Abstract
Dbp2p, a member of the large family of DEAD-box proteins and a yeast homolog of human p68, was shown to interact with Upf1p, an essential component of the nonsense-mediated mRNA decay pathway. Dbp2p:Upf1p interaction occurs within a large conserved region in the middle of Upf1p that is largely distinct from its Nmd2p and Sup35/45p interaction domains. Deletion of DBP2, or point mutations within its highly conserved DEAD-box motifs, increased the abundance of nonsense-containing transcripts, leading us to conclude that Dbp2p also functions in the nonsense-mediated mRNA decay pathway. Dbp2p, like Upf1p, acts before or at decapping, is predominantly cytoplasmic, and associates with polyribosomes. Interestingly, Dbp2p also plays an important role in rRNA processing. In dbp2Δ cells, polyribosome profiles are deficient in free 60S subunits and the mature 25S rRNA is greatly reduced. The ribosome biogenesis phenotype, but not the mRNA decay function, of dbp2Δ cells can be complemented by the human p68 gene. We propose a unifying model in which Dbp2p affects both nonsense-mediated mRNA decay and rRNA processing by altering rRNA structure, allowing specific processing events in one instance and facilitating dissociation of the translation termination complex in the other.
The degradation of eukaryotic mRNAs can be triggered by at least three distinct events: deadenylation, endonucleolytic cleavage, and aberrant translational termination (6, 31). The latter mechanism, also known as nonsense-mediated mRNA decay (32), is responsible for the rapid turnover of mRNAs containing premature stop codons (51) as well as unspliced pre-mRNAs that enter the cytoplasm (23), mRNAs with a poor translation initiation context (61), some mRNAs with upstream open reading frames (47), and transcripts with extended 3′-untranslated regions (46). In Saccharomyces cerevisiae, trans-acting factors involved in the nonsense-mediated mRNA decay pathway were initially identified as frameshift suppressors and are encoded by UPF1 (IFS2/SAL2/MOF4), NMD2 (UPF2/SUA1/IFS1), and UPF3 (SUA6) (7, 10, 38, 39). These genes have also been identified in screens for omnipotent suppressors as well as in two-hybrid interaction screens (9, 20, 21; D. Zuk, A. H. Brown, S. Liebman, and A. Jacobson, unpublished experiments). Mutations in, or deletions of, these genes generally cause stabilization of nonsense-containing mRNAs while having little or no effect on most wild-type mRNAs (51).
The UPF1 gene encodes a 109-kDa protein that contains two putative Zn2+ fingers near its N terminus and is a member of RNA helicase superfamily I. Upf1p, isolated from yeast, has been shown to possess nucleic acid binding, RNA helicase, and ATPase activities (13, 64) and to interact directly with the translational termination factors Sup35p and Sup45p (12). A two-hybrid screen using Upf1p as bait identified several genes, many of which may play important roles in mRNA decay, including NMD2, DCP2 (NMD1), NMD3, and DBP2 (21). NMD2 is an essential component of the nonsense-mediated mRNA decay pathway, and its encoded protein interacts with Upf3p in a two-hybrid assay (20). DCP2 is a high-copy suppressor of a temperature-sensitive allele of DCP1, the gene encoding the decapping enzyme (15). NMD3 has been identified as a high-copy suppressor of a mutation in GRC5, a ribosomal protein gene, and as a gene required for viability in the absence of XRN1, the gene encoding the major 5′-to-3′ exonuclease involved in mRNA decay (27, 33, 34). Nmd3p associates with the 60S ribosomal subunit and appears to also play a role in ribosome biogenesis (5, 25). DBP2 was initially identified as a yeast homolog of the human p68 gene (28), and the respective proteins encoded by these genes are members of the extensive helicase superfamily II, and more specifically the large DEAD-box family of proteins. p68 has ATP-dependent RNA helicase and RNA-dependent ATPase activities in vitro (29), localizes to the nucleus, and exhibits cell cycle-dependent shuttling to the nucleolus in vivo (28). Expression of the p68 gene in Escherichia coli protects mRNA from degradation, although the mechanism of this protection has not been elucidated (30). Interestingly, the DBP2 gene contains a large 3′ proximal intron, the position and size of which has been conserved from yeast to humans, and it has been demonstrated that splicing of the intron is autoregulated by Dbp2p (4). Although DBP2 is not an essential gene, cells harboring a disruption of this gene exhibit slow-growth and cold-sensitive phenotypes (4).
Here we report further characterization of the function of DBP2. We have mapped the respective Upf1p and Dbp2p interaction domains and shown that a dbp2Δ strain accumulates nonsense-containing transcripts. Like Upf1p, Dbp2p acts upstream of Dcp1p and Xm1p in the nonsense-mediated mRNA decay pathway. In addition, Dbp2p also plays a prominent role in the rRNA processing pathway, as indicated by the accumulation of 35S pre-rRNA in dbp2Δ cells. Expression of the human p68 gene in dbp2Δ cells reverses the rRNA processing defect of these cells but not their deficiency in nonsense-mediated mRNA decay. A role for Dbp2p in both nonsense-mediated mRNA decay and rRNA processing leads us to suggest a common mechanism by which this putative RNA helicase can influence ribosome maturation and function.
MATERIALS AND METHODS
General methods.
Preparation of standard yeast media and methods of cell culture were as described previously (54). Transformation of yeast was done by the rapid method (59), and DNA manipulations were performed according to standard techniques (55). All PCR amplifications were performed with Taq DNA polymerase (65) and confirmed, where appropriate, by DNA sequencing by the method of Sanger et al. (56) using a Sequenase 2.0 kit (USB Corp.). Plasmids were propagated in the E. coli strain DH5α.
Oligonucleotides.
The oligonucleotides used in this study were prepared by Operon, Inc., and are listed in Table 1.
TABLE 1.
Oligonucleotides
| Name | Sequence |
|---|---|
| DBP2–3 | 5′-CGGATTTCGCTCCAGGCGATGGACC-3′ |
| DBP2–4 | 5′-CGGAATTCATGTTAGATATGGGTTTTGAACC-3′ |
| DBP2–5 | 5′-GCCTCGAGTCAGGTTTGTCTATCAGGTCTG-3′ |
| DBP2–6 | 5′-GCCTCGAGTCAGGCGGCCACATCAGTAGCAACC-3′ |
| DBP2–7 | 5′-GCCTCGAGTCAGCCACCATAAGATCTCCTGTCG-3′ |
| DBP2–8 | 5′-GCCTCGAGTCAATAGTTTGAACGACCTCTGTTACCC-3′ |
| DBP2–9 | 5′-CGGAATTCGGTTCTCTAGAACTATCTGCCTCCC-3′ |
| DBP2-de15′ | 5′-TGAAACGACCGCGGTGGATGCTCTTCA-3′ |
| DBP2-de13′ | 5′-CTTCCAGAATAGCCCACACTCTTCTCC-3′ |
| K163R-1 | 5′-GCCACTGGTTCCGGTAGGACTTTGTCTTATAG-3′ |
| K163R-2 | 5′-CAATAAGACAAAGTCCTACCGGAACCAGTGGC-3′ |
| E268D-1 | 5′-CCTGGTTCTTGATGATGCTGATAGAATGTTAGATATGGG-3′ |
| E268D-2 | 5′-CCCATATCTAACATTCTATCAGCATCATCAAGAACCAGG-3′ |
| T300A-1 | 5′-CTTGATGTGGTCTGCCGCTTGGCCAAAGG-3′ |
| T200A-2 | 5′-CCTTTGGCCAAGCGGCAGACCACATCAAG-3′ |
| R447K-1 | 5′-GATTATGTTCACAAAATCGGTAGAACTGGTAGAGCAGG-3′ |
| R447K-2 | 5′-CCTGCTCTACCAGTTCTACCGATTTTGTGAACATAATC-3′ |
| P68-4 | 5′-GCGGAATTCATGCTTGATATGGGCTTTGAACCC-3′ |
| P68-8 | 5′-GCGCGCTCGAGTTATTGGGAATATCCTGTTGGC-3′ |
Strains.
The yeast strains used in this study include: (i) L40 (MATa ade2 his3Δ200 leu2-3,112 trp1-901 LYS::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ gal4 gal80), a kind gift from Stanley Hollenberg; (ii) BJ2168 (MATa ura3-52 leu2 trp1 pep4-3 prb1-1122 prc1-407 gal2), a kind gift from Elizabeth Jones; (iii) yIB12/4 (MATa ade2-1 ura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100 dbp2::URA3), a kind gift from Richard Iggo; (iv) HFY1200 (MATa ade2-1 ura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100), the UPF/NMD wild-type strain for this study (21); (v) yATB100, the dbp2Δ strain, constructed by PCR, using the oligonucleotide pair DBP2-del5′ and DBP2-del3′ to amplify a deletion construct from a genomic DNA preparation of yIB12/4 cells (the PCR product was recovered with a QIAquick PCR purification kit [Qiagen, Inc.] and transformed into strain HFY1200; transformants were selected on synthetic complete [SC]-ura plates, and colonies were screened by PCR for the presence of the deletion); (vi) yATB101-yATB104, yATB100 derivatives harboring plasmids with point mutations in DBP2, constructed as described below; and (vii) yATB200, a derivative of yATB100, containing the p68 gene on plasmid pIB13 (see below). HFY871 (20), HFY1081 (22), SJ21R (42), TP11B-4-1 (42) and SWP154− (51) were as described previously.
Plasmid constructs (see Fig. 1 for nomenclature). (i) Plasmids for directed two-hybrid experiments.
FIG. 1.
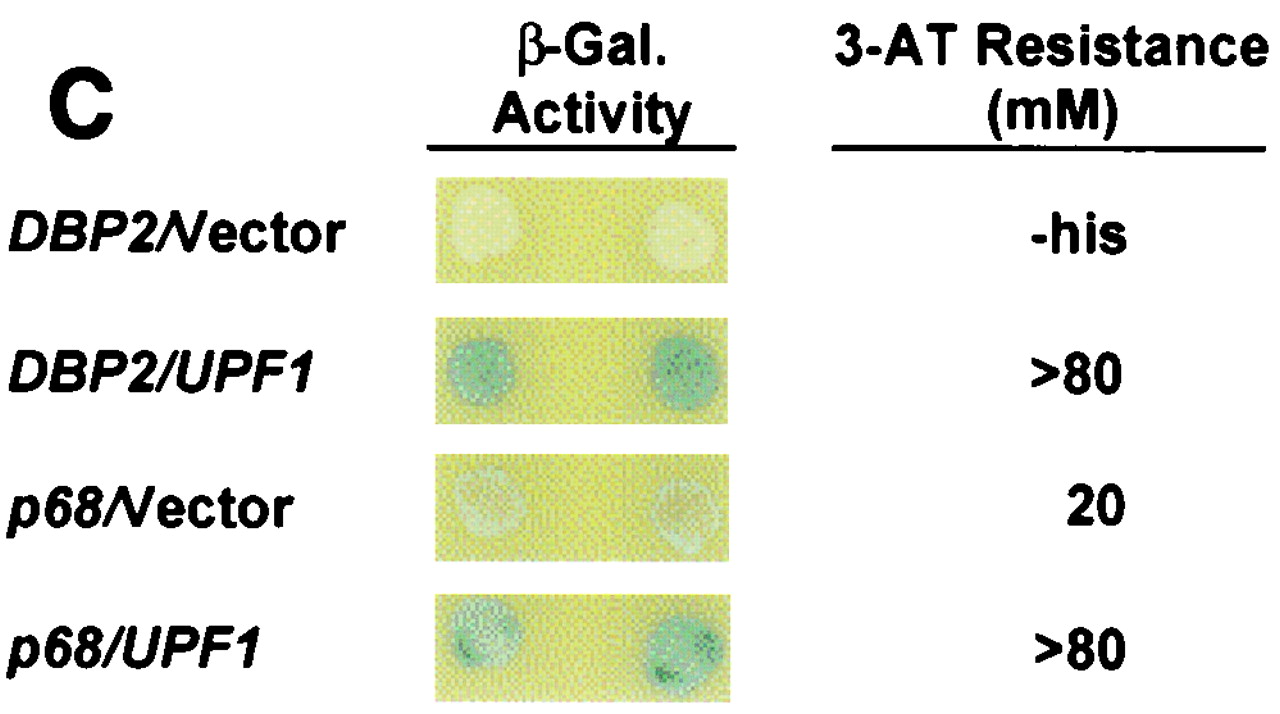
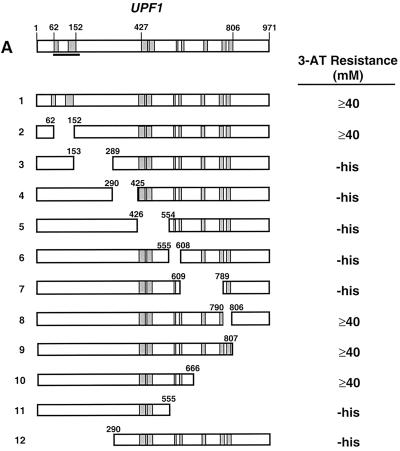
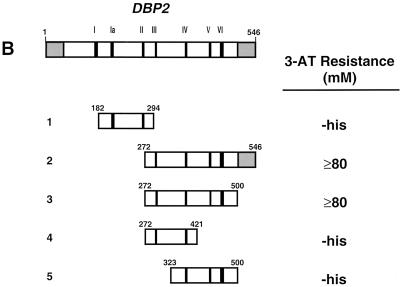
Mapping Upf1p-Dbp2p interaction domains. (A) Schematic representation of full-length Upf1p (amino acids 1 to 971) with known motifs denoted by shaded boxes and the Nmd2p interaction domain underlined. Interaction of full-length GAL4(AD)-UPF1, and fragments thereof, with lexA(DB)-DBP2 (amino acids 272 to 546) was assayed by measuring the extent of resistance to 3-AT. (B) Schematic representation of full-length Dbp2p (amino acids 1 to 546) with known RNA helicase domains and RGG motifs denoted by Roman numerals and shaded boxes. GAL4(AD)-UPF1 (full-length) interaction with lexA(DB)-DBP2 was assayed by measuring the extent of resistance to 3-AT. (C) GAL4(AD)-UPF1 or GAL4(AD) interaction with lexA(DB)-DBP2 (amino acids 272 to 546) or lexA(DB)-p68 (amino acids 253 to 614) assayed by determining the extent of resistance to 3-AT and by the production of β-galactosidase activity in individual clones analyzed on X-Gal plates. For the 3-AT assay, the highest concentration of 3-AT (on SC-his-leu-trp plates) that still allowed substantial cellular growth is noted; -his, cells could grow in the absence of histidine but were unable to grow in the presence of 5 mM 3-AT.
Plasmids containing UPF1 deletions and truncations fused with GAL4(AD) have been described previously (22, 62, 63). To create the lexA(DB)-DBP2 constructs, DNA fragments were amplified by PCR using the following pairs of oligonucleotides: DBP2-3 and DBP2-5 [lexA(DB)-DBP2(182–294)], DBP2-4 and DBP2-8 [lexA(DB)-DBP2(272–546)], DBP2-4 and DBP2-7 [lexA(DB)-DBP2(272–500)], DBP2-4 and DBP2-6 [lexA(DB)-DBP2(272–421)], and DBP2-9 and DBP2-7 [lexA(DB)-DBP2(323–500)]. The PCR products were digested with EcoRI and XhoI and subcloned into the plasmid pBTM116 (a generous gift from Stanley Hollenberg). To create the lexA(DB)-p68 construct, a fragment corresponding to the Dbp2p interaction domain was amplified by PCR using oligonucleotides P68-4 and P68-8 [lexA(DB)-p68(253–614)]. The PCR product was digested with EcoRI and XhoI and subcloned into pBTM116.
(ii) Plasmids for analysis of DBP2 function.
Plasmids containing different pgk1 alleles have been described previously (51). Plasmid pIB13, containing the human p68 gene, and plasmid pIG75, containing a wild-type copy of DBP2, have been described previously (4). Plasmids containing DBP2 alleles harboring point mutations were made with the Quick Change mutagenesis kit (Stratagene, Inc.) using plasmid pIG75 as a template and the oligonucleotide pairs K163R-1 and K163R-2, E268D-1 and E268D-2, T300A-1 and T300A-2, and R447K-1 and R447K-2 (see Table 1).
Two-hybrid assays.
The yeast two-hybrid tester strain L40 was used to assay interactions between Upf1p and Dbp2p. In each case a lexA(DB) fusion construct was cotransformed with a GAL4(AD) construct into the L40 strain (26, 41). Patches of cells were replica-plated onto SC-leu-trp-his plates containing increasing concentrations of 3-aminotriazole (3-AT; 0, 5, 10, 20, 40, and 80 mM) and incubated for 4 to 6 days at 30°C (41).
RNA extraction and Northern analysis.
RNA was isolated by the hot phenol method as described previously (24). Aliquots (10 μg) of each RNA sample were analyzed by Northern blotting using radiolabeled probes prepared by random priming (CYH2 and CAN1) with a kit from Boehringer Mannheim or end-labeled oligonucleotide probes (mini-PGK1 and PGK1) prepared as described previously (16). CAN1 mRNA was detected with a probe made from a 1.0-kbp EcoRI-SalI fragment of YEplac195-CAN1 (40). Total RNA used for analysis of nuclear pre-rRNA (see Fig. 9) was isolated by the glass bead/phenol method (43) and characterized by Northern blotting using equal amounts of total RNA per lane. Hybridization conditions were as described by Peltz et al. (52).
FIG. 9.
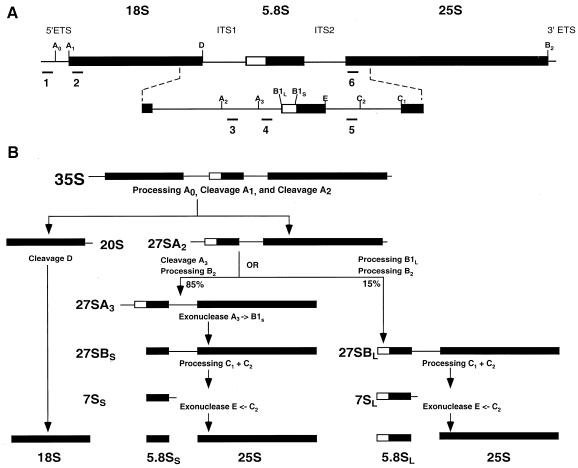
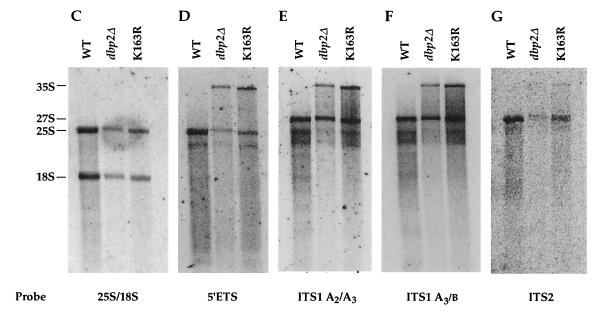
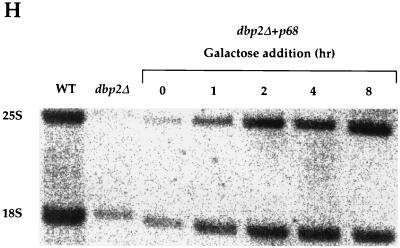
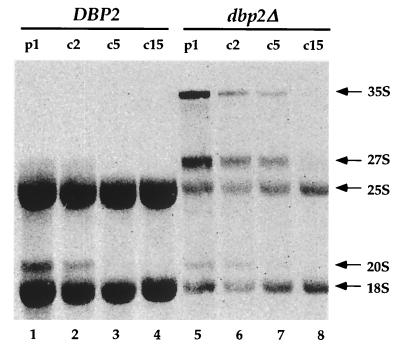
Accumulation of rRNA precursor species in dbp2Δ cells. (A) Schematic of the yeast 35S pre-rRNA and its principal processing sites, derived from a figure in reference 60. The locations of the oligonucleotide probes used in this study are numbered (1 to 6) and highlighted by black bars. (B) Schematic of the processing of the 35S pre-rRNA and its principal products. (C to G) Northern analysis of rRNAs. Total cellular RNA was isolated from HFY1200 (wild type [WT]), yATB100 (dbp2Δ), and yATB101 (K163R) cells and fractionated by Northern blotting, and the same blot was stripped and reprobed repeatedly. (C) Oligonucleotides 2 and 6, complementary to sites within the mature 18S and 25S rRNA sequences, respectively, were used to probe the blot. (D) The blot was probed with oligonucleotide 1, which is complementary to a portion of the 5′ ETS. (E) The blot was probed with oligonucleotide 3, which hybridizes between site A2 and A3 in ITS1. (F) The blot was probed with oligonucleotide 4, which hybridizes between site A3 and B1 in ITS1. (G) The blot was probed with oligonucleotide 5, which hybridizes upstream of C2 in ITS2. (H) Total cellular RNA was isolated from HFY1200 (wild type) and yATB100 (dbp2Δ) cells and from yATB200 (dbp2Δ+p68) cells at 0, 1, 2, 4, and 8 h after induction with galactose. Oligonucleotides 2 and 6, complementary to sites within the mature 18S and 25S rRNA sequences, respectively, were used to probe the blot.
Polyribosome analysis.
Cells utilized for the experiments of Fig. 6B through D and Fig. 8A through D were grown in yeast extract-peptone-dextrose medium. Temperature shifts utilized for Fig. 6C and D were as described previously (42). Cells analyzed in Fig. 8E and F were grown in SC-ura+raffinose medium at 30°C to an optical density at 600 nm (OD600) of 0.3, induced by adding galactose to a concentration of 2%, and shaken for 4 h to a final OD600 of 0.8 to 1.0. Cytoplasmic extracts were prepared and fractionated on 15 to 47% or 7 to 25% sucrose gradients as described previously (42, 52).
FIG. 6.
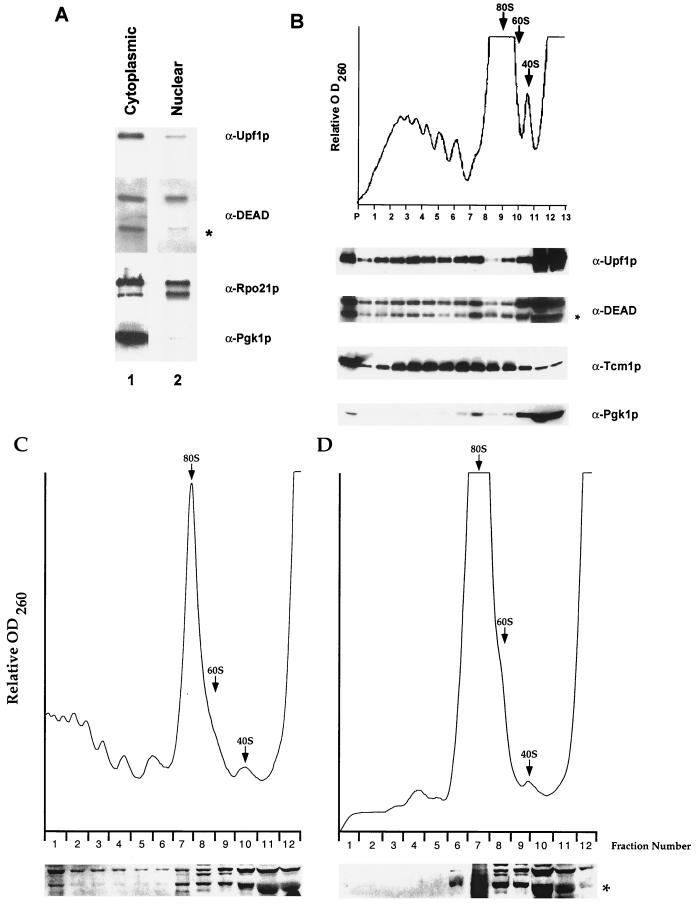
Dbp2p copurifies with nuclei and cofractionates with polyribosomes. (A) Western blot analysis of cytoplasm and purified nuclei isolated from strain BJ2168. Samples loaded represented equivalent numbers of cells. (B) An extract of SWP154− cells (51) was fractionated on a 15 to 50% sucrose gradient that was subsequently analyzed by Western blotting. (Top) Absorbance at 260 nm (OD260), with sedimentation proceeding from right to left. The 80S, 60S, and 40S ribosome peaks are indicated by arrows. (Bottom) Western blot analysis of gradient fractions 1 to 9 and the pellet fraction (P) included the entire sample, whereas fractions 10 to 13 included only one-fifth of the sample. The blots in both panels were serially stripped and reprobed with the indicated antibodies. The asterisk denotes Dbp2p. The upper band in the α-DEAD panels is a mixture of the comigrating proteins Dbp1p and Ded1p (4). (C and D) Cultures of SJ21R (PRT1) (C) and TP11B-4-1 (prt1-1) (D) were grown at 23°C and shifted to 37°C for 30 min. Extracts were prepared, fractionated, and analyzed as for panel B.
FIG. 8.
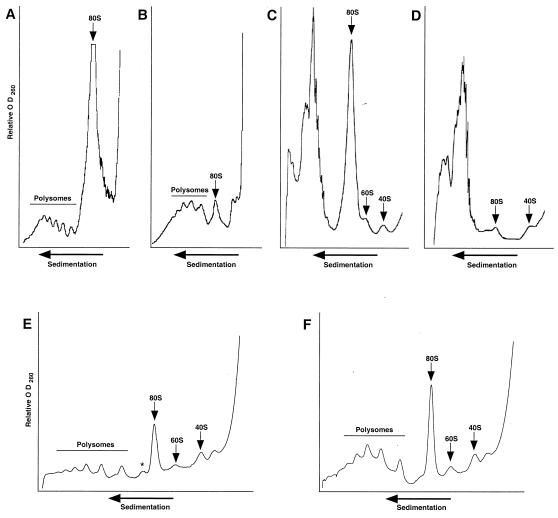
Polysome profiles are altered in dbp2Δ cells. Cytoplasmic extracts were prepared from wild-type (A and C) and dbp2Δ (B and D) cells and fractionated on either 15 to 47% (A and B) or 7 to 25% (C and D) sucrose gradients as described in Materials and Methods. The A260 trace of the gradients is shown, with the polysome and 80S, 60S, and 40S ribosome peaks indicated. Cytoplasmic extracts prepared from yATB200 (dbp2Δ+p68) cells, grown in raffinose (E) or induced for 4 h in galactose (F), were fractionated on 7 to 47% sucrose gradients.
Measurement of amino acid incorporation.
Cells were grown in 100 ml of SC-ura-met+raffinose medium at 30°C to an OD600 of 0.6 to 0.8, harvested by centrifugation, resuspended in 20 ml of fresh medium, and shaken for 10 min at 30°C. Galactose was then added to a final concentration of 2%. Incorporation of 35S-translabel (70% methionine and ∼15% cysteine; ICN) was monitored by trichloroacetic acid precipitation as described previously (68). Each experiment was repeated at least three times.
Subcellular fractionation.
Yeast nuclei were isolated by osmotic lysis of spheroplasts, followed by banding two times on Ficoll gradients (17). The purity of the nuclei was monitored by Western blotting using enrichment for a nucleus-associated protein (Rpo21p) and the loss of a cytoplasmic protein (Pgk1p) as standards.
Pulse-chase analysis of rRNA processing.
HFY1200 (wild type) and yATB100 (dbp2Δ) cells were grown to an OD600 of 0.8 in 40 ml of SC-met medium. Cells were centrifuged, resuspended in 1 ml of pre-warmed SC-met medium, pulsed with 250 μCi of [methyl-3H]methionine (70 to 85 Ci/mmol; Amersham), and then chased by mixing 250-μl aliquots of cells with 1.75 ml of prewarmed SC medium containing 1 mg of methionine per ml. Samples were taken at several times during the chase, pelleted, washed in ice-cold water, repelleted, and frozen in liquid nitrogen. Total RNA was isolated by the glass bead-phenol method (43). For each sample, 20,000 cpm of labeled RNA was resolved on 1.2% agarose-formaldehyde gels and transfered to Zeta-Probe membrane. The membrane was sprayed with EN3HANCE (New England Nuclear) and then exposed to X-ray film for 3 days at −80°C with an intensifying screen.
RESULTS
Dbp2p interacts with an internal domain of Upf1p adjacent to the Nmd2p interaction domain.
Previously we identified the product of the DBP2 gene as a Upf1p-interacting protein (21). To better understand the functional consequences of this interaction, we utilized a directed yeast two-hybrid assay to map the domain on Upf1p that interacts with Dbp2p. A series of UPF1 truncations, fused in-frame to GAL4(AD), were tested for their ability to promote two-hybrid interactions with the minimal interacting fragment of DBP2 (Fig. 1A). The extent of interaction between the respective fusion proteins was assessed by monitoring the expression of (lexAop)4-HIS3 and (lexAop)8-lacZ reporters in cotransformants grown on plates containing increasing concentrations of 3-AT and by qualitative and quantitative β-galactosidase assays, respectively. Coexpression of lexA(DB)-DBP2(272–546) fusions with GAL4(AD)-UPF1 (full length) led to high levels of resistance to 3-AT (Fig. 1A, constructs 1, 2, and 8 to 10) and to high levels of β-galactosidase activity (data not shown). The high degree of 3-AT resistance indicates that the interaction between Upf1p and Dbp2p is most likely to be direct and not bridged by another protein or RNA moiety (20). Coexpression of lexA(DB)-DBP2 with GAL4(AD)-UPF1 fusions lacking sequences in an internal 515-amino-acid sequence of Upf1p (from amino acids 152 to 666) led to background levels of 3-AT resistance (Fig. 1A, constructs 3 to 7 and 11 and 12) and β-galactosidase activity (data not shown). The Dbp2p interaction domain of Upf1p contains the ATP binding, ATPase, and helicase motifs of the superfamily I group of helicases. Interestingly, the N-terminal 181 amino acids of Upf1p containing the Zn2+ finger domain, which is sufficient for Nmd2p and Sup35p interaction (19–21; F. He and A. Jacobson, unpublished experiments), is only minimally overlapping with the Dbp2p interaction domain.
To determine the regions of Dbp2p that are necessary for interaction with Upf1p, we constructed DBP2 amino- and/or carboxy-terminal truncations and tested their abilities to promote two-hybrid interaction with Upf1p (Fig. 1B). lexA(DB)-DBP2 fusions containing the 272 N-terminal amino acids of Dbp2p are toxic to E. coli and to the L40 yeast strain used in this experiment (data not shown); therefore, it was not possible to test full-length Dbp2p for interaction with Upf1p. Fragments of DBP2 were fused to lexA(DB), and full-length UPF1 was fused to GAL4(AD). The extent of interaction was monitored as above. Coexpression of GAL4(AD)-UPF1 with lexA(DB)-DBP2 fusions that encode an internal 229-amino-acid segment of Dbp2p led to increased resistance to 3-AT (Fig. 1B, constructs 2 and 3) and to detectable β-galactosidase activity (data not shown). Coexpression of GAL4(AD)-UPF1 with lexA(DB)-DBP2 fusions that lack 70 N-terminal amino acids or 80 C-terminal amino acids of this 229-amino-acid segment led to background levels of 3-AT resistance (Fig. 1B, constructs 4 and 5) and β-galactosidase activity (data not shown). The fragment of Dbp2p that is sufficient for Upf1p interaction is highly conserved (61% identity with human p68 protein) and contains the predicted helicase and RNA binding domains.
DBP2 was originally cloned as a yeast homolog to the human p68 gene (28) (the respective proteins share 57% identity and 72% similarity). To test whether the p68 protein could interact with Upf1p, a p68 gene fragment containing sequences homologous to the mapped DBP2 interaction domain was fused to the lexA(DB). A plasmid containing this construct was cotransformed with the GAL4(AD)-UPF1 plasmid, and two-hybrid interaction was monitored as before. Coexpression of the lexA(DB)-p68(253–614) and GAL4(AD)-UPF1 constructs promoted resistance to >80 mM 3-AT with the HIS3 reporter and increased expression of the lacZ reporter (Fig. 1C). Although the lexA(DB)-p68(253-614) construct showed some self-activation when coexpressed with the GAL4(AD) vector alone (3-AT resistance, 20 mM), at least a fourfold increase in reporter expression was seen in conjunction with the GAL4(AD)-UPF1 construct (3-AT resistance, >80 mM). These results suggest that the Dpb2p-Upf1p interaction has been conserved in p68, a conclusion supported by the 45% identity of the minimal Dbp2p-interacting domain of yeast Upf1p and the equivalent segment of human Upf1p (1, 53).
Nonsense-containing transcripts accumulate in a dbp2Δ strain.
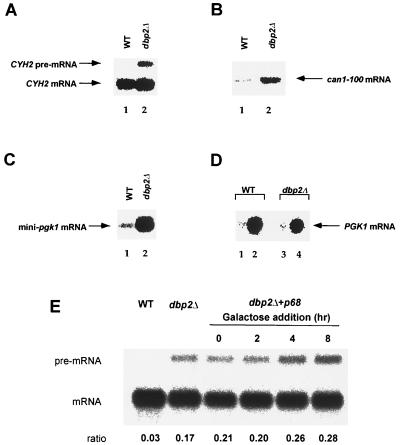
Since Dbp2p interacts with Upf1p we sought to determine whether a deletion of DBP2 affects nonsense-mediated mRNA decay or other aspects of mRNA decay. We assayed the abundance of two endogenous substrates of the nonsense-mediated mRNA decay pathway as well as that of wild-type and mutant mRNAs in strains that differed in their DBP2 genotypes. Five different nonsense-containing transcripts (CYH2pre-mRNA, can1-100 mRNA, mini-PGK1 mRNA, and full-length PGK1 mRNAs with early or late nonsense codons) were assayed by Northern analysis in wild-type and dbp2Δ strains. Cells harboring upf/nmd alleles typically demonstrate substantial increases in the abundance of transcripts containing early nonsense codons (23, 40, 51, 66). Deletion of DBP2 resulted in an eightfold increase in the abundance of the CYH2 pre-mRNA while having no effect on CYH2 mRNA levels (Fig. 2A, lanes 1 and 2). Similarly, the abundance of the can1-100 mRNA was increased sixfold in a dbp2Δ strain compared to the wild-type (Fig. 2B, lanes 1 and 2). The mini-PGK1 nonsense transcript, previously shown to be stabilized upon inactivation of the nonsense-mediated mRNA decay pathway (51), was also found to be significantly more abundant in dbp2Δ cells (Fig. 2C, lanes 1 and 2). The magnitude of these effects is comparable to that seen in upf1Δ cells. We also tested the abundance of full-length PGK1 transcripts, containing early and late nonsense codons (51), in the dbp2Δ strain. As expected, full-length PGK1 mRNA with a late nonsense codon was comparably abundant in wild-type and dbp2Δ cells (Fig. 2D). However, unlike results observed in upf1 cells (51), the full-length PGK1 transcript with an early nonsense codon was not stabilized in the dbp2Δ strain (Fig. 2D, lane 3). The levels of wild-type mRNAs of differing stabilities, including those encoded by the STE2 and PAB1 genes, were not affected by deletion of DBP2 (data not shown). These results suggest that DBP2 is involved in the nonsense-mediated mRNA decay pathway but functions in a manner different from that of UPF1, NMD2, or UPF3.
FIG. 2.
Nonsense-containing transcripts are stabilized in a dbp2Δ strain. Total RNA from yeast strains of the indicated genotypes was isolated and analyzed by Northern hybridization. The blots were hybridized with radioactive probes that detected the CYH2 pre-mRNA and mRNA (A and E); the can1–100 mRNA (B); the mini-pgk1 nonsense-containing mRNA (C); and full-length PGK1 mRNAs harboring either an early (panel D, lanes 1 and 3) or late (panel D, lanes 2 and 4) nonsense codon. The transcripts analyzed in panels C and D were expressed from plasmids. In panel E, galactose was added at t0 to cells growing in raffinose, and samples were taken every 2 h. The ratios of CYH2 pre-mRNA/mRNA are indicated below each lane. WT, wild-type.
Previous studies demonstrated that the human p68 gene was able to complement the slow-growth phenotype exhibited by dbp2Δ cells (4). Since Dbp2p and p68 both interact with yeast Upf1p (see above), we tested whether the p68 protein would complement the nonsense-mediated mRNA decay phenotype seen in a dbp2Δ strain. The p68 gene was placed under the control of the inducible GAL1,10 promoter transformed into dbp2Δ cells, and expression of p68 was induced by the addition of galactose. Samples were taken at several time points thereafter and subjected to Northern analysis using the relative abundance of the CYH2 pre-mRNA to assess the nonsense-mediated mRNA decay phenotype. These experiments showed that, although it did correct the previously observed slow-growth phenotype (data not shown), induction of p68 did not complement the nonsense-mediated mRNA decay phenotype. The abundance of CYH2 pre-mRNA was low in wild-type cells, high in the dbp2Δ strain containing the p68 gene under GAL1,10 control, and relatively unaffected by galactose induction of p68 (Fig. 2E). These results indicate that the human p68 gene does not complement the nonsense-mediated mRNA decay phenotype seen in a dbp2Δ strain and that the slow-growth phenotype must be attributable to a different function of Dbp2p.
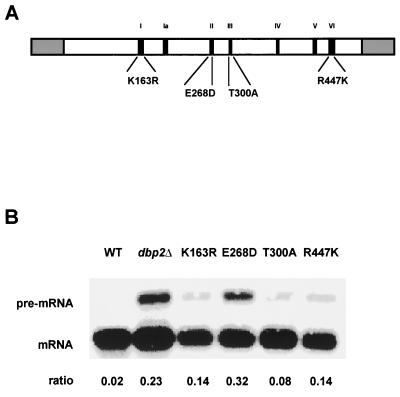
Point mutations in highly conserved motifs identify functionally important domains within Dbp2p.
Studies with other DEAD-box proteins have identified at least four functionally important domains (for a review see reference 14). By homology, Dbp2p also contains domains for ATP binding, ATPase activity, helicase activity, and RNA binding. To test whether these highly conserved motifs are functionally important for Dbp2p activity in the nonsense-mediated mRNA decay pathway, we created point mutations within each domain and determined the relative abundance of the CYH2 pre-mRNA and mRNA in dbp2Δ strains expressing one of the four dbp2 alleles from a CEN plasmid. Western analysis showed that the levels of DBP2 protein expressed from the different alleles did not differ from that expressed by the wild-type gene (data not shown). Each of the four mutations increased the abundance of the CYH2 pre-mRNA, albeit to varying degrees (Fig. 3B). The strain that exhibited the largest increase carried the E268D mutation, which resides within the putative ATPase domain of Dbp2p. The K163R mutation, within the highly conserved ATP binding motif, as well as the R447K mutation, within the putative RNA-binding domain, also increased the abundance of the CYH2 pre-mRNA, but not to levels approaching that of the dbp2Δ strain. The T300A mutation, which resides within the switch-helicase motif, had only a modest effect on the abundance of the CYH2 pre-mRNA.
FIG. 3.
Mutations in conserved domains of Dbp2p affect the abundance of nonsense-containing transcripts. (A) Schematic diagram of Dbp2p showing the amino acids that were changed. (B) Northern analysis of total RNA from wild-type, dbp2Δ, and dbp2Δ cells transformed with a CEN plasmid containing one of the four dbp2 alleles. Blots were hybridized with a CYH2 probe as described in the legend to Fig. 2. Ratios of CYH2 pre-mRNA/mRNA are indicated below each lane. WT, wild type.
To determine whether these mutations might have dominant-negative effects on nonsense-mediated mRNA decay, we transformed the wild-type strain with CEN plasmids containing wild-type DBP2, or one of its four mutant alleles, and assayed the abundance of the CYH2 pre-mRNA. Two of the alleles tested, those in the RNA binding and helicase domains, showed modest increases in the relative abundance of the CYH2 pre-mRNA (data not shown), while the two mutations in the ATP binding and ATPase domains had no significant effects on the level of this transcript (data not shown).
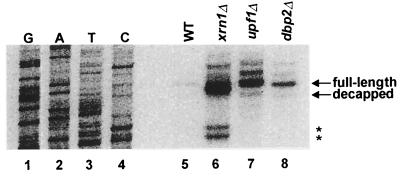
DBP2 acts before or at decapping in the nonsense-mediated mRNA decay pathway.
The general pathway of mRNA decay is ordered such that deadenylation is followed by decapping, which, in turn, is followed by 5′-to-3′ digestion of the transcript (45). The decapping and exonucleolytic activities are encoded by the DCP1 and XRN1 genes, respectively (18, 36, 37). Recent studies have shown that Upf1p, Nmd2p, and Upf3p act upstream of Dcp1p, regulating but not catalyzing mRNA decapping (22). To test whether Dbp2p is also upstream to the decapping reaction, we analyzed the 5′ end of the CYH2 pre-mRNA in wild-type, xrn1Δ, upf1Δ, or dbp2Δ strains. The decapped version of this transcript, readily seen in xrn1Δ cells, is two nucleotides shorter than its capped counterpart and is detectable by a primer extension assay (Fig. 4, lanes 5 and 6; see also reference 22). In upf1Δ and dbp2Δ strains, the CYH2 pre-mRNA is primarily full length (Fig. 4, lanes 7 and 8), leading us to conclude that Dbp2p, like Upf1p, acts concurrently with or prior to Dcp1p.
FIG. 4.
Nonsense-containing mRNAs are predominantly full-length in dbp2Δ cells. Total RNA was isolated from yeast strains of the indicated genotypes, and the 5′ end of the CYH2 pre-mRNA was analyzed by primer extension. DNA sequencing reactions with the same primer (run on lanes G, A, T, and C) were used to determine the positions of the primer extension products. The major transcriptional start sites for the CYH2 pre-mRNA and decapped mRNA are indicated by an arrow. The atypical extension products detected in the xm1Δ strain are marked by asterisks. WT, wild type.
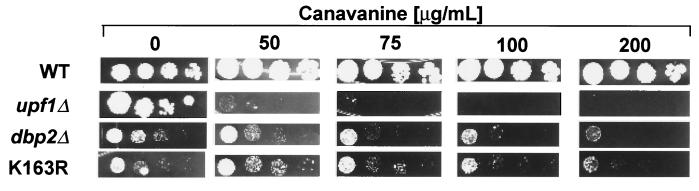
Translational fidelity is altered in a dbp2Δ strain.
All three of the UPF/NMD factors have been shown to play a role in translation termination, promoting nonsense suppression in their absence (39, 40, 62, 63). Using a recently described quantitative assay for nonsense suppression (40), we tested whether such suppression is affected in DBP2 mutant strains. This assay exploits the can1-100 nonsense allele. This mutation promotes efficient premature translational termination in wild-type cells, leading to resistance to high concentrations of the toxic drug canavanine. In upf/nmd cells, however, premature translation termination is less efficient, leading to canavanine sensitivity (40). As shown in Fig. 5, serial dilutions of wild-type cells harboring the can1-100 allele were completely resistant to all concentrations of the drug that were tested. In contrast, upf1Δ cells were sensitive to as little as 50 ug of canavanine per ml, whereas it required 200 ug of the drug per ml to affect dbp2 strains to a comparable degree (Fig. 5). Interestingly, in spite of their modest mRNA decay phenotype cells containing the K163R allele of DBP2 showed canavanine sensitivity similar to that of the dbp2Δ strain. These data indicate that nonsense suppression occurs in dbp2 cells, albeit at least fourfold less efficiently than in upf1Δ cells, and suggest that the mRNA decay and translation functions of Dbp2p can be separated.
FIG. 5.
Translational fidelity is altered in dbp2Δ cells. Cultures of the identified genotypes were diluted serially and spotted on SC-arg plates containing 0, 50, 75, 100, or 200 μg of canavanine per ml. Cells were analyzed after 7 days of growth at 30°C. WT, wild type.
To analyze further Dbp2p's role in translational fidelity, we assayed the sensitivity of wild-type and dbp2Δ strains to drugs affecting translation, including the antibiotics cycloheximide, which inhibits translational elongation, and paromomycin, which promotes translational misreading (48, 58, 68). Zones of growth inhibition surrounding filter discs containing different concentrations of cycloheximide or paromomycin were measured, and the results are presented in Table 2. The dbp2Δ strain was found to be hypersensitive to paromomycin, exhibiting much larger zones of inhibition than the wild-type strain. However, both strains showed similar sensitivities to cycloheximide. This experiment suggests that the absence of Dbp2p decreases the fidelity of translation.
TABLE 2.
Sensitivity to drugs affecting translation
| Strain | Zone of inhibition (cm) with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Paromomycin (mg)
|
Cycloheximide (μg)
|
|||||||
| 2.50 | 1.25 | 0.50 | 0.25 | 2.50 | 0.50 | 0.13 | 0.05 | |
| Wild type | <0.6 | <0.6 | <0.6 | <0.6 | 2.1 | 0.7 | <0.6 | <0.6 |
| dbp2Δ | 1.7 | 1.7 | 1.3 | 0.8 | 2.0 | 0.8 | <0.6 | <0.6 |
Dbp2p is predominantly cytoplasmic and associates with polysomes.
In S. cerevisiae, nonsense-mediated mRNA decay occurs in the cytoplasm. This conclusion stems from several observations, including those showing that the major factors and mRNA decay intermediates specific to the pathway are associated with polysomes (2, 3, 42) and that decay occurs while the transcripts remain associated with ribosomes (42, 67). To understand the role of Dbp2p in the nonsense-mediated mRNA decay pathway further, we examined its subcellular distribution by Western blotting of cellular fractions. As controls for these experiments, blots were probed with antibodies to a known nuclear protein (the α-subunit of RNA polymerase II, Rpo21p) and to a known cytoplasmic protein (phosphogycerate kinase, Pgk1p). The antibody used to detect Dbp2p was generated against the DEAD-box domain and thus cross-reacts with other DEAD-box proteins (4). We identified the specific Dbp2p band on Western blots by comparing extracts from wild-type and dbp2Δ strains (data not shown and R. Iggo, personal communication). In a comparison of cytoplasmic and nuclear extracts, Dbp2p, like Upf1p, shows a largely cytoplasmic profile, with a small amount of the protein localized to the nucleus (Fig. 6A). The relative absence of Pgk1p in the nuclear fraction underscores the purity of this fraction and the significance of the cytoplasmic localization of Dbp2p.
A higher resolution analysis of the localization of Dbp2p was obtained by determining whether it associated with polyribosomes. Extracts from wild-type cells were fractionated on sucrose gradients, and fractions were evaluated for the presence of Dbp2p by Western blotting. As seen with Upf1p, Dbp2p cofractionated with polysomes, although the majority of the protein was found in the lighter, nonpolysomal fractions (Fig. 6B). The polysomal association appears to be specific, since the ribosomal protein, Tcm1p, was also detected in the polysomal fractions while Pgk1p was absent from these fractions. To confirm the specificity of the Dpb2p association with polyribosomes, we repeated the analysis in a strain harboring the prt1-1 allele. This temperature-sensitive mutation in the gene encoding the p90 subunit of elF3 confers rapid inhibition of growth and translation initiation at 37°C (61). When extracts were prepared from cells shifted to the nonpermissive temperature, the prt1-1 sample showed a loss of Dbp2p from the polysomal fractions with which it remained associated in the wild-type sample (Fig. 6C and D). This suggests that a portion of cytoplasmic Dbp2p associates specifically with polysomes.
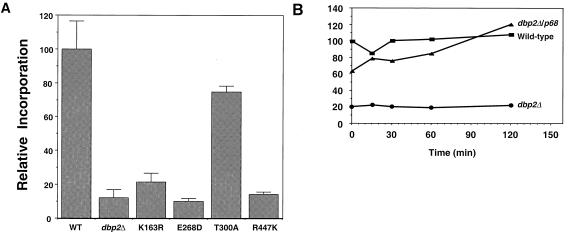
Cells harboring a dbp2Δ mutation show decreased levels of protein synthesis.
The slow growth rate and translational fidelity effects that accompanied deletion of DBP2 prompted us to determine the relative rates of protein synthesis in wild-type and dbp2 strains. This was monitored initially by assaying the incorporation of 35S-labeled amino acids (a mixture of methionine and cysteine). In this assay, the extent of protein synthesis in dbp2Δ cells was approximately 10-fold less than that of wild-type cells (Fig. 7A). Consistent with this result, cells harboring dbp2 alleles with mutations affecting the putative ATP binding (K163R), ATPase (E268D), and RNA binding (R447K) domains exhibited 5- to 10-fold reductions in amino acid incorporation (Fig. 7A). The only dpb2 allele with a modest effect on protein synthesis was that harboring the T300A mutation in the helicase-switch region.
FIG. 7.
Amino acid incorporation in wild-type (WT) and mutant strains. (A) Incorporation of 35S-labeled amino acids was measured in HFY1200 (wild type), yATB100 (dbp2Δ), yATB101 (K163R), yATB102 (E268D), yATB103 (T300A), and yATB104 (R447K) as described in Materials and Methods. The values are given as the percentage incorporation, with the wild type taken as 100%, and are the averages of at least five samples. The error bars denote the standard deviations of the five separate samples. (B) Cells were subjected to galactose induction for different lengths of time, and incorporation of 35S-labeled amino acids was measured as described in Materials and Methods. Data are expressed as the percentage of incorporation at t0 and are the averages of triplicate samples. Squares depict HFY1200 cells (wild type), circles depict yATB100 cells (dbp2Δ), and triangles depict yATB200 cells (dbp2Δ+p68).
Expression of the human p68 gene in a dbp2Δ strain fully complements the slow-growth phenotype but does not restore nonsense-mediated mRNA decay (see reference 4 and Fig. 2E). To consider whether the effects on growth rate were related to the effects on protein synthesis, we induced expression of the human p68 gene in a dbp2Δ strain and monitored incorporation of 35S-labeled amino acids. Induction of the p68 gene in dbp2Δ cells restored protein synthesis to wild-type levels in approximately 90 min (Fig. 7B). Furthermore, the presence of the galactose-inducible p68 gene was sufficient to cause the same cells to increase their level of amino acid incorporation approximately threefold when grown in glucose-containing medium, i.e., expression of the inducible gene must be leaky. These results demonstrate that expression of p68 complements the translational deficit of dbp2Δ and, more importantly, indicates that Dbp2p's role in translation is separate from the role it plays in mRNA decay. This conclusion is significant because there is a considerable body of evidence indicating that mRNA decay can be affected by perturbations in protein synthesis (31).
Protein synthesis effects in dbp2Δ cells are accompanied by reduced levels of polysomes, monosomes, and 60S ribosomal subunits.
To obtain insight into the specific step of translation affected by the absence of Dbp2p, we analyzed polysome profiles from wild-type and dbp2Δ cells. Cytoplasmic extracts from dbp2Δ cells, fractionated on 15 to 47% sucrose gradients, routinely showed fewer ribosomes per mRNA (an average of 6 in wild-type cells and 3 in dbp2Δ cells), lower levels of 80S subunits than wild-type cells (Fig. 8A and B), and, in higher-volume gradients, the presence of half-mers (data not shown). To obtain higher resolution of the fractions containing monosomes and ribosomal subunits, we repeated these experiments using 7 to 25% sucrose gradients and demonstrated again the relative dearth of 80S monosomes in the dbp2Δ strain (Fig. 8C and D). These experiments also showed that although the levels of 40S subunits were comparable in extracts from mutant and wild-type cells, there were substantially fewer free 60S subunits in the dbp2Δ cells (Fig. 8C and D). The decrease in free 60S ribosomal subunits in dbp2Δ cells is consistent with the occurrence of smaller polysomes, half-mers, and diminished translation.
To determine the means by which the human p68 gene is able to restore high levels of protein synthesis to dbp2Δ cells (see above), we assessed the effects of p68 gene expression on the levels of polysomes, 80S monosomes, and 60S ribosomes. Expression of the p68 gene was induced as before, and after 4 h of induction cytoplasmic extracts were prepared and fractioned over 15 to 47% sucrose gradients. As is evident in Fig. 8E and F, expression of p68 increased the amount of polysomes and the number of 60S and 80S ribosomes and eliminated the trace levels of half-mers present between the smaller polysome peaks. This result is consistent with the ability of the p68 gene to complement the translation defect of a dbp2Δ strain and implicates p68 in 60S ribosome biogenesis or function.
Dbp2p is involved in rRNA processing.
The dramatic decrease in 60S ribosomal subunits observed in the polysome analyses of dbp2Δ cells (Fig. 8) are comparable to those observed previously in strains harboring mutations that affect rRNA processing (reference 60 and references therein). This decrease, as well as the observation that total RNA isolated from dbp2Δ cells appears to contain reduced amounts of rRNA as detected by ethidium bromide staining (data not shown), suggests that Dbp2p, too, may have a role in rRNA processing. To consider this possibility, RNA from wild-type, dbp2Δ, and dbp2 K163R strains was analyzed by Northern blotting using oligonucleotide probes complementary to the 18S and 25S rRNAs or to selected regions of their common 35S precursor. Figures 9A and B illustrate the rRNA processing pathway (60) and the regions of the different transcripts complementary to the respective probes. Initially, these experiments confirmed the ethidium bromide staining results, demonstrating that the mature 25S and 18S rRNAs are considerably reduced in amount in both dbp2 mutant strains (Fig. 9C). To assess whether this reduction reflected altered levels of the 35S precursor, the blot was reprobed with a 5′ external transcribed spacer (ETS)-specific oligonucleotide. This experiment demonstrated that both dbp2 mutant strains accumulated significant levels of the 35S precursor, whereas wild-type cells do not (Fig. 9D). To determine whether the accumulation of the 35S precursor in mutant cells might be attributable to alterations in the downstream processing events that lead to the accumulation of 25S rRNA, the blot was sequentially reprobed with oligonucleotides capable of detecting the 27SA2 RNA, all 27SA species, or all categories of 27S rRNA precursor (Fig. 9E through G). Probing the blot with an oligonucleotide complementary to the internal transcribed spacer 1 (ITS1) region between A2 and A3 showed that dbp2Δ cells contain slightly less 27SA2 RNA than wild-type cells, but this effect is not manifested in the K163R mutant (Fig. 9E). Similar results were obtained when the blot was probed for overall 27SA by using an oligonucleotide specific to the A3-to-B region of ITS1 (Fig. 9F), indicating that the 27SA2 is the predominant species found with both probes. Using a probe specific for the ITS2 region between the 5.8S and 25S subunits that detects all species of 27S RNA, we observed that dbp2Δ cells contain substantially less 27S RNA than wild-type cells and that K163R mutant cells also contain decreased amounts of this RNA population (Fig. 9G). These results indicate that there may be a delay in processing between the 27SA2 and the 27SB precursor rRNAs and lead us to conclude that Dbp2p has a role in the biogenesis of the 25S rRNA.
Since the human p68 gene is able to complement the slow growth, protein synthesis, and 60S ribosome deficit of dbp2Δ cells (see above), we tested whether its expression would also complement the rRNA processing deficiency of the mutant cells. After inducing the regulatable p68 gene with galactose, RNA was isolated at different time intervals and analyzed by Northern analysis for the relative abundance of the 18S and 25S rRNA species (Fig. 9H). It is evident from this experiment that the defect leading to low levels of the mature rRNAs is complemented by expression of the human p68 gene.
Generation of mature 25S rRNA is reduced in the dbp2Δ strain.
To further dissect the role that Dbp2p plays in the formation of mature rRNAs, we performed a pulse-chase labeling of pre-rRNA with [methyl-3H]methionine. Processing of the 35S pre-rRNA, monitored during a 15-min chase, was considerably slower in the dbp2Δ strain than in the wild-type strain (Fig. 10; compare lanes 1 to 4 with 5 to 8). In addition, a delay in the formation of the 25S rRNA was evident. Interestingly, the processing of the 20S pre-rRNA to the mature 18S rRNA was not affected in the dbp2Δ strain. These results indicate that Dbp2p plays a specific role in 35S and 27S processing which leads to a decrease in the 60S ribosomal subunits.
FIG. 10.
The dbp2 null mutation leads to reduced synthesis of the mature 25S rRNA. Strains HFY1200 (wild type) and yATB100 (dbp2Δ) were grown in SC-met medium. Cells were pulse-labeled (p) for 1 min with [methyl-3H]methionine and then chased (c) for 2, 5, and 15 min with unlabeled methionine. Total RNA was isolated, and equal counts per minute were run on a 1.2% agarose-formaldehyde gel, transferred to a Zeta-probe membrane, and visualized by fluorography. The positions of the pre-rRNAs and mature rRNAs are indicated.
DISCUSSION
Dbp2p, a Upf1p-interacting DEAD-box protein, has a role in nonsense-mediated mRNA decay.
Many complex cellular processes that affect gene function and expression, including DNA recombination, pre-mRNA splicing, rRNA processing, and protein synthesis, have been shown to require the activities of the DNA and RNA helicases of superfamilies I and II (reviewed in references 14 and 44). Helicase function has also been implicated in mRNA decay. The UPF1 gene, for example, is required for the rapid turnover of transcripts containing premature translational termination codons (39, 51), and biochemical analyses of its encoded protein have shown that it binds RNA and has ATPase and helicase activities (13, 62–64). Further elucidation of the role of Upf1p in nonsense-mediated mRNA decay was sought by using a two-hybrid screen to identify its potential interactors. Six interacting proteins were identified in this screen, including two (Dcp2p/Nmd1p and Nmd2p) that were shown subsequently to be involved in mRNA decay (9, 15, 21) and one (Nmd3p) with a role in ribosome biogenesis that may also link Upf1p to the 60S ribosomal subunit (5). In this study, we have characterized another of the Upf1p-interacting proteins, the putative helicase Dbp2p.
Dbp2p interacts specifically with Upf1p, and the Dbp2p interaction domain on Upf1p is largely separate from that involved in interactions with Nmd2p or Sup35p (Fig. 1) (19, 22, and F. He and A. Jacobson, unpublished experiments). Upf1p:Dbp2p interaction appears to be evolutionarily conserved, since the Upf1p interaction domain of Dbp2p encompasses a highly conserved region in the p68 family of DEAD-box proteins and the corresponding region in the human p68 protein is able to interact with Upf1p (Fig. 1D). Interaction with Upf1p implicated Dbp2p in the nonsense-mediated mRNA decay pathway and the enhanced abundance of the CYH2 pre-mRNA, can1-100 mRNA, and mini-PGK1 nonsense transcript in dbp2Δ cells (Fig. 2A through D) confirmed this notion. Dbp2p's role in nonsense-mediated mRNA decay must differ from that of Upf1p, however, since a full-length PGK1 transcript harboring an early nonsense codon was not stabilized in dbp2Δ cells (Fig. 2D). This same phenomenon, stabilization of truncated but not full-length nonsense-containing transcripts, has been observed previously in cells harboring a mof2/sui1 allele and may reflect the ability of an impaired but not completely inactivated decay pathway to respond to the occurrence of multiple downstream elements within the larger transcript (8). Although the human p68 gene complemented other phenotypes of dbp2Δ cells, it was not capable of reversing the mRNA decay defect seen in these cells. This suggests that either Dbp2p has at least two discrete functions, one of which cannot be carried out by p68, or that p68 is limited in its ability to reproduce precisely the principal role of its yeast counterpart (see below).
Mutations within evolutionarily conserved DEAD-box family motifs inactivate Dbp2p.
Among the conserved motifs of the DEAD-box proteins, those designated I, II, III, and VI have been characterized by mutational analysis. Motif I has also been described as the Walker A motif of the large family of ATPases, and mutational analysis of elF-4A, a member of the DEAD-box family, has shown that it is essential for ATP binding and, accordingly, for ATPase and helicase activities (49). Using site-directed mutagenesis, we replaced the highly conserved lysine residue of this motif with an arginine residue. Although this conservative change retained the positive charge of the lysine residue, this allele was not able to complement the slow-growth or nonsense-mediated mRNA decay phenotypes of a dbp2Δ strain (Fig. 3B and data not shown). Motif II, the DEAD-box, is homologous to the Walker B motif of the ATPase family of proteins and mutational analyses of the conserved D-E residues have implicated this domain as the site of ATP hydrolysis (49, 57). Again, a conservative mutation within this site was sufficient to render the protein incapable of complementing the slow-growth (data not shown) and nonsense-mediated mRNA decay phenotypes of a dbp2Δ strain (Fig. 3B). Motif III within the DEAD-box family of helicases, known for its conserved S-A-T residues, has also been studied in elF-4A. Substitution of the conserved serine and threonine residues of this motif with alanines completely abolished elF-4A helicase activity, without effects on ATP binding, ATPase, or RNA binding activities (49, 50). This domain is sometimes referred to as the switch region because it is thought to act as a switch from ATP hydrolysis to RNA unwinding. Dbp2p containing a T300A mutation in this region is still able to complement the slow-growth phenotype (data not shown) and at least partially complement the nonsense-mediated mRNA decay phenotype of dbp2Δ cells (Fig. 3B). Finally, Motif VI is thought to be involved in protein-RNA interaction, since mutations within this domain of elF-4A reduce RNA binding and, therefore, RNA helicase activity (50). The conservative R447K mutation that we constructed in this domain of Dbp2p could not complement the slow-growth or the nonsense-mediated mRNA decay phenotypes of a dbp2Δ strain (Fig. 3B and data not shown) and may exert a dominant-negative effect (data not shown). Collectively, the mutational analyses of Dbp2p implicate ATP binding, ATP hydrolysis, and RNA binding as Dbp2p activities essential for nonsense-mediated mRNA decay, implying that this protein, like p68 (29), is likely to behave as an RNA helicase in vivo. If correct, this would indicate that, in yeast, nonsense codon recognition, translation termination, and subsequent mRNA destabilization may require the activity of at least three RNA helicases, i.e., Upf1p, Mtt1p, and Dbp2p (11, 13, 62–64).
Dbp2p also plays a role in ribosome biogenesis and function.
Sixteen of 20 DEAD-box proteins identified in S. cerevisiae appear to function in either ribosome biogenesis or translation (14). This observation, combined with the cold sensitivity, reduced growth rate, and poor yield of rRNA in dbp2Δ cells led us to investigate whether Dbp2p also had a role in these posttranscriptional processes. A Dbp2p effect on translation was indicated by several different experimental results, including those showing that cells with disruptions or point mutations within DBP2 had altered sensitivity to translation inhibitors, reduced rates of amino acid incorporation, fewer ribosomes per mRNA, fewer monosomes and 60S subunits, and more half-mers than wild-type cells (Fig. 7 and 8, Table 2, and data not shown).
The appearance of half-mers and the reduced yield of 60S subunits suggested that dbp2Δ cells might also have defects in rRNA processing, a hypothesis confirmed by Northern blotting and pulse-chase analyses. Cells harboring dbp2Δ or the dbp2 K163R mutation had elevated levels of the 35S rRNA precursor and significant decreases in the 25S/27SA2 ratio and the overall levels of 27S and 25S species (Fig. 9). Since 27SA2 precursor accumulation was only slightly affected (Fig. 9C), Dbp2p most likely plays a principal role in the formation of the 27SA3 precursor and a lesser role in the formation of the 27SA2 precursor. The lack of functional Dbp2p must, therefore, interfere with the major processing pathway that leads to the 27SA3 precursor and, ultimately, to the 5.8Ss and 25S rRNAs. The minor pathway leading to the 5.8SL and 25S rRNAs must compensate for this deficit (see Fig. 9B). This would explain why the levels of 25S rRNA are decreased to a greater degree than 18S rRNA in dbp2 strains and why the level of free 60S subunits is severely affected with little or no detectable change in the level of free 40S subunits. Additional evidence for an active role in rRNA processing is demonstrated by the kinetic delay in 27S and 25S rRNA formation, as seen in the pulse-chase experiment (Fig. 10). Interestingly, there is no delay in the processing of the 20S precursor, although the overall levels of 18S rRNA are diminished. These observations lead us to the conclusion that Dbp2p plays a specific role in the formation of the 27S pre-rRNA and the 25S rRNA.
Expression of the human p68 gene in dbp2Δ cells complements the translation defect of these cells as well as their reduced levels of 60S ribosomes, altered polysome profiles, and deficiencies in rRNA processing (Fig. 7 to 9). These results indicate that the translation defect of dbp2Δ cells is most likely to be directly attributable to reductions in, or alterations of, their ribosome pool and suggest that p68 may well have a role in rRNA processing in human cells as well. The latter conclusion is consistent with earlier studies showing that p68 shuttles between the nucleus and the nucleolus in a cell cycle-dependent manner (28). Expression of the p68 gene in dbp2Δ cells did not, however, complement the nonsense-mediated mRNA decay phenotype of these cells (Fig. 2E). Since there is substantial evidence that some degree of ongoing protein synthesis is a requirement for nonsense-mediated mRNA decay (32, 61, 67, 68), the fact that p68 did restore translation minimizes the likelihood that the failure to restore decay can be attributed to indirect effects.
A common function for Dbp2p in rRNA processing and mRNA decay?
Our data indicate that Dbp2p has a role in both rRNA processing and nonsense-mediated mRNA decay. While it appears that the mRNA decay phenotype of dbp2Δ cells cannot be ascribed to their translation defects (see above), the possibility remains that rRNA processing is altered slightly in p68-complemented cells and that such alterations may interfere with the ability of the ribosome to trigger mRNA destabilization upon nonsense codon recognition. However, Dbp2p appears to have a major role in the cytoplasm, since it accumulates in that compartment (Fig. 6A) as well as in nuclei (Fig. 6A) (35) and cofractionates with polysomes (Fig. 6B). Dbp2p also interacts with Upf1p, a largely cytoplasmic protein (2, 3, 32, 42), and affects mRNA decay, translational fidelity, and sensitivity to drugs that alter translational fidelity in the same manner as Upf1p (Fig. 4 and 5 and Table 2) (7, 22, 40, 48, 58).
Upf1p is thought to play a role in translational termination, and it may interact with factors 3′ of the stop codon to promote efficient ribosome release and possible recycling for additional rounds of initiation (32). At premature termination codons, factors normally proximal to the termination site may be absent and qualitative or quantitative changes in ribosome release and recycling may ensure. In turn, such changes may trigger rapid decay of the aberrant transcript (32). Dbp2p could fit into this scheme, as well as the rRNA processing pathway, if it unwound portions of rRNA structure, allowing specific processing events in one instance and facilitating dissociation of the termination complex in the other. The latter role is quite similar to that proposed for Nmd3p and Prt1p, two proteins that interact with Upf1p or selectively promote degradation of nonsense-containing transcripts, respectively (5, 61).
ACKNOWLEDGMENTS
This work was supported by a grant to A.J. (GM27757) from the National Institutes of Health and a postdoctoral fellowship to D.M. from the Medical Foundation/Charles A. King Trust.
We are indebted to Richard Iggo for helpful information, Stanley Hollenberg, Richard Iggo, Elizabeth Jones, David Lane, and Stuart Peltz for plasmids, antibodies, and yeast strains, and the members of the Jacobson lab for their experimental advice and editorial comments.
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jack H M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Schenkman L R, Eastman M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 4.Barta I, Iggo R. Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 1995;14:3800–3808. doi: 10.1002/j.1460-2075.1995.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belk J P, He F, Jacobson A. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA. 1999;5:1055–1070. doi: 10.1017/s1355838299990027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Y, Dinman J, Peltz S W. mof4–1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and -1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Y, Gonzalez C I, Kinzy T G, Dinman J D, Peltz S W. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA. 1999;5:794–804. doi: 10.1017/s1355838299982055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 10.Culbertson M R, Underbrink K M, Fink G R. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czaplinski K, Majlesi N, Banerjee T, Peltz S W. Mtt1 is a Upf1-like helicase that interacts with the translation termination factors and whose expression can modulate termination efficiency. RNA. 2000;6:730–743. doi: 10.1017/s1355838200992392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1p: a factor involved in mRNA turnover. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 14.De La Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 15.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg A, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink G, editors. Methods in enzymology: molecular biology of Saccharomyces cerevisiae. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 18.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He F, Brown A, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 20.He F, Brown A, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway using an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 22.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho J H, Johnson A W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu C, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iggo R D, Jamieson D J, MacNeill S A, Southgate J, McPheat J, Lane D P. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol. 1991;11:1326–1333. doi: 10.1128/mcb.11.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iggo R D, Lane D. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iost I, Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson A, Peltz S. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson A, Peltz S. Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae. In M. Mathews, J. Hershey, and N. Sonenberg (ed.), Translational control. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 827–847. [Google Scholar]
- 33.Johnson A W. Rat1p and Xm1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karl T, Onder K, Kodzius R, Pichova A, Wimmer H, Thur A, Hundsberger H, Loffler M, Klade T, Beyer A, Breitenbach M, Koller L. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr Genet. 1999;34:419–429. doi: 10.1007/s002940050416. [DOI] [PubMed] [Google Scholar]
- 35.Kumar A, Cheung K H, Ross-Macdonald P, Coelho P S, Miller P, Snyder M. TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res. 2000;28:81–84. doi: 10.1093/nar/28.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaGrandeur T, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larimer F W, Hsu C L, Maupin M K, Stevens A. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 38.Lee B S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2203–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 40.Maderazo A B, He F, Mangus D A, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangus D A, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangus D A, Jacobson A. Linking turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 43.Mangus D A, Jang S H, Jaehning J A. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J Biol Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- 44.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 45.Muhlrad D, Decker C, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 46.Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliveira C C, McCarthy J E. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 48.Palmer E, Wilhelm J, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 49.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor elF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peltz S, Brown A, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 52.Peltz S W, Donahue J L, Jacobson A. A mutation in tRNA nucleotidyltransferase stabilizes mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perlick H A, Medghalchi S M, Spencer F A, Kendzior R J, Jr, Dietz H C. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose M, Winston F, Heiter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 55.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid S R, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 59.Soni R, Carmichael J, Murray J. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simple procedure. Curr Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- 60.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 61.Welch E, Jacobson A. An internal open reading frames triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weng Y, Czaplinski K, Peltz S W. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol. 1996;16:5477–5590. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weng Y, Czaplinski K, Peltz S W. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weng Y, Czaplinski K, Peltz S W. ATP is a cofactor of the Upf1p protein that modulates its translation termination and RNA binding activities. RNA. 1998;4:205–214. [PMC free article] [PubMed] [Google Scholar]
- 65.White T, Arnheim N, Erlich H. The polymerase chain reaction. Trends Genet. 1989;5:185–189. doi: 10.1016/0168-9525(89)90073-5. [DOI] [PubMed] [Google Scholar]
- 66.Zhang S, Ruiz-Echevarria M J, Quan Y, Peltz S W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol Cell Biol. 1995;15:2231–2244. doi: 10.1128/mcb.15.4.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang S, Welch E M, Hogan K, Brown A H, Peltz S W, Jacobson A. Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA. 1997;3:234–244. [PMC free article] [PubMed] [Google Scholar]
- 68.Zuk D, Jacobson A. A single amino acid substitution in yeast elF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]