, Zhu D, Muratova O, et al. (2021). Pfs230 yields higher malaria transmission-blocking vaccine activity than Pfs25 in humans but not mice. J Clin Invest 131, e146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tonkin ML, Arredondo SA, Loveless BC, Serpa JJ, Makepeace KA, Sundar N, Petrotchenko EV, Miller LH, Grigg ME, and Boulanger MJ (2013). Structural and biochemical characterization of Plasmodium falciparum 12 (Pf12) reveals a unique interdomain organization and the potential for an antiparallel arrangement with Pf41. J Biol Chem 288, 12805–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker ML, Peng F, and Boulanger MJ (2015). The Structure of Plasmodium falciparum Blood-Stage 6-Cys Protein Pf41 Reveals an Unexpected Intra-Domain Insertion Required for Pf12 Coordination. PLoS One 10, e0139407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu SJ, Almagro-Garcia J, and McVean G (2018). Deconvolution of multiple infections in Plasmodium falciparum from high throughput sequencing data. Bioinformatics 34, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F, Doumbo OK, Greenwood B, Hall BF, Levine MM, et al. (2011). A research agenda to underpin malaria eradication. PLoS Med 8, e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graves PM, Carter R, Burkot TR, Quakyi IA, and Kumar N (1988). Antibodies to Plasmodium falciparum gamete surface antigens in Papua New Guinea sera. Parasite Immunol 10, 209–218. [DOI] [PubMed] [Google Scholar]
- 33.Ouedraogo AL, Roeffen W, Luty AJ, de Vlas SJ, Nebie I, Ilboudo-Sanogo E, Cuzin-Ouattara N, Teleen K, Tiono AB, Sirima SB, et al. (2011). Naturally acquired immune responses to Plasmodium falciparum sexual stage antigens Pfs48/45 and Pfs230 in an area of seasonal transmission. Infect Immun 79, 4957–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Healer J, McGuinness D, Hopcroft P, Haley S, Carter R, and Riley E (1997). Complement-mediated lysis of Plasmodium falciparum gametes by malaria-immune human sera is associated with antibodies to the gamete surface antigen Pfs230. Infect Immun 65, 3017–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickey TH, Tang WK, Butler B, Ouahes T, Orr-Gonzalez S, Salinas ND, Lambert LE, and Tolia NH (2022). Design of the SARS-CoV-2 RBD vaccine antigen improves neutralizing antibody response. Sci Adv 8, eabq8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauser BM, Sangesland M, St Denis KJ, Lam EC, Case JB, Windsor IW, Feldman J, Caradonna TM, Kannegieter T, Diamond MS, et al. (2022). Rationally designed immunogens enable immune focusing following SARS-CoV-2 spike imprinting. Cell Rep 38, 110561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caradonna TM, and Schmidt AG (2021). Protein engineering strategies for rational immunogen design. NPJ Vaccines 6, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLeod B, Mabrouk MT, Miura K, Ravichandran R, Kephart S, Hailemariam S, Pham TP, Semesi A, Kucharska I, Kundu P, et al. (2022). Vaccination with a structure-based stabilized version of malarial antigen Pfs48/45 elicits ultra-potent transmission-blocking antibody responses. Immunity 55, 1680–1692 e1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura K, Deng B, Tullo G, Diouf A, Moretz SE, Locke E, Morin M, Fay MP, and Long CA (2013). Qualification of standard membrane-feeding assay with Plasmodium falciparum malaria and potential improvements for future assays. PLoS One 8, e57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunbar J, Krawczyk K, Leem J, Marks C, Nowak J, Regep C, Georges G, Kelm S, Popovic B, and Deane CM (2016). SAbPred: a structure-based antibody prediction server. Nucleic Acids Res 44, W474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- 46.Miura K, Swihart BJ, Deng B, Zhou L, Pham TP, Diouf A, Burton T, Fay MP, and Long CA (2016). Transmission-blocking activity is determined by transmission-reducing activity and number of control oocysts in Plasmodium falciparum standard membrane-feeding assay. Vaccine 34, 4145–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azasi Y, Gallagher SK, Diouf A, Dabbs RA, Jin J, Mian SY, Narum DL, Long CA, Gaur D, Draper SJ, et al. (2020). Bliss’ and Loewe’s additive and synergistic effects in Plasmodium falciparum growth inhibition by AMA1-RON2L, RH5, RIPR and CyRPA antibody combinations. Sci Rep 10, 11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura K, Deng B, Wu Y, Zhou L, Pham TP, Diouf A, Wu CK, Lee SM, Plieskatt JL, Morin MJ, and Long CA (2019). ELISA units, IgG subclass ratio and avidity determined functional activity of mouse anti-Pfs230 antibodies judged by a standard membrane-feeding assay with Plasmodium falciparum. Vaccine 37, 2073–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structures have been deposited to the Protein Data Bank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. The nucleotide sequences from BCR sequencing have been deposited to the GenBank and are publicly available as of the date of publication. Accession number is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contacts upon request.


