Abstract
Background
We aimed to analyze the genotype-phenotype correlations of STXBP1 pathogenic variants, prognostic factors and the treatment choices in a case-series of STXBP1-related disorders from China.
Methods
The clinical data and genetic results of the children diagnosed with STXBP1-related disorders at Xiangya hospital from 2011 to 2019 were collected retrospectively, and analyzed. We divided our patients into groups for comparison purposes: patients with missense variants and nonsense variants, patients who are seizure-free and not seizure-free, patients with mild to moderate intellectual disability (ID) and severe to profound global developmental delay (GDD).
Results
Nineteen patients were enrolled: 17 (89.5%) unrelated and 2 (10.5%) familial. Twelve (63.2%) were females. Developmental epileptic encephalopathy (DEE) was observed in 18 (94.7%) patients and ID alone in 1 (5.3%) individual. Thirteen patients (68.4%) had profound ID/GDD, 4 (23.53%) severe, 1 (5.9%) moderate and 1 (5.9%) mild. Three patients (15.8%) with profound ID died. A total of 19 variants were detected: pathogenic (n = 15) and likely pathogenic (n = 4). Seven were novel variants: c.664-1G>-, M486R, H245N, H498Pfs*44, L41R, L410del, and D90H. Of the 8 previous reported variants, 2 were recurrent: R406C and R292C. Anti-seizure medications were used in combinations, and 7 patients became seizure-free, and most of them achieved seizure freedom within the first 2 years of life irrespective of the type of the mutation. Effective medications for the seizure-free individuals included adrenocorticotropic (ACTH) and/or levetiracetam and/or phenobarbital and/or sodium valproate and/or topiramate and/or vigabatrin and/or nitrazepam. There was no correlation between the types of pathogenic variants and the phenotypes.
Conclusion
Our case-series showed that there is no genotype-phenotype correlation in patients with STXBP1-related disorders. This study adds 7 novel variants which expand the spectrum of STXBP1-related disorders. Combinations of levetiracetam and/or sodium valproate and/or ACTH and/or phenobarbital and/or vigabatrin and/or topiramate and/or nitrazepam were more often associated with seizure freedom in our cohort within 2 years of life.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-023-01474-2.
Keywords: STXBP1-related disorders, Genotype-phenotype correlation, Intellectual disability, Global developmental delay, Epilepsy, Treatments
Background
Syntaxin-binding protein 1 (STXBP1) is located on chromosome 9q34.3, and it encodes for an important protein that regulates neurotransmitter release [1]. STXBP1-related disorders are spectrum of conditions characterized by neurodevelopmental abnormalities and epilepsy [2–4]. The STXBP1 spectrum comprises early onset epileptic encephalopathy (EOEE), neurodevelopmental disorders, developmental epileptic encephalopathy (DEE), West syndrome (WS), Ohtahara syndrome (OS), and atypical Rett syndrome [4]. The predicted incidence rate of the STXBP1-related disorders is 3.30–3.81 per 100 000 births [5]. A few studies from China have evaluated genotype-phenotype correlations of STXBP1 pathogenic variants [6–8]. Some studies from other parts of the world have also examined the topic [2, 4].
This study aims to analyze the genotype-phenotype correlations of STXBP1 pathogenic variants, prognostic factors and the treatment options. This study will expand the understanding of STXBP1-related disorders, clinical management and research for potential treatment.
Materials and methods
Ethical clearance
Written consents were obtained from the parents or guardians of the subjects which were approved by the Institutional Ethics Committee of Xiangya Hospital, Central South University, China.
Participants
The clinical data of the patients diagnosed with STXBP1-related disorders and referred to Xiangya hospital, Central South University from 2011 to 2019 were collected retrospectively. We included all patients with DEE/developmental encephalopathy (DE) pathogenic/likely pathogenic variants of STXBP1.
Data collection
Retrospective data was collected including demographics, onset age, medical history, seizure semiology, family history, growth and development record, physical examination (head circumference, malformations, muscle tone, other neurologic examinations), treatment provided, and treatment outcomes. Additional data that was collected included blood glucose, complete blood count, electrolytes, urea, creatinine, AST, ALT, creatine kinase, ammonia, lactate, homocysteine, serum ceruloplasmin, electroencephalograph (EEG), magnetic resonance imaging (MRI), and intelligence test (IQ/DQ).
Routine methods of intelligence assessment
The degree of intellectual disability (ID) or global developmental delay (GDD) was assessed using the diagnostic criteria of the DSM-5 for intellectual disabilities (Diagnostic and Statistical Manual of Mental Disorders, 5th Edition, American Psychiatric Association, 2013). Standardized age-related rating scales, clinical interview and observations were used for the assessment of adaptive functioning. However, diagnosis was often made by clinical judgment instead of using formal standardized assessments, especially for young patients [9]. Standardized age-related rating scales that were used included: Gesell Developmental Schedules for patients younger than 2–4 years, Wechsler Preschool and Primary Scale of Intelligence-Fourth Edition (WPPSI-IV) for patients between 4 and 6 years, and Wechsler Intelligence Scale for Children-Fourth Edition (WISC-IV) for patients who were 6 years old or above. More details regarding assessment of intelligence can be found in our previous studies [10, 11].
Grouping of the patients
We divided our patients into groups for comparison purposes: groups of patients with missense (including non-frameshift) and nonsense (including splice and frameshift) pathogenic variants, patients who were seizure-free and not seizure-free (including the deceased ones), and groups with mild to moderate ID/GDD and severe to profound ID/GDD (including the deceased ones).
Evaluation of the efficacy of anti-seizure medications
Patients were followed retrospectively to determine their response towards anti-seizure medications. Seizure freedom was defined as lack of seizures for more than one year. A medicine that led to the reduction of seizures frequency by 50% within the first month of the administration was considered to be effective. The last follow up was on November 2022.
Mutation analysis
For the children and parents who consented to genetic testing, 2 millimeters of blood were collected in EDTA anticoagulant tube, and the DNA was extracted within 72 h. Genetic tests included whole exome sequencing (WES), sub-whole exome sequencing (SWES), customized multigene panel and molecular inverted probe (MIP). Samples were tested at the precision medical genetic testing company (www.precisionmdx.com). Sanger sequencing was used to verify the parental origin of the identified variants. The genetic results were collected and interpreted according to the variant curation guidelines published in 2015 by the American College of Medical Genetics (ACMG) [12]. The pathogenicity of all detected variants was rated as: pathogenic (P), likely pathogenic (LP), variant of unknown significance (VUS), likely benign (LB) and benign (B).
T vector sequencing
For families with possible mosaicism, at least 100 T vectors from suspected mosaicism sources were sequenced to calculate the mutation ratio.
Statistical analysis
Statistical analysis was done using SPSS Version 25 (IBM, Armonk, NY). Categorical data was summarized in the form of frequencies and proportions. Univariate statistical analysis was carried out to compare different clinical variables (including age of onset, sex, EEG, brain imaging features, and severity of ID/GDD) among groups of the patients with missense and nonsense pathogenic variants. Similar comparison was done for patients who were seizure-free and not seizure-free, and patients with mild to moderate ID/GDD and severe to profound ID/GDD in order to identify the prognostic factors. The Chi-squared test or Fisher’s exact test was used when applicable. Results with a value of P ≤ 0.05 were considered to be statistically significant.
Results
Baseline characteristics of the whole cohort
A total of 19 patients were enrolled in this study, and 12 (63.2%) were females. Onset age of seizures ranged from 1 day to 1 year; and it was 3 months for 14 (73.7%) individuals. DEE was observed in 18 (94.7%) patients; whereas ID only was seen in 1 (5.2%) individual. Thirteen patients (68.4%) had profound ID/GDD, and 4 (23.53%) had severe ID/GDD (Table 1). Three children with profound GDD died: one at the age of 2 years, one at the age of 6 months and one at the age of 7 years. The causes of deaths were severe uncontrolled epilepsy for 2 patients and unknown for one child.
Table 1.
Baseline characteristics of the whole cohort
| Variable | Total number of cases (n = 19) | Percentage (%) |
|---|---|---|
| Sex | ||
| Male | 7 | 36.8% |
| Female | 12 | 63.2% |
| Ethnicity | ||
| Han | 19 | 100% |
| Non-Han | 0 | 0% |
| Age of onset of seizures | ||
| ≤ 3 m | 14 | 73.7% |
| 4–12 m | 4 | 21.1% |
| > 12 m | 1 | 5.3% |
| Presence of seizures | ||
| Yes | 18 | 94.7% |
| No | 1 | 5.3% |
| Intellectual disability/global developmental delay categories | ||
| Mild | 1 | 5.9% |
| Moderate | 1 | 5.9% |
| Severe | 4 | 23.53% |
| Profound | 13 | 68.4% |
| Presence of hypsarrhythmia in an EEG | ||
| Yes | 11 | 57.9% |
| No | 7 | 36.8% |
| No EEG results | 1 | 5.3% |
| Presence of burst-suppression in EEG | ||
| Yes | 7 | 36.8% |
| No | 11 | 57.9% |
| No EEG results | 1 | 5.3% |
| Brain MRI results | ||
| Normal | 13 | 68.4% |
| Abnormal | 5 | 26.3% |
| No MRI results | 1 | 5.3% |
Abbreviations: EEG: electroencephalograph, m; month, MRI; magnetic resonance imaging.
Genetic analysis results
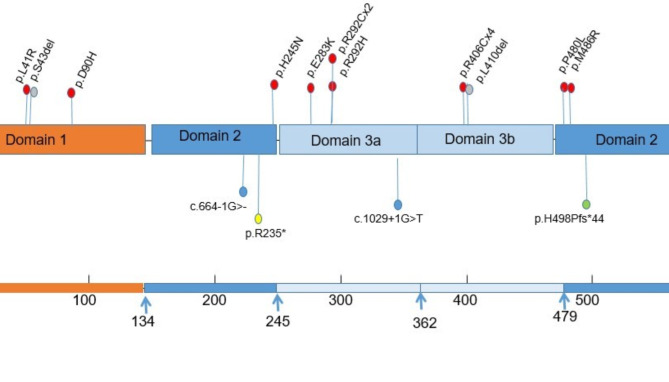
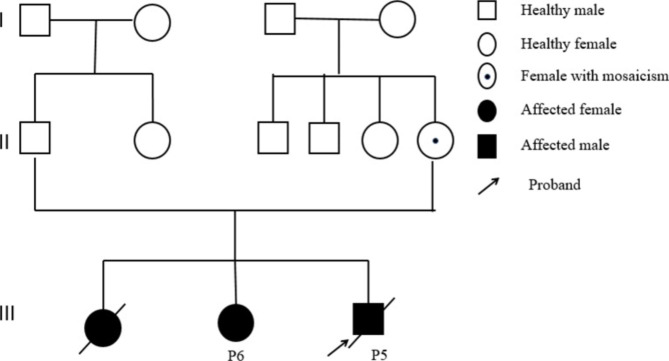
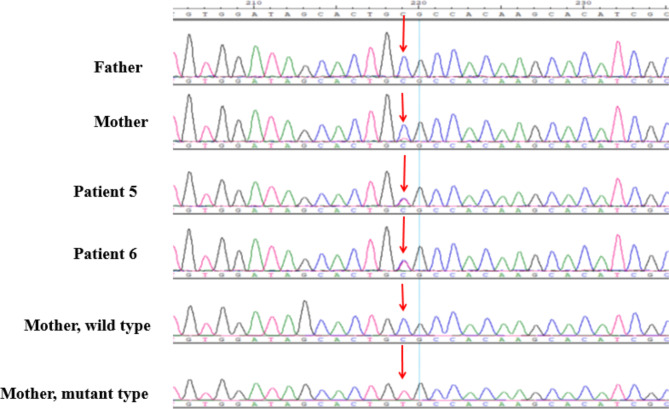
A total of 19 variants were detected in those 19 patients: pathogenic (n = 15) and likely pathogenic (n = 4). We found 15 different STXBP1 variants: 7 novel and 8 reported in previous studies. The novel variants were c.664-1G>-, M486R, H245N, H498Pfs*44, L41R, L410del, and D90H. Of the 6 previous reported variants [4, 17–20, 22, 24–30], 2 variants were recurrent (detected in ≥ 2 children): R406C, R292C. The details of the variants have been submitted to ClinVar public database (https://www.ncbi.nlm.nih.gov/clinvar). The pathogenic variants were de novo for 16 individuals and inherited from maternal side for 3 children (Table 2). The mother of the two non-twin siblings (P5 and P6) had DEE and the mother of P14 had history of febrile seizures but she is well now. The P5 and P6’ mother had mild ID and had a history of experiencing syncope. Figure 1 summarizes the locations of the identified variants. Notably, maternal mosaicism was discovered in mother of P5 and P6 (Fig. 2A and Fig. 2B). All variants were detected by the next generation sequencing (NGS), including 11 patients of WES, 2 patients of SWES, 3 individuals of epileptic gene panel, and 2 with MIP sequencing of nerve development related genes.
Table 2.
Genetic analysis results
| Case | Genomic position | Base alteration | Amino acid change | Exon | Domain | Pathogenic variant type | Parental origin | ACMG scoring | Score | Reported/Not reported |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | Chr9:130438188 | c.1216 C > T | p.R406C | 14 | 3b | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [24, 26, 27] |
| P2 | Chr9:130434297 | c.664-1G>- | - | 9 | 3a | Splice | De novo | PVS1 + PS2 + PM2 | P | No |
| P3 | Chr9:130430439 | c.875G > A | p.R292H | 10 | 3a | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [28, 29] |
| P4 | Chr9:130438188 | c.1216 C > T | p.R406C | 14 | 3b | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [24, 26, 27] |
| P5 | Chr9:130430438 | c.874 C > T | p.R292C | 10 | 3a | Missense | Maternal | PS1 + PS4 + PM2 + PP1 | P | [2] |
| P6 | Chr9:130430438 | c.874 C > T | p.R292C | 10 | 3a | Missense | Maternal | PS1 + PS4 + PM2 + PP1 | P | [2] |
| P7 | Chr9:130440807 | c.1457T > G | p.M486R | 16 | 2 | Missense | De novo | PS2 + PM2 | LP | No |
| P8 | Chr9:130438188 | c.1216 C > T | p.R406C | 14 | 3b | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [24, 26, 27] |
| P9 | Chr9:130428514 | c.733 C > A | p.H245N | 8 | 2 | Missense | De novo | PS2 + PM2 + PP2 | P | No |
| P10 | Chr9:130434396 | c.1029 + 1G > T | - | 12 | 3a | Splice | De novo | PS1 + PS2 + PM2 | P | [30] |
| P11 | Chr9:130442466 | c.1493_1505del | p.H498Pfs*44 | 17 | 2 | Frameshift | De novo | PVS1 + PS2 + PM2 | LP | No |
| P12 | Chr9:130440789 | c.1439 C > T | p.P480L | 17 | 2 | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [4, 17–20] |
| P13 | Chr9:130438188 | c.1216 C > T | p.R406C | 14 | 3b | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | [24, 26, 27] |
| P14 | Chr9:130430411 | c.847G > A | p.E283K | 10 | 3a | Missense | Maternal | PS1 + PM2 + PP1 | P | [19] |
| P15 | Chr9:130416028 | C.122T > G | p.L41R | 3 | 1 | Missense | De novo | PS1 + PS2 + PS4 + PM2 | P | No |
| P16 | Chr9:130438197 | c.1227_1229del | p.L410del | 14 | 3b | Non frameshift | De novo | PS2 + PM2 + PM4 | LP | No |
| P17 | Chr9:130422330 | c.268G > C | p.D90H | 5 | 1 | Missense | De novo | PS2 + PS4 + PM2 | P | No |
| P18 | Chr9:130416030 | c.128_130del | p.S43del | 3 | 1 | Non frameshift | De novo | PS2 + PM2 | LP | [22] |
| P19 | Chr9:130428484 | c.703 C > T | p.R235* | 10 | 2 | Nonsense | De novo | PVS1 + PS1 + PM2 | P | [25] |
Abbreviations: P; pathogenic, LP: likely pathogenic.
Fig. 1.
This figure shows the distributions of the identified STXBPI variants. Red circles represent missense pathogenic variants, grey circles represent non-frameshift pathogenic variants, blue circles represent splice pathogenic variants, yellow circles represent nonsense pathogenic variant, and green circles represent frameshift pathogenic variant
Fig. 2A.
Pedigree map of patient 5 (P5) and patient 6 (P6) with p.R292C pathogenic variant. Squares represent males, circles represent females, filled squares and circles represent individuals affected, and circle with a small dot inside represent maternal mosaicism of the p.R292C pathogenic variant
Fig. 2B.
p.R292C Sanger sequencing map and mother T vector sequencing peak map. Arrows indicate pathogenic variant sites
Phenotypes, treatment and outcomes of the patients with missense and non-frameshift pathogenic variants
Fifteen patients (78.95%) carried missense and non-frameshift pathogenic variants. The onset age ranged from 2 days to 3 months, and the mean current age is 7.7 years. Fourteen patients (93.3%) had DEE while 1 (6.7%) had ID only. Eleven patients (73.3%) had severe to profound ID/GDD. Five patients (33.3%) were hypertonic, 6 (40%) had normal muscle tone, 3 (20%) were hypotonic, and 1 (6.7%) was hypotonic while young but changed to hypertonic as she grew up. Of the 14 children with DEE, 5 (35.7%) had OS initially but later evolved to WS; 3 (21.4%) had EOEE initially but evolved to WS later; and 3 (21.4%) had WS from the beginning. The commonest initial seizure classification of those 14 individuals were as follows: 5 (35.7%) presented with focal seizures at the beginning, 1 (7.1%) with tonic-clonic, 6 (42.8%) with spasms, and 5 (35.7%) with tonic seizures. Nevertheless, the seizure semiology changed later. Five (35.7%) of 14 children with brain imaging results information had abnormal conventional brain MRI. Of those 5 patients, 1 (20%) had thin corpus callosum, 1 (20%) had arachnoid cyst on the right side, 1 (20%) had thickened parietal cortex, 1 (20%) had widened cerebral lateral ventricle mostly on the left side, and 1 (20%) had increased T1WI signal in bilateral basal ganglia, bilateral thalamus and brainstem dentate nucleus.
Out of the 15 individuals that presented with missense and non-frameshift pathogenic variants, 14 patients (93.3%) had anti-seizure medications information; and 1 patient (7.1%) who had ID was therefore exempted. Two patients (14.2%) received one anti-seizure medication, 2 patients (14.2%) received 2 anti-seizure medications, 3 patients (21.4%) received 3 anti-seizure medications 3 patients (21.4%) received 4 anti-seizure drugs, and 3 patients (21.4%) received ≥ 5 anti-seizure drugs. Of those 14 individuals, 6 (42.9%) became seizure-free, 6 (42.9%) were not seizure-free and 2 (14.2%) died. Five (83.3%) of the 6 seizure-free cases achieved seizure freedom within 2 years of life. Efficient drugs for seizure-free patients included the combination of ACTH and/or levetiracetam and/or phenobarbital and/or sodium valproate and/or topiramate and/or vigabatrin. Table 3 summarizes this information.
Table 3.
Phenotypes, treatment and outcomes of the patients with missense and non-frameshift pathogenic variants
| Patient number | Pathogenic variant type | Sex | Current age (y) | Diagnosis | Seizure onset age | Initial seizure semiology (s) | Latest seizure semiology (s) | EEG manifestations | MRI findings | Previous medicine (s) | Recent medicine (s) | Efficient medicine (s) | Seizure outcome (age) | Muscle tone | Degree of ID/GDD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | Missense | F | 5.4 | OS evolved to WS | 2m10d | FS | TS, S, FMS, FCS | H/BS/M | A | TPM,VPA, ACTH and PRD | LEV | ACTH and LEV | NSF | High | Profound |
| P3 | Missense | F | 5.3 | EOEE evolved to WS | 16d | FS | FS, S | M | N | PB and LEV | LEV | PB and LEV | SF (0.3 y) | Normal | Severe |
| P4 | Missense | M | 11.3 | WS | 2 m | TCS, S | TCS, S | ESE/H/ M | A | VPA, TPM, ACTH and PRD | No medication | VPA and ACTH | SF (1 y) | High | Profound |
| P5 | Missense | M | Died | OS evolved to WS | 2d | FS, S | TS, S, FS, | H/BS | A | LEV | Died | - | Died (2 y) | High | Died |
| P6 | Missense | F | 9.8 | ID, EP | 7 m | TS | TS | No results | No results | No medicine | No medicine | No medicine | NSF | Low/high | Profound |
| P7 | Missense | M | 4.8 | OS evolved to WS | 1m19d | S | S, FS, FS-GTS | H/BS/W/F | N | LEV, TPM, ACTH, CZP, and NZP | LEV | ACTH | SF (0.6 y) | Normal | Severe |
| P8 | Missense | M | Died | OS evolved to WS | 2m28d | TS, S | S, TS | BS/F | A | VPA and ACTH | Died | None | Died (0.5 y) | Normal | Died |
| P9 | Missense | F | 9 | OS evolved to WS | 0.5 m | S, TS | S, FS, T | H/BS/M/W | N | TPM, ACTH, PRD, LEV, NZP, and VNS | LEV, TPM | None | NSF | High | Profound |
| P12 | Missense | M | 7.8 | ID | 10 m | Not applicable | Not applicable | Slow back ground | N | Not applicable | Not applicable | Not applicable | Not applicable | High | Profound |
| P13 | Missense | F | 13 | ID, EP | 5 m | TS | TS | W | A | LEV | No medication | LEV | SF (10y) | Normal | Moderate |
| P14 | Missense | F | 4.1 | GDD, EP | 1y3m | TS | TS, FS | M | N | VPA and LEV | LEV | LEV | NSF | Normal | Mild |
| P15 | Missense | F | 5.6 | WS | 2d | FS | FS, S | M | N | TPM and VPA | TPM, VPA | TPM and VPA | SF (2y) | Low | Severe |
| P16 | Non frameshift | F | 5.8 | WS | 7d | S | S, TS | H/W | N | VPA, ACTH, and VGB | VPA | ACTH and VGB | NSF | Low | Severe |
| P17 | Missense | F | 5.4 | EOEE evolved to WS | 1m23d | FS | FS, TS, S | W | N | LEV, TPM, and VPA | LEV and VPA | LEV and VPA | SF (2y) | Normal | Profound |
| P18 | Non frameshift | F | 5.4 | EOEE evolved to WS | 4 m | TS | TS, S, FS, | W/H/F | N | LEV, VPA, ACTH, KD, VGB, and CLB | VGB | LEV, VPA and VGB | NSF | Low | Profound |
Abbreviations: A; abnormal, BS; burst-suppression, d; day, EOEE; early onset epileptic encephalopathy, EEG; electroencephalography, ESE; epileptic status epilepticus, F; female, FCS; focal-clonic seizures, FS; focal seizures, FS-GTCS; focal seizures secondary to generalized tonic-clonic seizures, FS-S; focal spasm seizures, AAS; atypical aphasia, GDD; global developmental delay, H; hypsarrhythmia, ID; intellectual disability, m; month, M; male; MRI; magnetic resonance imaging, M; multifocal discharge, N; normal, OS- ohtahara syndrome, S; spasms seizures, TS; tonic seizures, TCS; tonic clonic seizures, W; widespread discharge, WS; west syndrome, ACTH; adrenocorticotropic hormone, CZP; clonazepam, KD; ketogenic diet, LEV; levetiracetam, NZP; nitrazepam, PRD; prednisone tablets, PB; phenobarbital, P; patient, TPM; topiramate, VPA; sodium valproate, VGB; vigabatrin, CLB; clobazam, VNS; vagus nerve stimulation, SF; seizure free, NS; not seizure-free
Phenotypes, treatment and outcomes of the patients with nonsense pathogenic variants
Four patients (21%) out of 19 patients had nonsense pathogenic variants. The onset age ranged from 1 day to 2 months. Two patients (50%) had OS that evolved to WS and 2 patients (50%) had EOEE that evolved to WS. Two patients (50%) presented with focal seizures at the beginning. Two patients (50%) were hypertonic, 1 (25%) was hypotonic and 1 (25%) had alternating muscle tone (high/low) as she grew up. All 4 patients (100%) had profound ID/GDD. At the end of follow up, 1 patient (25%) became seizure-free at the age of 8 years, and 2 patients (50%) were not seizure-free and 1 patient (25%) died. ACTH and levetiracetam were effective for the patient who achieved seizure freedom (Table 4).
Table 4.
Phenotypes, treatment and outcomes of the patients with nonsense (including splice and frameshift) pathogenic variants
| Patient number | Pathogenic variant type | Sex | Current age | Diagnosis | Seizure onset age | Initial seizure semiology (s) | Latest seizure semiology (s) | EEG manifestations | MRI findings | Previous drug (s) | Current drug (s) | Efficient drug (s) | Seizure outcome | Muscle tone | Degree of ID/GDD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P2 | Splice | F | 6.8y | OS evolved to WS | 8d | FS | FS, TS, S | H/BS/W | N | ACTH, PRD, NZP, and LEV | VPA, TPM, and CBZ | ACTH and LEV | NSF | High | Profound |
| P10 | Splice | F | 9.6y | EOEE evolved to WS | 2 m | FS | S, FS | H | N | ACTH, PRD, VPA, and NZP | None | ACTH and NZP | SF (8y) | High/Low | Profound |
| P11 | Frameshift | M | Died (7y) | EOEE evolved to WS | 1d | TS | TS, S, AAS | H/W/M | N | LEV, ACTH, PRD, VPA, TPM, and NZP | Died(7y) | LEV, ACTH and TPM | Died (7y4m) | High | Profound |
| P19 | Nonsense | M | 7.6y | OS evolved to WS | 2 m | TS, FS | TS, FS, S | BS/H/M | N | TPM, VPA, VGB, and LEV | LEV and VPA | VPA, VGB and LEV | NSF | Low | Profound |
Abbreviations: d; days, EP; epilepsy, EOEE; early onset epileptic encephalopathy, F; female, FS; focal seizure, GDD; global developmental delay, ID; intellectual disability, M; male, m: months, OS; ohtahara syndrome, S; spasms seizure, TS; tonic seizure, TCS; tonic clonic seizure, W: widespread spike, y; years.
The overall seizure outcomes for both groups of patients with missense and non-frameshift pathogenic variants as well as patients with nonsense (including splice and frameshift) pathogenic variants
At the end of follow up, 7 patients (38.9%) became seizure-free and most of them achieved seizure freedom at the age of ≤ 2 years. Few individuals had early seizure onset but delayed offset. Patients whose current age ranged from 6 to 9 years, lacked seizure freedom in comparison to the younger age (< 6 years) and older age (> 9 years).
Comparison of phenotypes between groups of patients with missense against those with nonsense pathogenic variants
The comparison between the groups of patients who carried missense against those with nonsense pathogenic variants was carried out. However, there was no statistically significant difference observed between the groups (Supplementary Table 1).
Factors associated with seizure-freedom according to univariate analysis.
The presence of epileptic spasms, onset age, hypsarrhythmia, burst suppression pattern, type of the pathogenic variant, and the utilization of ACTH, sodium valproate, levetiracetam and topiramate did not show any association with seizure outcome (Supplementary Table 2).
Factors associated with the severity of the intellectual disability/global developmental delay according to univariate analysis.
The presence or absence of burst suppression pattern, presence or absence of hypsarrhythmia, abnormal brain imaging results, seizure outcome and the type of the pathogenic variant did not show any association with the degree of severity of ID/GDD (Supplementary Table 3).
Discussion
This case series encompassed 17 unrelated and 2 familial patients in which the genotypes and phenotypes of STXBP1-related disorders were analyzed. A total of 15 variants were detected, of which 7 were novel and 2 were recurrent. The initial clinical diagnosis included DEE and ID only. All individuals had different degrees of ID/GDD, mainly severe to profound. Majority of the patients presented with epileptic spasms and focal seizures. We could not find genotype-phenotype correlation. Some patients presented with abnormal movements. Seizure-free patients in both groups of patients with missense and nonsense pathogenic variants received the combination of ACTH and/or levetiracetam and/or phenobarbital and/or sodium valproate and/or topiramate and/or vigabatrin and/or nitrazepam.
The proportion of missense pathogenic variants was the highest in our study which is similar to the recent study [3]. A recent study has revealed that splice site, frameshift variants, whole gene deletions and partial gene deletions were linked with WS, infantile spasms, and ataxia; while patients with missense variants were more likely to have other DEE [3]. Nevertheless, our study and two other studies did not find a genotype-phenotype correlation [2, 13]. Lack of the genotype-phenotype correlations in our study can be due to the small sample size to obtain spontaneous clusters of sub-phenotypes and too heterogeneous for a holistic approach.
Our study revealed 4 patients with p.R406C variant and EOEE, WS, and DEE; which corroborates the recent findings of recent publications that genetic hotspots for STXBP1 include p.R406C (n = 40) followed by p.R292C /H/L/P (n = 30)[3]. Although our study showed that the pathogenic variant sites of P5 and P6 were the same (p.R292C), and they were siblings with the same genetic background; however, their phenotypes were different. One female sibling had similar phenotype as P5, however, the genetic results were missing. Nevertheless, their mother had mild phenotype. Notably, maternal mosaicism was discovered in the mother of P5 and P6, which is similar to previous reports [14–16]. In this study, the pathogenic variant site of P12 was p. P480L but his phenotype differ from the previous reported individuals; the phenotype of this child was a profound ID complicated with ataxia while the 2 previously reported individuals presented with WS [17–20].
All of our patients presented with different degrees of ID/GDD, mainly severe to profound, and all except one presented with DEE which is similar to the review [2]. A recent study conducted in adults suggested that severe cognitive impairments and movement disorders involving multiple systems are often present in STXBP1-DEE; which correlates with our findings [21]. In addition, it has been revealed that the age of seizure onset is correlated with severity of ID/GDD; a later seizure onset is associated with better developmental outcome [22]. Burst suppression pattern in EEG has been reported to relate with worse seizure prognosis [23], and it is linked with p.R406C/H variants [4]. Nevertheless, we could not establish such a relationship in our study due to a small sample size.
Most of our patients presented with seizures within the first year of life. It has been reported that seizure frequency is highest in the first year of life, but decreases dramatically by the age of 7 years [4]. Seizure freedom was achieved within 2 years of life for the majority of our patients, but we have several patients who continued to have seizures at 6–9 years. It has been reported that STXBP- related disorders can have prolonged seizure-free periods but never achieving permanent seizure control [21]. We have observed this pattern in three of our patients: P6, P11 and P16. Consequently, we are not certain whether the seizure freedom among the remained cases will be sustainable. It is difficult to control epileptic seizures in STXBP1-related disorders, and therapies are not helpful for ID/GDD. Most patients need to be treated with two or more anti-seizure medications. In this study, individuals who received the combinations of the ACTH and/or levetiracetam and/or phenobarbital and/or sodium valproate and/or topiramate and/or vigabatrin and/or nitrazepam achieved seizure freedom within the first 2 years of life which is similar to one study which reported that anti-seizure medications response were limited to the first 2 years of life [4]. Notably, ACTH and phenobarbital were also reported recently to have beneficial effect in seizure frequency reduction in WS and focal seizures, respectively [4]. In our cohort, none of the medicines seem to be superior to others. One of our patients was treated with VNS, but it aggravated seizures. There is no previous report of patients treated with VNS, indicating that further studies are required about the therapeutic effect of VNS for this condition.
Conclusion
STXBP1-related disorders is a spectrum of disease whose major clinical manifestations include DEE, ID, and abnormal movements. It begins from the age of 1 month and presents with different types of seizures. Most of the patients present with WS as initial or final epileptic syndrome. It is difficult to control seizures and improve cognition. However, the combination of the ACTH and/or levetiracetam and/or phenobarbital and/or sodium valproate and/or topiramate and/or vigabatrin and/or nitrazepam seem to be effective in seizures management especially in the first 2 years of life. Some cases may have a temporary seizure freedom that continues later in life. This study adds 7 novel variants which expand the mutational spectrum of STXBP1-related disorders.
Limitations
This study involves a small sample size and it was retrospective in nature. We could not find genotype-phenotype correlations since the sample size was too small to obtain spontaneous clusters of sub-phenotypes, and too heterogeneous for a holistic approach as many of our patients evolved from one condition to another. The retrospective nature of the study makes the revision of instrumental (EEG/MRI) and laboratory (e.g., genetic) investigations quite approximate. It could not provide the details of the efficacy of the individual drugs used. Future prospective studies are invited to expand the understanding of this devastating condition.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the participating patients and their families.
List of Abbreviations
- ACTH
Adrenocorticotropic hormone
- B
Benign
- DSM-5
Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition, American Psychiatric Association, 2013
- EEG
Electroencephalograph
- MRI
Magnetic resonance imaging
- ID
Intellectual disability
- GDD
Global developmental delay
- DEE
Developmental epileptic encephalopathy
- DE
Developmental encephalopathy
- EOEE
Early onset epileptic encephalopathy
- VNS
Vagus nerve stimulation
- STXBP1-E
STXBP1-encephalopathy
- WES
Whole exome sequencing
- SWES
Sub-whole exome sequencing
- MIP
Customized multigene panel and molecular inverted probe
- NGS
Next generation sequencing
- WS
West syndrome
- OS
Ohtahara syndrome
- P
Pathogenic
- LP
Likely pathogenic
- VUS
Variant of unknown significance
- LB
Likely benign
Author Contribution
M.K is the first author who drafted and revised the manuscript. B.C, L.S collected data and carried out the initial analyses. Y.W, L.Y, F.Y, F.H and J.P coordinated and supervised data collection. G.W is the corresponding author who conceptualized and designed the study, coordinated and supervised data collection, and critically reviewed the manuscript for important intellectual content. All authors reviewed the manuscript and approved the submitted version (and any substantially modified version that involves the author’s contribution to the study) and have agreed both to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
Funding
We are grateful for the support we received from the National Natural Science Foundation of China [No. 81801297, No.81771408], and the Hunan Key Research and Development Program [No.2019SK2081].
Data Availability
The datasets generated and/or analysed during the current study are available in the ClinVar public database (clinvar@ncbi.nlm.nih.gov) repository.
Declarations
Ethics approval and consent to participate
The study including all methods adhered to the tenets of the Declaration of Helsinki and received approval from the Institutional Review Board and Research Ethics Committee of Xiangya Hospital, Central South University, Changsha, Hunan. Written consents were obtained from the parents/guardians of the subjects which were approved by the Institutional Ethics Committee of Xiangya Hospital, Central South University.
Consent for publication
Written informed consents for publication of clinical details and clinical images were obtained from participants or the parents of the participants under the age of 16.
Competing interests
None of the authors has any conflict of interest to disclose. The manuscript has been read and approved by all the authors, the requirements for authorship as stated in the journal guideline have been met, and each author believes that the manuscript represents honest work.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swanson DA, Steel JM, Valle D. Identification and characterization of the human ortholog of rat STXBP1, a protein implicated in vesicle trafficking and neurotransmitter release. Genomics. 1998;48:373–6. doi: 10.1006/geno.1997.5202. [DOI] [PubMed] [Google Scholar]
- 2.Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, Baier H, et al. STXBP1 encephalopathy: a neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–62. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- 3.Xian J, Parthasarathy S, Ruggiero SM, Balagura G, Fitch E, Helbig K, et al. Assessing the landscape of STXBP1-related disorders in 534 individuals. Brain. 2022;145:1668–83. doi: 10.1093/brain/awab327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xian J, Parthasarathy S, Ruggiero SM, Balagura G, Fitch E, Helbig K et al. Assessing the landscape of STXBP1-related disorders in 534 individuals.Brain. 2021. [DOI] [PMC free article] [PubMed]
- 5.López-Rivera JA, Pérez-Palma E, Symonds J, Lindy AS, McKnight DA, Leu C, et al. A catalogue of new incidence estimates of monogenic neurodevelopmental disorders caused by de novo variants. Brain. 2020;143:1099–105. doi: 10.1093/brain/awaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Cheng M, Wang J, Hong S, Li M, Liao S, et al. De novo mutations of STXBP1 in chinese children with early onset epileptic encephalopathy. Genes Brain Behav. 2018;17:e12492. doi: 10.1111/gbb.12492. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Wang L, Cai XT, Zhou H, Yu D, Wang Z. Therapeutic benefits of ACTH and levetiracetam in STXBP1 encephalopathy with a de novo mutation: a case report and literature review. Med (Baltim) 2018;97:e0663. doi: 10.1097/MD.0000000000010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao JJ, Ji XN, Mao YY, Zhang PP, Liu WT, Zhang HZ, et al. [Clinical and genetic characteristics of children with STXBP1 encephalopathy]. Zhonghua er ke za zhi = Chinese. J Pediatr. 2020;58:493–8. doi: 10.3760/cma.j.cn112140-20191028-00683. [DOI] [PubMed] [Google Scholar]
- 9.Shevell M. Global developmental delay and mental retardation or intellectual disability: conceptualization, evaluation, and etiology. Pediatr Clin North Am. 2008;55:1071–84. doi: 10.1016/j.pcl.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Kessi M, Xiong J, Wu L, Yang L, He F, Chen C, et al. Rare Copy Number Variations and Predictors in Children with Intellectual Disability and Epilepsy. Front Neurol. 2018;9:947. doi: 10.3389/fneur.2018.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arafat A, Jing P, Ma Y, Pu M, Nan G, Fang H, et al. Unexplained early infantile epileptic Encephalopathy in Han Chinese Children: next-generation sequencing and phenotype enriching. Sci Rep. 2017;7:46227. doi: 10.1038/srep46227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramov D, Guiberson NGL, Burré J. STXBP1 encephalopathies: clinical spectrum, disease mechanisms, and therapeutic strategies. J Neurochem. 2021;157:165–78. doi: 10.1111/jnc.15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uddin M, Woodbury-Smith M, Chan A, Brunga L, Lamoureux S, Pellecchia G, et al. Germline and somatic mutations in STXBP1 with diverse neurodevelopmental phenotypes. Neurol Genet. 2017;3:e199. doi: 10.1212/NXG.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Møller RS, Liebmann N, Larsen LHG, Stiller M, Hentschel J, Kako N, et al. Parental mosaicism in epilepsies due to alleged de novo variants. Epilepsia. 2019;60:e63–6. doi: 10.1111/epi.15187. [DOI] [PubMed] [Google Scholar]
- 16.Saitsu H, Hoshino H, Kato M, Nishiyama K, Okada I, Yoneda Y, et al. Paternal mosaicism of an STXBP1 mutation in OS. Clin Genet. 2011;80:484–8. doi: 10.1111/j.1399-0004.2010.01575.x. [DOI] [PubMed] [Google Scholar]
- 17.Di Meglio C, Lesca G, Villeneuve N, Lacoste C, Abidi A, Cacciagli P, et al. Epileptic patients with de novo STXBP1 mutations: key clinical features based on 24 cases. Epilepsia. 2015;56:1931–40. doi: 10.1111/epi.13214. [DOI] [PubMed] [Google Scholar]
- 18.Milh M, Villeneuve N, Chouchane M, Kaminska A, Laroche C, Barthez MA, et al. Epileptic and nonepileptic features in patients with early onset epileptic encephalopathy and STXBP1 mutations. Epilepsia. 2011;52:1828–34. doi: 10.1111/j.1528-1167.2011.03181.x. [DOI] [PubMed] [Google Scholar]
- 19.Tso WWY, Kwong AKY, Fung CW, Wong VCN. Folinic acid responsive epilepsy in Ohtahara syndrome caused by STXBP1 mutation. Pediatr Neurol. 2014;50:177–80. doi: 10.1016/j.pediatrneurol.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Shen Q, Zheng G, Guo H, Lu X, Wang X, et al. Gene and phenotype expansion of unexplained early infantile epileptic Encephalopathy. Front Neurol. 2021;12:633637. doi: 10.3389/fneur.2021.633637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamberger H, Crosiers D, Balagura G, Bonardi CM, Basu A, Cantalupo G, et al. Natural history study of STXBP1-Developmental and epileptic Encephalopathy Into Adulthood. Neurology. 2022;99:e221–33. doi: 10.1212/WNL.0000000000200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balagura G, Xian J, Riva A, Marchese F, Ben Zeev B, Rios L, et al. Epilepsy Course and Developmental Trajectories in STXBP1-DEE. Neurol Genet. 2022;8:e676. doi: 10.1212/NXG.0000000000000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes ML, Giraldes MM, Pinho AP, Costa JC. da. Prognostic value of non-reactive burst suppression EEG pattern associated to early neonatal seizures. Arq Neuropsiquiatr. 2005;63:14–9. [DOI] [PubMed]
- 24.Sajan SA, Jhangiani SN, Muzny DM, Gibbs RA, Lupski JR, Glaze DG, et al. Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet Med. 2017;19:13–9. doi: 10.1038/gim.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitsu H, Kato M, Okada I, Orii KE, Higuchi T, Hoshino H, et al. STXBP1 mutations in early infantile epileptic encephalopathy with suppression-burst pattern. Epilepsia. 2010;51:2397–405. doi: 10.1111/j.1528-1167.2010.02728.x. [DOI] [PubMed] [Google Scholar]
- 26.Vidal S, Brandi N, Pacheco P, Gerotina E, Blasco L, Trotta J-R, et al. The utility of Next Generation sequencing for molecular diagnostics in Rett syndrome. Sci Rep. 2017;7:12288. doi: 10.1038/s41598-017-11620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen NM, Conroy J, Shahwan A, Lynch B, Correa RG, Pena SDJ, et al. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016;57:e12–7. doi: 10.1111/epi.13250. [DOI] [PubMed] [Google Scholar]
- 28.Michaud JL, Lachance M, Hamdan FF, Carmant L, Lortie A, Diadori P, et al. The genetic landscape of infantile spasms. Hum Mol Genet. 2014;23:4846–58. doi: 10.1093/hmg/ddu199. [DOI] [PubMed] [Google Scholar]
- 29.Trump N, McTague A, Brittain H, Papandreou A, Meyer E, Ngoh A, et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J Med Genet. 2016;53:310–7. doi: 10.1136/jmedgenet-2015-103263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deprez L, Weckhuysen S, Holmgren P, Suls A, Van Dyck T, Goossens D, et al. Clinical spectrum of early-onset epileptic encephalopathies associated with STXBP1 mutations. Neurology. 2010;75:1159–65. doi: 10.1212/WNL.0b013e3181f4d7bf. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the ClinVar public database (clinvar@ncbi.nlm.nih.gov) repository.