Abstract
T/EBP/NKX2.1, a member of the NKX family of homeodomain-containing transcription factors, regulates the expression of a number of genes in lung and thyroid. Here we describe the isolation and characterization of a novel target gene, termed claudin-18, that is down-regulated in the lungs of T/ebp/Nkx2.1-null mouse embryos. The gene product exhibits an amino acid sequence similar to those of the claudin multigene family of proteins that constitute tight junction strands in epithelial cells. The gene was localized by fluorescence in situ hybridization to mouse chromosome 9 at region 9E3-F1 and to human chromosome 3 at region 3q21–23. The claudin-18 gene has two promoters, each with its own unique exon 1 that is spliced to common exons 2 through 5. Alternative usage of these promoters leads to production of lung and stomach-specific transcripts. The downstream lung-specific promoter contains two T/EBP/NKX2.1 binding sites responsible for trans activation of the gene by T/EBP/NKX2.1 in lung cells. Only claudin-18 was down-regulated in T/ebp/Nkx2.1-null embryo lungs among 11 claudin transcripts examined. Furthermore, the claudin-18 transcript has an alternative 12-bp insertion derived from the 5′ end of intron 4, which produces a C-terminally truncated isoform in lung and stomach. Immunohistochemistry demonstrated complete membrane localization of claudin-18 with small focal dots in the lung and stomach epithelial cells. Immunogold electron microscopy analysis revealed that claudin-18 is concentrated at the cell-cell borders of epithelial cells. These unique features suggest a potentially important role for claudin-18 in the structure and function of tight junctions in lung and stomach.
T/EBP/NKX2.1, also known as TTF-1, is a homeodomain-containing transcription factor that is expressed in lung, thyroid, and a part of the brain (15, 21, 22, 28). T/EBP/NKX2.1 was originally characterized as a transcription factor that regulates expression of thyroid-specific genes such as those encoding thyroglobulin (7), thyroid peroxidase (7, 11, 20), TSH receptor (6, 37), and the Na/I symporter (10). In the lung, T/EBP/NKX2.1 is expressed in all epithelial cells early during pulmonary morphogenesis, but the expression becomes progressively restricted to alveolar type II and Clara cells towards the end of gestation and in postnatal days (46). T/EBP/NKX2.1 activates transcription of genes specifically expressed in lung, including the genes for surfactant proteins A (2), B (1), and C (19) and Clara cell secretory protein (also called uteroglobin) genes (34, 35). Targeted disruption of the T/ebp/Nkx2.1 locus was shown to result in immediate postnatal death due to respiratory failure caused by profoundly hypoplastic lungs (21, 26). These mice also lack the thyroid, pituitary, and parts of the ventral forebrain such as the hypothalamus and basal ganglia (21, 41, 42). Detailed analyses of the T/ebp/Nkx2.1-null mouse respiratory system revealed that T/EBP/NKX2.1 may function in the establishment of pattern formation of early pulmonary structure and pulmonary morphogenesis during embryonic development (26). A role for T/EBP/NKX2.1 in pulmonary morphogenesis was also suggested by in vitro experiments in which a T/EBP/NKX2.1 antisense oligonucleotide inhibited normal branching morphogenesis in lung organ culture (25). Based on these results, it is hypothesized that lung branching morphogenesis must be related to the ability of T/EBP/NKX2.1 to activate and/or suppress specific downstream target genes. One such category of target genes in lung consists of the surfactant proteins A (2), B (1), and C (19) and Clara cell secretory protein genes (34, 35). However, they are not known to have morphoregulatory function.
Extensive studies have shown that epithelial-mesenchymal interaction plays an instructive role in lung branching morphogenesis (3, 17, 24, 45). In this context, a possible function for T/EBP/NKX2.1 may be in activation of epithelial cell pathways that are necessary for receiving and/or interpreting the instructive signals that originate from the mesenchyme. In this role, downstream target genes for T/EBP/NKX2.1 would potentially include those encoding cell surface receptors, components of the signal transduction pathway, and/or a variety of other factors connecting the cell surface to changes in gene expression and cellular behavior. In T/ebp/Nkx2.1-null embryo lungs, expression of some extracellular matrix proteins and their cellular receptors, including collagen type IV and α integrins, and some growth factors such as VEGF3 and BMP4 is reduced or absent (46). Whether the abnormal phenotype in T/ebp/Nkx2.1-null embryo lungs is entirely or partially due to the reduction or absence of expression of these genes remains to be examined.
Among possible target genes are those encoding elements of tight junctions (TJs), a specialized membrane domain at the most apical region of polarized epithelial cells that creates a primary barrier to prevent paracellular transport of solutes and restrict the lateral diffusion of membrane lipids and proteins to maintain cellular polarity (4, 5, 23, 27, 40, 44). Claudins, products of a recently identified multigene family, are components of TJ strands and have four transmembrane domains and two extracellular loops with both NH2 and COOH termini in the cytoplasm (29, 43). Claudins have several functional characteristics consistent with a role in barrier formation and dependent on the specific claudin species that exhibit tissue specificity (27, 29). Claudin-1 and -2 have the ability to induce the formation of networks of strands and grooves at cell-cell contact sites when introduced into fibroblasts lacking TJs (12). Both claudin-3 and -4 are receptors for a cytotoxic enterotoxin (CPE) produced by the bacterium Clostridium perfringens, and the interaction with CPE results in increased membrane permeability by forming small pores in plasma membrane (39). A claudin-11 (oligodendrocyte-specific protein) knockout mouse showed the absence of TJ strands in central nervous system myelin and Sertoli cells in testis (14). Mutations in human claudin-16 (paracellin-1) cause renal hypomagnesemia with hypercalciuria and nephrocalcinosis, suggesting that it creates a channel that allows magnesium to diffuse through renal TJs (38). To date, at least 20 members of the claudin gene family have been identified (27). However, many of these claudins have not yet been examined in detail, and the functional differences are largely unknown.
Here, we isolated and characterized a novel member of claudin gene family, claudin-18, which has two isoform transcripts produced by alternative splicing that exhibit lung- and stomach-specific expression. Further, claudin-18 has a splicing variant lacking the C-terminal cytoplasmic domain. Analysis of the promoter function of the mouse claudin-18 gene suggests that the lung-specific form is a downstream target gene regulated by the T/EBP/NKX2.1 homeodomain transcription factor.
MATERIALS AND METHODS
Identification of claudin-18 by SSH.
T/ebp/Nkx2.1+/− mice were bred to generate T/ebp/Nkx2.1-null embryos (21). The embryos were obtained by dissection of pregnant mice at E16.5, with noon on the day when the vaginal plug was detected designated stage E0.5. The embryos were individually microdissected to isolate lungs, and genotyping was performed by PCR using yolk sacs. Total RNA, isolated from lungs of null mutant (driver) and wild-type (tester) embryos using the ULTRASPEC RNA isolation system (Biotecx Laboratories, Houston, Tex.), was used as a template to synthesize double-stranded cDNAs using the SMART PCR cDNA synthesis kit (Clontech Laboratories, Palo Alto, Calif.). Suppressive subtractive hybridization (SSH) was performed using the PCR-Select cDNA subtraction kit (Clontech) according to the manufacturer's instructions. For differential screening, subcloned cDNAs were isolated by colony PCR amplification, and dot blot hybridization was performed using forward- and reverse-subtracted cDNAs as probes with the PCR-Select differential screening kit (Clontech). Clones that hybridized with only the forward-subtracted probe were selected for virtual Northern blot analyses as follows. Double-stranded cDNAs synthesized with the SMART PCR cDNA synthesis kit were electrophoresed on a 0.8% agarose gel and transferred onto a GeneScreen Plus nylon membrane (NEN Life Science Products, Boston, Mass.). The membrane was prehybridized at 60°C in ExpressHyb hybridization solution (Clontech) for 30 min and hybridized in fresh buffer with denatured random-primer-labeled probe at 60°C for 3 h. After hybridization, the blot was washed twice in 2× SSC (2× SSC is 0.3 M NaCl and 30 mM Na citrate, pH 7.0) containing 0.1% sodium dodecyl sulfate (SDS) at room temperature for 10 min, followed by 0.1× SSC containing 0.1% SDS at 50°C for 20 min. The filter was then exposed to a PhosphorImager screen overnight. Signal intensities were analyzed using the ImageQuant program (Molecular Dynamics, Inc., Sunnyvale, Calif.). Differentially expressed clones were subjected to DNA sequencing analysis.
Cloning and DNA sequencing.
An adult mouse lung cDNA library in the λZAPII vector (Stratagene, La Jolla, Calif.) was screened by plaque hybridization using cDNAs isolated from SSH as probes. Hybridization was carried out at 65°C in 6× SSC–0.5% SDS–5× Denhardt solution–0.1 mg of denatured salmon sperm DNA per ml for 16 h. Positive plaques were subjected to secondary and tertiary screenings. In order to amplify a missing 3′ sequence, we performed 3′ rapid amplification of the cDNA ends (RACE) with the SMART RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions using 2 μg of adult lung total RNA.
The amino acid sequence of mouse claudin-18A1.1 was used to conduct TBLASTN searches in order to isolate a human ortholog of claudin-18. One human genomic DNA clone (GenBank database accession number AC010810) that exhibited sequence homology to mouse claudin-18 was found. Human claudin-18A1∗1 cDNA was amplified by reverse transcription-PCR (RT-PCR) using human adult lung RNA as a template and the primer pair, 5′-TGGGTGCCATTGGCCTCCTG-3′ and 5′-TGGAAGGATAAGATTGTACC-3′. Human claudin-18A2∗1 cDNA was amplified by RT-PCR using human adult stomach RNA as a template and the primer pair, 5′-TGTGCGCCACCATGGCCGTG-3′ and 5′-GTGCTGAGAGGTCTTAGAGC-3′. Three independent clones of amplified cDNAs from each RT-PCR were subjected to DNA sequencing analysis.
Mouse claudin-18a2 was initially identified by 5′ RACE with the Smart RACE cDNA amplification kit (Clontech) using 2 μg of mouse adult stomach total RNA. In order to isolate mouse claudin-18a2∗1 cDNA containing an entire open reading frame, PCR was performed using mouse adult stomach RNA as a template and the primer pair 5′-GCCTGTCTCTTGTCCTCTCC-3′ and 5′-CAGGTTGGCGGGTCTTAGAGC-3′.
Genomic DNA was isolated from a mouse embryonic stem cell lambda genomic library (Stratagene) and a mouse BAC DNA library (Incyte Genomics, St. Louis, Mo.) using labeled cDNAs as probe. Genomic DNA fragments containing the mouse claudin-18 gene were digested with restriction enzymes, subcloned into pBluescript II (Stratagene), and sequenced. All DNA sequencing was carried out using an ABI prism dye terminator cycle sequencing Ready Reaction kit and a model 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.).
Determination of the transcription start site.
The transcription start sites of the mouse claudin-18a1 and -18a2 transcripts were determined using the SMART RACE cDNA amplification kit (Clontech). Briefly, first-strand cDNA was synthesized from 2 μg of adult lung or stomach total RNAs using 5′ cDNA synthesis primer, SMART II oligonucleotide, and Superscript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, Calif.). After the first-strand cDNA synthesis, PCR was performed with a gene-specific antisense primer (5′-GTCAGAGTCATCTTGGCCTTGGCAG-3′) and a universal primer mix. Sequencing of the resultant cloned PCR products indicated the presence of multiple transcription start sites. Since 5 out of 12 cDNAs for lung and 10 out of 14 cDNAs for stomach had the exact sequence, we designated this site the major transcription start site.
Chromosomal mapping.
A human claudin-18 probe of ∼100 kb (whole BAC clone sequence) and a mouse claudin-18 probe of ∼20 kb (whole phage clone sequence) of genomic DNA labeled with biotin or digoxigenin was used for fluorescence in situ hybridization of chromosomes derived from methotrexate-synchronized normal peripheral lymphocytes and from mouse spleen cultures, respectively. The conditions of hybridization; the detection of fluorescence signals; digital-image acquisition, processing, and analysis; and direct localization of signals on banded chromosomes were carried out as previously described (32, 47). To confirm the identity of mouse chromosomes, preparations were rehybridized with mouse chromosome painting probe (Cambio) and previously observed labeled metaphases were recorded.
RT-PCR analyses.
For RT of mRNAs, 2 μg of total RNA was pretreated with DNase I, incubated for 10 min at 70°C, and chilled on ice. The reactions were carried out in a final volume of 20 μl containing RNA, 4 μl of 5× first strand synthesis buffer (Invitrogen Life Technologies), 1 μl of a mixture of four deoxynucleotide triphosphates (2.5 mM each), 2 μl of 0.1 M dithiothreitol (DTT), and 100 ng of random primers. After incubation at 37°C for 2 min, 200 U of Superscript II reverse transcriptase (Invitrogen Life Technologies) was added, and the incubation was continued for 60 min at 37°C. Single-stranded cDNAs in 0.1 μl of the reaction mixture were amplified by PCR using AmpliTaq DNA polymerase (PE Applied Biosystems) under the following conditions: denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min for 35 cycles. Oligonucleotide primers used for RT-PCR were as follows: mouse claudin-1, 5′-CTGGGTTTCATCCTGGCTTC-3′ and 5′-TTGATGGGGGTCAAGGGGTC-3′; mouse claudin-3, 5′-AAAGAATTCTGCTCCTGCCCACCGCGCGAC-3′ and 5′-AAACTCGAGAAGTAGCTGCAGTGGCCACC-3′; mouse claudin-4, 5′-TAGGGGGCAAGTGCACCAAC-3′ and 5′-CCCCAGCAAGCAGTTAGTGG-3′; mouse claudin-6, 5′-ATCTTGGGGATCGTCCTGAC-3′ and 5′-TTTGAGCATCAGCCACCAAG-3′; mouse claudin-7, 5′-TCCCTCAGTGGCAGATGAGC-3′ and 5′-AAGGACCAGAGCAGACCCTG-3′; mouse claudin-8, 5′-GCCTCAGTGGAGAGTGTCTG-3′ and 5′-AGAACAGTGCTCCTCCAGCG-3′; mouse claudin-10, 5′-ACACTGCCCACCGACTACTG-3′ and 5′-AGAGAAGCTCCTGCCCATCC-3′; mouse claudin-12, 5′-TAACTGGAGGAAACTGCGGC-3′ and 5′-CCCCTGAGCTAGCAATAGTG-3′; mouse claudin-13, 5′-TGCCATTGTGAGCTGCGTGC-3′ and 5′-CGGGGAAAGTCTCTGCATAC-3′; mouse claudin-15, 5′-ATGTCGGTAGCTGTGGAGAC-3′ and 5′-TCCCTGCAATGGCCAGCAGC-3′; and mouse claudin-18, P1 (5′-ACCTTCCCAGCAAGAGGGTG-3′), P2 (5′-TGATTGCACAGATGCCGGAG-3′), P3 (5′-GTCTGTGTTTGCCAACATGC-3′), P4 (5′-AGACACAGCTTTGAAGTTGC-3′), and P5 (5′-CTTTGAAGGGGAGGACTCAC-3′) (for details, see Fig. 1).
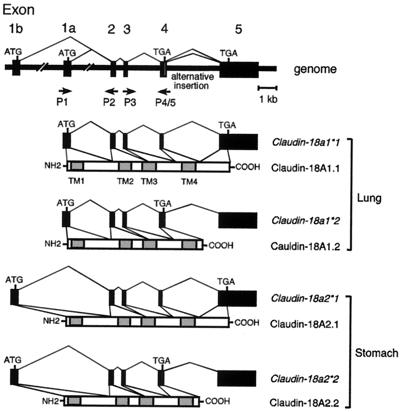
FIG. 1.
Schematic representation of four types of mouse claudin-18 transcripts and polypeptides. Organization of exons and introns and predictions of how the four types of transcripts are produced are shown. Solid boxes represent exons. The positions of the translation initiation and termination codons defining open reading frames are indicated. The alternatively inserted 12-bp sequence after exon 4 is depicted by an open box. Arrows indicate the positions of primers used for RT-PCR analysis (P1 to P5). Shaded boxes represent four transmembrane domains (TM1 to TM4). The nomenclature for the genes and their respective polypeptides is shown on the right for lung and stomach forms.
Northern blot analysis.
Total RNAs (10 μg) from adult mouse lung and stomach were electrophoresed on a 1.2% agarose gel containing 0.22 M formaldehyde and blotted onto GeneScreen Plus nylon membranes. Filters were serially hybridized with mouse claudin-18 exon 1a and exon 1b and β-actin as a probe. A mouse multiple-tissue Northern blot (Clontech) was hybridized with a full-length mouse claudin-18a1∗1 cDNA as a probe. A human multiple-tissue expression array (MTE; Clontech) was hybridized with a full-length human claudin-18A1∗1 cDNA as a probe. Hybridization was performed in ExpressHyb hybridization solution at 68°C for 2 h. The membrane was washed twice with 2× SSC containing 0.1% SDS at room temperature for 10 min and twice with 0.1× SSC containing 0.1% SDS at 55°C for 20 min, followed by exposure to X-ray film at −80°C.
Luciferase reporter plasmid construction and site-directed mutagenesis.
A 4-kb EcoRI fragment containing 3.5 kb of 5′ flanking sequences of the mouse claudin-18a1 genomic DNA was subcloned into the EcoRI site of pBluescript II, followed by PCR amplification with a T7 primer (5′-GTAATACGACTCACTATAGGGC-3′) and a claudin-18a1 gene-specific primer (5′-GGGCGCAGGCTTCCAGGAGC-3′). The PCR product was subcloned into pCR2.1 (Invitrogen Life Technologies), and an SpeI-XhoI fragment from this plasmid was inserted into the NheI-XhoI site of the pGL3-Basic luciferase reporter vector (Promega, Madison, Wis.) to generate plasmid pGL3-3500. This construct was further digested with KpnI and MluI for construction of deletion plasmids using exonuclease III (New England Biolabs, Beverly, Mass.) and S1 nuclease (Invitrogen Life Technologies). Five deletion constructs (pGL3-70, -111, -148, -212, and -259) were sequenced to determine the exact sequences.
Site-directed mutations of two potential T/EBP/NKX2.1 binding sites were introduced into the pGL3-259 plasmid by using a QuikChange site-directed mutagenesis kit (Stratagene). The following primers were used to make the plasmids (mutations are in boldface): pGL3-259 mut 1 plasmid, 5′-GCTCAACCAGACCCGTGCCTCTTGGGGC-3′ and 5′-GCCCCAAGAGGCACGGGTCTGGTTGAGC-3′ (complementary strand); pGL3-259 mut 2 plasmid, 5′-GACAAGTGCCTCGGGGGGCTGCTTCCCC-3′ and 5′-GGGGAAGCAGCCCCCCGAGGCACTTGTC-3′ (complementary strand); and pGL3-259 mut 3 plasmid, 5′-CAAATGAAAACCATGCCAGTGAAGCAGGAGCGAAACTG-3′ and 5′-CAGTTTTCGCTCCTGCTTCACTGGCATGGTTTTCATTTG-3′ (complementary strand).
Transfection and reporter gene assays.
A human lung adenocarcinoma cell line, NCI-H441, was maintained in RPMI 1640 medium containing 10% fetal calf serum. HeLa cells were cultured in minimum essential medium containing 10% fetal calf serum. Cells in 12-well plates at 50 to 70% confluency were transfected by using the Effectene transfection reagent (Qiagen, Valencia, Calif.) with 250 ng of reporter plasmid, 25 ng of expression vector, and 25 ng of pCH110 (Amersham Pharmacia Biotech) as an internal control. For dose-dependent reporter activity assays, increasing amounts of pCMV4-T/EBP/NKX2.1 or pCMV4 (0 to 1 μg) were transfected into NCI-H441 cells together with 0.2 μg of pGL3-259 and 25 ng of pCH110. After 48 h, the cells were harvested in reporter lysis buffer (Promega), and the lysates were assayed for β-galactosidase and luciferase activities using the high-sensitivity β-galactosidase assay kit (Stratagene) and luciferase assay system (Promega), respectively. To correct for transfection efficiency, luciferase activity was normalized to β-galactosidase activity. Relative luciferase activities of various mouse claudin-18a1 promoter constructs were expressed on the basis of the activity of pGL3-Basic in the presence of the same trans-activating plasmid, which was given a value of 1. Dose-dependent luciferase activities were expressed on the basis of the activity of pGL3-259 without any cotransfected vector, which was given a value of 1. Data are expressed as mean values from at least three experiments (duplicate samples) ± standard deviations (SD). P values were obtained using the Student t test.
Preparation of nuclear extracts.
Nuclear extracts of NCI-H441 cells were prepared as described previously (9). All procedures were performed on ice and in ice-cold reagents. Briefly, confluent cell monolayers in 10 15-cm-diameter dishes were washed twice with phosphate-buffered saline (PBS), harvested by scraping in PBS, and centrifuged at 500 × g for 5 min. The pellets were washed once with PBS and repelleted as before. The cell pellets were suspended in 5 packed-cell volumes of lysis buffer (10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 2 μg of pepstatin A per μl, and 2 μg of leupeptin per μl, and the cells were lysed on ice for 10 min. The cell lysates were then centrifuged at 500 × g for 5 min, and the pellet was suspended in 3 packed-cell volumes of lysis buffer, followed by homogenization in a Dounce homogenizer with pestle B (Wheaton, Millville, N.J.). The homogenate was centrifuged at 500 × g for 5 min, and the nuclear pellet was suspended in 0.5 ml of extraction buffer (50 mM HEPES-KOH [pH 7.9], 420 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 20% (vol/vol) glycerol, 1 mM DTT, 0.5 mM PMSF, 2 μg of pepstatin A per μl, and 2 μg of leupeptin per μl). The nuclear suspension was rotated for 30 min and was centrifuged at 24,000 × g for 30 min. The supernatant recovered was saved as nuclear extracts and stored at −80°C until use.
Electrophoretic mobility shift assays.
Single-stranded oligonucleotides were annealed at a concentration of 10 μM in annealing buffer (1 mM Tris [pH 7.5], 1 mM MgCl2, and 5 mM NaCl) at 95°C for 5 min and then slowly cooled to room temperature. Double-stranded DNA was end labeled with [α-32P]dCTP and DNA polymerase Klenow fragment (Invitrogen Life Technologies). Labeled DNA was separated from free [α-32P]dCTP by filtration through a ProbeQuant G-50 Micro Column (Amersham Pharmacia Biotech Inc.).
Nuclear extracts (15 μg) and, when indicated, unlabeled oligonucleotide competitor DNAs were preincubated in 23 μl of gel mobility shift assay buffer [10 mM HEPES-KOH (pH 7.9), 50 mM KCl, 0.6 mM EDTA, 5mM MgCl2, 10% glycerol, 5 mM DTT, 0.7 mM PMSF, 2 μg of pepstatin A per μl, 2 μg of leupeptin per μl, and 87 μg of poly(dI-dC) (Amersham Pharmacia Biotech) per μl] for 10 min on ice. An oligonucleotide probe (105 cpm) was added to the mixture and incubated for an additional 30 min at room temperature. For antibody supershift analyses, 1 μl of anti-TTF1 monoclonal antibody (Lab Vision Corporation, Fremont, Calif.) was added, and the incubation was continued for an additional 1 h. DNA-protein complexes were separated from free probe by 5% nondenaturing polyacrylamide gel electrophoresis. After electrophoresis, the gel was blotted onto Whatman no. 3MM paper, dried, and exposed to X-ray film.
Immunohistochemistry.
A polypeptide, DGGARTEDDEQSHPTKYDYV, corresponding to the COOH-terminal cytoplasmic domain of mouse claudin-18A1.1 and -18A2.1 (amino acids 245 to 264) was synthesized and coupled via cysteine to keyhole limpet hemocyanin. The conjugated peptide was used as an antigen to generate a polyclonal antibody in rabbits (Macromolecular Resources, Fort Collins, Colo.). The antibody specificity was examined by Western blotting using bacterially expressed mouse claudin-1, which shares the last three amino acid residues with claudin-18. It was reported that antibody against claudin-5 cross-reacts with claudin-6, which share the last four amino acid residues (31). Immunohistochemistry was carried out using an 800-fold dilution of claudin-18 antibody and a Vectastain ABC Rabbit Elite Kit (Vector Laboratories, Burlingame, Calif.).
Postembedding IEM.
Details of tissue sample preparation for postembedding immunogold electron microscopy (IEM) were described previously (16). Briefly, mouse embryonic tissue was fixed in 4% paraformaldehyde (Tousimis, Rockville, Md.) in PBS for 2 h at 4°C and dehydrated through graded cold ethanol. Infiltration was made in 1:1 and 1:2 mixtures of 100% ethanol and L.R. Gold resin (Polysciences, Warrington, Pa.) for 1 h each and in pure resin overnight. The tissue was embedded in L.R. Gold resin containing initiator (0.1% [wt/vol] benzil [Ladd Research Industries, Burlington, Vt.]), and the resin was cured in a −20°C UV Cryo Chamber (Ted Pella, Redding, Calif.) for 24 h. Thin sections were cut and mounted on meshed nickel grids. Nonspecific binding sites were blocked by incubation of the sections in PBS containing 10% normal goat serum, 0.1% bovine serum albumin, and 0.01% Tween 20. Primary incubation was done with dilutions of anti-claudin-18 antibody followed by 15-nm-diameter-immunogold (Amersham Life Science, Arlington Heights, Ill.)-conjugated secondary antibody (1:100 dilution). The sections were stained in uranyl acetate and lead citrate and examined and photographed with an H7000 electron microscope (Hitachi, Tokyo, Japan) operated at 75 kV.
Nucleotide sequence accession numbers.
The nucleotide sequences are available from the GenBank nucleotide sequence database under the following accession numbers: mouse claudin-18a1∗1 cDNA, AF221068; mouse claudin-18a1∗2 cDNA, AF349450; mouse claudin-18a2∗1 cDNA, AF349451; mouse claudin-18a2∗2 cDNA, AF349453; human claudin-18A1∗1 cDNA, AF221069, and human claudin-18A2∗1 cDNA, AF349452.
RESULTS
Isolation of T/ebp/Nkx2.1 downstream target gene.
To isolate putative T/EBP/NKX2.1 downstream target genes, an SSH method was used to generate a cDNA library of clones enriched for E16.5 wild-type versus T/ebp/Nkx2.1-null embryos. The latter lungs are severely hypoplastic and appear to consist solely of proximal pulmonary structures (46). One hundred ninety-two clones were initially picked and probed with forward-subtracted cDNAs (wild type) or reverse-subtracted cDNAs (mutant). Twenty-seven clones that gave strong signals with the forward-subtracted probe compared to the reverse probe were subjected to Northern blotting analyses. Five clones were found to be differentially expressed. Sequence analyses revealed that two of them encode a polypeptide exhibiting sequence similarity to the claudin family of proteins (29). The predicted polypeptide sequence of the cDNA is 264 amino acids in length and possesses four transmembrane domains, typical of all members of the claudin family (29, 44). We refer to this gene as claudin-18 (Fig. 1). At present, 20 members are registered in the GenBank database (27). The gene nomenclature that we used (see below) is in agreement with the recommendation of the nomenclature committee.
Structure of claudin-18 gene and encoded polypeptides.
To isolate a full-length claudin-18 cDNA, a mouse adult lung cDNA library was screened using the cDNA clone obtained by SHH. Ten clones were identified in 106 recombinant phage. Among them, one clone possessed an in-frame stop codon upstream of the authentic stop codon by an alternative 12-bp nucleotide insertion (see below) (Fig. 2A).
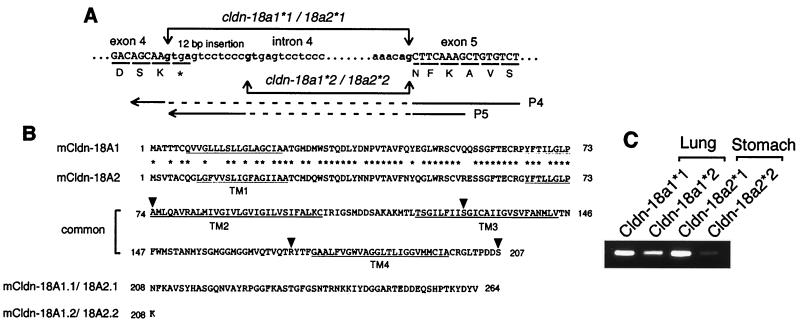
FIG. 2.
(A) Alternative splicing of the mouse claudin-18 gene between exons 4 and 5. The intron 4,5′-splice donor gt, and 3′-splice acceptor ag are in boldface. Amino acid residues are shown below each underlined codon, and the termination codon is marked with an asterisk. The claudin (cldn)-18a1∗1/18a2∗1 gene encodes the amino acid sequence DSNFK at the exon-intron junction, whereas the claudin-18a1∗2/18a2∗2 gene encodes the amino acid sequence DSK. The P4 primer has a continuous sequence of exons 4 and 5 without alternative insertion, and P5 is an alternative insertion-specific primer. The primer sequences are indicated by solid lines. Primer pairs P3-P4 and P3-P5 (Fig. 1) produce fragments specific to claudin-18a1∗1/2∗1 and claudin-18a1∗2/2∗2, respectively. (B) Deduced amino acid sequences of four types of claudin-18 polypeptides. The locations of the putative transmembrane domains are underlined and indicated as TM1 to TM4. Amino acids that are identical in claudin-18A1 and -18A2 are indicated by asterisks. The exon-intron junctions are indicated by arrowheads. (C) RT-PCR confirmation of expression of the four types of transcripts. Products obtained with primer pairs P3-P4 and P3-P5 using mouse adult lung and stomach mRNAs are shown.
In order to analyze the claudin-18 gene structure and to define the origin of the truncated form of cDNA, a mouse genomic library was screened using a full-length cDNA as a probe. The mouse claudin-18 gene is composed of five exons and four introns (Fig. 1). All exon-intron boundaries matched the consensus sequence for RNA splicing. Interestingly, the first 12 bp in intron 4 can be alternatively retained, which appears to be responsible for the truncated form of the transcript (Fig. 2A). Since in-frame stop codon TGA is present within the 12-bp sequence, the resulting polypeptide lacks almost the entire C-terminal cytoplasmic domain, including possible ZO-1/ZO-2/ZO-3 binding sequences (Fig. 2B) (see Discussion) (18).
The amino acid sequence of mouse claudin-18 was used to conduct TBLASTN searches in order to isolate a human ortholog of claudin-18. One human genomic DNA clone (GenBank database accession number AC010810) that exhibits sequence homology to mouse claudin-18 was identified. The sequence of this clone was used to isolate human claudin-18 cDNA by RT-PCR using human lung RNAs. The human claudin-18 demonstrated 88% amino acid sequence identity to mouse claudin-18. The presence of a truncated version of the human claudin-18 was not confirmed.
While searching for the human claudin-18 ortholog gene, we found another first exon 11 kb upstream of exon 1. To determine whether this exon is actually transcribed, RT-PCR was performed with exon-specific primers using human lung and stomach RNAs as templates, since strong signals were observed in human lung and stomach by dot blot analysis (see Fig. 4B). The results revealed that the upstream exon 1 (exon 1b) is stomach specific and that the downstream exon 1 (exon 1a) is lung specific. In order to examine whether stomach-specific exon 1b is also present in mouse, 5′ RACE was carried out using mouse stomach RNA. Some of the clones isolated had a sequence at their 5′ ends that shared homology with human exon 1b but not with mouse exon 1a. It appears that each exon 1 is spliced to common exons 2 through 5, resulting in two transcripts sharing identical C-terminal domains (Fig. 1). PCR analysis using mouse genomic DNA as a template revealed that exon 1b is 6 kb upstream of exon 1a. The predicted amino acid sequences of exons 1a and 1b share 73% sequence identity (Fig. 2B). Thus, the mouse claudin-18 gene has two alternately used exon 1s, each producing lung- and stomach-specific isoforms (see below). In addition, a C-terminally truncated form of the transcript was also found in stomach, indicating that the mouse claudin-18 gene produces four types of transcripts (Fig. 1). PCRs using mouse lung and stomach RNAs confirmed the presence of four types of transcripts, although the stomach-specific truncated form of mRNA (claudin-18a2∗2) appears to be underrepresented (Fig. 2C).
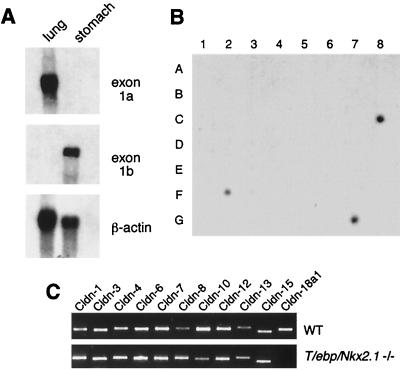
FIG. 4.
Expression of claudin-18 transcripts. (A) Northern blot analysis using exon-specific probes in adult mouse lung and stomach. Ten micrograms of total RNA from the indicated tissues was consecutively hybridized with each probe shown on the right. (B) Dot blot analysis of claudin-18 mRNA in human tissues. RNA sources are as follows: A1, whole brain; A2, amygdala; A3, caudate nucleus; A4, cerebellum; A5, cerebral cortex; A6, frontal lobe; A7, hippocampus; A8, medulla oblongata; B1, occipital lobe; B2, putamen; B3, substantia nigra; B4, temporal lobe; B5, thalamus; B6, subthalamic nucleus; B7, spinal cord; C1, heart; C2, aorta; C3, skeletal muscle; C4, colon; C5, bladder; C6, uterus; C7, prostate; C8, stomach; D1, testis; D2, ovary; D3, pancreas; D4, pituitary gland; D5, adrenal gland; D6, thyroid gland; D7, salivary gland; D8, mammary gland; E1, kidney; E2, liver; E3, small intestine; E4, spleen; E5, thymus; E6, peripheral leukocyte; E7, lymph node; E8, bone marrow; F1, appendix; F2, lung; F3, trachea; F4, placenta; G1, fetal brain; G2, fetal heart; G3, fetal kidney; G4, fetal liver; G5, fetal spleen; G6, fetal thymus; and G7, fetal lung. (C) Differential expression of claudin mRNAs in wild-type and T/ebp/Nkx2.1-null embryo lungs. Total RNAs prepared from E12.5 embryonic lungs of wild-type (WT) or T/ebp/Nkx2.1-null mutant (−/−) mice were analyzed by RT-PCR using various claudin primers as indicated. Primer pair P1-P2 (shown in Fig. 1) was used for claudin-18a1.
In summary, the mouse lung-specific claudin-18a1∗1 gene encodes a protein of 264 amino acids (termed claudin-18A1.1) with a calculated molecular mass of 28,120 kDa, and its C-terminally truncated type claudin-18a1∗2 gene encodes a protein of 208 amino acids (claudin-18A1.2) with a calculated molecular mass of 21,977 kDa. The stomach-specific claudin-18a2∗1 gene encodes a protein of 264 amino acids (claudin-18A2.1) with a calculated molecular mass of 28,142 kDa, and its C-terminally truncated type claudin-18a2∗2 gene encodes a protein of 208 amino acids (claudin-18A2.2) with a calculated molecular mass of 21,999 kDa.
Chromosomal localization of claudin-18 gene.
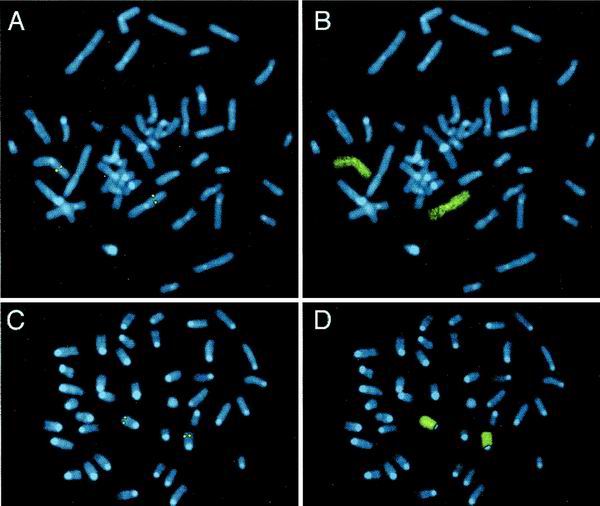
In two fluorescence in situ hybridization experiments with human lymphocytes from different individuals, specific fluorescent signals at identical sites on both chromatids of the long arm of chromosome 3 were detected in approximately 90% of the 25 metaphases randomly selected in each sample. Double fluorescent signals were not observed at any other chromosome, and nonspecific brighter fluorescent spots were sporadic. The locations of the signals were determined directly in 20 metaphases with DAPI (4′,6′-diamidino-2-phenylindole)-enhanced G-like banding at region 3q21–23, where we assign the locus of the human claudin-18 gene (Fig. 3). Mouse chromosome spreads hybridized with a digoxigenin-labeled genomic DNA probe had specific fluorescent signals at identical sites on both homologs of a medium-size chromosome in 40 out of 50 metaphases randomly selected for analysis. Chromosomes of similar size with double symmetrical fluorescent signals were not observed, while single fluorescent spots were randomly distributed. Twenty complete metaphases without overlapping chromosomes were analyzed by imaging of DAPI-enhanced G-like banding. The banding pattern generated by DAPI banding was consistent with mouse chromosome 9, and the signal localized at region 9E3-F1, a syntenic region for human 3q21-23 (8, 36).
FIG. 3.
Chromosomal localization. Human (A) and mouse (C) chromosome spreads after hybridization with species-specific biotin-labeled claudin-18 gene probes are shown. Both chromosomes 3 in human and chromosomes 9 in mouse exhibit symmetrical fluorescent signals at regions 3q21-23 and 9E3-F1, respectively, as depicted by enhanced DAPI-induced chromosome banding. To confirm the identity of chromosomes with specific signals, metaphases from panels A and C were rehybridized with human chromosome painting probe for human chromosome 3 (B) and mouse chromosome 9 (D).
Expression of claudin-18.
Claudin-18 expression was examined in adult mouse tissues by Northern blot analysis using the full coding region of the claudin-18a1∗1 cDNA as a probe. A single 2.8-kb transcript was detected only in the lung among the tissues examined, including heart, brain, spleen, skeletal muscle, liver, kidney, and testis (data not shown). Next, the expression of claudin-18 in mouse lung and stomach was analyzed by Northern analysis using exon 1a- and 1b-specific probes (Fig. 4A). The exon 1a-specific probe identified a band only in lung, whereas the exon 1b-specific probe detected a band only in stomach. RNA dot blot analysis of various human tissues indicated that human claudin-18 is expressed only in lung and stomach (Fig. 4B). These results indicate that both mouse and human claudin-18 genes produce lung-and stomach-specific transcripts.
Claudin-18a1 expression was examined along with that of other claudin family members, using RT-PCR and E12.5 embryonic lung RNAs derived from wild-type and T/ebp/Nkx2.1-null embryos (Fig. 4C). In wild-type lungs, at least 10 other claudin family members in addition to claudin-18a1 were expressed. In contrast, claudin-18a1 was not expressed in T/ebp/Nkx2.1-null lungs, while the expression of all other claudins was detectable, raising the possibility that claudin-18 is the only member of the claudin family of genes that is regulated by T/EBP/NKX2.1.
claudin-18a1 promoter is trans activated by T/EBP/NKX2.1.
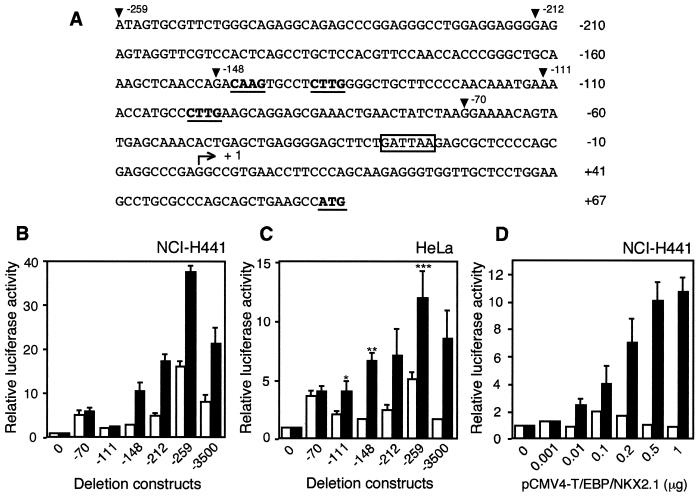
In order to determine whether the lung-specific claudin-18a1 promoter is responsive to activation by T/EBP/NKX2.1, a DNA fragment containing the 5′ flanking region of the mouse claudin-18a1 gene was isolated and sequenced (Fig. 5A). A major transcription initiation site was determined by the 5′ RACE method using adult mouse lung RNA as a template. A TATA-like box (GATTAA) was found at position −25 bp.
FIG. 5.
T/EBP/NKX2.1 activates transcription of the mouse claudin-18a1 gene. (A) Sequence of the mouse claudin-18a1 gene promoter. Arrowheads indicate the positions of the pGL3-259, -212, -148, -111 and -70 deletion constructs. The minimal T/EBP/NKX2.1 binding consensus sequences (5′-CAAG-3′) (19) and ATG initiation codon are shown in boldface and underlined. A TATA-like box is boxed. The bent arrow with + 1 indicates the major transcription start site. (B and C) Deletion analysis of the mouse claudin-18a1 gene promoter. The relative luciferase activities in NCI-H441 cells (B) or HeLa cells (C) transiently transfected with the indicated deletion constructs are shown. Activities obtained in the presence of cotransfected pCMV4-T/EBP/NKX2.1 (black bars) or pCMV4 (white bars) are relative to the activity of pGL3-Basic (construct 0). Values represent the means ± SD from three separate experiments. ∗, significantly different from the control pCMV4 vector at a P value of <0.05, ∗∗, significantly different from construct pGL3-111 with the expression plasmid at a P value of <0.01; ∗∗∗, significantly different from construct pGL3-212 with the expression plasmid at a P value of <0.05. (D) Dose-dependent increase of claudin-18a1 reporter activity with pCMV4-T/EBP/NKX2.1. Increasing concentrations of pCMV4-T/EBP/NKX2.1 (black bars) or pCMV4 (white bars) were cotransfected with the construct pGL3-259 in NCI-H441 cells. Luciferase activities are relative to the activity of construct pGL3-259 without any cotransfected vectors. Values represent the means ± SD from three separate experiments.
To map regions responsible for claudin-18a1 transcriptional activity and to identify potential T/EBP/NKX2.1-responsive regions, successive deletions of the claudin-18a1 promoter were linked to a luciferase reporter gene (pGL3-70, -111, -148, -212, -259, and -3500). These constructs were cotransfected into human lung NCI-H441 cells with the expression plasmid pCMV4-T/EBP/NKX2.1 or control pCMV4 vector (Fig. 5B). Cotransfection of pCMV4-T/EBP/NKX2.1 resulted in significant increases (two to threefold) in the claudin-18a1 reporter activities of constructs pGL3-148, -212, -259, and -3500 in NCI-H441 cells. Similar induction was observed in non-lung-derived HeLa epithelial cells for these deletion constructs (Fig. 5C). However, an induction was also observed for construct pGL3-111 in HeLa cells, which could be related to the presence of sufficient levels of endogenous T/EBP/NKX2.1 in NCI-H441 cells to mediate trans activation of construct pGL3-111, a possibility which remains to be examined. These results suggest that T/EBP/NKX2.1 is likely to be directly involved in the transcriptional control of the mouse claudin-18a1 gene and that elements that are necessary to activate claudin-18a1 transcription may be located between −148 and −70 bp. The nucleotide sequences in this region contain three minimal consensus sequences for potential T/EBP/NKX2.1 binding sites (CAAG) (19) at positions −134, −143, and −98 bp. Basal reporter activity (in the absence of T/EBP/NKX2.1 cotransfection) was also affected by deletion of the claudin-18a1 gene promoter. In both cell lines tested, basal activity was increased upon deletion of the claudin-18a1 promoter from −3500 to −259 bp. Furthermore, there was a significant decrease in reporter activity upon deletion from position −259 to −212 of the claudin-18a1 promoter. These results point to the presence of elements within the claudin-18a1 promoter and/or protein factors that mediate the constitutive transcriptional activity of claudin-18a1 independent of T/EBP/NKX2.1.
A dose-dependent increase in claudin-18a1 reporter activity in NCI-H441 cells was observed with increasing amounts of T/EBP/NKX2.1 DNA (0.005- to 5-fold relative to the amount of construct pGL3-259). A plateau appears to be reached when an approximately fivefold excess amount of expression plasmid is cotransfected. Under these transfection conditions, however, expression of the endogenous human claudin-18A1 gene was not detectable by RT-PCR in NCI-H441 cells suggesting that T/EBP/NKX2.1 may not be required for transcription of the human claudin-18A1 gene or that other factors that may be required for the expression of the gene are deficient in this cell line. Alternatively, the claudin-18A1 mRNA is inherently unstable in NCI-H441 cells.
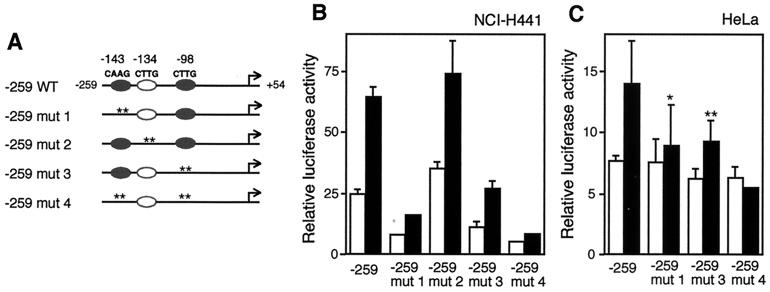
Site-directed mutagenesis of the putative T/EBP/NKX2.1 binding sites (at positions −143 bp [pGL3-259 mut 1] and −98 bp [pGL3-259 mut 3]) (Fig. 6A) showed incomplete but significant reduction of trans activation by T/EBP/NKX2.1 in both NCI-H441 and HeLa cells (Fig. 6B and C, respectively). Mutations at position −134 bp (pGL3-259 mut 2) had no discernible effect. Mutations of both T/EBP/NKX2.1 binding sites (at positions −143 and −98 bp [pGL3-259 mut 4]) resulted in profound reductions of trans activation in both NCI-H441 and HeLa cells. These results indicate that the putative T/EBP/NKX2.1 binding sites at positions −143 and −98 participate in trans activation of the mouse claudin-18a1 promoter by T/EBP/NKX2.1.
FIG. 6.
Mutation of T/EBP/NKX2.1 binding sites reduces trans activation by T/EBP/NKX2.1. (A) Schematic representation of wild-type (−259 WT) and mutant (−259 mut 1 to 4) constructs. Closed ovals represent potential T/EBP/NKX2.1 binding sites. Open ovals depict functionless T/EBP/NKX2.1 binding consensus sites. Site-specific mutations are indicated by asterisks (each asterisk corresponds to a nucleotide). The mutated sequences are shown in Fig. 7A. (B and C) The deletion −259 construct with or without various site-specific mutations was transiently transfected into NCI-H441 cells (B) or HeLa cells (C) in the presence of coexpressed pCMV4-T/EBP/NKX2.1 (black bars) or pCMV4 (white bars). Values represent the means ± SD from three separate experiments. ∗, significantly different from construct −259 with the expression vector at a P value of <0.01; ∗∗, significantly different from construct −259 with the expression vector at a P value of <0.05.
T/EBP/NKX2.1 protein binds to sequences of the claudin-18a1 promoter.
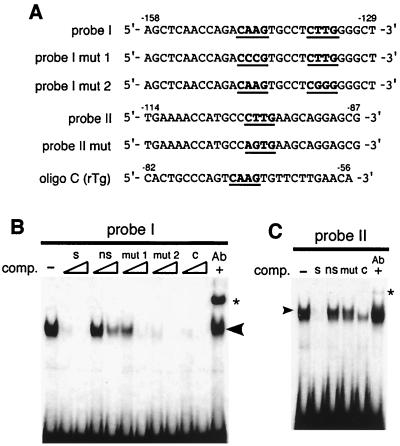
In order to further define the function of these elements, electromobility shift assays were performed with nuclear extracts from NCI-H441 cells (Fig. 7). Migration of the 32P-labeled oligonucleotide containing two T/EBP/NKX2.1 consensus binding sites (probe I; −158 to −129 bp) (Fig. 7A) was slower in the presence of NCI-H441 cell nuclear extracts (Fig. 7B). Formation of the specific protein-DNA complex was inhibited by the addition of 100-and 500-fold excesses, of unlabeled specific oligonucleotide but not by nonspecific oligonucleotide. An oligonucleotide containing one of the two T/EBP/NKX2.1 binding sites mutated (at −143 bp; probe I mut 1) did not compete with formation of the specific protein-DNA complex, whereas an oligonucleotide containing a mutated site at position −134 (probe I mut 2) did. These results demonstrate that T/EBP/NKX2.1 does not bind to the minimum consensus site at position −134. An oligonucleotide containing the T/EBP/NKX2.1 binding site identified in the rat thyroglobulin gene promoter (7) completely prevented complex formation. The specific protein-DNA complex was supershifted by T/EBP/NKX2.1 antibody (Fig. 7B, lane Ab +). Radiolabeled oligonucleotides containing the third T/EBP/NKX2.1 binding site at position −98 (probe II; −114 to −87 bp) exhibited results similar to those for probe I (Fig. 7C). Thus, the elements that mediate transcriptional activation of T/EBP/NKX2.1 are located at positions −143 and −98 bp in the 5′ region of the mouse claudin-18a1 gene.
FIG. 7.
The T/EBP/NKX2.1 minimal consensus sequence in the mouse claudin-18a1 gene promoter specifically binds to NCI-H441 nuclear proteins. (A) Sequences of probes and competitors used in electrophoretic gel mobility shift analysis. Oligo C is taken from the rat thyroglobulin promoter (rTG) (−82 to −56 bp), which had been identified as a T/EBP/NKX2.1 binding site (7). Putative T/EBP/NKX2.1 consensus sequences (19) are shown in boldface and underlined. (B and C) Electrophoretic mobility shift assay of the T/EBP/NKX2.1 binding site. NCI-H441 nuclear extracts were incubated with 32P-labeled probe I (B) or probe II (C). The specifically retarded complex is shown by an arrowhead. Competition assays were performed with 100-fold (C) or 100- and 500-fold (B) excesses of unlabeled specific (lanes s), nonspecific (lanes ns), mutated (lanes mut 1 and mut 2), or oligo C (lanes c) oligonucleotides. For antibody supershift analysis, TTF1-specific monoclonal antibody was added to the reaction mixture (lanes Ab +). Asterisks indicate the position of the supershifted complex.
Cellular localization of claudin-18.
To analyze the subcellular localization of caludin-18, we examined the distribution of claudin-18 in mouse lung and stomach by immunohistochemistry (Fig. 8). Since the claudin-18 antibody was raised against synthetic C-terminal peptides, this antibody recognizes both claudin-18A1.1 and -18A2.1. In lung bronchus and bronchioles (Fig. 8A) (data on bronchioles not shown) and glandular stomach (Fig. 8B and C), immunoreactivity was seen along the cell membrane diffusely, in the cytoplasm of some cells, and in punctate spots that may represent TJs. In particular, dramatic complete membrane staining was observed in adult mouse glandular stomach around the entire surface of epithelial cells (Fig. 8C).
FIG. 8.
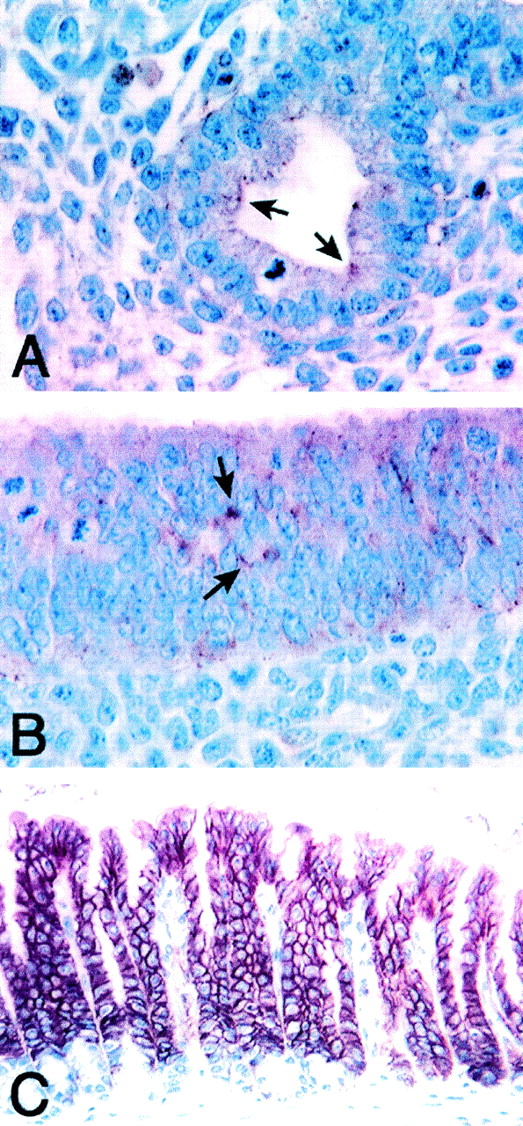
Immunolocalization of claudin-18 in mouse lung and stomach. E13.5 whole normal mouse embryo (A and B) and normal adult mouse stomach (C) were fixed in Bouin's fixatives and were subjected to immunohistochemistry. (A) Lung bronchial epithelial cells. (B) Glandular stomach epithelial cells. Positive staining is shown by arrows. (C) Glandular stomach epithelial layers. Magnification, ×300.
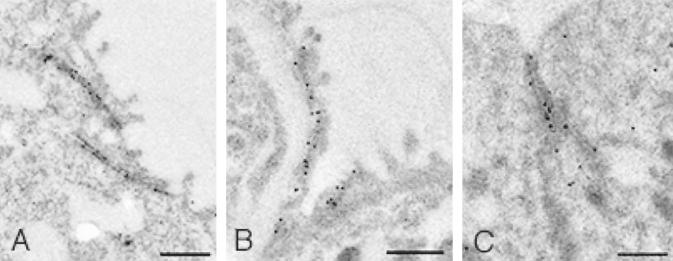
In order to examine at which cellular organelles claudin-18A1.1 is present, IEM was performed using adult mouse stomach and lungs from E15.5 and E18.5 embryos and adults. All of the tissues examined clearly demonstrated that claudin-18 is localized at TJs in epithelial cells (Fig. 9). No other organelles had immunogold spots. T/ebp/Nkx2.1-null embryonic lungs of the same gestational ages were also examined for claudin-18 protein expression. As expected, no positive staining was obtained, further demonstrating the specificity of the antibody for claudin-18 (data not shown). Interestingly, T/ebp/Nkx2.1-null embryo lungs as early as E12.5 clearly demonstrated the presence of TJs (data not shown). However, the latter observation can be explained by the presence of claudins other than claudin-18 in the mutant lungs (Fig. 4).
FIG. 9.
Ultrastructural localization of claudin-18A1.1 and claudin-18A2.1 in mouse lung and stomach, respectively. IEM of E15.5 embryo (A) and adult (B) normal mouse lungs and of adult stomach (C) is shown. Note that the immunogold labeling is localized at TJs. Bars, 0.5 μm.
DISCUSSION
We have identified a novel T/EBP/NKX2.1 target gene, claudin-18, which is a member of the claudin multigene family. Unlike other claudin gene family members, claudin-18 encodes two types of lung- and stomach-specific polypeptides by alternative splicing. An alternative 12-bp insertion at the 3′ end of exon 4 produces an isoform that lacks the C-terminal cytoplasmic domain. Most claudins have tyrosine-valine (Y-V) amino acid sequences at their C termini, which directly bind to the membrane-associated guanylate kinase family of proteins, ZO-1, ZO-2, and ZO-3, through their first PDZ (postsynaptic protein-95 [PSD-95]/discs large [DLG]/zonula occludens-1 [ZO-1]) domains (18, 29). These proteins are believed to function as scaffolds of the TJ plaque to cross-link TJ strands to the actin-based cytoskeleton, and they are involved in the regulation of the barrier function of TJs by modulating actin filament-TJ strand association (18, 23). Considering these hypotheses and the fact that claudins can be polymerized to form TJ strands without interacting with ZO-1 (13), the C-terminally truncated isoform may have a dominant-negative-type function in regulating the barrier function of TJs. Determination of whether this is the case or not must await further studies.
Alternative usage of two promoters and their own first exons leads to production of lung- and stomach-specific claudins. Although detailed descriptions of most claudins have not yet been reported, most claudins have wide tissue distribution patterns, except claudins-11 and -16 (27, 29; our unpublished observation). Claudin-11 is the only claudin expressed in the myelin sheaths of oligodendrocytes in the central nervous system and Sertoli cells in the testis (14, 30). Claudin-16 expression is mainly restricted to the thick ascending limb of Henle in the kidney, and it is assumed that claudin-16 creates a magnesium-selective channel through the TJs (38). In our analyses by RT-PCR, more than 10 claudins are expressed in mouse lung (this study) and stomach (unpublished observation). However, lung- and stomach-restricted expression of claudin-18 suggests that claudin-18 may have an important role such as a channel-like activity as seen in claudin-16.
The results of transient-transfection studies indicate that the homeodomain-containing transcription factor T/EBP/NKX2.1 is required to trans activate expression of the mouse claudin-18a1 promoter even though the level of induction is low (two to threefold). To our knowledge, this is the first report that describes functional analysis of the claudin gene promoter. Cotransfection experiments using a plasmid expressing T/EBP/NKX2.1 with claudin-18a1–luciferase reporter constructs delineated the minimal region of the mouse claudin-18a1 gene promoter that is sufficient to activate transcription of the gene. This region contains two consensus T/EBP/NKX2.1 binding elements, 5′-CAAG-3′ (19). Mutation of each motif interfered equally with T/EBP/NKX2.1 binding to the site and reduced claudin-18a1 promoter activity by approximately one-half. Mutation of both sites nearly abolished the transcriptional activity of the gene. Data obtained from electrophoretic mobility shift assays were consistent with these results. The dose-dependent increase in reporter activity in response to cotransfected T/EBP/NKX2.1 is also consistent with the hypothesis that the mouse claudin-18a1 gene is a direct target of T/EBP/NKX2.1. Interestingly, claudin-18a1 is the only gene that appears to be activated by T/EBP/NKX2.1, although at least 10 claudins are expressed in mouse embryonic lungs (27, 29). T/EBP/NKX2.1 not only functions as a transcriptional regulator of surfactant protein (1, 2, 19) and Clara cell secretory protein (34, 35) genes in lung but also is proposed to be one of the key regulators of early lung development (26). Recently, it was demonstrated that morphogenesis and cellular differentiation of the distal lung compartments are strictly dependent on the activity of T/EBP/NKX2.1 (26, 46). Whether claudin-18a1 has a role in lung morphogenesis remains to be studied.
claudin-18a2 is a stomach-specific isoform of the claudin-18 gene that has a promoter distinct from that of claudin-18a1. High levels of expression in adult mouse stomach were demonstrated by immunostaining. Of interest is its immunolocalization, where claudin-18 appears to be located on entire cell membranes and in some cases cytoplasm rather than just TJs. This pattern of expression is very similar to those of caludin-3 and -5 in stomach (33). Diffuse cytoplasmic staining of claudin-18 may be the result of using thicker paraffin sections (4 to 6 μm) for immunostaining instead of very thin sections such as the 50- to 100-nm sections used for EM. However, IEM demonstrated claudin-18 immunogold spots only at TJs and not within any other cellular organelles. For claudin-3 and -5, IEM has yet to be performed (33). Thus, it is possible, as proposed for claudin-3 and -5 (33), that claudin-18 may be localized along the lateral and basolateral intercellular space without forming direct cell-to-cell contacts that can influence ion or solute movement along the lateral or basolateral membranes. It is possible that these claudins are recruited into TJs to quickly establish new paracellular transport properties in response to acute physiological challenge. Our data on claudin-18 argue for the proposal that some, if not all, of the TJ barriers may be regulated by both transcriptional and subcellular localization mechanisms (33).
Our results demonstrated that the stomach-specific form of the transcript is present in both mouse and human. In preliminary experiments, there was no cis-regulatory element for stomach-specific expression within 1.8 kb upstream of the mouse claudin-18a2 gene when a human stomach adenocarcinoma-derived AGS cell line was used in transfection assays. Initially, the expression of the human claudin-18A2 gene in this cell line was confirmed by using RT-PCR. To our knowledge, no stomach-specific transcription factor has been reported. Further sequences upstream of 1.8 kb of the mouse claudin-18a2 gene may contain a cis-regulatory element for stomach-specific expression. It is not yet known whether the two different tissue-specific forms of claudin-18 possess distinct functions at their specific expression sites. In this regard, it is interesting that claudin-18al is the only claudin that is missing in T/ebp/Nkx2.1-null mouse lungs. Determination of whether claudin-18A1 plays a role in lung development requires further experiments.
ACKNOWLEDGMENTS
We thank Frank Gonzalez and Taro Akiyama for helpful discussions and critical review of the manuscript and Barbara Kasprzak and Donna Butcher for immunohistochemistry.
REFERENCES
- 1.Bohinski R J, Di Lauro R, Whitsett J A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruno M D, Bohinski R J, Huelsman K M, Whitsett J A, Korfhagen T R. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem. 1995;270:6531–6536. doi: 10.1074/jbc.270.12.6531. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso W V. Lung morphogenesis revisited: old facts, current ideas. Dev Dyn. 2000;219:121–130. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1053>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Cereijido M, Shoshani L, Contreras R G. Molecular physiology and pathophysiology of tight junctions. I. Biogenesis of tight junctions and epithelial polarity. Am J Physiol Gastrointest Liver Physiol. 2000;279:G477–G482. doi: 10.1152/ajpgi.2000.279.3.G477. [DOI] [PubMed] [Google Scholar]
- 5.Cereijido M, Valdes J, Shoshani L, Contreras R G. Role of tight junctions in establishing and maintaining cell polarity. Annu Rev Physiol. 1998;60:161–177. doi: 10.1146/annurev.physiol.60.1.161. [DOI] [PubMed] [Google Scholar]
- 6.Civitareale D, Castelli M P, Falasca P, Saiardi A. Thyroid transcription factor 1 activates the promoter of the thyrotropin receptor gene. Mol Endocrinol. 1993;7:1589–1595. doi: 10.1210/mend.7.12.8145764. [DOI] [PubMed] [Google Scholar]
- 7.Civitareale D, Lonigro R, Sinclair A J, Di Lauro R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBry R W, Seldin M F. Human/mouse homology relationships. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Endo T, Kaneshige M, Nakazato M, Ohmori M, Harii N, Onaya T. Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/l− symporter gene. Mol Endocrinol. 1997;11:1747–1755. doi: 10.1210/mend.11.11.0012. [DOI] [PubMed] [Google Scholar]
- 11.Francis-Lang H, Price M, Polycarpou-Schwarz M, Di Lauro R. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanisms for thyroid-specific gene expression. Mol Cell Biol. 1992;12:576–588. doi: 10.1128/mcb.12.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gow A, Southwood C M, Li J S, Pariali M, Riordan G P, Brodie S E, Danias J, Bronstein J M, Kachar B, Lazzarini R A. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell. 1999;99:649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- 15.Guazzi S, Price M, De Felice M, Damante G, Mattei M G, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayat M A. Colloidal gold, Principles, methods, and applications. 1 to 3. San Diego, Calif: Academic Press Inc.; 1989. [Google Scholar]
- 17.Hogan B L. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 18.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly S E, Bachurski C J, Burhans M S, Glasser S W. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem. 1996;271:6881–6888. doi: 10.1074/jbc.271.12.6881. [DOI] [PubMed] [Google Scholar]
- 20.Kikkawa F, Gonzalez F J, Kimura S. Characterization of a thyroid-specific enhancer located 5.5 kilobase pairs upstream of the human thyroid peroxidase gene. Mol Cell Biol. 1990;10:6216–6224. doi: 10.1128/mcb.10.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox C H, Ward J M, Gonzalez F J. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 22.Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 23.Madara J L. Regulation of the movement of solutes across tight junctions. Annu Rev Physiol. 1998;60:143–159. doi: 10.1146/annurev.physiol.60.1.143. [DOI] [PubMed] [Google Scholar]
- 24.Metzger R J, Krasnow M A. Genetic control of branching morphogenesis. Science. 1999;284:1635–1639. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- 25.Minoo P, Hamdan H, Bu D, Warburton D, Stepanik P, deLemos R. TTF-1 regulates lung epithelial morphogenesis. Dev Biol. 1995;172:694–698. doi: 10.1006/dbio.1995.8080. [DOI] [PubMed] [Google Scholar]
- 26.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1 (−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- 27.Mitic L L, Van Itallie C M, Anderson J M. Molecular physiology and pathophysiology of tight junctions. I. Tight junction structure and function: lessons from mutant animals and proteins. Am J Physiol Gastrointest Liver Physiol. 2000;279:G250–254. doi: 10.1152/ajpgi.2000.279.2.G250. [DOI] [PubMed] [Google Scholar]
- 28.Mizuno K, Gonzalez F J, Kimura S. Thyroid-specific enhancer-binding protein (T/EBP): cDNA cloning, functional characterization, and structural identity with thyroid transcription factor TTF-1. Mol Cell Biol. 1991;11:4927–4933. doi: 10.1128/mcb.11.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol. 1999;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popescu N, Zimonjic D, Hatch C, Bonner W. Chromosomal mapping of the human histone gene H2AZ to 4q24 by fluorescence in situ hybridization. Genomics. 1994;20:333–335. doi: 10.1006/geno.1994.1182. [DOI] [PubMed] [Google Scholar]
- 33.Rahner C, Mitic L L, Anderson J M. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology. 2001;120:411–422. doi: 10.1053/gast.2001.21736. [DOI] [PubMed] [Google Scholar]
- 34.Ray M K, Chen C Y, Schwartz R J, DeMayo F J. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transcription factor 1 and cardiac muscle-specific homeobox protein (CSX) Mol Cell Biol. 1996;16:2056–2064. doi: 10.1128/mcb.16.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawaya P L, Stripp B R, Whitsett J A, Luse D S. The lung-specific CC10 gene is regulated by transcription factors from the AP-1, octamer, and hepatocyte nuclear factor 3 families. Mol Cell Biol. 1993;13:3860–3871. doi: 10.1128/mcb.13.7.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Searle A G, Peters J, Lyon M F, Hall J G, Evans E P, Edwards J H, Buckle V J. Chromosome maps of man and mouse. IV. Ann Hum Genet. 1989;53:89–140. doi: 10.1111/j.1469-1809.1989.tb01777.x. [DOI] [PubMed] [Google Scholar]
- 37.Shimura H, Okajima F, Ikuyama S, Shimura Y, Kimura S, Saji M, Kohn L D. Thyroid-specific expression and cyclic adenosine 3′,5′-monophosphate autoregulation of the thyrotropin receptor gene involves thyroid transcription factor-1. Mol Endocrinol. 1994;8:1049–1069. doi: 10.1210/mend.8.8.7997232. [DOI] [PubMed] [Google Scholar]
- 38.Simon D B, Lu Y, Choate K A, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton R P. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 39.Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 1999;147:195–204. doi: 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spring K R. Routes and mechanism of fluid transport by epithelia. Annu Rev Physiol. 1998;60:105–119. doi: 10.1146/annurev.physiol.60.1.105. [DOI] [PubMed] [Google Scholar]
- 41.Sussel L, Marin O, Kimura S, Rubenstein J L. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 42.Takuma N, Sheng H Z, Furuta Y, Ward J M, Sharma K, Hogan B L, Pfaff S L, Westphal H, Kimura S, Mahon K A. Formation of Rathke's pouch requires dual induction from the diencephalon. Development. 1998;125:4835–4840. doi: 10.1242/dev.125.23.4835. [DOI] [PubMed] [Google Scholar]
- 43.Tsukita S, Furuse M. Occludin and claudins in tight-junction strands: leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 44.Tsukita S, Furuse M, Itoh M. Structural and signalling molecules come together at tight junctions. Curr Opin Cell Biol. 1999;11:628–633. doi: 10.1016/s0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 45.Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson K D, Cardoso W V. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 46.Yuan B, Li C, Kimura S, Engelhardt R T, Smith B R, Minoo P. Inhibition of distal lung morphogenesis in Nkx2.1 (−/−) embryos. Dev Dyn. 2000;217:180–190. doi: 10.1002/(SICI)1097-0177(200002)217:2<180::AID-DVDY5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 47.Zimonjic D B, Rezanka L, DiPaolo J A, Popescu N C. Refined localization of the erbB-3 proto-oncogene by direct visualization of FISH signals on LUT-inverted and contrast-enhanced digital images of DAPI-banded chromosomes. Cancer Genet Cytogenet. 1995;80:100–102. doi: 10.1016/0165-4608(94)00161-4. [DOI] [PubMed] [Google Scholar]