Abstract
The UV-sensitive V-H1 cell line has a T46I substitution mutation in the Walker A box in both alleles of XPD and lacks DNA helicase activity. We characterized three partial revertants that curiously display intermediate UV cytotoxicity (2- to 2.5-fold) but normal levels of UV-induced hprt mutations. In revertant RH1-26, the efficient removal of pyrimidine (6-4) pyrimidone photoproducts from both strands of hprt suggests that global-genomic nucleotide excision repair is normal, but the pattern of cyclobutane pyrimidine dimer removal suggests that transcription-coupled repair (TCR) is impaired. To explain the intermediate UV survival and lack of RNA synthesis recovery in RH1-26 after 10 J of UV/m2, we propose a defect in repair-transcription coupling, i.e., the inability of the cells to resume or reinitiate transcription after the first TCR event within a transcript. All three revertants carry an R658H suppressor mutation, in one allele of revertants RH1-26 and RH1-53 and in both alleles of revertant RH1-3. Remarkably, the R658H mutation produces the clinical phenotype of trichothiodystrophy (TTD) in several patients who display intermediate UV sensitivity. The XPDR658H TTD protein, like XPDT46I/R658H, is codominant when overexpressed in V-H1 cells and partially complements their UV sensitivity. Thus, the suppressing R658H substitution must restore helicase activity to the inactive XPDT46I protein. Based on current knowledge of helicase structure, the intragenic reversion mutation may partially compensate for the T46I mutation by perturbing the XPD structure in a way that counteracts the effect of this mutation. These findings have implications for understanding the differences between xeroderma pigmentosum and TTD and illustrate the value of suppressor genetics for studying helicase structure-function relationships.
The nucleotide excision repair (NER) pathway in eukaryotic cells is required for the error-free removal from DNA of lesions such as pyrimidine (6-4) pyrimidone photoproducts (PPs), cyclobutane pyrimidine dimers (CPDs), and bulky chemical adducts (1, 9, 37, 48). The molecular mechanism of NER is a complex process requiring about 25 proteins in humans. NER can be subdivided into global-genomic repair and transcription-coupled repair (TCR). These subpathways differ with respect to damage recognition and the rate at which some forms of DNA damage are repaired. For many lesions, including CPDs, the global-genomic pathway operates in all parts of the genome but acts relatively slowly. In this pathway, the XPA, RPA, and XPC-hHR23B proteins participate in damage recognition (41, 61, 62). Removal of CPDs and chemical adducts during TCR occurs more rapidly in the transcribed strand of active genes (30). Repair is initiated when an RNA polymerase II elongation complex is blocked by a lesion and requires the CSA and CSB proteins but not XPC-hHR23B (9). In both repair pathways, repair or transcription factor TFIIH is required to unwind the DNA duplex surrounding the lesion to facilitate incision by the XPG and XPF-ERCC1 single-strand or double-strand junction-specific endonucleases (10, 11, 34, 35, 38). This unwinding reaction absolutely requires the 3′ to 5′ and 5′ to 3′ helicase activities of the TFIIH subunits XPB/ERCC3 (28) and XPD/ERCC2 (42, 66), respectively.
Along with its role in NER, TFIIH (reviewed in reference 15) is necessary for promoter opening during basal transcription initiation by RNA polymerase II. This nine-member protein complex has several enzymatic activities, including a cyclin-dependent kinase activity comprising subunits Cdk7, cyclin H, and Mat1. The XPB and XPD subunits of TFIIH possess single-stranded DNA-dependent helicase-associated ATPase activities. Interestingly, the helicase activity of only XPB is required for transcription. Although XPD is required for transcription, its role appears structural rather than catalytic (7, 16, 32, 51). A K48R mutation in the canonical GKS/T ATP binding motif of XPD or the Rad3 homolog abolishes repair but does not compromise transcription (43, 66).
Mutations in the XPD/ERCC2 gene result in three distinct diseases (reviewed in references 3, 5, and 48): cancer-prone xeroderma pigmentosum (XP), XP in combination with Cockayne syndrome (CS; a developmental disorder), and trichothiodystrophy (TTD), which involves neurological and developmental abnormalities distinct from those of CS. Unlike the situation with most other XP genes, XPD mutations exhibit variable and complex clinical phenotypes due to the fact that XPD functions in both transcription and repair. Mutations in XPD resulting in simple XP appear to affect only the NER pathway and probably result from defective helicase activity. XPD-CS mutations show a more complex phenotype that includes defective TCR of oxidative lesions from ionizing radiation and hydrogen peroxide and increased mutations from an 8-oxo-7,8-dihydroguanine lesion in the transcribed strand in a shuttle vector (26). Mechanistically, TTD defects have been ascribed to an altered enzyme structure that reduces stability and/or interaction with other TFIIH members, thereby impairing transcription initiation (4, 7, 15). However, it is becoming apparent in the case of some XPD mutants that the clinical phenotypes require a more complex explanation than either lack of activity or lack of native XPD conformation. For example, it was reported that the R683W XPD mutant has a reduced interaction with the TFIIH p62 subunit, which normally stimulates XPD helicase activity (7). It is clear that understanding the structure-function relationships of wild-type (WT) and mutant XPD proteins will increase insight into the molecular bases of these diseases.
A step in this direction was taken with the isolation of the hamster V-H1 mutant (68) and its partially UV-resistant revertants (69). The T46I substitution mutation in both alleles of V-H1 resides in helicase motif I (21), which contains an ATP binding motif found in all helicases (12). V-H1 cells are about sixfold more UV sensitive to killing than parental cells (69) and are defective in XPD helicase activity (19). In this study, we characterize three phenotypic revertants by demonstrating that these revertants have partially restored the NER activity of XPD. In each revertant, the suppressing mutation(s) was found to be R658H. Remarkably, this substitution is identical to the mutation seen in three UV-sensitive TTD patients. These results suggest that TTD mutations do in fact change the structure of XPD and that suppressor genetics is a useful tool for studying the gross structural features of the XPD helicase.
MATERIALS AND METHODS
Cell strains and culture conditions.
The UV-sensitive V-H1 mutant was derived from the V79 cell line (68). The revertants RH1-3, RH1-26, and RH1-53 were isolated from a single culture of V-H1 after UV mutagenesis with a dose of 3 J/m2 on each of three consecutive days (69). Culture conditions were previously defined (64). The doubling times (mean and standard error of the mean) in hours of V-H1, RH1-26, and RH1-3 were 16.2 ± 0.72, 17.4 ± 1.4, and 18.3 ± 1.5, respectively.
UV survival curves and mutagenesis.
UV-C survival curves were obtained as previously described (49). V79, V-H1, RH1-3, RH1-26, RH1-53, and various transformed cell lines were grown in mass cultures with continued neo selection for transformants. Cells were then grown in normal α minimal essential medium for 1 to 3 days before use in the survival experiments. Cells were plated on 10-cm dishes in triplicate at each UV dose. Dishes were incubated at 37°C for 2 to 5 h and exposed to UV at various doses (49).
In the mutation experiment, cells were thawed and grown in regular medium for 2 days and then grown in hypoxanthine-aminopterin-thymidine medium for 96 to 120 h to kill preexisting hprt mutants. They then were placed in regular medium for 48 to 72 h before being irradiated at 0, 2.5, and 5.0 J/m2. Aliquots were plated for survival, and then 0.8 × 107 to 1.0 × 107 cells were inoculated into an 850-cm2 plastic roller bottle (350 ml). The WT was diluted on day 3 and revertant cultures were diluted on day 4 to 6 × 106 cells per bottle. The WT was plated for mutations on day 6 of expression, RH1-26 was plated on day 8, and RH1-3 was plated on day 9 because of differences in growth rates. In each instance, cells were plated in triplicate for plating efficiency (300 cells/dish) and for 6-thioguanine selection (5 μg/ml) on eight dishes (unirradiated cells) or four dishes (irradiated cells) at 4.8 × 105 cells per 15-cm dish. Selection dishes were incubated for 10 to 15 days.
Plasmid transfection and cDNA cloning.
Plasmid pXPD/R658H was constructed by digesting pXPD/RH1-53 with SfiI and XhoI to release a 1.66-kb fragment containing the suppressing mutation. After gel purification, this fragment was ligated to the complementary 6.15-kb fragment isolated from SfiI/XhoI-digested pXPD/WT.
The XPD cDNAs from revertants RH1-3, RH1-26, and RH1-53 were produced from total RNAs isolated from 2 × 107 to 5 × 107 cells using a total RNA Maxi or Midi kit from Qiagen. First-strand cDNA synthesis was accomplished using an oligo(dT)12–18 primer and a Life Technologies preamplification system for first-strand cDNA synthesis by following the manufacturer's recommendations with the following exceptions. After the addition of reverse transcriptase, reaction mixtures were incubated for 70 to 90 min at 50°C; after the addition of RNase H, reaction mixtures were incubated at 37°C for 20 to 30 min.
With the first-strand product, cDNA was produced by use of an Opti-Prime PCR optimization kit (Stratagene). This reaction was performed with a 100-μl mixture containing 10 mM Tris-HCl (pH 8.8), 3.5 mM MgCl2, 25 mM KCl, 0.2 mM each deoxynucleoside triphosphate (dNTP), 100 μg of bovine serum albumin (BSA)/ml, 10 mM NH4SO4, 7.5% dimethyl sulfoxide, 2.5 U of Pfu DNA polymerase, 1 U of Perfect Match DNA polymerase enhancer, 10 μl of reverse transcriptase reaction mixture, and 0.2 mM each primers JK105 and JK109 (see below). Pfu DNA polymerase was added last to initiate the reaction at 80°C. Reaction mixtures were heated to 94°C for 2.5 min followed by 31 to 36 cycles of PCR (1 cycle is 1 min at 94°C, 1 min at 62°C, 2 min at either 68 or 72°C, and then a 6-min extension in the last cycle). To subclone the cDNAs into pcDNA3 (Invitrogen), vector DNA was digested with EcoRV and ligated to gel-purified phosphorylated PCR product DNA.
Stable cDNA transformants were obtained by electroporation followed by selection for Geneticin resistance (64, 65). Transformants were chosen for detailed study based on their level of UV sensitivity in pilot experiments and/or on their level of overexpressed XPD protein, as measured by Western blotting.
RNA synthesis.
Exponentially growing cells were prelabeled for 20 h with 0.01 μCi of 14C-thymidine/ml (56 mCi/mmol) (44). Cells (106) were seeded in 6-cm dishes with fresh medium 1 day before UV irradiation. At various times after irradiation (10 J/m2), the medium was replaced with fresh medium containing 2 μCi of 3H-uridine/ml (39 Ci/mmol), after which cells were incubated for 30 min at 37°C. Subsequently, cells were washed with phosphate-buffered saline, and the incorporation of radioactivity in trichloroacetic acetic acid-precipitated material was determined (54). The ratio of 3H to 14C incorporation was taken as a measure of RNA synthesis.
Analysis of gene-specific repair.
The method used to analyze gene-specific repair was essentially that described previously (59). Exponentially growing cells were prelabeled for 20 h in the presence of 3H-thymidine (0.012 μCi/ml; 82 Ci/mmol) and 0.1 μM thymidine. Prior to irradiation, the 3H-thymidine-containing medium was removed and the cells were rinsed with phosphate-buffered saline. Exponentially growing cells were irradiated with UV-C at 10 or 30 J/m2 and either lysed immediately or incubated for 2, 4, 8, or 24 h in the presence of 10 μM 5-bromodeoxyuridine and 1 μM 5-fluorodeoxyuridine. DNA was isolated and purified by phenol-chloroform extraction and digested with EcoRI. Restricted DNA was centrifuged to equilibrium in neutral CsCl gradients to allow separation of replicated and parental DNAs (59). The frequency of UV-induced CPDs per restriction fragment was determined by incision with T4 endonuclease V. For analysis of 6-4 PP frequency, CPDs were removed by use of a photolyase and visible light, and then the DNA was incubated with the UvrABC excinuclease complex of Escherichia coli. After electrophoresis of samples in alkaline agarose gels, DNA was transferred to Hybond N+ membranes and hybridized with gene-specific probes (59). Filters were scanned using InstantImagerTM (Packard Instrument Co.). The number of CPDs or 6-4 PPs per restriction fragment was calculated from the relative band intensities of full-size restriction fragments in the lanes containing mock-treated DNA or DNA treated with either T4 endonuclease V or UvrABC excinuclease, assuming the Poisson distribution. Correction for nonspecific activity of UvrABC was performed as described previously (53).
DNA probes.
hprt cDNA fragments containing exons 3 to 5 or exons 6 to 9 were used to prepare specific probes. Strand-specific single-stranded probes were radioactively labeled with [32P]dATP by linear PCR using a single primer recognizing one strand (36).
Excision repair assay.
Hamster cell extracts were prepared from 1 × 109 to 3 × 109 cells as previously described (29). The excision repair substrate is a 140-bp DNA duplex containing a cholesterol adduct instead of a base in the middle of the duplex. The 32P label was positioned five nucleotides 5′ from the cholesterol adduct, and the substrate was prepared from six oligonucleotides as previously described (17). Reactions were performed at 30°C for 60 min with 50 μg of cell extract in mixtures containing 3 fmol of substrate, 40 mM HEPES (pH 7.9), 80 mM KCl, 8 mM MgCl2, 2 mM rATP, 20 μM each dNTP, 1 mM dithiothreitol, 7% glycerol (vol/vol), and 100 μg of BSA/ml. After the reactions were complete, the substrate and product DNAs were processed, resolved by denaturing polyacrylamide gel electrophoresis (PAGE), and quantitated as previously described (33).
DNA sequencing.
The XPD cDNAs from RH1-3, RH1-26, and RH1-53 were sequenced from supercoiled plasmids using the dye termination procedure (20). Primers for sequencing both strands were as follows (written 5′ to 3′): JK105 (TATTCAAGAGGCGGGCGAGCGG), JK72 (GCGGAAAAGCCCTCTGGTAG), JK107 (GCTCAAGAAAGAACCTGTGCATTC), JK74 (GTATCGAGCCAGGAAGTAGG), JK02 (CAGACCTGGTGTCCAAAGAG), JK76 (TGCTCGTCTGTCTCCTTGAT), JK77 (TGCCAGGCTCCATCCGCACT), JK85 (TAAGTGCTGACGAGAGTGGC), JK35 (GTCCCCACTGGACATCTACC), JK82 (TACTTCTCCAGGGCCACACT), JK78 (GGGGCAAAGTCTCAGAAGGG), JK80 (CAGATGGCTCAACCCTTCCA), JK37 (TTGCCCCTGATGGCACGACC), and JK109 (TTTCAGTACCTCCTGGTGCCACAGACGTCA).
Identification of genomic mutations.
Hamster genomic DNA was prepared from 1.6 × 107 to 4 × 107 cells using a blood and cell culture DNA Maxi kit (Qiagen). To determine the XPD genotype of the revertants, a 954-bp DNA fragment was PCR amplified from genomic DNA by two cycles of PCR using the following conditions and primers in a 50-μl reaction volume: 10 mM Tris-HCl (pH 9.2), 1.5 mM MgCl2, 25 mM KCl, 0.2 mM each dNTP, 100 μg of BSA/ml, 10 mM NH4SO4, 7.5% dimethyl sulfoxide, 2.5 U of Pfu DNA polymerase, 1 U of Perfect Match DNA polymerase enhancer, 300 to 400 ng of genomic DNA, and primers JK78 (see above) and JK81 (5′-TAGACTGAGCAGTGACAGGC-3′). First-round PCR was performed as described above, except that annealing was done at 60°C and the final extension reaction was carried out for 8 min. Second-round PCR was performed with 2 μl of the first-round product under the conditions described above for cDNA synthesis. Second-step PCR products were purified using a QIAquick PCR purification kit (Qiagen) and then HhaI digested. Reaction products were resolved on a 15% polyacrylamide gel and visualized by staining with 0.5 μg of ethidium bromide/ml and illuminating on a standard UV-B light box.
Western blot analysis.
Cell extracts were prepared as previously described (19). Fifty micrograms of protein from each extract line was resolved by sodium dodecyl sulfate (SDS)-PAGE. The resolved proteins were electroblotted onto a nitrocellulose membrane (Amersham) and probed with a rabbit anti-XPD polyclonal antibody (19) in blotting buffer containing 2% dry milk, 2% BSA, 10 mM Tris (pH 7.5), 150 mM NaCl, and 0.20% Tween 20. Antibody binding was visualized by film autoradiography using a goat anti-rabbit alkaline phosphatase-linked secondary antibody (Santa Cruz Biotechnology, Inc.) and an ECL kit Western blotting detection reagent (Amersham).
RESULTS
UV survival and mutagenesis responses of V-H1 revertants.
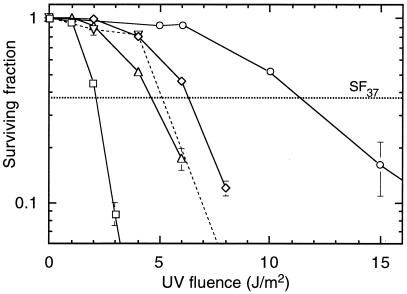
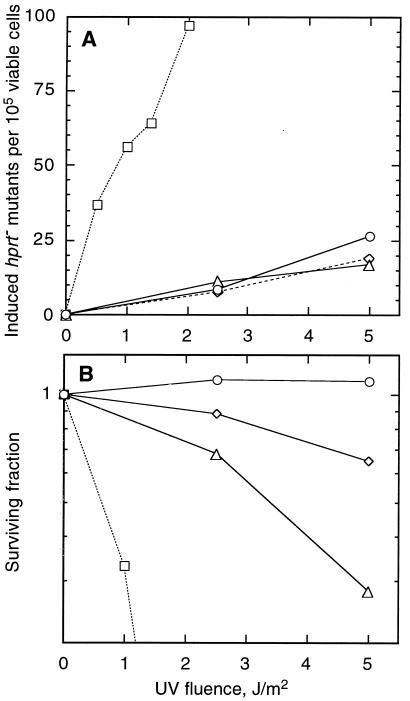
Partial revertants were previously isolated from UV-mutagenized V-H1 cells (69). We chose revertants RH1-3, RH1-26, and RH1-53 for further characterization to determine the molecular basis of their phenotypic reversion. UV survival experiments confirmed that all three revertants were restored only partially compared to the WT level of resistance (Fig. 1). Mutant V-H1 exhibited ∼5.5-fold higher sensitivity (based on D37 values) than the WT, whereas RH1-26 and RH1-53 were ∼2.5-fold more sensitive than the WT. RH1-3 was consistently more resistant to UV than the other two revertants. The sensitivity of RH1-3 and RH1-26 cells to UV-induced mutations at the hprt locus was compared with that of WT cells using a large-culture format to minimize variations caused by sampling (see Materials and Methods). V-H1 cells were previously shown to have a sevenfold higher rate of UV-induced mutagenesis than WT cells (69). The data show two important findings (Fig. 2). First, in contrast to their differential cell survival responses, RH1-3 and RH1-26 gave virtually identical mutation responses. Second, neither revertant differed significantly from the WT. Since the mutation responses suggest that repair is normal, the results prompt the question as to why survival is impaired.
FIG. 1.
UV survival based on colony-forming ability of revertants and parental cell lines. Cell lines tested were WT V79 (○), V-H1 (□), and the phenotypic revertants RH1-3 (⋄), RH1-26 (▵), and RH1-53 (▿). Lines intersecting the abscissa indicate data points below the axis. Each survival curve represents the average of two experiments. Error bars represent SEMs.
FIG. 2.
UV-induced hprt mutations based on thioguanine-resistant cells for WT, mutant, and revertant cell lines. (A) Mutations were induced by irradiation with 254-nm UV light in parental V79 (○), V-H1 (□), RH1-3 (◊), and RH1-26 (▵) cell lines. Data for V-H1 were taken from reference 69. The spontaneous mutation frequencies that were subtracted were as follows: WT, 0.4 × 10−5; V-H1, 1.8 × 10−5; RH1-3, 1.6 × 10−5; and RH1-26, 1.0 × 10−5. (B) Survival curves measured immediately after UV exposure for the cultures shown in panel A.
In vivo and in vitro repair in revertant RH1-26.
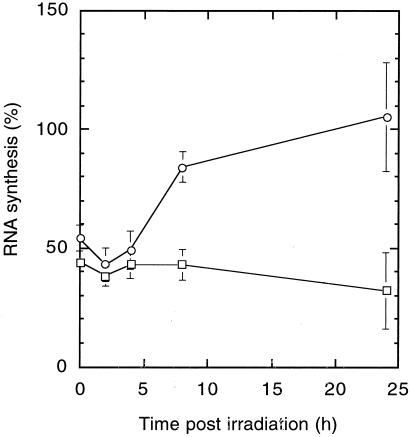
Previous studies showed that RH1-26 cells removed 6-4 PPs from the genome with normal kinetics overall, based on a radioimmunoassay, but showed very little removal of CPDs in the hprt gene (70). To assess TCR, we first determined whether RH1-26 cells were able to recover from UV-inhibited RNA synthesis after a UV dose of 10 J/m2. No RNA synthesis recovery was observed in RH1-26 cells after this dose, whereas WT cells recovered RNA synthesis by 24 h postirradiation (Fig. 3).
FIG. 3.
Rates of RNA synthesis after UV irradiation. Values are expressed relative to those for untreated cells following irradiation with 10 J/m2. V79 (○) and RH1-26 (□) cells were tested. Error bars represent SEMs.
The removal of CPDs was analyzed with the 13- and 18-kb EcoRI fragments of the hprt gene in WT and RH1-26 cells after irradiation with 10 J/m2 using T4 endonuclease V and quantitative Southern blot analysis (60). The initial CPD frequencies in the two cell lines were similar. No removal of dimers from the nontranscribed strand of the 18-kb fragment of the hprt gene was observed after 24 h in the WT or RH1-26 (data not shown). Most CPDs (87%) were removed from the transcribed strand in the hprt gene in WT cells, whereas little or no removal occurred in the same fragment in RH1-26 cells (Table 1). However, assay of the transcribed strand in an upstream 13-kb EcoRI fragment which contains hprt exons 3 to 5 indicated that the level of repair of CPDs after 24 h was high compared to that in the downstream 18-kb fragment, i.e., 56 and 9%, respectively. In WT cells, the removal of CPDs from the transcribed strand of the hprt gene was somewhat faster in the 13-kb fragment than in the 18-kb fragment during the first 6 h after irradiation; in both fragments, the transcribed strand was completely repaired after 24 h (60).
TABLE 1.
CPD removal from the transcribed strand of the EcoRI fragment of the hprt gene
| Cell line (exons) | Mean ± SEM CPD frequency per fragment at h:
|
% Repair after 24 h | |
|---|---|---|---|
| 0 | 24 | ||
| V79 (6–9) | 0.78 ± 0.06 | 0.10 ± 0.12 | 87 |
| RH1-26 (3–5) | 0.71 ± 0.12 | 0.31 ± 0.04 | 56 |
| RH1-26 (6–9) | 0.75 ± 0.17 | 0.68 ± 0.23 | 9 |
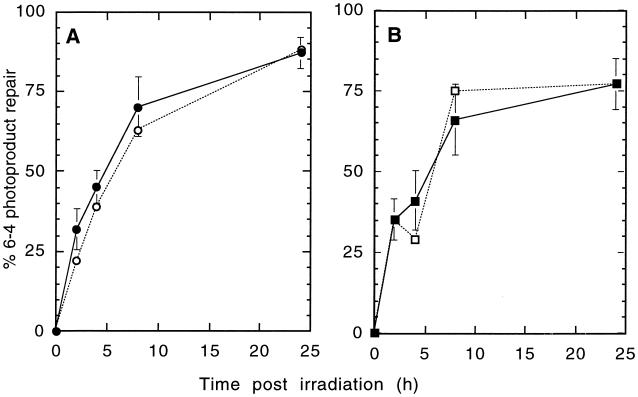
The removal of 6-4 PPs was measured with the same 18-kb EcoRI hprt fragment using the UvrABC excinuclease method, which avoids a potential limitation of the radioimmunoassay for detecting nonremoved 6-4 PPs that have become refractory to antibody binding. Since the level of induction of 6-4 PPs is ∼3-fold lower than that of CPDs (59), a UV dose of 30 J/m2 was used to obtain sufficient numbers of 6-4 PPs for accurate analysis. Prior to incubation with UvrABC, CPDs were removed by treatment with photolyase and visible light. (When photoreactivated DNA was incubated in the presence of T4 endonuclease V, no incisions were observed in the fragments analyzed, indicating complete removal of CPDs.) Photoreactivated DNA was incubated with UvrABC and then analyzed by alkaline gel electrophoresis. 6-4 PPs were removed from both strands of the hprt gene with the same kinetics, and there was no detectable difference in repair kinetics between the WT and RH1-26 (Fig. 4). Thus, the normal removal of 6-4 PPs from hprt in RH1-26 agrees with previous antibody data showing efficient removal of 6-4 PPs from the genome overall (70) but contrasts sharply with the lack of removal of CPDs in this particular hprt fragment (see Discussion).
FIG. 4.
Removal of 6-4 PPs from the hprt gene after UV irradiation with 30 J/m2. (A) WT cells, transcribed strand (●) and nontranscribed strand (○). (B) RH1-26, transcribed strand (▪) and nontranscribed strand (□). Error bars represent SEMs.
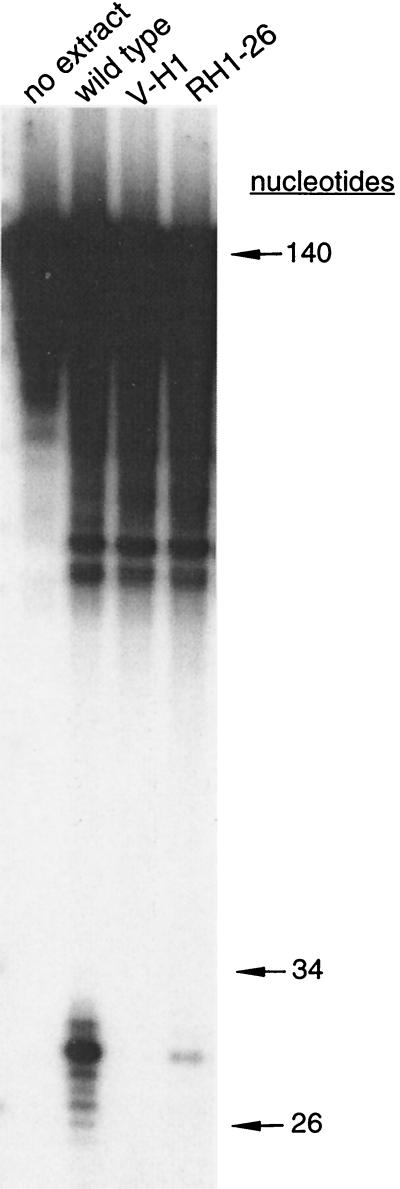
To further examine cellular NER capacity, Manley (29) whole-cell extracts were prepared, and NER activity was measured using a 140-bp linear DNA duplex substrate containing a cholesterol “adduct” in the middle of the substrate (see Materials and Methods). To record NER activity, a 32P-labeled base was positioned five nucleotides 5′-ward of the cholesterol moiety. In this assay, activity is detected when dual incisions occur on opposite sides of the lesion, thereby producing excised oligonucleotides 24 to 32 bases long.
In reactions lacking cell extracts, no detectable incision was observed (Fig. 5, first lane). Reactions containing WT extracts converted 5.2% ± 0.4% (standard error of the mean) of the substrate to product (Fig. 5, second lane). In contrast, no activity was detected in reactions containing V-H1 extracts (Fig. 5, third lane), although the extracts were shown to be active by complementation using XPG-deficient extracts (data not shown). Reactions containing RH1-26 extracts incised 1.1% ± 0.7% of the substrate constituting, on average, 22% WT activity (Fig. 5, fourth lane). In another experiment, RH1-3 exhibited 29% WT activity (data not shown; based on triplicate measurements of each cell line). These results are qualitatively consistent with the differential UV sensitivity of the two revertants, but the quantitative levels of repair are clearly lower than one would expect from in vivo survival and mutagenesis. The lack of activity in the V-H1 extracts is in agreement with the high UV sensitivity of this mutant. The partial restoration of in vitro NER activity in RH1-26 suggested that the suppressing mutation resided in XPD or another NER protein.
FIG. 5.
Partial restoration of in vitro NER activity in revertant RH1-26. Cell extracts were incubated with a 140-bp duplex DNA containing a cholesterol moiety in the middle of the substrate. The substrate was internally labeled with 32P 5 bp 5′-ward of the adduct on the same strand of DNA. Reactions were performed at 30°C for 60 min. After protein removal, the substrate was separated from the product by denaturing PAGE and visualized by film autoradiography.
The unwinding activity of XPD is essential for NER (66), and the XPDT46I protein of V-H1 lacks helicase activity, as recently reported (19). Thus, it seemed plausible that the suppressing mutation(s) in the revertants may have occurred within XPD. Alternatively, the mutation may have occurred in an XPD-interacting protein within TFIIH, such as p44 (8), to restore helicase activity by reestablishing an essential protein-protein interaction that was disrupted by the T46I substitution.
XPD suppressor mutations in V-H1 revertants.
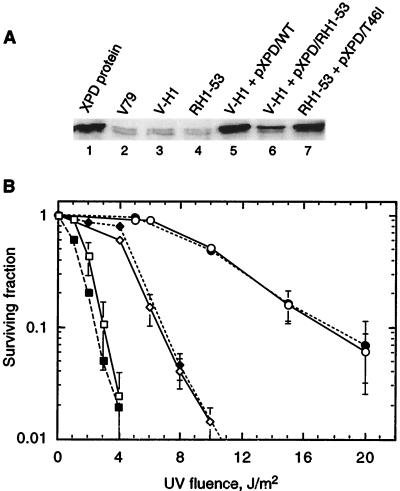
To test directly whether a suppressing mutation was intragenic, the XPD cDNA from RH1-53 was subcloned into expression vector pcDNA3, and several plasmid clones were individually transfected into V-H1. Individual G418-resistant transformants were then examined for overexpression of XPD by Western analysis (Fig. 6A) and for increased UV resistance (Fig. 6B). Since V-H1 cells harbor two identical mutant xpd alleles (21), initial experiments were directed at identifying transformants that had increased UV resistance and increased levels of XPD protein (data not shown). Overexpressing transformants showing increased UV resistance were presumed to have integrated the cDNA of a revertant allele of RH1-53 (e.g., Fig. 6A, lane 6, and Fig. 6B). These clones showed a level of UV resistance identical to that of RH1-53 cells. Transformant cultures from some cDNA clones of RH1-53 did not show any UV-resistant cells. This overexpression-competition experiment suggested that the suppressing mutation in RH1-53 resided in an xpd allele. As a positive control for complementation, V-H1 transformants overexpressing wild-type XPD were complemented to the WT level of UV resistance (Fig. 6A, lane 5, and Fig. 6B).
FIG. 6.
Evidence that a suppressing mutation resides in an xpd allele. (A) Fifty micrograms of soluble protein from each extract was resolved by SDS-PAGE, and XPD protein was identified by Western blotting using an affinity-purified anti-XPD polyclonal antibody. In some lanes, the XPD protein appears as a doublet, for reasons not understood. (B) UV survival curves for cells of the WT (○), V-H1 (□), revertant RH1-53 (◊), V-H1 transfected with XPDWT (●), V-H1 transfected with XPDRH1-53 (⧫), and RH1-53 transfected with XPDT46I (▪). Each survival curve represents the average of two experiments. Error bars represent SEMs.
As a confirmatory approach to the transfection of V-H1 with XPDRH1-53 cDNA, RH1-53 cells were transfected with XPDV-H1 cDNA to determine whether the overexpression of XPDT46I could convert RH1-53 to a UV-sensitive phenotype. As shown in Fig. 6A, lane 7, and Fig. 6B, an RH1-53 transformant expressing the XPDT46I protein was at least as sensitive to UV as V-H1. Taken together, these results demonstrated that the suppressing mutation mapped to one of the xpd alleles in RH1-53.
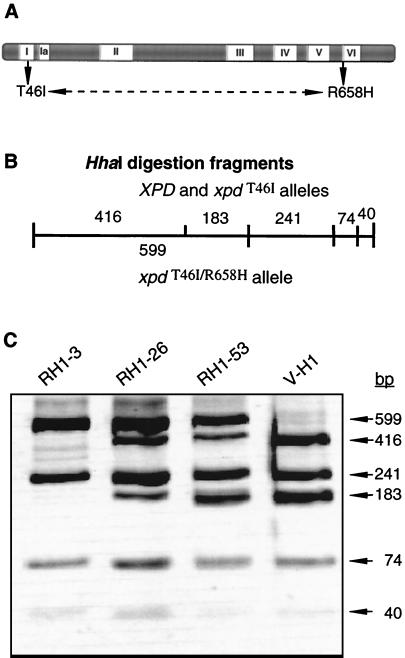
Nucleotide sequencing of UV resistance-producing XPD cDNA clones from the revertants was performed to identify the secondary mutations. The same CGC-to-CAC substitution at nucleotide 1973 of the coding sequence was found in all three revertants. This change represents a R658H amino acid substitution in the XPD protein which is quite distant from the original T46I mutation and resides just outside of helicase motif VI (Fig. 7A). These results suggest that these two regions of the protein normally interact to form a functional domain. Especially remarkable and curious is the fact that R658H is the causative mutation in three TTD patients having intermediate UV sensitivity (45, 46). An R658C substitution was present in another TTD patient (45).
FIG. 7.
Analysis of the XPD intragenic suppressor mutation(s) in revertants. (A) The positions of the conserved helicase motifs in XPD, as described by Koonin for the Rad3 helicase family (23), are shown. The original mutation in the V-H1 cell line is a homozygous substitution in the Walker A box of motif I (GT46GKT) of the protein that results in a T46I change. The suppressing mutation in all three revertant cell lines changed arginine at position 658 to histidine. Arginine 658 is positioned on the immediate N-terminal side of motif VI. (B) HhaI digestion of the genomic PCR products from WT and V-H1 DNAs produces DNA fragments of 416, 241, 183, 74, and 40 bp. In the revertant DNA, the suppressing mutation eliminates an HhaI site. (C) Genomic DNAs from V-H1 and the revertants were isolated, and a portion of the XPD gene corresponding to nucleotide positions 13113 to 14066 was PCR amplified and purified. PCR products were digested with HhaI and resolved by PAGE. The 40-bp bands are faint. The image was black-white inverted for ease of viewing.
Since the suppressing mutation eliminates an HhaI cleavage site, restriction enzyme analysis was used to determine the genotypes of the revertants with respect to each allele. A 954-bp genomic fragment surrounding the position of the suppressing mutation was amplified by PCR from V-H1 and the revertants. The predicted restriction patterns for the alleles are diagrammed in Fig. 7B. The loss of the HhaI site in a revertant allele changes the restriction map in two ways: a unique 599-bp HhaI digestion fragment is predicted along with the loss of the 416- and 183-bp fragments seen with the WT and V-H1 alleles. The HhaI restriction patterns are shown in Fig. 7C. The amplified and digested V-H1 DNA contains the five predicted fragments. In contrast, the RH1-3 digest suggests that this revertant is homozygous for the XPDT46I/R658H suppressing allele. In comparison, both RH1-26 and RH1-53 are heterozygous for this allele. Confirmatory analysis was done on cDNA of each revertant by performing HhaI digestion of reverse transcriptase PCR-derived products (data not shown). All the restriction digestion data were also consistent with the idea that RH1-3 expresses two revertant alleles. Table 2 summarizes these XPD genotypes.
TABLE 2.
XPD intragenic suppressor mutations in revertants of V-H1
| Cell line | Amino acid at the following position in the indicated allele:
|
|||
|---|---|---|---|---|
| 46
|
658
|
|||
| 1 | 2 | 1 | 2 | |
| WT V79 | Thr | Thr | Arg | Arg |
| V-H1 | Ile | Ile | Arg | Arg |
| RH1-26 | Ile | Ile | Arg | His |
| RH1-53 | Ile | Ile | Arg | His |
| RH1-3 | Ile | Ile | His | His |
Since the R658H substitution partially restored the activity of the XPDT46I mutant protein in V-H1, it was of interest to determine how well the XPDR658H allele itself would complement V-H1 cells. This information would help us to assess how much effect the T46I substitution had on the XPDR658H protein. Since TTD with the R658H mutation shows mild UV sensitivity, we would not expect the XPDR658H allele to fully complement V-H1.
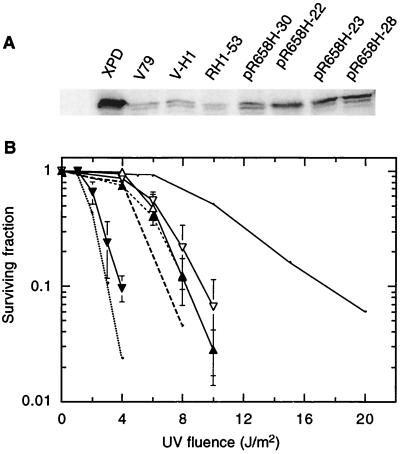
V-H1 cells transfected with a plasmid containing the XPDR658H cDNA were selected for resistance to G418 and then screened for XPDR658H overexpression by Western blot analysis. As shown in Fig. 8A, four transformants showed various levels of XPD overexpression. UV survival curves for these transformants are compared to those for V-H1, RH1-53, and the WT in Fig. 8B. Transformant clone 30, which appeared to express the smallest amount of XPDR658H protein, minimally complemented V-H1. Transformant clones 22 and 23, showing about the same level of expression, complemented V-H1 to UV resistance slightly higher than that of RH1-53. Finally, transformant 28 (arguably producing the highest level of XPDR658H) repeatedly showed the highest level of resistance, which was appreciably above that of RH1-53 and approximately equivalent to that of RH1-3 (compare Fig. 1 with Fig. 8B). There are two possible explanations for this partial restoration of UV resistance. First, the XPDR658H protein may be moderately helicase deficient and adequately competitive with the XPDT46I protein at these expression levels. Alternatively, the XPDR658H protein may be fully active enzymatically but not highly competitive with XPDT46I, even at a very high expression level, because of defective interactions with partner proteins.
FIG. 8.
UV survival of V-H1 clones overexpressing XPDR658H cDNA. (A) Fifty micrograms of soluble protein extract from each cell line resolved by SDS-PAGE and probed for XPD using anti-XPD antibody from a rabbit as described in Materials and Methods. (B) UV survival curves for V-H1 (dotted line), RH1-53 (dashed line, no symbols), WT (solid line, no symbols), and VH-1/R658H transformant clone 30 (▾), clone 22 (▴), clone 23 (▵) and clone 28 (▿). The curves for V-H1, RH1-53, and WT are taken from Fig. 6.
DISCUSSION
Partial restoration of in vitro NER capacity in revertants.
Like that of other XPD mutants (18), the phenotype of V-H1 cells has been paradoxical because of their high sensitivity to UV killing and UV-induced hprt mutation but their retention of both high unscheduled DNA synthesis (69) and, ostensibly, intermediate levels of 6-4 PP removal (31). However, recent studies examining in vitro incision and XPD helicase activity lead to the conclusion that V-H1 is completely defective in NER and that the apparent repair is an artifact arising from unproductive incision and associated “repair” synthesis (19). The helicase-motif I synthetic XPDK48R mutation shows similar properties, such as aberrant repair synthesis (66). Our in vitro data show that the suppressor mutations in RH1-3 and RH1-26 partially restore incision activity (Fig. 5 and unpublished results), inferentially by producing a mutant protein that has partially regained helicase activity, which is essential for repair (42, 66). Both revertants show less in vitro repair activity than expected from the in vivo repair and survival data. One possible explanation for this discrepancy is that the XPDT46I/R658H protein is relatively inefficient or labile in vitro, like, for example, temperature-sensitive mutants that lack detectable in vitro activity.
Nature of the revertant phenotype.
The phenotype of RH1-26 (and the other revertants), like that of its parent mutant, also presented a paradox because sensitivity to UV killing was intermediate, whereas UV-induced hprt mutation was reported to be below normal (70). Our data show an essentially WT level of mutagenesis under mutation expression conditions that compensated for the reduced rate of growth of revertant cells compared with WT cells (Fig. 2). Examination of the kinetics of removal of CPDs and 6-4 PPs in the hprt gene was intended to provide insight into the previous finding that the efficiency and spectrum of UV-induced mutations were normal in RH1-26 (70). Almost all mutations produced at 2 J/m2 were due to photolesions in the nontranscribed strand. Our data (Fig. 4) suggest that 6-4 PP removal is normal in both strands of hprt, even at the high dose of 30 J/m2. The possibility of abortive incision (as discussed above for V-H1) does not seem to apply to RH1-26 because we did not observe any break induction that would be indicative of this occurring (unpublished results). However, normal UV mutagenesis may seem at first to be inconsistent with inhibition of recovery of transcription, as measured by RNA synthesis after 10 J/m2 (Fig. 3). This inhibition is likely to be the cause of the substantial UV cytotoxicity of RH1-26 (increased ∼2.5-fold).
The lack of recovery of RNA synthesis implies that (i) TCR per se is defective or that (ii) transcription fails to resume or reinitiate after the removal of the first CPD in an active gene (average of about two CPDs in the 18-kb fragment at 10 J/m2). Since TCR normally removes DNA photolesions in a sequential way (53), CPDs in the 5′ region of the hprt gene (i.e., the 13-kb fragment) will be removed first. After the first TCR event has occurred in the gene, either transcription elongation can resume (if the polymerase is still bound) or transcription can reinitiate (if the polymerase is released). RH1-26 appears to be defective in whichever of these steps normally occurs, causing failure of RNA synthesis to recover at 10 J/m2 (Fig. 3). However, after low doses, such as 2 J/m2 (the dose used to determine the mutation spectrum), less than one CPD, on average, is present in hprt; CPDs will be removed by ongoing transcription or after initiation of transcription, thus leading to a normal mutation frequency and spectrum.
Therefore, to explain the intermediate UV sensitivity and lack of recovery of RNA synthesis in RH1-26, we favor a model in which only cells in the population that are able to restore RNA synthesis can survive. These surviving cells have approximately WT levels of hprt mutations (Fig. 2) because of efficient 6-4 PP removal and intermediate CPD repair. In the remainder of the cells, suppression of transcription may lead to cell cycle arrest and/or apoptosis (2, 27). It is well established that sensitivity to killing varies during the cell cycle, with early S phase being the most sensitive. In contrast, mutagenesis shows a flat age response (67). In asynchronous RH1-26 populations, survival of hprt+ cells after irradiation could be preferentially limited to a portion of the cell cycle (67).
Moreover, it is instructive to compare the phenotype of RH1-26 with that of the CHO CSB/ERCC6 mutant UV61, which shows efficient repair of 6-4 PPs, little repair of CPDs in the transcribed strand, no recovery of transcription, and enhanced mutagenesis (58). UV61 is also somewhat more UV sensitive than RH1-26 (50). Whereas RH1-26 appears to be partially defective in CPD removal and reinitiation of transcription after gene repair, UV61 is defective in TCR per se, resulting in an elevated level of UV mutagenesis. The phenotype of RH1-26 also differs from that of highly UV-sensitive human XPD-CS mutants, which show defective RNA synthesis recovery but also greatly reduced 6-4 PP repair (52).
Mechanism of phenotypic suppression of XPD helicase mutation.
The fact that RH1-3 is homozygous for the suppressing mutation agrees with its UV resistance being significantly higher than that of RH1-26 and RH1-53 (Fig. 1 and 2). These data, along with the XPDT46I/R658H overexpression data shown in Fig. 6, indicate that this revertant allele is codominant. The homozygosity of the revertant allele in RH1-3 cells is surprising, just as is the homozygosity of the XPDT46I allele in parental V-H1 cells. In both instances, homozygosity presumably arose through a process of gene conversion or chromosome nondisjunction. The chromosomes of V79 cells show multiple rearrangements (47). It is possible that one of the chromosomes carrying XPD is particularly prone to such events. If the initial reversion event had improved the growth rate, there might have been selection in favor of a second event producing homozygosity in RH1-3. However, this scenario is unlikely because RH1-3 grows more slowly than RH1-26 and RH1-26 grows more slowly than V-H1 (see Materials and Methods). These slower doubling times for revertant cells than for V-H1 and parental V-79 cells are consistent with the idea that the R658H mutation results in less efficient transcription.
Suppression of the XPDT46I mutant phenotype by the R658H substitution is especially intriguing in light of the fact that this mutation causes TTD (45, 46). The transcription hypothesis proposes that TTD patients have a defect in basal transcription due to impaired transcription initiation (7, 15, 57). There is now considerable evidence that mutations underlying TTD alter the structure of XPD and destabilize TFIIH. The xpd R658C mutation present in several TTD patients confers a temperature-sensitive phenotype for repair, transcription, and cell growth (56) and explains the hair loss occurring in patients after fevers. Moreover, this mutation destabilizes TFIIH, since the level of the p62 subunit is also temperature sensitive. The XPDR722W protein, the result of a recurring TTD mutation, is unable to interact with the p44 TFIIH subunit in cell extracts, unlike WT XPD (8). Also, the TTD mutation in the TTD-A complementation group causes a reduction in the TFIIH level to ∼25% the normal level by an unknown mechanism (55). We propose that the R658H substitution in V-H1 revertants causes a subtle structural change that compensates for a structural perturbation arising from the T46I substitution in the ATP binding site.
The two heterozygous revertants showed similar, intermediate levels of UV resistance, indicating that the XPDT46I/R658H revertant protein competes effectively with the XPDT46I protein for incorporation into TFIIH. To begin to understand the mechanism underlying the suppressing R658H substitution, the XPDR658H protein was overexpressed at different levels in V-H1 cells and was also found to compete effectively and produce more UV resistance at higher levels of expression (Fig. 8). The intermediate UV sensitivity associated with the human R658H mutation (46) is consistent with the intermediate sensitivity of the V-H1 transformants overexpressing XPDR658H. We assume that this mutation acts identically in hamster and human proteins because the amino acid sequences are 99% identical.
Helicase domains and intragenic suppression.
Based on sequence homology, five RNA/DNA helicase superfamilies were defined (12, 13). Superfamily II contains the Rad3 family, to which XPD belongs (23). Despite the overall weak similarity among the various families, conserved motifs have been defined (13, 14). The limited homology that does exist across superfamilies is restricted to motifs I and II, which are involved in nucleotide triphosphate binding (14). However, these motifs are shared with other enzymes that hydrolyze nucleotide triphosphates (referred to as Walker A and B motifs) (63).
X-ray crystal structures for several helicases, including the PcrA and Rep DNA helicases and the hepatitis C virus NS3 RNA helicase, are known (22, 24, 40). Although the PcrA and Rep proteins belong to superfamily I and the NS3 protein belongs to superfamily II, they are structurally similar, implying the conservation of folds and relationships among helicase motifs across these superfamilies (14, 25). The tertiary structures of all three proteins place the helicase motifs within two domains forming a cleft that contains motifs I and II on one side and motif VI on the other. In each structure, the GxGKT/S residues of motif I are responsible for binding the β-phosphate of ATP, while the aspartic acid residue of motif II binds Mg2+. These bonds help orient the ATP-Mg2+ complex for hydrolysis. Upon binding ATP, the gap between the cleft closes, positioning the arginine residues of motif VI such that they interact with the phosphate groups of ATP. Upon hydrolysis, protein translocation and DNA unwinding occur, ADP is released, and the cleft opens, thus completing a reaction cycle (39).
An intimate spatial relationship between residues T46 and R658 in XPD is suggested based on our reversion data and the general picture of helicase structure gained from the crystal structures mentioned above. We speculate that because residue T46 is positioned between invariant residues of motif I (GT46GKT), the T46I change may distort ATP binding in such a way that the arginine residues of motif VI are unable to interact properly with the phosphate groups of ATP to facilitate hydrolysis. The normal R658 residue may influence the position of the arginine residues of motif VI, since R658 is positioned seven residues away from the highly conserved Gly and Arg residues in this motif (R658HAAQCVGRAIR). In suppressing the effect of the T46I substitution, the R658H substitution may position the arginine residues of motif VI in a conformation that improves the efficiency of hydrolysis, albeit not to the normal rate.
To our knowledge, this study provides the first instance in which intragenic suppressor mutations have been identified in a DNA helicase. Reversion mutations in the yeast Prp28 RNA helicase identified several putative motif interactions, including a remarkably analogous suppression of T223I in motif I by a substitution in motif IV (6). Thus, suppressor genetics may help determine helicase structure and function by identifying interacting regions or motifs. It is of particular interest to understand exactly how TTD mutations such as R658H alter the structure of XPD to confer various degrees of UV sensitivity.
ACKNOWLEDGMENTS
We thank Anita Avery for technical assistance.
This research was supported by NIH grant CA52679 and was performed under the auspices of the U.S. Department of Energy at the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405-Eng-48. Other support was provided by grants GM32833 (to A.S.), EU-FIGH-CT1999-00010 (to M.Z.Z.), and Dutch Cancer Society grant IKW 92-32 (to L.H.F.M.).
REFERENCES
- 1.Araujo S J, Wood R D. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 2.Balajee A S, DeSantis L P, Brosh Jr R M, Selzer R, Bohr V A. Role of the ATPase domain of the Cockayne syndrome group B protein in UV induced apoptosis. Oncogene. 2000;19:477–489. doi: 10.1038/sj.onc.1203372. [DOI] [PubMed] [Google Scholar]
- 3.Berneburg M, Lehmann A R. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma D, Hoeijmakers J H. The molecular basis of nucleotide excision repair syndromes. Mutat Res. 1994;307:15–23. doi: 10.1016/0027-5107(94)90273-9. [DOI] [PubMed] [Google Scholar]
- 5.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In: Vogelstein B, Kinzler K, editors. The genetic basis of human cancer. New York, N.Y: McGraw-Hill Book Co.; 1998. pp. 245–274. [Google Scholar]
- 6.Chang T H, Latus L J, Liu Z, Abbott J M. Genetic interactions of conserved regions in the DEAD-box protein Prp28p. Nucleic Acids Res. 1997;25:5033–5040. doi: 10.1093/nar/25.24.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coin F, Bergmann E, Tremeau-Bravard A, Egly J M. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coin F, Marinoni J C, Rodolfo C, Fribourg S, Pedrini A M, Egly J M. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 9.de Laat W L, Jaspers N G, Hoeijmakers J H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 10.Evans E, Fellows J, Coffer A, Wood R D. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans E, Moggs J G, Hwang J R, Egly J M, Wood R D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A E, Koonin E V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- 13.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall M C, Matson S W. Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoeijmakers J H J, Egly J M, Vermeulen W. TFIIH: a key component in multiple DNA transactions. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 16.Holstege F C P, van der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J C, Hsu D S, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R T, Squires S. The XPD complementation group. Insights into xeroderma pigmentosum, Cockayne's syndrome and trichothiodystrophy. Mutat Res. 1992;273:97–118. doi: 10.1016/0921-8777(92)90072-b. [DOI] [PubMed] [Google Scholar]
- 19.Kadkhodayan S, Coin F, Salazar E, George J, Egly J, Thompson L. Codominance associated with overexpression of certain XPD mutations. Mutat Res. 2001;485:153–168. doi: 10.1016/s0921-8777(00)00077-x. [DOI] [PubMed] [Google Scholar]
- 20.Kadkhodayan S, Salazar E P, Lamerdin J E, Weber C A. Construction of a functional cDNA clone of the hamster ERCC2 DNA repair and transcription gene. Somat Cell Mol Genet. 1996;22:453–460. doi: 10.1007/BF02369437. [DOI] [PubMed] [Google Scholar]
- 21.Kadkhodayan S, Salazar E P, Ramsey M J, Takayama K, Tucker J D, Zdzienicka M Z, Weber C A. Molecular analysis of ERCC2 mutations in the repair deficient hamster mutant UVL-1 and V-H1. Mutat Res. 1997;385:47–57. doi: 10.1016/s0921-8777(97)00030-x. [DOI] [PubMed] [Google Scholar]
- 22.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 23.Koonin E V. Escherichia coli dinG gene encodes a putative DNA helicase related to a group of eukaryotic helicases including Rad3 protein. Nucleic Acids Res. 1993;21:1497. doi: 10.1093/nar/21.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 25.Korolev S, Yao N, Lohman T M, Weber P C, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 27.Ljungman M, Zhang F. Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 28.Ma L, Siemssen E D, Noteborn H M, van der Eb A J. The xeroderma pigmentosum group B protein ERCC3 produced in the baculovirus system exhibits DNA helicase activity. Nucleic Acids Res. 1994;22:4095–4102. doi: 10.1093/nar/22.20.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell D L, Zdzienicka M Z, van Zeeland D, Nairn R S. Intermediate (6-4) photoproduct repair in Chinese hamster V79 mutant V-H1 correlates with intermediate levels of DNA incision and repair replication. Mutat Res. 1989;226:43–47. doi: 10.1016/0165-7992(89)90091-2. [DOI] [PubMed] [Google Scholar]
- 32.Moreland R J, Tirode F, Yan Q, Conaway J W, Egly J M, Conaway R C. A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J Biol Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- 33.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 34.Mu D, Wakasugi M, Hsu D S, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 35.O'Donovan A, Davies A A, Moggs J G, West S C, Wood R D. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 36.Ruven H J, Seelen C M, Lohman P H, van Kranen H, van Zeeland A A, Mullenders L H. Strand-specific removal of cyclobutane pyrimidine dimers from the p53 gene in the epidermis of UVB-irradiated hairless mice. Oncogene. 1994;9:3427–3432. [PubMed] [Google Scholar]
- 37.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 38.Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G J, Hoeijmakers J H J, Wood R D. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 39.Soultanas P, Wigley D B. DNA helicases: ‘inching forward’. Curr Opin Struct Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 40.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 41.Sugasawa K, Ng J M, Masutani C, Iwai S P, van der Spek J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 42.Sung P, Guzder S N, Prakash L, Prakash S. Reconstitution of TFIIH and requirement of its DNA helicase subunits, Rad3 and Rad25, in the incision step of nucleotide excision repair. J Biol Chem. 1996;271:10821–10826. doi: 10.1074/jbc.271.18.10821. [DOI] [PubMed] [Google Scholar]
- 43.Sung P, Higgins D, Prakash L, Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988;7:3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tagder K, Scheuermann W. Estimation of absorbed doses in the cell nucleus after incorporation of 3H- or 14C-labeled thymidine. Radiat Res. 1970;41:202–216. [PubMed] [Google Scholar]
- 45.Takayama K, Salazar E P, Broughton B C, Lehmann A R, Sarasin A, Thompson L H, Weber C A. Defects in the DNA repair and transcription gene ERCC2 (XPD) in trichothiodistrophy. Am J Hum Genet. 1996;58:263–270. [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor E M, Broughton B C, Botta E, Stefanini M, Sarasin A, Jaspers N G J, Fawcett H, Harcourt S A, Arlett C F, Lehmann A R. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci USA. 1997;94:8658–8663. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thacker J. The chromosomes of a V79 Chinese hamster line and a mutant subline lacking HPRT activity. Cytogenet Cell Genet. 1981;29:16–25. doi: 10.1159/000131547. [DOI] [PubMed] [Google Scholar]
- 48.Thompson L H. Nucleotide excision repair: its relation to human disease. In: Nickoloff J, Hoekstra M, editors. DNA damage and repair. 2. DNA repair in higher eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 335–393. [Google Scholar]
- 49.Thompson L H, Fong S, Brookman K. Validation of conditions for efficient detection of HPRT and APRT mutations in suspension-cultured Chinese hamster cells. Mutat Res. 1980;74:21–36. doi: 10.1016/0165-1161(80)90188-0. [DOI] [PubMed] [Google Scholar]
- 50.Thompson L H, Mitchell D L, Regan J D, Bouffler S D, Stewart S A, Carrier W L, Nairn R S, Johnson R T. CHO mutant UV61 removes (6-4) photoproducts but not cyclobutane dimers. Mutagenesis. 1989;4:140–146. doi: 10.1093/mutage/4.2.140. [DOI] [PubMed] [Google Scholar]
- 51.Tirode F, Busso D, Coin F, Egly J M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 52.van Hoffen A, Kalle W H, de Jong-Versteeg A, Lehmann A R, van Zeeland A A, Mullenders L H. Cells from XP-D and XP-D-CS patients exhibit equally inefficient repair of UV-induced damage in transcribed genes but different capacity to recover UV-inhibited transcription. Nucleic Acids Res. 1999;27:2898–2904. doi: 10.1093/nar/27.14.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoffen A, Venema J, Meschini R, van Zeeland A A, Mullenders L H. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Oosterwijk M F, Versteeg A, Filon R, van Zeeland A A, Mullenders L H. The sensitivity of Cockayne's syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol Cell Biol. 1996;16:4436–4444. doi: 10.1128/mcb.16.8.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vermeulen W, Bergmann E, Auriol J, Rademakers S, Frit P, Appeldoorn E, Hoeijmakers J H, Egly J M. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat Genet. 2000;26:307–313. doi: 10.1038/81603. [DOI] [PubMed] [Google Scholar]
- 56.Vermeulen W, Rademakers S, Jaspers N G, Appeldoorn E, Raams A, Klein B, Kleijer W J, Hansen L K, Hoeijmakers J H. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- 57.Vermeulen W, van Vuuren A J, Chipoulet M, Schaeffer L, Appeldoorn E, Weeda G, Jaspers N G J, Priestley A, Arlett C F, Lehmann A R, Stefanini M, Mezzina M, Sarasin A, Bootsma D, Egly J-M, Hoeijmakers J H J. Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH). Evidence for the existence of a transcription syndrome. Cold Spring Harbor Symp Quant Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- 58.Vreeswijk M P, Overkamp M W, Westland B E, van Hees-Stuivenberg S, Vrieling H, Zdzienicka M Z, van Zeeland A A, Mullenders L H. Enhanced UV-induced mutagenesis in the UV61 cell line, the Chinese hamster homologue of Cockayne's syndrome B, is associated with defective transcription coupled repair of cyclobutane pyrimidine dimers. Mutat Res. 1998;409:49–56. doi: 10.1016/s0921-8777(98)00038-x. [DOI] [PubMed] [Google Scholar]
- 59.Vreeswijk M P, van Hoffen A, Westland B E, Vrieling H, van Zeeland A A, Mullenders L H. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in Chinese hamster cells. J Biol Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 60.Vrieling H, Venema J, van Rooyen M L, van Hoffen A, Menichini P, Zdzienicka M Z, Simons J W, Mullenders L H, van Zeeland A A. Strand specificity for UV-induced DNA repair and mutations in the Chinese hamster HPRT gene. Nucleic Acids Res. 1991;19:2411–2415. doi: 10.1093/nar/19.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci USA. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 63.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber C A, Salazar E P, Stewart S A, Thompson L H. Molecular cloning and biological characterization of a human gene, ERCC2, that corrects the nucleotide excision repair defect in CHO UV5 cells. Mol Cell Biol. 1988;8:1137–1146. doi: 10.1128/mcb.8.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber C A, Salazar E P, Stewart S A, Thompson L H. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 1990;9:1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winkler G S, Araujo S J, Fiedler U, Vermeulen W, Coin F, Egly J M, Hoeijmakers J H, Wood R D, Timmers H T, Weeda G. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 67.Wood R D, Burki H J. Repair capability and the cellular age response for killing and mutation induction after UV. Mutat Res. 1982;95:505–514. doi: 10.1016/0027-5107(82)90281-0. [DOI] [PubMed] [Google Scholar]
- 68.Zdzienicka M Z, Simons J W I M. Mutagen-sensitive cell lines are obtained with a high frequency in V79 Chinese hamster cells. Mutat Res. 1987;178:235–244. doi: 10.1016/0027-5107(87)90274-0. [DOI] [PubMed] [Google Scholar]
- 69.Zdzienicka M Z, Van der Shans S, Westerveld A, van Zeeland D, Simons J W I M. Phenotypic heterogeneity within the first complementation group of UV-sensitive mutants of Chinese hamster cells. Mutat Res. 1988;193:31–41. doi: 10.1016/0167-8817(88)90005-3. [DOI] [PubMed] [Google Scholar]
- 70.Zdzienicka M Z, Venema J, Mitchell D L, van Hoffen A, van Zeeland A A, Vrieling H, Mullenders L H, Lohman P H, Simons J W. (6-4) photoproducts and not cyclobutane pyrimidine dimers are the main UV-induced mutagenic lesions in Chinese hamster cells. Mutat Res. 1992;273:73–83. doi: 10.1016/0921-8777(92)90051-4. [DOI] [PubMed] [Google Scholar]