Abstract
Islet-brain 1 (IB1) is the human and rat homologue of JIP-1, a scaffold protein interacting with the c-Jun amino-terminal kinase (JNK). IB1 expression is mostly restricted to the endocrine pancreas and to the central nervous system. Herein, we explored the transcriptional mechanism responsible for this preferential islet and neuronal expression of IB1. A 731-bp fragment of the 5′ regulatory region of the human MAPK8IP1 gene was isolated from a human BAC library and cloned upstream of a luciferase reporter gene. This construct drove high transcriptional activity in both insulin-secreting and neuron-like cells but not in unrelated cell lines. Sequence analysis of this promoter region revealed the presence of a neuron-restrictive silencer element (NRSE) known to bind repressor zinc finger protein REST. This factor is not expressed in insulin-secreting and neuron-like cells. By mobility shift assay, we confirmed that REST binds to the NRSE present in the IB1 promoter. Once transiently transfected in β-cell lines, the expression vector encoding REST repressed IB1 transcriptional activity. The introduction of a mutated NRSE in the 5′ regulating region of the IB1 gene abolished the repression activity driven by REST in insulin-secreting β cells and relieved the low transcriptional activity of IB1 observed in unrelated cells. Moreover, transfection in non-β and nonneuronal cell lines of an expression vector encoding REST lacking its transcriptional repression domain relieved IB1 promoter activity. Last, the REST-mediated repression of IB1 could be abolished by trichostatin A, indicating that deacetylase activity is required to allow REST repression. Taken together, these data establish a critical role for REST in the control of the tissue-specific expression of the human IB1 gene.
Islet-brain 1 (IB1) is the rat and human homologue of JIP-1, a murine inhibitor of the c-Jun amino-terminal kinase (JNK). It was termed IB1 since its expression is mostly detectable in pancreatic islets and in the brain (2). IB1 was identified by studying the transcriptional mechanisms responsible for the pancreatic β-cell-specific control of glucose transporter gene GLUT2 (2, 4, 24, 36). Subsequently, the human MAPK8IP1 gene, encoding IB1, was established as a candidate gene for diabetes mellitus (35). Indeed, the direct sequencing of this gene in human type 2 diabetes patients revealed the presence of a missense mutation (resulting in protein mutation S59N) which cosegregated with a rare form of monogenic type 2 diabetes. Ex vivo, this mutation was shown to induce an accelerated apoptosis in pancreatic β cells (35). These observations identified IB1 as a key regulator for β-cell survival since it modulates the activation of the JNK signaling pathway, a system which plays an essential role in maturation, differentiation, and/or apoptosis (8, 16). For example, when the IB1 protein content is decreased, β cells are more sensitive to cytokine-induced apoptosis by increasing the JNK activity (3). Thus, the IB1 expression level is critical for β-cell function.
The goal of the present study was to understand how MAPK8IP1 gene expression is controlled in a tissue-specific manner. Work by Atouf and coauthors has correlated the selective presence of several gene transcripts in pancreatic β and neuronal cells with the absence in these cells of a transcription factor named REST (RE-1 silencing transcription factor, also termed as NRSF) (1). This protein is a zinc finger transcriptional repressor found to be widely expressed during embryogenesis in all tissues, except in endocrine pancreas and mature neuronal tissues (1, 5). The REST gene displays modular organization conserved across humans, rats, and mice within the protein-coding region and is regulated by alternative splicing of REST pre-mRNA. It was reported that the REST isoform with nine zinc fingers is the most predominant isoform found in various tissues (25). Alternatively spliced REST protein isoforms differ in their DNA-binding domains and transrepression domains (26), suggesting different functions of REST. For example, the REST4 isoform, which is a truncated protein, inhibits REST activity by acting as a dominant-negative form of REST (DNREST) (26, 32). REST binds to a 21-bp cis element called the RE-1 silencer element (NRSE), also known as repressor element RE-1, to negatively regulate in nonneuronal tissues several genes preferentially expressed in neuronal cells such as the rat SCG10, the rat type II sodium channel, the human synapsin I, and the rat N-methyl-d-aspartate (NMDA) receptor 1 genes (6, 18, 20, 33). The repression effect induced by REST required the interaction of REST with the corepressor mSin3 and histone deacetylase I (HDACI) to form a complex which induces hypoacetylation of histone (15). These authors proposed that a remodeling of the chromatin structure is required for gene control by REST. Others models where the deacetylase activity may not always be required for REST-mediated repression were proposed. Indeed, REST repression is mediated by recruiting corepressors mSin3 and coREST by a mechanism independent of HDAC (12).
Although little is known about the effect of REST on the transcription of pancreatic β-cell-specific genes, some of the mentioned neuronal genes, such as the rat type II sodium channel and the rat NMDA receptor 1 genes, have been also detected in pancreatic β cells (1). Consistent with this, by transient transfection, the NRSE located in the regulatory regions of these genes is unable to silence gene reporter activity in neuronal and β cells (1, 33). Thus, the absence of REST expression would allow the same selective expression of some genes in both neuronal and pancreatic β tissues. Based on these observations, we hypothesized that REST could be a regulator for tissue-specific expression of IB1.
Herein, we report the sequence of the human IB1 promoter and the presence in this region of an NRSE. Moreover, we show that REST binds to this NRSE. Once transfected in an insulin-secreting line, the expression vector encoding REST repressed the transcriptional activity of the IB1 promoter. The introduction of a mutated NRSE into the human IB1 promoter abolished this silencing effect mediated by REST in insulin-secreting cells and relieved the low transcriptional activity observed in unrelated cell lines expressing REST. DNREST derepressed IB1 promoter activity in unrelated cells. Finally, REST-mediated repression is dependent on HDAC activity since IB1 transcriptional activity was relieved in trichostatin-treated cell lines. Taken together, these observations indicate that transcriptional repressor REST, through a deacetylase activity, controls the selective expression of the IB1 gene in both neuronal and β pancreatic cells.
MATERIALS AND METHODS
Screening of a BAC library of human genomic DNA and plasmid construction.
A BAC library of human genomic DNA (Research Genetics, Rockville, Md.) was screened using human IB1 cDNA as previously described (22). By sequencing, one positive clone (CIT304M9) was found to contain a 731-bp fragment of the IB1 promoter. This fragment was then subcloned at KpnI/XhoI sites into the promoterless expression vector pGL3 basic (Promega). To generate plasmid RIP REST containing the full-length cDNA of human REST under the control of the rat insulin promoter (RIP), the EcoRI-digested REST cDNA from the REEX1 expression vector (a kind gift from R. Scharfmann, Robert Debré Hospital, Paris, France) was ligated into pBS RIP vector (a gift from P. Herrera, Geneva University, Geneva, Switzerland) at EcoRI sites. Mutation in the NRSE sequence of the IB1 promoter was generated by PCR–site-directed mutagenesis using high-fidelity Pfu DNA polymerase according to the manufacturer's protocol (QuikChange; Stratagene, La Jolla, Calif.). In vitro mutagenesis was carried out on full-length IB1LUC from double-stranded 5.6-kb plasmid DNA using two oligonucleotide primers, each complementary to opposite strands of the vector (forward: 5′-GGCTTCAGCACCGCTTAGAGCGCCATCTCC-3′; reverse: 5′-CCGGAGATGGCGCTCTAAGCGGTGCTGAAG-3′). The mutated nucleotides are underlined. To construct luciferase reporter plasmids containing two copies of the IB1 NRSE sequence, wild-type (forward: 5′-GGCTTCAGCACCGCGGAGAGCGCCATCTCC-3′; reverse: 5′-CCGGAGATGGCGCTCTCCGCGGTGCTGAAG-3′) and mutated (indicated above) primers were hybridized, ligated, and filled in with Klenow fragments of DNA polymerase I according to a standard protocol before blunt-ended insertion into SmaI sites of a simian virus 40 (SV40) constitutive promoter containing the luciferase gene (SV40LUC) (Promega). The plasmid encoding DNREST consisted of a REST gene fragment (1.6 kb) encoding the DNA-binding domain; the fragment was amplified by PCR from the REEX1 expression vector using degenerate primers as described previously (7). The PCR product was cloned at HindIII/XbaI sites into corresponding sites of the pCDNA3 eukaryotic expression vector. All constructs were verified by DNA sequencing to confirm the integrity and orientation of the cloning and the introduction of the desired mutations.
Cell lines, transient transfection, and luciferase assays.
Human carcinoma HeLa and mouse NIH 3T3 (fibroblast) cells were cultured as previously described (27). Human lymphoma Jurkat and mouse macrophage-like RAW cells were routinely grown in RPMI 1640 (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS). Mouse βTC3 and rat INS1 pancreatic β cells were maintained as described previously (4, 37). The rat pheochromocytoma PC12 cell line, derived from neural-crest-derived tissue, was cultured in 10% CO2 in Dulbecco's minimal essential medium supplemented with 10% donor horse serum and 5% FBS. All cells were transfected using cationic DOTAP reagent in solution according to the manufacturer's procedure (Roche Diagnostics). On the day of transfection, 2 μg of total DNA was mixed with 15 μl of DOTAP solution and incubated at room temperature for 10 min. The trypsinized cells (4 × 104) were added to the DNA-DOTAP solution and then plated in each of 12 wells. Cells were incubated for 48 h and harvested with 100 μl of the passive lysis buffer (Promega). Luciferase activities (from IB1LUC and normalization Renilla pRLCMV [pRLCMVrenilla] vectors) were measured with 50 μl of protein extract solution by using the Dual-Luciferase reporter assay system (Promega). All experiments were repeated at least three times in triplicate.
RNA preparation, RT-PCR, and Northern blot analysis.
Total RNA extraction from cell lines was conducted exactly as described previously (4). For reverse transcription-PCR (RT-PCR), 5 μg of total RNA was reverse transcribed with superscript II reverse transcriptase using random hexamers [pd(N)6] as primers. PCRs were carried out with 1/10 volume of the reverse-transcribed product in a final volume of 50 μl using recombinant Taq DNA polymerase (Gibco-BRL) and were performed in a GeneAmp 9700 PCR machine (Perkin-Elmer). Each PCR cycle consisted of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a 5-min extension at 72°C. PCR products were separated in a 1.5% agarose gel with Tris-borate-EDTA buffer. Primer sets used for human, rat, and mouse REST mRNA isoforms (26) are as follows. For human isoforms, primers were h-p2s (5′-GTGACCGCTGCGGCTACAATACTAA-3′; primer 1) and h-p8as (5′-GGACAAGTAGGATGCTTAGATTTGA-3′; primer 2). The human isoform encoding a truncated REST protein with a 4-bp insertion of exon N (hREST-N4) was amplified with primer set pN4s (5′-GCGTACTCATTCAGTGGGGTGA-3′) and pN4as (5′-CACATTTAAATGGCTTCTCACCCCACT-3′). For rat and mouse REST, the primers were mrp2s (5′-CTACATGGCACACCTGAAGCACCAC-3′; primer 3) and mrp8as (5′-GCGTAGTCACACACGGGGCAGTTGAAC-3′; primer 4). The rat and mouse REST4 isoform was amplified with primer set p2s (5′-CTACATGGCACACCTGAAGCACCAC-3′) and pR4as (5′-GGCTTCCTCACCCAACTAGATCACACT-3′). For detection of ubiquitous cytochrome oxidase 4 (COX4) mRNA, 24 cycles of PCR were performed using forward and reverse primers 5′-ATGTTGGCTTCCAGAGCGCTGA-3′ and 5′-CTTCTTCCACTCATTCTTGTCATAG-3′. For IB1, the primer set used was 5′-GCGTCGCCTCCCAATTTCAG-3′ and 5′-CAGGTCCATCTGCAGCATCTC-3′. Northern blot analysis from different cell lines was performed as described previously (4).
RACE.
For 5′ rapid amplification of cDNA ends (RACE), 5 μg of total RNA from human insulinoma tissue was reverse transcribed by avian myeloblastosis virus reverse transcriptase according to the 5′/3′ RACE kit procedure (Roche Diagnostics) using an IB1-specific primer (IBSP1) (5′-ACGCCGGCTGGTGGCCGGACTCGGCCTT-3′). The purified cDNA was then subjected to a terminal transferase tailing reaction, and the deoxyribosyladenosine-tailed cDNA was then used as a template for PCR using an oligo(dT) anchor primer provided by the 5′/3′ RACE kit plus IBSP2 (5′-ATCAGGTCCATCTGCAGCATCT-3′). A second nested-PCR round reaction was then performed using the PCR anchor primer (5′-GACCACGCGTATCGATGTCGAC-3′) and IBSP3 (5′-GCCACACTCATCAGTGATC-3′). The RACE products were subcloned into the T-Easy vector (Promega) according to the manufacturer's protocol, and individual clones were sequenced.
Preparation of nuclear extracts and electrophoretic mobility shift assays (EMSA).
Nuclear extracts from cells were prepared as previously described (1). Sequences of the IB1 NRSE oligonucleotides used in the gel retardation are those described above. Complementary sense and antisense oligonucleotides were hybridized and then filled in by the Klenow fragment of DNA polymerase I (Roche Diagnostics) in the presence of deoxycytosine [α-32P]triphosphate (Amersham). Free nucleotides were separated by centrifugation through a G-50 column. For binding, 10 μg of nuclear proteins was preincubated on ice in the presence or absence of an excess of unlabeled competitor DNA for 10 min in 20 μl of a solution containing 20 mM HEPES (pH 7.6), 0.1% Nonidet P-40, 10% glycerol, 1 mM dithiothreitol, 2.5 mM MgCl2, 250 mM KCl, and 2 μg of poly(dI-dC) · poly(dI-dC). Approximately 100 fmol of double-stranded labeled oligonucleotides was mixed with nuclear proteins, and the mixture was incubated for 20 min on ice. For supershift assays, 5 to 10 μg of nuclear proteins was preincubated on ice with or without a polyclonal rabbit anti-REST antibody (a generous gift from G. Mandel, Howard Hughes Medical Institute, New York, N.Y.) for 30 min before addition of the labeled probe. Samples were loaded onto a 6% nondenaturing polyacrylamide gel with 0.25× Tris-borate-EDTA buffer. The gels were fixed in a solution of 10% acetic acid and 30% methanol, dried, and exposed to Hyperfilm-MP (Amersham).
Nucleotide sequence accession numbers.
The sequence of the 731-bp fragment of the IB1 promoter found in clone CIT304M9 was assigned GenBank accession no. AJ304445. The mRNA sequence for the REST isoforms expressed in HeLa, NIH 3T3, Jurkat, and RAW cells and that for REST isoforms expressed in humans and rats have been assigned GenBank accession no. U22314 and AF037199, respectively.
RESULTS
Identification of the IB1 promoter region.
Subclones from a BAC clone containing most of the human MAPK8IP1 gene (CIT304M9) were subjected to sequencing analysis (22). One clone containing the first exon and part of the putative 5′ regulatory region of the gene was identified. The nucleic acid sequence of this region is shown in Fig. 1. The initiation start site was determined using a 5′ RACE reaction with several primers as described in Materials and Methods. For this analysis, we used as the template total RNA isolated from a human insulinoma known to express IB1 (35). The products of the 5′ RACE reaction were cloned and sequenced. The most prominent 5′ nucleotide identified the start site (bp +1 of the sequence shown in Fig. 1). Numerous putative sequences for a variety of transcription factors were identified within the 731 bp by computer analysis (http://bioinformatics.weizmann.ac.il/transfac/), in particular a RE-1 silencer element (NRSE). As shown in Table 1, the 21-bp NRSE cis element identified (−229 to −209 bp) is almost identical to the consensus NRSE sequence (29).
FIG. 1.
Nucleic acid sequence of a fragment of the human IB1 promoter. The transcription start site (asterisk, +1) was determined by a 5′RACE reaction and computer analysis. A putative NRSE binding site is underlined. The ATG in boldface is the codon of translation initiation of the IB1 protein.
TABLE 1.
Sequence comparison of the Islet-brain 1 NRSE with functional known NRSEs
| NRSEa | Sequenceb |
|---|---|
| Consensus | TTCAGCACCACGGACAGCGCC |
| Rat SCG10 | TTCAGCACCACGGAGAGTGCC |
| Human synapsin | TTCAGCACCGCGGACAGTGCC |
| Rat NMDA rec 1 | TTCAGCACCTCGGACAATGCC |
| WTIB1 | TTCAGCACCGCGGAGAGCGCC |
| mutIB1 | TTCAGCACCGCTTAGAGCGCC |
WTIB1, wild-type IB1; mutIB1, mutant IB1.
Nucleotides that differ from the consensus are in boldface; mutated nucleotides are underlined.
IB1 transcript is detected in insulin-secreting and neuron-like cell lines but not in REST-expressing cell lines.
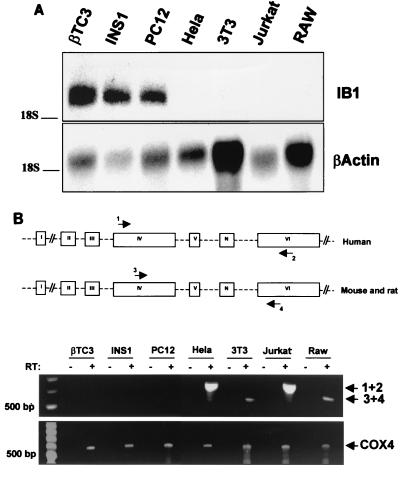
Insulin-secreting cell lines βTC3 (mouse) and INS1 (rat), neuron-like PC12 (rat) cells, fibroblast-derived NIH 3T3 (mouse) cells, carcinoma HeLa (human) cells, lymphocyte Jurkat (human) cells, and macrophage-like RAW (mouse) cells were used as models to study the mechanism controlling the cell-specific expression of IB1. Total RNA was isolated and subjected to Northern blot analysis to detect the presence of the IB1 transcript. As shown in Fig. 2A, IB1 transcripts were detected in βTC3, INS1, and PC12 cells but not in HeLa, NIH 3T3, Jurkat, and RAW cell lines. All these cells were also analyzed for expression of REST transcript isoforms by RT-PCR using different primer sets (see Materials and Methods). Primers were designed in order to discriminate the most abundant REST mRNA isoforms in humans, rats, and mice (26). RT-PCR analysis of RNA from HeLa, NIH 3T3, Jurkat, and RAW cell lines yielded only one detectable PCR fragment (Fig. 2B). Sequence analysis of this PCR product revealed the sequences of the REST isoforms expressed in HeLa, NIH 3T3, Jurkat, and RAW cells to be the same as the human and rat REST mRNA sequences. The specific expression of the human and mouse REST mRNA encoding truncated proteins was also investigated by RT-PCR using specific primer pairs (see Materials and Methods) designed to span exon N. No PCR fragment was detected in RNA from HeLa, NIH 3T3, Jurkat, and RAW cells (data not shown), indicating that the main transcript detected in these cells is the REST mRNA encoding the nine-zinc-finger protein. RT-PCR analysis of RNA from insulin-secreting βTC3, INS1, and PC12 cells failed to detect any transcript, as anticipated (1).
FIG. 2.
Expression of the IB1 and REST genes in several cell lines. (A) Northern blot analysis of IB1 and β-actin mRNAs in insulin-secreting cells (βTC3 and INS1), in rat neuron-like PC12 cells, in human carcinoma HeLa cells, in mouse fibroblast-derived NIH 3T3 cells, in human lymphoma Jurkat cells, and in mouse macrophage-like RAW cells. Total RNA (15 μg) was hybridized with the human IB1 cDNA probe as described previously (4). β-Actin was used to control the quantity of the total RNA loaded. (B) Analysis of REST expression in βTC3, INS1, PC12, HeLa, NIH 3T3, Jurkat, and RAW cells. RT-PCR analysis of RNA from different cell lines yielded one detectable PCR fragment using specific primer pairs for human and mouse and rat REST mRNA (primer pairs comprising primers 1 and 2 and 3 and 4, respectively). These primers, designed as described previously (26), were between exon 4 and exon 6 to span the region which differs in REST mRNA isoforms. Sequence analysis of the amplified fragment showed that the PCR product of HeLa, NIH 3T3, Jurkat, and RAW cells is the REST full-length cDNA fragment. This band was not detectable in βTC3, INS1, and PC12 cells. (Bottom) Ubiquitous cytochrome oxidase 4 (COX4) was also amplified to control the quality of the reverse transcription for each mRNA sample. PCR was performed as described in Materials and Methods. −, negative control for which no reverse transcriptase was added to the reaction.
The promoter region of IB1 drives high transcriptional activity in insulin-secreting and neuron-like cell lines but not in unrelated cell lines.
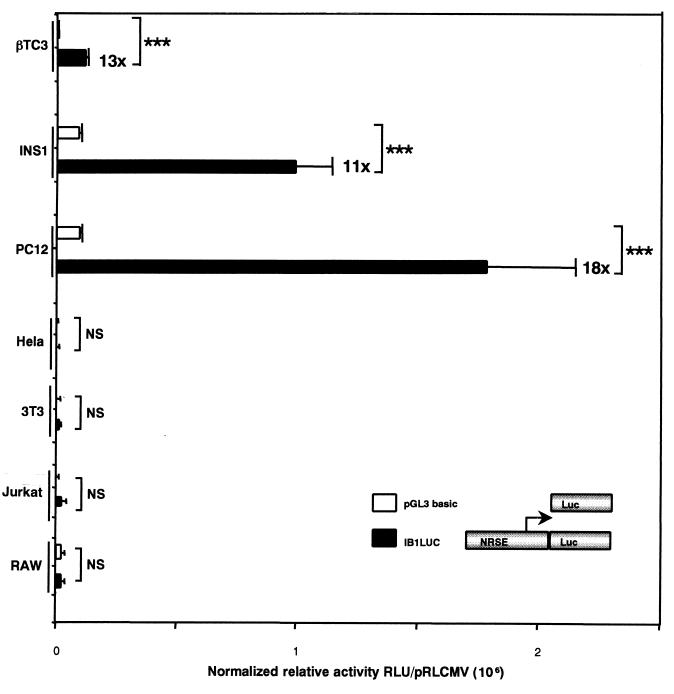
We next evaluated the transcriptional activity of the fragment comprising bp −691 to + 41 of the promoter region of the MAPK8IP1 gene in insulin-secreting and neuron-like cells and in unrelated cells. The IB1 promoter region was cloned into the eukaryotic expression vector pGL3 basic (IB1LUC) and transiently transfected into βTC3, INS1, PC12, HeLa, NIH 3T3, Jurkat, and RAW cells, together with a vector encoding the Renilla protein (pRLCMVrenilla) for normalization. As shown in Fig. 3, this construct drove high transcriptional activity in βTC3, INS1, and PC12 cells (13-, 11-, and 18-fold higher than that for the promoterless pGL3 basic vector, respectively) whereas no significant reporter gene activities were detected in HeLa, NIH 3T3, Jurkat, and RAW cells. These data indicate that important cis regulatory elements are present within the human IB1 promoter to confer the β-cell-specific expression of the reporter gene.
FIG. 3.
Transcriptional activity of the IB1 promoter region. The region of the IB1 promoter comprising bp −691 to + 41 (IB1LUC) drove a high luciferase activity in the insulin-secreting βTC3, INS1, and PC12 cells (13-, 11-, and 18-fold higher, respectively, than that for the promoterless vector pGL3 basic) but not in unrelated HeLa, NIH 3T3, Jurkat, and RAW cells. Each experiment was repeated at least three times in triplicate. Luciferase activities were normalized using pRLCMVrenilla, and results are expressed as means ± standard errors of the means (triple asterisk, P < 0.001).
REST binds to the NRSE of the human IB1 promoter.
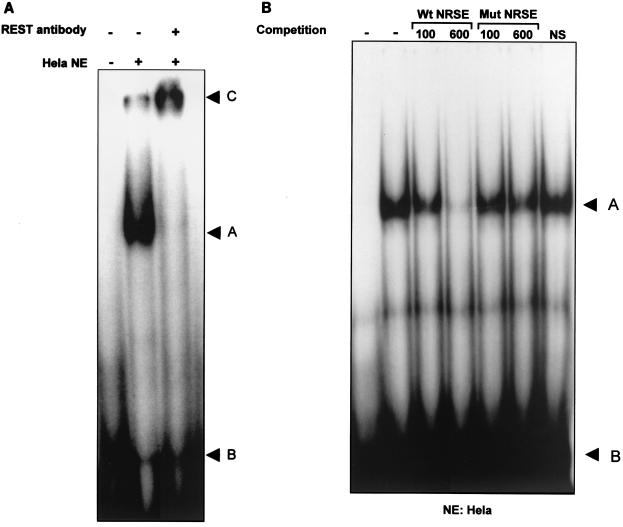
To examine the REST-binding activity to the newly identified human IB1 NRSE, we performed an EMSA using nuclear extracts prepared from βTC3, INS1, PC12, HeLa, NIH 3T3, Jurkat, and RAW cells. We also designed a mutated NRSE oligonucleotide, described in Table 1, in which two guanine residues in the core of the element were replaced with two thymine residues. Mutation of these two bases was previously shown to alter the activity of the binding of REST to its preferential NRSE binding site (20). As shown in Fig. 4A, a DNA-binding complex from HeLa nuclear extracts is observed using the NRSE of the IB1 promoter as the labeled probe and this complex was supershifted in the presence of polyclonal REST antibody. Competition experiments were conducted using a 100- or 600-fold molar excess of unlabeled wild-type NRSE or mutated NRSE (Fig. 4B). The wild-type oligonucleotide competed for protein-DNA interactions, whereas the mutated NRSE did not decrease the binding of REST to the human NRSE. This DNA-binding complex was not detected in nuclear extracts obtained from βTC3, INS1, and PC12 cells, while this binding pattern was detected in nuclear extracts obtained from NIH 3T3, Jurkat, and RAW cells (data not shown). These data indicate that REST, present in HeLa, NIH 3T3, Jurkat, and RAW cells, is able to bind to the human IB1 NRSE and that the endogenous REST is unable to bind to the mutated NRSE.
FIG. 4.
Sequence-specific binding activity of REST to the human IB1 NRSE. (A) EMSA with 32P-labeled IB1 NRSE using nuclear extracts from HeLa cells. A slow-migrating complex (arrowhead A) was detected, compared to free-probe migration (arrowhead B). This pattern was supershifted by using REST antibodies (arrowhead C). (B) The DNA-binding activity with IB1 NRSE was competed by adding a 100- or 600-fold molar excess of unlabeled wild-type NRSE (Wt) but not by adding unlabeled mutated NRSE (Mut). NS, nonspecific competitor.
Transcriptional activation of the IB1 promoter mutated in the NRSE in non-β cells.
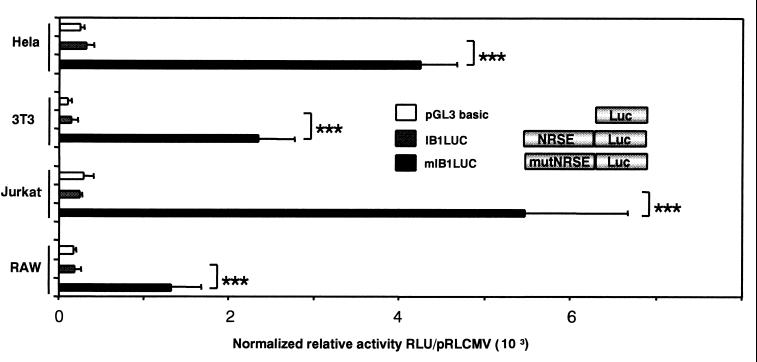
Using site-directed mutagenesis, the NRSE of the human IB1 promoter was modified to the mutated NRSE described in Table 1. Substitution of the two nucleotides caused a loss of activity of REST binding to mutated NRSE as shown in Fig. 4B. In transient transfection, the reporter gene activity mediated by the mutated NRSE in the IB1 promoter (mIB1LUC) was completely relieved compared to IB1LUC activity in HeLa cells (Fig. 5). This implies that the NRSE contributes to the repression of the IB1 promoter activity in non-β pancreatic and nonneuronal HeLa, NIH 3T3, Jurkat, and RAW cells.
FIG. 5.
Mutation of the NRSE motif in the human IB1 promoter relieved transcriptional activity in REST-expressing cell lines. The IB1 promoter containing mutated or wild-type NRSE, mIB1LUC and IB1LUC, respectively, was transiently transfected into REST-expressing HeLa, NIH 3T3, Jurkat, and RAW cells. The luciferase activity was relieved in all these cells with mIB1LUC. Each experiment was performed at least three times in triplicate. Luciferase activities were normalized using pRLCMVrenilla, and results are expressed as means ± standard errors of the means (triple asterisk, P < 0.001). RLU, relative light units.
REST represses IB1 promoter activity in β cells.
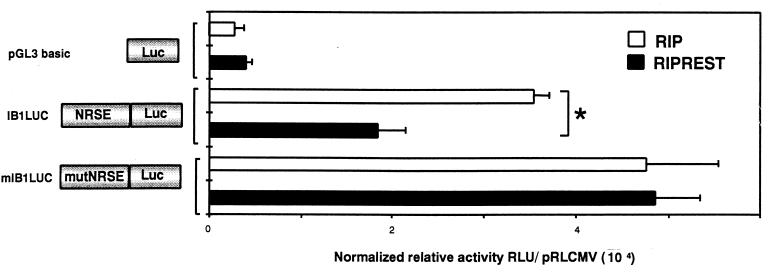
We then evaluated whether REST could repress IB1 promoter activity in β cells. An expression vector encoding or not encoding REST under the control of the RIP was transiently transfected into the insulin-secreting βTC3 cells in the presence of the promoterless pGL3 basic vector or IB1LUC, which is wild type or mutated (mIB1LUC) in NRSE. REST significantly decreased the IB1LUC relative activity by 50% (Fig. 6), whereas it had no effect on the promoterless pGL3 basic vector and mIB1LUC relative activities. These data established that REST is able to repress the human IB1 promoter in non-REST-expressing cell lines and that this effect is mediated through the identified NRSE.
FIG. 6.
REST represses IB1 promoter activity. IB1LUC and mIB1LUC constructs were cotransfected in the insulin-secreting cell line (βTC3), with REST expression vector RIP REST (under the control of the RIP) or the empty RIP vector as a control. The luciferase activity of IB1LUC was decreased by 50% in REST-expressing cells, whereas REST expression had no effect on mIB1LUC activity. Each experiment was performed at least three times in triplicate. Luciferase activities were normalized using pRLCMVrenilla, and results are expressed as means ± standard errors of the means (asterisk, P < 0.05). RLU, relative light units.
The IB1 NRSE acts as a silencer in a heterologous promoter.
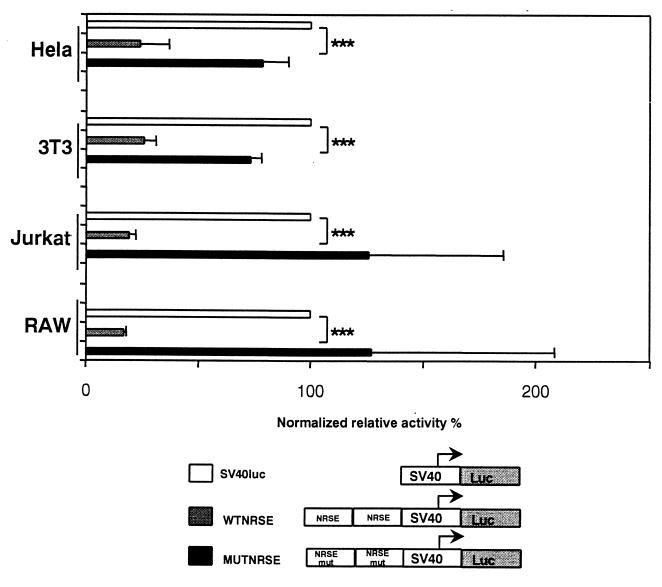
Mutational analysis of the human IB1 NRSE suggested that the NRSE motif plays a critical role in β-cell-specific expression of IB1. To assess whether this identified motif is also able to act as a silencer in REST-expressing cell lines, two copies of the human wild-type or mutated NRSE motifs were cloned upstream of a viral SV40 promoter linked to a luciferase gene. These constructs were then transfected into HeLa, NIH 3T3, Jurkat, and RAW cells. As shown in Fig. 7, the wild-type human NRSE motif decreased the heterologous promoter activity by 76, 74, 81, and 83% compared to SV40LUC relative activity in HeLa, NIH 3T3, Jurkat, and RAW cells, respectively. The promoter activity was partially restored in HeLa, NIH 3T3, Jurkat, and RAW cells when the NRSE motif was mutated.
FIG. 7.
Human NRSE silences the activity of the heterologous SV40 promoter. The wild-type and mutated NRSEs of the IB1 promoter, WTNRSE and MUTNRSE, respectively, were cloned as dimers upstream an SV40 promoter. Constructs were then transiently transfected into HeLa, NIH 3T3, Jurkat, and RAW cells. WTNRSE activity is markedly decreased in all these cells compared to the basal activity of SV40LUC. The mutation in NRSE partially restored the activity of the MUTNRSE promoter. Luciferase activities were normalized using pRLCMVrenilla. Each experiment was performed at least three times in triplicate. All values are expressed as percentages of the SV40LUC activity and are means ± standard errors of the means (triple asterisk, P < 0.001).
Expression of DNREST in unrelated cells.
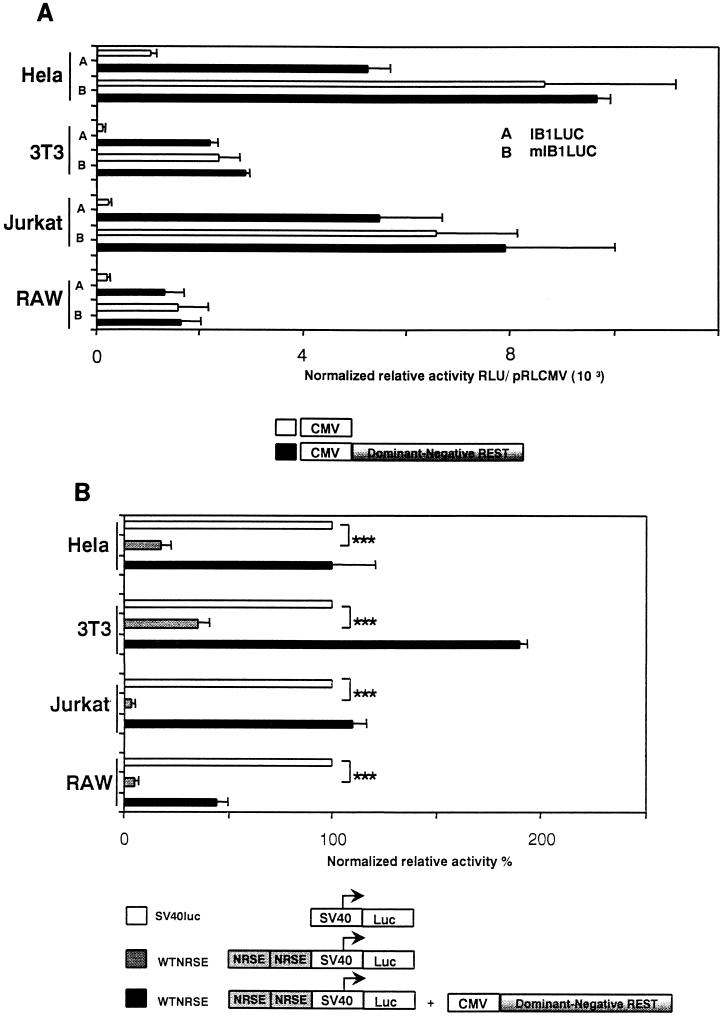
We constructed an expression vector encoding DNREST to evaluate whether the repression mediated by REST could be relieved. DNREST corresponds to human cDNA encoding only the DNA-binding domain of REST without the two repressor domains of the protein. As shown in Fig. 8A, the IB1LUC construct was transiently transfected into HeLa, NIH 3T3, Jurkat, and RAW cells in the presence or the absence of DNREST. Cotransfection of DNREST derepressed the IB1 promoter relative activity in transfected cells, and this effect was absent when using the mutated NRSE (mIB1LUC). These data indicated that the repression of IB1 promoter activity in non-β and nonneuronal cells is mediated by REST and that mutation of the NRSE or the use of DNREST allowed a complete derepression of the IB1 promoter activity. Furthermore, cotransfection of DNREST relieved the repression driven by multimerized NRSE in the heterologous SV40 promoter (WTNRSE) in HeLa, NIH 3T3, Jurkat, and RAW cells (Fig. 8B). This indicated that the derepression is mediated through the NRSE. It has been described that DNREST mediates its derepression by binding to the NRSE (7). To determine whether this mechanism occurs for the NRSE present in the IB1 promoter, HeLa cells were transfected with the DNREST or with the control vector. By EMSA, a binding pattern was observed with nuclear extracts from DNREST-transfected cells. This binding was fully competed with a 600-fold excess of unlabeled wild-type NRSE but did not compete with unlabeled mutated NRSE (data not shown). This result indicated that DNREST binds specifically to the NRSE of the IB1 promoter.
FIG. 8.
Expression of DNREST derepressed the IB1 promoter and heterologous SV40 promoter activities in HeLa cells. (A) IB1LUC and mIB1LUC vectors were transiently transfected with or without DNREST into HeLa, NIH 3T3, Jurkat, and RAW cells. The IB1LUC activity was relieved with the expression of DNREST. Triple asterisk, P < 0.001. RLU, relative light units. (B) The heterologous construction WTNRSE was transfected in HeLa, NIH 3T3, Jurkat, and RAW cells with or without DNREST. The WTNRSE activity was dramatically decreased compared to the SV40LUC basal activity. The WTNRSE activity was completely restored upon DNREST expression. Triple asterisk, P < 0.001.
The IB1 transcriptional repression is TSA sensitive through the NRSE.
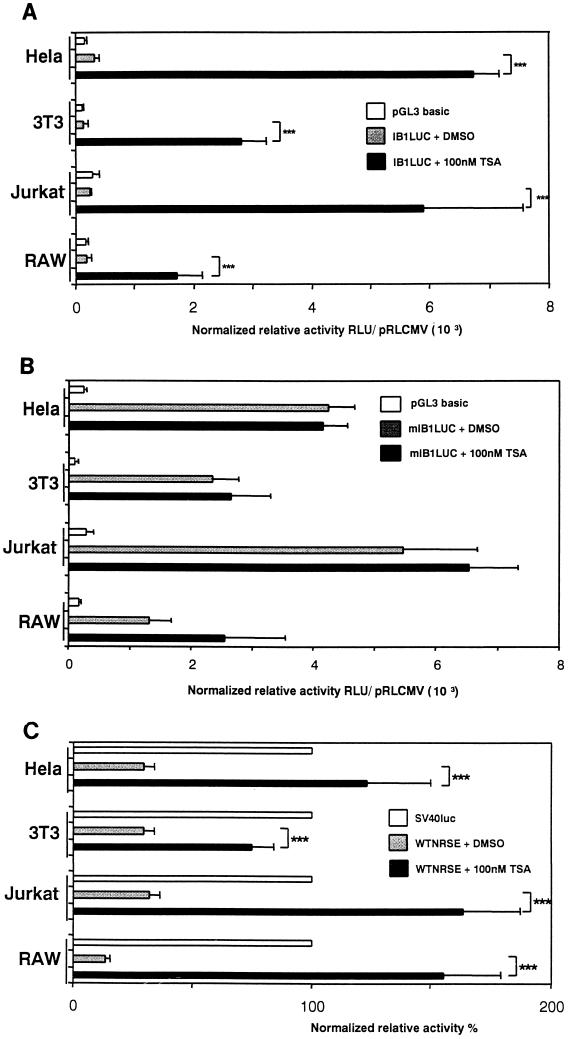
To assess whether the repression of IB1 promoter activity is HDAC dependent, HeLa, NIH 3T3, Jurkat, and RAW cells were transiently transfected with IB1LUC and incubated for 24 h with 100 nM trichostatin A (TSA), a specific inhibitor of HDAC (40). The luciferase activity was significantly relieved in all TSA-treated cell lines, HeLa, NIH 3T3, Jurkat, and RAW cells (Fig. 9A), indicating that the transcriptional repression is TSA sensitive. We investigated whether the effect of the TSA is mediated through the NRSE within the promoter. Then, HeLa, NIH 3T3, Jurkat, and RAW cells were also transiently transfected with mIB1LUC in the presence of dimethyl sulfoxide (DMSO) or TSA. As a result, we did not observe any significant differences in luciferase activity between TSA-treated and DMSO-treated HeLa, NIH 3T3, Jurkat, and RAW cells when NRSE is mutated (Fig. 9B). This suggested that the derepression induced by TSA is mediated through the newly identified NRSE. To verify that the NRSE-mediated repression is TSA sensitive, we transiently transfected the heterologous NRSE-multimerized SV40 promoter in HeLa, NIH 3T3, Jurkat, and RAW cells for 24 h with 100 nM TSA. The activity of the heterologous promoter was restored in all TSA-treated HeLa, NIH 3T3, Jurkat, and RAW cells (Fig. 9C). No significant effect of TSA on the SV40 or cytomegalovirus promoter activities which are not pancreatic or neuron selective was observed (data not shown). These data indicate that the IB1 NRSE is a TSA-sensitive regulatory element and that REST transcriptional repression of the IB1 promoter in nonpancreatic β and nonneuronal cells involves a TSA-sensitive mechanism that is mediated through the NRSE.
FIG. 9.
IB1 promoter activity is repressed through NRSE in a TSA-sensitive manner. (A) IB1 promoter construct IB1LUC was transiently transfected into HeLa, NIH 3T3, Jurkat, and RAW cells. TSA (100 nM) or DMSO as a control was added to medium 24 h after transfection. The IB1LUC activity was relieved in all TSA-treated cells. (B) The mIB1LUC construct was also transfected in the REST-expressing cells with TSA or DMSO. No significant differences in mIB1Luc activity between TSA-treated cells and DMSO-treated cells were observed. (C) The heterologous WTNRSE construct was also transiently transfected in the presence of TSA or DMSO. The WTNRSE activity was restored in all TSA-treated cells. Each experiment was performed at least three times in triplicate. Triple asterisk, P < 0.001. RLU, relative light units.
Repression of the endogenous IB1 transcriptional activity is relieved in TSA-treated cells.
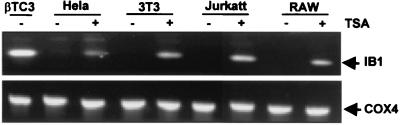
We also investigated whether repression of the transcription of the endogenous IB1 gene could be relieved in REST-expressing cells after TSA treatment since IB1 promoter activity was relieved in TSA-treated cells. To perform this experiment, total RNAs from HeLa, NIH 3T3, Jurkat, and RAW cells treated or not with TSA were analyzed by RT-PCR for expression of the IB1 transcript using an IB1-specific primer set. As shown in Fig. 10, the reaction yielded a PCR fragment detectable only in cells treated with TSA, not in untreated cells. This observation was consistent with previous observations on the IB1 promoter performed by transient transfection assays, indicating that deacetylase activity is required for the repression of endogenous MAPK8IP1 gene transcription.
FIG. 10.
Endogenous IB1 gene expression is relieved in TSA-treated cells. Total RNAs from HeLa, NIH 3T3, Jurkat, and RAW cells treated in the presence of DMSO (−) or 100 nM TSA (+) were analyzed for IB1 gene expression by RT-PCR using specific primer pairs as described in Materials and Methods. RNA from βTC3 cells was used as a positive control for IB1 gene expression. In REST-expressing cells, the PCR yielded one PCR fragment detected only in the presence of TSA-treated cells.
DISCUSSION
The present study provides new insight into the transcriptional mechanisms which control the cell-specific expression of the IB1 gene. First, we identified a fragment of the human IB1 promoter which drives high transcriptional activity in a pancreatic β and neuron-like cell lines but not in unrelated cell lines. Second, we identified an NRSE within this fragment of promoter which binds transcription factor REST. Third, REST was shown to silence IB1 promoter activity in non-β cell lines. Fourth, expression of DNREST relieved the repression of IB1 transcriptional activity driven by REST. Fifth, trichostatin relieved endogenous IB1 transcriptional activity through the NRSE in REST-expressing cells. The results of our study establish a critical role for REST in the control of the preferential pancreatic and neuronal expression of the IB1 gene.
It has been described that REST silences, in nonneuronal and nonpancreatic β cells, a subset of neuronal genes which are also detected in pancreatic β cells such as SCG10, synapsin I, the brain type II voltage-dependent sodium channel, and the dopamine β-hydroxylase genes (1). These authors (1) proposed that identical mechanisms control the expression of neuronal genes in pancreatic β cells. Herein, we confirm that REST is a potent repressor that contributes to the preferential expression of IB1 in both tissues. One possible mechanism would be that REST silences IB1 gene transcription through histone acetylation, suggesting that a mechanism involving chromatin remodeling regulates the expression of IB1 in both neuronal and β-pancreatic cells. In our model, this seems likely since the IB1 transcriptional activity was relieved in all TSA-treated cells. These data showed that deacetylase activity is required for the repression of IB1 gene transcription by REST. To mediate the repression, REST may recruit the deacetylase activity and mSin3 by a mechanism similar to that previously described by others (15, 40). Thus, REST-induced hypoacetylation around the NRSE could change the local nucleosomal structure and therefore could have a direct effect on TFIID-RNA polymerase II holocomplex access to the IB1 promoter. Indeed, the NRSE at the IB1 promoter is located only 229 bp upstream of the transcription start site, and so local changes to nucleosomal structure may affect the basal trancriptional complex.
However, REST repression may be insufficient to control the cell-specific expression of IB1. Consistent with this, we showed that cotransfection of the IB1 promoter containing mutated NRSE or the use of DNREST was unable to relieve entirely the luciferase activity to a level comparable to that seen in β cells. This was also confirmed by the fact that the IB1 transcript was not detectable by Northern blotting in TSA-treated cells (data not shown). These data suggest that non-β and nonneuronal cells are lacking (or express at very low levels) some trans-acting factors which are present in β and neuronal cells and are critical to achieve high transcriptional activity of the IB1 gene.
Although the expression of the IB1 gene has been shown to be mainly detected in neuronal and pancreatic β cells, several reports, including ours, have described low levels of IB1 transcripts in some tissues such as testis and kidney tissues (2, 9, 22, 34), where REST transcripts have been detected (25). One explanation for this observation would be that these cells display a low level of REST expression which is sufficient to decrease IB1 gene expression. In accordance with this model, it was described that REST, which controls synapsin I gene expression, is coexpressed in neuroblastoma cell lines and that the levels of REST expression were inversely proportional to the levels of synapsin I mRNA. In this model, an increased expression of synapsin I was directly correlated with decreased expression of REST (23). Alternatively, the MAPK8IP1 gene could be regulated by other REST isoforms since different REST alternative transcripts could reduce the repressor effect of REST upon a cell context (32).
By controlling IB1 gene transcription, REST may contribute indirectly to the temporal and spatial regulation of apoptosis, proliferation, or differentiation of neuronal and pancreatic β cells. IB1 was shown to interact with several components of JNK signaling, such as mitogen-activated protein kinase 7 (MAPK7), mixed-lineage 3 (MLK3), dual leucine zipper-bearing kinase (DLK), and JNK1 and JNK2 (3, 38, 39). By acting as a scaffold protein, IB1 modulates JNK activity. As mentioned, this activated cascade could promote either cell survival, cell death, or differentiation (8, 16). We propose that REST controls IB1 gene transcription to maintain a high or low basal JNK activity. Interestingly, using DNREST, Lawinger and coauthors have recently shown that medulloblastoma cells are more sensitive to apoptosis and undergo a more pronounced differentiation when REST is inactivated (19). This observation could be related to a modulation of IB1 gene expression. In vivo, the selective disruption of REST in mice leads to malformation in several nonneuronal tissues, as well as apoptosis and embryonic lethality. In addition, expression of several REST target genes was derepressed in nonneuronal tissues and in neuronal progenitors in these mice. IB1 could be one of these derepressed genes which may in turn contribute to the observed phenotype.
Among genes which are linked to a direct pancreatic β-cell function, the IB1 gene is the first found to be regulated by REST. Indeed, the function of neuronal genes regulated by REST in pancreatic β cells remains unclear. However, genes such as BETA2/NEUROD1, Islet-1, Pax4, Pax6, Nkx2.2, Brn4, and neurogenin 3 genes, which are all essential for endocrine pancreas development, are also found to be predominantly expressed in neuronal cells (10, 11, 13, 14, 21, 28, 30, 31). Although we failed to identify any NRSE in the known sequences of these genes by computer analysis because the promoter regions of these genes were not (or partially) identified or not published, a REST-mediated regulation of the expression of these genes remains possible. Moreover, REST is able to mediate repression through an NRSE, even one located far upstream in the promoter or conversely in intronic sequences (29). A fine characterization of the coding, intronic, and regulatory regions of these genes is required to allow the identification of some putative NRSEs. If functional NRSEs are identified, REST could contribute to endocrine cell fate as observed for the transcriptional repressor hairy of split1 (HES1), a general repressor of basic helix-loop-helix factors (17). The mouse deficient in HES1 displayed a severe pancreatic hypoplasia caused by depletion of pancreatic epithelial precursors (17). Subsequent work will be required to evaluate whether REST contributes to the pancreatic development. The understanding of the mechanism leading to the development of mature insulin-secreting cells is critical for the engineering of precursor endocrine cells, which could be an unlimited source of insulin-secreting cells for transplantation purposes.
In conclusion, we show that REST is a critical transcriptional factor that contributes to the cell-specific expression of the human IB1 gene. We propose that this modulation might be implicated in the differentiation of the endocrine pancreas and/or the central nervous system and/or in tumorogenesis.
ACKNOWLEDGMENTS
We are grateful to Philippe Dupraz for the kind help in constructing DNREST. We are indebted to Gail Mandel for the REST antibody and Fouad Atouf for useful discussions. We thank Eric Bernardi for the critical reading of the manuscript.
G.W., J.A.H., C.B., and V.M. are supported by the Swiss National Science Foundation (grants 32-48916.96, 31-56689.99, 32-94471.95, and 32-54119.98), the Juvenile Diabetes Research Foundation (grant 1-2001-555 to G.W.), and the Placide Nicod and Octav Botnar Foundations.
REFERENCES
- 1.Atouf F, Czernichow P, Scharfmann R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/repressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem. 1997;272:1929–1934. doi: 10.1074/jbc.272.3.1929. [DOI] [PubMed] [Google Scholar]
- 2.Bonny C, Nicod P, Waeber G. IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. J Biol Chem. 1998;273:1843–1846. doi: 10.1074/jbc.273.4.1843. [DOI] [PubMed] [Google Scholar]
- 3.Bonny C, Oberson A, Steinmann M, Schorderet D F, Nicod P, Waeber G. IB1 reduces cytokine-induced apoptosis of insulin-secreting cells. J Biol Chem. 2000;275:16466–16472. doi: 10.1074/jbc.M908297199. [DOI] [PubMed] [Google Scholar]
- 4.Bonny C, Thompson N, Nicod P, Waeber G. Pancreatic-specific expression of the glucose transporter type 2 gene: identification of cis-elements and islet-specific trans-acting factors. Mol Endocrinol. 1995;9:1413–1426. doi: 10.1210/mend.9.10.8544849. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z F, Paquette A J, Anderson D J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 6.Chin L S, Li L, Greengard P. Neuron-specific expression of the synapsin II gene is directed by a specific core promoter and upstream regulatory elements. J Biol Chem. 1994;269:18507–18513. [PubMed] [Google Scholar]
- 7.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 8.Davis R J. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 9.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 10.Edlund H. Transcribing pancreas. Diabetes. 1998;47:1817–1823. doi: 10.2337/diabetes.47.12.1817. [DOI] [PubMed] [Google Scholar]
- 11.Edlund H. Pancreas: how to get there from the gut? Curr Opin Cell Biol. 1999;11:663–668. doi: 10.1016/s0955-0674(99)00033-2. [DOI] [PubMed] [Google Scholar]
- 12.Grimes J A, Nielsen S J, Battaglioli E, Miska E A, Speh J C, Berry D L, Atouf F, Holdener B C, Mandel G, Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 13.Huang H-P, Liu M, El-Hodiri H M, Chu K, Jamrich M, Tsai M-J. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol Cell Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H P, Tsai M J. Transcription factors involved in pancreatic islet development. J Biomed Sci. 2000;7:27–34. doi: 10.1007/BF02255915. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Myers S J, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 16.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 17.Jensen J, Pedersen E E, Galante P, Hald J, Heller R S, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen O D. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 18.Kraner S D, Chong J A, Tsay H J, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 19.Lawinger P, Venugopal R, Guo Z S, Immaneni A, Sengupta D, Lu W, Rastelli L, Marin Dias C A, Levin V, Fuller G N, Echelard Y, Majumder S. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci USA. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Pereira F A, Price S D, Chu M, Shope C, Himes D, Eatock R A, Brownell W E, Lysakowski A, Tsai M J. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooser V, Maillard A, Bonny C, Steinmann M, Shaw P, Yarnall D P, Burns D K, Schorderet D F, Nicod P, Waeber G. Genomic organization, fine-mapping, and expression of the human islet-brain 1 (IB1)/c-Jun-amino-terminal kinase interacting protein-1 (JIP-1) gene. Genomics. 1999;55:202–208. doi: 10.1006/geno.1998.5641. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura E, Sasaki K, Maruyama K, Tsukada T, Yamaguchi K. Decrease in neuron-restrictive silencer factor (NRSF) mRNA levels during differentiation of cultured neuroblastoma cells. Neurosci Lett. 1996;211:101–104. doi: 10.1016/0304-3940(96)12722-1. [DOI] [PubMed] [Google Scholar]
- 24.Ogawa Y, Noma Y, Davalli A M, Wu Y J, Thorens B, Bonner-Weir S, Weir G C. Loss of glucose-induced insulin secretion and GLUT2 expression in transplanted beta-cells. Diabetes. 1995;44:75–79. doi: 10.2337/diab.44.1.75. [DOI] [PubMed] [Google Scholar]
- 25.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palm K, Metsis M, Timmusk T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res Mol Brain Res. 1999;72:30–39. doi: 10.1016/s0169-328x(99)00196-5. [DOI] [PubMed] [Google Scholar]
- 27.Richardson R T, Batova I N, Widgren E E, Zheng L X, Whitfield M, Marzluff W F, O'Rand M G. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J Biol Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 28.Sander M, German M S. The beta cell transcription factors and development of the pancreas. J Mol Med. 1997;75:327–340. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- 29.Schoenherr C J, Paquette A J, Anderson D J. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab M H, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya F J, Zhao S, Frotscher M, Tsai M J, Nave K A. Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwitzgebel V M, Scheel D W, Conners J R, Kalamaras J, Lee J E, Anderson D J, Sussel L, Johnson J D, German M S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- 32.Shimojo M, Paquette A J, Anderson D J, Hersh L B. Protein kinase A regulates cholinergic gene expression in PC12 cells: REST4 silences the silencing activity of neuron-restrictive silencer factor/REST. Mol Cell Biol. 1999;19:6788–6795. doi: 10.1128/mcb.19.10.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapia-Ramirez J, Eggen B J, Peral-Rubio M J, Toledo-Aral J J, Mandel G. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc Natl Acad Sci USA. 1997;94:1177–1182. doi: 10.1073/pnas.94.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson N A, Haefliger J A, Senn A, Tawadros T, Magara F, Ledermann B, Nicod P, Waeber G. Islet-brain1/JNK-interacting protein-1 is required for early embryogenesis in mice. J Biol Chem. 2001;276:27745–27748. doi: 10.1074/jbc.C100222200. [DOI] [PubMed] [Google Scholar]
- 35.Waeber G, Delplanque J, Bonny C, Mooser V, Steinmann M, Widmann C, Maillard A, Miklossy J, Dina C, Hani E H, Vionnet N, Nicod P, Boutin P, Froguel P. The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nat Genet. 2000;24:291–295. doi: 10.1038/73523. [DOI] [PubMed] [Google Scholar]
- 36.Waeber G, Pedrazzini T, Bonny O, Bonny C, Steinmann M, Nicod P, Haefliger J A. A 338-bp proximal fragment of the glucose transporter type 2 (GLUT2) promoter drives reporter gene expression in the pancreatic islets of transgenic mice. Mol Cell Endocrinol. 1995;114:205–215. doi: 10.1016/0303-7207(95)96801-n. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Antinozzi P A, Hagenfeldt K A, Maechler P, Wollheim C B. Molecular targets of a human HNF1 alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J. 2000;19:4257–4264. doi: 10.1093/emboj/19.16.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda J, Whitmarsh A J, Cavanagh J, Sharma M, Davis R J. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]