Abstract
Nascent pre-mRNAs associate with hnRNP proteins in hnRNP complexes, the natural substrates for mRNA processing. Several lines of evidence indicate that hnRNP complexes undergo substantial remodeling during mRNA formation and export. Here we report the isolation of three distinct types of pre-mRNP and mRNP complexes from HeLa cells associated with hnRNP A1, a shuttling hnRNP protein. Based on their RNA and protein compositions, these complexes are likely to represent distinct stages in the nucleocytoplasmic shuttling pathway of hnRNP A1 with its bound RNAs. In the cytoplasm, A1 is associated with its nuclear import receptor (transportin), the cytoplasmic poly(A)-binding protein, and mRNA. In the nucleus, A1 is found in two distinct types of complexes that are differently associated with nuclear structures. One class contains pre-mRNA and mRNA and is identical to previously described hnRNP complexes. The other class behaves as freely diffusible nuclear mRNPs (nmRNPs) at late nuclear stages of maturation and possibly associated with nuclear mRNA export. These nmRNPs differ from hnRNPs in that while they contain shuttling hnRNP proteins, the mRNA export factor REF, and mRNA, they do not contain nonshuttling hnRNP proteins or pre-mRNA. Importantly, nmRNPs also contain proteins not found in hnRNP complexes. These include the alternatively spliced isoforms D01 and D02 of the hnRNP D proteins, the E0 isoform of the hnRNP E proteins, and LRP130, a previously reported protein with unknown function that appears to have a novel type of RNA-binding domain. The characteristics of these complexes indicate that they result from RNP remodeling associated with mRNA maturation and delineate specific changes in RNP protein composition during formation and transport of mRNA in vivo.
Formation of mature cytoplasmic mRNAs in eukaryotic cells involves extensive processing of their corresponding pre-mRNAs in the nucleus, resulting in mature mRNAs that are subsequently exported across the nuclear envelope to the cytoplasm (for recent reviews on RNA export, see references 27 and 51). The natural substrates for nuclear events in mRNA maturation are ribonucleoprotein (RNP) complexes formed by the persistent association of pre-mRNAs and mRNAs with specific proteins. Prominent among these is a group of pre-mRNA- and mRNA-binding proteins collectively known as hnRNP proteins. The association of hnRNP proteins with RNA begins as the nascent pre-mRNA emerges from the RNA polymerase II transcription machinery and remains through processing and export of mRNA (17).
In human cells, the hnRNP proteins comprise a family of ca. 24 different polypeptides, termed hnRNP A1 (ca. 35 kDa) through hnRNP U (ca. 120 kDa), which are among the most abundant components of the cell nucleus (17, 59). hnRNP proteins are recruited to different transcripts in different relative amounts (43, 61, 76) and, rather than being passive components of the substrate, several hnRNP proteins have been shown to have specific roles in many different aspects of mRNA formation (17, 33). Furthermore, the protein composition of hnRNP complexes is not temporally fixed. There is substantial evidence that maturation and nuclear export of mRNA are accompanied by changes in the protein composition of hnRNP complexes, as described below.
Under normal growth conditions, hnRNP proteins are concentrated in the nucleus, where they are apparently excluded from the nucleolus (57). A subset of the hnRNP proteins (e.g., hnRNPs A1 and K) shuttle constantly between the nucleus and the cytoplasm, whereas others (e.g., hnRNP C1/C2 and hnRNP U) do not shuttle and are retained in the nucleus (60). Nuclear export of hnRNP A1 is mediated by a specific amino acid sequence termed M9, which functions as a bona fide nuclear export signal (NES) (46). M9 also functions as the hnRNP A1 nuclear location signal by mediating binding of its nuclear import receptor, transportin (62). hnRNP A1 retains its ability to bind mRNA at least transiently in the cytoplasm and probably also during its passage through the nuclear pore complex (NPC) (60). In contrast to hnRNP A1, hnRNP C1 and hnRNP C2 are retained in the nucleus, and this retention is mediated by a specific amino acid sequence in the C proteins that functions as a nuclear retention sequence (NRS). Importantly, this NRS can override NESs (50), and therefore it is likely that removal of NRS-containing hnRNP proteins from mRNA is a prerequisite for nuclear export of mRNA.
Based on the information described above, it was proposed that shuttling hnRNP proteins accompany mRNAs during their passage through NPCs, while nonshuttling hnRNP proteins are removed prior to or concomitant with mRNA export (57, 60). Support for this was provided by studies of the giant Balbiani ring (BR) RNP complex in Chironomus tentans. BR pre-mRNA- and mRNA-containing RNPs can be observed directly by electron microscopy due to their large size and abundance, and specific morphological changes can be monitored as their RNA matures and is exported through NPCs (15). Early electron microscopy observations provided evidence that mRNA is indeed exported to the cytoplasm through NPCs as an mRNP complex (69). Immunoelectron microscopy studies have shown that C-hrp36, which resembles vertebrate hnRNP A/B proteins, associates with nascent BR pre-mRNAs and remains in BR granules following their release into the nucleoplasm. Importantly, this association persists while BR granules are transported through NPCs and continues transiently in the cytoplasm (73). Proteins of the cap-binding complex also remain associated with BR RNPs during their nuclear export, while others, such as hrp45 and hrp23, do not appear to shuttle and dissociate at various stages prior to or upon association of transport RNPs with NPCs (15).
The specific factors and mechanisms involved in mRNP export through NPCs are not well understood. Nuclear export of mRNA is a facilitated process that requires energy (27, 51). The present models for mRNA export are that the mRNP, rather than the mRNA itself, is the actual recognition substrate and that mRNP proteins mediate the interaction of the bound mRNA with a nuclear protein export machinery (27, 51). A number of candidate factors specific for mRNA export in metazoans have emerged through similarities with mRNA export factors identified in yeast and through studies of retroviral RNA export. One of the best characterized is TAP, a homolog of the essential Saccharomyces cerevisiae mRNA export factor Mex67p, which also binds the constitutive transport element (CTE) of type D retroviruses. TAP promotes export of unspliced CTE-containing pre-mRNA, and an excess of CTE RNA inhibits cellular mRNA export (22, 30, 55). The participation of TAP in export of cellular mRNAs is probably effected through its interaction with an hnRNP-like protein, termed REF or Aly, that is similar to another essential S. cerevisiae mRNA export factor, Yra1p (4, 70, 71). Recent studies have indeed shown that REF can stimulate nuclear export of mRNA derived from intron-containing as well as intronless RNAs (65, 77). Association of REF with some RNAs appears to result from pre-mRNA splicing, and the present thinking is that TAP associates with cellular mRNPs at some stage after recruitment of REF (65). Other mammalian proteins similarly implicated in mRNA export include hGLE1, RAE1 (or mrnp41) (a homolog of Gle2p), and Dbp5, a DEAD-box protein (32, 63, 67, 75).
Processing itself of pre-mRNA into mRNA plays a central role in mRNA export. It has long been known that, for intron-containing genes, the presence of an intron (and its subsequent removal) is required for efficient gene expression (21, 23). Some naturally intronless transcripts (e.g., herpes simplex virus thymidine kinase mRNA and histone mRNA) contain specific sequences that can mediate intron-independent expression when placed on otherwise intron-dependent RNAs (25, 39). In the case of thymidine kinase mRNA, one such sequence provides a high-affinity binding site for the hnRNP L protein (39). Introns themselves prevent RNA export, and this is likely a result of retention by the spliceosome (12, 36). In vitro splicing and oocyte microinjection studies showed that for at least some mRNAs produced from intron-containing pre-mRNAs, mRNPs assembled in vitro through splicing are different from those assembled on fully spliced mRNA and are exported from the nucleus more efficiently (41). Several proteins are recruited to mRNPs as a result of splicing, including DEK, SRm160p, RNPS1, Y14, and REF (31, 37, 38, 45, 77). This results at least in part from the deposition of a specific protein complex at or near splice junctions (37, 38). Among these, Y14 persists with the mRNA in the cytoplasm (31).
It is apparent that nuclear processing and export of mRNA are accompanied by multiple rearrangements in the RNP complexes with which pre-mRNA and mRNA are associated. These rearrangements would include formation of an export-competent nuclear mRNP (nmRNP) intermediate that contains mature mRNA with bound shuttling hnRNP proteins, as well as specific proteins (such as REF) recruited through pre-mRNA splicing and from which nonshuttling hnRNP proteins have been removed (57, 60). Such an RNP assembled in vivo has not yet been isolated. Besides splicing, it is likely that other cellular events in mRNA formation also contribute to this remodeling. Shuttling hnRNP proteins associated with mRNA may contribute to mRNA export through their NESs (46, 47). Indeed, microinjection experiments with Xenopus laevis oocytes have implicated hnRNP A1 in mRNA export (28). Further remodeling of this mRNP would then occur following nuclear export, as the nmRNP proteins are exchanged for cytoplasmic mRNP components (16).
In the work presented here, we set out to identify and isolate RNP complexes from human cells containing shuttling hnRNP proteins associated with pre-mRNA and mRNA at different stages of maturation. At least three distinct complexes can be separated, one of which exhibits characteristics of a nuclear mRNP (nmRNP) intermediate. We have identified nmRNP-specific proteins that include specific alternatively spliced isoforms of the hnRNP D and E proteins, as well as a novel RNA-binding protein. This novel nmRNP complex is likely to represent a late nuclear stage of mRNA formation and as such is a candidate substrate for nuclear export of mRNA.
MATERIALS AND METHODS
Cell culture and labeling.
HeLa cells were grown at 37°C in monolayer culture to subconfluent densities in Dulbecco modified Eagle medium containing 10% fetal calf serum and supplemented with penicillin and streptomycin. Where indicated, cells were labeled with [35S]methionine at 20 μCi/ml for 20 h in Dulbecco modified Eagle medium containing 5% fetal calf serum and 1/10 the normal concentration of methionine.
Subcellular fractionation.
All fractionation steps were carried out on ice. Cells grown in monolayer culture were rinsed three times with phosphate-buffered saline and collected with a cell scraper in RSB-100 (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 2.5 mM MgCl2) (0.75 to 1.0 ml/plate) containing digitonin (Calbiochem) at a final concentration of 40 μg/ml. The cells were then incubated on ice for 5 min, and the soluble cytosolic fraction was separated from the nuclear and digitonin-insoluble fractions by centrifugation at 2,000 × g for 8 min. The supernatant fraction was collected, and the remaining pellet was resuspended in RSB-100 containing 0.5% (vol/vol) Triton X-100. Following incubation on ice for 5 min, the Triton-extracted material was separated by centrifugation at 2,000 × g for 8 min. The supernatant was collected, and the remaining pellet was resuspended in the same buffer and disrupted by two 5-s exposures to sonication on ice, using a microtip sonicator (model XL2015; Heat Systems, Farmingdale, N.Y.) set at scale 2.5. The sonicated material was then layered onto a 30% (wt/vol) sucrose cushion in RSB-100 and centrifuged at 4,000 × g for 15 min, and the supernatant was collected.
Immunopurification of RNP complexes.
RNP complexes were immunopurified from the different subcellular fractions with the indicated anti-hnRNP antibodies essentially as previously described (13, 59), except that protease inhibitors were omitted. Bound complexes were eluted from protein A-Sepharose beads with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer or non-equilibrium pH gradient gel electrophoresis (NEPHGE) sample buffer for analysis by SDS-PAGE and two-dimensional gel electrophoresis, respectively.
RNA analysis.
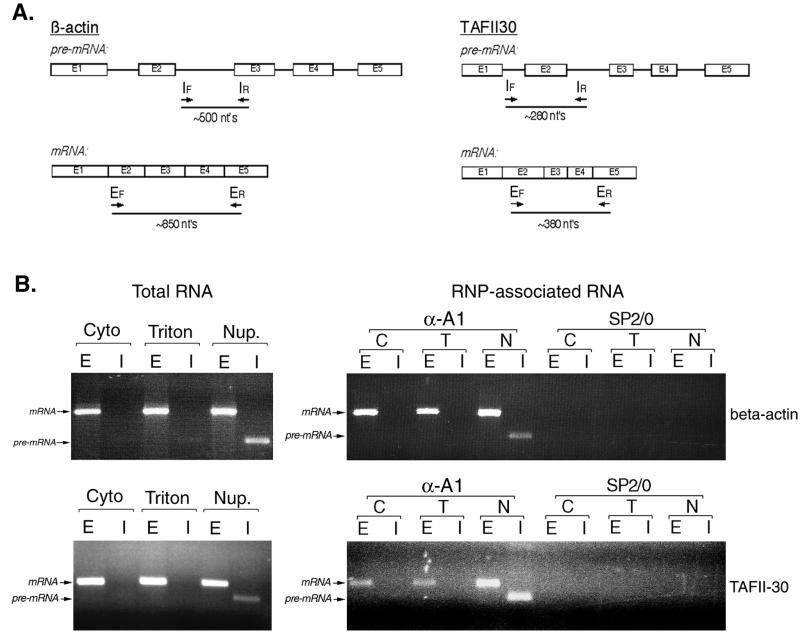
Total RNA was prepared from subcellular fractions using the Trizol reagent (GIBCO-BRL) according to the manufacturer's instructions. For analysis of RNP-associated RNA, immunopurified complexes were heated to 65°C for 5 min in Tris-EDTA containing 1% SDS. RNA was then extracted from the samples by extraction with phenol and precipitated with ethanol according to standard protocols. RNA samples were incubated with 4 U of RQ1 DNase (Promega) for 40 min, and RNA was recovered by phenol extraction and ethanol precipitation. RNP-associated RNAs or ≈1.5 μg of total RNA was used in reverse transcription (RT) reactions (5 mM MgCl2, 0.6 mM deoxynucleoside triphosphates, and 2.5 μM random hexamer primers) with murine leukemia virus reverse transcriptase (Perkin-Elmer) for 15 min at 45°C, followed by 5 min at 95°C. PCRs (with 0.5 pmol of the indicated primers per μl, 1.5 mM MgCl2, and 2.5 U of AmpliTaq polymerase [Perkin-Elmer]) included 30 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C. PCR products were resolved by agarose gel electrophoresis. The positions of the primers used in each reaction are indicated in Fig. 3. Sequences of the β-actin-specific primers were as follows: EF, 5′ GAAAATCTGGCACCACACCT; ER, 5′ GGCCGGACTCGTCATACTC; IF, 5′ CGCTACCTCTTCTGGTGGC; and IR, 5′ ACCATGTCACACTGGGGAAG. Sequences of the TAFII30-specific primers were as follows: EF, 5′ AGGGGGCCATATCTAACGG; ER, 5′ AGTAGTGCGGCTTCTTCACATT; IF, 5′ GGGTGAGGGCAGAGGGTATAG; and IR, 5′ TTTGTCAGCAGGCTAGGTGG.
FIG. 3.
Subcellular distribution and RNP association of specific pre-mRNAs and mRNAs. (A) Diagram depicting the positions in the actin and TAFII30 pre-mRNA and mRNA of the primers used for RT-PCR analysis. Subscripts F and R refer to the forward and reverse primers, respectively, used in the reactions. (B) Left panel, RT-PCR analysis of the distribution of pre-mRNA and mRNA for the transcripts shown in panel A, using total RNA from the various subcellular fractions as a template. Right panel, analysis of hnRNP A1-associated RNA in complexes isolated from the different subcellular fractions. Lanes E, RT-PCR products when all primers correspond to exon sequences. Lanes I, use of at least one intron-specific primer in the RT-PCR. Abbreviations are as in Fig. 1 and 2.
Gel electrophoresis and immunoblot analysis.
SDS-PAGE and immunoblot analyses were carried out essentially as previously described (59). For SDS-PAGE, the separating gel had an acrylamide concentration of 12.5%. Two-dimensional gel electrophoresis was carried out as described by O'Farrell et al. (53). Separation in the first dimension was by NEPHGE, using pH 3 to 10 ampholites (Bio-Rad, Richmond, Calif.). After electrophoresis of [35S]methionine-labeled proteins, the gel was stained with Coomassie blue and impregnated with 2,5-diphenyloxazole for fluorography (35). The following antibodies were used for immunoblot analysis: 4B10 (anti-hnRNP A1) (59), 4F4 (anti-hnRNP C1/C2) (14), 5B9 (anti-hnRNP D) (26), 5H3 (anti-hnRNP E) (26), 12G4 (anti-hnRNP K/J) (44), anti-PABP1 (19), antitransportin (Transduction Laboratories, Lexington, Ky.), and anti-REF (kindly provided by E. Izaurralde, European Molecular Biology Laboratory).
UV light-induced cross-linking of proteins to RNA in living cells and analysis of cross-linked RNP complexes.
Cross-linking of proteins to RNA in vivo by UV light irradiation of cells, followed by selection of cross-linked complexes by oligo(dT) chromatography, was carried out essentially as previously described (16, 58), except that following UV irradiation cells were fractionated as described above. Proteins were released from the cross-linked complexes by digestion with RNase A at 50 μg/ml for 1 h at 30°C, resolved by SDS-PAGE, and visualized by autoradiography.
RESULTS
Differential subcellular fractionation of complexes containing shuttling and nonshuttling hnRNP proteins.
In order to isolate RNP complexes at different stages of maturation in association with the shuttling hnRNP protein A1, a subcellular fractionation approach was devised to allow their separation. To isolate complexes in transit through the cytoplasm, the plasma membrane was first permeabilized with low concentrations of digitonin that leave the nuclear envelope intact (1), and nuclei and associated structures were then removed through centrifugation. The supernatant fraction from this step is expected to contain primarily soluble cytoplasmic material, and we refer to it as cytosol. The second fraction was obtained by extracting the nuclear pellet with 0.5% Triton X-100, which partially solubilizes the nuclear envelope, and again pelleting the extracted nuclei. The resulting supernatant contains soluble nuclear components that are selectively released from the nucleus (4, 20; F. Triolo and S. Piñol-Roma, unpublished data), as well as organellar material that is solubilized by Triton X-100. We refer to this as the Triton-extracted fraction. The remaining nuclear pellet was disrupted by sonication and clarified by centrifugation, yielding a soluble fraction that is operationally defined as nucleoplasm and that was used previously for isolation of hnRNP complexes (13, 56, 59).
Initial immunoblotting experiments revealed small amounts of hnRNP A1 in the cytosol, consistent with its nucleocytoplasmic shuttling. In addition, detectable amounts of A1 were selectively extracted with Triton X-100 from the nuclear fraction, as compared to hnRNP C1/C2 (not shown, but see Fig. 4). To address whether A1 is in RNP complexes in these fractions, we carried out immunopurifications using an antibody against hnRNP A1 (4B10) under conditions that preserve most protein-protein and protein-RNA interactions (13, 59). Immunopurifications from the nucleoplasmic fraction yielded a set of proteins similar to that of previously described hnRNP complexes (Fig. 1A). This was confirmed by two-dimensional gel electrophoresis (see, e.g., Fig. 5A) and by the fact that virtually identical complexes were immunopurified from this fraction with an antibody against hnRNP C1/C2 (Fig. 1A). Notably, immunopurifications from the other two fractions yielded distinct sets of proteins associated with hnRNP A1. In the cytosol, the most prominent proteins migrated at ca. 70 and ca. 90 kDa (Fig. 1A). In the Triton-extracted fraction, A1 was associated with a prominent protein of ca. 130 kDa as well as with other proteins of lower relative abundance, many of which appear to be unique to this complex (Fig. 1A). By contrast, no detectable complexes were recovered from the cytosol or Triton-extracted fractions with an anti-hnRNP C antibody (Fig. 1). The presence of proteins specific to each of the different hnRNPA1-associated complexes shows that these are indeed distinct complexes. This also indicates that the differential fractionation of these complexes reflects differences in their subcellular localization and/or association with subcellular structures, rather than cross-contamination of the fractions or disruption of hnRNP complexes during fractionation.
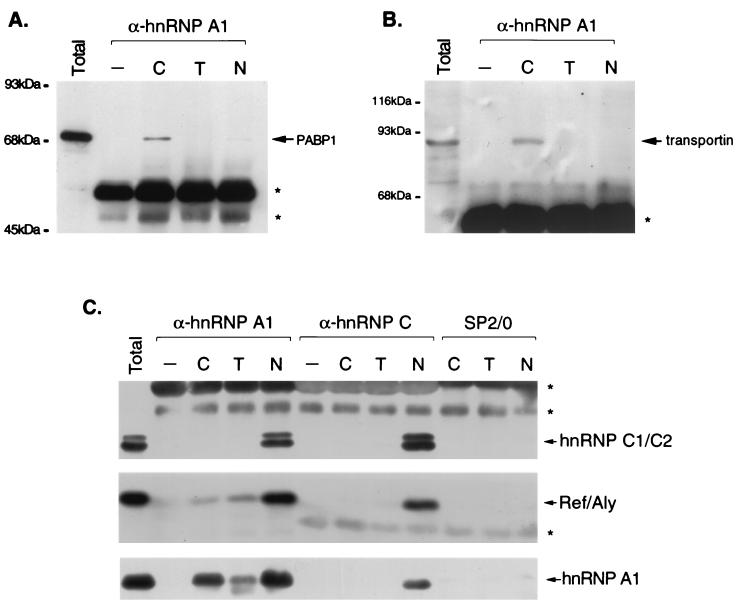
FIG. 4.
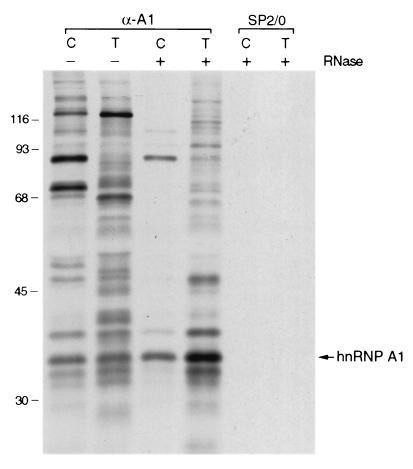
Immunoblot analysis of immunopurified RNPs. HeLa cells were fractionated and RNP complexes were immunopurified from each fraction with monoclonal antibodies to hnRNP A1 (4B10) or hnRNP C1/C2 (4F4) or nonimmune antibodies (SP2/0). The proteins in the complexes were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with monoclonal antibodies to PABP1 (A) or to transportin (B) or with antibodies against hnRNP A1 (4B10), hnRNP C (4F4), and REF (C). Lanes C, T, and N, cytosol, Triton-extracted, and nucleoplasmic fractions, respectively. Asterisks denote the positions of the heavy or light chains from antibodies used for immunopurification. Lanes −, mock immunopurifications in which cellular material was omitted.
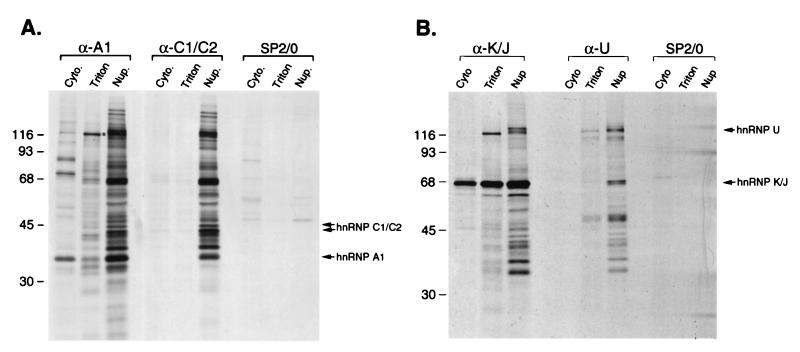
FIG. 1.
Immunopurification of complexes containing hnRNP proteins from different subcellular fractions. (A) HeLa cells were labeled with [35S]methionine and fractionated as described in the text. Immunopurifications were carried out from each fraction with monoclonal antibodies to hnRNP A1 (4B10) or hnRNP C1/C2 (4F4) or nonimmune parent myeloma immunoglobulins (SP2/0). The positions of hnRNP A1 and hnRNP C1/C2 in the gel are indicated on the right. The asterisk indicates the 130-kDa protein discussed in the text. Cyto., cytosol; Triton, Triton-extracted fraction; Nup., nucleoplasmic fraction. (B) Immunopurifications carried out under conditions identical to those shown in panel A, using monoclonal antibodies to hnRNP K/J (12G4) and hnRNP U (3G6). The positions of hnRNP K/J and hnRNP U in the gel are indicated on the right. Positions of molecular mass standards (in kilodaltons) are indicated on the left.
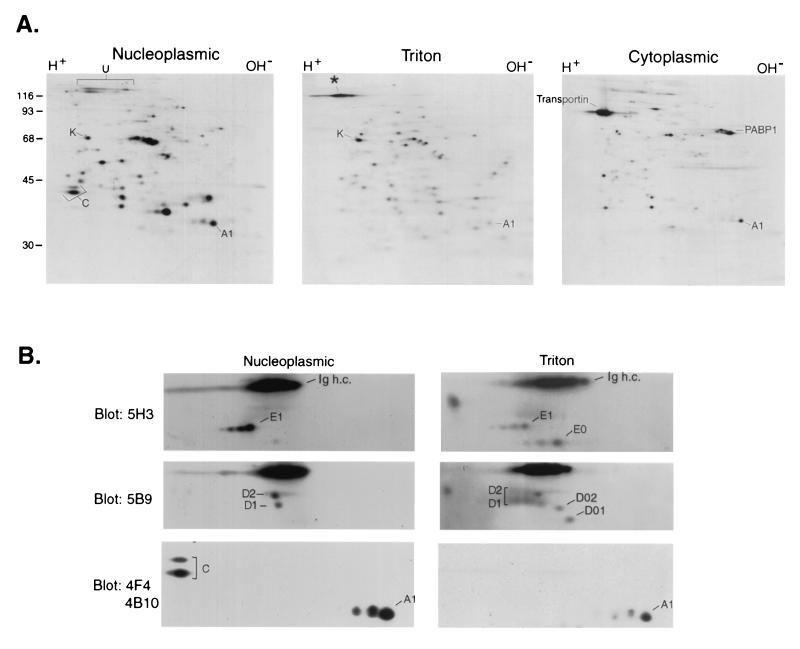
FIG. 5.
Two-dimensional gel electrophoresis of hnRNP A1-associated RNP complexes. (A) Proteins in complexes immunopurified with 4B10 from each subcellular fraction of [35S]methionine-labeled cells were resolved by two-dimensional gel electrophoresis as described in the text and visualized by autoradiography. The positions of proteins discussed in the text are indicated. (B) Immunoblot analysis. Nucleoplasmic and Triton-soluble nuclear RNPs resolved by two-dimensional gel electrophoresis as for panel A were immunoblotted with the indicated monoclonal antibodies. The positions of the hnRNP D, E, A1, and C1/C2 proteins and corresponding alternatively spliced isoforms are indicated. Ig h.c., position of the heavy chains from antibodies used for immunopurification.
The presence of complexes associated with C proteins only in the nucleoplasm, in contrast to the presence of hnRNP A1-associated complexes also in the cytosol and Triton-extracted fractions, suggested that this differential fractionation reflects different properties of RNP complexes associated with shuttling compared to nonshuttling hnRNP proteins. Indeed, immunopurifications with antibodies against two other shuttling proteins, hnRNP K/J (Fig. 1B) and hnRNP A2 (data not shown), yielded similar complexes from the Triton-extracted fraction, in which the 130-kDa protein was again a prominent component (Fig. 1B). By contrast, an antibody against hnRNP U (a nonshuttling protein) immunopurified the hnRNP complex from nucleoplasm, as observed previously (13), but only small detectable amounts of hnRNP U from the other two fractions (Fig. 1B). Release from the nuclear fraction with Triton X-100 therefore appears to be a property of complexes associated with shuttling hnRNP proteins that do not contain nonshuttling hnRNP proteins. The specificity of the immunopurifications was confirmed by the absence of detectable proteins when nonimmune SP2/0 myeloma immunoglobulins were used (Fig. 1). The specificity of the anti-hnRNP antibodies in immunopurification experiments has been reported previously (13, 44, 59). Therefore, the additional proteins observed in the various complexes are coimmunopurified due to interactions (direct or indirect) with the respective hnRNP proteins.
The complexes of hnRNP A1 in different fractions are RNP complexes.
Previous studies showed that copurification of the >20 hnRNP proteins in nucleoplasmic hnRNP complexes requires RNA (13). To address whether RNA is also required for the association of proteins with hnRNP A1 in the cytosol and Triton-extracted fractions, the fractions were digested with RNase prior to immunopurification. As shown in Fig. 2, RNase digestion disrupts the complexes in both fractions. Specifically, association of the 130-kDa protein and of most of the other proteins with A1 in the Triton-extracted fraction is completely disrupted by RNase treatment (Fig. 2). In the cytosol fraction, whereas the 70-kDa protein does not copurify with A1 after RNase treatment, association of the 90-kDa protein is resistant to digestion of the RNA (Fig. 2). These results indicate that, as is the case with nucleoplasmic hnRNPs, the complexes isolated from cytosol and Triton-extracted fractions contain RNA. Furthermore, the associations between most of these proteins and hnRNP A1 are likely to be mediated primarily by their binding to the same RNA molecules.
FIG. 2.
RNase sensitivity of hnRNP A1-associated complexes. Complexes associated with hnRNP A1 were isolated from the soluble cytosolic fraction (lanes C) or Triton-soluble nuclear fraction (lanes T), as shown in Fig. 1, without (lanes −) or with (lanes +) prior digestion of the fractions with RNase A. The position of hnRNP A1 in the gel is indicated on the right. Identical immunopurifications were carried out with nonimmune parent myeloma immunoglobulins (SP2/0). Positions of molecular mass standards (in kilodaltons) are indicated on the left.
The hnRNP A1-associated complexes contain RNAs at different stages of maturation.
Previous work showed that immunopurified hnRNP complexes contain a heterogeneous population of rapidly labeled, RNA polymerase II-transcribed RNAs (13). Furthermore, hnRNP A1 is bound transiently to poly(A)+ RNA in the cytoplasm and probably also during its nucleocytoplasmic transport (60). In agreement with this, RNA polymerase II-transcribed RNA is coimmunopurified with hnRNP A1 in all three fractions and can be detected as a heterogeneous population of RNAs after labeling with [3H]uridine (data not shown).
The fractionation properties of the RNPs in the different fractions could be explained in at least two ways. First, each fraction could contain different subsets of transcripts with different extraction properties. Alternatively, the different fractionation of the RNPs may reflect sequential stages in maturation of their associated pre-mRNAs and mRNAs. In the absence of a unifying feature of pre-mRNAs that would allow us to distinguish them experimentally as a family from mRNAs, we addressed these possibilities by focusing our attention on specific transcripts for analysis by RT-PCR. Two different constitutively expressed RNAs were examined, corresponding to β-actin (49) and TAFII-30 (66). These RNAs were selected because they encode proteins with very different properties: a cytoplasmic abundant cytoskeletal protein (β-actin) and a nuclear protein of relatively low abundance involved in transcription (TAFII-30). Furthermore, both RNAs encode intracellular proteins, and therefore their translation should not be carried out in association with detergent-soluble structures, such as the endoplasmic reticulum. This property would allow us to distinguish between selectively extracted nuclear RNAs and those that are preferentially associated with the endoplasmic reticulum. Primers specific for exon or intron sequences were designed so that pre-mRNA and spliced mRNA for each transcript could be distinguished based on the size of the resulting PCR product (Fig. 3A). RT-PCR analysis of total RNA from each fraction showed that spliced mRNA for both transcripts is readily detected in all three fractions, and therefore it is unlikely that each fraction merely contains different subsets of transcripts (Fig. 3B). More importantly, intron-containing precursors for both RNAs were readily detected in the nucleoplasmic fraction, whereas little (if any) pre-mRNA was detected in the cytosol and Triton-extracted fractions. This is consistent with the second scenario raised above, namely, that the different fractions contain RNAs at different stages of maturation. The amplified PCR products do not result from DNA contamination of the RNA preparations, since identical reactions where reverse transcriptase was omitted did not yield any detectable amplified products (data not shown).
In order to determine whether RNA associated with hnRNP A1 in the different fractions showed a similar distribution of pre-mRNA versus mRNA, RNP complexes were immunopurified with 4B10, and RNAs extracted from the complexes were used as templates in identical RT-PCRs. Both pre-mRNA and mRNA were found associated with hnRNP A1 in the nucleoplasmic fraction, whereas only mature spliced mRNA was associated with hnRNP A1 in the Triton-extracted and cytosol fractions (Fig. 3B). Similar results were observed for both transcripts. The RNAs were specifically immunopurified because of their association with hnRNP A1, since no RNA was detected in immunopurifications with nonimmune SP2/0 myeloma immunoglobulins (Fig. 3B). The fact that pre-mRNA associated with hnRNP A1 was found only in the nucleoplasm indicates that hnRNP complexes represent an earlier stage of mRNA maturation than RNP complexes isolated from the Triton-extracted and cytosol fractions.
Identification of common as well as specific proteins associated with hnRNP A1 in different RNP complexes.
The apparent molecular mass, isoelectric point (see Fig. 5A), and RNase-resistant association in the cytosol with hnRNP A1 of the 90-kDa protein suggested that it might correspond to transportin, which associates with A1 in the cytoplasm and mediates its nuclear import (62). In agreement with this, immunoblot analysis (Fig. 4B) revealed that transportin is indeed associated with hnRNP A1 preferentially in the cytosolic complexes. This is consistent with the observation that coimmunopurification of the major 90-kDa band with hnRNP A1 persists after RNase digestion (Fig. 2), indicating that it associates with hnRNP A1 by direct protein-protein interactions. The presence of transportin in hnRNP A1-associated complexes in the cytoplasmic fraction is consistent with the hypothesis that these complexes represent a bona fide soluble cytoplasmic pool. This is further supported by the finding that the cytoplasmic poly(A)-binding protein (PABP1) is also specifically enriched in the cytosolic complexes (Fig. 4A) and likely corresponds to the other major band, of ca. 70 kDa, that also associates with A1 in this fraction, as suggested by its mobility on two-dimensional gels (Fig. 5A). These characteristics suggest that the hnRNP A1-associated complexes from the cytoplasmic fraction correspond to soluble cytoplasmic intermediates following nuclear export of hnRNP A1 with its bound mRNA.
The fractionation properties of the other two hnRNP A1-associated RNPs and the characteristics of their associated RNAs raised the likely possibility that these complexes represent distinct nuclear stages in the nucleocytoplasmic shuttling cycle of hnRNP A1 with its bound mRNAs. At least one protein directly implicated in mRNA export, REF, (70, 71, 77), is associated with all three RNPs (Fig. 4). Notably, however, while the amounts of REF detected in the Triton-extracted RNPs are smaller than those in hnRNPs, the amount of hnRNP A1 (and its associated complexes) recovered from the Triton-extracted fraction is also always smaller than that recovered from the nucleoplasm (Fig. 4C). We therefore conclude that REF is associated with hnRNP A1 in at least similar relative amounts in the complexes isolated from the nucleoplasm and from the Triton-extracted fraction. Consistent with its ability to shuttle between the nucleus and the cytoplasm (77), a relatively smaller amount of REF is also present in the cytosolic A1-associated RNPs.
A comparison of the pattern of proteins in the nucleoplasmic and Triton-extracted complexes resolved by two-dimensional gel electrophoresis revealed that in addition to REF, other shuttling hnRNP proteins (e.g., hnRNP K and hnRNP A2/B1/B2) are also present in both complexes (Fig. 5A). On the other hand, apart from the different association of the nonshuttling hnRNP C1/C2 proteins, which, as observed previously (Fig. 4C), were not detected by immunoblot analysis in the Triton-extracted complexes, the nonshuttling hnRNP U protein (60) was also not present in any detectable amounts in the Triton-extracted complexes (Fig. 5A). Therefore, a unique characteristic of these mRNA-containing complexes from the Triton fraction that distinguishes them from the pre-mRNA- and mRNA-containing hnRNP complex is the absence of nonshuttling hnRNP proteins.
A second unique feature of the Triton-extracted complexes was the presence of proteins that are not found in hnRNP complexes. Among them, the most prominent migrates at ca. 130 kDa (Fig. 5A). Differences in protein composition were also apparent in the region of the gel between 35 and 60 kDa (Fig. 5A, compare panels Nucleoplasmic and Triton). Some of these proteins were identified using antibodies against known hnRNP proteins. An antibody against hnRNP D (5B9) reacts with four alternatively spliced isoforms of this protein (D1, D2, D01, and D02) (26). Immunoblotting experiments revealed that whereas only D1 and D2 are present in the nucleoplasmic hnRNP complex, all four isoforms are associated with A1 in the Triton-extracted complexes (Fig. 5B). Similarly, an anti-hnRNP E antibody (5H3) recognizes one of the hnRNP E proteins, E1, as well as an immunologically related protein, E0 (26). Only the E1 protein is detected in the hnRNP complex, whereas both E1 and E0 are present in the Triton complex (Fig. 5B). Therefore, the mRNA-containing RNP complexes from the Triton-extracted fraction contain a subset of hnRNP proteins (namely, shuttling hnRNP proteins), as well as additional proteins that include alternatively spliced isoforms of hnRNP proteins.
Identification of the 130-kDa protein as LRP130, a novel RNA-binding protein.
Since the 130-kDa protein was the most prominent one associated with shuttling hnRNP proteins in the Triton-extracted fraction and because its association appeared to be specific to these mRNA-containing complexes, it was chosen for further characterization. For this purpose, immunopurification from the Triton-extracted fraction with the 4B10 antibody was scaled up to enable analysis of the 130-kDa protein by mass spectrometry (MS). Tandem MS (MS-MS) (5) was used to derive sequence information (64) from tryptic peptides of the 130-kDa protein. The resulting peptide masses and corresponding daughter fragments were used to identify proteins by searching the NCBI nonredundant protein sequence database and EST sequence database with the program PepFrag (18). This analysis yielded unambiguous identification of a peptide sequence, ADAVWNKIQEENVIPR, unique to a previously reported 130-kDa leucine-rich protein (LRP130) (24). The same identification was obtained by MS-MS analysis of a different tryptic fragment (not shown). Matrix-assisted laser desorption ionization–time of flight MS analysis of the 130-kDa band confirmed its identity with LRP130. The function of this protein was previously unknown, although it was found to be overexpressed in hepatoblastoma cells (24). Extensive computer searches of databases (2) showed no regions in LRP130 with significant or obvious similarity to any known RNA-binding domains (8). The only recognizable amino acid sequence motif in LRP130 is the recently described PPR motif, a TPR-related motif that has been suggested to mediate protein-protein or protein-RNA interactions (68). LRP130 contains 11 such PPR motifs, which are clustered in the N terminus and in the middle region of the protein (68).
Association of LRP130 with A1 is readily disrupted by RNase treatment (Fig. 2), indicating that the interaction between these two proteins is mediated by their association with the same RNA molecules. The absence of known RNA-binding motifs in LRP130 raised the question of whether it is bound directly to RNA or whether this association is indirect (for example, through interaction with an RNA-bound protein). To address this directly, RNA-protein cross-links were induced in vivo by exposure of living cells to UV irradiation. Under the conditions used here, only proteins that are in direct contact with the RNA in vivo can be covalently cross-linked to it. RNA with its cross-linked proteins can then be isolated under protein-denaturing conditions in order to eliminate adventitious association of proteins with RNA during fractionation (see, e.g., references 16 and 72). HeLa cells were exposed to UV light, and polyadenylated RNA with its cross-linked proteins was selected from cytosol, Triton-extracted, and nucleoplasmic fractions by oligo(dT) chromatography under denaturing conditions that allow coisolation of only those proteins covalently cross-linked to the RNA. Proteins in the cross-linked complexes were then released by digestion with RNase and resolved by SDS-PAGE (Fig. 6). As previously observed (see, e.g., references 16 and 72), even at this level of resolution one can observe differences in the sets of proteins cross-linked to RNA in nucleoplasmic and cytoplasmic fractions. Importantly, the most prominent cross-linked protein in the Triton-extracted fraction migrates at ca. 130 kDa (lane T). The specificity of this cross-linking is underscored by the absence of detectable proteins in samples prepared from cells that were not exposed to UV light (Fig. 6, lane −UV). Matrix-assisted laser desorption ionization–time of flight MS and MS-MS analyses of the 130-kDa cross-linked band in the Triton-extracted fraction identified it also as LRP130 (not shown), which demonstrates that LRP130 is indeed bound to poly(A)-containing mRNA in living cells.
FIG. 6.
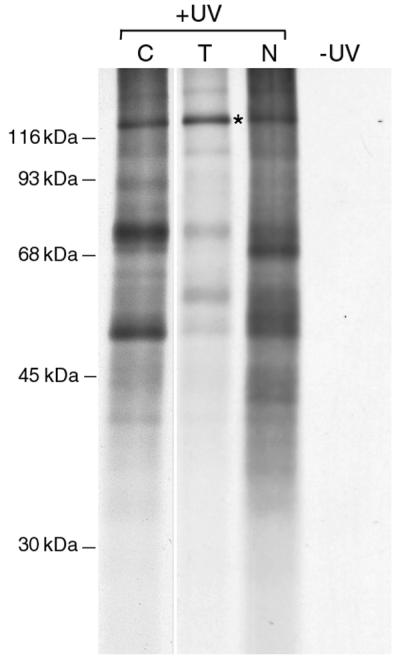
Selection of proteins bound to poly(A)+ RNA in vivo in the different subcellular fractions. HeLa cells were exposed to UV light to induce covalent protein-RNA cross-links, and the cells were fractionated into cytoplasmic (lane C), Triton-extracted (lane T), and nucleoplasmic (lane N) fractions. The cross-linked complexes were selected from each fraction by oligo(dT) chromatography under protein-denaturing conditions. Bound proteins were released from the cross-linked complexes by digestion with RNase, resolved by SDS-PAGE, and visualized by autoradiography. The position of LRP130 is indicated with an asterisk. A shorter exposure of the lane corresponding to cytoplasmic cross-linked proteins is shown here for clarity.
DISCUSSION
We have dissected the nucleocytoplasmic shuttling pathway of hnRNP A1 into at least three distinct classes of RNPs with characteristics of sequential stages in mRNA formation. Among these, we identified a likely intermediate nuclear mRNP (nmRNP) complex (or set of complexes) that contains mRNA with associated shuttling hnRNP proteins but no pre-mRNA or nonshuttling hnRNP proteins. The strategy for the isolation of these complexes is based on the different nucleocytoplasmic traffic characteristics of hnRNP proteins and exploits differences in subcellular associations of RNP complexes at different stages of maturation in combination with specific immunopurification of RNPs. The immunopurification approach taken here has been used successfully in the past to isolate hnRNP complexes under conditions that minimize disruption and rearrangements of the complex (13, 59). Therefore, the complexes described in this study are likely to represent endogenous RNPs that were assembled in vivo.
The three classes of RNP complexes described here can be distinguished from each other by their RNA and protein compositions and by their association with different subcellular fractions. One class corresponds to the previously described nucleoplasmic hnRNP complexes (13, 59), which contain shuttling and nonshuttling hnRNP proteins and which we show here contain both pre-mRNA and mRNA. This is distinct from a second class of RNPs, also associated with nuclei, which display characteristics of mature nuclear mRNPs (nmRNPs). Specifically, they contain mRNA but no detectable pre-mRNA. They also contain shuttling hnRNP proteins as well as the nuclear mRNA export factor REF, as is the case with hnRNP complexes, but no nucleus-retained hnRNPs such as C1/C2 and U. In addition, there are several proteins specifically associated with nmRNPs that are not found in hnRNP complexes. RNP complexes associated with hnRNP A1 are also found in the cytosol, where A1 is associated with mRNA as well as with the major cytoplasmic mRNP protein PABP1 (19) and with transportin, the nuclear transport receptor for hnRNP A1 (62). Importantly, all three types of complexes contain proteins in common, as well specific proteins that are not found in the other complexes.
The existence of distinct RNP complexes of hnRNP proteins was suggested by a number of previous studies, which indicated that nuclear formation of mRNAs from pre-mRNAs, as well as nucleocytoplasmic transport of mRNAs, is accompanied by substantial changes in the proteins associated with these RNAs (see the introduction). In addition, the major proteins bound to mRNA in the cytoplasm at steady-state levels are different from those associated with pre-mRNA in the nucleus (16, 72), indicating a wholesale exchange of mRNP for hnRNP proteins as the mRNA is exported from the nucleus to the cytoplasm. Taking these previous observations together with our findings, the distinct RNP complexes that we have isolated can best be fit into a temporal sequence of events in which the hnRNP complexes represent the initial pre-mRNPs as well as early postsplicing mRNPs in which RNA polymerase II-transcribed transcripts are found (17). In agreement with this, pre-mRNA and mRNA for both β-actin and TAFII30 are detected in these complexes (Fig. 3). In addition, the mRNA export factor REF is also present in hnRNP complexes. The presence of spliced mRNA in hnRNP complexes indicates that many of the subsequent changes in RNP protein composition represented by the additional mRNPs described here occur after processing of pre-mRNA to mRNA.
In contrast to hnRNPs, the fractionation properties of the cytosolic complexes and their association with transportin and PABP1 (a primarily cytosolic mRNP protein [19]) indicate that they represent the last stage(s) in the nucleocytoplasmic shuttling of hnRNP A1 during its transit in the cytoplasm. These results also indicate that PABP1 can bind mRNA prior to complete release of hnRNP A1 from the same mRNA. By contrast, no significant amounts of PABP1 copurify with hnRNP K in the cytosolic fraction (Fig. 1B). This suggests that hnRNP K dissociates from mRNA prior to binding of PABP1 and therefore prior to release of hnRNP A1. Indeed, electron microscopy studies of BR mRNP export in C. tentans have also shown different proteins dissociating from the mRNP in the cytoplasm at different stages following its nuclear export (for a review, see reference 15). We cannot determine from these results whether both transportin and PABP1 coexist simultaneously in the same complexes with hnRNP A1 or whether they interact with hnRNP A1 in distinct complexes. The specificity of the interaction of hnRNP A1 with transportin is underscored by the absence of transportin in association with hnRNP K/J (Fig. 1B), in agreement with previous findings that hnRNP K does not require transportin for its nuclear import (47). It is noteworthy that the number of proteins that can be cross-linked to mRNA in the cytoplasm is substantially larger than the number of proteins associated with hnRNP A1 (16) (Fig. 6). This would be consistent with a transient nature of the cytosolic hnRNP A1-containing mRNPs (supported by immunofluorescence microscopy data [60]), indicating that additional proteins associate with cytoplasmic mRNA once hnRNP A1 is released from the complex (and therefore such proteins would not be coimmunoprecipitated with hnRNP A1).
The third class of RNP complexes, which are associated with hnRNP A1 in the Triton-extracted fraction, is of particular interest because their properties are consistent with those hypothesized for nuclear mRNPs at late stages of mRNA formation and possibly as substrates for nuclear export of mRNA. This conclusion is supported both by their protein and RNA compositions, as described above, and by their subcellular fractionation properties. Specifically, these nmRNPs are associated with the nuclear fraction and are not solubilized with digitonin under conditions that retain the integrity of the nuclear envelope (1). Therefore, they are unlikely to represent soluble cytoplasmic complexes. On the other hand, they are readily released by Triton X-100 treatment of the digitonin-insoluble fraction, which contains nuclei as well as insoluble cytoplasmic structures. Treatment of nuclei with nonionic detergents is known to result in selective release of some nuclear contents, including RNA export factors, and we have observed this to also be the case for the nucleoplasmic pool of another abundant nuclear protein, nucleolin, with its associated rRNA (4, 20; Triolo and Piñol-Roma, unpublished data). It is unlikely that the complexes released by treatment with Triton X-100 originate from an otherwise insoluble cytoplasmic pool of mRNPs associated with the cytoskeleton. It has been reported that translated mRNA associates with the cytoskeleton, and there are conflicting reports as to the sensitivity of cytoskeleton-associated mRNA to treatment with detergents (6, 11, 54). While we have not completely ruled out a cytoplasmic origin of these complexes, we consider this unlikely since they are not released into the soluble cytosolic fraction by treatment of cells with a variety of cytoskeleton-disrupting conditions (S. Mili and S. Piñol-Roma, unpublished observations). The properties of these RNPs, therefore, indicate that they are precursors to the cytosolic mRNPs described here and that they correspond to a later stage in mRNA formation than (and are a product of) the pre-mRNA-containing hnRNP complexes. This suggests strongly that these nmRNP complexes are a novel intermediate in the pathway of mRNA formation.
An important finding presented here is that several of the proteins in the nmRNPs are specific to these complexes and are not found in hnRNP complexes. The most prominent among them is LRP130, to which no specific function had been attributed (24) and which our results indicate binds specifically to mRNA. Importantly, a protein with electrophoretic mobility similar to that of LRP130 is associated with all shuttling hnRNP proteins that we have examined thus far, including hnRNP A1 and hnRNP K/J (this work) and hnRNP A2/B1/B2 (Mili and Piñol-Roma, unpublished observations). Analysis of the LRP130 amino acid sequence revealed no readily apparent RNA-binding motifs. However, we show here that it binds poly(A)+ RNA in vivo, as it is readily cross-linked to mRNA by UV irradiation of living cells (Fig. 6). The only recognizable amino acid sequence motif in LRP130 is the recently described PPR motif (68). Other proteins with PPR motifs have also been shown to bind RNA and/or participate in mRNA metabolism, raising the possibility that the PPR motif itself is an RNA-binding motif (68). Therefore, LRP130 is an RNA-binding protein with none of the known RNA-binding motifs, suggesting that it contains a novel type of RNA-binding domain. A recent study with Drosophila melanogaster has shown that BSF, a protein highly similar to LRP130, has RNA-binding activity and is involved in regulating the stability of bicoid mRNA (42).
Other proteins associated with this intermediate and not present in nucleoplasmic hnRNP complexes are the D0 and E0 proteins. There are four alternatively spliced isoforms of hnRNP D: D2 (p45), D1 (p42), D02 (p40), and D01 (p37) (74). Of these, D2 and D1 isoforms are preferentially associated with hnRNP complexes (Fig. 5B). D1 and D2 contain a specific amino acid sequence that is encoded by an alternatively spliced exon that is absent from the D01 and D02 isoforms. By contrast to D1 and D2, D01 and D02 are specifically associated with nmRNPs. While all D protein isoforms are predominantly nuclear at steady-state levels, they differ in their ability to shuttle between the nucleus and the cytoplasm. Interestingly, the amino acid sequence specific to the D1/D2 isoforms has been proposed to be responsible for retaining them in the nucleus by mediating their interaction with the nuclear matrix-associated factor SAF-B. D01 and D02, which lack this sequence, do not associate with SAF-B and are able to shuttle (3). These properties are consistent with our observation that D1- and D2-containing hnRNP complexes are resistant to extraction with nonionic detergent, whereas D01- and D02-containing nmRNP complexes seem to be relatively freely diffusible in the nucleoplasm, since they are readily extracted by mild detergent treatment. This distribution of hnRNP D protein isoforms is reminiscent of the Drosophila RNA-binding protein How, which exists in two isoforms with different subcellular distributions. It was proposed that the shorter isoform can compete directly with the longer nuclear isoform for binding to target RNAs, thereby releasing inhibition of nuclear export in a developmentally regulated manner (48). We speculate that a similar mechanism could operate in the case of the hnRNP D proteins, with the D01 and D02 isoforms possibly displacing the nucleus-retained D1 and D2 isoforms at a specific stage prior to nuclear RNA export. The specific relationship between E1 and E0 is not known, but their immunological relatedness, together with precedent from other hnRNP proteins (9), suggests that they may also be produced by alternative splicing from the same pre-mRNAs.
It is noteworthy that recruitment of a number of RNA-binding proteins onto mRNA in the nucleus has been shown or hypothesized to be required for subsequent function of these proteins in the cytoplasm, e.g., in RNA localization, stability, and regulation of translation (see, e.g., references 7, 34, and 40). For example, the D. melanogaster hrp40 protein, which is similar to hnRNP D, mediates cytoplasmic localization of a number of pair-rule transcripts, and this function requires recruitment of Hrp40 to the mRNA in the nucleus (34). Furthermore, contributions of hrp40 to regulation of Gurken localization during oogenesis vary among specific hrp40 isoforms (52). In vertebrate cells, recruitment of specific hnRNP D protein isoforms (also known as AUF1) to mRNA in the nucleus has been proposed to determine the subsequent function of these proteins in cytoplasmic regulation of mRNA stability (40). These observations, therefore, are in agreement with our finding that specific isoforms of hnRNP proteins initiate their association with the mRNA in nuclear mRNP complexes. RNP complexes containing β-actin and TAFII30 pre-mRNA and mRNA exhibit similar fractionation properties, suggesting that the overall characteristics of the observed complexes are a general feature of most transcripts in the cell. However, it is likely that the actual relative amounts of different proteins in the RNPs vary among specific transcripts (43, 61, 76), depending on the sequence characteristics and ultimate fate of the mRNA.
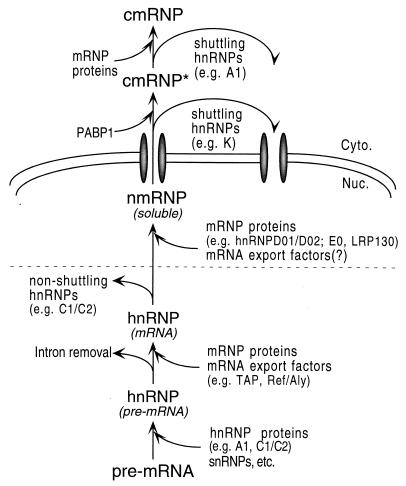
Based on these findings, we propose the model shown in Fig. 7 for remodeling of pre-mRNP and mRNP complexes during nuclear maturation and export of mRNA. As previously shown, most hnRNP proteins associate with nascent transcripts produced by RNA polymerase II, in pre-mRNA-containing hnRNP complexes. In addition to hnRNP proteins, other components of the pre-mRNA processing machinery (including snRNPs) associate transiently with hnRNP complexes as the reactions leading to formation of mature mRNA take place. For transcripts derived from intron-containing genes, the RNAs are retained in the nucleus as long as spliceosomes can be formed on these RNAs (12, 36). In addition, RNA-bound proteins with NRSs such as hnRNP C1/C2 and D1/D2 may also mediate nuclear retention of the bound RNAs (50). Completion of splicing leads to recruitment of a subset of mRNP-specific proteins, such as Y14, DEK, SRm160, and the mRNA export factor REF. Subsequent removal of NRS-containing proteins would release this mRNP from a “nuclear anchor,” possibly rendering the mRNP freely diffusible in the nucleoplasm. The mechanism for removal of NRS-containing proteins is not known. By analogy to the D. melanogaster RNA-binding protein How (48), this dissociation could result from displacement by alternatively spliced isoforms (as could be the case for hnRNP D and E) or by a different mechanism (e.g., through the action of helicases [29, 67]). This complex would also acquire additional mRNA-specific proteins such as LRP130 and hnRNP D01/D02, leading to formation of an nmRNP intermediate that is subsequently exported to the cytoplasm. Following export from the nucleus, shuttling hnRNP proteins remain associated in a transient cytosolic mRNP (cmRNP*). An exchange of cytoplasmic mRNP proteins for nmRNP proteins takes place, ultimately resulting in a distinct cytoplasmic mRNP (cmRNP) that serves as a substrate for cytoplasmic mRNA metabolism. Shuttling hnRNP proteins dissociate from the mRNA at different stages and are reimported into the nucleus. Therefore, RNP remodeling during mRNA maturation and export would involve stage-specific release of bound proteins, as well as stage-specific recruitment of additional RNA-binding proteins. Any of the stages depicted in this model are potential targets for regulation.
FIG. 7.
Model for the sequential changes in protein composition of complexes associated with pre-mRNA and mRNA during maturation and nuclear export of mRNA. See text for details.
At least two of the proteins in this nmRNP, namely, hnRNP A1 and hnRNP K, contain sequences that can mediate nuclear protein export (46, 47), and thus multiple NESs are found on individual mRNAs. Our results show that at least one of the mRNA export factors identified thus far in vertebrate cells, REF (71, 77), is also associated with the nmRNP. We have been unable to determine unambiguously whether TAP is also associated with the nmRNP. It is possible that the interaction of TAP with the nmRNP is too unstable or weak to survive the RNP isolation procedure, as suggested also by results from other laboratories (see, e.g., reference 77). Experiments are now in progress to determine which (if any) additional known RNA export factors and/or NPC components, as well as other proteins recruited to mRNA through splicing (see the introduction), are present in or interact with this nmRNP. Additional nmRNP components may include members of the SR family of splicing factors, some of which also shuttle between the nucleus and the cytoplasm (10). Experiments are also in progress to determine whether mRNAs derived from intronless genes also associate in similar complexes and with similar proteins.
ACKNOWLEDGMENTS
We thank Audrey Marcu and Rosalie Perez for excellent technical assistance, Soojin Kim for assistance with the initial RT-PCR experiments, and Fabio Triolo for sharing his initial observations on selective solubility of mature nuclear RNPs and for helpful discussions and suggestions throughout the course of this work. We also thank Elisa Izaurralde, Maria Carmo-Fonseca, and Angelo Calado for antibodies and Kathy Borden, Avrom Caplan, Jeanne Hirsch, Fabio Triolo, and Paul Wassarman for critical reading of the manuscript.
This work was supported by a grant (GM-53468) from the NIH to S.P.-R.
REFERENCES
- 1.Adam S A, Sterne-Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arao Y, Kuriyama R, Kayama F, Kato S. A nuclear matrix-associated factor, SAF-B, interacts with specific isoforms of AUF/hnRNP D. Arch Biochem Biophys. 2000;380:228–236. doi: 10.1006/abbi.2000.1938. [DOI] [PubMed] [Google Scholar]
- 4.Bachi A, Braun I C, Rodrigues J P, Panté N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Görlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biemann K. Mass spectrometry of peptides and proteins. Annu Rev Biochem. 1992;61:977–1010. doi: 10.1146/annurev.bi.61.070192.004553. [DOI] [PubMed] [Google Scholar]
- 6.Bird R C, Sells B H. Cytoskeleton involvement in the distribution of mRNP complexes and small cytoplasmic RNAs. Biochim Biophys Acta. 1986;868:215–225. doi: 10.1016/0167-4781(86)90057-6. [DOI] [PubMed] [Google Scholar]
- 7.Bouvet P, Wolffe A P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 8.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 9.Burd C G, Swanson M S, Görlach M, Dreyfuss G. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA-binding proteins is generated by small peptide inserts. Proc Natl Acad Sci USA. 1989;86:9788–9792. doi: 10.1073/pnas.86.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervera M, Dreyfuss G, Penman S. Messenger RNA is translated when associated with the cytoskeletal framework in normal and VSV-infected HeLa cells. Cell. 1981;23:113–120. doi: 10.1016/0092-8674(81)90276-2. [DOI] [PubMed] [Google Scholar]
- 12.Chang D D, Sharp P A. Regulation of HIV depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 13.Choi Y D, Dreyfuss G. Isolation of the heterogeneous nuclear ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci USA. 1984;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi Y D, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 16.Dreyfuss G, Choi Y D, Adam S A. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984;4:1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. HnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 18.Fenyo D, Zhang W, Chait B T, Beavis R C. Internet-based analytical chemistry resource: a model project. Anal Chem. 1996;68:721A–726A. [Google Scholar]
- 19.Görlach M, Burd C G, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 20.Gotzmann J, Eger A, Meissner M, Grimm R, Gerner C, Sauermann G, Foisner R. Two-dimensional electrophoresis reveals a nuclear matrix-associated nucleolin complex of basic isoelectric point. Electrophoresis. 1997;18:2645–2653. doi: 10.1002/elps.1150181421. [DOI] [PubMed] [Google Scholar]
- 21.Gruss P, Lai C J, Dhar R, Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci USA. 1979;76:4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 23.Hamer D H, Leder P. Splicing and the formation of stable RNA. Cell. 1979;18:1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- 24.Hou J, Wang F, McKeehan W L. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HEPG2) In Vitro Cell Dev Biol. 1994;30A:111–114. doi: 10.1007/BF02631402. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Carmichael G G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 28.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jankowsky E, Gross C H, Shuman S, Pyle A M. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 30.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka N, Yong J, Kim V N, Velazquez F, Perkinson R A, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 32.Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krecic A M, Swanson M S. hnRNP complexes: composition, structure and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 34.Lall S, Francis-Land H, Flament A, Norvell A, Schüpbach T, Ish-Horowicz D. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell. 1999;98:171–180. doi: 10.1016/s0092-8674(00)81012-0. [DOI] [PubMed] [Google Scholar]
- 35.Laskey R A, Mills A D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 36.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 37.Le Hir H, Izaurralde E, Maquat L E, Moore M J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 40.Loflin P, Chen C Y, Shyu A-B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;15:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo M-J, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mancebo R, Zhou X, Shillinglaw W, Henzel W, Macdonald P M. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol Cell Biol. 2001;21:3462–3471. doi: 10.1128/MCB.21.10.3462-3471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matunis E L, Matunis M J, Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matunis M J, Michael W M, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992;12:164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson J A, Awrey D, Greenblatt J, Grosveld G, Blencowe B J. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 47.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nabel-Rosen H, Dorevitch N, Reuveny A, Volk T. The balance between two isoforms of the Drosophila RNA-binding protein How controls tendon differentiation. Mol Cell. 1999;4:573–584. doi: 10.1016/s1097-2765(00)80208-7. [DOI] [PubMed] [Google Scholar]
- 49.Nakajima-Ijima S, Hamada H, Reddy P, Kakunaga T. Molecular structure of the human cytoplasmic beta-actin gene: interspecies homology of sequences in the introns. Proc Natl Acad Sci USA. 1985;82:6133–6137. doi: 10.1073/pnas.82.18.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 52.Norvell A, Kelley R L, Wehr K, Schüpbach T. Specific isoforms of Squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 1999;13:864–876. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Farrell P Z, Goodman H M, O'Farrell P H. High resolution two-dimensional gel electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 54.Ornelles D A, Fey E G, Penman S. Cytochalasin releases mRNA from the cytoskeletal framework and inhibits protein synthesis. Mol Cell Biol. 1986;6:1650–1662. doi: 10.1128/mcb.6.5.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pederson T. Proteins associated with heterogeneous nuclear RNA in eukaryotic cells. J Mol Biol. 1974;83:163–183. doi: 10.1016/0022-2836(74)90386-6. [DOI] [PubMed] [Google Scholar]
- 57.Piñol-Roma S. HnRNP proteins and the nuclear export of mRNA. Semin Cell Dev Biol. 1997;8:57–63. doi: 10.1006/scdb.1996.0122. [DOI] [PubMed] [Google Scholar]
- 58.Piñol-Roma S, Adam S A, Choi Y D, Dreyfuss G. Ultraviolet-induced crosslinking of RNA to proteins in vivo. Methods Enzymol. 1989;28:410–418. doi: 10.1016/0076-6879(89)80114-4. [DOI] [PubMed] [Google Scholar]
- 59.Piñol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals and assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 60.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 61.Piñol-Roma S, Swanson M S, Gall J G, Dreyfuss G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J Cell Biol. 1989;109:2575–2587. doi: 10.1083/jcb.109.6.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 63.Pritchard C E J, Fornerod M, Kasper L H, van Deursen J M A. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–253. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qin J, Herring C J, Zhang X. De novo peptide sequencing in an ion trap mass spectrometer with 18O labeling. Rapid Commun Mass Spectrom. 1998;12:209–216. doi: 10.1002/(SICI)1097-0231(19980314)12:5<209::AID-RCM141>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues J P, Rode M, Gatfield D, Blencowe B J, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheer E, Mattei M-G, Jacq X, Chambon P, Tora L. Organization and chromosomal localization of the gene (TAF2H) encoding the human TBP-associated factor II 30 (TAFII30) Genomics. 1995;29:269–272. doi: 10.1006/geno.1995.1243. [DOI] [PubMed] [Google Scholar]
- 67.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues J P, Boscheron C, Rigaut G, Wilm M, Séraphin B, Carmo-Fonseca M, Izaurralde E. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Small I D, Peters N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 69.Stevens B J, Swift H. RNA transport from nucleus to cytoplasm in Chironomus salivary glands. J Cell Biol. 1966;31:55–77. doi: 10.1083/jcb.31.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stutz F, Bachi A, Doerks T, Braun I C, Séraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.vanEekelen C A, Riemen T, vanVenrooij W. Specificity in the interaction of hnRNP and mRNA with proteins as revealed by in vivo cross linking. FEBS Lett. 1981;130:223–226. doi: 10.1016/0014-5793(81)81125-8. [DOI] [PubMed] [Google Scholar]
- 73.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 74.Wagner B J, DeMaria C T, Sun Y, Wilson G M, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 75.Watkins J L, Murphy R, Emtage J L T, Wente S R. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc Natl Acad Sci USA. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wurtz T, Kiseleva E, Nacheva A, Alzhanova-Ericsson A, Rosen A, Daneholt B. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1996;16:1425–1435. doi: 10.1128/mcb.16.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Z, Luo M-J, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]