Abstract
Background
The prevalence of multidrug-resistant hypervirulent K. pneumoniae (MDR-hvKP) has gradually increased. It poses a severe threat to human health. However, polymyxin-resistant hvKP is rare. Here, we collected eight polymyxin B-resistant K. pneumoniae isolates from a Chinese teaching hospital as a suspected outbreak.
Results
The minimum inhibitory concentrations (MICs) were determined by the broth microdilution method. HvKP was identified by detecting virulence-related genes and using a Galleria mellonella infection model. Their resistance to serum, growth, biofilm formation, and plasmid conjugation were analyzed in this study. Molecular characteristics were analyzed using whole-genome sequencing (WGS) and mutations of chromosome-mediated two-component systems pmrAB and phoPQ, and the negative phoPQ regulator mgrB to cause polymyxin B (PB) resistance were screened. All isolates were resistant to polymyxin B and sensitive to tigecycline; four were resistant to ceftazidime/avibactam. Except for KP16 (a newly discovered ST5254), all were of the K64 capsular serotype and belonged to ST11. Four strains co-harbored blaKPC-2, blaNDM-1, and the virulence-related genes prmpA, prmpA2, iucA, and peg344, and were confirmed to be hypervirulent by the G. mellonella infection model. According to WGS analysis, three hvKP strains showed evidence of clonal transmission (8–20 single nucleotide polymorphisms) and had a highly transferable pKOX_NDM1-like plasmid. KP25 had multiple plasmids carrying blaKPC-2, blaNDM-1, blaSHV-12, blaLAP-2, tet(A), fosA5, and a pLVPK-like virulence plasmid. Tn1722 and multiple additional insert sequence-mediated transpositions were observed. Mutations in chromosomal genes phoQ and pmrB, and insertion mutations in mgrB were major causes of PB resistance.
Conclusions
Polymyxin-resistant hvKP has become an essential new superbug prevalent in China, posing a serious challenge to public health. Its epidemic transmission characteristics and mechanisms of resistance and virulence deserve attention.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-023-02808-x.
Keywords: Klebsiella pneumoniae, Polymyxin B, ST11-K64, Hypervirulence, Clonal transmission, Whole-genome sequencing
Background
Klebsiella pneumoniae (KP) is a common clinical opportunistic pathogen causing various infections in immunocompromised hosts [1]. According to the China Antimicrobial Surveillance Network (https://www.chinets.com/), in 2021, K. pneumoniae ranked the second most prevalent isolate (14.12%). Carbapenems are essential antibiotics to treat severe K. pneumoniae infections. However, the resistance rates of K. pneumoniae to meropenem and imipenem have significantly increased from 2.9% and 3% in 2005 to 26.3% and 25% in 2018 [2]. The global spread of carbapenemases such as Klebsiella pneumoniae carbapenemase-2 (KPC-2) and New Delhi metallo-β-lactamase-1 (NDM-1) has exacerbated the health hazards worldwide [3, 4]. With the emergence of carbapenem-resistant isolates, polymyxins have been used in clinical treatment. Polymyxins, mainly including polymyxin B (PB) and polymyxin E (also called colistin), are a group of cationic, basic peptides produced by Bacillus polymyxa and have broad-spectrum antibacterial activity against Gram-negative bacteria [5]. Although polymyxins were disfavored due to their potential nephrotoxicity, they are currently used in hospitals and are known as the last line of defense for the clinical treatment of multidrug-resistant Gram-negative bacterial infections. However, as treatment with polymyxins is becoming more common, reports of polymyxin resistance (including chromosomal and plasmid-mediated resistance) are increasing, especially in areas with high polymyxin use [6, 7].
Hypervirulent Klebsiella pneumoniae (hvKP) is a new variety of K. pneumoniae that can cause severe infections to immunocompetent hosts. According to previous studies, hvKP is usually sensitive to most antibiotics, including cephalosporins and carbapenems [8]. However, recent years the emergence of multidrug-resistant hvKP (MDR-hvKP) has been seen, and the latest research has reported the increasing emergence of colistin-resistant hvKP [9]. Such multidrug-resistant and hypervirulent strains have brought great challenges to clinical anti-infective treatment.
Polymyxin B-resistant hvKP (PBR-hvKP) was rarely reported in previous research. Therefore, to identify the characteristics of PBR-hvKP and to explore its mechanisms of multidrug resistance and hypervirulence, we collected PB-resistant K. pneumoniae isolates and conducted in-depth investigations of their clinical and molecular characteristics, including information about their resistance and virulence.
Results
Clinical characteristics of eight patients infected with Polymyxin B-resistant K. pneumoniae
A total of eight non-repetitive clinical isolates of PB-resistant K. pneumoniae were collected. Clinical characteristics of patients infected with PB-resistant K. pneumoniae are summarised in Table 1. Six (75%) patients were male, with a mean age of 58.3 ± 15.0 years. The patients came from six different wards, and three (infected with KP17, KP21, and KP25) came from the same intensive care unit (ICU). Main underlying conditions included pneumonia (87.5%, 7/8), hypertension (37.5%, 3/8), abnormal liver function (37.5%, 3/8), tumor (37.5%, 3/8), and cerebral haemorrhage (25%, 2/8). Three patients had a disturbance of consciousness or were in a coma at admission. All the patients had prior antibiotic exposure before detecting PB-resistant K. pneumoniae. All the patients underwent invasive procedures, and six patients were admitted to ICU. The main antibiotics used within 72 h after infection included TZP (37.5%, 3/8), MEM (37.5%, 3/8), and TGC (25%, 2/8). Five patients were discharged without recovery. None of the patients died.
Table 1.
Clinical characteristics of patients infected with PB-resistant K. pneumoniae
| Patient | Strain | Residence time | Age/sex | Department | Underlying conditions | ICU stay time | Prior antibiotic exposure | Source of infection | Treated antibiotics in 72 h | Discharge status |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | KP25 | 2020.11.4–2020.12.20 | 65/Male | ICU | Multiple fractures, Severe pneumonia, Acute renal injury, Hypertension | 47 | SCF, LZD, TGC, TZP | Wound | TGC, TZP | Unhealed |
| 2 | KP14 | 2020.11.17–2020.12.4 | 57/Female | Recovery unit | Pneumonia, Rupture of intracranial aneurysm, Abnormal liver function | 8 | TZP | Respiratory tract | TZP | critically ill |
| 3 | KP17 | 2021.12.14–2021.2.20 | 70/Male | ICU | Severe pneumonia, Lung cancer surgery, Hypertension | 47 | TZP, MEM, PB, VA, CAZ | Respiratory tract | SCF | Improved |
| 4 | KP16 | 2021.1.5–2021.1.15 | 37/Male | Pancreato-biliary surgery unit | Acute severe pancreatitis | None | CDZ | Digestive tract | CDZ | Improved |
| 5 | KP18 | 2021.1.13–2021.3.15 | 38/Male | ICU, Neurosurgery unit | Pneumonia, Intracranial infection, Abnormal liver function | 71 | MEM, PB | Respiratory tract | MEM, TZP | critically ill |
| 6 | KP21 | 2021.2.18–2021.3.26 | 77/Female | ICU | Hepatocellular carcinoma, Pneumonia, Hepatitis B | 8 | LVX, CN, SCF | Liver | MEM | Unhealed |
| 7 | KP20 | 2021.3.8–2021.3.19 | 70/Male | ICU, Respiratory unit | Lymphoma, Severe pneumonia, Abnormal liver function, Type II diabetes | 14 | MEM, PB | Respiratory tract | MEM, PB | Unhealed |
| 8 | KP24 | 2021.3.25–2021.5.27 | 52/Male | Integrated traditional Chinese and western medicine unit | Brainstem hemorrhage, Pneumonia, Hypertension, Type II diabetes | None | PB, TGC | Urinary tract | TGC | Improved |
Abbreviations: ICU Intensive care unit, SCF Cefoperazone/sulbactam, LZD Linezolid, TGC Tigecycline, TZP Piperacillin/tazobactam, MEM Meropenem, PB Polymyxin B, VA Vancomycin, CDZ Cefodizime, LVX Levofloxacin, CN Gentamicin
Antimicrobial susceptibility and antibiotic resistance genes
All isolates were resistant to PB and sensitive to TGC; four were resistant to CZA (Table 2). All eight PB-resistant isolates carried carbapenemase gene blaKPC-2 and fosfomycin-resistance gene fosA; four isolates carried carbapenemase gene blaNDM-1; five isolates carried extended-spectrum β-lactamase gene blaSHV-12; and three isolates carried blaSHV-182, blaCTX-M-65, and blaTEM-1B (Fig. 1). The plasmid-borne colistin resistance gene mcr-1 was not detected. All isolates were found to have D150G amino acid substitutions in the phoQ gene and R256G amino acid substitution in the pmrB gene. Four isolates had different amino acid substitutions in the pmrB gene, including T157P and S85R. ISKpn74-like and IS903B-like insertion mutations in the mgrB gene were detected in two isolates (Table 2).
Table 2.
Antimicrobial susceptibility results of PB-resistant K. pneumoniae and the E. coli transconjugants and substitution mutations of chromosomal regulators in PB-resistant isolates
| Minimum inhibitory concentrations (μg/mL) | Mutations of chromosomal regulators | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Category | CAZ | FEP | CZA | ATM | TZP | NIT | IPM | MEM | AMK | LVX | TGC | PB | mgrB | phoQ | pmrB |
| KP14 | XDR | 64 | > 128 | ≤ 1/4 | > 128 | > 512/4 | 512 | > 32 | > 32 | > 512 | > 16 | 1 | > 16 | - | D150G | R256G |
| KP16 | MDR | > 128 | > 128 | ≤ 1/4 | > 128 | > 512/4 | 256 | 32 | 32 | ≤ 4 | 16 | 0.5 | 16 | - | D150G | R256G |
| KP17 | XDR | > 128 | > 128 | > 128/4 | > 128 | > 512/4 | 256 | > 32 | > 32 | > 512 | > 16 | 0.5 | 16 | - | D150G | T157P, R256G |
| KP18 | XDR | 64 | > 128 | ≤ 1/4 | > 128 | > 512/4 | 512 | > 32 | > 32 | > 512 | > 16 | 1 | > 16 | ISKpn74 | D150G | R256G |
| KP20 | XDR | 128 | > 128 | 1 | > 128 | > 512/4 | 256 | > 32 | > 32 | > 512 | 16 | 0.5 | 16 | - | D150G | S85R, R256G |
| KP21 | XDR | > 128 | > 128 | > 128/4 | > 128 | > 512/4 | 128 | > 32 | > 32 | > 512 | > 16 | 1 | 16 | - | D150G | T157P, R256G |
| KP24 | XDR | > 128 | > 128 | > 128/4 | > 128 | > 512/4 | 512 | > 32 | > 32 | > 512 | > 16 | 2 | > 16 | IS903B | D150G | R256G |
| KP25 | XDR | > 128 | > 128 | > 128/4 | > 128 | > 512/4 | 256 | > 32 | > 32 | > 512 | 16 | 0.5 | 16 | - | D150G | T157P, R256G |
| EC600 | - | ≤ 1 | ≤ 1 | ≤ 1/4 | ≤ 1 | 8/4 | 8 | ≤ 0.25 | ≤ 0.25 | ≤ 4 | ≤ 0.125 | ≤ 0.125 | 0.5 | - | - | - |
| KP17-T | MDR | > 128 | 64 | > 128/4 | ≤ 1 | 256/4 | 8 | 16 | 16 | > 512 | ≤ 0.125 | ≤ 0.125 | 0.5 | - | - | - |
| KP21-T | MDR | > 128 | 64 | > 128/4 | ≤ 1 | 512/4 | 8 | 8 | 16 | > 512 | 0.25 | ≤ 0.125 | 0.5 | - | - | - |
| KP25-T | MDR | > 128 | 32 | > 128/4 | ≤ 1 | 256/4 | 8 | 16 | 16 | > 512 | ≤ 0.125 | ≤ 0.125 | 0.5 | - | - | - |
KP17-T, KP21-T, KP25-T, Transconjugants
Abbreviations: CAZ Ceftazidime, FEP Cefepime, CZA Ceftazidime/avibactam, ATM Aztreonam, TZP Piperacillin/tazobactam, NIT Nitrofurantoin, IPM Imipenem, MEM Meropenem, AMK Amikacin, LVX Levofloxacin, TGC Tigecycline, PB Polymyxin B
Fig. 1.
Phylogenetic relationship, resistance genes, and virulence genes of 8 PB-resistant strains (4 PBR-hvKP strains with red background, 4 PB-resistant strains with purple background) in this study and 13 ST11 PBR-hvKP strains from other provinces in China. The darker blue box represents the presence of genes. K. pneumoniae HS11286 was used as the reference for core SNPs calling
Virulence phenotype and genetic characteristics
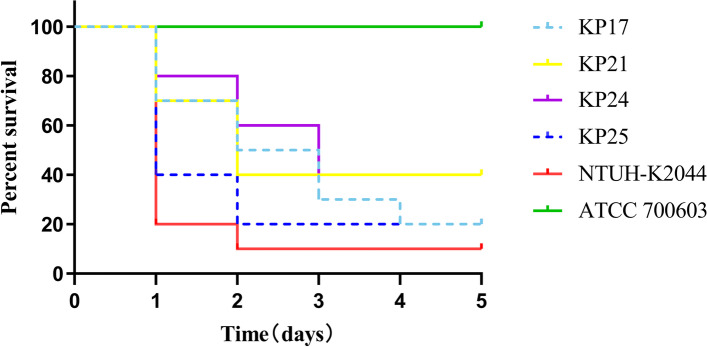
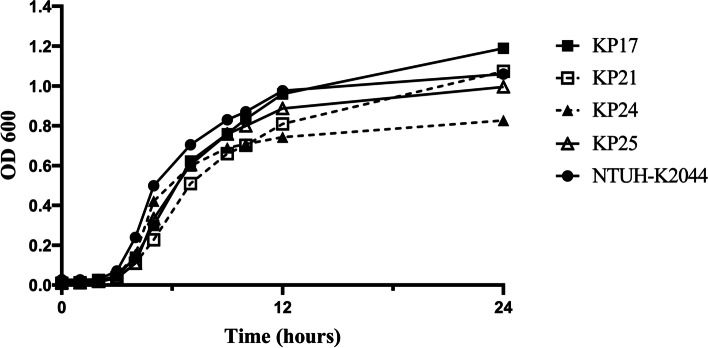
All isolates were non-mucoviscous according to the string test. Four isolates (KP17, KP21, KP24, and KP25) carried hypervirulent biomarker genes rmpA, rmpA2, peg344, and iucA (Fig. 1). It was worth noting that KP17, KP21, and KP25 had in-frame truncations in rmpA2, which resulted in a 636-bp sequence variation. Results of the G. mellonella infection model showed that the five-day mortality rates of these isolates were greater than or equal to 60% (Fig. 2), suggesting that they were PBR-hvKP. Except for KP24, almost all isolates could form biofilms. Six were classified as strong film-forming isolates, and one was classified as a moderate film-forming isolate. All PBR-hvKP isolates were sensitive to serum. However, the growth abilities of four PBR-hvKP differed (Fig. 3). After 8–12 h of monitoring, the growth of KP17 was the strongest, while the growth of KP24 was weak (P < 0.05).
Fig. 2.
Four PBR-hvKP infections of G. mellonella larvae. The survival rate of larvae infected with KP17, KP21, KP24, and KP25 was 20.0%, 40.0%, 40.0%, and 20.0%. NTUH-K2044 and ATCC 700603 were used as high and low virulence control strains
Fig. 3.
Growth curves of four PBR-hvKP. NTUH-K2044 was used as a high virulence control strain
MLST genotyping, serotypes, and phylogenetic analysis
Features of genomes of eight PB-resistant K. pneumoniae isolates are included in Supplementary Table S1. Table 3 shows the origin, types, major resistance and virulence characteristics, capsular serotypes, and sequence types (STs) of eight PB-resistant isolates. ST11 was the predominant ST (7/8, 87.5%), except for KP16, which belonged to ST5254. ST5254 is a newly discovered ST type, a single-locus (phoE) variant of ST11, with the phoE515 gene in ST5254 differing from phoE1 in ST11 (Supplementary Table S2). All PB-resistant isolates belonged to the K64 serotype. In order to further clarify the relationship between our PB-resistant strains and those reported in China, we reviewed colistin/PB-resistant hypervirulent ST11 strains reported previously. A total of 13 genomes of colistin/PB-resistant hypervirulent ST11 strains were downloaded from NCBI and used for phylogenetic analysis together with genomes in this study (Fig. 1). The results showed that strains in this study had specific differences in affinity with those reported before and did not belong to the same cluster. Notably, three PBR-hvKP KP17, KP21, and KP25 were classified as the same cluster. The SNPs ranged from 8 to 20 (Supplementary Figure S1), indicating probable clonal transmission [10]. Another cluster included KP14, KP20, and KP18. KP16 and KP24 were individual isolates. The distribution of colistin/PB-resistant hypervirulent ST11 K. pneumoniae in those clades indicated that PBR-hvKP KP17, KP21, and KP25 belonged to a new clade with blaNDM-1.
Table 3.
Characteristics of eight PB-resistant K. pneumoniae
| Strains | Patient | Specimen | STs | Capsular type | Strain type | Carbapenem resistance genes | String test | Virulence genes | Biofilm assay | Serum resistance |
|---|---|---|---|---|---|---|---|---|---|---|
| KP14 | 2 | Sputum | 11 | K64 | XDR-KP | blaKPC-2 | - | - | + + + | 0.73% |
| KP16 | 4 | Drainage | 5254 | K64 | MDR-KP | blaKPC-2 | - | - | + + | 0.08% |
| KP17 | 3 | Sputum | 11 | K64 | XDR-hvKP | blaNDM-1, blaKPC-2 | - | rmpA, rmpA2, peg344, iucA | + + + | 0.00% |
| KP18 | 5 | Sputum | 11 | K64 | XDR-KP | blaKPC-2 | - | - | + + + | 0.62% |
| KP20 | 7 | Sputum | 11 | K64 | XDR-KP | blaKPC-2 | - | - | + + + | 40.00% |
| KP21 | 6 | Blood | 11 | K64 | XDR-hvKP | blaNDM-1, blaKPC-2 | - | rmpA, rmpA2, peg344, iucA | + + + | 0.00% |
| KP24 | 8 | Urine | 11 | K64 | XDR-hvKP | blaNDM-1, blaKPC-2 | - | rmpA, rmpA2, peg344, iucA | - | 0.00% |
| KP25 | 1 | Wound | 11 | K64 | XDR-hvKP | blaNDM-1, blaKPC-2 | - | rmpA, rmpA2, peg344, iucA | + + + | 1.15% |
Abbreviations: XDR-KP Extensively drug-resistant K. pneumoniae, MDR-KP Multidrug-resistant K. pneumoniae, XDR-hvKP Extensively drug-resistant hypervirulent K. pneumoniae
Complete genomic and comparative analysis of KP25
The KP25 genome contained one chromosome with a length of 4,588,001 bp and seven plasmids. Table 4 summarises the genome characteristics of KP25. The chromosome carried type 3 fimbriae gene mrkABCDFHJ, and yersiniabactin siderophore genes ybtPQSX–ybtAUTE. The major plasmids included ColRNAI (plasmid 2), IncFII (Yp) (plasmid 3), IncFII (pHN7A8) (plasmid 4), IncFII (pCRY) (plasmid 6), and repB (plasmid 7). Drug resistance genes blaNDM-1 (in plasmid 3), blaKPC-2 (in plasmid 4), tet(A) (in plasmid 6), and virulence genes rmpA, rmpA2, iutA, iucB, iucC, and peg344 (in plasmid 7) were distributed in different plasmids.
Table 4.
Genome characteristics of isolate KP25
| Genetic context | Length (bp) | GC content (%) | Plasmid type | Closest plasmid match (identity) | #MEGs | Resistance genes | Virulence genes |
|---|---|---|---|---|---|---|---|
| Chromosome | 4,588,001 | 57.39 | - | - | 37 | - | mrkABCDFHJ, ybtPQSX–ybtAUTE |
| Plasmid 1 | 743,487 | 57.38 | - | - | 3 | fosA5 | - |
| Plasmid 2 | 23,917 | 55.57 | ColRNAI | pKP8695-p4 (99.76%) | 0 | - | - |
| Plasmid 3 | 110,782 | 54.83 | IncFII (Yp) | pRJF866 (99.92%) | 10 | blaNDM-1, sul1, rmtC | - |
| Plasmid 4 | 92,836 | 55.21 | IncFII (pHN7A8) | p12085-KPC (99.92%) | 12 | blaKPC-2, blaSHV-12 | - |
| Plasmid 5 | 91,870 | 56.99 | - | - | 0 | - | - |
| Plasmid 6 | 83,552 | 54.00 | IncFII (pCRY) | p16ZR-187-IncFII-83-R (99.99%) | 2 | tet(A), catA2, sul2, qnrS1, blaLAP-2 | - |
| Plasmid 7 | 219,442 | 49.93 | repB | pK2044 (99.29%) | 21 | - | rmpA, rmpA2, iutA, iucB, iucC, peg344 |
#MEGs, the number of mobile genetic elements
The most similar genomes to KP25 were previously reported carbapenem-resistant K. pneumoniae (CR-KP) strains KP-C76 [11] and KP20194b2 [12] isolated respectively in Hangzhou and Hengyang in China, both containing a blaKPC-2-carrying plasmid (Fig. 4B), a tet(A)-carrying plasmid, and a pLVPK-like virulence plasmid (Fig. 4C). In the blaKPC-2-carrying plasmid (pKP25-4), the genetic environment of blaKPC-2 and blaSHV-12 was IS26-ΔTn3-ISKpn27-blaKPC-2-ISKpn6-korC-klcA-ΔrepB-tnpR-ΔTn3-blaSHV-12-deoR-ygbJ-ygbK-IS26 (Fig. 5), which was a composite transposon consisting of Tn1722 in pKPHS2 (GenBank no. CP003224.1) and a truncated IS26-blaSHV-12-IS26 unit [13]. The tet(A)-carrying plasmid (pKP25-6) carried an MDR region consisting of multiple resistance genes sul2, catA2, tet(A), blaLAP-2, qnrS1, and transposases IS26, IS5075, ISVsa3, and ISKpn19. β-lactamase gene blaLAP-2 was flanked upstream by ΔTn1721 and presumed to exist in a truncated IS26-blaLAP-2-qnrS1-IS26 unit. The virulence plasmid (pKP25-7) had a high similarity (99.29%) with the virulence plasmid pK2044 carried by the reference strain NTUH-K2044. It was worth noting that rmpA and peg344 were flanked upstream by ISKpn26 and downstream by IS903B. The stop codon TAG in rmpA2 led to a fragment deletion. Interestingly, the blaNDM-1-carrying plasmid (pKP25-3) was not present in previous similar genomes of KP-C76 and KP20194b2. It shared 99.92% nucleotide identity with the published plasmid pRJF866 (GenBank no. KF732966) found in a CR-KP isolate from a blood specimen from a hospital in Shanghai (Fig. 4A), which was a pKOX_NDM1-like plasmid [14]. Sequence analysis showed that blaNDM-1, sul1, and rmtC formed an MDR region flanked upstream by IS5 and downstream by ISKpn26 (Fig. 5). The genetic environment was IS5-ΔISEhe3-groEL-groES-cutA-dsbD-trpF-bleMBL-blaNDM-1-rmtC-ΔTn3- DDE-type integrase/transposase/recombinase-tniB-tniQ-sul1-copG-ISKpn26.
Fig. 4.
Genetic comparison of blaNDM-1-carrying plasmid (A), blaKPC-2-carrying plasmid (B), and virulence plasmid (C) from KP25, including the best matching plasmids; plasmids from KP-C76 [11] and plasmids from KP20194b2 [12]
Fig. 5.
The blaNDM-1 region from plasmid 3 and the blaKPC-2 region from plasmid 4 in KP25. Genes are depicted as arrows according to the direction of transcription. Resistance genes are in red, mobile elements are in blue, and other traits are in grey
Conjugation assays showed that the plasmid carrying blaNDM-1 could be successfully transferred to E. coli EC600 from KP17, KP21, and KP25. The transconjugants were resistant to CAZ, FEP, CZA, TZP, IPM, MEM, and AMK (Table 2), whereas the plasmid-carrying blaKPC-2 was not transferred under experimental conditions.
Discussion
The resistance rates of hvKP were generally lower than those of classical K. pneumoniae (cKP) reported in previous studies [15]. However, recent studies in many countries have reported the emergence of MDR-hvKP. The fatality rate of MDR-hvKP infected patients was 56.3%-66.7% in China, and the rate of isolation of MDR-hvKP strains is increasing [16–18]. Polymyxins are essential antibiotics for Gram-negative bacteria exhibiting MDR. In this study, we collected eight strains of PB-resistant K. pneumoniae and analyzed the clinical and molecular characterization of these strains.
A clonal transmission of ST11-K64 PB-resistant K. pneumoniae in the hospital and two clusters among these isolates were investigated. Epidemiological information showed that patients were admitted to different ICU wards, and patients 1, 3, and 6 had overlapping stays (Fig. 6). Following the fatal outbreak of ST11-K47 CR-KP in ICU wards [19], this study indicated that ST11 PBR-hvKP formed a new clade in China and spread within a hospital setting. Most of the strains isolated in this study were ST11, except KP16, which was first discovered ST5254. ST307, ST512 and ST147 have been shown to be hyperepidemic CR-KP clones in Europe [20–22]. In China, ST11 and ST23 have been the main ST types of MDR-KP, and the KPC-2-producing ST11 was the dominant clone of CR-KP [23]. In the present study, we reported ST5254 for the first time and demonstrated that this new ST is a high-risk clone resistant to PB and carries MDR genes such as blaKPC-2, which requires close attention. The identification of clinically isolated K. pneumoniae by wzc or wzi gene sequencing has revealed that K64 is one of the major serotypes of CR-KP [24]. ST11-K64 CR-hvKP carried MDR genes such as blaKPC-2 and blaNDM-1 together with pLVPK-like virulence plasmid, expressing high virulence in a G. mellonella infection model, and the fatality rate of patients was high [12, 25, 26].
Fig. 6.
Epidemiology of eight PB-resistant K. pneumoniae cases. The rectangles on the timeline represent the duration of the patient’s admission. Antibiotics used before and after the isolation of PB-resistant strains were marked
All PB-resistant isolates in this study carried at least two common mutations, including D150G in phoQ and R256G in pmrB [27–29]. Homologous PBR-hvKP strains showed consistent mutation patterns, whereas new mutations were derived in other strains. T157P in pmrB caused overexpression of the operons pmrCAB and pmrHFIJKLM, resulting in the increase of MICs of PB in clinical isolates [30]. S85R in pmrB was predicted to affect protein function by PROVEAN (http://provean.jcvi.org/seq_submit.php) (Supplementary Figure S2). Insertion mutation and inactivation of mgrB led to overexpression of the phoPQ operon, which activated the pmrHFIJKLM operon and produced 4-amino-4-deoxy-L-aminoarabinose (L-Ara4N), leading to resistance [31]. Resistance to polymyxins can be influenced by external environmental stimuli such as antibiotics. Choi et al. [32] showed that under the stimulation of colistin, mutations in phoP, phoQ, and pmrB were generated in PB-sensitive ST23-KP. The acquisition of drug resistance occurs at the cost of reducing capsular polysaccharides and virulence. Murtha et al. [33] studied Enterobacterales and found that meropenem could induce PhoPQ and increase modifications of L-Ara4N. Exposure to different antibiotics can lead to changes in bacterial tolerance. Long-term hospitalization and the use of β-lactams increase the risk of PB-resistant infections.
WGS and comparative analysis revealed the molecular characterization of PB-resistant MDR-hvKP. PBR-hvKP strains in this study were related to isolates previously reported in Hangzhou (KP-C76) and Hengyang (KP20194b2), China [11, 12]. Similarly, KP-C76 was also found to develop colistin resistance due to the insertion mutation of mgrB. However, it differed from mutations in KP25, suggesting that the resistant mutations of polymyxins were relatively independent in different strains. Another difference was that KP25 obtained a pKOX_NDM1-like plasmid, which was firstly discovered in Taiwan and spread in eastern China, such as Shanghai and Jiangxi [14, 34, 35]. The plasmid carrying blaNDM-1 made PBR-hvKP resistant to CZA. Class B metallo-β-lactamase and mutations in blaKPC are the main mechanisms of CZA resistance [36, 37]. Conjugation assays showed that the pKOX_NDM1-like plasmid demonstrated a stronger transfer ability than the blaKPC-2-carrying plasmid [14, 38]. pKP25-3 carries the entire tra region (Fig. 4A), which encodes the highly conserved gene products of the type IV secretion system, while in pKP25-4, the region was inserted by IS26 and led to gene deletion (Fig. 4B), which explains the low conjunction rate of the blaKPC-2 plasmid [39]. The conjugation and expression of the plasmid carrying blaNDM-1 and rmtC endowed E. coli EC600 with resistance to CAZ, FEP, CZA, TZP, IPM, MEM, and AMK, while the chromosome-mediated resistant genes and qnrS1-carrying plasmid were not conjugated. Strains co-harboring blaKPC-2 and blaNDM-1 were able to spread stably in the environment. The blaNDM-1 gene is highly transferable between different species in Enterobacterales, expanding the drug resistance spectrum [38].
Virulence-related genes mrk coding for type 3 fimbriae and ybt coding for yersiniabactin in the chromosome, aerobactin genes iuc/iutA and salmochelin gene iro in pLVPK-like virulence plasmid were found in PBR-hvKP [40]. Four PBR-hvKP isolates in this study carried rmpA and rmpA2. However, the rmpA2 was a truncated 636-bp fragment with a frameshift mutation, which may be responsible for the loss of mucus phenotype and weak serum resistance [41]. In fact, frameshift rmpA2 was not uncommon in ST11-K64 isolates. Zhou et al. [42] reported the presence of frameshift mutations in 52.2% (48/92) rmpA2-positive ST11-K64 isolates. The virulence of strains with mutant rmpA2 was not significantly different from that of wild strains in the G. mellonella infection model. However, the environmental survival rate was significantly increased and had the advantage of hospital transmissibility. On the other hand, loss of hypermucoviscosity and weak serum resistance may be associated with the acquisition of PB resistance [32]. Stimulation of PB led to enhanced resistance in mutant strains, accompanied by the reduction in virulence-related phenotypes. Strong film-forming ability and low mucus phenotype increased the ability of strains to colonize and spread in hospitals, leading to persistent chronic infections in patients [43].
Conclusions
In conclusion, we studied the clonal transmission and molecular characterization of co-harboring blaNDM-1 and blaKPC-2 PBR-hvKP in a tertiary hospital in China. PBR-hvKP not only carried multiple resistance genes but also had hypervirulence. Mutations in chromosomal genes phoQ and pmrB, and insertion mutations in mgrB were major causes of PB resistance. In addition, we discovered a novel ST type, ST5254, which broadened the types of pan-resistant K. pneumoniae. ST11-K64 is a new type of PBR-hvKP epidemic in China. Rapid remodeling and diversification of genomes and promoting mobile genetic elements make it a new crucial super bacterium. Therefore, strengthening the clinical monitoring of ST11-K64 strains and taking effective measures to avoid transmission of resistant bacteria is significant for preventing and treating MDR-hvKP infections.
Methods
Polymyxin B-resistant K. pneumoniae strains and data collection
We conducted a retrospective study at Xiangya Hospital, Central South University. K. pneumoniae isolates were collected from clinical specimens processed routinely between November 2020 and April 2021; these isolates were chosen based on being resistant to PB as involved in a suspected outbreak. Only the first culture was included for patients with two or more PB-resistant K. pneumoniae strains. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics GmbH, Bremen, Germany) and the Vitek 2 Compact System (BioMérieux, Marcy l'Etoile, France) were used for identification and antimicrobial susceptibility testing. In addition, patients' clinical data were collected from hospital computer databases.
Antimicrobial susceptibility test
Antimicrobial susceptibility testing was performed using the broth microdilution method. The antibiotics (Wenzhou Kangtai Biotechnology Company, Wenzhou, China) included ceftazidime (CAZ), cefepime (FEP), ceftazidime/avibactam (CZA), aztreonam (ATM), piperacillin/tazobactam (TZP), nitrofurantoin (NIT), imipenem (IPM), meropenem (MEM), amikacin (AMK), levofloxacin (LVX), tigecycline (TGC), and PB. The minimum inhibitory concentrations (MICs) of the antibiotics, except that of TGC, were in accordance with those defined by the Clinical and Laboratory Standards Institute (CLSI) 2021 breakpoints [44]. Susceptibility to TGC was interpreted according to the US Food and Drug Administration (US-FDA) breakpoints (https://www.fda.gov/drugs/development-resources/tigecycline-injection-products). E. coli ATCC 25922 was used for quality control.
Detection of capsular type, hypermucoviscosity, and virulence-related genes prmpA, prmpA2, iroB, peg344, and iucA
Capsular type of K. pneumoniae was detected by polymerase chain reaction (PCR), and the wzi locus was sequenced as previously described [45]. Mucoviscous phenotype was evaluated using the string test [46]. Isolates were grown on blood agar plates overnight at 37 °C, and colonies were gently touched and lifted with an inoculation loop. Hypermucoviscosity was defined as a macroscopically observed mucus string > 5 mm in length. Virulence-related genes plasmid-borne rmpA (prmpA), prmpA2, iroB, peg344, and iucA were detected by PCR to identify hypervirulent strains as previously [44]. In short, DNA was extracted by boiling method and specific primers, and PCR conditions were according to the previous study [47]. PCR was performed using an Applied Biosystems thermal cycler (Applied Biosystems, Foster City, CA) as follows: denaturation for 5 min at 94 °C, 30 cycles of 30 s at 95 °C, 30 s at primer-specific annealing temperature, and 30 s at 72 °C, and a final extension for 10 min at 72 °C.
Galleria mellonella infection model
The virulence of the strain was evaluated using G. mellonella larvae (Tianjin Huiyude Biotechnology Company, Tianjin, China). The experiment was modified as described in a previous study [48]. First, the strains to be tested were cultured overnight; then, a single colony was selected and used to prepare 1.0 × 107 colony forming units (CFU)/mL bacterial solution with phosphate-buffered saline (PBS). Each larva was injected with 10 μL of the bacterial suspension through the last left proleg, then placed in disposable plates and incubated at 37 °C in the dark for 5 d. The number of deaths was recorded every 24 h. NTUH-K2044 and K. pneumoniae ATCC 700603 (National Clinical Laboratory Center, Beijing, China) were used as high- and low-virulence control strains, respectively. Tests were performed in triplicate.
Serum resistance assay
The serum resistance assay was conducted as previously described [49]. Strains were cultured in Luria–Bertani (LB) medium overnight, then washed and suspended with PBS. Then, 100 μL of bacterial suspension was added to 300 μL of healthy human serum and incubated at 37 °C for 3 h. Before and after co-cultivation, 100 μL of suspension was taken to determine the CFUs. The bacterial killing effect was defined as the percentage of CFU after culture in serum compared with the bacterial CFU before culture. Tests were performed in triplicate.
Growth curves and biofilm assay
The growth curves were made following a previous study [50]. First, a single colony of bacteria was standardized to match a 0.5 McFarland followed by 1:100 dilution in LB broth at 37 °C with medium shaking. Then, 200 μL of the bacterial solution was taken out, and the optical density at 600 nm (OD600) was measured every hour during the first 12 h. Finally, multiple t-test was used to compare growth at various time points.
Biofilm formation was assessed by the crystal violet staining method [51]. The strains were prepared into 1.0 × 108 CFU/mL solution with PBS, diluted to 1:100 with LB broth, and cultured in a 96-well plate (3 parallel wells for each strain) at 37 °C for 24 h. The plate was then washed with PBS, stained with 1% crystal violet for 15 min, and then decolorized with 95% ethanol for 10 min. Absorbance was measured at OD570. The classification standard followed the previous study [52]. Growth curves and biofilm assay were performed in triplicate.
Whole-genome sequencing (WGS) and data analysis
Genomic DNA was isolated using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and submitted to next-generation high-throughput sequencing (NGS) on a HiSeq 2000 platform (Illumina Inc., San Diego, CA, USA) with 2 × 100-bp paired-end reads. Pacbio sequel II and DNBSEQ platform (Beijing Genomics Institute, Shenzhen, China) were used for sequencing of the Genomic DNA of KP25. The Canu program was used for self-correction. GATK (https://www.broadinstitute.org/gatk/) was used to make single-base corrections. The whole-genome sequence was annotated by the RAST tool version 2.0 (https://rast.nmpdr.org/). The multilocus sequence typing (MLST) profiles were determined with the MLST database (https://bigsdb.pasteur.fr/klebsiella/). Snippy was applied to run core single-nucleotide polymorphisms (SNPs) calling (https://github.com/tseemann/snippy) and generate a phylogenetic tree based on the maximum-likelihood method with K. pneumoniae HS11286 (GenBank no. CP003200.1) as the reference. ChiPlot (https://www.chiplot.online/) was used for the visualization of the phylogenetic tree. The antibiotic resistance genes and virulence loci of the assembled genome sequences were identified using ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and the MLST database. PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to predict the plasmid types. ISfinder database (https://www-is.biotoul.fr/index.php) was used to determine the insert sequences. A comparison of sequences of plasmids was conducted using BRIG [53]. The sequences of PB-resistant strains are available in the National Center for Biotechnology Information (NCBI) database under the BioProject accession number PRJNA824787.
Bacterial conjugation
PB-resistant strains were used as the donors (meropenem resistant), and E. coli EC600 (rifampicin resistant) was used as the recipient strain for plasmid-binding assays. The donor and recipient strains cultured in LB broth overnight were mixed and spotted on sterile filter paper, then incubated on a blood plate at 37 °C for 18 h. Transconjugants were selected on MacConkey agar supplemented with 1 mg/L meropenem and 600 mg/L rifampicin. blaNDM-1 PCR and antimicrobial susceptibility test of meropenem were performed to confirm the transconjugants.
Supplementary Information
Additional file 1: Table S1. Features of genomes of eight PB-resistant K. pneumoniae isolates. Table S2. Allelic profiles of eight PB-resistant K. pneumoniae isolates. Figure S1. Paired SNP distance of eight PB-resistant K. pneumoniae isolates. Figure S2. PROVEAN result shows the effect of S85R in PmrB.
Acknowledgements
We want to express our gratitude to the doctors from the clinical laboratory of Xiangya Hospital, Central South University, China, for their support in sample collection. In addition, we thank Medjaden Inc. for the scientific editing of this manuscript.
Abbreviations
- MDR
Multidrug-resistant
- hvKP
Hypervirulent K. pneumoniae
- MICs
Minimum inhibitory concentrations
- WGS
Whole-genome sequencing
- KP
Klebsiella pneumoniae
- KPC-2
Klebsiella pneumoniae carbapenemase-2
- NDM-1
New Delhi metallo-β-lactamase-1
- PB
Polymyxin B
- PBR-hvKP
Polymyxin B-resistant hypervirulent K. pneumoniae
- MALDI-TOF MS
Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry
- CAZ
Ceftazidime
- FEP
Cefepime
- CZA
Ceftazidime/avibactam
- ATM
Aztreonam
- TZP
Piperacillin/tazobactam
- NIT
Nitrofurantoin
- IPM
Imipenem
- MEM
Meropenem
- AMK
Amikacin
- LVX
Levofloxacin
- TGC
Tigecycline
- CLSI
Clinical and Laboratory Standards Institute
- US-FDA
US Food and Drug Administration
- PCR
Polymerase chain reaction
- CFU
Colony forming units
- PBS
Phosphate-buffered saline
- LB
Luria–Bertani
- cgMLST
Core genome multilocus sequence typing
- MLST
Multilocus sequence typing
- NCBI
National Center for Biotechnology Information
- ICU
Intensive care unit
- CR-KP
Carbapenem-resistant K. pneumoniae
- cKP
Classical K. pneumoniae
- XDR
Extensively drug-resistant
- L-Ara4N
4-Amino-4-deoxy-L-aminoarabinose
Authors’ contributions
MT and JL analyzed the data and wrote and edited the manuscript. ZL, FX, and CM carried out the experiments. YH and HW contributed to the methodology and data curation. MZ designed the project and revised the manuscript. All authors contributed to the article and approved the submitted version. The author(s) read and approved the final manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities of Central South University under Grant (No. 2021zzts1043); the Undergraduate Free Exploration Foundation of Central South University under Grant (No. XCX2022031); the Natural Science Foundation of Hunan Province under Grant (No. 2020JJ4886, 2022JJ70084); and the Science Foundation of Hunan Health Commission in Hunan Province under Grant (No. 202111000066).
Availability of data and materials
The data supporting this study's findings are openly available in the NCBI BioProject repository at https://www.ncbi.nlm.nih.gov/bioproject/824787 under BioProject accession number PRJNA824787.
Declarations
Ethics approval and consent to participate
This study was conducted using a protocol approved by the Central South University Ethics Committee (Changsha, Hunan Province, China) with ID 201703302 and according to the principles of the Declaration of Helsinki and its later amendments or comparable ethical standards. All participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengli Tang and Jun Li contributed equally to this work.
References
- 1.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu F, Guo Y, Yang Y, Zheng Y, Wu S, Jiang X, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38:2275–2281. doi: 10.1007/s10096-019-03673-1. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trimble MJ, Mlynárčik P, Kolář M, Hancock RE. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb Perspect Med. 2016;6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ah YM, Kim AJ, Lee JY. Colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2014;44:8–15. doi: 10.1016/j.ijantimicag.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Narimisa N, Goodarzi F, Bavari S. Prevalence of colistin resistance of Klebsiella pneumoniae isolates in Iran: a systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2022;21:29. doi: 10.1186/s12941-022-00520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YT, Cheng YH, Chuang C, Chou SH, Liu WH, Huang CH, et al. Molecular and Clinical Characterization of Multidrug-Resistant and Hypervirulent Klebsiella pneumoniae Strains from Liver Abscess in Taiwan. Antimicrob Agents Chemother. 2020;64:e00174–20. doi: 10.1128/AAC.00174-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Wu Y, Zhu Y, Jia P, Li X, Jia X, et al. Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKP) in China. Emerg Microbes Infect. 2022;11:648–661. doi: 10.1080/22221751.2022.2036078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Zeng Y, Zhang R, Cai J. In vivo Emergence of Colistin and Tigecycline Resistance in Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae During Antibiotics Treatment. Front Microbiol. 2021;12:702956. doi: 10.3389/fmicb.2021.702956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Ouyang J, He W, Zeng T, Liu B, Jiang H, et al. Co-occurrence of Rapid Gene Gain and Loss in an Interhospital Outbreak of Carbapenem-Resistant Hypervirulent ST11-K64 Klebsiella pneumoniae. Front Microbiol. 2020;11:579618. doi: 10.3389/fmicb.2020.579618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford PJ, Avison MB. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHVmobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother. 2004;54:69–75. doi: 10.1093/jac/dkh251. [DOI] [PubMed] [Google Scholar]
- 14.Qu H, Wang X, Ni Y, Liu J, Tan R, Huang J, et al. NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3-type plasmids may contribute to the dissemination of blaNDM-1. Int J Infect Dis. 2015;34:8–13. doi: 10.1016/j.ijid.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 16.Huang QS, Liao W, Xiong Z, Li D, Du FL, Xiang TX, et al. Prevalence of the NTE(KPC)-I on IncF Plasmids Among Hypervirulent Klebsiella pneumoniae Isolates in Jiangxi Province. South China Front Microbiol. 2021;12:622280. doi: 10.3389/fmicb.2021.622280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. 2020;11:1215–24. doi: 10.1080/21505594.2020.1809322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Wu Q, He L, Zhang H, Xu M, Yuan B, et al. Clinical and Molecular Characteristics of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Isolates in a Tertiary Hospital in Shanghai. China Infect Drug Resist. 2021;14:2697–2706. doi: 10.2147/IDR.S321704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 20.Rossi M, Chatenoud L, Gona F, Sala I, Nattino G, D'Antonio A, et al. Characteristics and clinical implications of carbapenemase-producing klebsiella pneumoniae colonization and infection. Italy Emerg Infect Dis. 2021;27:1416–1426. doi: 10.3201/eid2705.203662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cañada-García JE, Moure Z, Sola-Campoy PJ, Delgado-Valverde M, Cano ME, Gijón D, et al. CARB-ES-19 multicenter study of carbapenemase-producing klebsiella pneumoniae and escherichia coli from all spanish provinces reveals interregional spread of high-risk clones such as ST307/OXA-48 and ST512/KPC-3. Front Microbiol. 2022;13:918362. doi: 10.3389/fmicb.2022.918362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin MJ, Corey BW, Sannio F, Hall LR, MacDonald U, Jones BT, et al. Anatomy of an extensively drug-resistant Klebsiella pneumoniae outbreak in Tuscany, Italy. Proc Natl Acad Sci U S A. 2021;118:e2110227118. doi: 10.1073/pnas.2110227118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global Dissemination of carbapenemase-producing klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan YJ, Lin TL, Lin YT, Su PA, Chen CT, Hsieh PF, et al. Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob Agents Chemother. 2015;59:1038–1047. doi: 10.1128/AAC.03560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Gu Y, Li X, Liu Y, Ye Y, Guan S, et al. Identification and Characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med. 2019;39:167–175. doi: 10.3343/alm.2019.39.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Shi Q, Shen S, Huang C, Wu H. Clinical and Bacterial Characteristics of Klebsiella pneumoniae Affecting 30-Day Mortality in Patients With Bloodstream Infection. Front Cell Infect Microbiol. 2021;11:688989. doi: 10.3389/fcimb.2021.688989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother. 2015;59:2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Lin Y, Wang Z, Hu N, Liu Q, Zhou W, et al. Molecular Mechanisms of Colistin Resistance in Klebsiella pneumoniae in a Tertiary Care Teaching Hospital. Front Cell Infect Microbiol. 2021;11:673503. doi: 10.3389/fcimb.2021.673503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aires CA, Pereira PS, Asensi MD, Carvalho-Assef AP. mgrB Mutations Mediating Polymyxin B Resistance in Klebsiella pneumoniae Isolates from Rectal Surveillance Swabs in Brazil. Antimicrob Agents Chemother. 2016;60:6969–6972. doi: 10.1128/AAC.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. 2014;58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi MJ, Ko KS. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother. 2015;59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murtha AN, Kazi MI, Schargel RD, Cross T, Fihn C, Cattoir V, et al. High-level carbapenem tolerance requires antibiotic-induced outer membrane modifications. PLoS Pathog. 2022;18:e1010307. doi: 10.1371/journal.ppat.1010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang TW, Wang JT, Lauderdale TL, Liao TL, Lai JF, Tan MC, et al. Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob Agents Chemother. 2013;57:4072–4076. doi: 10.1128/AAC.02266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L, Zhong Q, Shang Y, Wang H, Ning C, Li Y, et al. The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol Infect. 2014;142:1972–1977. doi: 10.1017/S0950268813002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schillaci D, Spanò V, Parrino B, Carbone A, Montalbano A, Barraja P, et al. Pharmaceutical approaches to target antibiotic resistance mechanisms. J Med Chem. 2017;60:8268–8297. doi: 10.1021/acs.jmedchem.7b00215. [DOI] [PubMed] [Google Scholar]
- 37.Hemarajata P, Humphries RM. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother. 2019;74:1241–1243. doi: 10.1093/jac/dkz026. [DOI] [PubMed] [Google Scholar]
- 38.Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi: 10.1016/j.ebiom.2019.102599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dmowski M, Gołębiewski M, Kern-Zdanowicz I. Characteristics of the Conjugative Transfer System of the IncM Plasmid pCTX-M3 and Identification of Its Putative Regulators. J Bacteriol. 2018;200:e00234–18. doi: 10.1128/JB.00234-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai P, Hu D. The making of hypervirulent Klebsiella pneumoniae. J Clin Lab Anal. 2022;36:e24743. [DOI] [PMC free article] [PubMed]
- 41.Lin TH, Wu CC, Kuo JT, Chu HF, Lee DY, Lin CT. FNR-Dependent RmpA and RmpA2 regulation of capsule polysaccharide biosynthesis in klebsiella pneumoniae. Front Microbiol. 2019;10:2436. doi: 10.3389/fmicb.2019.02436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou K, Xiao T, David S, Wang Q, Zhou Y, Guo L, et al. Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility. China Emerg Infect Dis. 2020;26:289–297. doi: 10.3201/eid2602.190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bengoechea JA, Sa PJ. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 31th ed. Pennsylvania: Clin Lab Stand Inst; 2021.
- 45.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, et al. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med. 2006;259:606–614. doi: 10.1111/j.1365-2796.2006.01641.x. [DOI] [PubMed] [Google Scholar]
- 47.Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56:e00776–18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Insua JL, Llobet E, Moranta D, Pérez-Gutiérrez C, Tomás A, Garmendia J, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun. 2013;81:3552–3565. doi: 10.1128/IAI.00391-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin J, Ko KS. Effect of plasmids harbouring blaCTX-Mon the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents. 2015;46:214–218. doi: 10.1016/j.ijantimicag.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 50.Sun L, Sun L, Li X, Hu X, Wang X, Nie T, et al. A Novel Tigecycline Adjuvant ML-7 Reverses the Susceptibility of Tigecycline-Resistant Klebsiella pneumoniae. Front Cell Infect Microbiol. 2021;11:809542. doi: 10.3389/fcimb.2021.809542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffey BM, Anderson GG. Biofilm formation in the 96-well microtiter plate. Methods Mol Biol. 2014;1149:631–641. doi: 10.1007/978-1-4939-0473-0_48. [DOI] [PubMed] [Google Scholar]
- 52.Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123:1003–1018. doi: 10.1111/jam.13533. [DOI] [PubMed] [Google Scholar]
- 53.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Features of genomes of eight PB-resistant K. pneumoniae isolates. Table S2. Allelic profiles of eight PB-resistant K. pneumoniae isolates. Figure S1. Paired SNP distance of eight PB-resistant K. pneumoniae isolates. Figure S2. PROVEAN result shows the effect of S85R in PmrB.
Data Availability Statement
The data supporting this study's findings are openly available in the NCBI BioProject repository at https://www.ncbi.nlm.nih.gov/bioproject/824787 under BioProject accession number PRJNA824787.