Abstract
Oscillations of the period (per) and timeless (tim) gene products are an integral part of the feedback loop that underlies circadian behavioral rhythms in Drosophila melanogaster. Resetting this loop in response to light requires the putative circadian photoreceptor cryptochrome (CRY). We dissected the early events in photic resetting by determining the mechanisms underlying the CRY response to light and by investigating the relationship between CRY and the light-induced ubiquitination of the TIM protein. In response to light, CRY is degraded by the proteasome through a mechanism that requires electron transport. Various CRY mutant proteins are not degraded, and this suggests that an intramolecular conversion is required for this light response. Light-induced TIM ubiquitination precedes CRY degradation and is increased when electron transport is blocked. Thus, inhibition of electron transport may “lock” CRY in an active state by preventing signaling required either to degrade CRY or to convert it to an inactive form. High levels of CRY block TIM ubiquitination, suggesting a mechanism by which light-driven changes in CRY could control TIM ubiquitination.
In all organisms examined thus far, a feedback loop comprising cycling gene products that negatively regulate their own synthesis lies at the heart of the circadian clock (7). In Drosophila melanogaster, two of the genes that autoregulate in this fashion are period (per) and timeless (tim). The per and tim mRNA levels cycle with a circadian rhythm, such that RNA levels are high at the end of the day (or beginning of the night) and low at the end of the night (13, 36, 37). The two encoded proteins (PER and TIM) also cycle, with protein accumulation starting in the early evening and peaking in the middle of the night (8, 15, 26, 47). As they accumulate in the cytoplasm PER and TIM associate to form heterodimers (20, 47). This association stabilizes PER and permits nuclear entry of both proteins (30, 31, 34). Thus, while TIM does not require PER for stability, it is dependent on PER for nuclear transport. In the nucleus, either one or both proteins repress the synthesis of the per and tim mRNAs. Turnover of the two proteins, TIM in the late night and PER in the early morning, allows RNA levels to rise once again and the cycle continues.
Light acts as the primary zeitgeber, or timegiver, to synchronize an organism to its environment. At a molecular level the effect of light is to reduce levels of the TIM protein, an effect that appears to mediate entrainment of the molecular loop and thereby that of behavioral rhythms (15, 20, 26, 47). In fact, in all organisms examined (Neurospora crassa, Drosophila, and mammals), light changes levels of a clock component, indicating that this is a conserved general mechanism (7, 32, 41). The photoreceptors used by the circadian clock are still a subject of considerable debate, but in Drosophila it is clear that the visual system is not required although it is a redundant pathway that can mediate entrainment (14, 40, 46). The dedicated circadian photoreceptor in Drosophila as well as in Arabidopsis thaliana is the flavin-binding photoreceptor, cryptochrome (CRY) (1, 9–11, 39). In Drosophila, levels of cry RNA and protein cycle such that RNA levels peak at the end of the day and protein levels are high at night (9). Flies containing a mutant cry gene (cryb) display free-running behavioral rhythms but show deficits in entrainment that are especially pronounced when cryb is coupled with a visual-system mutation (14, 39). In addition, cryb mutant flies are rhythmic in constant light while wild-type flies are arrhythmic under such conditions, further demonstrating that their circadian system is defective in its ability to perceive light (10).
We recently found that TIM degradation in response to light is mediated by the proteasome and that TIM is phosphorylated and ubiquitinated prior to its degradation (27). Ubiquitination of TIM in response to light can be observed in cultured Drosophila S2 cells. To analyze the upstream events in the photic input pathway, we investigated the response of CRY to light and the effect of CRY signaling on TIM ubiquitination in the S2 cell culture system. Our data support a model in which CRY undergoes a light-induced conformation change which leads to its degradation by the proteasome. Blocking the transport of electrons from reduced flavin inhibits CRY degradation and increases TIM ubiquitination, indicating a causal relationship between the two events. However, overexpression of CRY attenuates TIM ubiquitination. Based on these data we propose a model according to which CRY controls TIM ubiquitination by actively blocking it in the dark and promoting it in the presence of light.
MATERIALS AND METHODS
Plasmid construction.
The pIZ-cry expression plasmid was constructed by ligating the entire coding region of dcry (nucleotides 1 to 1629; GenBank accession no. AB018050) into expression vector pIZ/His-V5 (Invitrogen), which uses the OpIE2 promoter. The design inserted the V5 epitope at the carboxy terminus of CRY. CRY mutants CRY-N (amino acids 1 to 423 of CRY), CRY-C (amino acids 244 to 542), and CRYb (39) were also subcloned into pIZ/His-V5. Expression plasmids hs-tim, hs-cry and hs-Ub were constructed by cloning full-length cDNAs for tim (28), cry, and an hemagglutinin (HA)-tagged ubiquitin octamer (42), respectively, into hsp70 promoter-driven expression vector pCasPer-hs (27). All constructs were verified by restriction enzyme digestion and sequencing.
Fly genetics.
The UAS-cry transgenic line, P{UAS-dcry}, was generously provided by T. Tanimura (Kyushu University, Ropponmatsu, Japan) (16). To obtain CRY-overexpressing flies, the P{UAS-dcry} line was crossed with the act5C-GAL4 effector line.
Cell transfections and pharmacological treatment.
All plasmids, hs-tim (1 μg/well), hs-Ub (1 μg/well), hs-cry, and wild-type and mutant pIZ-cry (amounts specified in each figure legend) were transfected using the calcium phosphate coprecipitation method. The calcium phosphate precipitate was mixed with 5 × 106 cells, and the mixture was incubated for 16 h. The cells were grown for an additional 24 h in fresh media. To induce the expression of transfected genes in the pCasPer-hs vector, cells were incubated for 30 min at 37°C and allowed to recover at room temperature for the times indicated in the figure legends. For light treatment (at the bench top, ∼600 lx) the cells were exposed to a 10-min or 2- or 5-h light pulse after recovery from heat shock (see figure legends for details). For transfections where no heat shock was involved, light treatment was initiated 40 to 48 h after transfection. Diphenylene iodonium (DPI; Sigma) and MG115 (Z-Leu-Leu-Nva-H; Peptides International) dissolved in dimethyl sulfoxide and lactacystin (BIOMOL) dissolved in water were used at a final concentration of 20 μM. After the treatments required for each experiment, cells were harvested and lysed as described below.
Coimmunoprecipitations.
S2 cells were washed once with phosphate-buffered saline (PBS) and lysed in Triton immunoprecipitation buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 10 mM EDTA [pH 8.0], 0.2% Triton X-100, 10 mM dithiothreitol, protease inhibitor cocktail; Boehringer Mannheim). Cell extracts were clarified by centrifugation (16,181 × g, 20 min at 4°C), and protein concentrations were determined using the Bio-Rad DC protein assay. One milligram of total protein from each clarified supernatant was incubated with 1 μl of an antibody to TIM (UPR8) overnight at 4°C. The immune complex was bound to protein G beads by incubation at 4°C for 1 h. Beads were then washed three times with PBS, mixed with sodium dodecyl sulfate (SDS) gel loading buffer (40 μl), and boiled, and the supernatant was loaded onto SDS-polyacrylamide gel electrophoresis gels. To assay ubiquitination, Western blots of the precipitates were probed with an anti-HA antibody as described below (the transfected ubiquitin is tagged with HA).
Western blots.
Fly head extracts were obtained as follows. Heads from frozen flies were homogenized in the Triton immunoprecipitation buffer described above. Extracts were clarified by centrifugation (16,181 × g, 20 min at 4°C), and the protein concentration was determined. One hundred to 150 μg of protein from the clarified supernatant was loaded onto SDS-polyacrylamide gel electrophoresis gels and transferred to a nitrocellulose membrane. For S2 cell Western blots, equivalent amounts of protein from S2 cells were loaded (amount used in each experiment is indicated in the figure legends). After being blocked in 1% bovine serum albumin and 3% nonfat dry milk in PBS, the blot was incubated with either a 1:1,000 dilution of rabbit anti-HA antibody (Clontech; see Fig. 5 and 7A), a 1:300 dilution of mouse anti-HA antibody (provided by Jeffrey Field; see Fig. 6) (12), a 1:1,000 dilution of rat anti-TIM antibody UPR8 (15), a 1:1,000 dilution of mouse anti-V5 antibody (Invitrogen), a 1:200 dilution of rat anti-CRY (raised to a full-length glutathione S-transferase–CRY fusion protein; see Fig. 7A), or a 1:5,000 dilution of rabbit anti-MAPK (mitogen-activated protein kinase; Sigma) in blocking solution for 1 h at room temperature. Subsequently, the blot was washed three times for 10 min each in PBS and then incubated with horseradish peroxidase-conjugated secondary antibody (1:1,000; Amersham). The signal was visualized with the ECL kit (Amersham). To ensure equal loading on each lane, the blot was stripped in stripping buffer (62.5 mM Tris-HCl [pH 7.6], 100 mM 2-mercaptoethanol, 5% SDS) at 55°C for 30 min, rinsed in PBS, blocked, and processed for detecting other bound proteins. In CRY experiments, overall levels of MAPK were quantitated to control for protein concentration and loading (see Fig. 1 and 3), whereas immunoprecipitated TIM levels were used as a control in TIM ubiquitination experiments (see Fig. 6 and 7). The Western blotting data were quantified on a densitometer (Molecular Dynamics), and the relative optical density (ROD) of the protein of interest was then determined. The means ± standard errors of the means (SEM) of the experiments were plotted, and Student's t test was used to make comparisons between control and experimental groups.
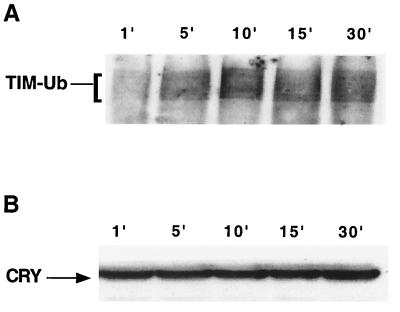
FIG. 5.
TIM ubiquitination precedes CRY degradation. We determined the time course of light-induced TIM ubiquitination and of CRY degradation in S2 cells. Cells were cotransfected with hs-tim, hs-Ub (this expresses an HA-tagged ubiquitin octamer), and pIZ-cry (2 μg/well). (A) Expression of TIM and HA-tagged ubiquitin was induced through heat shock treatment (see Materials and Methods). After recovery from heat shock, cells were exposed to light for the indicated times and harvested immediately. TIM ubiquitination was assayed by probing TIM immunoprecipitates with an anti-HA antibody. Similar results were obtained in two independent experiments. (B) Samples from panel A were run on a separate gel and probed with anti-V5 to determine the level of CRY after light treatment.
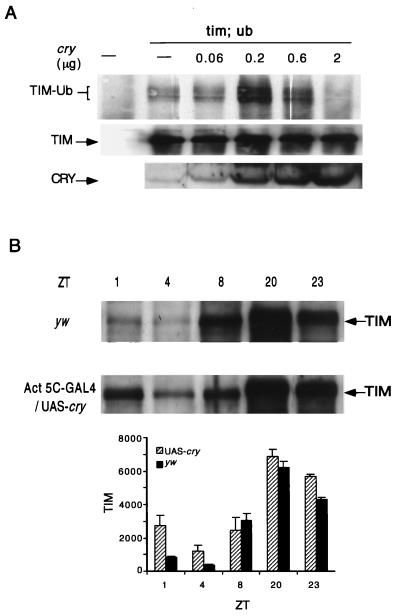
FIG. 7.
Effect of CRY overexpression on light-induced TIM ubiquitination and TIM expression. (A) hs-tim, hs-Ub, and different amounts of the hs-cry plasmid were transfected into S2 cells where indicated. (Top) To analyze ubiquitination, TIM immunoprecipitates were probed with an anti-HA antibody. (Middle) The blots were reprobed with an anti-TIM antibody to evaluate overall levels of TIM. (Bottom) CRY levels were verified by probing the same lysates (100 μg) with an anti-CRY antibody. The differential effects of low and high concentrations of CRY were observed in two independent experiments with the hs-cry plasmid. Blocking by high concentrations of CRY was also seen in multiple experiments with the pIZ-cry plasmid (see text). (B) Light-induced degradation of TIM protein is delayed in flies that overexpress CRY. Flies were collected at the indicated zeitgeber times (ZT) during the third day of a 12-h–12-h LD cycle. (Top) Western blots of yw and Act5C-GAL4/UAS-cry adult head extracts (100 μg/lane) were probed with an anti-TIM antibody. (Bottom) Data from two or three independent experiments (the data for all the yw time points and UAS-cry ZT23 are based on two experiments; data for UAS-cry time points other than ZT23 are based on three) were quantified on a densitometer. Means ± SEM are plotted. A more detailed comparison of early day time points (ZT1, -2, and -4 to -6) between the two genotypes confirmed that TIM levels were higher in UAS-cry flies in the early part of the day (data not shown).
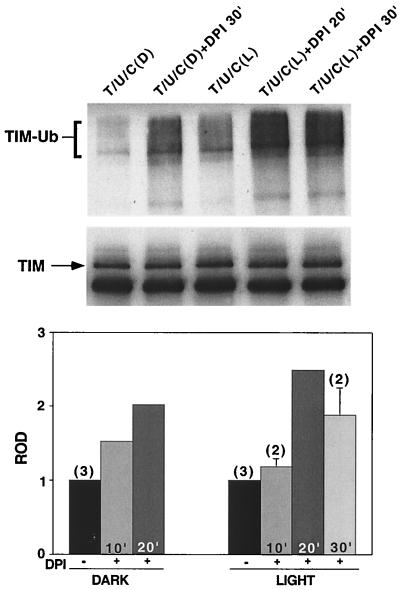
FIG. 6.
TIM ubiquitination is increased by blocking electron transport. Cells were transfected with hs-tim (T), hs-Ub (U), and pIZ-cry (C) (0.2 μg/well). After a 30-min heat shock, the cells were maintained in the dark for 2 h. DPI (20 μM) was added to the cultures 10, 20, or 30 min prior to the initiation of a 10-min light pulse (L). Controls were maintained in the dark (D) during this time. (Top) TIM ubiquitination was assayed as described for Fig. 5. (Middle) The blot was reprobed with an anti-TIM antibody to visualize overall TIM levels. TIM ubiquitination was increased in the DPI-treated groups. (Bottom) Quantitation of data from three independent experiments. In all cases, the ROD of the ubiquitinated TIM species in DPI-treated groups was quantified on a densitometer and normalized to the ROD of ubiquitinated TIM in untreated cells. The duration of DPI treatment prior to the light pulse is indicated within each bar. The numbers of times an experimental condition was repeated are in parentheses.
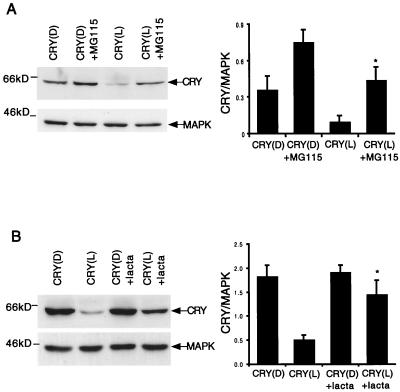
FIG. 1.
CRY is degraded by the proteasome in response to light. S2 cells were transfected with pIZ-cry (2 μg/well) and then left in the dark for 40 h. A proteasome inhibitor, either MG115 (A) or lactacystin (lacta) (B), was added to the culture media at 20 μM. The cells were subsequently kept in light (L) or dark (D) for 5 h prior to Western blot analysis. (Left) Western blots (150 μg/lane) were probed first with an anti-V5 antibody to detect pIZ-cry and then with an anti-MAPK antibody to control for loading. (Right) For each proteasome inhibitor, data from three independent experiments were quantified on a densitometer (Molecular Dynamics). The ROD of the CRY signal was normalized to that of MAPK, and the means ± SEM are graphed. ∗, significantly different from the light-treated control assayed in the absence of the proteasome inhibitor (t = −3.05 and P < 0.04 for MG115, and t = −3.22 and P = 0.03 for lactacystin). MG115 and lactacystin did not produce significant changes in CRY expression in the samples that were maintained in the dark (t = −2.66 and P = 0.056, and t = −0.32 and P = 0.77, respectively).
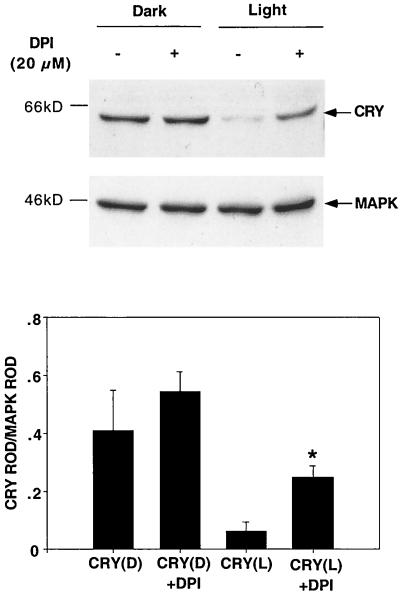
FIG. 3.
CRY degradation requires electron transport. The CRY response to light was tested in transfected S2 cells in the presence of electron transport inhibitor DPI. Cells were transfected with pIZ-cry (2 μg/well), and DPI was added to the culture media immediately before a 5-h light pulse. (Top) CRY expression was assayed through Western blots using an anti-V5 antibody. The blots were then stripped and reprobed with an anti-MAPK antibody. Three independent experiments were performed and analyzed. (Bottom) The ROD of the CRY signal was determined and normalized to that of MAPK. ∗, significantly different from the light-treated sample assayed in the absence of DPI (t = −3.65, P < 0.03).
Reverse transcription-PCR.
mRNA from S2 cells was isolated using the MicroPoly(A)Pure kit and transcribed to first-strand cDNA with an oligo(dT) primer using the Superscript preamplification system (Life Technologies). To ensure that the PCR product was derived from mRNA and not genomic DNA, a negative control in which reverse transcriptase was omitted was included. Subsequent PCR amplification was carried out with a primer pair that amplifies nucleotides 654 to 1089 of cry.
RESULTS
CRY is degraded by the proteasome in response to light.
The profile of CRY protein expression in light-dark (LD) cycles suggested that the protein is unstable in the presence of light (9). Light-induced instability of CRY was supported by experiments in which CRY was expressed in S2 cells (4). We transfected cells with a pIZ-cry construct, in which CRY is tagged with a V5 epitope, and noticed that levels of the protein were reduced by light treatment. To identify the mechanisms that degrade CRY, we treated CRY -transfected cells with light in the presence of proteasome inhibitors MG115 and lactacystin. Both these agents were effective in blocking CRY degradation (Fig. 1). This effect of light on both TIM (27) and CRY suggests that the action of a ubiquitin/proteasome degradation pathway may be one of the first events in photic resetting in Drosophila.
CRY mutants are not degraded by light.
We previously reported light-induced ubiquitination of TIM in S2 cells (further discussed below). The photoreceptor that transduced photic signals to TIM was not known, but, based on data implying a role for CRY in circadian photoreception (11, 39) together with the reported light-activated direct interaction of TIM and CRY (4), we suspected that S2 cells expressed endogenous cry. As shown in Fig. 2A, we confirmed the presence of cry RNA in S2 cells. Consistent with the presence of endogenous CRY in our S2 cells, we found that light-dependent inhibition of PER-TIM feedback activity, which is known to be CRY dependent (4), occurred to a significant extent in the absence of transfected CRY (S. Sathyanarayanan, F. Lin, and A. Sehgal, unpublished data). The presence of endogenous photic signaling mechanisms in S2 cells provided us with a system in which we could assay the degradation of transfected CRY regardless of its ability to transduce a photic signal.
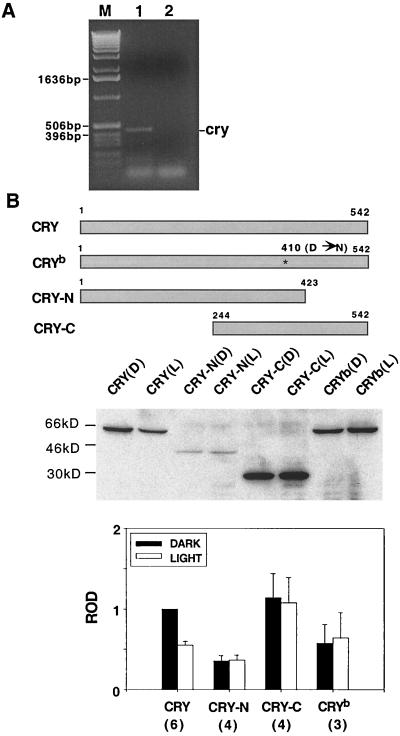
FIG. 2.
Effect of light on CRY mutants. (A) Expression of endogenous cry in S2 cells. A 435-bp band corresponding to dcry (bp 654 to 1089) was detected through reverse transcription-PCR with S2 mRNA (lane 1). There was no band when reverse transcriptase was eliminated from the reaction, indicating that the product is derived from RNA (lane 2). Lane M, molecular weight markers (Life Technologies). (B) (Top) Schematic representation of CRY mutants. The CRYb mutant carries a missense mutation (asterisk) that substitutes an asparagine (N) for an aspartic acid (D) in the flavin-binding site. Both CRY-N and CRY-C contain the 14 highly conserved residues required for flavin binding but lack sequences at the C and N termini, respectively. (Middle) Western analysis of S2 cells transfected with the mutant cry DNA (2 μg/well). Light-treated cells were exposed to a 2-h light pulse. Equal amounts of light-treated (L) and dark control (D) lysates (150 μg/lane) were loaded onto the gel, and the blot was probed with an anti-V5 antibody. (Bottom) The Western blotting data from this experiment and from several others were quantified on a densitometer (Molecular Dynamics). The ROD of the signal obtained under different conditions was normalized to that of wild-type CRY in the dark, and the means ± SEM were plotted. The numbers of independent experiments are in parentheses.
To determine whether a specific region of CRY mediates its degradation, we transfected different CRY mutants and assayed their response to light. As shown in Fig. 2B, CRY-N (amino acids 1 to 423) has the C-terminal 119 amino acids deleted and CRY-C (amino acids 244 to 542) has the N-terminal 243 amino acids deleted. The cryb mutation is a missense mutation within the sequence that encodes the highly conserved flavin-binding region of Drosophila CRY. It corresponds to the original cry mutation isolated through a genetic screen of Drosophila (39). Treatment with light did not reduce the levels of any of these mutant CRY proteins (Fig. 2B). For CRY-N the levels were consistently low with and without light treatment, indicating a general instability of the protein. CRY-C was expressed at high levels, and levels of CRYb were equivalent to those of the wild type. However, none of these proteins showed a response to light. Since there is no common sequence that is deleted in all these constructs, this indicates either that CRY degradation requires more than one part of the molecule or that the overall conformation of the molecule is important for its recognition by the degradation system. The large deletions in CRY-N and CRY-C may prevent such a conformation change. For CRYb, the mutation is thought to prevent association with flavin, which may be required for a redox-mediated conformation change (see below).
CRY degradation requires redox activity.
CRY signaling in plants requires redox activity and is mediated, at least in part, by the flavin moiety bound to CRY (23, 45). This is based on the finding that DPI, which inhibits the transport of electrons from reduced flavin, was effective in blocking CRY-mediated photic signaling in Arabidopsis (23). In addition, flavins participate in electron transport in other systems, the most relevant system being that of the photolyases, which are homologous to CRYs and which are known to repair DNA through an electron transfer mechanism (2, 6).
Based on the data implicating electron transport in Arabidopsis CRY signaling, we determined the effect of electron transport inhibitors on degradation of Drosophila CRY. As shown in Fig. 3, DPI attenuated CRY degradation. Thus, photic signaling by Drosophila CRY involves redox activity, most likely mediated by the flavin. Note that the doses of DPI used here have not been associated with any toxic effects in cell culture systems (5, 35). Ferricyanide, which blocks electron transport in the membrane, attenuated CRY degradation to a limited extent (data not shown).
Relationship between light responses of CRY and TIM.
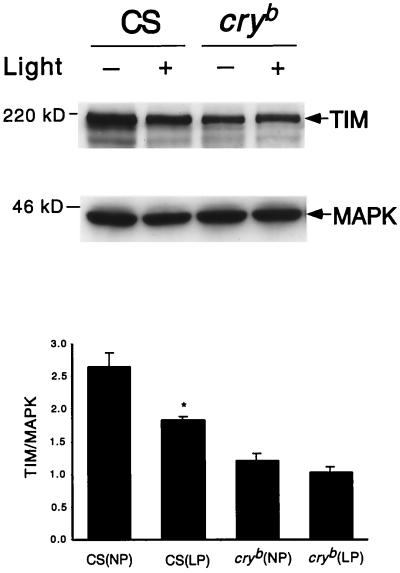
The presence of endogenous CRY in our S2 cells supported the idea that the light-dependent TIM ubiquitination we reported previously (27) was mediated by CRY. As mentioned above, the data of Ceriani et al., demonstrating a light-dependent interaction between TIM and CRY, also suggested that CRY regulates TIM (4). To determine if the TIM response to light requires CRY, we assayed TIM levels in light-pulsed and unpulsed cryb flies. As shown in Fig. 4, while wild-type flies showed the characteristic decrease in TIM levels with light treatment, this response was lacking in cryb flies. Using a histochemistry assay, Ivachenko et al. also recently reported that TIM does not respond to light in larval lateral neurons and adult Malpighian tubules of cryb flies (17).
FIG. 4.
TIM is not degraded by light in cryb flies. Wild-type (Canton S) and cryb flies were treated with a 1-h light pulse at zeitgeber time 20 (ZT20; ZT0, lights on; ZT12, lights off [12-h–12-h LD cycle]). (Top) At the end of 1 h, light-pulsed flies (LP) and unpulsed controls (NP) were harvested and head extracts (100 μg/lane) were assayed for TIM expression. The Western blots were stripped and reprobed with an anti-MAPK antibody to control for loading (in other experiments we determined that levels of MAPK do not cycle in adult fly heads [J. Williams and A. Sehgal, unpublished data]). (Bottom) The data were quantified on a densitometer, and TIM levels were normalized relative to those of MAPK. The means ± SEM of three experiments are shown for both genotypes in light and dark. TIM levels were significantly reduced by light in wild-type flies (t = −3.78; P = 0.019) but not in the cryb mutant (t = −1.31; P = 0.26).
We used the S2 cell system to determine the relationship between light-induced CRY degradation and TIM ubiquitination and degradation. One possibility we considered was that CRY was required for TIM stability. In this model, light-induced CRY degradation would lead to TIM degradation, perhaps by exposing relevant sites on TIM to phosphorylation and ubiquitination events. Although TIM and CRY do not bind each other in the dark in the yeast two-hybrid system, they can be coimmunoprecipitated from S2 cells, suggesting that they are present in the same complex (4). Thus, removal of CRY in response to light could affect TIM processing. Alternatively, light exposure may lead to some conformational and/or redox changes in CRY which trigger downstream events including TIM ubiquitination and CRY degradation. To distinguish between these two possibilities, we examined the time course of TIM ubiquitination and that of CRY degradation in S2 cells. We detected an increase in TIM ubiquitination within 5 min of light exposure (Fig. 5A), while CRY levels in the same extracts remained unchanged up to the end of a 30-min light pulse (Fig. 5B). Thus, overall degradation of CRY does not appear to be required for TIM ubiquitination. Although we cannot exclude the possibility that CRY is removed from a complex with TIM, it is more likely that in response to light CRY transmits a signal that leads to TIM ubiquitination.
We failed to detect significant degradation of TIM in S2 cells in response to light. This may be due, in part, to the HA tag on the ubiquitin, which could interfere with proteasomal digestion. However, other researchers have also noted that TIM is not turned over upon light exposure in S2 cells (4). Extended incubation (up to 6 h post-TIM induction) of transfected cells resulted in TIM degradation in both dark- and light-treated cells (data not shown).
We next examined TIM ubiquitination in the presence of electron transport inhibitor DPI. TIM ubiquitination was increased by DPI (Fig. 6), although CRY degradation was blocked, which is consistent with the idea that TIM ubiquitination does not require degradation of CRY (Fig. 5). In fact, the increased TIM ubiquitination is most likely due to the accumulation of activated CRY, effected through a block either in degradation or in the reconversion of CRY to an inactive form.
Effect of CRY overexpression on the TIM light response.
As noted above, we believe that light-induced TIM ubiquitination in S2 cells is mediated by endogenous CRY. To test the effects of increasing CRY levels on light-induced TIM ubiquitination, we cotransfected S2 cells with hs-tim, hs-Ub (27), and different concentrations of either pIZ-cry or hs-cry and assayed TIM ubiquitination 2 h after light exposure.
High concentrations of both hs-cry and pIZ-cry decreased TIM ubiquitination (Fig. 7A and data not shown). However, ubiquitination of TIM was enhanced when <0.2 μg of hs-cry/well was transfected (Fig. 7A). pIZ-cry did not increase TIM ubiquitination when transfected at low concentrations, most likely because this plasmid yielded higher levels of CRY expression. Taken together these observations indicate that small increases in CRY promote TIM ubiquitination after 2 h of light exposure but that high levels attenuate it. However, even in the presence of high levels of CRY, TIM ubiquitination increased during the first 10 to 15 min of light treatment (Fig. 5A). The block at later time points in CRY-overexpressing cells (Fig. 7A) is indicative of a deficit in the maintenance of TIM ubiquitination, which may be due to enhanced deactivation of CRY (see Discussion).
Our data on the differential effects of low and high CRY concentrations are supported by results of CRY overexpression in transgenic flies. Flies that overexpress CRY under control of the tim promoter show enhanced resetting, while those that express CRY under the actin 5c promoter show a reduction of light-induced phase delays (16). The difference in the phenotypes of these two overexpression strains may lie in the level of overexpression. To determine whether the reduced resetting in the actin 5c line correlated with reduced TIM degradation in response to light, we crossed flies carrying a UAS-cry construct to others carrying an actin 5c promoter-GAL4 transgene and assayed the resulting progeny for TIM expression. TIM expression was examined at different times of day by Western blotting of adult fly head extracts (Fig. 7B). In CRY overexpression flies TIM levels were considerably higher than wild-type levels at time points 1 and 4 but equivalent to wild-type levels at all other time points. Thus, the effect was specific for the early part of the day, when TIM is normally turned over in response to light.
DISCUSSION
Degradation of CRY by light invokes analogies with the plant photoreceptors, phytochrome (PHY) and CRY, both of which are degraded in response to light (22, 38). Thus, it may be a common mechanism to control levels of the photoreceptor and thereby the strength of the photic response. Moreover, as noted here for CRY, PHY is known to be degraded by the proteasome (38).
The role of the proteasome in degradation of both CRY and TIM also underscores similarities with the cell cycle. The cell cycle is characterized by cycling proteins that undergo phosphorylation and subsequent degradation, in many cases by the proteasome (18). We now know that both PER and TIM are cyclically phosphorylated and that phosphorylation plays a role in turnover of both proteins (27, 30). For TIM, light-induced degradation is effected through an increase in phosphorylation and ubiquitination (27). Thus, as for the cell cycle, multiple proteins in the circadian cycle are turned over by the ubiquitin-proteasome pathway. However, PER turnover may utilize a different pathway since ubiquitination of PER has not been observed (27).
The mechanisms that lead to CRY degradation in response to light are not clear, but we hypothesize that a conformation change in CRY is required. A light-induced conformation change is supported by the following lines of evidence. (i) In the yeast two-hybrid system CRY interacts with full-length TIM in the presence of light but not in the dark (4). (ii) Sequences that mediate CRY degradation do not appear to map to a unique part of the molecule, suggesting that the tertiary structure is important (Fig. 2). (iii) The CRYb protein is not degraded by light (Fig. 2). All these mutants were tested in the presence of endogenous CRY, and so their ability to signal was dissociated from their degradation. Although the single amino acid mutated in the flavin-binding region in CRYb could play a direct role in the degradation process, it is far more likely that it affects a flavin-mediated conformation change. The fact that CRYb does not associate with TIM in the yeast two-hybrid system (4) is consistent with an inability to undergo a conformation change.
Ceriani et al. showed recently that CRY blocks PER and TIM autoregulation of their own RNA synthesis in a light-dependent manner in S2 cells (4). As TIM degradation is not detectable in S2 cells, they suggested that the inhibition of TIM activity, rather than its degradation, by CRY is the primary response to light. We believe that this block in PER-TIM activity may be the immediate response of the clock to light. Presumably this block persists as long as the photic signals are present and CRY is not degraded. However, a phase change of several hours, which can be produced with a pulse of <1 min of light, must require an irreversible change in a clock component. We show that TIM is ubiquitinated in S2 cells within 5 min of light treatment. In flies, TIM degradation (which presumably follows ubiquitination) occurs within 30 to 60 min of light treatment (15) and is apparently critical for resetting the clock (40, 46). As shown in Fig. 4 and as reported by Ivachenko et al., a CRY molecule with a functional flavin-binding domain is required for this response (17).
Signaling by flavins frequently involves a redox change (2, 23, 24). In fact, we show here that a reagent that blocks the transfer of electrons from reduced flavin prevents CRY degradation by light. At the same time, it increases TIM ubiquitination. Based on the recently proposed models for Arabidopsis CRY (45), DPI may block either intramolecular electron transport required for a change in CRY conformation or intermolecular transport to a signaling pathway that effects degradation. Assuming that active CRY, which promotes TIM ubiquitination, is produced by a conformation change, we suggest that the DPI-sensitive step occurs after the conformation change. It should be noted that DPI can also block the activity of other flavoproteins, such as NADPH oxidase and nitric oxide synthase, that play a role in redox processes (21, 44).
We show here that high levels of CRY block TIM ubiquitination. Our data on the effects of CRY in S2 cells are supported by in vivo fly data. Transgenic flies that overexpressed CRY under control of the tim promoter showed enhanced resetting, indicating that within a certain range increasing levels of the photoreceptor amplify the photic signal (9). Likewise, at very low concentrations in cultured cells we observed increased TIM ubiquitination (Fig. 7A). However, when CRY was overexpressed by the actin 5c promoter in flies (16) and also in the cultured cells when it increased beyond a certain level (Fig. 7A), the photic signal is reduced. The transgenic lines that expressed CRY under the control of the actin 5c promoter showed smaller phase shifts than the wild type, perhaps because the actin promoter drives a high level of expression at all times of day, as distinct from the tim promoter, which is rhythmically transcribed. In addition, the actin 5c line that expressed the highest levels of CRY showed decreased sensitivity to light such that a higher intensity of light was required to produce shifts equivalent to that for the wild type. Our data demonstrate that in these flies levels of TIM in the early part of the day are increased. This could be due to the effect of CRY overexpression on the feedback mechanism; since CRY is known to attenuate negative feedback by PER and TIM in a light-dependent manner (4), CRY overexpression could result in reduced feedback and thereby increased tim RNA expression. However, one would expect the increased RNA to also result in high levels of TIM at night, which was not observed here (Fig. 7B). Given that daytime levels are preferentially affected, together with the behavioral phenotype of these flies, we favor the alternative explanation that the TIM response to light is reduced due to attenuation of photic signaling by overexpressed CRY.
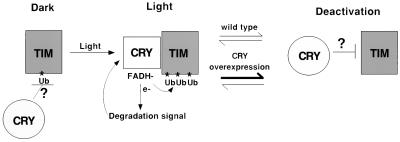
We infer that, in the actin 5c-CRY flies as well as in the S2 cells, less CRY is present in an active (light-induced) conformation. Overexpression may result in rapid deactivation of CRY in continuous light, which would account for the decrease in TIM ubiquitination observed at later time points (compare Fig. 5 and 7). We note that photoreceptors are known to be deactivated in the presence of continuous light (25), a process that may be augmented by overexpression. An attractive possibility is that the inactive form of CRY prevents TIM ubiquitination while the light-induced form, which binds TIM directly, promotes TIM ubiquitination (Fig. 8). In the CRY overexpression situation the high levels of inactive CRY during both the light and the dark attenuate TIM ubiquitination. Since the inactive form is normally found in the dark, this would be indicative of a role for CRY in blocking TIM ubiquitination in the dark (Fig. 8). It may be possible to test this model by generating mutations that lock CRY in a particular conformation. It would also be interesting to determine the effects of acute CRY induction on overall TIM levels and on light-induced TIM degradation in flies. A function for CRY in stabilizing TIM may also have relevance for its mechanism of action within the clock. To date, there is no evidence to suggest that CRY is involved in generating free-running behavioral rhythms in Drosophila, but it apparently participates in clock function in peripheral tissues (17, 19). Interestingly, in Malpighian tubules, where CRY appears to be part of the clock, TIM levels are low in cryb flies (17). In addition, mammalian CRY is part of the clock that controls behavioral rhythms and is thought to stabilize mPER2 (33). It is conceivable that, as a clock component, CRY controls free-running degradation of clock proteins, thereby promoting molecular oscillations.
FIG. 8.
Model for the effect of CRY on TIM. The light-induced conformation of CRY promotes TIM ubiquitination, perhaps through direct interaction with TIM. This conformation of CRY also signals to a degradation pathway through a DPI-sensitive electron transport mechanism. As a result, both CRY and TIM are degraded (although TIM degradation is not observed in S2 cells, light-induced TIM degradation in flies is CRY dependent [17] [Fig. 4]). We assume that, like other photoreceptors, CRY is eventually deactivated. Overexpression of CRY may favor its deactivation, resulting in decreased TIM ubiquitination in S2 cells and decreased degradation in flies (Fig. 7). We suggest that the inactive CRY actively inhibits TIM ubiquitination, a mechanism by which CRY could also control TIM ubiquitination in the dark (see Discussion). FADH, reduced flavin adenine dinucleotide.
The lack of TIM degradation in S2 cells suggests that some components of the pathway are absent or expressed at low levels in these cells. Nevertheless, it is clear that S2 cells are capable of circadian photoreception and support all the events that lead up to the first response of a clock protein. Given that the S2 cell line is an embryonic line that has not differentiated completely, this suggests that photoreceptors, and in some cases perhaps even circadian oscillators, are found in undifferentiated embryonic cells, which give rise to specific organs. In this context, note that some mammalian cells display cyclic expression of clock genes when serum shocked (3). In addition, circadian oscillators that can be entrained by light have been described in isolated organs from both vertebrates and invertebrates (29, 43).
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Yifeng Chen for raising the CRY antibody used here, Teiichi Tanimura for flies carrying the UAS-cry construct, J. D. Alvarez and J. Field for comments on the manuscript, Tony Cashmore and Peter Schotland for comments on a previous version, and other members of the laboratory for useful discussions.
The work was supported in part by grants from the NIH and NSF. A.S. is an Associate Investigator in the HHMI.
REFERENCES
- 1.Ahmad M, Cashmore A R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 2.Aubert C, Ves M H, Mathis P, Eker A P M, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 3.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 4.Ceriani M F, Darlington T K, Staknis D, Mas P, Petti A A, Weitz C J, Kay S A. Light-dependent sequestration of Timeless by cryptochrome. Science. 1999;285:553–556. doi: 10.1126/science.285.5427.553. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y-C, Lin-Shiau S-Y, Lin J-K. Involvement of reactive oxygen species and caspase 3 activation in arsenite-induced apoptosis. J Cell Physiol. 1998;177:324–333. doi: 10.1002/(SICI)1097-4652(199811)177:2<324::AID-JCP14>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Deisenhofer J. DNA photolyases and cryptochromes. Mutat Res. 2000;460:143–149. doi: 10.1016/s0921-8777(00)00023-9. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap J C. Molecular bases for circadian biological clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 8.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emery P, So W, Kaneko M, Hall J C, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 10.Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall J C, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 11.Emery P, Stanewsky R, Rosbash M, Hall J C. dCRY is a unique Drosophila circadian photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 12.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardin P E, Hall J C, Rosbash M. Feedback of the Drosophila period gene on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 14.Helfrich-Forster C, Winter C, Hofbauer A, Hall J C, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 15.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–686. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa T, Matsumoto A, Kato T, Jr, Togashi S, Ryo H, Ikenaga M, Takeshi T, Ueda R, Tanimura T. DCRY is a Drosophila photoreceptor protein implicated in light entrainment of circadian rhythm. Genes Cells. 1999;4:57–65. doi: 10.1046/j.1365-2443.1999.00237.x. [DOI] [PubMed] [Google Scholar]
- 17.Ivachenko M, Stanewsky R, Giebultowicz J M. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- 18.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 19.Krishnan B, Levine J D, Lynch M K S, Dowse H B, Funes P, Hall J C, Hardin P E, Dryer S E. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- 20.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Trush M A. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem Biophys Res Commun. 1998;253:295–299. doi: 10.1006/bbrc.1998.9729. [DOI] [PubMed] [Google Scholar]
- 22.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore A R. Enhancement of blue-light sensitivity of arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long J C, Jenkins G I. Involvement of plasma membrane redox activity and calcium homeostasis in the UV-B and UV-A/blue light induction of gene expression in Arabidopsis. Plant Cell. 1998;10:2077–2086. doi: 10.1105/tpc.10.12.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda H, Iyanagi T. Calmodulin activates intramolecular electron transfer between the two flavins of neuronal nitric oxide synthase flavin domain. Biochim Biophys Acta. 1999;1473:345–355. doi: 10.1016/s0304-4165(99)00193-2. [DOI] [PubMed] [Google Scholar]
- 25.Mendez A, Burns M E, Roca A, Lem J, Wu L W, Simon M I, Baylor D A, Chen J. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 2000;28:153–164. doi: 10.1016/s0896-6273(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 26.Myers M P, Wager-Smith K, Rothenflugh A, Young M W. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 27.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 28.Ousley A, Zafarullah K, Chen Y, Emerson M, Hickman L, Sehgal A. Conserved regions of the timeless (tim) clock gene in Drosophila analyzed through phylogenetic and functional studies. Genetics. 1998;148:815–825. doi: 10.1093/genetics/148.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plautz J D, Kaneko M, Hall J C, Kay S A. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 30.Price J L, Blau J, Rothenflugh A, Abodeely M, Kloss B, Young M W. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 31.Price J L, Dembinska M E, Young M W, Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–4049. doi: 10.1002/j.1460-2075.1995.tb00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reppert S M. A clockwork explosion. Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 33.Reppert S M, Weaver D. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 34.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins Period and Timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 35.Schubert D, Behl C, Lesley R, Brack A, Dargusch R, Sagara Y, Kimura H. Amyloid peptides are toxic via a common oxidative mechanism. Proc Natl Acad Sci USA. 1995;92:1989–1993. doi: 10.1073/pnas.92.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sehgal A, Price J L, Man B, Young M W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 37.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M P, Young M W. Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 38.Shanklin J, Jabben M, Vierstra R D. Red light-induced formation of ubiquitin-phytochrome conjugates: identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci USA. 1987;84:359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay S A, Rosbash M, Hall J C. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 40.Suri V, Zuwei Q, Hall J C, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 41.Tamaru T, Isojima Y, Yamada T, Okada M, Nagai K, Takamatsu K. Light and glutamate-induced degradation of the circadian oscillating protein BMAL1 during the mammalian clock resetting. J Neurosci. 2000;20:7525–7530. doi: 10.1523/JNEUROSCI.20-20-07525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 43.Whitmore D, Foulkes N, Strahle U, Sassone-Corsi P. Zebrafish clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- 44.Xia Y, Roman L J, Masters B S S, Zweier J L. Inducible nitric-oxide synthase generates superoxide from the reductase domain. J Biol Chem. 1998;273:22635–22639. doi: 10.1074/jbc.273.35.22635. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Wu Y, Tang R, Liu D, Liu Y, Cashmore A R. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z, Emerson M, Su H S, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a non-visual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/s0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 47.Zeng H, Qian Z, Myers M P, Rosbash M. A light entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]