Abstract
Ubiquitination is one of the most important post‐translational modifications which plays a significant role in conserving the homeostasis of cellular proteins. In the ubiquitination process, ubiquitin is conjugated to target protein substrates for degradation, translocation or activation, dysregulation of which is linked to several diseases including various types of cancers. E3 ubiquitin ligases are regarded as the most influential ubiquitin enzyme owing to their ability to select, bind and recruit target substrates for ubiquitination. In particular, E3 ligases are pivotal in the cancer hallmarks pathways where they serve as tumour promoters or suppressors. The specificity of E3 ligases coupled with their implication in cancer hallmarks engendered the development of compounds that specifically target E3 ligases for cancer therapy. In this review, we highlight the role of E3 ligases in cancer hallmarks such as sustained proliferation via cell cycle progression, immune evasion and tumour promoting inflammation, and in the evasion of apoptosis. In addition, we summarise the application and the role of small compounds that target E3 ligases for cancer treatment along with the significance of targeting E3 ligases as potential cancer therapy.
Keywords: cancer, E3 ubiquitin ligases, targeted therapy, ubiquitination
E3 ubiquitin ligases are important players in cellular processes by ubiquitinating substrate proteins in disease progression such as cancer. Owing to this, scientists attempt to identify safe and bioavailable compounds targeting E3 ligases. In this review, we summarise the roles of E3 ligases in cancer‐related pathways, highlight the application and clinical significance of small molecule inhibitors and discuss further directions
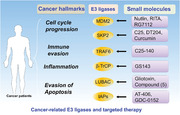
.
1. INTRODUCTION
Cancer is a heterogeneity disease characterised by diverse dysregulated biological processes. 1 Normal cell growth is controlled by extracellular and intracellular cues that direct the determination of various cellular processes such as cell signalling, cell cycle, DNA repair, transcriptional regulation and apoptosis. 2 The regulation of these cellular processes is greatly influenced by post‐translational modifications. In principle, these are achieved by regulating the activity of proteins involved in the various cellular processes with the dysregulated post‐translational modification associated with aberrant cell function and the manifestation of diseases such as cancer. 3 , 4 Ubiquitination is one of the most important post‐translational modification that involves the transfer of ubiquitin, a small regulatory protein, to specific substrates for proteasome degradation. 5 Non‐proteolytic functions of ubiquitin have also been reported including transcription and translation regulation, DNA repair, protein trafficking, signalling activation and suppression. 6 , 7 , 8
Ubiquitin‐substrate linkage is an isopeptide bond between the C‐terminal (glycine 76) of ubiquitin and ɛ‐amino group of the lysine residue of substrate protein. 9 Ubiquitin can also be conjugate to threonine, serine or cysteine residue as well as the free N‐terminal residue (α‐NH2 group) of the substrate. 10 , 11 , 12 Substrates are monoubiquitinated, multi‐monoubiquitinated or polyubiquitinated. Monoubiquitination is mostly implicated in non‐proteolytic function as exemplified in histone monoubiquitination implicated in chromatin modification, DNA damage response and repair signalling. 13 Histone 2A (H2A) ubiquitination at lysine 119 (K119) by polycomb repressor complex 1 (PRC1) and histone 2B (H2B) ubiquitination at lysine 120 (K120) by RING finger complexes (RNF20/40) are the well‐known histone monoubiquitination whose deregulation has been identified in several malignancies. 13 , 14 In polyubiquitination, the first ubiquitin binds the specific lysine residue of the protein substrate while the incoming ubiquitin binds the internal lysine of the previously bound ubiquitin forming Ub–Ub chain. There are seven lysine residues and an N terminal methionine (K6, K11, K27, K29, K33, K48, K63 and M) within ubiquitin moiety for polyubiquitination chain assembly. 15 Polyubiquitin chains via K48 and K11 are known to target substrate for degradation by downstream 26S proteasome. 16 , 17 K63‐ and M1‐linked linear ubiquitin chains perform non‐proteolytic functions and are critical players in immune response and inflammatory signalling. 17 , 18 , 19 Other ubiquitin linkages are not fully characterised although they have been shown to function in immune response, DNA damage and repair response, mitophagy and cellular stress response. 20 , 21 , 22
Three component enzymes are engaged in the ubiquitination process including ubiquitin‐activating enzyme (E1), ubiquitin‐conjugating enzyme (E2) and ubiquitin ligase (E3). 23 The specificity of the ubiquitin system is determined by E3 ligases which specifically select proteins for ubiquitination. 24 E3 ligases can select target proteins by identifying a specific peptide motif termed degron in the substrate. 25 Such degrons include N‐terminal and C‐terminal degrons, 26 proline‐rich motif (PPRX) typically recognised by WW domain containing E3 ligase, 27 phospho‐degron common for WD40 containing E3 ligase 28 and D box and Ken box found commonly in APC/C substrates. 29 Plethora of studies have shown that E3 ligases are an important player in cancer progression explainable by their regulatory effect on proteins involved in various cellular processes. Owing to this fact, there has been a growing attempt to identify safe and bioavailable compounds that target E3 ligases with high specificity for cancer therapy. Herein, we summarise the roles of E3 ligases in cancer promoting pathways like cell cycle progression, immune evasion, inflammatory signalling and apoptosis escape. In addition, we highlight the application and clinical significance of small molecule inhibitors of E3 ligase as well as small molecule degraders, recruiting E3 ligase for target protein degradation.
2. CLASSIFICATION OF E3 UBIQUITIN LIGASES
There are over 600 E3 ligases identified hitherto, each targeting specific substrate proteins. All E3 ligases can be grouped into three families; the really interesting new gene (RING) finger family, the homologous to E6AP C terminus (HECT) family and the RING between RING (RBR) family.
2.1. RING finger family E3 ligases
The RING finger E3 ligases make up the largest E3 family that contains the RING or U‐box catalytic domain. The canonical RING finger is a cysteine‐rich domain with the sequence of Cys‐X2‐Cys‐X(9‐39)‐Cys‐X(1‐3)‐His‐X(2‐3)‐Cys‐X2‐Cys‐X(4‐48)‐Cys‐X2‐Cys (where X represents any amino acid). It uses the cysteine and histidine residues to coordinate two zinc ions in an eight ligands cross based structure. 30 , 31 RING finger E3 ligases do not bind ubiquitin directly but mediate the transfer of ubiquitin from bound E2 (E2‐Ub) to the target substrate. 32 Ring finger E3 ligases may function independent of any auxiliary protein, as a monomer or homodimer, such as TRIM (Tripartite Motif containing E3 ligases), TRAF6 (TNF Receptor Associated Factors 6), cIAP (cellular Inhibitor of Apoptosis), XIAP (X‐linked inhibitor of apoptosis), RING finger (RNF) containing proteins and MDM2 (Murine Double Minute 2) 33 , 34 , 35 , 36 , 37 or associate with another protein and function as a heterodimer or multisubunit complex. Heterodimeric ring finger E3 ligases include TRAF heterodimers, 38 MDM2/MDMX, 39 BRCA1/BARD1 40 and RING1B/BMI‐1. 41 MDMX, BARD1 and BMI‐1 in the above heterodimers have ring domain, but do not possess E3 ligase activity therefore they associate with the E3 ligase ring domain counterpart to promote substrate ubiquitination.
Multisubunit ring finger E3 ligases include the Cullin Ring Finger ligases (CRL) and Anaphase Promoting Complex/cyclosome (APC/C). The CRL is a large family, containing Cullin scaffold protein, E2 binding RING‐box protein (Rbx1 and Rbx2), adaptor protein and substrate recognition protein. 42 The substrate recognition proteins bind adaptors while the Cullin forms a central scaffold that bridges Rbx1/2 and the adaptor‐substrate recognition protein. Differences in Cullin type (Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, Cul7 and Cul9) form the basis for different groups of the CRL subfamily. 43 The specificity of substrate recruitment is determined by substrate recognition proteins with over 400 kinds identified. 44 SCF (SKP1‐Cullin1‐F‐box) E3 ligase is the largest CRL family with SKP1 as the adaptor and F‐box proteins as the substrate recognition unit. 45 There are 69 F‐box proteins encoded in human genome which can be grouped into three subfamilies based on their substrate recruiting domain: F‐box with WD‐40 domain (FBXW), F‐box with leucine rich repeat (FBXL) and F‐box with other domains (FBXO). 46 , 47 F‐box proteins are important regulators of diverse cell functions; an example is SKP2 (also called FBXL1), a popular oncoprotein that mediates the degradation of CDK inhibitors to enable G1 to S phase cell cycle progression. 48 The other ring finger complex, APC/C, functions in cell cycle mitotic progression. They contain 19 subunits that are grouped into three sub‐complexes: scaffolding platform, ring finger containing catalytic core and the tetratricopeptide repeat (TPR). 49 APC/C requires co‐activator proteins, CDC20 and CDH1 for substrate recruitment to the APC/C complex. The co‐activator enters the APC/C complex by binding APC3 and APC8 in the TPR sub‐complex, 50 , 51 recognises and recruits substrate containing KEN‐box or D‐box to the APC/C complex for ubiquitination. 29
2.2. HECT family E3 ligases
The HECT families of E3 ligase bear HECT catalytic domain at their Carboxyl terminal lobe. A flexible link with a cysteine active site connects the N terminal and C terminal lobes. 52 This cysteine active site, confers on them a distinct mechanism of ubiquitination from the ring finger families. In HECT‐mediated ubiquitination, ubiquitin from E2 first forms an intermediate thioester bond with the cysteine before being transferred to the target substrate. 53 Twenty‐eight HECT E3 ligases have been identified in human and are grouped into three different subfamilies based on N terminal domain including the NEDD4 family containing N‐terminal WW and C2 domain, the HERC family containing N‐terminal RCC‐like domain (RLD) and other HECT with variable N‐terminal domain. HECT members are actively involved in several cellular processes that drive cancer progression. For example, E6AP, a member of other HECT family drives cervical cancer by associating with E6 protein of human papillomavirus to promote proteasome degradation of p53 54 , 55 ; the NEDD4 family members WWP1 and NEDD4 promote PI3K/AKT signalling by catalysing the ubiquitin‐mediated degradation of PTEN 56 ; ITCH, another NEDD4 member, is an important regulator of immune response. 57 , 58
2.3. RBR family E3 ligases
The RBR ligases are considered the smallest E3 family with only 14 members among which some well‐known E3 ligases such as Parkin, HHARI, TRIAD1, HOIP, HOIL‐IL and RNF144 are included. 59 , 60 Canonical RBRs consist of three components: RING1 with an E2 binding domain, RING2 with a catalytic cysteine residue and an In‐between Ring domain (IBR). 61 RBRs are described as RING–HECT hybrid because the RING1 is structurally similar to Ring type E3 ligase, while the RING2 bears an exposed catalytic cysteine residue that forms intermediate thioester linkage with ubiquitin during substrate ubiquitination just like the HECT family. 62 Traditional RBRs exist in the form of N‐RING1–IBR–RING2‐C; however, they can contain additional domains at the N terminal, middle or C terminal. These outside domains confer a characteristic auto‐inhibitory action on RBR, for example, the C‐terminal Ariadne domain of HHARI blocks RING2 active site leading to HHARI autoinhibition. 60 Parkin, a tumour suppressor RBR implicated in neurodegeneration disease and innate immune response, is kept in an autoinhibition state by three outside domains, ubiquitin like domain (Ubl), Ring0 domain and a REP domain. 63 , 64 REP and Ubl mask E2 binding site on RING1, while Ring0 binds to RING2 and blocks its catalytic cysteine residue thereby keeping Parkin in an autoinhibition state. 65 , 66 Phosphorylation of the Ubl domain by PTEN‐induced kinase 1 (PINK1) and the binding of phosphorylated ubiquitin to Parkin disrupt the autoinhibition state, releasing Parkin for activity. 67 , 68 LUBAC (Linear ubiquitin chain assembly complex), another RBR ubiquitin ligase active in innate immune response, is composed of HOIP, HOIL‐IL and SHARPIN that generates linear polyubiquitin chains. 19 , 60 HOIP bears ubiquitination catalytic activity but is kept in the inactive auto‐inhibited state by the ubiquitin‐associated (UBA) domain at its N terminus. To distort the autoinhibition, the ubiquitin like domain of either HOIL‐IL or SHARPIN interacts and forms a complex with the UBA domain of HOIP thereby activating HOIP for M1 linear ubiquitination. 69 , 70 In addition to the established E3 ligase catalytic activity of HOIP, HOIL‐1L has been identified as E3 ligase that catalyses oxyester linked self‐monoubiquitination and monoubiquitination of protein components of Toll‐like receptor (TLR). 12
3. ROLES OF E3 LIGASES IN CANCER HALLMARKS
Hanahan and Weinberg 71 described ten hallmarks of cancer which are acquired strategies by cancer cells that enable their survival, growth and metastasis. Significant numbers of evidences have demonstrated the participation of ubiquitination in cancer hallmark pathways. For brevity, we will particularly summarise recent findings on the implication of E3 ubiquitin ligases in cell cycle progression, immune evasion and inflammation and the evasion of apoptosis.
3.1. Role of E3 ubiquitin ligases in cell cycle progression
Cancer cells abrogate cell cycle regulation to sustained proliferation and progression. The cell cycle is a series of incidents that progress through four phases: gap1 phase (G1 phase), DNA synthesis phase (S phase), gap2 phase (G2 phase) and mitosis phase (M phase). The cyclin‐dependent kinases (CDKs) are key regulators of the cell cycle that drive cell division by forming complex with cyclins with distinct CDK/cyclin complex operating at different phases. 72 , 73 Cyclins are short‐lived proteins degraded during cell cycle by E3 ligases, 74 while CDKs are relatively stable; however, their activities can be inhibited by other cell cycle regulatory proteins among which E3 ligases represent specific functions. 75 The classical E3 ligases regulating cell cycle include APC/C and SCF containing the substrate recognition proteins such as SKP2, β‐TrCP and FBXW7 (Figure 1 and Table 1). 76 , 77
FIGURE 1.
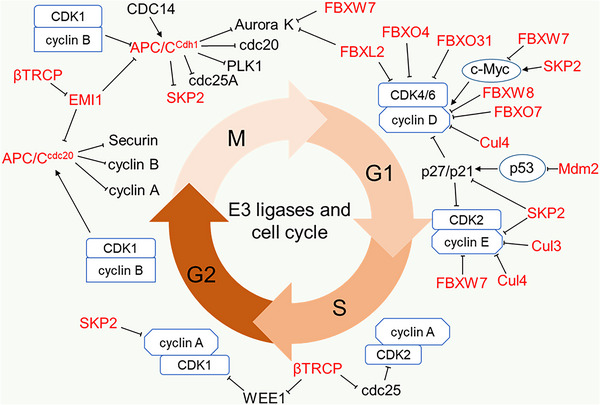
E3 ligases in cell cycle regulation. E3 ligases regulate cell cycle progression via regulating the CDK and cyclin activity at the four phases of the cell cycle. E3 ligases majorly act directly on Cyclins and mediate their ubiquitination and degradation, but for CDKs, E3 ligases act on the primary CDK regulators such as CDK inhibitors (p27, p21, p57), and mediate their ubiquitination and degradation.
TABLE 1.
E3 ligases implicated in cell cycle.
| E3 ligase | Target | Modification | Function of E3 ligase | Role in cancer | Reference |
|---|---|---|---|---|---|
| APC/CCDC20 | Cyclin A | Degradation | Promote anaphase onset | Oncogene | 49 , 314 |
| Cyclin B | Degradation | Promote anaphase onset | |||
| Securin | Degradation | Promote sister chromatid separation | |||
| APC/CCDH1 | CDC20 | Degradation | Promote mitotic exit | Tumour suppressor | 85 , 315 , 316 , 317 , 318 |
| Plk1 | Degradation | Promote mitotic exit | |||
| Aurora A/B | Degradation | Promote mitotic exit | |||
| CDC25A | Degradation | Prevent CDK2 accumulation | |||
| SKP2 | Degradation | Prevent CDK2 accumulation | |||
| SCFSKP2 | p21, p27, p57 | Degradation | Promote G1/S transition | Oncogene | 48 , 94 , 97 |
| Cyclin E | Degradation | Foster S phase progression | |||
| c‐MYC | Stabilisation | Cell cycle progression | |||
| SCFβ‐TrCP | EMI1 | Degradation | Activate APC/C | Oncogene /Tumour suppressor | 87 , 90 , 91 , 319 |
| CDC25A | Degradation | Prevent CDK2 activation | |||
| WEE1 | Degradation | Promote CDK1 activation | |||
| FOXO3 | Degradation | Promote cell cycle progression | |||
| SCFFBXW7 | Cyclin E | Degradation | Inhibit cell cycle progression | Tumour suppressor | 95 , 98 |
| c‐MYC | Degradation |
Prevent c‐MYC expression Inhibit cell proliferation |
|||
| CUL3RhoBTB3 | Cyclin E | Degradation | Inhibit cell cycle progression | Tumour suppressor | 320 , 321 |
|
Cul4CDT2 Cul4DCAF11 |
p21 | Degradation | Promote S phase progression | Oncogene | 101 , 284 |
| CuL4DDB2 | p27 | Degradation | Promote cell proliferation | Oncogene | 100 |
| CUL4AMBRA1 | Cyclin D | Degradation | Inhibit cell proliferation | Tumour suppressor | 322 |
| MDM2 | p53 | Degradation | Promote cell proliferation | Oncogene | 223 |
APC/C mainly mediates K11 polyubiquitination and degradation of its substrate. It requires its co‐activators CDC20 and CDH1 to regulate the cell cycle between the M phase and early G1 phase. 51 APC/CCDC20 is activated in the M phase by CDK1‐mediated phosphorylation while APC/CCDH1 is simultaneously phosphorylated and inhibited until the late M phase when there is a low mitotic kinase level. 78 , 79 , 80 The activated APC/CCDC20 is inhibited by the mitotic checkpoint complex at metaphase. This induces metaphase arrest to ensure the correct attachment of sister chromatids to bipolar spindle before transitioning into anaphase. 50 , 81 Upon mitotic checkpoint complex satisfaction, APC/CCDC20 catalyses the ubiquitination and proteasomal degradation of securin (a separase inhibitor) to promote chromatid segregation, as well as the degradation of cyclin A and cyclin B to promote anaphase onset. 49 The degradation of cyclin B weakens CDK1 and abates CDK1‐mediated inhibition of CDH1 during anaphase. This activity coupled with CDC14 phosphatase‐induced dephosphorylation of CDH1 promotes the activation of APC/CCDH1. 82 , 83 Active APC/CCDH1 then mediates the proteasome degradation of CDC20, Polo‐like kinase 1 (Plk1) and Aurora kinases (Aurora A and B) at late mitosis to ensure mitotic exit and the degradation of CDC25A (a phosphatase activator of CDK2) and SKP2 at early G1 to lower CDK accumulation. 84 , 85 In late G1 phase, early mitotic inhibitor 1 (EMI1) inhibits APC/C and disrupts APC/CCDH1‐mediated degradation of its substrates. Although EMI1 is an F‐box protein, its cell cycle regulating activity is F‐box domain independent, to this end, EMI1 regulates APC/CCDH1 by antagonising the APC/CCDH1 generation the of K11 polyubiquitin chain. 86 This inhibitory action of EMI1 continues through the S phase and G2 phase until it is degraded by SCFβ‐TrCP to allow cell cycle progression through mitosis. 87 Additionally, SCFβ‐TrCP promotes the degradation of CDC25A to suppress cell cycle progression and degrades WEE1 (CDK1 inhibitor) and transcription factor FOXO3 for cell cycle progression. 88 , 89 , 90 , 91
SKP2 is activated in the late G1 phase by CDK2, which phosphorylates and protects it from APC/CCDH1‐mediated degradation. 92 Once activated, SCFSKP2 facilitates G1/S transition by promoting the ubiquitination and degradation of Kip/Cip members of CDK inhibitors (p21CIP1, p27KIP1 and p57KIP2). 48 , 93 During the G1 phase, both SCFSKP2 and SCFFBXW7 target cyclin E for proteasomal degradation 94 , 95 , 96 and act as E3 ligases of the oncogenic transcription factor c‐MYC but with distinct functions. SCFSKP2 stabilises and promotes c‐MYC transcriptional activity, while SCFFBXW7 mediates proteasome degradation of c‐MYC. 97 , 98
In addition to the classical regulators mentioned above, many other E3 ligases including Cul3 E3 ligase complex, Cul4 E3 ligase complex and the SCF containing FBXW8, FBXO4, FBXO7, FBXO31 and FBXL2 have been identified as cell cycle regulators that directly target cyclins and CDK inhibitors as shown in Figure 1. 99 , 100 , 101 More important also is MDM2, the key E3 ligase targeting p53 for proteasome degradation. 102
Deregulation of cell cycle‐related E3 ligases has been reported in several cancers. SCFSKP2 complex is a positive regulator of cell cycle considered as proto‐oncogene because it targets CDK inhibitors and other tumour suppressors. The oncogenic role of SKP2 reconciles with its overexpression in various human cancers including ovarian adenocarcinoma, breast cancer, lung cancer, colorectal cancer, prostate cancer, leukaemia and squamous cell carcinoma. 103 , 104 , 105 , 106 Interestingly, deletion of SKP2 has been shown to compensate for anti‐tumour and cell safeguarding deficiency in p53 deleted cancer cells due to elevated p27 levels. Zhao and co‐workers demonstrated this using SKP2 knockout mice and found that loss of p53 and pRB in these mice blocked tumorigenesis due to cell cycle arrest mediated by accumulated p27, suggesting that SKP2 could be a promising target for p53 mutant cancer therapy. 107 Likewise, APC/C co‐activator CDC20 has been identified as an oncoprotein that is highly expressed in several human cancers. 108 , 109 , 110 , 111 , 112 Tumorigenic role of CDC20 is linked to its involvement in diverse cellular pathways where it targets tumour suppressors. CDC20 aberrant expression or the disruption of SAC‐mediated inhibition of CDC20 leads to tumorigenesis due to aneuploidy. 113 Conversely, APC/CCDH1 has been associated with a tumour suppressive role as majority of its substrates including SKP2 are known oncoproteins. APC/CCDH1‐mediated regulation of SKP2 turnover in cell cycle significantly contributes to its control of tumorigenesis in cells. Studies have shown that inhibition of CDH1 resulted in high cellular proliferation while an induced overexpression in solid tumours is associated with patients’ survival and low histological tumour grade in solid tumours. 114 , 115 Nonetheless, SCFβ‐TrCP performs dual roles in the cell cycle, promoting both cell cycle progression and cell cycle arrest. This suggests both oncogenic and tumour‐suppressive roles. Hence, the expression of β‐TrCP in cancers is not clearly defined; however, the expression could be linked to a context‐dependent role. The Cul4 E3 ligase complex is also a key regulator of cell cycle that targets CDK inhibitors p21 and p27 as well as cyclin E for degradation. Aberrant expression of Cul4 has been indicated in breast cancer, lung cancer, ovarian cancer and squamous cell cancer. 116 Studies show that Cul4 depletion reduces the proliferation of lung squamous cell carcinoma and small cell lung cancer. 117 Moreover, conditional knockdown of Cul4A in mice reduces sensitivity to UV‐induced skin cancer in a mechanism linked to its regulation of p21. 118 MDM2 is the primary inhibitor of p53 and is amplified in most cancers, especially p53WT cancer types. 119 , 120 FBXW7, on the other hand, is an established tumour suppressor that functions as SCF substrate recognition protein targeting several oncogenic proteins including cyclin E and c‐MYC for proteasome degradation. Concomitant with its tumour‐suppressive role, FBXW7 is frequently underexpressed or inactivated in most human cancers. 121 Taken together, these studies show that E3 ligases are critical regulators of cell cycles, and their cell cycle‐dependent oncogenic function could be considered in the development of therapies that selectively target E3 ligases to induce the cell cycle arrest and corresponding death of cancer cells with considerably lesser toxicity.
3.2. Roles of E3 ubiquitin ligases in immune response and inflammation
Evasion of immune response and stimulation of tumour‐promoting inflammation are among the several strategies adopted by cancer cells to sustain proliferation and progression. Host cells through the innate and adaptive immune response, release proinflammatory cytokines that activate macrophages, dendritic cells and natural killer (NK) cells for the destruction of tumour cells and infectious agent that threatens cell immune homeostasis. During immune surveillance, growing cancer cells escape from immune destruction and colonise the infected tissue. They achieve this by promoting the activation of tumour suppressive cells such as the regulatory T cell (Treg), myeloid‐derived suppressor cell and regulatory B cell which suppress anti‐tumour immunity within the tumour microenvironment (TME). 122 , 123 , 124 , 125 Strikingly, cancer cells promote uncontrollable inflammation which becomes chronic, causing a damaging effect that increases cancer progression. 125 Evidence of chronic inflammation progression into cancer is exemplified in the study where Mdr2‐knockout mice developed liver inflammation that subsequently progressed to liver cancer. 126 , 127 E3 ligases are actively involved in immune response and inflammatory signalling either as promoters or suppressors and could thus be considered novel therapeutic targets for improving anti‐tumour immunity and blocking tumour‐promoting inflammation.
To achieve immune tolerance, cancer cells increase the expression of E3 ligases that promote the activity of Treg and decrease anti‐tumour immunity. Among these E3 ligases, the TRAF6 ligase promotes the activity of Treg within TME by mediating K63‐linked polyubiquitination and activation of FoxP3, a transcription factor for Treg. TRAF6‐deficient Treg cells resist growth of implanted tumours, abolish immune tolerance in cancer cells and enhance anti‐tumour immunity. 128 The effect of TRAF6 inhibition for immunotherapy was investigated in Hela 1–6 tumour model and the result showed that TRAF6 inhibitors accelerated T cell‐mediated anti‐tumour immunity and blocked Treg infiltration to reduce the Treg tumour population. 129 Therefore, TRAF6 represents a promising targeting candidate for cancer immunotherapy. Elsewhere, it has also been reported that the E3 ligase ITCH positively regulates Treg by catalysing the monoubiquitination of the transcription factor TIEG1. Monoubiquitination promotes the nuclear translocation of TIEG1 necessary for FoxP3 expression. 58 Interestingly, ITCH can also mediate IL‐6‐dependent K27‐linked polyubiquitination of TIEG1 which in opposite to monoubiquitination abrogates TIEG1 nuclear translocation thereby preventing FoxP3 expression. 130 Inhibition of Tregs in this case is found to correlate with an increase in Th17 response and enhance anti‐tumour immunity. Given these contrasting functions of ITCH‐mediated mono‐ and poly‐ubiquitination, targeting ITCH might not be a best option for increasing anti‐tumour immunity but the driving mechanism involved in each case could be considered. Another E3 ligase casitas B‐lineage lymphoma proto‐oncogene‐b (Cbl‐b) has been identified as an important immune checkpoint regulator of CD8+ T‐cell and NK cells. 131 , 132 Findings from a recent study showed that depletion of Cbl‐b in tumours restores the effector activity of CD8+ tumour‐infiltrating lymphocytes and prevents chimeric antigen receptor T‐cell exhaustion. 133 Given this immune suppressive role, APN401, a small interfering RNA‐based cellular immunotherapy that specifically targets and silences Cbl‐b is currently in clinical studies in patients with advanced solid tumours. 134
Tumour‐promoting inflammation is mainly regulated via nuclear factor‐kappa B (NF‐κB) signalling. NF‐κB is the key mediator of downstream inflammatory response and is composed of five members (RelA/p65, RelB, c‐Rel, NF‐κB1/p50 and NF‐κB2/p52), from which p52 and p50 are generated from p105 and p100 precursors, respectively. 70 Both canonical and non‐canonical NF‐κB signalling pathways are regulated by several E3 ligases. For instance, SCFβ‐TrCP via a non‐canonical pathway mediates ubiquitination and partial degradation of NF‐κB precursor p100 to generate p52 which then translocates to the nucleus for target gene expression. 135 In the physiological state, the non‐canonical pathway is inhibited by the E3 ligases cIAP, TRAF2 and TRAF3. These E3 ligases form a destruction complex with NF‐κB‐inducing kinase (NIK) where cIAP conjugates K48‐linked polyubiquitin on NIK to induce its proteasome degradation. 136 , 137 , 138 , 139 , 140 When released from the destruction complex, NIK activates downstream IκB kinase alpha (IKKα), which in turn phosphorylates p100 for its partial degradation to p52. 137 , 141 Under stimulation, TRAF2 induces cIAP to catalyse proteasome degradation of TRAF3 leading to destruction complex disassembly and consequent release of NIK. 142 , 143 Consistent with negative regulation, TRAF2, TRAF3 and cIAPs inactivation mutation has been detected in B‐cell malignancy and their tumour suppressive function is mainly associated with non‐canonical NF‐κB inhibition. 137 , 144 – 147
Activation of the canonical NF‐κB signalling pathway promotes nuclear translocation of NF‐κB dimer (RelA‐p50 heterodimer), where they activate target genes. Under normal physiological conditions, NF‐κB inhibitor alpha (IκBα) arrests NF‐κB dimer in the cytoplasm to prevent their nuclear translocation and consequent expression of target genes. 148 However, with inflammation signalling, IκBα is phosphorylated by IκB kinase (IKK) complex (containing NEMO, IKKα and IKKβ), to promote its ubiquitination and degradation by SCFβ‐TrCP E3 ligase. 149 , 150 , 151
Upstream signalling activators of canonical NF‐κB such as tumour necrosis factor receptor 1 (TNFR1) and pattern recognition receptors (PRR) including TLR, RIG‐I‐like receptor (RLR) and NOD‐like receptor (NLR) are regulated by E3 ligases. Upon binding of inflammatory effector ligands (TNFα and cytokines) to their cell surface receptors, TRADD (TNFR‐associated death domain protein) and RIP1 (receptor‐interacting protein 1) are recruited via their death domain to the TNFR1 death domain. 152 , 153 TRADD then recruits TRAF2 E3 ligase which further recruits another E3 ligase cIAP to form complex I, undergoing self‐polyubiquitination and K63, K11 and K48 ubiquitination of RIP1 (Figure 2). 154 , 155 These polyubiquitin chains act as scaffolds for the recruitment of another ubiquitin ligase LUBAC for M1 linear polyubiquitination of RIP1 and NEMO. 156 , 157 , 158 , 159 , 160 TGF‐β activated Kinase 1 (TAK1) is also recruited by the K63 polyubiquitin scaffold, promoting downstream signalling by inducing the phosphorylation and activation of the IKK complex as shown in Figure 2. 161 , 162 Key E3 ligases involved in canonical NF‐κB signalling are listed in Table 2. Other E3 ligases regulating NF‐κB signalling include Makorin ring finger 2 (MKRN2) E3 ligase which suppresses NF‐κB signalling via proteasome degradation of p65 and the E3 ligase ITCH which cooperates with deubiquitinating enzyme cylindromatosis tumour suppressor protein (CYLD) to mediate deubiquitination and degradation of TAK1. 163 Although TRAF2 exhibits anti‐tumour activity via noncanonical NF‐κB, it has also been shown to elicit protooncogenic function by activating canonic NF‐κB signalling. 164 As TRAF2 is required to interact with cIAP1 or cIAP2 to promote TNFR1‐induced NF‐κB signalling and suppress TNF‐induced cell death, inhibition of TRAF2 or cIAPs is necessary to sensitise tumour cells to TNF‐induced cell death. To this end, several IAP antagonists have been developed for inducing anti‐tumour immunity and have proven effective as a monotherapy or a combination therapy with immune checkpoint blockers such as programmed death 1. 165 , 166
FIGURE 2.
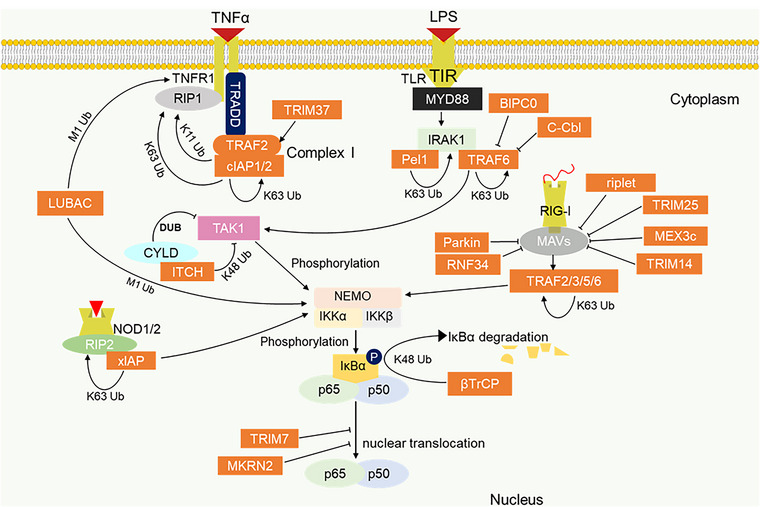
E3 ligase promoting inflammation pathway. Progression of the NF‐κB inflammation pathway via TNFR1 and the pattern recognition receptors: TLR, RIG‐I and NLR, is actively regulated by E3 ligases. K63 and M1 ubiquitin linkages serve as scaffold for the recruitment of downstream activators of NF‐κB. The K48 ubiquitination of IκBα promotes nuclear translocation of p65/p50 for the target gene expression.
TABLE 2.
Key E3 ligases involved in canonical NF‐κB signalling.
| E3 ligase | Ubiquitination mode | Signalling receptor | Function | NF‐κB‐associated role in cancer | References |
|---|---|---|---|---|---|
| SCFβ‐TrCP | K48 polyubiquitination | N/A |
Degrade IκBα, promote nuclear translocation of NF‐κB Degrade β‐catenin, promote NF‐κB‐β‐catenin crosstalk |
Oncogenic | 135 |
| TRAF2 | K63 polyubiquitination | TNFR1 | Polyubiquitinate RIP1, promote recruitment and activation of downstream NF‐κB activators | Oncogenic/tumour suppressor | 323 |
| cIAPs |
K63, K11, K48 polyubiquitination |
TNFR1 |
Polyubiquitinate RIP1, promote recruitment and activation of downstream NF‐κB activators | Oncogenic/tumour suppressor | 231 , 324 |
| K63 | NLR | Polyubiquitinate RIP2, promote recruitment and activation of downstream NF‐κB activators | Oncogenic/tumour suppressor | 178 | |
| XIAP | K63 | NLR | Polyubiquitinate RIP2, promote recruitment and activation of downstream NF‐κB activators | Oncogenic | 176 , 325 |
| LUBAC | M1‐linear polyubiquitination | TNFRI, TLR, RLR | Polyubiquitinate RIP1 and NEMO, recruit IKK complex for activation | Oncogenic | 69 |
| TRAF6 | K63 polyubiquitination | TLR, RLR | Self‐polyubiquitination, promote recruitment and activation of downstream NF‐κB activators | Oncogenic | 168 , 326 |
| Pel 1 | K63 polyubiquitination | TLR(MD88) | Polyubiquitinate IRAK1 and TAK1 and promote recruitment and activation of downstream NF‐κB activators | Oncogenic/tumour suppressor | 169 , 327 , 328 |
| K63 polyubiquitination | TRL(TRIF) | Polyubiquitinate RIP1, promote recruitment and activation of downstream NF‐κB activator | Oncogenic/tumour suppressor | ||
|
TRIM25,TRIM7 MEX3c, Riplet |
K63 polyubiquitination | RLR | Activation of RIG‐I |
Not identified TRIM7 is oncogenic |
179 |
| TRAFs (TRAF2/3/5/6) | K63 polyubiquitination | RLR | Bind MAVs, mediate polyubiquitination for recruitment and activation of IKK complex | Oncogenic/tumour suppressor | 180 , 181 |
TLRs play essential roles in the innate immune system, which include both cell surface and intracellular receptor types. TLRs contain TIR domain through which they recruit adaptor proteins MyD88 (myeloid differentiating factor 88) and TRIF (TIR domain‐containing adaptor‐inducing IFN‐β) for both NF‐κB and/or interferon (IFN) induction. 167 TRAF6 and Pellino 1 (Pel1) are the key E3 ligases regulating TLR‐induced NF‐κB signalling. They mediate K63 polyubiquitination for induction and activation of TAK1 complex and IKK complex necessary for downstream NF‐κB activation. 168 , 169 K63 polyubiquitin is conjugated on TAB2 and TAB3 which complexes with TAK1 (TAK1–TAB1–TAB2 or TAB3 complexes) to aid TRAF6–TAK1 interaction and activation of TAK1. 170 , 171 Besides, a ring finger containing E3 ligase BICP0 (bovine herpes virus‐encoded protein, infected cell protein 0), functions as a negative regulator of NF‐κB signalling by mediating K48 polyubiquitination and degradation of TRAF6. 172 Besides, c‐Cbl has also been reported to inhibit NF‐κB signalling by promoting K48‐linked ubiquitination of TRAF6. 173 TRAF members particularly TRAF6 are known key regulators of tumour‐promoting inflammation and immune response along TLR–NF‐κB pathway, therefore, small molecule inhibitors of TRAF6‐induced NF‐κB activating inflammation have been identified. Such inhibitor like the TRAF–CD40 inhibitor has been shown to suppress breast cancer metastasis effectively as either a single agent or in combination therapy with conventional chemotherapy. 174 NLRs and RLRs are cytosolic PRR activated following recognition by bacteria cell walls and cytosolic RNA virus, respectively. These two receptors are master regulators of the innate immune response against pathogens and are considered relevant for inducing immunogenic cell death and anti‐tumour immunity. However, their constitute activation due to dysregulation may elicit proinflammatory signals and chronic inflammation that predisposes to malignancies as such they could be considered a double‐edged sword in inflammation and cancer. NOD1 and NOD2 are the major NLR family that drives NF‐κB signalling. They contain caspase activating and recruiting domain (CARD) for interaction with RIP2. 175 This complex promotes the recruitment of E3 ligases such as XIAP, cIAPs and LUBAC for recruiting downstream NF‐κB activating cascade. 176 , 177 , 178
Polyubiquitination of RIG‐I by the E3 ligases TRIM25, TRIM14, MEX3c and Riplet (RNF135) is essential for activation and initiation of RIG‐I‐mediated signalling. 179 RLRs on the other hand include RIG‐I, MDA5 and LGP2 are known to mediate anti‐viral immunity by binding the mitochondria anti‐viral proteins (MAVs). This promotes MAVs interactions with TRAFs (TRAF2/3/5/6) for K63 polyubiquitination and subsequent IKK complex induction. 180 , 181 The regulation of NLR and RLR‐driven NF‐κB signalling by E3 ligases has not been clearly described in cancer; however, a few studies have demonstrated the cancer implication of E3 ligase regulation of NOD1/2‐driven NF‐κB signalling. For instance, overexpression of TRIM22 has been shown to decrease the proliferation and migration of endometrial cancer cells as knockdown of TRIM22 was found to accelerate cancer progression via NOD‐NF‐κB pathway. 182 More so, NOD1 can attenuate Helicobacter pylori infection‐induced caudal‐related homeobox 2 (Cdx2) expression and intestinal metaplasia via induction of TRAF3. Since, H. pylori infection‐induced Cdx2 expression is dependent on NF‐κB activation, NOD1 induction of TRAF3 inhibits NF‐κB signalling and protects the intestinal cells from precancerous changes. 183
Wnt/β‐catenin is a significant pathway in carcinogenesis and embryonic development, which has been shown to regulate inflammatory response via cross‐talking with the NF‐κB pathway. 184 , 185 SCFβ‐TrCP is a key regulator of both Wnt/β‐catenin and NF‐κB signalling pathways. SCFβ‐TrCP negatively regulates Wnt/β‐catenin by catalysing K48‐polyubiquitin‐induced proteasome degradation of β‐catenin, subsequently inhibiting its nuclear translocation and target gene expression. It is shown that Wnt/β‐catenin signalling induces high β‐TrCP expression which in turn activates NF‐κB signalling. 186 Strikingly, an integrated association of β‐TrCP, β‐catenin and NF‐κB is detected in colorectal cancer and is considered important for tumour metastasis and apoptosis inhibition. 187 In addition, β‐TrCP promotes lymphocytic leukaemia cell proliferation through concomitant activation of NF‐κB and β‐catenin/TCF signalling pathways, suggesting that β‐TrCP–NF‐κB–β‐catenin pathway could be considered a potential target for cancer therapy. 188
Active Rel/NF‐κB act as a transcription factor for hundreds of genes involved in various biological process including cytokines/chemokines, immunoreceptors, apoptotic regulators, growth factors and transcription factors. 189 It is reported that increasing NF‐κB signalling is central to driving cancer cell proliferation. As a result, genetic alterations of E3 ligases regulating this pathway have been identified in several human cancers. Oncogenic functions of immunomodulating E3 ligases are mostly attributed to the activation of NF‐κB signalling. For instance, TRAF6 is overexpressed in colon cancer and lung cancer and investigation of its oncogenic function shows the activation of NF‐κB signalling pathway. 190 , 191 , 192 TRAF6‐induced NF‐κB activation promotes multiple myeloma cell adhesion to bone marrow stroma. 193 Similarly, TRAF2 functions as a NF‐κB activating oncogene that promotes epithelial cancers and skeletal tumour growth in osteotropic breast cancer. 164 , 194 Furthermore, mutational activation of BRAF in melanoma promotes β‐TrCP expression and induces NF‐κB activation and melanoma cell growth. 195 Constitutively, enhanced expression of β‐TrCP in pancreatic carcinoma cells positively correlates with NF‐κB expression and chemoresistance in pancreatic carcinoma. 196 On the other hand, the U‐box containing E3 ligase CHIP (carboxyl terminus of Hsc70‐interacting protein) as a tumour suppressor weakly expressed in colon cancer and gastric cancer and negatively regulates NF‐κB signalling. 197 , 198 CHIP was found to repress the growth of colorectal cancer cells by mediating ubiquitination and degradation of p65 and its expression correlated with TNM stages with the lowest expression in stage 3 and 4 patients. 198 Further a similar result revealed that TRIM7 E3 ligase negatively regulates NF‐κB signalling by promoting the ubiquitination and proteasomal degradation of p65 in lung cancer. 199 In this work, Jin and colleagues not only detected low expression levels of TRIM7 in tumours compared to normal cells, but also found that tumour size decreases with stable expression of TRIM7. 199 With an increasing understanding of the target substrates and mechanisms of action, RNF40 could be considered a target for colorectal cancer treatment. Conversely, TRIM37, another member of the TRIM family, is reported to activate NF‐κB signalling and promote non‐small cell lung cancer aggressiveness due to its K63‐mediated polyubiquitination and activation of TRAF2. 200 These studies verify the active involvement of E3 ligases in cancer‐promoting inflammatory signalling presenting them as excellent targets for inducing tumour regression via inhibition of NF‐κB tumour‐promotion inflammation.
3.3. Evasion of apoptosis by E3 ubiquitin ligases
Cell death is a natural suicidal event for destroying malignant and potentially harmful cells. Of all modes of cell death, programmed cell death (apoptosis) is considered the most significantly inhibited type by cancer cells. This is primarily because it does not elicit adverse effects, thus, diverse therapies targeting apoptotic pathways have been developed for cancer treatment. 201 , 202 Table 3 lists significant E3 ubiquitin ligases associated with apoptotic regulation.
TABLE 3.
E3 ligases of apoptotic signalling.
| Intrinsic pathway | |||
|---|---|---|---|
| E3 ligase | Activity | Role in apoptosis | References |
| TRIM17 | Degrade Mcl‐1 by ubiquitination | Promote apoptosis | 329 |
| SCFβ‐TrCP |
Degrade Mcl‐1 by ubiquitination Degrade Bim‐EL by ubiquitination |
Context dependent | 219 , 220 |
| SCFFbxw7 | Degrade Mcl‐1 by ubiquitination | Promote apoptosis | 330 |
| MULE | Degrade Mcl‐1 by ubiquitination | Promote apoptosis | 331 |
| APC/CCDC20 | Degrade Mcl‐1 by ubiquitination | Promote apoptosis | 332 |
| APC/CCDH1 | Degrade MOAP1 by ubiquitination | Inhibit apoptosis | 207 |
| TRIM39 | Prevent APC/CCDH1 degradation | Promote apoptosis | 207 |
| XIAP |
Inactivate caspase 3/7/9 Ubiquitinate and degrade ccaspase3 |
Inhibit apoptosis | 209 , 222 , 333 |
| cIAP1/2 | Ubiquitinate caspase 3/7, Selfubiquitination, Ubiquitinate Ring IAPs | Inhibit apoptosis | 210 , 211 , 212 |
| AREL1 | Ubiquitinate and degrade SMAC | Inhibit apoptosis | 218 |
| Extrinsic pathway | |||
|---|---|---|---|
| cIAPs | Ubiquitinate RIP1, Prevent complex II formation | Inhibit apoptosis | 231 , 232 |
| TRAF2 | Promote RIP1 ubiquitination and prevent complex II formation | Inhibit apoptosis | 229 , 334 |
| Ubiquitinate and degrade caspase 8 | Inhibit apoptosis | 235 , 236 | |
| LUBAC | Stabilise ubiquitinated RIP1, prevent complex II formation | Inhibit apoptosis | 233 |
| ITCH | Degrade FLIP | Promote apoptosis | 234 |
| TRIM9 | Upregulate BCL‐2, downregulate caspase 7/9, decrease activity of caspase 3 | Inhibit apoptosis | 246 , 335 |
| SIAH2 and POSH | Downregulate caspase 8 | Inhibit apoptosis | 237 |
| MKRN1 | Degrade FADD | Inhibit apoptosis | 238 |
3.3.1. Intrinsic apoptotic pathway
Apoptosis can be propagated through two pathways: intrinsic pathway and extrinsic pathway. The intrinsic pathway is mediated from mitochondria by intrinsic stimuli arising from cellular stress and DNA damage that induce mitochondrial outer membrane permeabilisation (MOMP). There are two regulators of MOMP, the anti‐apoptotic Bcl‐2 members including Bcl‐2, Mcl‐1, Bcl‐XL and Bcl‐w antagonise the formation of MOMP and the pro‐apoptotic Bcl‐2 members (Bax, Bad, Bak, Bid, Noxa, Puma and Bim) which prompt MOMP to release downstream signalling caspase activating proteins, cytochrome c and SMAC (Second mitochondria‐derived activator of caspases) for apoptosis. 203 , 204 Cytochrome c recruits an apoptotic peptidase activating factor‐1 to form an apoptosome complex responsible for activating caspase 9 and downstream effectors, caspases 3 and caspases 7 for apoptosis. 205 Interestingly, E3 ligases play a critical regulatory role in intrinsic pathways. For instance, Mcl‐1 is found to be polyubiquitinated and degraded by the E3 ligases TRIM17, MULE, SCFβ‐TrCP, SCFFBXW7 and APC/CCDC20. 206 Likewise, TRIM39 E3 ligase inhibits APC/CCDH1‐mediated degradation of MOAP1 (Modulator of Apoptosis 1), a tumour suppressor that activates the pro‐apoptotic Bax protein. 207 These E3 ligases function in the promotion of apoptosis.
In contrast, the IAP family proteins, most importantly cIAPs and XIAP are notable apoptosis inhibitors. XIAPs via their BIR3 domain inhibit apoptosis by binding and inactivating caspases and their BIR2 domain inhibit the catalytic activity of caspases 7 and 3. 208 , 209 Utilising E3 ligase activity in apoptosis inhibition, cIAPs mediate ubiquitination of caspase 3/7, self‐ubiquitination and the ubiquitination of other ring finger IAPs, 210 , 211 , 212 while XIAP mediates the ubiquitination and degradation of caspase 3. Unlike XIAP, cIAPs binding to caspases do not inhibit caspase apoptotic function, 213 therefore, the anti‐apoptotic function of cIAPs could be explained in part by their interaction with TRAFs to activate NF‐κB signalling and prevent TNFα‐induced apoptosis, ubiquitin ligase activity and/or neutralisation of IAP antagonist. 214 , 215 SMAC/DIABLO, a natural IAP antagonist from permeabilised mitochondria contains an amino‐terminal tetrapeptide motif with which it binds the BIR domain of IAPs disrupting their anti‐apoptotic function. 216 SMAC analogues (SMAC mimetics) have currently been developed for promoting the apoptosis of cancer cells. 217 The SMAC mimetics were originally thought to bind XIAP but subsequent studies reveal that they primarily target cIAPs inducing their autoubiquitination and proteasome degradation and TNFα‐dependent apoptosis, suggesting that cIAPs stabilise the binding and inhibition of caspases by XIAP. 138 , 139 Another anti‐apoptotic E3 ligase AREL1 (Apoptosis Resistance E3 Ubiquitin Ligase 1) in turn regulates SMAC by catalysing the ubiquitination and degradation of cytosolic SMAC. 218 SCFβ‐TrCP functions as an anti‐apoptotic E3 ligase promoting the ubiquitination and degradation of pro‐apoptosis protein, Bim‐EL, following Rsk1/2‐induced Bim‐EL phosphorylation. Therefore, inhibition of either β‐TrCP or Rsk1/2 facilitates apoptosis in non‐small cell lung cancer cells. 219 On the contrary, SCFβ‐TrCP also functions as an apoptosis promoter by mediating the degradation of Mcl‐1 following Mcl‐1 phosphorylation by GSK3. 220 This promiscuous nature of SCFβ‐TrCP, similar to what was found in cell cycle regulation indicates that the function of SCFβ‐TrCP in cancer varies with tissue type. The situation of unrepaired DNA damage triggers p53 to activate the expression of pro‐apoptotic BH3‐only proteins for intrinsic apoptotic signalling. 221 , 222 P53‐mediated apoptosis is inhibited by the E3 ligase MDM2 which catalyses the polyubiquitination and degradation of p53 as well as monoubiquitination resulting in cytoplasmic translocation of p53. 223
3.3.2. Extrinsic apoptotic pathway
TNFR family containing death receptors (DR) interact with effector ligands such as Fas ligand (Fasl), TNFα and TNF‐related apoptosis‐inducing ligand (TRAIL) and recruit death domain‐containing adaptor molecules (TRADD and FADD) to effectuate intracellular signalling that led to cell death. 224 Unlike other ligands which trigger only death response, TNFα associates with the receptor TNFR1 and induces either pro‐survival/inflammatory signalling via complex I or apoptotic signalling via complex II. 225 , 226 E3 ligases are important regulators of TNFα–TNFR1‐mediated switch from pro‐survival/inflammatory signalling to apoptosis signalling. To induce apoptosis signalling under TNFα stimulation, TNFR1 adaptor TRADD recruits FADD which in turn recruits pro‐caspase 8 to form complex II for activation of downstream apoptosis effector. 227 TRADD recruitment of the E3 ligases TRAF2 and cIAP on the other hand inhibits apoptosis and promotes complex I formation for pro‐survival/inflammatory signalling. 153 , 155 RIP1 can also recruit FADD to form another complex II that comprises RIP1, FADD and caspase 8, so RIP1 interplays between pro‐survival/inflammation and apoptosis signalling. 228 , 229 , 230 Ubiquitination of RIP1 by cIAPs and TRAF2 inhibits RIP1 induction of apoptotic signalling and favours pro‐survival signalling. 229 , 231 , 232 In the absence of cIAP and TRAF2 or in deubiquitinated form, RIP1 dissociated from membrane‐bound complex I to recruit FADD for the formation of apoptotic complex II signalling (Figure 3). 224 LUBAC is another E3 ligase that impedes RIP1‐dependent apoptosis signalling by mediating the linear ubiquitination of RIP1. This ubiquitination is necessary for recruiting a hybrid protein A20 that blocks CYLD from deubiquitinating RIP1thereby stabilising RIP1 in pro‐survival response. 233 Cellular FLICE inhibitory protein (c‐FLIP) which interferes with pro‐caspase 8 activation in complex II is inhibited by ITCH that degrades c‐FLIP to promote apoptosis. 234 In addition, the apoptotic inhibitor, TRAF2 promotes polyubiquitination and degradation of caspase 8 and this contributes to insensitivity to TRAIL ligand‐induced apoptosis in gastric cancer. 235 , 236 Likewise, two E3 ligases, Siah2 (Seven in absentia homolog) and POSH (plenty of SH3s), interact together to abrogate TRAIL or FAS ligand‐induced apoptosis by inhibiting downstream caspase 8 activity. 237 The stability of FADD is impaired in the presence of the E3 ligase MKRN1 which promotes FADD proteasomal degradation. 238
FIGURE 3.
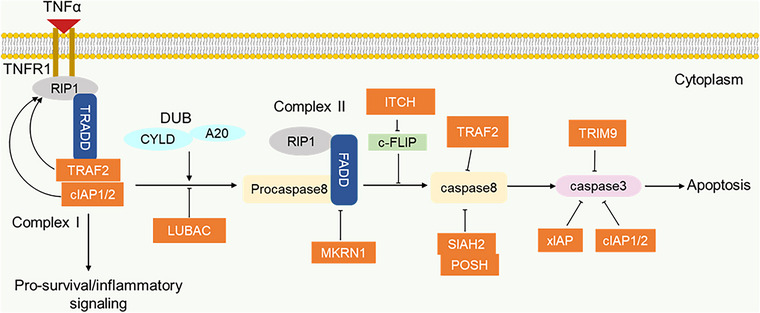
E3 ligases in the regulation of pro‐survival/apoptosis switch. The E3 ligases TRAF2 and cIAP mediate ubiquitination in favour of pro‐survival complex I. The presence of deubiquitinases (CYLD and A20) promote the switch from pro‐survival complex I to apoptotic complex II. LUBAC stabilises the ubiquitin chain in complex I and prevents complex II formation. Several E3 ligases function as negative regulators of apoptotic caspases.
The expression levels of E3 ligases that negatively affect apoptosis are high in most cancers. An example is seen in XIAP, which is abundantly expressed in anaplastic thyroid carcinoma (ATC). Most notably, this overexpression is associated with proliferation, migration, invasion and chemoresistance in ATC cells. 239 Additionally, XIAP overexpression has also been reported in breast cancer, renal cancer, colon cancer and leukaemia and correlated with poor overall survival, thus, positioning it as a potential therapeutic for cancer treatment. 240 , 241 , 242 , 243 , 244 In a related study, overexpression of TRIM9 is opined to correlate with the upregulation of Bcl‐2 and downregulation of caspases 9/7 leading to apoptosis inhibition in lung cancer. 245 Similarly, Yang et al. 246 reported a correlation between TRIM9 overexpression and decreased caspase 3 activity in uterine leiomyoma. TRAF2 shows high expression in prostate cancer cells which is associated with inhibition of TRAIL‐induced apoptosis. In vitro study in prostate DU‐145 cells shows that TRAF2 affects caspase‐8, cFLIP and SIRT1 expression. 247 Altogether, E3 ligases represent an attractive target that could be considered in clinical application for promoting apoptosis in tumour cells.
4. TARGETING E3 UBIQUITIN LIGASE IN THE CANCER HALLMARK PATHWAY
Since the success of the proteasome inhibitor Bortezomib in treating multiple myeloma, there has been a growing attempt towards targeting the ubiquitin–proteasome system for cancer therapy. 248 , 249 The new generation therapy is geared toward targeting ubiquitin pathway enzymes (E1, E2 and E3) which act on selected substrates unlike the proteasome inhibitors with broader effects and more associated side effects. 250 E3 ligases have attracted more attention owing to their specificity, as a result, an increasing number of small molecules targeting E3 ligases have been developed and are currently under clinical trials. 251 Small molecules targeting E3 ligases of the cell cycle, apoptosis and NF‐κB signalling pathway are elaborated in Table 4 and discussed herein.
TABLE 4.
Small molecules targeting E3 ligases in cancer hallmark pathways.
| Small compound | Targeted E3 ligase | Mode of action | Targeted cancer conditions | Clinical trial phase | References |
|---|---|---|---|---|---|
| Nutlin | MDM2 | Bind p53 binding pocket in MDM2 and competitively inhibit MDM2–p53 interaction | Osteosarcoma xenograft | N/A | 254 |
| RG7112 | MDM2 | Bind MDM2 and inhibit MDM2–p53 interaction. | Leukaemia, advanced solid tumours, liposarcoma | Phase I | 256 , 258 , 336 |
| RG7388 (Nutlin derivative) | MDM2 | Bind MDM2 and inhibit MDM2–p53 interaction. | AML, solid tumours, neuroblastoma, breast cancer | Phase I/II (Phase III terminated) | |
| AMG‐232 | MDM2 | Bind MDM2 with high potency and inhibit MDM2–p53 interaction | AML, advanced solid tumours, glioblastoma metastatic melanoma, multiple myeloma | Phase I | 261 , 263 |
| RITA | MDM2 | Specifically bind p53 and induce a conformational change that inhibits MDM2–p53 interaction | Fibro sarcoma and lymphoma cell lines and cervical carcinoma xenograft | N/A | 265 , 266 |
| SAR405838 (MI‐773‐01) | MDM2 | Bind MDM2 with high affinity and inhibit MDM2–p53 interaction | Malignant neoplasm, lymphoma advanced solid tumours | Phase I | 268 |
| APG‐115 (AA‐115) | MDM2 | Bind MDM2 blocking its inhibitory effect on p53 | Advanced solid tumours, lymphoma, advanced liposarcoma, AML T‐prolymphocytic leukaemia | Phase I/II | |
| Milademetan (DS‐3032b) | MDM2 | Disrupts MDM2–p53 interaction | Advanced solid tumours, lymphomas, AML, dedifferentiated liposarcoma | Phase I/II/III |
NCT0501237 |
| CGM097 | MDM2 | Bind MDM2 inhibiting MDM2–p53 interaction | Advanced solid tumours | Phase I | NCT01760525 |
| Siremadlin (HDM201) | MDM2 | Bind MDM2 preventing MDM2–p53 interaction | Liposarcoma, AML, Advanced/metastatic soft tissue sarcoma, colorectal cancer | Phase I/II | |
| BI 907828 | MDM2 | Disrupts MDM2–p53 interaction | Advanced solid tumours, glioblastoma, pancreatic neoplasm dedifferentiated liposarcoma | Phase I/II | |
| MK‐8242 | MDM2 | Bind to MDM2 and prevent HDM2‐p53 interaction | Solid tumours, AML | Phase I (terminate) | |
| Compound A | SKP2 | Prevent SKP2 association in SCF complex | Multiple myeloma cell lines | N/A | 269 |
| Compound #25 (C25) | SKP2 | Prevent SKP2 interaction with adaptor SKP1 and inhibit E3ligase activity of SKP2 | Liver, lungs, prostate and osteosarcoma cell lines | N/A | 270 |
| DT204 | SKP2 | Prevent SKP2 interaction with Cullin1 and Commd1 | Myeloma tumours (murine model | N/A | 271 |
| C series compound (C1, C2, C16, C20) | SKP2 | Inhibit Cks1 activity to destabilise SKP2‐p27 interaction | Melanoma cell lines | N/A | 272 |
| Curcumin | SKP2 | Downregulate SKP2 expression | Prostate cancer, pancreatic cancer | Phase I/II/III | 274 , 275 , 276 |
| Dioscin | SKP2 | Promote SKP2‐CDH1 interaction necessary for CDH1‐medaited degradation of SKP2 | Colorectal cancer cell lines | N/A | 277 |
| Oridonin | FBXW7 | Fbxw7 agonist, promotes degradation of c‐MYC | Leukaemia and lymphoma cell lines | N/A | 278 |
| Pevonedistat (MLN4924) | CRLs | Inhibit NAE to prevent cullin ring neddylation | Advanced solid tumours, AML, MDS, lymphoma | Phase I/II/III | 282 , 283 , 344 |
| NSC1892 | CRL4DCAF4 | Disrupt Cul4A/B‐DDB1 interaction and prevent CRL4DCAF4 formation. Lead to accumulation of ST7, p21 and p27 | Colorectal cancer cell lines | N/A | 284 |
| Pro‐Tame | APC/C | Bind APC/C and inhibit IR tail dependent recruitment of CDC20 and CDH1 | NA | N/A | 285 |
| Apcin | APC/CCDC20 | Bind the D‐box binding site of CDC20 and prevent recruitment of APC/CDC20 substrate | NA | N/A | 286 |
| GS143 | β‐TrCP | Inhibit β‐TrCP ubiquitination of IKBα | NA | N/A | 287 |
| C25‐140 | TRAF6 | Inhibit TRAF6‐Ubc13 interaction to reduce TRAF6 E3 ligase activity | A study conducted in Autoimmune condition | N/A | 288 |
| Gliotoxin | LUBAC | Bind the RBR domain of HOIP and inhibit linear ubiquitin chain formation | NA | N/A | 290 |
| Compound 5 | LUBAC | Bind active cysteine of HOIP and inhibit thioester bond formation | NA | N/A | 291 |
| HOIPINs | LUBAC | Bind active Cys885 and other associated residue of HOIP and inhibit linear ubiquitin chain formation | NA | N/A | 292 |
| SMAC mimetic: AT‐406 (DEBIO1143) | IAPs | Bind XIAP and cIAPs to induce cIAP1 degradation and caspase activation | Solid tumours, AML, lymphoma, squamous cell carcinoma | Phase I/II/III | 294 , 295 , 345 |
| SMAC mimetic: GDC‐0152 (Compound 1) | IAPs | Bind BIR3 domains of XIAP and CIAP and BIR domain of ML‐IAP and induce caspase 3/7 activation and degradation of cIAP | Solid cancers | Phase I (Terminated) | |
| SMAC mimetic: LCL161 | CIAPs | Bind the BIR3 domain of cIAPs and induce their autoubiquitination and degradation | Solid tumours, multiple myeloma, breast cancers, small cell lung cancer | Phase I/II | 347 , 348 |
| SMAC mimetic: Birinapant (TL327711) | CIAPs | Bind BIR3 domain of cIAPs to induce their autoubiquitination and degradation | Solid tumours, MDS, ovarian cancers, Neck squamous cell carcinoma | Phase I/II | 296 , 349 , 350 |
| PRT4165 | PRC1 | Inhibit PRC1‐induced H2A and H2AX ubiquitination | NA | N/A | 298 |
| GW‐516 | PRC1 | Catalytically inhibits RNF2 component of PRC1 | Prostate cancer cell lines | N/A | 351 |
| RB‐3 | PRC1 | Binds RING1B/BMI1 complex and induces conformational changes that disrupt their interaction with nucleosomes | Leukaemia cell lines | N/A | 352 |
4.1. Small molecules targeting cell cycle E3 ligases
To promote cell cycle arrest and inhibit cellular proliferation in cancer, several small molecule compounds have been developed to target cell cycle E3 ligases such as MDM2, SKP2, FBXW7, APC/C and Cullin4.
4.1.1. MDM2
MDM2 is implicated in both cell cycle and apoptosis due to the broad functions of its substrate, p53, in regulating diverse cellular processes including cell cycle, apoptosis, senescence and DNA repair. 252 A critical study of the structural basis of MDM2–p53 interaction revealed that the N terminal transactivation domain with conserved amino acids Phe19, Trp23 and Leu26 in p53 is required for interaction with the hydrophobic pocket of MDM2. 253 This finding prompted the development of small compounds that mimic and bind MDM2 with high affinity. The cis‐imidazole‐containing compound, Nutlin is the first potent MDM2 inhibitor which antagonises MDM2–p53 interaction by acting as a competitive inhibitor that binds to the p53 binding pocket on MDM2. 254 Nutlin‐3, a derivative of Nutlin, was subsequently developed and it is shown to exhibit anti‐tumour efficacy against tumour cell lines harbouring wild‐type p53. 255 Accordingly, Nutlin‐3 activates p53‐dependent cell cycle arrest but shows reduced apoptosis induction ability. 255 Further optimisation of Nutlin‐3 led to the development of RG7112, the first classical MDM2 inhibitor to enter clinical trial. RG7112 effectively induced cell cycle arrest and apoptosis in wild‐type p53 harbouring cancer cells especially solid tumours. 256
In its phase I clinical trial, RG7112 produced a moderate response but with associated adverse side effects like neutropenia and thrombocytopenia in about half of the patients. 257 , 258 For this reason, an improved second‐generation analogue of Nutlin, Idasanutlin (RG7388), was developed and entered clinical trial with results that prove it to be more potent, tolerable and selective than RG7112. 259 , 260 However, the phase III clinical trial assessing the overall survival of Idasanutlin as a combination therapy with Cytarabine in patients with relapsed acute myeloid leukaemia (AML) in comparison with only cytarabine and placebo, was terminated for futility based on efficacy results at the interim analysis (ClinicalTrials.gov: NCT02545283). Another MDM2–p53 inhibitor AMG‐232 is a piperidinone‐based compound that also binds the p53 binding pocket in MDM2. AMG‐232 binding with MDM2 showed higher potency than p53 due to additional interaction with glycine58 ‘shelf region’ in MDM2 hydrophobic pocket. 261 In a preclinical study using tumour xenograft model, AMG‐232 effectively inhibited tumour growth and completely repressed MDM2‐amplified SJSA‐1 via cell cycle arrest and apoptosis. 262 AMG‐232 progressed into a clinical trial showing modest safety and tolerability with similar but milder side effects than previous MDM2 inhibitors. 263 The Furan‐derived compound RITA (Reactivation of p53 Induction of Tumour Apoptosis) is another inhibitor of MDM2–p53. Unlike other small compound inhibitors, RITA specifically binds to the p53 N‐transactivation domain rather than MDM2 and induces a conformational change that blocks not only MDM2–p53 interaction but also p53 interaction with other E3 ligase inhibitors. 264 , 265 , 266 RITA is reported to accelerate p53‐dependent apoptosis and inhibit the growth of cancer cells that retain both wild‐type and mutant p53. 265 Additional important MDM2 inhibitors are the members of the Spiro‐oxindoles class which include the MI series of compounds that mimics p53 binding residue and binds p53 binding pocket in MDM2 to inhibit tumour growth. 267 Among the M1 series, MI‐773‐01 (SAR405838) has very high binding affinity and specificity for MDM2 and has progressed into clinical trials in patient with solid tumour. 268 Other important MDM2–p53 inhibitors in clinical studies have been listed in Table 4.
4.1.2. SKP2
Given that SKP2 substrates including p27 and p21 are popular cell cycle inhibitors, small molecule inhibitors of SKP2 inhibit cell growth by inducing cell cycle arrest and cell death. Compound A prevents SKP2 from associating with its SCF complex, leading to G1/S cell cycle arrest and cell death. 269 Similarly, the small compound inhibitor, Compound #25 (C25) prevents SKP2 interaction with adaptor molecule SKP1 and inhibits E3 ligase activity of SKP2 thereby promoting cell cycle arrest and cell death. 270 Another SKP2 antagonist of this type is DT204, which overcomes Bortezomib resistance in multiple myeloma by reducing the interaction of SKP2 with Cullin1 and Commd1 adaptor thereby disrupting SCFSKP2 complex. 271 Wu and colleagues identified the C‐series compounds (C1, C2, C16 and C20), which are SKP2 inhibitors that prevent SKP2‐mediated ubiquitination of p27. The C‐series compounds target and inhibit the activities of Cks1, a SKP2‐p27 interacting bridge that promotes SKP2 ubiquitination of p27. 272 Additionally, the naturally occurring compounds, Curcumin, lycopene and quercetin exhibit anti‐cancer effects by down‐regulating SKP2 expression leading to cell cycle arrest. 273 Curcumin derived from curcuma longa rhizome (Turmeric) has passed human phase I and phase II clinical trials for cancer and its high safety and no dose‐limiting toxicity enabled progression to phase III clinical in patients with metastatic colon cancer. 274 , 275 , 276 Another natural plant‐derived steroid saponin, dioscin, has been shown to promote CDH1‐mediated polyubiquitination and degradation of SKP2 and a significant reduction in the cell growth of colorectal cancer. 277
4.1.3. FBXW7 and CRL
FBXW7 as a tumour suppressor is mutated or inhibited in human cancers with high frequency. Oridonin, a naturally occurring compound, is employed as an FBXW7 agonist to promote proteasome degradation of c‐MYC. 278
CRLs are inhibited by the pevonedistat (MLN4924), a small compound that prevents Cullin neddylation. In the preclinical study using mice model, pevonedistat selectively and efficiently suppressed the growth of human tumour xenograft which facilitated their clinical evaluation. 279 , 280 The result of the phase I dose‐escalation and pharmacodynamics study in patients with advanced solid tumours showed that pevonedistat is well tolerated and functions effectively as an NEDD8‐activating enzyme inhibitor. 281 The phase II and III studies evaluated pevonedistat as a combination therapy with azacitidine in comparison with azacitidine alone in patients with MDS and AML. Here, pevonedistat and azacitidine combination treatment produced a better outcome in phase II study than azacitidine single agent; however, the phase III study failed to meet the prespecified primary endpoint as there was no significant difference between the outcome of the combined therapy and monotherapy. 282 , 283 A Cullin scaffold–adaptor interaction is also employed as a target region, by small molecule inhibitor small molecule NSC1892 which specifically disrupts CUL4 interaction with the DDB1 adaptor molecule. 284
4.1.4. APC/C
The role of APC/C in mitotic progression makes it an important target for inducing mitotic arrest and subsequent tumour death. APC/C co‐activators, CDC20 and CDH1, contain a C‐terminal IR tail and N‐terminal C‐box motif for recruitment to the APC/C complex. 50 To inhibit APC/C activity, Pro‐Tame (Pro drug of Tosyl‐l‐Arginine Methyl Ester), a small compound that induces spindle checkpoint‐dependent mitotic arrest in human cells, binds APC/C to inhibit IR tail‐dependent recruitment of CDC20 and CDH1. 285 Another small molecule inhibitor Apcin binds to the D‐box binding site of CDC20 to competitively inhibit APCCDC20 ubiquitination of D‐box containing substrates. 286
4.2. Small molecules targeting E3 ligase along inflammatory NF‐κB pathway and apoptotic pathway
Among the targeted pathways for cancer treatment are the tumour promoting inflammation pathway and apoptotic pathway. The strong involvement of E3 ligases in NF‐κB and apoptotic pathways, present them as an important target for inducing apoptosis and inhibiting inflammation in cancer cells. Hence, small compounds targeting the concerned E3 ligases such as SCFβ‐TrCP, TRAF6, LUBAC and IAPs have been developed for cancer treatment.
β‐TrCP is targeted by the small molecule GS143 for the inhibition of NF‐κB‐mediated inflammation in cancer. GS143 inhibits β‐TrCP‐mediated ubiquitination of IκBα to prevent activation of NF‐κB. 287
TRAF6 in association with the E2 ubiquitin‐conjugating enzyme Ubc13 mediates K63 polyubiquitination for NF‐κB inflammation signalling. 35 The small molecule inhibitor of TRAF6, C25‐140, inhibits TRAF6–Ubc13 interaction to reduce TRAF6 E3 ligase activity. 288 The interaction of CD40 with TRAFs particularly TRAF6 has been shown to activate inflammatory signalling via the NF‐κB pathway. This finding prompted the identification of a small molecule inhibitor of TRAF6–CD40, 6877002 that binds TRAF6 and blocks TRAF6 interaction with CD40 thereby suppressing NF‐κB inflammation signalling. 174 , 289
Most small molecule inhibitors of LUBAC, target HOIP, the catalytic subunit in LUBAC. The small molecule, Gliotoxin, inhibits LUBAC by binding to the Ring‐IBR‐Ring domain of HOIP to prevent linear ubiquitin chain formation. 290 Johansson et al. 291 designed α, β‐unsaturated methyl ester‐linked compound (Compound 5) which binds the active cysteine to inhibit HOIP E3‐UB thioester bond formation thereby preventing NF‐κB activation. Another similar LUBAC antagonist, HOIPINs bind active Cys885 and other associated residues in HOIP to suppress the RING–HECT‐hybrid reaction of HOIP, thus hindering LUBAC‐mediated linear ubiquitination. 292
IAP contains a BIR domain that binds caspases in the cell to inhibit cell apoptosis. 208 Natural IAP antagonist mimetics (SMAC mimetics) are currently developed and under clinical evaluation in cancer subjects. 217 Small compounds AT‐406 (DEBIO1143), GDC‐0152, LCL161 and Birinapant (TL32711), are SMAC mimetics under clinical evaluation (Table 4), but the clinical trial for GDC‐0152 was terminated after only one phase I study of safety and pharmacokinetics in patients with advanced malignancy although the reason for the termination is not related to patients’ safety or anti‐tumour activity (ClinicalTrials.gov: NCT00977067). 293 , 294 , 295 NF‐κB inflammation signalling and the extrinsic apoptotic pathway crosstalk between complex I and complex II via TNFR1 signalling, and this point could be considered a potential target for cancer therapy. 226 For instance, TRAF2 recruits cIAP to complex I and prevents the formation of apoptotic promoting complex II. In this case, Birinapant with a high affinity for the BIR3 domain of cIAP binds cIAPs that are associated with TRAF2 to induce their autoubiquitination and degradation. 296 By this means, Birinapant abrogates complex I‐induced NF‐κB activation and promotes RIP1‐mediated complex II formation for apoptosis of tumour cells. 296
Besides the polyubiquitinating E3 ligase, small molecule inhibitors of monoubiquitinating E3 ligase such as PRC1 responsible for H2A monoubiquitination have been developed. The catalytic components of PRC1 such as RING1 A or B/BMI1 complex are associated with several cancers and correlate with poor prognosis, thus PRC1 represents an attractive therapeutic target for cancer. PRT4165 was initially designed as small molecule inhibitor of BMI1/Ring1A‐mediated ubiquitination and degradation of topoisomerase II α (Top2α)/drug complex and was later found to inhibit PRC1‐induced H2A and H2AX ubiquitination at DNA damage site. PRT4165 abrogates DNA double‐strand break repair thereby promoting cell cycle arrest and death of DNA‐damaged cells. 297 , 298 Su and co‐workers 299 designed GW‐516, an inhibitor of H2A ubiquitination that catalytically inhibits the RNF2 component of PRC1 to truncate immunosuppression and prevent metastasis of prostate cancer cells. Recently, Grembecka and Cierpicki's laboratory developed the small molecule RB‐3 that inhibits RING1B/BMI1‐mediated H2A ubiquitination in cancer cells. RB‐3 binds RING1B/BMI1 complex and induces conformational changes that disrupt their interaction with nucleosomes. RB‐3 was also shown to induce differentiation in leukaemia cell lines and AML, establishing that RB‐3 may act as a promising therapeutic compound for leukemic cells. 300
In summary, these studies are convincing evidence that E3 ligases are credible and attractive targets for cancer therapy. The studies also show that small molecule compounds can be directed at E3 ligases, E3 ligase–substrate interaction point or E3 ligase substrates as in the case of RITA to inhibit E3 ligase‐mediated substrate degradation. With a complete understanding of the E3 ligase mechanism within ubiquitin–proteasome system, more efficient and potent E3 ligase inhibitors will be developed for clinical assessment.
5. TARGETED E3 LIGASE SUBSTRATES DEGRADATION BY PROTAC IN CANCER THERAPY
Translational therapeutics based on E3 ligase research have evolved beyond small molecule inhibition to induce E3 ligase for the targeted degradation of disease‐causing proteins. This therapeutic approach is accomplished through a class of targeting protein degraders known as the Proteolysis Targeting Chimera (PROTAC) molecule. PROTAC enables chemical hijacking of the endogenous proteasome system to degrade the protein of interest (POI). Since it was first proposed in 2001, 301 the PROTAC technology has attracted significant interest from academia and industry due to its demonstrated advantages over small molecule inhibitors. Significant advantages of the PROTAC molecule include its event‐driven pharmacology, reversible catalytic mechanism, selectivity and ability to modulate enzyme–non‐enzyme proteins and transcription factors that are considered non‐digestible by traditional small inhibitor molecules. 302 , 303
PROTACs are heterobifunctional small molecules composed of two ligand warheads (target protein–ligand and E3 ligase ligands) and a linker connecting them. This design facilitates the recruitment of POI and E3 ligase nearby within a ternary complex for target protein ubiquitination and proteasome degradation. 301 , 304 The mechanism of PROTAC‐mediated degradation of POI is catalytic and allows for iteration, since after induction of ubiquitination, PROTAC dissociates from the ternary complex to recruit new targets as shown in Figure 4. Unlike occupancy‐driven small molecule inhibitors that modulate protein functions by binding to active sites, PROTACs eliminate target proteins by binding alternative non‐active sites. This modality enhances the pharmacodynamic action of PROTACs due to the need to resynthesise new proteins and limits the potential for mutational drug resistance. 305 , 306 While the binding affinity for the target protein is the key determinant of the potency of small molecule inhibitors, the degradation efficiency of PROTACs has other influencing factors such as the cooperativity of the ternary complex, the linker size and the choice of E3 ligase, all of which are highly considerable factors in the design and optimisation of PROTACs for efficiency. 307 , 308 , 309 , 310 , 311 , 312 An excellent review by Rao's group discussed in detail the strategies in the design and optimisation of PROTACs for potency. 313
FIGURE 4.
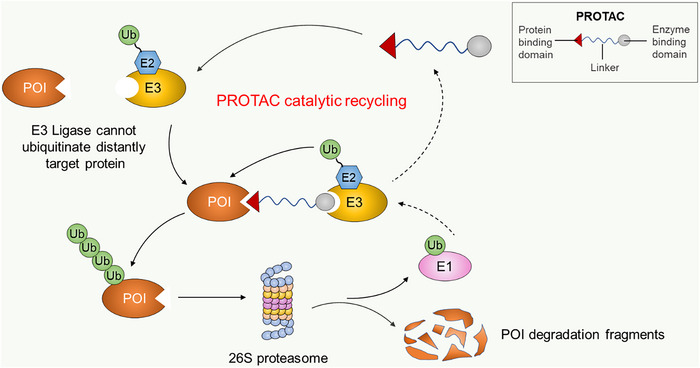
The mechanism of PROTAC‐mediated target protein degradation. PROTACs simultaneously recruit POI and E3 ligase into a ternary complex where ubiquitin is transferred from the E2–E3 complex to POI. Ubiquitinated proteins are attracted to the 26S proteasome system for degradation while PROTAC is recycled to recruit new targets.
6. CONCLUSION AND FUTURE PERSPECTIVE
In previous studies, E3 ligases are known to mediate substrate monoubiquitination or polyubiquitination through their substrate recognition function and without doubt play a classic role in the ubiquitination process. However, the mechanism underlying monoubiquitination and polyubiquitination of one substrate by the same E3 ligase remains to be fully understood. For instance, MDM2 catalyses both monoubiquitination and polyubiquitination of p53 with different effects, as such insight into the mechanism that prompts mono‐ versus poly‐ubiquitination for one substrate as well as other ubiquitin chains including branched chains and heterotypic chains would be useful in defining E3 ligase specificity and optimising the design of small molecule E3 ligase‐based therapies.
The specificity of E3 ubiquitin ligases as well as their critical involvement in both normal cell function and cancer hallmark pathways have attracted much attention for possible application in cancer therapy. The understanding of E3 ligase–substrate interaction has facilitated the design of small molecule compounds targeting E3 ligases as inhibitors, modulators or agonists. Oncogenic E3 ligases are overexpressed in most cancers and are targeted by inhibitors while tumour‐suppressing E3 ligases are underexpressed or mutated in some cancer types and are targeted by agonists. Some substrates are targeted for ubiquitination by more than one E3 ligase exemplified in p53 which serves as a substrate for over five E3 ubiquitin ligases. In this case, targeting and inhibiting just one of these E3 ligases might not effectively control tumour progression as other E3 ligases would likely act on the substrate to counteract the effect of the inhibitor. In addition, some E3 ligases play a contrasting function in cell processes as both oncogene and tumour suppressors, therefore, therapies regarding this class of E3 ligases would be more effective at targeting E3 ligase–substrate interaction than targeting the E3 ligases.
The majority of the small molecule compounds targeting E3 ligases are inhibitor compounds that inhibit oncogenic E3 ligases leading to an increased level of their substrates. However, the growing interest in more recent years is towards harnessing E3 ligase machinery for degrading oncogenic proteins. The target protein degradation technology PROTAC has opened a new phase in drug design with its overwhelming advantages over small molecule inhibitors. Although this new technology has seen remarkable clinical success within a few years of development, there are still concerning challenges that need to be addressed and they include structural, kinetics and dynamics understanding of ternary complex, computational approach for studying ternary complex interactions, effective screening for easy identification of ligand, effecting approach for expanding the scope of hijackable E3 ligase and for expanding the library of undruggable targets. To this end, high throughput techniques that address these challenges would aid a complete understanding of the PROTAC‐mediated degradation mechanism for improved PROTAC design and optimisation.
In the past few years, E3 ligase has gained substantial research interest which has fostered a rising concern in E3 ligase‐based therapeutic intervention. Several E3 ligase‐based therapies are under clinical evaluation for cancer treatment and some like the E3 ligase modulators (thalidomide, lenalidomide and pomalidomide) have been approved by FDA in patients with multiple myeloma. The emergence of target protein degraders particularly PROTACs has strikingly increased the number of E3 ligase‐based therapies in clinics and with the increasing successful clinical outcomes, the coming years will have more FDA‐approved E3‐ligase‐based therapies for cancer treatment.
CONFLICT OF INTEREST STATEMENT
All authors declare no competing interest.
ACKNOWLEDGEMENTS
This study is supported by National Natural Science Foundation of China grants (No. 81972625), Dalian Science and Technology Innovation Funding (2019J12SN52) and Liaoning Revitalization Talents Program (XLYC2002035).
Sampson C, Wang Q, Otkur W, et al. The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin Transl Med. 2023;13:e1204. 10.1002/ctm2.1204
Chibuzo Sampson and Qiuping Wang contributed equally to this work.
Contributor Information
Yun Lu, Email: yyida2009@126.com.
Xiaolong Liu, Email: xlliu@dicp.ac.cn.
Hai‐long Piao, Email: hpiao@dicp.ac.cn.
REFERENCES
- 1. Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giancotti FG. Deregulation of cell signaling in cancer. FEBS Lett. 2014;588(16):2558‐2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunphy K, Dowling P, Bazou D, O'Gorman P. Current methods of post‐translational modification analysis and their applications in blood cancers. Cancers (Basel). 2021;13(8):1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma B, Prabhakaran V, Desai A, Bajpai J, Verma R, Swain P. Post‐translational modifications (PTMs), from a cancer perspective: an overview. Oncogen. 2019;2:12. [Google Scholar]
- 5. Dantuma NP, Masucci MG. Stabilization signals: A novel regulatory mechanism in the ubiquitin/proteasome system. FEBS Lett. 2002;529(1):22‐26. [DOI] [PubMed] [Google Scholar]
- 6. Hirayama S, Sugihara M, Morito D, et al. Nuclear export of ubiquitinated proteins via the UBIN‐POST system. Proc Natl Acad Sci USA. 2018;115(18):E4199‐e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dougherty SE, Maduka AO, Inada T, Silva GM. Expanding role of ubiquitin in translational control. Int J Mol Sci. 2020;21(3):1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, Qiu Z, Wu Y. Ubiquitin regulation: the histone modifying enzyme's story. Cells. 2018;7(9):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chau V, Tobias JW, Bachmair A, et al. A multiubiquitin chain is confined to specific lysine in a targeted short‐lived protein. Science. 1989;243(4898):1576‐1583. [DOI] [PubMed] [Google Scholar]
- 10. Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N‐terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. Embo j. 1998;17(20):5964‐5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDowell GS, Philpott A. Non‐canonical ubiquitylation: Mechanisms and consequences. Int J Biochem Cell Biol. 2013;45(8):1833‐1842. [DOI] [PubMed] [Google Scholar]
- 12. Kelsall IR, Zhang J, Knebel A, Arthur JSC, Cohen P. The E3 ligase HOIL‐1 catalyses ester bond formation between ubiquitin and components of the Myddosome in mammalian cells. Proc Natl Acad Sci USA. 2019;116(27):13293‐13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marsh DJ, Dickson KA. Writing histone monoubiquitination in human malignancy—the role of RING finger E3 ubiquitin ligases. Genes (Basel). 2019;10(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cole AJ, Clifton‐Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocr Relat Cancer. 2015;22(1):T19‐33. [DOI] [PubMed] [Google Scholar]
- 15. Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921‐926. [DOI] [PubMed] [Google Scholar]
- 16. Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin‐chain formation by the human anaphase‐promoting complex. Cell. 2008;133(4):653‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ziv I, Matiuhin Y, Kirkpatrick DS, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics. 2011;10(5):M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayakawa M. Role of K63‐linked polyubiquitination in NF‐κB signalling: which ligase catalyzes and what molecule is targeted? J Biochem. 2012;151(2):115‐118. [DOI] [PubMed] [Google Scholar]
- 19. Spit M, Rieser E, Walczak H. Linear ubiquitination at a glance. J Cell Sci. 2019;132(2). [DOI] [PubMed] [Google Scholar]
- 20. Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu Y, Zheng Q, Erramilli SK, et al. K29‐linked ubiquitin signaling regulates proteotoxic stress response and cell cycle. Nat Chem Biol. 2021;17(8):896‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu X, Lei C, Xia T, Zhong X, Yang Q, Shu HB. Regulation of TRIF‐mediated innate immune response by K27‐linked polyubiquitination and deubiquitination. Nat Commun. 2019;10(1):4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neutzner M, Neutzner A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012;52:37‐50. [DOI] [PubMed] [Google Scholar]
- 24. Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15‐30. [DOI] [PubMed] [Google Scholar]
- 25. Lucas X, Ciulli A. Recognition of substrate degrons by E3 ubiquitin ligases and modulation by small‐molecule mimicry strategies. Curr Opin Struct Biol. 2017;44:101‐110. [DOI] [PubMed] [Google Scholar]
- 26. Varshavsky A. N‐degron and C‐degron pathways of protein degradation. Proc Natl Acad Sci USA. 2019;116(2):358‐366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang J, Wang N, Jiang Y, et al. Characterization of substrate binding of the WW domains in human WWP2 protein. FEBS Lett. 2015;589(15):1935‐1942. [DOI] [PubMed] [Google Scholar]
- 28. Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730‐738. [DOI] [PubMed] [Google Scholar]
- 29. Barford D. Structural insights into anaphase‐promoting complex function and mechanism. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3605‐3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399‐434. [DOI] [PubMed] [Google Scholar]
- 31. Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional analysis of the RING‐type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137(1):13‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING‐type E3 ligases: master manipulators of E2 ubiquitin‐conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843(1):47‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Campbell SJ, Edwards RA, Leung CC, et al. Molecular insights into the function of RING finger (RNF)‐containing proteins hRNF8 and hRNF168 in Ubc13/Mms2‐dependent ubiquitylation. J Biol Chem. 2012;287(28):23900‐23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fiorentini F, Esposito D, Rittinger K. Does it take two to tango? RING domain self‐association and activity in TRIM E3 ubiquitin ligases. Biochem Soc Trans. 2020;48(6):2615‐2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu TM, Shen C, Li Q, Zhang P, Wu H. Mechanism of ubiquitin transfer promoted by TRAF6. Proc Natl Acad Sci USA. 2018;115(8):1783‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mace PD, Linke K, Feltham R, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin‐conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283(46):31633‐31640. [DOI] [PubMed] [Google Scholar]
- 37. Nakatani Y, Kleffmann T, Linke K, Condon SM, Hinds MG, Day CL. Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase. Biochem J. 2013;450(3):629‐638. [DOI] [PubMed] [Google Scholar]
- 38. Das A, Middleton AJ, Padala P, Ledgerwood EC, Mace PD, Day CL. The structure and ubiquitin binding properties of TRAF RING heterodimers. J Mol Biol. 2021;433(8):166844. [DOI] [PubMed] [Google Scholar]
- 39. Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15(5):841‐848. [DOI] [PubMed] [Google Scholar]
- 40. Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1‐BARD1 heterodimeric RING‐RING complex. Nat Struct Biol. 2001;8(10):833‐837. [DOI] [PubMed] [Google Scholar]
- 41. Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. Structure of a Bmi‐1‐Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281(29):20643‐20649. [DOI] [PubMed] [Google Scholar]
- 42. Jang SM, Redon CE, Aladjem MI. Chromatin‐bound cullin‐ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy. Front Mol Biosci. 2018;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Z, Sui J, Zhang F, Zhang C. Cullin family proteins and tumorigenesis: genetic association and molecular mechanisms. J Cancer. 2015;6(3):233‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y, Sun Y. Cullin‐RING Ligases as attractive anti‐cancer targets. Curr Pharm Des. 2013;19(18):3215‐3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fouad S, Wells OS, Hill MA, D'Angiolella V. Cullin ring ubiquitin ligases (CRLs) in cancer: responses to ionizing radiation (IR) treatment. Front Physiol. 2019;10:1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heo J, Eki R, Abbas T. Deregulation of F‐box proteins and its consequence on cancer development, progression and metastasis. Semin Cancer Biol. 2016;36:33‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tekcham DS, Chen D, Liu Y, et al. F‐box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics. 2020;10(9):4150‐4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frescas D, Pagano M. Deregulated proteolysis by the F‐box proteins SKP2 and beta‐TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8(6):438‐449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou Z, He M, Shah AA, Wan Y. Insights into APC/C: From cellular function to diseases and therapeutics. Cell Div. 2016;11:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bodrug T, Welsh KA, Hinkle M, Emanuele MJ, Brown NG. Intricate regulatory mechanisms of the anaphase‐promoting complex/cyclosome and its role in chromatin regulation. Front Cell Dev Biol. 2021;9:687515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang LF, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase‐promoting complex. Nature. 2014;513(7518):388‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weber J, Polo S, Maspero E. HECT E3 ligases: a tale with multiple facets. Front Physiol. 2019;10:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sluimer J, Distel B. Regulating the human HECT E3 ligases. Cell Mol Life Sci. 2018;75(17):3121‐3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Masuda Y, Saeki Y, Arai N, et al. Stepwise multipolyubiquitination of p53 by the E6AP‐E6 ubiquitin ligase complex. J Biol Chem. 2019;294(41):14860‐14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shai A, Pitot HC, Lambert PF. E6‐associated protein is required for human papillomavirus type 16 E6 to cause cervical cancer in mice. Cancer Res. 2010;70(12):5064‐5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee YR, Chen M, Lee JD, et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC‐WWP1 inhibitory pathway. Science. 2019;364(6441). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense‐mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20(7):406‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Venuprasad K, Huang H, Harada Y, et al. The E3 ubiquitin ligase itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9(3):245‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smit JJ, Sixma TK. RBR E3‐ligases at work. EMBO Rep. 2014;15(2):142‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reiter KH, Klevit RE. Characterization of RING‐between‐RING E3 ubiquitin transfer mechanisms. Methods Mol Biol. 2018;1844:3‐17. [DOI] [PubMed] [Google Scholar]
- 61. Spratt DE, Walden H, Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458(3):421‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17(8):1221‐1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Seirafi M, Kozlov G, Gehring K. Parkin structure and function. Febs j. 2015;282(11):2076‐2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang X, Lin C, Song J, et al. Parkin facilitates proteasome inhibitor‐induced apoptosis via suppression of NF‐κB activity in hepatocellular carcinoma. Cell Death Dis. 2019;10(10):719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaugule VK, Burchell L, Barber KR, et al. Autoregulation of Parkin activity through its ubiquitin‐like domain. Embo j. 2011;30(14):2853‐2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. Embo j. 2013;32(15):2099‐2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kamienieva I, Duszyński J, Szczepanowska J. Multitasking guardian of mitochondrial quality: Parkin function and Parkinson's disease. Transl Neurodegener. 2021;10(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kumar A, Aguirre JD, Condos TE, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. Embo j. 2015;34(20):2506‐2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fiil BK, Gyrd‐Hansen M. The Met1‐linked ubiquitin machinery in inflammation and infection. Cell Death Differ. 2021;28(2):557‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu J, Wang Y, Gong Y, et al. Structural insights into SHARPIN‐mediated activation of HOIP for the linear ubiquitin chain assembly. Cell Rep. 2017;21(1):27‐36. [DOI] [PubMed] [Google Scholar]
- 71. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐674. [DOI] [PubMed] [Google Scholar]
- 72. Hochegger H, Takeda S, Hunt T. Cyclin‐dependent kinases and cell‐cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9(11):910‐916. [DOI] [PubMed] [Google Scholar]
- 73. Malumbres M. Cyclin‐dependent kinases. Genome Biol. 2014;15(6):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349(6305):132‐138. [DOI] [PubMed] [Google Scholar]
- 75. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev. 1999;13(12):1501‐1512. [DOI] [PubMed] [Google Scholar]
- 76. Teixeira LK, Reed SI. Ubiquitin ligases and cell cycle control. Annu Rev Biochem. 2013;82:387‐414. [DOI] [PubMed] [Google Scholar]
- 77. Zou T, Lin Z. The involvement of ubiquitination machinery in cell cycle regulation and cancer progression. Int J Mol Sci. 2021;22(11):5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kernan J, Bonacci T, Emanuele MJ. Who guards the guardian? Mechanisms that restrain APC/C during the cell cycle. Biochim Biophys Acta Mol Cell Res. 2018;1865(12):1924‐1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11(5):1555‐1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Qiao R, Weissmann F, Yamaguchi M, et al. Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc Natl Acad Sci USA. 2016;113(19):E2570‐2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517(7536):631‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Enquist‐Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC‐dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30(4):437‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203‐232. [DOI] [PubMed] [Google Scholar]
- 84. Borg NA, Dixit VM. Ubiquitin in cell‐cycle regulation and dysregulation in cancer. 2017.
- 85. Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164(2):233‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase‐promoting complex. Nat Cell Biol. 2013;15(7):797‐806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Margottin‐Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4(6):813‐826. [DOI] [PubMed] [Google Scholar]
- 88. Busino L, Donzelli M, Chiesa M, et al. Degradation of Cdc25A by beta‐TrCP during S phase and in response to DNA damage. Nature. 2003;426(6962):87‐91. [DOI] [PubMed] [Google Scholar]
- 89. Dang F, Nie L, Wei W. Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 2021;28(2):427‐438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Smith A, Simanski S, Fallahi M, Ayad NG. Redundant ubiquitin ligase activities regulate wee1 degradation and mitotic entry. Cell Cycle. 2007;6(22):2795‐2799. [DOI] [PubMed] [Google Scholar]
- 91. Tsai WB, Chung YM, Zou Y, et al. Inhibition of FOXO3 tumor suppressor function by betaTrCP1 through ubiquitin‐mediated degradation in a tumor mouse model. PLoS One. 2010;5(7):e11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rodier G, Coulombe P, Tanguay PL, Boutonnet C, Meloche S. Phosphorylation of Skp2 regulated by CDK2 and Cdc14B protects it from degradation by APC(Cdh1) in G1 phase. Embo j. 2008;27(4):679‐691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cerqueira A, Martín A, Symonds CE, et al. Genetic characterization of the role of the Cip/Kip family of proteins as cyclin‐dependent kinase inhibitors and assembly factors. Mol Cell Biol. 2014;34(8):1452‐1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nakayama K, Nagahama H, Minamishima YA, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. Embo j. 2000;19(9):2069‐2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Koepp DM, Schaefer LK, Ye X, et al. Phosphorylation‐dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173‐177. [DOI] [PubMed] [Google Scholar]
- 96. Yeh KH, Kondo T, Zheng J, Tsvetkov LM, Blair J, Zhang H. The F‐box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin‐dependent degradation of cyclin E. Biochem Biophys Res Commun. 2001;281(4):884‐890. [DOI] [PubMed] [Google Scholar]
- 97. Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11(5):1177‐1188. [DOI] [PubMed] [Google Scholar]
- 98. Welcker M, Orian A, Jin J, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation‐dependent c‐Myc protein degradation. Proc Natl Acad Sci USA. 2004;101(24):9085‐9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cheng J, Guo J, North BJ, Tao K, Zhou P, Wei W. The emerging role for Cullin 4 family of E3 ligases in tumorigenesis. Biochimica et Biophysica Acta (BBA) ‐ Reviews on Cancer. 2019;1871(1):138‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Higa LA, Yang X, Zheng J, et al. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5(1):71‐77. [DOI] [PubMed] [Google Scholar]
- 101. Zou Y, Mi J, Cui J, et al. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem. 2009;284(48):33320‐33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hao Q, Chen Y, Zhou X. The Janus face of p53‐targeting ubiquitin ligases. Cells. 2020;9(7):1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Gatti V, Bernassola F, Talora C, Melino G, Peschiaroli A. The impact of the ubiquitin system in the pathogenesis of squamous cell carcinomas. Cancers (Basel). 2020;12(6):1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112(7):1415‐1424. [DOI] [PubMed] [Google Scholar]
- 105. Shigemasa K, Gu L, O'Brien TJ, Ohama K. Skp2 overexpression is a prognostic factor in patients with ovarian adenocarcinoma. Clin Cancer Res. 2003;9(5):1756‐1763. [PubMed] [Google Scholar]
- 106. Traub F, Mengel M, Lück HJ, Kreipe HH, von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res Treat. 2006;99(2):185‐191. [DOI] [PubMed] [Google Scholar]
- 107. Zhao H, Bauzon F, Fu H, et al. Skp2 deletion unmasks a p27 safeguard that blocks tumorigenesis in the absence of pRb and p53 tumor suppressors. Cancer Cell. 2013;24(5):645‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Celebi G, Kesim H, Ozer E, Kutlu O. The effect of dysfunctional ubiquitin enzymes in the pathogenesis of most common diseases. Int J Mol Sci. 2020;21(17):6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non‐small cell lung cancer patients. J Surg Oncol. 2012;106(4):423‐430. [DOI] [PubMed] [Google Scholar]
- 110. Mondal G, Sengupta S, Panda CK, Gollin SM, Saunders WS, Roychoudhury S. Overexpression of Cdc20 leads to impairment of the spindle assembly checkpoint and aneuploidization in oral cancer. Carcinogenesis. 2007;28(1):81‐92. [DOI] [PubMed] [Google Scholar]
- 111. Vriend J, Nachtigal MW. Ubiquitin proteasome pathway transcriptome in epithelial ovarian cancer. Cancers (Basel). 2021;13(11):2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wu WJ, Hu KS, Wang DS, et al. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J Transl Med. 2013;11:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint‐mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185(6):983‐994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Fujita T, Liu W, Doihara H, Wan Y. Regulation of Skp2‐p27 axis by the Cdh1/anaphase‐promoting complex pathway in colorectal tumorigenesis. Am J Pathol. 2008;173(1):217‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fujita T, Liu W, Doihara H, Wan Y. An in vivo study of Cdh1/APC in breast cancer formation. Int J Cancer. 2009;125(4):826‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sang Y, Yan F, Ren X. The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications. Oncotarget. 2015;6(40):42590‐42602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Li T, Wu S, Jia L, et al. CUL4 E3 ligase regulates the proliferation and apoptosis of lung squamous cell carcinoma and small cell lung carcinoma. Cancer Biol Med. 2020;17(2):357‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu L, Lee S, Zhang J, et al. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34(4):451‐460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Argentini M, Barboule N, Wasylyk B. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene. 2000;19(34):3849‐3857. [DOI] [PubMed] [Google Scholar]
- 120. Oliner JD, Saiki AY, Caenepeel S. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb Perspect Med. 2016;6(6):a026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565‐1570. [DOI] [PubMed] [Google Scholar]
- 123. Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117(5):1137‐1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Whiteside T. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904‐5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhao H, Wu L, Yan G, et al. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct Target Ther. 2021;6(1):1‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Katzenellenbogen M, Mizrahi L, Pappo O, et al. Molecular mechanisms of liver carcinogenesis in the mdr2‐knockout mice. Mol Cancer Res. 2007;5(11):1159‐1170. [DOI] [PubMed] [Google Scholar]
- 127. Pikarsky E, Porat RM, Stein I, et al. NF‐kappaB functions as a tumour promoter in inflammation‐associated cancer. Nature. 2004;431(7007):461‐466. [DOI] [PubMed] [Google Scholar]
- 128. Ni X, Kou W, Gu J, et al. TRAF6 directs FOXP3 localization and facilitates regulatory T‐cell function through K63‐linked ubiquitination. Embo j. 2019;38(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wu X, Xue R, Peng H, et al. Traf6 inhibitor boosts antitumor immunity by impeding regulatory T cell migration in Hepa1‐6 tumor model. Int Immunopharmacol. 2019;77:105965. [DOI] [PubMed] [Google Scholar]
- 130. Peng DJ, Zeng M, Muromoto R, et al. Noncanonical K27‐linked polyubiquitination of TIEG1 regulates Foxp3 expression and tumor growth. J Immunol. 2011;186(10):5638‐5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Paolino M, Choidas A, Wallner S, et al. The E3 ligase Cbl‐b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507(7493):508‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Venuprasad K. Cbl‐b and itch: key regulators of peripheral T‐cell tolerance. Cancer Res. 2010;70(8):3009‐3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kumar J, Kumar R, Kumar Singh A, et al. Deletion of Cbl‐b inhibits CD8(+) T‐cell exhaustion and promotes CAR T‐cell function. J Immunother Cancer. 2021;9(1):e001688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Thell K, Urban M, Harrauer J, et al. Master checkpoint Cbl‐b inhibition: anti‐tumour efficacy in a murine colorectal cancer model following siRNA‐based cell therapy. Ann Oncol. 2019;30:v503‐v504. [Google Scholar]
- 135. Sun SC. Non‐canonical NF‐κB signaling pathway. Cell Res. 2011;21(1):71‐85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF‐kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12(2):115‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF‐kappaB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Varfolomeev E, Blankenship JW, Wayson SM, et al. IAP antagonists induce autoubiquitination of c‐IAPs, NF‐kappaB activation, and TNFalpha‐dependent apoptosis. Cell. 2007;131(4):669‐681. [DOI] [PubMed] [Google Scholar]
- 139. Vince JE, Wong WW, Khan N, et al. IAP antagonists target cIAP1 to induce TNFalpha‐dependent apoptosis. Cell. 2007;131(4):682‐693. [DOI] [PubMed] [Google Scholar]
- 140. Zarnegar BJ, Wang Y, Mahoney DJ, et al. Noncanonical NF‐kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9(12):1371‐1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Senftleben U, Cao Y, Xiao G, et al. Activation by IKKalpha of a second, evolutionary conserved, NF‐kappa B signaling pathway. Science. 2001;293(5534):1495‐1499. [DOI] [PubMed] [Google Scholar]
- 142. Liao G, Zhang M, Harhaj EW, Sun SC. Regulation of the NF‐kappaB‐inducing kinase by tumor necrosis factor receptor‐associated factor 3‐induced degradation. J Biol Chem. 2004;279(25):26243‐26250. [DOI] [PubMed] [Google Scholar]
- 143. Vallabhapurapu S, Matsuzawa A, Zhang W, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK‐dependent alternative NF‐kappaB signaling. Nat Immunol. 2008;9(12):1364‐1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Döppler H, Liou GY, Storz P. Downregulation of TRAF2 mediates NIK‐induced pancreatic cancer cell proliferation and tumorigenicity. PLoS One. 2013;8(1):e53676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gardam S, Turner VM, Anderton H, et al. Deletion of cIAP1 and cIAP2 in murine B lymphocytes constitutively activates cell survival pathways and inactivates the germinal center response. Blood. 2011;117(15):4041‐4051. [DOI] [PubMed] [Google Scholar]
- 146. Lalani AI, Moore CR, Luo C, et al. Myeloid cell TRAF3 regulates immune responses and inhibits inflammation and tumor development in mice. J Immunol. 2015;194(1):334‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Moore CR, Edwards SK, Xie P. Targeting TRAF3 downstream signaling pathways in B cell neoplasms. J Cancer Sci Ther. 2015;7(2):67‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zinatizadeh MR, Schock B, Chalbatani GM, Zarandi PK, Jalali SA, Miri SR. The nuclear factor kappa B (NF‐kB) signaling in cancer development and immune diseases. Genes Dis. 2021;8(3):287‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Courtois G, Fauvarque M‐O. The many roles of ubiquitin in NF‐κB signaling. Biomedicines. 2018;6(2):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yamamoto Y, Gaynor RB. IκB kinases: key regulators of the NF‐κB pathway. Trends Biochem Sci. 2004;29(2):72‐79. [DOI] [PubMed] [Google Scholar]
- 151. Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF‐κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct Target Ther. 2020;5(1):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cockram PE, Kist M, Prakash S, Chen S‐H, Wertz IE, Vucic D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021;28(2):591‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wajant H, Scheurich P. TNFR1‐induced activation of the classical NF‐κB pathway. FEBS J. 2011;278(6):862‐876. [DOI] [PubMed] [Google Scholar]
- 154. Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor α‐induced NF‐κB activation. J Biol Chem. 2006;281(19):13636‐13643. [DOI] [PubMed] [Google Scholar]
- 155. Vince JE, Pantaki D, Feltham R, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate NF‐κB and to prevent TNF‐induced apoptosis. J Biol Chem. 2009;284(51):35906‐35915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gerlach B, Cordier SM, Schmukle AC, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471(7340):591‐596. [DOI] [PubMed] [Google Scholar]
- 157. Rahighi S, Ikeda F, Kawasaki M, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF‐κB activation. Cell. 2009;136(6):1098‐1109. [DOI] [PubMed] [Google Scholar]
- 158. Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J Biol Chem. 2013;288(44):31728‐31737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Witt A, Vucic D. Diverse ubiquitin linkages regulate RIP kinases‐mediated inflammatory and cell death signaling. Cell Death Differ. 2017;24(7):1160‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wu C‐J, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63‐linked polyubiquitination by NEMO is a key event in NF‐κB activation. Nat Cell Biol. 2006;8(4):398‐406. [DOI] [PubMed] [Google Scholar]
- 161. Adhikari A, Xu M, Chen ZJ. Ubiquitin‐mediated activation of TAK1 and IKK. Oncogene. 2007;26(22):3214‐3226. [DOI] [PubMed] [Google Scholar]
- 162. Xu Y‐R, Lei C‐Q. TAK1‐TABs complex: a central signalosome in inflammatory responses. Front Immunol. 2021;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Shin C, Ito Y, Ichikawa S, Tokunaga M, Sakata‐Sogawa K, Tanaka T. MKRN2 is a novel ubiquitin E3 ligase for the p65 subunit of NF‐κB and negatively regulates inflammatory responses. Sci Rep. 2017;7(1):46097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Shen RR, Zhou AY, Kim E, et al. TRAF2 is an NF‐κB‐activating oncogene in epithelial cancers. Oncogene. 2015;34(2):209‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Chesi M, Mirza NN, Garbitt VM, et al. IAP antagonists induce anti‐tumor immunity in multiple myeloma. Nat Med. 2016;22(12):1411‐1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kearney CJ, Lalaoui N, Freeman AJ, Ramsbottom KM, Silke J, Oliaro J. PD‐L1 and IAPs co‐operate to protect tumors from cytotoxic lymphocyte‐derived TNF. Cell Death Differ. 2017;24(10):1705‐1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kawasaki T, Kawai T. Toll‐like receptor signaling pathways. Front Immunol. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Gohda J, Matsumura T, Inoue J. Cutting edge: TNFR‐associated factor (TRAF) 6 is essential for MyD88‐dependent pathway but not toll/IL‐1 receptor domain‐containing adaptor‐inducing IFN‐beta (TRIF)‐dependent pathway in TLR signaling. J Immunol. 2004;173(5):2913‐2917. [DOI] [PubMed] [Google Scholar]
- 169. Murphy M, Xiong Y, Pattabiraman G, Qiu F, Medvedev AE. Pellino‐1 positively regulates toll‐like receptor (TLR) 2 and TLR4 signaling and is suppressed upon induction of endotoxin tolerance. J Biol Chem. 2015;290(31):19218‐19232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ishitani T, Takaesu G, Ninomiya‐Tsuji J, Shibuya H, Gaynor RB, Matsumoto K. Role of the TAB2‐related protein TAB3 in IL‐1 and TNF signaling. Embo j. 2003;22(23):6277‐6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Jiang Z, Ninomiya‐Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin‐1 (IL‐1) receptor‐associated kinase‐dependent IL‐1‐induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22(20):7158‐7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Cao C, An R, Yu Y, et al. BICP0 negatively regulates TRAF6‐mediated NF‐κB and interferon activation by promoting K48‐linked polyubiquitination of TRAF6. Front Microbiol. 2019;10:3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Jang HD, Hwang HZ, Kim HS, Lee SY. C‐Cbl negatively regulates TRAF6‐mediated NF‐κB activation by promoting K48‐linked polyubiquitination of TRAF6. Cell Mol Biol Lett. 2019;24:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zarzycka B, Seijkens T, Nabuurs SB, et al. Discovery of small molecule CD40‐TRAF6 inhibitors. J Chem Inf Model. 2015;55(2):294‐307. [DOI] [PubMed] [Google Scholar]
- 175. Goncharov T, Hedayati S, Mulvihill MM, et al. Disruption of XIAP‐RIP2 association blocks NOD2‐mediated inflammatory signaling. Mol Cell. 2018;69(4):551‐65.e7. [DOI] [PubMed] [Google Scholar]
- 176. Krieg A, Correa RG, Garrison JB, et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Natl Acad Sci USA. 2009;106(34):14524‐14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Damgaard RB, Nachbur U, Yabal M, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell. 2012;46(6):746‐758. [DOI] [PubMed] [Google Scholar]
- 178. Bertrand MJ, Doiron K, Labbé K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30(6):789‐801. [DOI] [PubMed] [Google Scholar]
- 179. Okamoto M, Kouwaki T, Fukushima Y, Oshiumi H. Regulation of RIG‐I activation by K63‐linked polyubiquitination. Front Immunol. 2017;8:1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Fang R, Jiang Q, Zhou X, et al. MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 2017;13(11):e1006720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Liu S, Chen J, Cai X, et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2:e00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zhang L, Zhang B, Wei M, et al. TRIM22 inhibits endometrial cancer progression through the NOD2/NF‑κB signaling pathway and confers a favorable prognosis. Int J Oncol. 2020;56(5):1225‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Asano N, Imatani A, Watanabe T, et al. Cdx2 expression and intestinal metaplasia induced by H. pylori infection of gastric cells is regulated by NOD1‐mediated innate immune responses. Cancer Res. 2016;76(5):1135‐1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Bognar MK, Vincendeau M, Erdmann T, et al. Oncogenic CARMA1 couples NF‐κB and β‐catenin signaling in diffuse large B‐cell lymphomas. Oncogene. 2016;35(32):4269‐4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ma B, Hottiger MO. Crosstalk between Wnt/β‐catenin and NF‐κB signaling pathway during inflammation. Front Immunol. 2016;7:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Spiegelman VS, Slaga TJ, Pagano M, Minamoto T, Ronai Z, Fuchs SY. Wnt/beta‐catenin signaling induces the expression and activity of betaTrCP ubiquitin ligase receptor. Mol Cell. 2000;5(5):877‐882. [DOI] [PubMed] [Google Scholar]
- 187. Ougolkov A, Zhang B, Yamashita K, et al. Associations among beta‐TrCP, an E3 ubiquitin ligase receptor, beta‐catenin, and NF‐kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96(15):1161‐1170. [DOI] [PubMed] [Google Scholar]
- 188. Wang L, Feng W, Yang X, et al. Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of NF‐κB and β‐catenin/TCF signaling pathways. Cell Death Dis. 2018;9(4):427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Pahl HL. Activators and target genes of Rel/NF‐kappaB transcription factors. Oncogene. 1999;18(49):6853‐6866. [DOI] [PubMed] [Google Scholar]
- 190. Zhang T, Wang H, Han L. Expression and clinical significance of tumor necrosis factor receptor‐associated factor 6 in patients with colon cancer. Iran Red Crescent Med J. 2016;18(1):e23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhang XL, Dang YW, Li P, et al. Expression of tumor necrosis factor receptor‐associated factor 6 in lung cancer tissues. Asian Pac J Cancer Prev. 2014;15(24):10591‐10596. [DOI] [PubMed] [Google Scholar]
- 192. Zhu G, Cheng Z, Huang Y, et al. TRAF6 promotes the progression and growth of colorectal cancer through nuclear shuttle regulation NF‐kB/c‐jun signaling pathway. Life Sci. 2019;235:116831. [DOI] [PubMed] [Google Scholar]
- 193. Morgan JJ, McAvera RM, Crawford LJ. TRAF6 silencing attenuates multiple myeloma cell adhesion to bone marrow stromal cells. Int J Mol Sci. 2019;20(3):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Peramuhendige P, Marino S, Bishop RT, et al. TRAF2 in osteotropic breast cancer cells enhances skeletal tumour growth and promotes osteolysis. Sci Rep. 2018;8(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Liu J, Suresh Kumar KG, Yu D, et al. Oncogenic BRAF regulates beta‐Trcp expression and NF‐kappaB activity in human melanoma cells. Oncogene. 2007;26(13):1954‐1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Müerköster S, Arlt A, Sipos B, et al. Increased expression of the E3‐ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor‐kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Res. 2005;65(4):1316‐1324. [DOI] [PubMed] [Google Scholar]
- 197. Dai H, Chen H, Xu J, et al. The ubiquitin ligase CHIP modulates cellular behaviors of gastric cancer cells by regulating TRAF2. Cancer Cell Int. 2019;19:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wang Y, Ren F, Wang Y, et al. CHIP/Stub1 functions as a tumor suppressor and represses NF‐κB‐mediated signaling in colorectal cancer. Carcinogenesis. 2014;35(5):983‐991. [DOI] [PubMed] [Google Scholar]
- 199. Jin J, Lu Z, Wang X, et al. E3 ubiquitin ligase TRIM7 negatively regulates NF‐kappa B signaling pathway by degrading p65 in lung cancer. Cell Signal. 2020;69:109543. [DOI] [PubMed] [Google Scholar]
- 200. Li Y, Deng L, Zhao X, et al. Tripartite motif‐containing 37 (TRIM37) promotes the aggressiveness of non‐small‐cell lung cancer cells by activating the NF‐κB pathway. J Pathol. 2018;246(3):366‐378. [DOI] [PubMed] [Google Scholar]
- 201. Carneiro BA, El‐Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. 2020;17(7):395‐417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Liu L, Wang M, Li X, Yin S, Wang B. An overview of novel agents for cervical cancer treatment by inducing apoptosis: emerging drugs ongoing clinical trials and preclinical studies. Front Med (Lausanne). 2021;8:682366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Chin HS, Fu NY. Physiological functions of Mcl‐1: insights from genetic mouse models. Front Cell Dev Biol. 2021;9:704547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL‐2 family proteins. Nat Rev Mol Cell Biol. 2019;20(3):175‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Gómez‐Díaz C, Ikeda F. Roles of ubiquitin in autophagy and cell death. Semin Cell Dev Biol. 2019;93:125‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Gupta I, Singh K, Varshney NK, Khan S. Delineating crosstalk mechanisms of the ubiquitin proteasome system that regulate apoptosis. Front Cell Dev Biol. 2018;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Huang NJ, Zhang L, Tang W, Chen C, Yang CS, Kornbluth S. The Trim39 ubiquitin ligase inhibits APC/CCdh1‐mediated degradation of the Bax activator MOAP‐1. J Cell Biol. 2012;197(3):361‐367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Berthelet J, Dubrez L. Regulation of apoptosis by inhibitors of apoptosis (IAPs). Cells. 2013;2(1):163‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Obexer P, Ausserlechner MJ. X‐linked inhibitor of apoptosis protein ‐ a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front Oncol. 2014;4:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Cheung HH, Plenchette S, Kern CJ, Mahoney DJ, Korneluk RG. The RING domain of cIAP1 mediates the degradation of RING‐bearing inhibitor of apoptosis proteins by distinct pathways. Mol Biol Cell. 2008;19(7):2729‐2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Choi YE, Butterworth M, Malladi S, Duckett CS, Cohen GM, Bratton SB. The E3 ubiquitin ligase cIAP1 binds and ubiquitinates caspase‐3 and ‐7 via unique mechanisms at distinct steps in their processing. J Biol Chem. 2009;284(19):12772‐12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin‐protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem. 2000;275(35):26661‐26664. [DOI] [PubMed] [Google Scholar]
- 213. Eckelman BP, Salvesen GS. The human anti‐apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J Biol Chem. 2006;281(6):3254‐3260. [DOI] [PubMed] [Google Scholar]
- 214. Wilkinson JC, Wilkinson AS, Scott FL, Csomos RA, Salvesen GS, Duckett CS. Neutralization of Smac/Diablo by inhibitors of apoptosis (IAPs). A caspase‐independent mechanism for apoptotic inhibition. J Biol Chem. 2004;279(49):51082‐51090. [DOI] [PubMed] [Google Scholar]
- 215. Vince JE, Pantaki D, Feltham R, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate NF‐κB and to prevent TNF‐induced apoptosis. J Biol Chem. 2009;284(51):35906‐35915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Verhagen AM, Vaux DL. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis. 2002;7(2):163‐166. [DOI] [PubMed] [Google Scholar]
- 217. Martinez‐Ruiz G, Maldonado V, Ceballos‐Cancino G, Grajeda JP, Melendez‐Zajgla J. Role of Smac/DIABLO in cancer progression. J Exp Clin Cancer Res. 2008;27(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Kim JB, Kim SY, Kim BM, et al. Identification of a novel anti‐apoptotic E3 ubiquitin ligase that ubiquitinates antagonists of inhibitor of apoptosis proteins SMAC, HtrA2, and ARTS. J Biol Chem. 2013;288(17):12014‐12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Dehan E, Bassermann F, Guardavaccaro D, et al. betaTrCP‐ and Rsk1/2‐mediated degradation of BimEL inhibits apoptosis. Mol Cell. 2009;33(1):109‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Ding Q, He X, Hsu JM, et al. Degradation of Mcl‐1 by beta‐TrCP mediates glycogen synthase kinase 3‐induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27(11):4006‐4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. How does p53 induce apoptosis and how does this relate to p53‐mediated tumour suppression? Cell Death Differ. 2018;25(1):104‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin‐protein ligase activity of X‐linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase‐3 and enhances its anti‐apoptotic effect in Fas‐induced cell death. Proc Natl Acad Sci USA. 2001;98(15):8662‐8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Li M, Brooks CL, Wu‐Baer F, Chen D, Baer R, Gu W. Mono‐ versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972‐1975. [DOI] [PubMed] [Google Scholar]
- 224. Wertz IE, Dixit VM. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ. 2010;17(1):14‐24. [DOI] [PubMed] [Google Scholar]
- 225. Kokolakis G, Sabat R, Krüger‐Krasagakis S, Eberle J. Ambivalent effects of tumor necrosis factor alpha on apoptosis of malignant and normal human keratinocytes. Skin Pharmacol Physiol. 2021;34(2):94‐102. [DOI] [PubMed] [Google Scholar]
- 226. Micheau O, Tschopp J. Induction of TNF receptor I‐mediated apoptosis via two sequential signaling complexes. Cell. 2003;114(2):181‐190. [DOI] [PubMed] [Google Scholar]
- 227. Wang L, Du F, Wang X. TNF‐alpha induces two distinct caspase‐8 activation pathways. Cell. 2008;133(4):693‐703. [DOI] [PubMed] [Google Scholar]
- 228. Amin P, Florez M, Najafov A, et al. Regulation of a distinct activated RIPK1 intermediate bridging complex I and complex II in TNFα‐mediated apoptosis. Proc Natl Acad Sci USA. 2018;115(26):E5944‐e5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. O'Donnell MA, Legarda‐Addison D, Skountzos P, Yeh WC, Ting AT. Ubiquitination of RIP1 regulates an NF‐kappaB‐independent cell‐death switch in TNF signaling. Curr Biol. 2007;17(5):418‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat Rev Mol Cell Biol. 2013;14(11):727‐736. [DOI] [PubMed] [Google Scholar]
- 231. Bertrand MJ, Milutinovic S, Dickson KM, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30(6):689‐700. [DOI] [PubMed] [Google Scholar]
- 232. Gentle IE, Wong WW, Evans JM, et al. In TNF‐stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non‐canonical NF‐κB and activation of caspase‐8. J Biol Chem. 2016;291(5):2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Priem D, Devos M, Druwé S, et al. A20 protects cells from TNF‐induced apoptosis through linear ubiquitin‐dependent and ‐independent mechanisms. Cell Death Dis. 2019;10(10):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Chang L, Kamata H, Solinas G, et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha‐induced cell death by inducing c‐FLIP(L) turnover. Cell. 2006;124(3):601‐613. [DOI] [PubMed] [Google Scholar]
- 235. Gonzalvez F, Lawrence D, Yang B, et al. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase‐8 with a ubiquitin shutoff timer. Mol Cell. 2012;48(6):888‐899. [DOI] [PubMed] [Google Scholar]
- 236. Xu L, Zhang Y, Qu X, et al. DR5‐Cbl‐b/c‐Cbl‐TRAF2 complex inhibits TRAIL‐induced apoptosis by promoting TRAF2‐mediated polyubiquitination of caspase‐8 in gastric cancer cells. Mol Oncol. 2017;11(12):1733‐1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Christian PA, Fiandalo MV, Schwarze SR. Possible role of death receptor‐mediated apoptosis by the E3 ubiquitin ligases Siah2 and POSH. Mol Cancer. 2011;10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Woo SM, Kwon TK. E3 ubiquitin ligases and deubiquitinases as modulators of TRAIL‐mediated extrinsic apoptotic signaling pathway. BMB Rep. 2019;52(2):119‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Liu Y, Zhang B, Shi T, Qin H. Inhibition of X‐linked inhibitor of apoptosis protein suppresses tumorigenesis and enhances chemosensitivity in anaplastic thyroid carcinoma. Oncotarget. 2017;8(56):95764‐95772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Devi GR, Finetti P, Morse MA, et al. Expression of X‐linked inhibitor of apoptosis protein (XIAP) in breast cancer is associated with shorter survival and resistance to chemotherapy. Cancers (Basel). 2021;13(11):2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Hussain AR, Siraj AK, Ahmed M, et al. XIAP over‐expression is an independent poor prognostic marker in Middle Eastern breast cancer and can be targeted to induce efficient apoptosis. BMC Cancer. 2017;17(1):640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Mizutani Y, Nakanishi H, Li YN, et al. Overexpression of XIAP expression in renal cell carcinoma predicts a worse prognosis. Int J Oncol. 2007;30(4):919‐925. [PubMed] [Google Scholar]
- 243. Tamm I, Kornblau SM, Segall H, et al. Expression and prognostic significance of IAP‐family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000;6(5):1796‐1803. [PubMed] [Google Scholar]
- 244. Tu H, Costa M. XIAP's profile in human cancer. Biomolecules. 2020;10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Wang X, Shu Y, Shi H, et al. TRIM9 is up‐regulated in human lung cancer and involved in cell proliferation and apoptosis. Int J Clin Exp Med. 2016;9(6):10461‐10469. [Google Scholar]
- 246. Yang F, Liu H, Yu Y, Xu L. TRIM9 overexpression promotes uterine leiomyoma cell proliferation and inhibits cell apoptosis via NF‐κB signaling pathway. Life Sci. 2020;257:118101. [DOI] [PubMed] [Google Scholar]
- 247. Wei B, Ruan J, Mi Y, et al. Knockdown of TNF receptor‐associated factor 2 (TRAF2) modulates in vitro growth of TRAIL‐treated prostate cancer cells. Biomed Pharmacother. 2017;93:462‐469. [DOI] [PubMed] [Google Scholar]
- 248. Richardson PG, Hideshima T, Anderson KC. Bortezomib (PS‐341): a novel, first‐in‐class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control. 2003;10(5):361‐369. [DOI] [PubMed] [Google Scholar]
- 249. Yerlikaya A, Okur E, Eker S, Erin N. Combined effects of the proteasome inhibitor bortezomib and Hsp70 inhibitors on the B16F10 melanoma cell line. Mol Med Rep. 2010;3(2):333‐339. [DOI] [PubMed] [Google Scholar]
- 250. Wu HQ, Baker D, Ovaa H. Small molecules that target the ubiquitin system. Biochem Soc Trans. 2020;48(2):479‐497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. LaPlante G, Zhang W. Targeting the ubiquitin‐proteasome system for cancer therapeutics by small‐molecule inhibitors. Cancers (Basel). 2021;13(12):3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Feroz W, Sheikh AMA. Exploring the multiple roles of guardian of the genome: P53. Egypt J Med Hum Genet. 2020;21(1):1‐23. [Google Scholar]
- 253. Shangary S, Wang S. Targeting the MDM2‐p53 interaction for cancer therapy. Clin Cancer Res. 2008;14(17):5318‐5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small‐molecule antagonists of MDM2. Science. 2004;303(5659):844‐848. [DOI] [PubMed] [Google Scholar]
- 255. Tovar C, Rosinski J, Filipovic Z, et al. Small‐molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103(6):1888‐1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Tovar C, Graves B, Packman K, et al. MDM2 small‐molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73(8):2587‐2597. [DOI] [PubMed] [Google Scholar]
- 257. Andreeff M, Kelly KR, Yee K, et al. Results of the phase I trial of RG7112, a small‐molecule MDM2 antagonist in leukemia. Clin Cancer Res. 2016;22(4):868‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Ray‐Coquard I, Blay JY, Italiano A, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2‐amplified, well‐differentiated or dedifferentiated liposarcoma: an exploratory proof‐of‐mechanism study. Lancet Oncol. 2012;13(11):1133‐1140. [DOI] [PubMed] [Google Scholar]
- 259. Ding Q, Zhang Z, Liu JJ, et al. Discovery of RG7388, a potent and selective p53‐MDM2 inhibitor in clinical development. J Med Chem. 2013;56(14):5979‐5983. [DOI] [PubMed] [Google Scholar]
- 260. Yee K, Martinelli G, Vey N, et al. Phase 1/1b study of RG7388, a potent MDM2 antagonist, in acute myelogenous leukemia (AML) patients (Pts). Blood. 2014;124(21):116. [Google Scholar]
- 261. Rew Y, Sun D. Discovery of a small molecule MDM2 inhibitor (AMG 232) for treating cancer. J Med Chem. 2014;57(15):6332‐6341. [DOI] [PubMed] [Google Scholar]
- 262. Canon J, Osgood T, Olson SH, et al. The MDM2 inhibitor AMG 232 demonstrates robust antitumor efficacy and potentiates the activity of p53‐inducing cytotoxic agents. Mol Cancer Ther. 2015;14(3):649‐658. [DOI] [PubMed] [Google Scholar]
- 263. Gluck WL, Gounder MM, Frank R, et al. Phase 1 study of the MDM2 inhibitor AMG 232 in patients with advanced P53 wild‐type solid tumors or multiple myeloma. Invest New Drugs. 2020;38(3):831‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Espinoza‐Fonseca LM. Targeting MDM2 by the small molecule RITA: towards the development of new multi‐target drugs against cancer. Theor Biol Med Model. 2005;2:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53‐HDM‐2 interaction and activates p53 function in tumors. Nat Med. 2004;10(12):1321‐1328. [DOI] [PubMed] [Google Scholar]
- 266. Zhao CY, Szekely L, Bao W, Selivanova G. Rescue of p53 function by small‐molecule RITA in cervical carcinoma by blocking E6‐mediated degradation. Cancer Res. 2010;70(8):3372‐3381. [DOI] [PubMed] [Google Scholar]
- 267. Gupta AK, Bharadwaj M, Kumar A, Mehrotra R. Spiro‐oxindoles as a promising class of small molecule inhibitors of p53‐MDM2 interaction useful in targeted cancer therapy. Top Curr Chem (Cham). 2017;375(1):3. [DOI] [PubMed] [Google Scholar]
- 268. de Jonge M, de Weger VA, Dickson MA, et al. A phase I study of SAR405838, a novel human double minute 2 (HDM2) antagonist, in patients with solid tumours. Eur J Cancer. 2017;76:144‐151. [DOI] [PubMed] [Google Scholar]
- 269. Chen Q, Xie W, Kuhn DJ, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27‐ and Skp2‐mediated cell‐cycle arrest and activation of autophagy. Blood. 2008;111(9):4690‐4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Chan CH, Morrow JK, Li CF, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154(3):556‐568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Malek E, Abdel‐Malek MA, Jagannathan S, et al. Pharmacogenomics and chemical library screens reveal a novel SCF(SKP2) inhibitor that overcomes Bortezomib resistance in multiple myeloma. Leukemia. 2017;31(3):645‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2‐mediated p27 degradation. Chem Biol. 2012;19(12):1515‐1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Huang HC, Lin CL, Lin JK. 1,2,3,4,6‐penta‐O‐galloyl‐β‐D‐glucose, quercetin, curcumin and lycopene induce cell‐cycle arrest in MDA‐MB‐231 and BT474 cells through downregulation of Skp2 protein. J Agric Food Chem. 2011;59(12):6765‐6775. [DOI] [PubMed] [Google Scholar]
- 274. Cheng AL, Hsu CH, Lin JK, et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high‐risk or pre‐malignant lesions. Anticancer Res. 2001;21(4b):2895‐2900. [PubMed] [Google Scholar]
- 275. Dhillon N, Aggarwal BB, Newman RA, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14(14):4491‐4499. [DOI] [PubMed] [Google Scholar]
- 276. Mahammedi H, Planchat E, Pouget M, et al. The new combination docetaxel, prednisone and curcumin in patients with castration‐resistant prostate cancer: a pilot phase II study. Oncology. 2016;90(2):69‐78. [DOI] [PubMed] [Google Scholar]
- 277. Zhou L, Yu X, Li M, et al. Cdh1‐mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine. 2020;51:102570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Huang HL, Weng HY, Wang LQ, et al. Triggering Fbw7‐mediated proteasomal degradation of c‐Myc by oridonin induces cell growth inhibition and apoptosis. Mol Cancer Ther. 2012;11(5):1155‐1165. [DOI] [PubMed] [Google Scholar]
- 279. Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8‐activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732‐736. [DOI] [PubMed] [Google Scholar]
- 280. Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018;44:92‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Sarantopoulos J, Shapiro GI, Cohen RB, et al. Phase I study of the investigational NEDD8‐activating enzyme inhibitor pevonedistat (TAK‐924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. 2016;22(4):847‐857. [DOI] [PubMed] [Google Scholar]
- 282. Adès L, Girshova L, Doronin VA, et al. Pevonedistat plus azacitidine vs azacitidine alone in higher‐risk MDS/chronic myelomonocytic leukemia or low‐blast‐percentage AML. Blood Adv. 2022;6(17):5132‐5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Sekeres MA, Watts J, Radinoff A, et al. Correction to: randomized phase 2 trial of pevonedistat plus azacitidine versus azacitidine for higher‐risk MDS/CMML or low‐blast AML. Leukemia. 2021;35(12):3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Yang C, Wu J, He H, Liu H. Small molecule NSC1892 targets the CUL4A/4B‐DDB1 interactions and causes impairment of CRL4(DCAF4) E3 ligases to inhibit colorectal cancer cell growth. Int J Biol Sci. 2020;16(6):1059‐1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Zeng X, Sigoillot F, Gaur S, et al. Pharmacologic inhibition of the anaphase‐promoting complex induces a spindle checkpoint‐dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18(4):382‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Sackton KL, Dimova N, Zeng X, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514(7524):646‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Nakajima H, Fujiwara H, Furuichi Y, Tanaka K, Shimbara N. A novel small‐molecule inhibitor of NF‐kappaB signaling. Biochem Biophys Res Commun. 2008;368(4):1007‐1013. [DOI] [PubMed] [Google Scholar]
- 288. Brenke JK, Popowicz GM, Schorpp K, et al. Targeting TRAF6 E3 ligase activity with a small‐molecule inhibitor combats autoimmunity. J Biol Chem. 2018;293(34):13191‐13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Bishop RT, Marino S, Carrasco G, et al. Combined administration of a small‐molecule inhibitor of TRAF6 and Docetaxel reduces breast cancer skeletal metastasis and osteolysis. Cancer Lett. 2020;488:27‐39. [DOI] [PubMed] [Google Scholar]
- 290. Sakamoto H, Egashira S, Saito N, et al. Gliotoxin suppresses NF‐κB activation by selectively inhibiting linear ubiquitin chain assembly complex (LUBAC). ACS Chem Biol. 2015;10(3):675‐681. [DOI] [PubMed] [Google Scholar]
- 291. Johansson H, Isabella Tsai YC, Fantom K, et al. Fragment‐based covalent ligand screening enables rapid discovery of inhibitors for the RBR E3 ubiquitin ligase HOIP. J Am Chem Soc. 2019;141(6):2703‐2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Oikawa D, Sato Y, Ohtake F, et al. Molecular bases for HOIPINs‐mediated inhibition of LUBAC and innate immune responses. Commun Biol. 2020;3(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Chang Y‐C, Cheung CHA. An updated review of smac mimetics, LCL161, birinapant, and GDC‐0152 in cancer treatment. Applied Sciences. 2021;11(1):335. [Google Scholar]
- 294. Hurwitz HI, Smith DC, Pitot HC, et al. Safety, pharmacokinetics, and pharmacodynamic properties of oral DEBIO1143 (AT‐406) in patients with advanced cancer: results of a first‐in‐man study. Cancer Chemother Pharmacol. 2015;75(4):851‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Sun XS, Tao Y, Le Tourneau C, et al. Debio 1143 and high‐dose cisplatin chemoradiotherapy in high‐risk locoregionally advanced squamous cell carcinoma of the head and neck: a double‐blind, multicentre, randomised, phase 2 study. Lancet Oncol. 2020;21(9):1173‐1187. [DOI] [PubMed] [Google Scholar]
- 296. Benetatos CA, Mitsuuchi Y, Burns JM, et al. Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2‐associated cIAPs, abrogates TNF‐induced NF‐κB activation, and is active in patient‐derived xenograft models. Mol Cancer Ther. 2014;13(4):867‐879. [DOI] [PubMed] [Google Scholar]
- 297. Alchanati I, Teicher C, Cohen G, et al. The E3 ubiquitin‐ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex ‐ a potentially new drug target. PLoS One. 2009;4(12):e8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Ismail IH, McDonald D, Strickfaden H, Xu Z, Hendzel MJ. A small molecule inhibitor of polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double‐strand breaks. J Biol Chem. 2013;288(37):26944‐26954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Su W, Han HH, Wang Y, et al. The polycomb repressor complex 1 drives double‐negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell. 2019;36(2):139‐55.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Shukla S, Ying W, Gray F, et al. Small‐molecule inhibitors targeting Polycomb repressive complex 1 RING domain. Nat Chem Biol. 2021;17(7):784‐793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1‐Cullin‐F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98(15):8554‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Burslem GM, Smith BE, Lai AC, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol. 2018;25(1):67‐77.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Samarasinghe KTG, Crews CM. Targeted protein degradation: a promise for undruggable proteins. Cell Chem Biol. 2021;28(7):934‐951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Fisher SL, Phillips AJ. Targeted protein degradation and the enzymology of degraders. Curr Opin Chem Biol. 2018;44:47‐55. [DOI] [PubMed] [Google Scholar]
- 305. Alabi S, Jaime‐Figueroa S, Yao Z, et al. Mutant‐selective degradation by BRAF‐targeting PROTACs. Nat Commun. 2021;12(1):920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Mares A, Miah AH, Smith IED, et al. Extended pharmacodynamic responses observed upon PROTAC‐mediated degradation of RIPK2. Commun Biol. 2020;3(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Chan KH, Zengerle M, Testa A, Ciulli A. Impact of target warhead and linkage vector on inducing protein degradation: comparison of bromodomain and extra‐terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I‐BET726) BET inhibitor scaffolds. J Med Chem. 2018;61(2):504‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Gadd MS, Testa A, Lucas X, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol. 2017;13(5):514‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Hughes SJ, Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure‐guided design of chemical degraders. Essays Biochem. 2017;61(5):505‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Lai AC, Toure M, Hellerschmied D, et al. Modular PROTAC design for the degradation of oncogenic BCR‐ABL. Angew Chem Int Ed Engl. 2016;55(2):807‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Nowak RP, DeAngelo SL, Buckley D, et al. Plasticity in binding confers selectivity in ligand‐induced protein degradation. Nat Chem Biol. 2018;14(7):706‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Steinebach C, Ng YLD, Sosič I, et al. Systematic exploration of different E3 ubiquitin ligases: an approach towards potent and selective CDK6 degraders. Chem Sci. 2020;11(13):3474‐3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Cao C, He M, Wang L, He Y, Rao Y. Chemistries of bifunctional PROTAC degraders. Chem Soc Rev. 2022;51(16):7066‐7114. [DOI] [PubMed] [Google Scholar]
- 314. Zhang S, Tischer T, Barford D. Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C. Nat Commun. 2019;10(1):3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APC(Cdh1) form of the anaphase‐promoting complex persists until S phase and prevents the premature expression of Cdc20p. J Cell Biol. 2001;154(1):85‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. Control of the SCFSkp2–Cks1 ubiquitin ligase by the APC/CCdh1 ubiquitin ligase. Nature. 2004;428(6979):190‐193. [DOI] [PubMed] [Google Scholar]
- 317. Lindon C, Grant R, Min M. Ubiquitin‐mediated degradation of aurora kinases. Front Oncol. 2015;5:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Li M, Zhang P. The function of APC/CCdh1 in cell cycle and beyond. Cell Div. 2009;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Busino L, Donzelli M, Chiesa M, et al. Degradation of Cdc25A by β‐TrCP during S phase and in response to DNA damage. Nature. 2003;426(6962):87‐91. [DOI] [PubMed] [Google Scholar]
- 320. Davidge B, Rebola KGO, Agbor LN, Sigmund CD, Singer JD. Cul3 regulates cyclin E1 protein abundance via a degron located within the N‐terminal region of cyclin E. J Cell Sci. 2019;132(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Lu A, SRJJoCB Pfeffer. Golgi‐associated RhoBTB3 targets cyclin E for ubiquitylation and promotes cell cycle progression. J Cell Biol. 2013;203(2):233‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Chaikovsky AC, Li C, Jeng EE, et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature. 2021;592(7856):794‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Alvarez SE, Harikumar KB, Hait NC, et al. Sphingosine‐1‐phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465(7301):1084‐1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Bertrand MJ, Lippens S, Staes A, et al. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1‐4). PLoS One. 2011;6(9):e22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Witt A, Vucic D. Diverse ubiquitin linkages regulate RIP kinases‐mediated inflammatory and cell death signaling. Cell Death Differ. 2017;24(7):1160‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Dainichi T, Matsumoto R, Mostafa A, Kabashima K. Immune control by TRAF6‐mediated pathways of epithelial cells in the EIME (epithelial immune microenvironment). Front Immunol. 2019;10:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Chang M, Jin W, Sun SC. Peli1 facilitates TRIF‐dependent Toll‐like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10(10):1089‐1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Moynagh PN. The roles of Pellino E3 ubiquitin ligases in immunity. Nat Rev Immunol. 2014;14(2):122‐131. [DOI] [PubMed] [Google Scholar]
- 329. Magiera MM, Mora S, Mojsa B, Robbins I, Lassot I, Desagher S. Trim17‐mediated ubiquitination and degradation of Mcl‐1 initiate apoptosis in neurons. Cell Death Differ. 2013;20(2):281‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Inuzuka H, Shaik S, Onoyama I, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Zhong Q, Gao W, Du F, Wang X. Mule/ARF‐BP1, a BH3‐only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl‐1 and regulates apoptosis. Cell. 2005;121(7):1085‐1095. [DOI] [PubMed] [Google Scholar]
- 332. Allan LA, Skowyra A, Rogers KI, Zeller D, Clarke PR. Atypical APC/C‐dependent degradation of Mcl‐1 provides an apoptotic timer during mitotic arrest. EMBO J. 2018;37(17):e96831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Berthelet J, Dubrez L. Regulation of apoptosis by inhibitors of apoptosis (IAPs). Cells. 2013;2(1):163‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14(3):400‐410. [DOI] [PubMed] [Google Scholar]
- 335. Wang X, Shu Y, Shi H, et al. TRIM9 is up‐regulated in human lung cancer and involved in cell proliferation and apoptosis. Int J Clin Exp Med. 2016;9(6):10461‐10469. [Google Scholar]
- 336. Andreeff M, Kelly KR, Yee K, et al. Results of the Phase I Trial of RG7112, a small‐molecule MDM2 antagonist in leukemia phase I trial of RG7112 in leukemia. Clin Cancer Res. 2016;22(4):868‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Yee K, Giovanni M, Vey N, et al. Phase 1/1b study of RG7388, a potent MDM2 antagonist, in acute myelogenous leukemia (AML) patients (Pts). Blood. 2014;124:116‐. [Google Scholar]
- 338. Rasco DW, Lakhani NJ, Li Y, et al. A phase I study of a novel MDM2 antagonist APG‐115 in patients with advanced solid tumors. American Society of Clinical Oncology; 2019. [Google Scholar]
- 339. Takahashi S, Fujiwara Y, Nakano K, et al. Safety and pharmacokinetics of milademetan, a MDM2 inhibitor, in Japanese patients with solid tumors: a phase I study. Cancer Sci. 2021;112(6):2361‐2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Sekiguchi N, Kasahara S, Miyamoto T, et al. Phase I dose‐escalation study of milademetan in patients with relapsed or refractory acute myeloid leukemia. 2022:1‐10. [DOI] [PMC free article] [PubMed]
- 341. Stein EM, DeAngelo DJ, Chromik J, et al. Results from a first‐in‐human phase I study of siremadlin (HDM201) in patients with advanced wild‐type TP53 solid tumors and acute leukemia. Clin Cancer Res. 2022;28(5):870‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. LoRusso P, Gounder M, Patel MR, et al. A Phase I dose‐escalation study of the MDM2‐p53 antagonist BI 907828 in patients with advanced solid tumors. 2021. [DOI] [PMC free article] [PubMed]
- 343. Ravandi F, Gojo I, Patnaik MM, et al. A phase I trial of the human double minute 2 inhibitor (MK‐8242) in patients with refractory/recurrent acute myelogenous leukemia (AML). Leuk Res. 2016;48:92‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Swords RT, Erba HP, DeAngelo DJ, et al. Pevonedistat (MLN4924), a first‐in‐class NEDD8‐activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br J Haematol. 2015;169(4):534‐543. [DOI] [PubMed] [Google Scholar]
- 345. Cai Q, Sun H, Peng Y, et al. A potent and orally active antagonist (SM‐406/AT‐406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J Med Chem. 2011;54(8):2714‐2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Flygare JA, Beresini M, Budha N, et al. Discovery of a potent small‐molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC‐0152). J Med Chem. 2012;55(9):4101‐4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Infante JR, Dees EC, Olszanski AJ, et al. Phase I dose‐escalation study of LCL161, an oral inhibitor of apoptosis proteins inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2014;32(28):3103‐3110. [DOI] [PubMed] [Google Scholar]
- 348. Bardia A, Parton M, Kümmel S, et al. Paclitaxel with inhibitor of apoptosis antagonist, LCL161, for localized triple‐negative breast cancer, prospectively stratified by gene signature in a biomarker‐driven neoadjuvant trial. J Clin Oncol. 2018;Jco2017748392 36(31):3126‐3133. [DOI] [PubMed] [Google Scholar]
- 349. Senzer NN, LoRusso P, Martin LP, et al. Phase II clinical activity and tolerability of the SMAC‐mimetic birinapant (TL32711) plus irinotecan in irinotecan‐relapsed/refractory metastatic colorectal cancer. American Society of Clinical Oncology; 2013. [Google Scholar]
- 350. Amaravadi RK, Schilder RJ, Martin LP, et al. A phase I study of the SMAC‐mimetic birinapant in adults with refractory solid tumors or lymphoma phase I trial of birinapant. Mol Cancer Ther. 2015;14(11):2569‐2575. [DOI] [PubMed] [Google Scholar]
- 351. Su W, Han HH, Wang Y, et al. The polycomb repressor complex 1 drives double‐negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell. 2019;36(2):139‐55.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Shukla S, Ying W, Gray F, et al. Small‐molecule inhibitors targeting Polycomb repressive complex 1 RING domain. Nat Chem Biol. 2021;17(7):784‐793. [DOI] [PMC free article] [PubMed] [Google Scholar]


