Abstract
We monitored longitudinal changes in bovine milk IgG in samples from four cows at 9 time points in between 0.5 and 28 days following calving. We used peptide-centric LC–MS/MS on proteolytic digests of whole bovine milk, resulting in the combined identification of 212 individual bovine milk protein sequences, with IgG making up >50 percent of the protein content of every 0.5 d colostrum sample, which reduced to ≤3 percent in mature milk. In parallel, we analyzed IgG captured from the bovine milk samples to characterize its N-glycosylation, using dedicated methods for bottom-up glycoproteomics employing product ion-triggered hybrid fragmentation; data are available via ProteomeXchange with identifier PXD037755. The bovine milk IgG N-glycosylation profile was revealed to be very heterogeneous, consisting of >40 glycoforms. Furthermore, these N-glycosylation profiles changed substantially over the period of lactation, but consistently across the four individual cows. We identified NeuAc sialylation as the key abundant characteristic of bovine colostrum IgG, significantly decreasing in the first days of lactation, and barely detectable in mature bovine milk IgG. We also report, for the first time to our knowledge, the identification of subtype IgG3 in bovine milk, alongside the better-documented IgG1 and IgG2. The detailed molecular characteristics we describe of the bovine milk IgG, and their dynamic changes during lactation, are important not only for the fundamental understanding of the calf’s immune development, but also for understanding bovine milk and its bioactive components in the context of human nutrition.
Keywords: bovine milk immunoglobulins, bovine colostrum, glycoproteomics, NeuGc, passive immunity
Introduction
In mammals, the mammary gland secretion produced up to 3 days following birth is considered to be colostrum (McGrath et al. 2016). Colostrum is compositionally and functionally very different from mature milk (McGrath et al. 2016). One of the main functions of colostrum, next to providing essential nutrition, is to transfer passive immunity from the mother to the offspring (Weström et al. 2020). Following the production of colostrum, the mammary secretion gradually transitions into mature milk. The composition and functionality of colostrum and milk are species-specific. With human milk constituting the golden standard to which; e.g. bovine milk-based infant formula is designed, it is important to study and understand bovine milk at the molecular level, to investigate how it differs from human milk, and how it can be used to as closely as possible match its nutritional and functional qualities. While the protein fraction of human colostrum is made up by up to 20 percent by immunoglobulins, the majority of which consist of secretory IgA (sIgA) (Atyeo and Alter 2021), bovine colostrum has much higher proportions of immunoglobulins, i.e. up to 80 percent of total protein, consisting primarily of IgG (Marnila and Korhonen 2011). In the last 2 to 3 weeks before calving, plasma levels of IgG, particularly IgG1, decrease in the cow; at the same time, IgG1 is thought to be actively transported and accumulates into the mammary gland, with maximum transfer occurring 1–3 days before calving (Delouis 1978). The active transfer of IgG and serum proteins into bovine milk greatly diminishes and ultimately ceases over the first days after calving (Delouis 1978), but IgG remains present in the bovine milk throughout the entire lactation period, presumably through passive transfer or local production in the mammary gland.
IgG contains an N-glycosylation motif in the CH2 domain of the heavy chain, which is conserved between the different mammalian species (Raju et al. 2000). Protein glycosylation is a common and complex post-translational modification that controls many biological pathways (Schjoldager et al. 2020). Glycosylation can determine protein folding, stability, function, localization, trafficking, and half-life (Schjoldager et al. 2020). Furthermore, the diversity and complexity of glycosylation can be a hallmark of health or disease (Reily et al. 2019), pregnancy (Jansen et al. 2016), and lactation (Goonatilleke et al. 2019).
Common methodologies for studying bovine IgG glycosylation include lectin microarray profiling (Yu et al. 2020), the hydrolysis of glycans and quantification of monosaccharides (Feeney et al. 2019), and the release and analysis of intact glycans by mass spectrometry (van Leeuwen et al. 2012). Each of these strategies, however, has its limitations. Lectin microarray profiling can recognize and quantify glycan features (Hirabayashi et al. 2013), such as glycan type (i.e. complex, hybrid or high-mannose), glycan substructures or types of linkages in glycan substructures, without providing information on glycan composition, complete structure and localization in the protein. Quantification of the monosaccharides gives an average picture of the sample glycome (Rohrer et al. 2013), but does not provide specific information on the individual glycoforms and their localization on the proteins. Analysis of the released glycans does provide information on the composition, heterogeneity, and structure of the glycoforms (Rojas-Macias et al. 2019), but does not indicate which protein or where on the protein they originate from, and what the occupancy of the glycosylation sites on the proteins is. Emerging technologies in the constantly developing field of glycoproteomics have made possible the analysis of glycopeptides by mass spectrometry, providing compositional and structural information on both the peptide and the glycan, site localization of the glycan, and occupancy of the glycation site (Hinneburg et al. 2016; Reiding et al. 2018; Ruhaak et al. 2018).
Here, we designed a study to investigate how bovine milk IgG changes longitudinally during lactation in relation to the rest of the mammary secretion proteome. While the decrease in abundance of IgG during the first days of lactation is well documented, we combined careful experimental design and state-of-the-art glycoproteomics technology for the in-depth investigation of bovine milk IgG N-glycosylation. Next to the dynamic changes during lactation, we studied the samples of several individual cows to assess how variable the findings are between animals. We observed a considerable decrease of IgG abundance during the first days following calving, and a further gradual decrease towards the end of the first month post-partum. Next to the better-documented IgG1 and IgG2, we report here, for the first time to our knowledge, the identification of subtype IgG3 in the bovine milk of all four cows and at every investigated time point. We found bovine milk IgG glycosylation to be highly diverse and complex, seemingly much more so than human IgG glycosylation, and we identified biological features of the bovine milk IgG N-glycosylation, such as NeuAc sialylation, that are a hallmark of bovine colostrum, greatly diminishing in mature bovine milk.
Results
To study the longitudinal changes in the bovine milk proteome, IgG relative abundances, and N-glycosylation, we analyzed a total of 39 individual bovine milk samples collected from 4 individual Dutch Friesian-Holstein cows referred to as Cow 1, Cow 2, Cow 3, and Cow 4. The time scale of the bovine milk sampling is schematically illustrated in Fig. 1A.
Fig. 1.
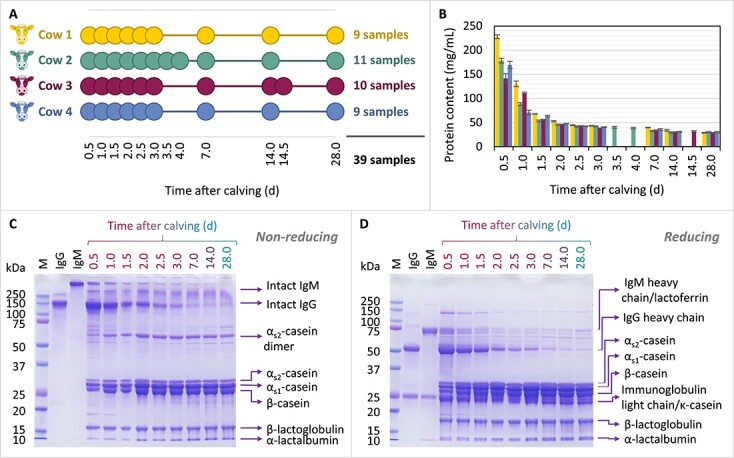
Insights into the protein content and composition of the bovine milk samples collected during lactation from the four individual cows, i.e. cow 1, cow 2, cow 3 and cow 4. (A) Chart depicting the sampling time points for each individual cow following calving. (B) Total protein content of the bovine milk samples, determined using a bicinchoninic acid (BCA) assay. The error bars represent the standard deviation of triplicate measurements. The colour codes are consistent between panels (A) and (B), and are specific for each individual cow. (C) and (D) SDS-PAGE gel images of the samples collected from cow 4 analyzed under non-reducing (C) and reducing (D) conditions. All bovine milk samples were loaded on the gel at 20 μg protein/well. The first 3 gel lanes display a protein marker panel: M = precision plus protein™ dual color standards, IgG = bovine serum IgG, IgM = bovine serum IgM, the latter two loaded on the gel at 4 μg protein/well.
Protein content and 1D gel electrophoresis of the bovine milk samples
The protein content of all collected samples was determined by the BCA method, and the samples were further analyzed by SDS-PAGE to gain a first overview of changes occurring during lactation in the bovine milk proteome. Figure 1B illustrates the longitudinal changes in protein content between the four individual cows. Considerable decreases in protein content occurred during the first few days of lactation in all four cows, with the most notable change occurring between the first two sampling time points. Substantial differences in the protein content between the individual cows were only observed in the first two days of lactation, after which the protein content of all samples stabilized to comparable levels for the rest of the sampled time points. The highest protein content was observed in bovine colostrum in the first sampled time point, i.e. 0.5 d, at (mean ± standard deviation) 228.0 ± 3.8 mg protein/mL for Cow 1, followed by 178.5 ± 4.8 and 169.4 ± 7.4 mg/mL for Cow 2 and Cow 4, respectively, and 141.5 ± 9.9 mg/mL for Cow 3. For all four cows, the protein content of the mature bovine milk stabilized to 28.7–30.2 mg/mL at 28 d after calving.
Next to the marked decrease in protein concentration during lactation, we also found considerable longitudinal changes in the protein composition of the samples. Bovine milk samples were separated on SDS-PAGE gels, from which a gradual shift in composition could be observed, as illustrated in Fig. 1C and D for the samples collected from Cow 4. At 0.5 d, the first sampled time point, the proteome was dominated by immunoglobulins, IgG making up a considerable proportion thereof, but this changed to a casein-dominated proteome over the course of approximately a week. While Fig. 1 shows the changes in protein composition for one cow (Cow 4), similar changes were seen in the bovine milk of the other three cows (Supplementary Fig. S1).
Bovine milk proteome composition
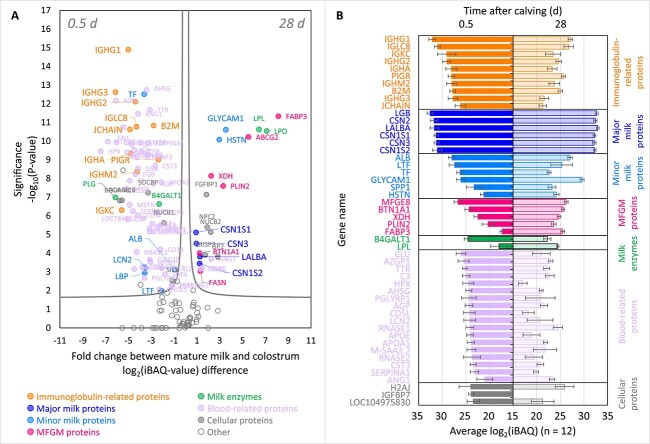
We next analyzed each individual sample by bottom-up proteomics, performing for each sample triplicate LC–MS/MS methods, accumulating to a total of 117 runs. This approach resulted in the identification of 212 individual bovine milk protein sequences across all samples analyzed. The abundances of the identified proteins were determined using label-free intensity Based Absolute Quantification (iBAQ) value. Some of the most notable differences between the bovine colostrum proteome at 0.5 d and the mature bovine milk proteome at 28 d, averaged over all cows, are depicted in Fig. 2A and B. The source data behind these graphs can be found in Supplementary Table S1. The volcano plots and bar charts per individual cows are provided in Supplementary Figs. S2 and S3, respectively.
Fig. 2.
Comparison between the proteomes of the samples collected at 0.5 and 28 d after calving averaged across all four cows, as determined by bottom-up mass spectrometry. (A) Volcano plot depicting the proteins with significantly different abundances in the proteomes of samples collected at 0.5 and 28 d after calving. Similar volcano plots are shown for each individual cow in Supplementary Fig. S2. (B) Differences in intensity based absolute quantification abundance (log2(iBAQ)) between the 50 most abundant protein sequences present in the proteomes of samples collected at 0.5 (left) and 28 (right) d after calving. For the selection of the 50 most abundant protein sequences, firstly the highest iBAQ value was selected for each individual protein sequence between the iBAQ values at 0.5 and 28 d. Next, the protein sequences with the 50 highest iBAQ values were established as the 50 most abundant proteins of bovine colostrum and mature milk proteomes combined. The error bars indicate the standard deviation of each protein sequence iBAQ value measured in 12 samples (i.e. triplicate measurements of samples from four cows). The colour codes highlighting the different protein sequence categories are consistent between panels (A) and (B). Similar bar charts are shown for each individual cow in Supplementary Fig. S3.
The major findings of the dynamic bovine milk proteome determined by proteomics proved to be in line with the results from SDS-PAGE (Fig. 1C and D, and Supplementary Fig. S4). All immunoglobulin-related proteins were found to be very abundant in the bovine colostrum, and significantly decreased in abundance during lactation. Next to the immunoglobulins, a majority of blood serum-related proteins were found to be significantly more abundant in the bovine colostrum than in the mature bovine milk. This included minor bovine milk proteins such as serotransferrin (TF), lactoferrin (LTF) and serum albumin (ALB), and endogenous bovine milk enzymes, such as plasmin(ogen) (PLG) and β-1,4-galactosyltransferase 1 (B4GALT1). Unlike the IgG1-rich bovine colostrum, the proteome of the mature bovine milk was dominated by the six major bovine milk proteins, i.e. the four caseins, α-lactalbumin (LALBA) and β-lactoglobulin (BLG). Alongside the six major bovine milk proteins, minor proteins such as lactophorin (GLYCAM1) and histatherin (HSTN), the majority of milk fat globule membrane (MFGM) proteins, and milk enzymes lipoprotein lipase (LPL) and lactoperoxidase (LPO), all showed significant increases in abundance in mature bovine milk. For a complete picture of the proteome changes at each time point during lactation and for each individual cow, please consult the heatmap in the Supplementary Fig. S4 and the source data provided in Supplementary Table S1.
Having identified immunoglobulin classes IgG, IgM, IgA as key components of the bovine colostrum proteome, Fig. 3A and B provides more detailed insight into their abundances during lactation. The abundance of the immunoglobulins relative to the rest of the bovine milk proteome was found to decrease up to ~50 times during lactation from 0.5 to 28 d after calving (Fig. 3A). Despite the decrease in concentration of the immunoglobulins during lactation, the relative ratios of the different immunoglobulins (i.e. IgG, IgM, IgA) to each other remained alike, showing higher similarity within each cow than between the four individual cows (Fig. 3B). While longitudinal and individual differences could be observed in the relative ratios of the different immunoglobulins to each other, the bovine milk immunoglobulin repertoire shared many similarities between all samples: IgG was found to be the dominant isotype, followed by IgM and IgA. All three bovine IgG subtypes (i.e. IgG1, IgG2, and IgG3) were detected in all of the analyzed samples, with IgG1 always showing the highest abundance, followed by IgG2, and to a much smaller extent also by IgG3;, this protein has not been previously reported to be present in bovine milk. The data shown in Fig. 3 also display that not all cows have a similar milk immunoglobulin profile, whereby in particular the Cow 4 appears to represent an outlier, with relatively high levels of IgG2, IgM, and IgA in the mature bovine milk.
Fig. 3.
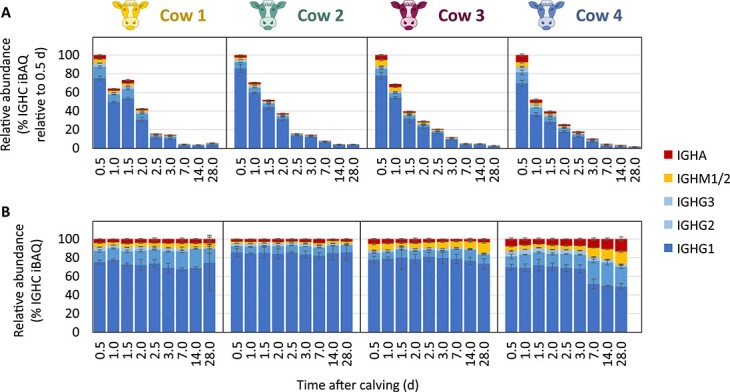
Changes in the relative abundances (% iBAQ) of IGHG1 (dark blue), IGHG2 (medium blue), IGHG3 (light blue), IGHM1/2 (yellow), and IGHA (red) in the bovine milk proteome of cow 1, cow 2, cow 3 and cow 4, between 0.5 and 28 d after calving. (A) The decrease of immunoglobulin concentration during lactation is shown by expressing heavy chain constant abundances during lactation relative to those in the first sampled time point. (B) Depicted are the relative abundances of the heavy chain constant regions to each other, irrespective of their abundance in the bovine milk proteome. The error bars represent the standard deviations of triplicate measurements.
N-glycosylation of IgG captured from bovine milk
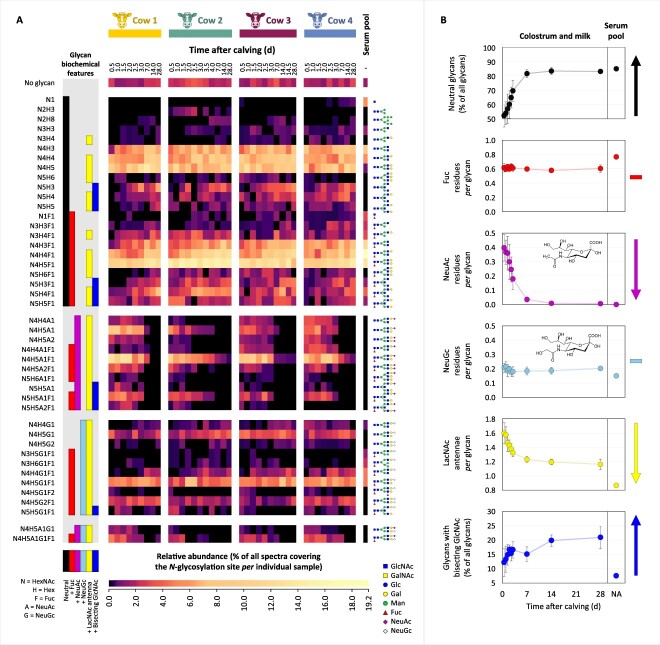
The data above show that the immunoglobulins, and particularly IgG, represented to a large extent the major differences between the proteome of bovine colostrum and mature milk. To further investigate whether not only the total concentration, but also the molecular characteristics, of IgG varied over lactation, we next affinity-captured IgG from all the bovine milk samples by making use of Protein G-based enrichment. Isolated IgG was subsequently analyzed by bottom-up mass spectrometry techniques employing product ion-dependent hybrid fragmentation methods optimized for the analysis of glycopeptides (Reiding et al. 2018; Dingess et al. 2021). The analysis of captured IgG from 39 samples and one bovine serum pool IgG using triplicate LC–MS/MS measurements amounted to a total of 120 runs, resulting in the identification of a repertoire of 44 different N-glycoforms occurring on the CH2 domain of IgG, identified across all samples, time points and cows. The heatmap in Fig. 4A depicts the combined information of glycan macro- and microheterogeneity, determined based on spectral counts for the N-glycosylation site in the CH2 domain of IgG from each individual sample.
Fig. 4.
Changes in the N-glycosylation profiles of bovine IgG in the interval 0.5–28 d after calving in the bovine milk of the four cows, i.e. cow 1, cow 2, cow 3 and cow 4. Additionally, in the last column, for comparison, the N-glycosylation profile of bovine serum IgG is depicted (pooled sample). (A) Heatmap depicting the macro- and microheterogeneity of the IgG CH2 domain N-glycosylation determined based on spectral counts. Normalization was performed relative to all spectra covering the glycosylation site, whereby the sum of all glycoforms and the non-glycosylated site amounts to 100 percent, and the relative quantitation of the non-glycosylated site indirectly indicates site occupancy. Clustering was performed based on the biochemical features of the N-glycans: neutral (black), fucosylated (red), sialylated with NeuAc (magenta), sialylated with NeuGc (light blue), containing LacNAc antennae (yellow) or bisecting GlcNAc (blue), respectively. The glycan composition is indicated to the left of the heatmap. To the right of the heatmap are proposed glycan structures corresponding to each glycan composition. (B) Illustrative lactation dynamics of the N-glycan biochemical features averaged across the four cows and compared to their corresponding level in the bovine serum IgG, with the panels from top to bottom describing the changes in neutral glycans, fucosylation, sialylation with NeuAc or NeuGc, containing LacNAc antennae or bisecting GlcNAc, respectively. The error bars represent the standard deviation of values from the four cows. The colour codes of the biochemical features are consistent between panels (A) and (B). Abbreviations: Gal = galactose; Glc = glucose; man = mannose; hex = hexose; GalNAc = N-acetylgalactosamine; GlcNAc = N-acetylglucosamine; HexNAc = N-acetylhexosamine; Fuc = fucose; NeuAc = N-acetylneuraminic acid; NeuGc = N-glycolylneuraminic acid.
Very high similarity was observed between the corresponding samples of the individual cows. The IgG N-glycosylation site was found to be occupied to an average proportion of 98 percent, mostly with complex N-glycans, although a very small proportion of non-glycosylated peptides occurred in all samples as well. The N-glycans were clustered in Fig. 4A based on their biochemical features, with Fig. 4B showing the changes thereof across lactation. Representative spectra of the non-glycosylated peptide, and glycopeptides exhibiting each selected biochemical feature, were manually inspected and are shown in Supplementary Figs. S5-S12. Of all IgG N-glycans in bovine colostrum at 0.5 d after calving, 48 percent (based on spectral count) were found to be sialylated, with the remaining 52 percent corresponding to neutral glycans. Preliminary screening of the raw files revealed strong oxonium ion evidence for the occurrence of sialylation with N-acetylneuraminic acid (NeuAc) and N-glycolylneuraminic acid (NeuGc), but not for the occurrence of acetylated sialic acid residues, or of deaminated neuraminic acid (Kdn) (Supplementary Fig. S15). Interestingly, sialylation with NeuAc, NeuGc, and also with the combination of NeuAc and NeuGc within the same glycan, were all detected in the bovine milk samples, particularly in the first days following parturition. NeuAc sialylation was found to be an exclusive characteristic of bovine colostrum, decreasing to nearly undetectable levels in mature bovine milk (Fig. 4B, third panel from the top). Comparably to the mature bovine milk IgG, no NeuAc sialylation was detected in the IgG sample acquired from pooled bovine serum. The level of NeuGc sialylation, on the other hand, remained fairly constant throughout lactation (Fig. 4B, fourth panel from the top). As a potential consequence of the decrease in NeuAc sialylation, the proportion of neutral glycans increased during lactation, reaching an average level of 83 percent (based on spectral count) in mature bovine milk samples. Fucosylation was present among both neutral and sialylated glycans, and remained relatively constant across time (Fig. 4B, second panel from the top). Branching of the complex N-glycans occurred with LacNAc antennae. The average number of LacNAc antennae per glycan gradually decreased from 1.60 ± 0.14 in bovine colostrum to 1.16 ± 0.07 in the mature bovine milk (Fig. 4B, fifth panel from the top). This appeared to be the result of the decrease in complexity and branching of the glycans during lactation. A small proportion of glycans with bisecting GlcNAc was also observed, whose abundance increased during lactation (Fig. 4B, sixth panel from the top). The N-glycosylation of the pooled bovine serum IgG was found to most closely resemble that of the mature bovine milk, and exhibited less resemblance to that of bovine colostrum or transitional bovine milk.
Discussion
In this study, we monitored the changes in bovine milk proteome composition, with emphasis on IgG, following its content and biochemical properties during lactation. Changes occurred most dramatically within the first days after calving, followed by a more gradual transition from bovine colostrum to mature bovine milk.
Changes in the bovine milk proteome during lactation converge around the immunoglobulins
The six main bovine milk proteins, i.e. the four caseins, α-lactalbumin and β-lactoglobulin (Gazi et al. 2022), were found to be abundant at all analyzed time points (Fig. 2, Supplementary Fig. S4). Next to these well-known constituents, and in line with the literature (McGrath et al. 2016), we found several immunoglobulins and related proteins, making up a great proportion of in particular the bovine colostrum proteome. Amongst the immunoglobulin-related proteins, we highlighted in our results (Fig. 2, Supplementary Fig. S4) the heavy and light chains of immunoglobulins IgG, IgA and IgM, the immunoglobulin joining chain (JCHAIN), the polymeric immunoglobulin receptor (PIGR), and β2-microglobulin (B2M). We found all these proteins to be very abundant in bovine colostrum and significantly decreased during lactation. The JCHAIN plays a role in the oligomerization of IgM and IgA, leading to the formation of IgM pentamers and IgA dimers, trimers and tetramers (Johansen et al. 2000; Dingess et al. 2022). PIGR is the precursor of the secretory component, the protein whose role it is to transport polymeric immunoglobulins across epithelial barriers (Mostov 1994), and in this case, into the bovine milk. B2M, initially isolated from bovine milk as the minor whey protein lactollin (Groves et al. 1963; Groves and Greenberg 1977), is the light chain component of the neonatal Fc receptor (FcRn) heterodimer that binds, transports, and protects IgG and serum albumin from degradation (Baumrucker et al. 2022). The purpose of its presence in bovine milk, particularly at higher concentration in the bovine colostrum, is as of yet uncertain. It can be speculated that B2M is shed from the FcRn heterodimer into bovine milk following transcytosis of IgG via the mammary secretory epithelial cells. It is unlikely that B2M from bovine milk plays a role in the further transcytosis of bovine milk IgG from the calf’s intestine into its blood circulation, considering that endogenous B2M is necessary for the correct folding and surface expression of the FcRn heterodimer in the intestinal epithelial cells (Praetor and Hunziker 2002). Next to the FcRn/B2M complex prolonging the half-life of circulatory IgG, it has also been shown that B2M alone also exhibits a protective effect on IgG (Kim et al. 2008). The role of B2M in bovine colostrum and bovine milk may therefore be to protect the passive immunity the calf receives from the cow, until such time that the calf’s own immune system is sufficiently developed.
The bovine milk immunoglobulome was dominated by IgG, which we found to make up 51–73 percent of the total protein content of bovine colostrum in the first bovine milking (Supplementary Fig. S13B), in line with previous reports (Marnila and Korhonen 2011). Conversely to bovine milk, the immunoglobulin repertoire of human milk is dominated by secretory IgA (sIgA), with IgG constituting only a minor component human milk (Atyeo and Alter 2021; Dingess et al. 2022). The placental barrier is species-specific and determines whether immune transfer to the fetus can occur in utero through the placenta (Chucri et al. 2010). In humans, prenatal transfer of IgG from the mother via the placenta ensures that neonates have systemic immunity at birth (Renegar 2005). Due to the high number of placental barrier layers in ruminants, including bovine, prenatal transfer of IgG is not possible (Chucri et al. 2010). Consequently, calves are born agammaglobulinemic, and rely on the IgG from bovine colostrum for survival (Godden et al. 2019). The IgG from bovine colostrum is therefore meant to be taken up into circulation and provide the calf with passive systemic immunity, explaining the dominant position of IgG at the expense of sIgA in the bovine milk immunoglobulome that we described in our results (Fig. 3). Unlike bovines, humans do not need to transfer passive systemic immunity to the neonate through milk. This leads to the human milk immunoglobulome to be dominated by sIgA, an immunoglobulin active in the neonate’s intestines, providing passive mucosal immunity (Renegar 2005).
The transfer of milk immunoglobulins from the offspring’s gut into its circulation ceases at the time of gut closure. This takes place 2–3 days after birth, after the calf has consumed the IgG-rich bovine colostrum essential for survival in its first days of life (Weaver et al. 2000). In the case of humans, gut closure occurs before birth (Weström et al. 2020). Consequently, neonates lack the ability to absorb milk IgG into circulation, thereby also explaining the low IgG levels present in human milk (Atyeo and Alter 2021).
While bovine milk IgG and human milk sIgA are designed for different purposes, as described above, the consumption of bovine milk IgG by infants may still provide health benefits comparable to the human milk sIgA (Ulfman et al. 2018). Considering that bovine milk IgG can also not be absorbed into circulation due to the pre-natal infant gut closure (Weström et al. 2020), its function in human infant nutrition may instead be that of passive mucosal immunity in the intestine, as in the case of the human milk sIgA.
The secretory component in sIgA (i.e. derived from the PIGR precursor shown in Fig. 2, Supplementary Fig. S4), next to ensuring transport of the IgA across the epithelial barrier into milk, also protects the IgA from proteolytic degradation in the gastrointestinal tract (Lindh 1975). While IgG lacks the secretory component, bovine IgG is less susceptible to proteolytic degradation by digestive proteases than its human counterpart (Burton et al. 2020). Furthermore, considering the infant’s immature and developing digestive system (Bourlieu et al. 2014), recovery of bovine IgG from infant feces has been shown to be high (Jasion and Burnett 2015), confirming its high resistance to proteolytic degradation.
Changes in bovine IgG N-glycosylation during lactation converge around loss of NeuAc sialylation
The IgG N-glycosylation repertoire detected here reveals that bovine milk IgG glycosylation is more diverse and heterogeneous than that of its human milk IgG counterpart, although both N-glycosylation sites are located in conserved regions of their respective CH2 domains. While we revealed here a diverse repertoire of 44 glycoforms co-occurring on bovine milk IgG between 0.5 and 28 d post-partum (Fig. 4), Zhu et al. (2020), using comparable LC–MS/MS approaches identified only 6 glycoforms on human milk IgG analyzed between 1 and 16 weeks post-partum. These differences may in part result from differences in sample preparation; i.e. we here analyzed bovine milk IgG glycosylation in affinity-captured IgG, whereas Zhu et al. (2020) analyzed glycopeptides enriched from a whole human milk proteolytic digest. Other studies, such as the one of Trbojević Akmačić et al. (2015), have shown that the human IgG glycosylation repertoire can also be quite diverse. However, conceptually, the number of bovine IgG glycoforms can be higher due to the presence of NeuGc and its combinations with NeuAc. All six human milk IgG glycoforms identified by Zhu et al. (2020) contained core fucosylation, whereas we determined here the bovine milk IgG to contain an average of 0.6 fucose residues per glycan at all investigated lactation time points (Fig. 4B, second panel from the top). Zhu et al. (2020) found a single sialylated glycoform, i.e. N4H5A1F1, making up 21 percent of the abundance of all glycoforms at 1 week, and gradually decreasing to 12 percent at 16 week. In terms of similarities shared with the bovine IgG glycosylation, all 6 human milk N-glycoforms are amongst the most abundant ones in the repertoire of 44 bovine glycoforms (Fig. 4). The physiological reason for the differences between the inter-species diversity of glycoforms is still unknown. However, we speculate they may relate to the different functionality of the bovine milk IgG between the two species. With IgG only constituting a minor component of the human milk immunoglobulome (Atyeo and Alter 2021), and with infant gut closure having occurred prior to birth (Weström et al. 2020), it is unlikely that IgG is actively transported into human milk. Conversely, bovine milk IgG is actively transported into the bovine colostrum (Delouis 1978), it needs to be resistant to the digestive proteases in the calf’s gastrointestinal tract, it needs to be actively transported from the calf’s intestine into its blood circulation (Godden et al. 2019), and it needs to have a sufficiently long half-life in the calf’s circulation to provide passive immunity while the calf can develop its own system immunity. Therefore, it can be speculated that the high diversity and complexity of the bovine milk IgG glycoforms (Fig. 4A), and the differences in biochemical features of the glycans in bovine colostrum from mature bovine milk (Fig. 4B), are correlated with the binding affinity of the FcRn receptors that need to transport and protect the IgG from degradation, and also determine the proteolytic susceptibility of the IgG.
A very interesting feature of the bovine milk IgG glycosylation found was the fact that NeuAc and NeuGc sialylation occurred simultaneously in the samples, at times even both on the same glycoprotein (Fig. 4, Supplementary Fig. S12). We found here NeuAc sialylation to be a unique hallmark of bovine colostrum IgG, with the NeuAc/NeuGc ratio decreasing from 1.91 in the 0.5 d colostrum to 0.03 in the 28 d mature milk. This is interesting, particularly from a human nutrition perspective. While being a widespread and abundant sialic acid in the glycoconjugates of most mammalian species, NeuGc cannot be produced by the human body and is therefore a non-human sialic acid (Irie et al. 1998). Consequently, NeuGc is immunogenic to humans, with anti-NeuGc antibodies detectable even in healthy individuals (Zhu and Hurst 2002). The high NeuAc sialylation of the particularly abundant IgG in bovine colostrum renders its glycosylation more human-like, making bovine colostrum IgG furthermore interesting from the perspective of biological functionality in human nutrition. The function of high sialylation of bovine colostrum IgG is hypothesized to be the extension of the IgG half-life; non-sialylated proteins are removed from circulation by binding to asialoglycoprotein receptors (ASGRs) in the liver and are subsequently directed to degradation (Stockert 1995). The ASGRs have a high affinity for glycan-terminal galactoses in the absence of sialic acids (D'souza and Devarajan 2015). While ASGRs are primarily present in the liver, Stockert (1995) reported their presence to a lower extent also in the gastro-intestinal tract of rats. Species-specific differences may exist in the distribution and binding affinity of the ASGRs. However, by similarity it can be speculated that the higher sialylation of bovine colostrum IgG is meant to extend its half-life by protecting it from ASGR recognition in the gastro-intestinal tract and circulation of the calf.
Our research lays the foundation for future investigations into the biological functionality of the here described heterogeneity, diversity and dynamic lactational changes in bovine milk IgG N-glycosylation. These findings are not only important from the perspective of the healthy development of the calf’s own immunity in the context of bovine animal health management, but also from the perspective of understanding the bioactivity of bovine milk components on human health, and particularly the potential role of bovine milk IgG in e.g. providing passive mucosal immunity to formula-fed infants.
Materials and methods
Chemicals and reagents
Bovine serum IgG (I5506) and IgM (I8135), enriched from pooled bovine serum, dithiothreitol (DTT), Tris(2-carboxyethyl)phosphine (TCEP), Tris, chloroacetamide (CAA), sodium deoxycholate (SDC) and formic acid (FA) were purchased from Sigma-Aldrich (Darmstadt, Germany). The Pierce BCA Protein Assay Kit (23225) for the bicinchoninic acid (BCA) assay, Imperial Protein Stain (24615), Pierce Spin Columns (69705), Dulbelcco’s phosphate-buffered saline (DPBS), Pierce IgG elution buffer (1856202) and neutralization buffer (1856281) were sourced from Thermo Scientific, Rockford, Illinois, USA. Trifluoroacetic acid (TFA) was purchased from Fisher Scientific (Landsmeer, The Netherlands). PepMap Trap Cartridges (5 μm C18 300 μm × 5 mm) were sourced from Thermo Fisher Scientific (Germering, Germany). XT sample buffer, 18-well 12 percent Criterion™ XT Bis-Tris Precast Gels and XT MOPS running buffer and Precision Plus Protein™ Dual Color Standards used for sodium-dodecyl sulphate – polyacrylamide gel electrophoresis (SDS-PAGE) were all purchased from Bio-Rad (Veenendaal, The Netherlands). Ultrapure MilliQ water was prepared with the system sourced from Merck Millipore, Darmstadt, Germany. Sequencing-grade endoproteinase GluC and broad range protease inhibitor cocktail (cOmplete, EDTA-free) were sourced from Roche Diagnostics (Mannheim, Germany).
Sequencing-grade trypsin was sourced from Promega (Madison, Wisconsin, USA). Sequencing-grade lysyl endopeptidase (LysC) was sourced from Wako Chemicals (Richmond, Virginia, USA). Oasis PRiME HLB 96-well plates (10 mg sorbent per well) for solid phase extraction (SPE) were sourced from Waters (Etten-Leur, The Netherlands). C18 column material (Poroshell 120 EC-C18, 2.4 μm) was sourced from Agilent Technologies (Amstelveen, The Netherlands). Protein G slurry (Protein G Sepharose 4 Fast Flow, GE17–0618-02) was sourced from GE Healthcare (Chicago, Illinois, USA).
Bovine milk samples
Bovine milk samples from four individual Dutch Friesian-Holstein cows, i.e. referred to as Cow 1, Cow 2, Cow 3, and Cow 4, were obtained from a farm in The Netherlands. Sampling started within 12 h after calving and continued up until four weeks after calving, with samples taken at 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 7.0, 14.0, and 28.0 d post-partum, as indicated in Fig. 1A. Additional samples were taken from Cow 2 at 3.5 and 4.0 d, and Cow 3 at 14.5 d post-partum. Calving dates of all four cows were in the interval of 24–28 January 2022. Milking was carried out twice daily at fixed times. The collected samples were stored at −20 °C until further analyses. The samples collected up until 3 d will be further referred to as bovine colostrum, samples collected at 28 d as mature bovine milk, and samples collected in between 3 and 28 d as transitional bovine milk.
Protein content and gel electrophoresis of bovine milk samples
The protein content of the bovine milk samples was determined by the BCA assay using the Pierce BCA Protein Assay Kit and 96-well plate method, according to manufacturer’s instructions. SDS-PAGE under both reducing (with DTT) and non-reducing (without DTT) conditions was performed as previously described (Gazi et al. 2022). The bovine milk samples, and the bovine serum IgG and IgM, were treated with XT sample buffer, loaded onto 18-well 12 percent Criterion™ XT Bis-Tris Precast Gels, and electrophoresis was run in XT MOPS running buffer. The bovine milk and immunoglobulin samples were loaded at 20 and 4 μg protein/well, respectively. Precision Plus Protein™ Dual Color Standards were run on the gels in parallel with the samples for protein size reference. The gels were stained with Imperial Protein Stain according to manufacturer’s instructions, and destained in ultrapure MilliQ water.
Capturing IgG from the bovine milk samples
For capturing IgG from bovine milk, the milk samples were warmed for 5 min at 37 °C. Broad range protease inhibitor cocktail was added at a ratio of 1 tablet to 23 mL of bovine milk. Sodium azide (NaN3) was added to a final concentration of 0.02 percent to the bovine milk samples. Skimming of the bovine milk samples was performed by centrifugation at 1500 × g and 37 °C for 10 min. Volumes of 100 μL of diluted Protein G slurry were packed into Pierce Spin Columns. The columns were conditioned with DPBS. Volumes of 10, 20, 50, 150, 250, 350, 400, and 450 μL of skim bovine milk were loaded from lactation time points 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, and 4.0 d, respectively. DPBS was added to a total skim bovine milk + DPBS volume of 700 μL. The bovine milk samples from 7–28 d after calving were loaded directly at the volume of 700 μL. The bovine milk/Protein G slurries were gently mixed on a rotary shaker for 1 h at room temperature. Following capturing of the IgG, the beads were washed two times with 150 μL of DPBS, and one time with ultrapure MilliQ water. The captured IgG was then eluted with Pierce IgG elution buffer and treated with neutralization buffer. The samples were stored at 4 °C until further analysis. The protein content of the captured IgG samples was determined spectrophotometrically using a NanoDrop 2000 instrument (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at 280 nm with the pre-set method for IgG quantification. The capturing efficiency was corrected based on the capturing from the bovine serum IgG reference, and the IgG concentrations were calculated back for every bovine milk sample. To verify the composition of the captured sample material, the samples were analyzed by non-reducing SDS-PAGE as described above. Bovine serum IgG was used as reference, and all samples were loaded at 2 μg protein/well.
Analysis of bovine milk proteome and captured IgG CH2 N-glycosylation by mass spectrometry
Amounts of 10 μg of protein from each whole bovine milk sample, captured IgG from bovine milk, and bovine serum IgG were denatured, reduced and alkylated in a buffer containing 10 mM TCEP, 100 mM Tris, 40 mM CAA and 1 percent (m/v) SDC, as previously described by Gazi et al. (2022). The whole bovine milk protein samples were digested with a ratio of 1:50 (m/m) GluC:protein for 4 h at room temperature, followed by the addition of 1:50 (m/m) trypsin:protein and overnight digestion at 37 °C. Digestion of the IgG samples was performed overnight at 37 °C with a mixture of 1:70 (m/m) LysC:protein and 1:50 (m/m) trypsin:protein. The enzymatic reactions were stopped, and SDC was precipitated by adjusting the sample pH to the range of 1.5–2.0 with 10 percent TFA. The peptides were extracted by SPE using Oasis PRiME HLB 96-well plates according to the instructions of the manufacturer. Following extraction, the samples were dried using a vacuum concentrator and stored at −20 °C until further analysis.
The dried samples were reconstituted in 2 percent FA, and amounts of 800 ng of digested whole bovine milk protein or 100 ng digested IgG were injected per run. The samples were analyzed using an Ultimate 3000 UHPLC system (Thermo Fisher Scientific, Germering, Germany) coupled online to an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Reversed-phase separation was achieved using PepMap Trap Cartridges and C18 analytical columns (50 cm length, 50 μm inner diameter). Mobile-phase solvent A consisted of 0.1 percent FA in water, and mobile-phase solvent B consisted of 0.1 percent FA in 80 percent ACN. Total method runtime for the whole bovine milk protein digests was 120 min at a flow rate of 300 nL/min, where the peptides were loaded at 9 percent B, followed by 1 min ramp from 9 to 13 percent B, the elution of the peptides during a 100 min linear gradient of 13–44 percent B, 3 min ramp from 44 to 99 percent B, 4 min washing of the column with 99 percent B, 1 min decrease from 99 to 9 percent B and 10 min equilibration to 9 percent B. The IgG digests were analyzed by a similar method, with the following modifications: 60 min total method run time and 40 min gradient elution time. MS scans were recorded at a resolution of 120,000, with an automatic gain control (AGC) target of 400,000, 50 ms maximum injection time (IT) and a scan range of m/z 350–2,000. Data-dependent MS2 scans were recorded at a resolution of 60,000, AGC target of 50,000, 50 ms maximum IT and a scan range of m/z 120–4,000.
Each sample was injected three times with identical chromatography and MS method, but with different MS/MS methods; this provided triplicate information at precursor level, and complementary results at fragmentation level. The different fragmentation methods consisted of (i) higher-energy collisional dissociation (HCD) at a normalized collision energy of 29, (ii) oxonium ion-triggered stepped HCD at normalized collision energy steps of 10, 25 and 40 percent, (iii) oxonium ion-triggered electron-transfer/higher-energy collisional dissociation (EThcD). The oxonium ion-triggered hybrid fragmentation methods favoured the fragmentation of specifically glycopeptides. The list of oxonium ions used for triggering hybrid fragmentation is provided in Supplementary Table S2.
The mass spectrometry raw data and complete Byonic search results of the glycoproteomics analyses on the captured IgG have been deposited to the ProteomeXchange Consortium (Deutsch et al. 2020) via the PRIDE (Perez-Riverol et al. 2022) partner repository with the dataset identifier PXD037755.
Protein database optimization, database search and identification of bovine milk proteome
For optimal results, and overcoming the limitations of incomplete, poorly annotated and highly redundant bovine protein databases, a customized protein database was created for the searches. The one protein sequence per gene version of the Bos taurus (taxon ID 9913) reference proteome (ID UP000009136) of 8 January 2022 was downloaded from UniProt (https://www.uniprot.org/proteomes/UP000009136) in FASTA format on 3 March 2022. This database was further processed by removing residual duplicate sequences (i.e. protein sequences with the same gene name), adding complete sequence where only fragment sequences were present, and updating the FASTA headers to the current headers of the date of download, i.e. 3 March 2022. Considering that the bovine immunoglobulins in UniProt were listed under the names “uncharacterized protein” or “Ig domain-containing protein”, functional bovine immunoglobulin heavy and light chain, constant and variable sequences were downloaded from the reference directory of the international ImMunoGeneTics information system (IMGT RefSeq, https://www.imgt.org/vquest/refseqh.html). The constant immunoglobulin heavy chain sequences were assembled from the respective fragment sequences of the constant heavy (CH) domains and hinge (H) regions. A single representative allele for each immunoglobulin gene sequence was kept in the working protein database. The contaminants FASTA file from the installation folder of the MaxQuant software was processed to remove all bovine contaminants. Database searches were performed on MaxQuant v 1.5.3.30 against the three databases described above combined, using default settings unless otherwise specified. The built-in contaminants feature of MaxQuant was deactivated. Digestion mode was set to specific against Trypsin/P and GluC, allowing a maximum of two missed cleavages. Methionine oxidation, protein N-terminus acetylation, and serine and threonine phosphorylation were searched as variable modifications. Cysteine carbamidomethylation was set as a fixed modification. Minimum peptide length was allowed at five amino acids, and maximum peptide mass was limited at 10,000 Da. Protein quantification was performed on a minimum of two unique + razor peptides. Label-free quantification was carried out using the intensity based absolute quantification (iBAQ) values. FTMS recalibration was activated. Following the database search, the protein groups table was further post-processed to remove non-bovine contaminants, reverse identifications, variable immunoglobulin sequences, proteins identified by less than two unique peptides, and proteins identified with an Andromeda score below 20. Supplementary Fig. S14 illustrates the shortcomings of commonly-used SwissProt and UniProt bovine protein databases, and the advantages of using our optimized bovine protein database.
Protein and glycan database optimization, database search and identification of IgG CH2 domain N-glycopeptides
The database search was performed with Byonic v4.5.2 (Protein Metrics, Cupertino, California, USA), a search engine specialized in the identification of glycopeptides. Considering the high apparent purity of the captured IgG samples based on the SDS-PAGE results (Supplementary Fig. S13), the samples were searched against a simplified database containing only the immunoglobulin sequences from IMGT RefSeq as described above. Decoys were added to the protein database. The N-glycan database used was based on the Byonic built-in database of 309 mammalian N-glycans. The default database was expanded to include complex and hybrid glycan antennae of N, N′-diacetyllactosamine (LacdiNAc) and all combinations of LacdiNAc and N-acetyllactosamine (LacNAc). To confirm which types of sialic acid to include into the glycan database, we verified MS2 chromatographic traces and spectra of relevant 0.5 d colostrum and 28 d mature milk samples from raw files resulting from methods employing higher-energy collisional dissociation. Supplementary Fig. S15 shows oxonium ion evidence for various sialic acids, including unmodified and acetylated N-acetylneuraminic acid (NeuAc), unmodified and acetylated N-glycolylneuraminic acid (NeuGc), and unmodified deaminated neuraminic acid (Kdn). The occurrence of NeuAc and NeuGc was undeniably confirmed, and they were found to overlap with oxonium ions derived from HexNAc residues. Acetylated residues and Kdn, however, could not be detected above noise levels. Based on this information, the database was expanded to include sialylation with both NeuAc and the non-human NeuGc, as well as all combinations thereof. The resulting database containing a total of 2440 N-glycan compositions that were used for identifying the IgG N-glycopeptides, is shown in Supplementary Table S3. Default search parameters were used, unless otherwise specified. Tryptic cleavage sites were defined C-terminal of arginine and lysine residues with two missed cleavages, but digestion specificity was set to non-specific. Fragmentation type was set either to HCD for the samples analyzed with HCD or product ion-triggered stepping HCD, or to both HCD & EThcD for the samples analyzed with product ion-triggered EThcD. Cysteine carbamidomethylation was set as a fixed modification. Methionine and tryptophan oxidation, and N-terminal cyclization of glutamine and glutamic acid to pyroglutamic acid were all searched as rare variable modifications, whereas N-glycosylation was searched as a common variable modification. A maximum of one rare and one common variable modifications were allowed per peptide. Protein false discovery rate (FDR) was set to 1 percent or 20 reverse counts. Further post-processing included filtering of the data based on |Log Prob| ≥ 1 and score ≥ 150. Based on the identified glycan composition, proposed glycan structures were built using GlycoWorkBench 2.1 build 146, according to the symbol nomenclature for glycan representation of the Consortium for Functional Glycomics (Varki et al. 2009).
Supplementary Material
Contributor Information
Inge Gazi, Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, University of Utrecht, Padualaan 8, 3584 Utrecht, CH The Netherlands; Netherlands Proteomics Center, Padualaan 8, 3584 Utrecht, CH The Netherlands.
Karli R Reiding, Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, University of Utrecht, Padualaan 8, 3584 Utrecht, CH The Netherlands; Netherlands Proteomics Center, Padualaan 8, 3584 Utrecht, CH The Netherlands.
André Groeneveld, FrieslandCampina, Stationsplein 4, LE, 3818 Amersfoort, The Netherlands.
Jan Bastiaans, FrieslandCampina, Stationsplein 4, LE, 3818 Amersfoort, The Netherlands.
Thom Huppertz, FrieslandCampina, Stationsplein 4, LE, 3818 Amersfoort, The Netherlands; Department of Agrotechnology and Food Sciences, Wageningen University, Bornse Weilanden 9, 6708 Wageningen, WG, The Netherlands.
Albert J R Heck, Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute for Pharmaceutical Sciences, University of Utrecht, Padualaan 8, 3584 Utrecht, CH The Netherlands; Netherlands Proteomics Center, Padualaan 8, 3584 Utrecht, CH The Netherlands.
Author contributions
I.G., T.H. and A.J.R.H. conceptualized the project; I.G. performed all experiments; I.G. performed data curation and analysis with support from K.R.R.; I.G. was responsible for visualization of the results and writing of the manuscript; all authors contributed to the interpretation of the findings and provided feedback for editing; A.J.R.H. and A.G. secured the funding for the project. All authors have read and agreed to the final version of the manuscript.
Funding
We acknowledge support from the Dutch Research Council (NWO) in the framework of the Innovation Fund for Chemistry (SATIN project 731.017.202). K.R.R. acknowledges support from NWO Veni project VI.Veni.192.058.
Conflict of interest statement. No conflict of interest to declare.
References
- Atyeo C, Alter G. The multifaceted roles of breast milk antibodies. Cell. 2021:184:1486–1499. [DOI] [PubMed] [Google Scholar]
- Baumrucker CR, Macrina AL, Bruckmaier RM. Colostrogenesis: Role and mechanism of the bovine fc receptor of the neonate (FcRn). J Mammary Gland Biol Neoplasia. 2021:26:419–453, 10.1007/s10911-021-09506-2. [DOI] [PubMed] [Google Scholar]
- Bourlieu C, Ménard O, Bouzerzour K, Mandalari G, Macierzanka A, Mackie AR, Dupont D. Specificity of infant digestive conditions: Some clues for developing relevant in vitro models. Crit Rev Food Sci Nutr. 2014:54:1427–1457. [DOI] [PubMed] [Google Scholar]
- Burton RE, Kim S, Patel R, Hartman DS, Tracey DE, Fox BS. Structural features of bovine colostral immunoglobulin that confer proteolytic stability in a simulated intestinal fluid. J Biol Chem. 2020:295:12317–12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chucri TM, Monteiro J, Lima A, Salvadori M, Junior JK, Miglino MA. A review of immune transfer by the placenta. J Reprod Immunol. 2010:87:14–20. [DOI] [PubMed] [Google Scholar]
- Delouis C. Physiology of colostrum production. Ann Rech Vet. 1978:9:193–203, PMID: 371503. [PubMed] [Google Scholar]
- Deutsch EW, Bandeira N, Sharma V, Perez-Riverol Y, Carver JJ, Kundu DJ, García-Seisdedos D, Jarnuczak AF, Hewapathirana S, Pullman BS. The ProteomeXchange consortium in 2020: Enabling ‘big data’approaches in proteomics. Nucleic Acids Res. 2020:48:D1145–D1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingess KA, Gazi I, van den Toorn HW, Mank M, Stahl B, Reiding KR, Heck AJ. Monitoring human milk β-casein phosphorylation and O-glycosylation over lactation reveals distinct differences between the proteome and endogenous Peptidome. Int J Mol Sci. 2021:22:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingess, KA, Hoek, M, van Rijswijk, DMH. et al. Identification of common and distinct origins of human serum and breastmilk IgA1 by mass spectrometry-based clonal profiling. Cell Mol Immunol (2023). 20:26–37. 10.1038/s41423-022-00954-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'souza AA, Devarajan PV. Asialoglycoprotein receptor mediated hepatocyte targeting—Strategies and applications. J Control Release. 2015:203:126–139. [DOI] [PubMed] [Google Scholar]
- Feeney S, Gerlach JQ, Slattery H, Kilcoyne M, Hickey RM, Joshi L. Lectin microarray profiling and monosaccharide analysis of bovine milk immunoglobulin G oligosaccharides during the first 10 days of lactation. Food Sci Nutr. 2019:7:1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazi I, Franc V, Tamara S, van Gool MP, Huppertz T, Heck AJ. Identifying glycation hot-spots in bovine milk proteins during production and storage of skim milk powder. Int Dairy J. 2022:129:105340. [Google Scholar]
- Godden SM, Lombard JE, Woolums AR. Colostrum management for dairy calves. Vet Clin North Am Food Anim Pract. 2019:35:535–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonatilleke E, Huang J, Xu G, Wu L, Smilowitz JT, German JB, Lebrilla CB. Human milk proteins and their glycosylation exhibit quantitative dynamic variations during lactation. J Nutr. 2019:149:1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M, Greenberg R. Bovine homologue of β2-microglobulin isolated from milk. Biochem Biophys Res Commun. 1977:77:320–327. [DOI] [PubMed] [Google Scholar]
- Groves ML, Basch JJ, Gordon WG. Isolation, characterization, and amino acid composition of a new crystalline protein, lactollin, from milk. Biochemistry. 1963:2:814–817. [DOI] [PubMed] [Google Scholar]
- Hinneburg H, Stavenhagen K, Schweiger-Hufnagel U, Pengelley S, Jabs W, Seeberger PH, Silva DV, Wuhrer M, Kolarich D. The art of destruction: Optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J Am Soc Mass Spectrom. 2016:27:507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi J, Yamada M, Kuno A, Tateno H. Lectin microarrays: Concept, principle and applications. Chem Soc Rev. 2013:42:4443–4458. [DOI] [PubMed] [Google Scholar]
- Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. 1998:273:15866–15871. [DOI] [PubMed] [Google Scholar]
- Jansen BC, Bondt A, Reiding KR, Lonardi E, De Jong CJ, Falck D, Kammeijer GS, Dolhain RJ, Rombouts Y, Wuhrer M. Pregnancy-associated serum N-glycome changes studied by high-throughput MALDI-TOF-MS. Sci Rep. 2016:6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasion VS, Burnett BP. Survival and digestibility of orally-administered immunoglobulin preparations containing IgG through the gastrointestinal tract in humans. Nutr J. 2015:14:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F, Braathen R, Brandtzaeg P. Role of J chain in secretory immunoglobulin formation. Scand J Immunol. 2000:52:240–248. [DOI] [PubMed] [Google Scholar]
- Kim J, Bronson C, Wani MA, Oberyszyn TM, Mohanty S, Chaudhury C, Hayton WL, Robinson JM, Anderson CL. β2-microglobulin deficient mice catabolize IgG more rapidly than FcRn-α-chain deficient mice. Exp Biol Med. 2008:233:603–609. [DOI] [PubMed] [Google Scholar]
- Lindh E. Increased resistance of immunoglobulin a dimers to proteolytic degradation after binding of secretory component. J Immunol. 1975:114:284–286. [PubMed] [Google Scholar]
- Marnila P, Korhonen H. Milk proteins | immunoglobulins. In: Fuquay JW, editors. Encyclopedia of dairy sciences. San Diego, United States: Academic Press; 2011. pp. 807–815 [Google Scholar]
- McGrath BA, Fox PF, McSweeney PL, Kelly AL. Composition and properties of bovine colostrum: A review. Dairy Sci Technol. 2016:96:133–158. [Google Scholar]
- Mostov KE. Transepithelial transport of immunoglobulins. Annu Rev Immunol. 1994:12:63–84. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022:50:D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetor A, Hunziker W. β2-microglobulin is important for cell surface expression and pH-dependent IgG binding of human FcRn. J Cell Sci. 2002:115:2389–2397. [DOI] [PubMed] [Google Scholar]
- Raju TS, Briggs JB, Borge SM, Jones AJ. Species-specific variation in glycosylation of IgG: Evidence for the species-specific sialylation and branch-specific galactosylation and importance for engineering recombinant glycoprotein therapeutics. Glycobiology. 2000:10:477–486. [DOI] [PubMed] [Google Scholar]
- Reiding KR, Bondt A, Franc V, Heck AJ. The benefits of hybrid fragmentation methods for glycoproteomics. TrAC Trends Anal Chem. 2018:108:260–268. [Google Scholar]
- Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019:15:346–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renegar KB. Passive Immunization: Systemic and Mucosal. In: Mestecky J, Lamm ME, McGhee JR, Bienenstock J, Mayer L, Strober W, editors. Mucosal Immunology 3rd ed. Academic Press, 2005. p. 841–851. [Google Scholar]
- Rohrer J, Basumallick L, Hurum D. High-performance anion-exchange chromatography with pulsed amperometric detection for carbohydrate analysis of glycoproteins. Biochemistry (Mosc). 2013:78:697–709. [DOI] [PubMed] [Google Scholar]
- Rojas-Macias MA, Mariethoz J, Andersson P, Jin C, Venkatakrishnan V, Aoki NP, Shinmachi D, Ashwood C, Madunic K, Zhang T. Towards a standardized bioinformatics infrastructure for N-and O-glycomics. Nat Commun. 2019:10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Xu G, Li Q, Goonatilleke E, Lebrilla CB. Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem Rev. 2018:118:7886–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol. 2020:21:729–749. [DOI] [PubMed] [Google Scholar]
- Stockert RJ. The asialoglycoprotein receptor: Relationships between structure, function, and expression. Physiol Rev. 1995:75:591–609. [DOI] [PubMed] [Google Scholar]
- Trbojević Akmačić I, Ventham NT, Theodoratou E, Vučković F, Kennedy NA, Krištić J, Nimmo ER, Kalla R, Drummond H, Štambuk J. Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm Bowel Dis. 2015:21:1237–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfman LH, Leusen JH, Savelkoul HF, Warner JO, Van Neerven RJ. Effects of bovine immunoglobulins on immune function, allergy, and infection. Front Nutr. 2018:5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen SS, Schoemaker RJ, Timmer CJ, Kamerling JP, Dijkhuizen L. N-and O-glycosylation of a commercial bovine whey protein product. J Agric Food Chem. 2012:60:12553–12564. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Marth JD, Bertozzi CR, Hart GW, Etzler ME. Symbol nomenclature for glycan representation. Proteomics. 2009:9:5398–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DM, Tyler JW, VanMetre DC, Hostetler DE, Barrington GM. Passive transfer of colostral immunoglobulins in calves. J Vet Intern Med. 2000:14:569–577. [DOI] [PubMed] [Google Scholar]
- Weström B, Arévalo Sureda E, Pierzynowska K, Pierzynowski SG, Pérez-Cano F-J. The immature gut barrier and its importance in establishing immunity in newborn mammals. Front Immunol. 2020:11:1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Shu J, Li Z. Lectin microarrays for glycoproteomics: An overview of their use and potential. Expert Rev Proteomics. 2020:17:27–39. [DOI] [PubMed] [Google Scholar]
- Zhu A, Hurst R. Anti-N-glycolylneuraminic acid antibodies identified in healthy human serum. Xenotransplantation. 2002:9:376–381. [DOI] [PubMed] [Google Scholar]
- Zhu J, Lin Y-H, Dingess KA, Mank M, Stahl B, Heck AJ. Quantitative longitudinal inventory of the N-glycoproteome of human milk from a single donor reveals the highly variable repertoire and dynamic site-specific changes. J Proteome Res. 2020:19:1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.