Abstract
Telomeres are ribonucleoproteins that cap chromosome-ends and their DNA length is controlled by counteracting elongation and shortening processes. The budding yeast Saccharomyces cerevisiae has been a leading model to study telomere DNA length control and dynamics. Its telomeric DNA is maintained at a length that slightly varies between laboratory strains, but little is known about its variation at the species level. The recent publication of the genomes of over 1,000 S. cerevisiae strains enabled us to explore telomere DNA length variation at an unprecedented scale. Here, we developed a bioinformatic pipeline (YeaISTY) to estimate telomere DNA length from whole-genome sequences and applied it to the sequenced S. cerevisiae collection. Our results revealed broad natural telomere DNA length variation among the isolates. Notably, telomere DNA length is shorter in those derived from wild rather than domesticated environments. Moreover, telomere DNA length variation is associated with mitochondrial metabolism, and this association is driven by wild strains. Overall, these findings reveal broad variation in budding yeast's telomere DNA length regulation, which might be shaped by its different ecological life-styles.
Keywords: telomere, yeast, S. cerevisiae, natural variation, population genomics, evolution, domestication
Introduction
Telomeres are ribonucleoprotein structures located at the ends of chromosomes (Blackburn and Gall 1978). They comprise tandem repeats of a DNA sequence whose motif and length vary among species (Monaghan 2010; Fulnečková et al. 2013). They maintain chromosome structure by distinguishing natural chromosome-ends from accidental double-stranded breaks (DSBs) and by contributing to chromosome segregation during cell division. Telomere DNA length (TL) is determined by the opposite actions of elongation and degradation pathways: elongation is ensured by telomerase, a reverse transcriptase that uses RNA as a template (Greider and Blackburn 1985, 1987), or by alternative lengthening of telomeres (ALT) recombination pathways (Lundblad and Blackburn 1993; Bryan et al. 1995; Teng and Zakian 1999); degradation is linked to the semi-conservative replication of DNA extremities coupled to specific nuclease activities (Watson 1972; Olovnikov 1973; Lingner et al. 1995). In mammalian somatic cells, telomerase is not or only weakly expressed and telomeres shorten at each cell division (Gilson and Géli 2007). By contrast, telomerase is constitutively expressed in the budding yeast Saccharomyces cerevisiae allowing telomeric DNA to be maintained at an equilibrium length (Marcand et al. 1999).
TL differs broadly among species. Human telomeres can range between 5 and 15 kb while mice can have extremely long telomeres (even >100 kb; Monaghan 2010). In comparison, yeast telomeres are relatively short at around 300 bp (Teixeira and Gilson 2005). Yeast telomeric sequences are often flanked by the so-called telomere-associated sequences (TAS). These comprise repetitive sequences like the X and Y′ elements, and interstitial telomeric sequences (ITS), which are located between them (Louis 1995; Wellinger and Zakian 2012). TL is a complex trait: in budding yeast it is modulated by more than 400 telomere length maintenance (TLM) genes that were identified in large-scale systematic genetic screens (Askree et al. 2004; Gatbonton et al. 2006; Ungar et al. 2009; Harari and Kupiec 2014), and by various environmental factors, including heat, caffeine, and ethanol (Romano et al. 2013; Kupiec 2014; Harari and Kupiec 2018b; Harari et al. 2020). TL also varies among individuals of the same species, including yeasts, nematodes, plants, mammals, and birds (Zijlmans et al. 1997; Liti, Haricharan et al. 2009; Heidinger et al. 2012; Fulcher et al. 2014; Cook et al. 2016; Hansen et al. 2016; Foley et al. 2020).
TL can be inferred from next-generation sequencing data by estimating the fraction of reads containing a sufficiently high number of telomeric DNA repeated sequences. This approach was implemented for human genomes containing regular telomeric DNA repeats (T2AG3)n (Ding et al. 2014; Nersisyan and Arakelyan 2015; Hakobyan et al. 2016; Lee et al. 2017) as well as for S. cerevisiae, which contains irregular telomeric DNA repeated sequences (TG1–3)n (Puddu et al. 2019). These methods are now broadly applied (Cook et al. 2016; Hakobyan et al. 2016; Barthel et al. 2017; Nersisyan et al. 2019; Taub et al. 2020).
In humans, TL variation has been associated with multiple pathological conditions (Aviv and Shay 2018) and with selection to attenuate the risk of cancer (Mangino et al. 2015; Hansen et al. 2016). In birds, TL has been used to predict key life-history traits like growth, reproduction and lifespan, and individuals with shorter TL have a lower life expectancy (Haussmann et al. 2005; Heidinger et al. 2012). Examples of extreme TL variation among different subpopulations have also been reported in yeast, including S. cerevisiae and its closest wild relative S. paradoxus (Liti, Haricharan, et al. 2009). This raises the question whether TL variation plays a role in the adaptation of a strain to its environment or if it derives from genetic drift. The lack of an adequate number of strains and experimental follow-up has so far prevented this question from being addressed.
The recent publication of the genomes and phenomes of more than 1,000 S. cerevisiae isolates (Peter et al. 2018; De Chiara et al. 2020, 2022) enabled us to explore TL variation at an unprecedented scale. Here, we report the first comprehensive analysis of TL variation in yeast strains isolated from a wide range of ecological niches and geographical areas, including domesticated and wild environments. For this purpose, we developed a bioinformatic pipeline that estimates the average TL, ITS content, and Y′ copy number from whole-genome sequencing data. We described TL variation across 26 well-defined lineages and identified new candidate genetic variants involved in TLM. We compared TL with a series of phenotypes including growth using different carbon and nitrogen sources, mitochondrial metabolism, sporulation capacity, and chronological lifespan (CLS). The results revealed different TL regulation patterns in wild and domesticated yeasts that correlate with their mitochondrial functions.
Materials and methods
A bioinformatic pipeline for estimating TL, ITS content, and Y′ copy number
We developed YeaISTY (Yeast ITS, Telomeres and Y′ elements estimator) in order to estimate TL, ITS content, and Y′ copy number in the 1,011 S. cerevisiae collection.
We optimized its running parameters by applying it to simulated reads derived from modified genome assemblies in which we simultaneously replaced all the native telomeres with designed ones of known length. We tested several cutoffs for the minimum length of the telomeric repeats stretch contained in the reads and set a threshold of 40 bp based on the results of our simulations (see Supplementary Discussion 1). We ran YeaISTY on the 1,011 S. cerevisiae collection with the following command line:
perl find_motif_in_reads.pl -i $SAMPLE.fasta -m motif.txt -o $SAMPLE.fasta.readscan -l $SAMPLE.fasta.readlist -c INT
where “i” is the input file (in FASTA format), “m” is a file containing the telomeric motifs for pattern matching (C{1,3}A, TG{1,3}), “o” is an output file containing a list of the reads classified as telomeric or ITS-derived and the position and length of their telomeric motifs, “l” is another output file containing the names of the reads classified as telomeric or ITS-derived, and “c” represents the minimum number of bp covered by telomeric motifs that must be contained in a read in order to classify it as telomeric or ITS-derived. This value can be set by the user and must be an integer number (INT). In this study, we set “c” as 40.
Subsequently, we performed mapping on a modified SGD reference genome in which we masked all repetitive sequences, including telomeres, ITS and Y′ elements, by BWA (version 0.7.12; Li and Durbin 2009). The list of repetitive sequences in the SGD genome was generated and masked using RepeatMasker (http://www.repeatmasker.org/). A representative, long version Y′ element (from TEL09L) was appended to this reference genome as an additional chromosome entry.
After applying the pipeline, 93 isolates stood out from the rest because they exhibited extremely high TL. These 93 isolates were sequenced in previous studies. Given that these results were not comparable to the rest of the collection, we excluded these isolates and only considered the remaining 918 strains. After assessing the impact of aneuploidy variation on TL estimation through computational simulations, we decided to only retain a subset of 706 euploid strains for further analyses (see Supplementary Discussion 2).
We observed an underestimation rate of 4- and 8-fold for TL and ITS content, respectively, as compared to the average values measured by teloblot and the annotation of the 12 genome assemblies from the YPRP. We adjusted this bias by increasing TL and ITS content of 4- and 8-fold, respectively, in the estimations of the 706 S. cerevisiae collection. Values reported in Supplementary Table 5 are already adjusted. Estimates from the other datasets used in this study varied around a different median value respect to the one of the 706 S. cerevisiae collection, indicating that the underestimation rate differs across datasets and therefore estimates are not directly comparable between datasets, but only within them (Puddu et al. 2019; Tattini et al. 2019). Since we did not perform teloblots of representative strains from all the other datasets, their exact underestimation rates remained unknown and their estimates were not adjusted. The reasons of the underestimation are unknown but might depend on the combination of the sequencing technology, library preparation methods and coverage. For instance, previous studies have shown that the probability of a DNA region to be targeted by a double-strand-break during the fragmentation step of the sequencing library preparation is not equal along the genome, with GC-rich and terminal regions like telomeres being particularly affected. The GC content bias and the positional bias result in an under-representation of sequencing reads derived from the affected regions, leading to a nonuniform distribution of coverage along the genome. Telomeres are particularly prone to show low coverage values due to their high GC content and their terminal position (Grokhovsky et al. 2011; Benjamini and Speed 2012; Poptsova et al. 2014).
We tested whether the stringent parameters used by YeaISTY to select telomeric and ITS reads might cause the underestimation by recalculating TL and ITS content in 704 isolates of the S. cerevisiae collection using a lower cutoff (20 bp). The new estimations significantly correlated with the previous ones (r = 0.97 for TL, r = 0.99 for ITS content; P < 2.2e−16) but TL was still 3-fold lower than the value estimated by teloblot. At the 20 bp cutoff, the correlation with the teloblot was lower (r = 0.85 vs r = 0.87 for 20 and 40 bp cutoff, respectively; Supplementary Tables 3 and 5). We conclude that the parameters of our pipeline are only responsible of a fraction of the underestimation, while the major role is likely played by other factors. Due to the lower correlation obtained with the 20 bp cutoff, we used the results derived from the 40 bp cutoff.
While relative values can still indicate whether one strain carries longer/shorter telomeres than another one within the same dataset, a comparison with already known TL and ITS is needed to determine the exact underestimation rate of each dataset and obtain absolute TL and ITS estimates.
YeaISTY is freely available through github (https://github.com/mdangiolo89/Different-telomere-length-patterns-between-domestic-and-wild-S.-cerevisiae-isolates).
Simulations of sequencing runs from genomes carrying designed telomeres
To estimate the efficiency of YeaISTY, we applied it to artificially constructed datasets, consisting of simulated sequencing runs derived from genomes where telomere lengths were set at specific values. We masked the native telomeres of the 12 previously described genome assemblies by using the program maskfasta contained in the suite bedtools (version 2.17.0; Quinlan and Hall 2010). Subsequently, we replaced the masked native telomeres with 14 designed telomeres of known length, consisting of yeast telomeric TG{1,3} or C{1,3}A repeats. We created 168 (12 × 14) designed genomes in each of which single telomeres were all identical to each other. The lengths of the designed telomeres had the following values: 18, 49, 77, 99, 135, 174, 208, 296, 360, 416, 504, 553, 602, 651 bp. ITS were not modified. Illumina reads were then simulated from the 168 designed genomes using the software dwgsim (https://github.com/nh13/DWGSIM) with the following parameters: type of read = paired-end, read length = 100 bp, sequencing error rate = 0%, insert size = 300 bp, coverage = 30. To test the performance of YeaISTY upon ploidy and aneuploidy variation, we simultaneously replaced all the native telomeres of the S288C genome assembly with a designed telomere of 360 bp. We manually copy-pasted the full genome sequence to generate identical S288C genome assemblies with ploidies ranging from 1n to 5n. Next, we manually copy-pasted combinations of chromosomes in order to artificially insert aneuploidies into the haploid genome assembly. We generated a total of 32 genome assemblies with a variable number of aneuploid chromosomes, ranging from 1 to 14. To assess the performance of YeaISTY against increasing amounts of mutations in telomeric sequences we used another modified haploid S288C genome assembly in which we simultaneously replaced all the native telomeres with a designed telomere of 296 bp. We manually introduced single-base mismatches in the telomeric sequence used to replace the native telomeres, generating a total of 5 genome assemblies with a variable percentage of mismatches (0%, 1%, 3%, 5%, 10%). We simulated 2 Illumina paired-end sequencing runs for each ploidy, aneuploidy and mismatch background, except the euploid 1n for which we generated 3 sequencing runs, by using the software ART (Huang et al. 2012) and the following parameters: type of read = paired-end, read length = 100 bp, insert size = 300 bp, coverage = 30.
ITS content, Y′ copy number, and TL estimation in the yeast population reference panel
We estimated TL, ITS content, and Y′ copy number in 7 S. cerevisiae and 5 S. paradoxus strains sequenced with Pacbio technology and whose complete genome assemblies were already available as part of the yeast population reference panel (YPRP; Yue et al. 2017). We estimated the frequency and distribution of stretches of telomeric repeats in the 12 genome assemblies by using custom Perl scripts. We merged the repeats which were interspaced by only 1 bp into a unique telomeric repeat stretch by using the software mergeBed included in the suite bedtools (version 2.17.0) with the option “-d 1” (Quinlan and Hall 2010). Then, we filtered the list to keep only the stretches longer than 10 bp. The interstitial telomeric repeats stretches were annotated as Y′-associated ITS if they closely preceded an annotated Y′ element, while the others were discarded. The ITS content of a strain was determined as the sum of the lengths of its Y′-associated ITS annotations.
TL of these 12 yeast strains was determined by analyzing their already available Sanger-sequencing reads, included in the SGRP project, as described in the paragraph “TL estimation in the SGRP collection”.
TL estimation in the SGRP collection
We estimated the TL of Sanger-sequenced strains of the Saccharomyces Genome Resequencing Project (SGRP; Liti, Carter et al. 2009) by using YeaISTY’s first module followed by manual annotation. First, we filtered the Sanger-sequencing reads to retain only the ones containing abundant telomeric repeats. We classified reads as telomeric if they contained a stretch of telomeric repeats (either C{1,3}A or TG{1,3}) longer or equal to 40 bp and located at the extremities of the read. Then, TL was estimated from each telomeric read by manually counting the telomeric repeats. Each read was considered as a single telomere. Lineage assignments in Supplementary Fig. 10b reflect the phylogenetic clades of Liti, Carter et al. (2009). S. cerevisiae strains were classified as domesticated if they had been isolated from fermentation-related environments, while they were classified as wild if they were isolated in nature. S. cerevisiae strains whose substrate of isolation was unknown were classified as “Unknown”.
Southern blots
Yeast cell cultures from 7 YPRP strains and 17 representative isolates of the 1,011 S. cerevisiae collection were grown overnight at 30°C in 5 ml of YPD medium to early stationary phase. Genomic DNA was subsequently extracted using MasterPure Yeast DNA Purification Kit. DNA (1–2 μg) was digested with XhoI at 37°C for 5 h. The digested DNA was purified using phenol/chloroform and ethanol precipitation and separated by electrophoresis (16 h at 30 V using 0.9% agarose). A 200 bp DNA ladder was loaded in the first lane of the gel to provide a reference for terminal restriction fragments (TRF) size estimation. The gel was soaked in ethidium bromide and an image was taken with a Typhoon machine. The DNA was then depurinated by incubating the gel in 200 ml of 0.25 M HCl for 30 min, and denaturated in 200 ml of 0.5N NaOH +1 M NaCl for 20 min twice. Neutralization was performed by incubating the gel twice in 200 ml of 1 M ammonium acetate for 20 min. The content of the gel was then transferred to a Hybond-N membrane and crosslinked to the membrane through UV irradiation. The membrane was hybridized with a TG1–3 probe (270 bp) containing 32P-labeled dCTP by incubating it in 15 ml of a prehybridization solution (6× SSC, 0.5% SDS, 1% milk) for 1 h at 50°C, and then in 15 ml of hybridization solution (6× SSC, 0.1% SDS, 1% milk) overnight at 50°C. The membrane was subsequently washed to remove any unbound probe by incubating it in 20 ml of 2× SSC for 10 min at 50°C, then in 20 ml of 2× SSC + 1% SDS for 30 min at 50°C and finally twice in 20 ml of 0.2× SSC + 1% SDS for 30 min at 50°C. TRF were visualized using Phosphorimager after 24 h of exposure.
TRF signals were analyzed using ImageQuant. For each lane, a pixel analysis was performed and the distance between each TRF and the respective well was annotated, where the distance represents the position of maximum pixel intensity of the band. For the lane containing the DNA ladder, the distance between each band and the well was annotated and used to construct a linear regression model describing the relationship between the size of the DNA fragments and their migration pattern. The function representing this model was used to predict the size of the TRF for each strain given its distance from the well. Given that the restriction site for XhoI is located in a conserved position around 875 bp before the start of each Y′ telomere, additional 875 bp were subtracted from the TRF size in order to obtain the final telomere length.
Statistics and data reproducibility
We estimated TL in 706 already sequenced S. cerevisiae strains (Peter et al. 2018). Strains classifications, including phylogenetic lineages and domesticated/wild assignments were maintained as described in (De Chiara et al. 2022). We used the whole collection (n = 706) for most of the analyses performed in this study, except for the GWAS (see below) and the correlation analysis where we used the euploid diploid collection (n = 555; see paragraphs “Genome-wide association study and LOFs analysis” and “Correlation analysis”).
Statistical analyses were performed using R (version 3.5.3). Two-group comparisons were performed using the function wilcox.test, performing a 2-tailed Mann–Whitney test. Multiple-group comparisons were performed using the function kruskal.test, performing a nonparametric ANOVA test. Subsequent pairwise analyses were performed using the function dunnTest included in the package FSA.
Genome-wide association study and LOFs analysis
We used a subset of euploid diploid strains (n = 555) for the genome-wide association study (GWAS) in order to minimize the confounding effects of ploidy variation.
Genotype data and the matrix of between-strains co-similarity to adjust for population structure were already available from (Peter et al. 2018). Only variants with a minor allele frequency (MAF) ≥ 0.05 in the whole population were included in the analysis (n = 101,579). TL data were converted to a normal distribution using the function qqnorm included in the R software. GWAS was performed using fast-lmm with the following command line:
fastlmmc -bfile $genotypes_file -bfileSim $cosimilarity_matrix -pheno $phenotypes_file -out $output_file
Variants with corrected P < 0.05 were considered statistically significant and retained for further analyses. Narrow-sense heritability (h2) for TL was calculated by dividing the genetic variance of the null model by the total variance of the null model (genetic variance and residual variance), computed using fast-lmm. The fraction of the phenotypic variance explained by the significant GWAS variants was calculated by removing all the rest of the genotypes from the co-similarity matrix and rerunning a second GWAS. Subsequently, we repeated the same calculations on the new output file.
LOF variants were already available from (Peter et al. 2018). A LOF was considered as present in a strain if it was either in homozygous or heterozygous state. LOFs were annotated as present in a clade if their frequency in the clade was higher than 0.6 but their frequency in the 555 S. cerevisiae collection was lower than 0.2.
Phenotypic measurements and correlation analyses
The set of phenotypes used in the correlation analysis was already available from previous studies. 35 phenotypes relative to resilience in the presence of stressors were derived from (Peter et al. 2018). 114 phenotypes relative to growth in multiple carbon and nitrogen sources were derived from (De Chiara et al. 2022). Absolute and relative mtDNA copy number were derived from (De Chiara et al. 2022). Mitochondrial volume and activity were partially derived from (De Chiara et al. 2020) but, in this study, the measurements were extended to the euploid diploid 555 yeasts collection and were performed as described in (De Chiara et al. 2020). Only the euploid diploid strains (n = 555) were used in the correlation analysis. Correlation analyses were performed using the function cor.test included in the R software applying the Pearson’s model.
To correct for potential bias due to population structure, we repeated the correlation analysis by applying the phylogenetic generalized least squares (PGLS) method. We computed Pagel’s λ and corrected correlation coefficients and P-values by using the function pgls included in the R package caper. The tree representing the population structure was based on a subset of a vcf file derived from Peter et al. (2018), which contained genetic variants corresponding to the 555 euploid diploid strains, and was created with the software iqtree (Nguyen et al. 2015). The tree was subsequently rooted on the branch of the Taiwanese clade using iTOL (Letunic and Bork 2021) and uploaded in the R environment in Newick format by using the function read.tree included in the package ape (Paradis et al. 2004).
In both analyses, missing data were not considered for the calculation of coefficients and P-values. In PGLS, the isolates containing missing data were also removed from the tree by using the function drop.tip included in the package ape. P-values were subsequently recomputed using the FDR correction for multiple hypothesis testing and associations were considered statistically significant if their corrected P < 0.05.
Results
S. cerevisiae telomere analysis from whole-genome sequences
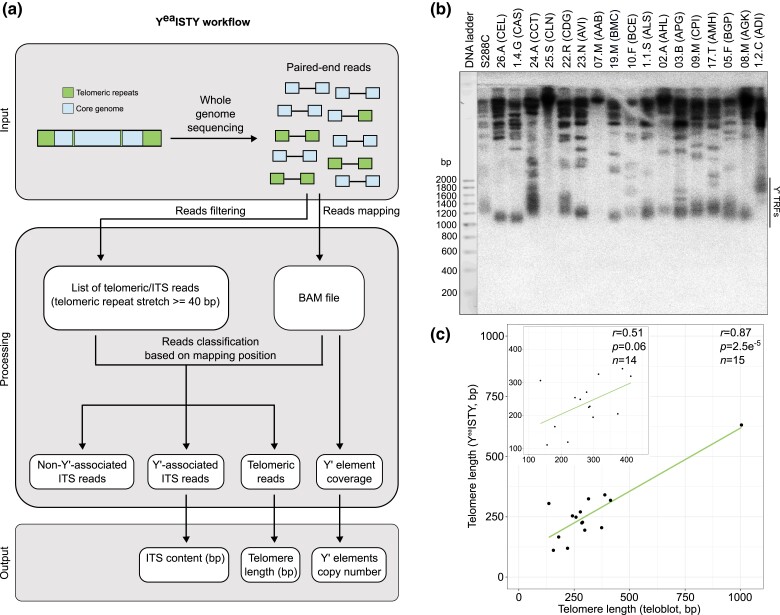
We developed YeaISTY (Yeast ITS, Telomeres and Y′ elements estimator) to estimate TL, ITS content and Y′ copy number from yeast whole-genome sequencing data (Methods). The performance of YeaISTY has been extensively benchmarked against simulated and experimental datasets and its running parameters have been optimized (see next paragraph and Supplementary Discussions 1–4). The YeaISTY workflow begins with a screening of Illumina paired-end reads. The pipeline detects single units of telomeric motifs (CA, CCA, CCCA, TG, TGG, TGGG). If units are consecutive to one another they are considered as part of the same telomeric repeats stretch and its start/end points are assigned, respectively, as the position of the first base of the first unit and the position of the last base of the last unit. The length of the telomeric repeats stretch is then calculated as end-start + 1. We allow the presence of only one mismatch of maximum 2 nucleotides inside the telomeric repeats stretch without changing the starting point (Supplementary Fig. 1a, panel 2, read 1). Instead, if the telomeric repeats stretch is interrupted by a mismatch of at least 3 bp or by multiple mismatches, the starting point is reassigned as the position of the next available telomeric unit in the read (Supplementary Fig. 1a, panel 2, reads 2 and 3). Reads are retained if they contain a stretch of telomeric repeats (either C{1,3}A or TG{1,3}) longer or equal to 40 bp (Fig. 1a and Supplementary Fig. 1a). Since telomeric and ITS-derived reads are virtually impossible to distinguish from each other based solely on their sequences, we introduced an additional step consisting in mapping all the reads to a modified reference genome in which all repetitive sequences, including telomeres, ITS and Y′ elements, have been masked. Moreover, a representative Y′ element was appended to this reference genome as an additional chromosome entry. The presence of a single representative Y′ element solves the problem of spurious mapping, provides a criterion to distinguish telomeric and ITS reads and enables us to reliably estimate Y′ elements copy number (Supplementary Fig. 1a). ITS reads are recognized based on the following assumptions: (1) if a read contains a stretch of C{1,3}A or TG{1,3} repeats and its pair maps on a nonsubtelomeric part of the genome; (2) if a read contains a stretch of TG{1,3} repeats and its pair maps on the representative Y′ element. ITS reads are further classified as non-Y′-associated or Y′-associated if they fit the first or the second criterion, respectively. All the other retained reads from the first filtering step are considered as telomeric. For simplicity, we refer to Y′-associated ITS content as “ITS content”, while the amount of non-Y′-associated ITS is not further analyzed in this study.
Fig. 1.
Overview of the YeaISTY workflow. a) Paired-end reads are scanned for telomeric repeats (either C{1,3}A or TG{1,3}). Reads containing a stretch of telomeric repeats longer or equal to 40 bp are retained. All the reads (retained and nonretained) are mapped to a modified reference genome. The mapping position of retained reads and their paired ones is used to classify them as ITS-derived or telomeric. The amount of ITS-derived and telomeric reads is then used to infer ITS content and telomere length, while coverage data are used to infer the copy number of Y′ elements. b) XhoI digestion of genomic DNA from representative strains of 17 S. cerevisiae lineages identified in Peter et al. (2018). Genomic DNA is probed with radioactively-labeled telomeric TG1–3 repeats. The black line denotes TRFs resulting from the digestion of a Y′ element. No TRFs were detected for the representatives of the Sake (25.S-CLN) and Mosaic beer (07.M-AAB) clades. The first lane shows a 200 bp DNA ladder used to derive the size of each TRF. c) Comparison of telomere length measured by teloblot and YeaISTY. Values in the plot are corrected for the 4-fold YeaISTY underestimation bias. Despite we estimated TL of S288C from sequencing data derived from Yue et al. (2017), we did not introduce this value in the plot as it derived from another sequencing batch and had a different underestimation rate. The Sake (CLN) and Mosaic beer (AAB) representatives were excluded from the comparison. The inset shows the correlation excluding the outlier with extremely long telomeres (1.2.C-ADI).
ITS content is calculated as 2(1Σn sITS)/c, while TL is calculated as (1Σn sTEL)/32c, where n is the number of reads assigned to each category (Y′-associated ITS or telomeric), s is the length of the telomeric repeats stretch contained in the reads assigned to each category, and c is the median coverage in regions with a GC content of 50–80%, which is similar to what is found in yeast telomeric repeats. ITS values are multiplied by 2 because we are only able to detect Y′-associated ITS reads containing the TG{1,3} motif, but we assume a similar amount of ITS reads containing the C{1,3}A motif. TL values are divided by 32 because this is the number of telomeres per haploid genome. In addition, Y′ elements copy number is estimated as cY′/c, where cY′ is the median coverage of the representative Y′ element and c is the median coverage along the whole genome (Fig. 1a and Supplementary Fig. 1a and Discussion 1). A more detailed description of the implementation of YeaISTY’s algorithm is presented in Supplementary Discussion 1.
Benchmarking of YeaISTY against simulated and experimental datasets
We assessed the efficiency of YeaISTY by using both experimental and simulated datasets. First, we used complete genome assemblies of 7 S. cerevisiae and 5 S. paradoxus strains belonging to the YPRP (Yue et al. 2017) and estimated their ITS and Y′ content (Supplementary Fig. 1b, c and Supplementary Table 1). Next, we replaced native telomeres of these genome assemblies with designed ones of designated length and simulated Illumina paired-end sequencing runs. Then, we applied YeaISTY to this dataset and compared the ITS content and Y′ copy number estimations to the real ones, while we compared the TL estimations to the length of the designed telomeres. Overall, we found very high and significant positive correlations (r = 0.98 and P < 2.2e−16 for TL, r = 0.97 and P < 2.2e−16 for ITS content, r = 0.89 and P < 2.2e−16 for Y′ copy number; Supplementary Discussions 1, 2, Supplementary Figs. 2, 3 and Supplementary Table 2).
Next, we tested the accuracy of YeaISTY in estimating TL and ITS/Y′ content from real sequencing datasets. We first estimated TL of the 12 YPRP strains by manually inspecting Sanger-sequencing reads containing telomeric repeats derived from already published studies involving these isolates (Liti, Carter, et al. 2009). In parallel, we applied YeaISTY to already available Illumina sequencing data derived from the same strains (Yue et al. 2017) and obtained significant positive correlations for all the measured variables (Pearson’s r = 0.63 and P = 0.04 for TL, r = 0.89 and P = 9.63e−5 for ITS content, r = 0.83 and P = 0.0008 for Y′ copy number; Supplementary Fig. 4a and Supplementary Table 3).
Finally, we used 7 strains from the YPRP and one representative strain for 17 clades of the 1,011 S. cerevisiae collection and measured their TL by southern blotting (teloblot). We correlated the teloblot results with the YeaISTY estimations and found an overall positive correlation (Pearson’s r = 0.78 and P = 0.06 for the YPRP, r = 0.87 and P = 2.5e−5 for the S. cerevisiae collection; Fig. 1b, c and Supplementary Fig. 4b–d and Supplementary Table 3). Two strains (25.S-CLN and 07.M-AAB) in the S. cerevisiae collection’s teloblot did not give any TRF and were not included in the statistical analysis. One strain (1.2.C-ADI) had extremely long telomeres, in accordance with previous reports showing that this strain has atypical TL and massive amplification of ITS and Y′ elements (Bergstrom et al. 2014; O’Donnell et al. 2022). Overall, TL resulting from YeaISTY was underestimated by 4-fold respect to the values of the S. cerevisiae collection’s teloblot. We therefore applied a 4-fold correction factor to the estimations of the S. cerevisiae collection.
Our TL estimations for the 15 representative strains of the S. cerevisiae collection (excluding 25.S-CLN and 07.M-AAB) also show positive correlation with TL measurements of the same strains derived from Nanopore sequencing (Pearson’s r = 0.69, P = 0.004 between the teloblot and Nanopore sequencing, Pearson’s r = 0.49, P = 0.06 between YeaISTY and Nanopore sequencing; Gilles Fischer, personal communication). Additional details on the benchmarking of YeaISTY are presented in Supplementary Discussions 1–4 (Supplementary Figs. 1–5 and Supplementary Tables 2–4). Taken together, these results show that YeaISTY estimations are reliable and can be used for further analyses.
Global telomere length variation in the S. cerevisiae population
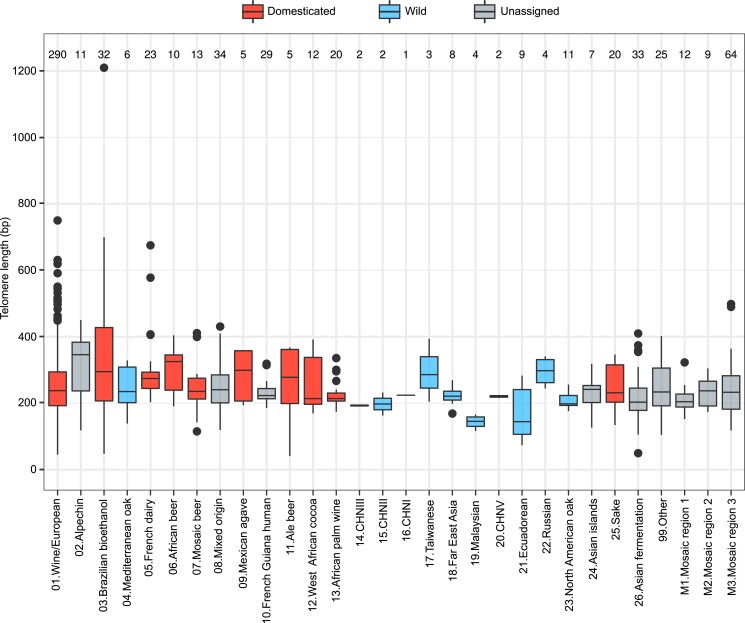
We applied YeaISTY to the 1,011 S. cerevisiae collection and estimated their TL. Strains derived from a former sequencing run (n = 93) were not included as their TL were not comparable to the rest of the dataset. Strains with aneuploidies (n = 212) were also excluded from this pool as our simulations showed that the number of aneuploidies can affect TL and ITS content estimations. We retained 706 strains and used these for further analyses (Supplementary Discussion 2 and Methods). TL was normally distributed with a median of 232 ± 98 bp, a value that is close to that of the S288C laboratory strain. Some isolates had either very short (minimum 41 bp) or very long (maximum 1210 bp) telomeres (Supplementary Fig. 6a and Supplementary Table 5). TL varied significantly among 26 previously described clades and 3 mosaic lineages (2-tailed Kruskal–Wallis test, P = 0.0002; Peter et al. 2018). TL was longer in the Alpechin and African beer clades, where the median exceeded 300 bp, and it was shorter in the Malaysian and Ecuadorean clades, where it remained below 150 bp (Fig. 2 and Supplementary Table 6). The Wine European clade’s TL had a distribution comparable to that of the entire collection, ranging from 45 to 750 bp (median = 238 ± 99 bp), consistent with the largest clade sample size (∼1/3 of the whole collection) and intraclade substructure consisting of 4 subclades (Semi-wild, Clinical/Y′amplification, Clinical/S. boulardii, Georgian; Supplementary Fig. 6b). The median TL varied significantly among the Wine European subclades (2-tailed Kruskal–Wallis test, P = 6.137e−8). It was highest in the Clinical/Y′ amplification subclade where it exceeded 600 bp, and lowest in the Clinical/S. boulardii subclade, where it remained below 200 bp (Supplementary Fig. 6c and Supplementary Table 6). Clinical/Y′ amplification strains are known to have aberrant TL and amplification of ITS and Y′ elements (Bergstrom et al. 2014; O’Donnell et al. 2022). No variation in TL was detected among strains with variable ploidy (2-tailed Kruskal–Wallis test, P = 0.14; Supplementary Fig. 6d).
Fig. 2.
TL variation in 706 S. cerevisiae isolates. Telomere length of the phylogenetic lineages in the 706 S. cerevisiae collection (Peter et al. 2018). Colors code represents the clade classification (domesticated, wild or unassigned), as reported in De Chiara et al. (2022). Clades are assigned as wild or domesticated based on the environmental origin of the isolates dominating (>66%) in the clade. They were labeled “Unassigned” if the threshold criterion was not passed. The order of clades on the x axis is the same as in Peter et al. (2018). In the box plots, horizontal lines denote the median, top and bottom hinges denote the IQR, whiskers denote maximum and minimum values within upper and lower hinges ± 1.5 × IQR. Numbers on top represent the number of isolates in each clade.
Finally, we measured the ITS content and Y′ copy number across the 706 isolates and found them to be highly variable but positively correlated with TL (Pearson’s r = 0.46 and P < 2.2e−16 between TL and ITS, r = 0.41 and P < 2.2e−16 between TL and Y′) and between themselves (Pearson’s r = 0.84, P < 2.2e−16; Supplementary Discussion 5, Supplementary Figs. 7–9, Supplementary Tables 7 and 8). The marked correlation between ITS and Y′ is consistent with the presence of short stretches of telomeric repeats between multiple Y′ copies (Louis and Haber 1992). Of note, ITS and Y′ content in the Wine European subclades followed the same trend of TL, being more abundant in the Clinical/Y′ amplification subclade and less abundant in the Clinical/S. boulardii subclade. Overall, our analyses show that TL and ITS exhibit both inter- and intraclade variation, but co-vary in the same direction.
Domesticated and wild yeast isolates exhibit different TLs
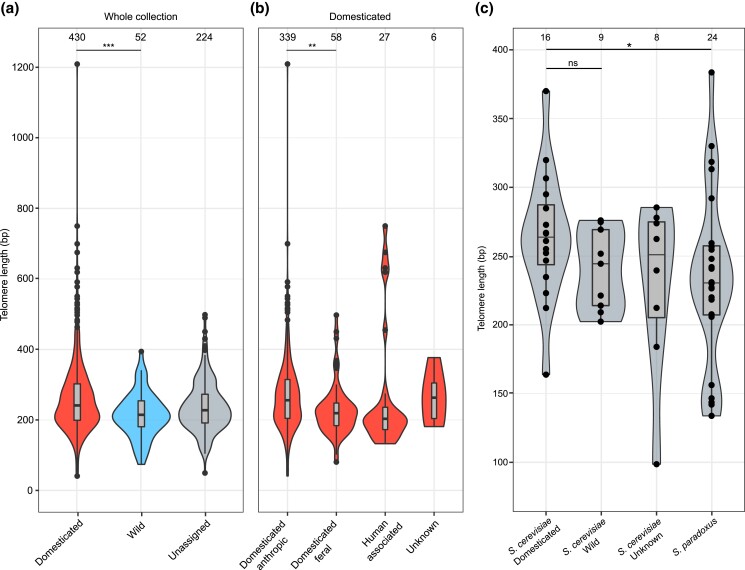
Since domestication represents a crucial life-style shift in S. cerevisiae (De Chiara et al. 2022), we investigated whether it shaped TL. Isolates were previously classified as wild (n = 52) or domesticated (n = 430) based on the dominant origin (>66%) in their clade (Peter et al. 2018; De Chiara et al. 2022). Isolates from clades that did not reach this threshold were classified as “Unassigned” (n = 224). Domesticated isolates had slightly longer telomeres than wild ones (median = 241 ± 111 bp and 215 ± 65 bp, respectively, 2-tailed Wilcoxon test, P = 0.0009), with a median difference of 26 bp (Fig. 3a and Supplementary Table 5). Consistently, TL was tendentially longer in domesticated clades than in wild ones (Fig. 2). This difference was significant even when using TL estimations obtained with a 20 bp cutoff (noncorrected TL = 95 ± 110 bp for domesticated, TL = 83 ± 24 bp for wild, P = 0.003; Supplementary Table 5), supporting the fact that the variation between the 2 groups is not due to differences in the presence of mismatches in telomeric sequences.
Fig. 3.
TL variation in domesticated and wild yeasts. a) Telomere length of domesticated, wild and unassigned isolates. Isolates classification is as in Fig. 2. b) Telomere length of domesticated isolates, divided in groups based on their substrate of isolation. “Domesticated-anthropic” are strains isolated from fermentation processes; “Domesticated-feral” are strains isolated from natural environments; “Human-associated” comprises strains isolated from the human body; “Unknown” comprises strains whose substrate of isolation is unknown. c) Telomere length of S. cerevisiae vs S. paradoxus strains from the SGRP. S. cerevisiae strains are further subdivided in groups based on their substrate of isolation. Box plots within in the violin plots are as in Fig. 2. Numbers on top represent the number of isolates in each group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = nonsignificant.
Within the domesticated group (n = 430), most isolates were indeed derived from fermentation-related processes in line with the dominant origin of the clade. We refer to these strains as “domesticated-anthropic” (n = 339), while others were isolated in nature and are defined here as “domesticated-feral” (n = 58). The remaining part of domesticated strains were either isolated from the human body (n = 27) or from unknown sources (n = 6; Supplementary Table 5). Feral strains have gone through domestication but have then been redispersed into a natural environment. We compared TL between the anthropic and feral groups and found that feral isolates had a shorter median TL than anthropic ones (domesticated-anthropic = 256 ± 108 bp, domesticated-feral = 219 ± 76 bp, 2-tailed Wilcoxon test, P = 0.001; Fig. 3b). This suggests that the long TL pattern is associated with ongoing domestication.
To further investigate the effects of domestication on TL, we compared the TL of S. cerevisiae to that of S. paradoxus, its closest related species which was never domesticated. We estimated the TL of S. cerevisiae (n = 33) and S. paradoxus (n = 24) strains from the SGRP collection using already available Sanger-sequencing data (Liti, Carter, et al. 2009). S. cerevisiae telomeres were slightly longer than their S. paradoxus counterparts with a median difference of 24 bp, although the comparison did not reach statistical significance (S. cerevisiae = 255 ± 49 bp, S. paradoxus = 231 ± 63 bp, 2-tailed Wilcoxon test, P = 0.18). The median difference in TL between the 2 species is comparable to what observed in the S. cerevisiae domesticated/wild dichotomy (Supplementary Fig. 10a). Therefore, we further examined this dataset by comparing the TL of SGRP S. cerevisiae strains isolated from fermentation-related environments (domesticated) vs the ones isolated in the wild. Telomeres were tendentially shorter in wild S. cerevisiae strains (264 ± 47 vs 244 ± 29 bp, respectively, 2-tailed Wilcoxon test, P = 0.17). Moreover, TL of domesticated S. cerevisiae strains was significantly higher than that of S. paradoxus ones (264 ± 47 vs 231 ± 63 bp, respectively, 2-tailed Wilcoxon test, P = 0.05; Fig. 3c). These results reinforce our previous observation that domestication might be associated with telomere lengthening.
S. paradoxus TL is known to be very different in its 3 continental subpopulations, with short telomeres in European isolates and very long ones in American isolates despite all of them being wild and isolated from similar ecological niches (Liti, Carter, et al. 2009; Liti, Haricharan, et al. 2009). While we observed a higher median TL in the American lineage, in agreement with previous findings, there was huge variation within both the S. paradoxus American and European lineages (Supplementary Fig. 10b and c). Therefore, the overall trend of S. paradoxus strains carrying shorter telomeres might also derive by a sampling bias toward European isolates (15/24) and the high variance in the S. paradoxus group suggests that genetic drift in geographic subpopulations can also result in large TL variation (Fig. 3c, Supplementary Fig. 10 and Supplementary Table 9).
We conclude that S. cerevisiae strains exhibit a broad distribution of TL patterns with medians that differ according to their life-styles: slightly longer for domesticated and shorter for wild strains. Remarkably, the domesticated pattern appears reversible, as exemplified in the feral isolates, showing that TL can change upon shifting life-style conditions and genetic drift.
Natural genetic variants underlie TL variation
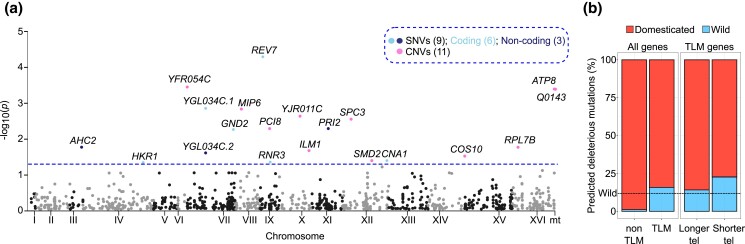
We searched for associations between common (MAF >5%) single nucleotide variants (SNV), copy number variants (CNV) and TL by conducting a GWAS across 555 euploid diploid strains. We identified 20 variants (false discovery rate (FDR) corrected α = 0.05), divided into 9 SNVs and 11 CNVs, including 2 CNVs encoded in the mitochondrial genome. Among the SNVs, 3 were in noncoding regions, while 6 were intragenic with 3 missense and 3 synonymous ones (Fig. 4a and Supplementary Table 10). Despite only one variant is in the intergenic region of a TLM gene previously identified in systematic genetic screens (AHC2;Askree et al. 2004; Gatbonton et al. 2006; Ungar et al. 2009), 5 others are part of telomere length regulation pathways or complexes (REV7, MIP6, PRI2, RPL7B, RNR3; Harari et al. 2020). The remaining 14 variants underlie associations with potential novel candidate TL regulators. The presence of intergenic hits suggests that part of TL regulation happens at the transcriptional level and these variants might act by regulating the expression of neighboring TLM genes.
Fig. 4.
TL genetic variants. a) Manhattan plot showing the position of the GWAS variants across the genome. 20 variants are beyond the genome-wide significance threshold (P < 0.05, dashed line). SNVs: single nucleotide variants; CNVs: copy number variants. Numbers in brackets denote the number of variants contained in that group. For simplicity, only the first 1,000 tested variants are shown in the plot. b) Fraction of TLM and non-TLM genes carrying predicted loss-of-function mutations in euploid diploid domesticated (n = 350) and wild (n = 49) isolates (total n = 399). Isolates classified as unassigned (n = 156) were not considered in this analysis. TLM genes are further subdivided into the ones conferring shorter vs longer telomeres when deleted. The dashed line indicates the fraction of wild strains in the 399 S. cerevisiae collection (12%).
In most nuclear CNVs, gene copy number amplification was associated with shorter TL, except for YFR054C, and in most cases TL variation was stratified in the Clinical/S. boulardii subclade and the Ecuadorean clade, which carry multiple gene copies (Supplementary Fig. 11a). Similarly, both mitochondrial CNVs inversely correlated with TL. Since accurate CN estimation of individual genes in the mitochondrial genome is difficult, we checked the relationship between TL and mtDNA copy number estimated using a set of 3 genes with nonproblematic mapping (De Chiara et al. 2020) and found an anticorrelation between the 2 variables (Supplementary Tables 12 and 13).
In most of the SNVs (except CNA1), the allele with the lowest frequency in the population (minor) was associated with longer telomeres, but no clades emerged as the main drivers of TL variation. The CNA1 minor allele was significantly enriched in domesticated clades (2-tailed X2 test, P = 7.68e−5) and associated with longer telomeres, therefore contributing to the TL difference between domesticated and wild isolates (Supplementary Fig. 11b and Supplementary Table 10). Overall, we estimated TL narrow-sense heritability to be 34%, with the significant GWAS variants explaining only 11% of the total phenotypic variance of the population. Therefore, we searched for candidate rare variants that could explain the rest of the TL variation. Rare genetic variants (MAF < 5%) evade GWAS detection, so we investigated their effect by selecting likely loss of function (LOF) variants in TLM genes (n = 383). TLM LOFs were significantly enriched in wild strains (2-tailed X2 test, P = 6.7e−12). Noteworthy, this enrichment was more pronounced in genes whose deletion causes telomere shortening (P = 7.7e−19) rather than lengthening (P = 0.001), in accordance with the trend of wild isolates to have shorter telomeres. This effect is specific to telomere length regulation, as LOFs in non-TLM genes (n = 5648) showed the opposite trend, being depleted in wild strains and enriched in domesticated ones (Fig. 4b and Supplementary Table 11). Some of the TLM LOFs were private to one or few clades and could contribute to explain the extreme TL observed in those lineages (Supplementary Discussion 6 and Supplementary Table 11).
In conclusion, we detected multiple genetic variants and LOF mutations that contribute to the TL variation observed in the S. cerevisiae collection segregating within specific clades.
Mitochondrial metabolism is associated with TL variation
We searched for associations between TL and other phenotypes. First, we measured mitochondrial volume and activity in the euploid diploid strains (n = 555) in either fermentative (YPD) or respiratory (YPEG) medium (Supplementary Table 12). Both mitochondrial activity and volume in YPD and YPEG were positively correlated (Pearson’s r = 0.48, P < 2.2e−16 for activity, r = 0.62, P < 2.2e−16 for volume, Supplementary Fig. 12a). While activity was higher in the glycerol-containing medium YPEG, in which fermentative metabolism is not active and respiration drives growth, volume was higher in YPD (2-tailed Wilcoxon test, P < 2.2e−16 for activity, P < 2.2e−16 for volume, Supplementary Fig. 12b and c).
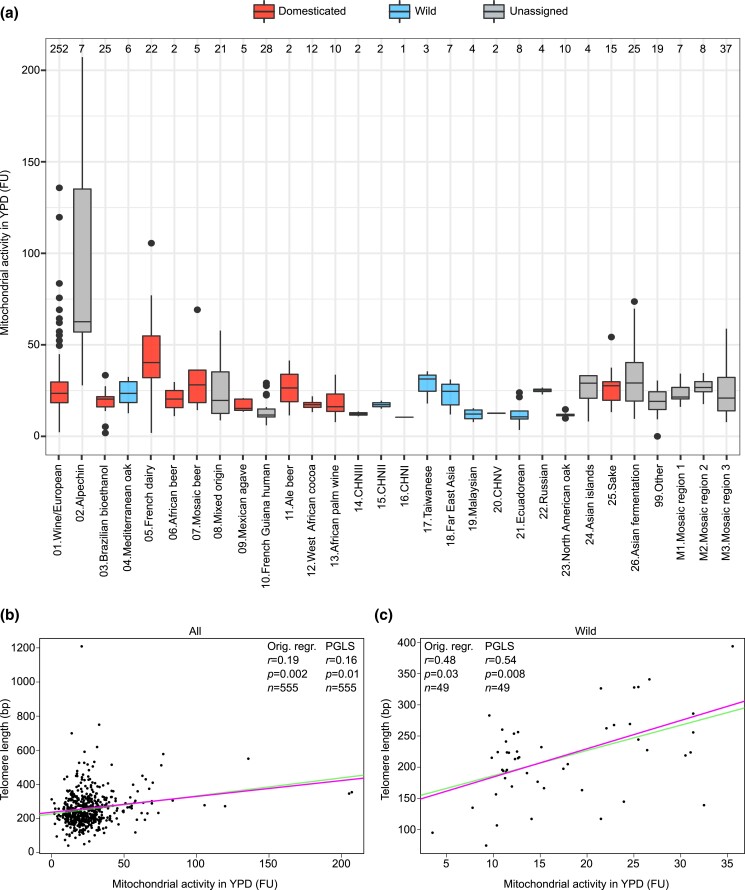
Mitochondrial activity and volume varied significantly among the clades (2-tailed Kruskal–Wallis test, P < 2.2e−16 for all the mitochondrial phenotypes; only mitochondrial activity in YPD is shown in Fig. 5a for simplicity). Mitochondrial activity in YPD was particularly high in the Alpechin clade (Fig. 5a). Wild isolates had lower mitochondrial activity and volume in both YPD and YPEG as compared to domesticated ones (2-tailed Wilcoxon test, P = 5.58e−7 and P = 2.93e−9 for mitochondrial volume and activity in YPD, respectively; P = 1.06e−14 and P < 2.2e−16 for mitochondrial volume and activity in YPEG, respectively; Supplementary Fig. 12d and e). We then examined the mitochondrial phenotypes of the feral strains and found they had a lower mitochondrial volume than anthropic ones (2-tailed Wilcoxon test, P = 0.0008 in YPD and P = 0.0008 in YPEG). Mitochondrial activity was also lower in feral strains but not significantly (2-tailed Wilcoxon test, P = 0.22 in YPD and P = 0.30 in YPEG).
Fig. 5.
TL and mitochondrial phenotypes. a) Mitochondrial activity in YPD of the phylogenetic lineages in the 555 S. cerevisiae collection. Isolates classification is as in Fig. 2. Box plots are as in Fig. 2. FU = fluorescence units. b, c) Associations between mitochondrial activity in YPD and telomere length in the 555 S. cerevisiae collection (left panel) and in the subset of wild isolates (n = 49; right panel). Each point represents a single isolate and the lines represent linear regression functions before (green) and after (pink) correction for phylogenetic signal. The association is significant when considering both the whole dataset (n = 555) and only the wild isolates. The other 3 mitochondrial phenotypes (mitochondrial activity and volume in YPD and YPEG) show similar trends but they are not shown for simplicity, while the copy number of mitochondrial DNA negatively correlates with telomere length (P < 0.05). Orig. regr = original regression.
There were positive correlations of TL with mitochondrial volume and activity in both YPD and YPEG (Pearson’s r = 0.16 and P = 0.02 for mitochondrial volume in YPEG, r = 0.18 and P = 0.006 for mitochondrial activity in YPEG, r = 0.13 and P = 0.08 for mitochondrial volume in YPD, r = 0.19 and P = 0.002 for mitochondrial activity in YPD). When switching to Spearman’s correlation, P-values were still significant, indicating that the association is robust (data not shown). Since domesticated and wild yeast isolates have different TL patterns, we also checked their associations with the mitochondrial phenotypes separately. The correlation coefficients of all mitochondrial phenotypes were higher when considering the wild isolates alone (Pearson’s r = 0.42 for mitochondrial volume in YPEG, r = 0.35 for mitochondrial activity in YPEG, r = 0.25 for mitochondrial volume in YPD, r = 0.48 for mitochondrial activity in YPD), although the P-value was only significant for the latter phenotype in this setting because of the reduced sample size (Fig. 5b and c, only mitochondrial activity in YPD is shown for simplicity, Supplementary Table 13). In domesticated strains we observed the opposite trend, with correlation coefficients close to 0, consistent with wild strains being the main drivers of the association between TL and mitochondria.
Closely related isolates might show more similar traits as a result of shared recent ancestry and this could bias the association analysis. We estimated a moderate phylogenetic signal for telomere length (λ = 0.54). Therefore, we reanalyzed our dataset by applying the PGLS method to correct for this bias. Telomere length and mitochondrial activity in YPD were still significantly correlated after correction (r = 0.16 and P = 0.01 for all, r = 0.54 and P = 0.009 for wild) and the linear model describing this association was only mildly affected (Fig. 5b and c). Upon correction, telomere length and mitochondrial copy number showed a significant anticorrelation that is mainly driven by the domesticated subgroup (r = 0.17 and P = 0.009 for all, r = 0.21 and P = 0.009 for domesticated).
Finally, we investigated the relationship between TL variation and a set of previously published phenotypes (n = 151) including growth in different carbon and nitrogen sources, in the presence of chemical compounds and environmental challenges, reproductive capacity and CLS under standard (SDC) and caloric-restricted conditions (Peter et al. 2018; De Chiara et al. 2022). No significant correlations were found between TL and other phenotypes except for growth yield in tryptophan when considering all the isolates or only the domesticated ones (Supplementary Table 13). This correlation was not anymore significant after PGLS correction.
Overall, these findings indicate that TL has only a limited impact on fitness traits, which is restricted to mitochondrial functions. The association between TL and mitochondrial metabolism is not spuriously driven by phylogenetic signal and it appears to be largely driven by wild isolates.
Discussion
By estimating the telomeric DNA length (TL) and ITS content of 706 S. cerevisiae isolates collected worldwide and associated with an extensive genomic and phenotyping dataset (Peter et al. 2018; De Chiara et al. 2020, 2022) we unveiled the species span of telomere length variation. We observed a wide spectrum of normally distributed TLs whose median value is close to what previously measured in the classical S288C laboratory strain. This variation is not explained by ploidy and instead is associated with several punctuated genetic variants.
We analyzed the association between TL and the ecological origin of the yeast isolates. Remarkably, we found that TL was shorter in wild strains than in domesticated ones, although the distributions within these 2 groups are broad. Noteworthy, within the domesticated group, the feral strains have shorter TL than anthropic ones, suggesting that the long TL pattern is associated with ongoing domestication while the return to wild-life is accompanied by TL shortening. The difference in TL between domesticated and wild strains further adds to the profound disparities in life-cycle and metabolism previously described, with wild strains growing better in stressful environments and uniformly having a more efficient sexual reproduction (De Chiara et al. 2022). Our findings, together with a recent report showing that wild mammals have shorter TL than domesticated ones (Le Pepke and Eisenberg 2021), unveil that domestication vs wild-life are important life-style traits that are associated with TL regulation.
These results raise the question of whether variations in TL underlie an adaptive strategy to different environments as proposed in various organisms (Young 2018; Casagrande and Hau 2019; Jacome Burbano and Gilson 2021). On one hand, a wild habitat imposes yeasts to cope with fluctuating environments and short telomeres could help survival through the transcriptional regulation of subtelomeric regions, which are enriched in stress response, nutrient utilization and cell wall composition genes (Stone and Pillus 1996; Wyrick et al. 1999; Ai et al. 2002; Robyr et al. 2002; Smith et al. 2011; Ye et al. 2014). In addition, short TL can lead to the release of transcription factors from telomeres, allowing them to bind and regulate their targets genome-wide (Maillet et al. 1996; Martin et al. 1999; Buck and Lieb 2006; Platt et al. 2013). This hypothesis agrees with the shorter telomere DNA length observed in feral strains, suggesting that the return to wild settings coincides with short telomeres. On the other hand, long telomeres could be favored in domesticated environments, where fermentative processes submit yeasts to previously unencountered stresses. Alternatively, TL may not be directly under selection but its variable pattern in domesticated/wild yeasts could be a corollary of the so-called “domestication syndrome” (De Chiara et al. 2022). A neutral scenario dictated by genetic drift could also explain the broad TL variability observed within the 2 classes of strains. For example, the highly diverged Asian lineages have different TL despite sharing the same life-style, habitat and some life-cycle traits, suggesting that TL was free to evolve after the split from their last common ancestor. The 706 S. cerevisiae collection is under-represented for wild isolates and a more extensive sampling of strains from natural environments will better elucidate the role of drift and/or selection in shaping TL. Further studies where fitness is measured in isogenic background with controlled TL and in culture conditions close to the wild and domestic environments are also needed to distinguish between these evolutionary scenarios.
Among a wide range of phenotypes tested, TL positively correlated with mitochondrial volume and activity while negatively correlated with mitochondrial DNA copy number. These results reinforce the view that telomeres and mitochondria are functionally connected (Nautiyal et al. 2002; Passos, Saretzki, Ahmed, et al. 2007; Passos, Saretzki, and Von Zglinicki 2007; Sahin et al. 2011; Qian et al. 2019; Robin et al. 2020). Instead, TL was generally not associated with growth in various conditions, in line with previous results showing that artificially shortened/elongated telomeres do not affect yeast fitness in laboratory conditions (Harari et al. 2017; Harari and Kupiec 2018a). Whether the association between mitochondria and telomeres in wild strains represents a fitness advantage is a possibility that warrants further investigation.
Overall, our analyses revealed extensive TL variation across a large population of S. cerevisiae isolates collected around the world, which can be explained, at least in part, by genetic variations, ecological origins and associations between telomere length and mitochondrial functions. The low narrow-sense heritability of TL in our population (34%) suggests that a substantial fraction of TL variation is affected by nonadditive genetic factors. Furthermore, isogenic S. cerevisiae backgrounds grown under defined environmental conditions (e.g. ethanol, acetic acid and caffeine) adjust TL through the modulation of TLM protein levels to a magnitude that overlaps the variation detected in the 1,011 collection, showing that this phenotype is highly plastic (Ungar et al. 2011; Kupiec and Weisman 2012; Harari et al. 2013; Romano et al. 2013). Future studies will quantify the role of genes, environment and their interactions in controlling telomere length in natural populations.
Supplementary Material
Acknowledgments
We thank Gilles Fischer for sharing TL estimations derived from Nanopore sequencing.
Contributor Information
Melania D’Angiolo, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France.
Jia-Xing Yue, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France; State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-sen University Cancer Center (SYSUCC), 651 Dongfeng Road East, China.
Matteo De Chiara, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France.
Benjamin P Barré, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France.
Marie-Josèphe Giraud Panis, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France.
Eric Gilson, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France; Department of Genetics, CHU, 06107 Nice, France.
Gianni Liti, Institute for Research on Cancer and Aging (IRCAN), Université Côte d’Azur, 28 Avenue de Valombrose, 06107 Nice, France.
Data Availability
The simulated reads of the designed genomes generated in this study are available at http://mdangiolo.ircan.org/sim_reads/. The phenotype data and other resources are available at: https://github.com/mdangiolo89/Different-telomere-length-patterns-between-domestic-and-wild-S.-cerevisiae-isolates.
Supplemental material available at GENETICS online.
Code availability
YeaISTY (Yeast ITS, Telomeres and Y′ elements estimator) and other custom scripts are available at: https://github.com/mdangiolo89/Different-telomere-length-patterns-between-domestic-and-wild-S.-cerevisiae-isolates.
Funding
We thank the French National Research Agency (ANR) LABEX SIGNALIFE ANR-11-LABX-0028-01 and the Fondation pour la Recherche Médicale (FDT201904008453) for supporting M.D. Ph.D. fellowship. This study was supported by ANR (ANR-18-CE12-0004, ANR-20-CE12-0020), Fondation pour la Recherche Médicale (EQU202003010413), CEFIPRA, Fondation ARC (No. PJA32020070002320) to GL. Work in EG laboratory is supported by grants from the Fondation ARC pour la recherche sur le cancer (Labelisation No. PGA20160203873) and the INSERM cross cutting program on aging (AGEMED).
Conflicts of interest
None declared.
Authors contributions
MD, JXY, and MDC designed and implemented YeaISTY; MD and MDC performed and analyzed GWAS; BB and MD performed and analyzed mitochondrial phenotyping; MD and MJGP performed teloblots; EG and GL conceived and supervised the project; MD, EG, and GL wrote the paper.
Literature cited
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XFS. Regulation of subtelomeric silencing during stress response. Mol Cell. 2002;10(6):1295–1305. doi: 10.1016/S1097-2765(02)00695-0. [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101(23):8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv A, Shay JW. Reflections on telomere dynamics and ageing-related diseases in humans. Philos Trans R Soc B Biol Sci. 2018;373(1741):20160436. doi: 10.1098/rstb.2016.0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel FP, Wei W, Tang M, Martinez-Ledesma E, Hu X, Amin SB, Akdemir KC, Seth S, Song X, Wang Q, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40(10):1–14. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom A, Simpson JT, Salinas F, Barre B, Parts L, Zia A, Nguyen Ba AN, Moses AM, Louis EJ, Mustonen V, et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol Biol Evol. 2014;31(4):872–888. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120(1):33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14(17):4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38(12):1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande S, Hau M. Telomere attrition: metabolic regulation and signalling function? Biol Lett. 2019;15(3):20180885. doi: 10.1098/rsbl.2018.0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Tanny RE, Seo B, Riccardi DD, Noble LM, Rockman MV, Alkema MJ, Braendle C, Kammenga JE, et al. The genetic basis of natural variation in caenorhabditis elegans telomere length. Genetics. 2016;204(1):371–383. doi: 10.1534/genetics.116.191148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara M, Barré BP, Persson K, Irizar A, Vischioni C, Khaiwal S, Stenberg S, Amadi OC, Žun G, Doberšek K, et al. Domestication reprogrammed the budding yeast life cycle. Nat Ecol Evol. 2022;6(4):448–460. doi: 10.1038/s41559-022-01671-9. [DOI] [PubMed] [Google Scholar]
- De Chiara M, Friedrich A, Barré B, Breitenbach M, Schacherer J, Liti G. Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC Biol. 2020;18(1):1–15. doi: 10.1186/s12915-020-00786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Mangino M, Aviv A, Spector T, Durbin R. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42(9):e75. doi: 10.1093/nar/gku181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Petit EJ, Brazier T, Finarelli JA, Hughes GM, Touzalin F, Puechmaille SJ, Teeling EC. Drivers of longitudinal telomere dynamics in a long-lived bat species, Myotis myotis. Mol Ecol. 2020;29(16):2963–2977. doi: 10.1111/mec.15395. [DOI] [PubMed] [Google Scholar]
- Fulcher N, Teubenbacher A, Kerdaffrec E, Farlow A, Nordborg M, Riha K. Genetic architecture of natural variation of telomere length in Arabidopsis thaliana. Genetics. 2014;199(2):625–635. doi: 10.1534/genetics.114.172163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulnečková J, Ševčíková T, Fajkus J, Lukešová A, Lukeš M, Vlček Č, Lang BF, Kim E, Eliáš M, Sýkorová E. A broad phylogenetic survey unveils the diversity and evolution of telomeres in eukaryotes. Genome Biol Evol. 2013;5(3):468–483. doi: 10.1093/gbe/evt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2(3):e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8(10):825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51(6):887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Grokhovsky SL, Il’icheva IA, Nechipurenko DY, Golovkin MV, Panchenko LA, Polozov RV, Nechipurenko YD. Sequence-specific ultrasonic cleavage of DNA. Biophys J. 2011;100(1):117–125. doi: 10.1016/j.bpj.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakobyan A, Nersisyan L, Arakelyan A. Quantitative trait association study for mean telomere length in the South Asian genomes. Bioinformatics. 2016;32(11):1697–1700. doi: 10.1093/bioinformatics/btw027. [DOI] [PubMed] [Google Scholar]
- Hansen MEB, Hunt SC, Stone RC, Horvath K, Herbig U, Ranciaro A, Hirbo J, Beggs W, Reiner AP, Wilson JG, et al. Shorter telomere length in Europeans than in Africans due to polygenetic adaptation. Hum Mol Genet. 2016;25(11):2324–2330. doi: 10.1093/hmg/ddw070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Gershon L, Alonso-Perez E, Klein S, Berneman Y, Choudhari K, Singh P, Sau S, Liefshitz B, Kupiec M. Telomeres and stress in yeast cells: when genes and environment interact. Fungal Biol. 2020;124(5):311–315. doi: 10.1016/j.funbio.2019.09.003. [DOI] [PubMed] [Google Scholar]
- Harari Y, Kupiec M. Genome-wide studies of telomere biology in budding yeast. Microb Cell. 2014;1(3):70–80. doi: 10.15698/mic2014.01.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Kupiec M. Do long telomeres affect cellular fitness? Curr Genet. 2018a;64(1):173–176. doi: 10.1007/s00294-017-0746-z. [DOI] [PubMed] [Google Scholar]
- Harari Y, Kupiec M. Mec1ATR is needed for extensive telomere elongation in response to ethanol in yeast. Curr Genet. 2018b;64(1):223–234. doi: 10.1007/s00294-017-0728-1. [DOI] [PubMed] [Google Scholar]
- Harari Y, Romano GH, Ungar L, Kupiec M. Nature vs nurture: interplay between the genetic control of telomere length and environmental factors. Cell Cycle. 2013;12(22):3465–3470. doi: 10.4161/cc.26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Zadok-Laviel S, Kupiec M. Long telomeres do not affect cellular fitness in yeast. mBio. 2017;8(4):e01314-17. doi: 10.1128/mBio.01314-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann MF, Winkler DW, Vleck CM. Longer telomeres associated with higher survival in birds. Biol Lett. 2005;1(2):212–214. doi: 10.1098/rsbl.2005.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger BJ, Blount JD, Boner W, Griffiths K, Metcalfe NB, Monaghan P. Telomere length in early life predicts lifespan. Proc Natl Acad Sci U S A. 2012;109(5):1743–1748. doi: 10.1073/pnas.1113306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28(4):593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome Burbano MS, Gilson E. The power of stress: the telo-hormesis hypothesis. Cells. 2021;10(5):1–21. doi: 10.3390/cells10051156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol Rev. 2014;38(2):144–171. doi: 10.1111/1574-6976.12054. [DOI] [PubMed] [Google Scholar]
- Kupiec M, Weisman R. TOR Links starvation responses to telomere length maintenance. Cell Cycle. 2012;11(12):2268–2271. doi: 10.4161/cc.20401. [DOI] [PubMed] [Google Scholar]
- Lee M, Napier CE, Yang SF, Arthur JW, Reddel RR, Pickett HA. Comparative analysis of whole genome sequencing-based telomere length measurement techniques. Methods. 2017;114:4–15. doi: 10.1016/j.ymeth.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Le Pepke M, Eisenberg DTA. On the comparative biology of mammalian telomeres: telomere length co-evolves with body mass, lifespan and cancer risk. Mol Ecol. 2021. 10.1111/mec.15870. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269(5230):1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- Liti G, Haricharan S, Cubillos FA, Tierney AL, Sharp S, Bertuch AA, Parts L, Bailes E, Louis EJ. Segregating YKU80 and TLC1 alleles underlying natural variation in telomere properties in wild yeast. PLoS Genet. 2009;5(9):e1000659. doi: 10.1371/journal.pgen.1000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Tsai IJ, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458(7236):337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11(16):1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- Louis EJ, Haber JE. The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics. 1992;131(3):559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73(2):347–360. doi: 10.1016/0092-8674(93)90234-H. [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 1996;10(14):1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Mangino M, Christiansen L, Stone R, Hunt SC, Horvath K, Eisenberg DTA, Kimura M, Petersen I, Kark JD, Herbig U, et al. DCAF4, A novel gene associated with leucocyte telomere length. J Med Genet. 2015;52(3):157–162. doi: 10.1136/jmedgenet-2014-102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18(12):3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97(5):621–633. doi: 10.1016/S0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Monaghan P. Telomeres and life histories: the long and the short of it. Ann N Y Acad Sci. 2010;1206(1):130–142. doi: 10.1111/j.1749-6632.2010.05705.x. [DOI] [PubMed] [Google Scholar]
- Nautiyal S, DeRisi JL, Blackburn EH. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99(14):9316–9321. doi: 10.1073/pnas.142162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersisyan L, Arakelyan A. Computel: computation of mean telomere length from whole-genome next-generation sequencing data. PLoS One. 2015;10(4):e0125201. doi: 10.1371/journal.pone.0125201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersisyan L, Nikoghosyan M; Genome of the Netherlands consortium, Arakelyan A. WGS-based telomere length analysis in Dutch family trios implicates stronger maternal inheritance and a role for RRM1 gene. Sci Rep. 2019;9(1):18758. doi: 10.1038/s41598-019-55109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell S, Yue JX, Saada OA, Agier N, Caradec C, Cokelaer T, De Chiara M, Delmas S, Dutreux F, Fournier T, et al. 142 telomere-to-telomere assemblies reveal the genome structural landscape in Saccharomyces cerevisiae. bioRxiv 510633. 10.1101/2022.10.04.510633, 4 October 2022, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41(1):181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, et al. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5(5):1138–1151. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Von Zglinicki T. DNA damage in telomeres and mitochondria during cellular senescence: is there a connection? Nucleic Acids Res. 2007;35(22):7505–7513. doi: 10.1093/nar/gkm893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue JX, Pflieger D, Bergström A, Sigwalt A, Barre B, Freel K, Llored A, et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556(7701):339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt JM, Ryvkin P, Wanat JJ, Donahue G, Ricketts MD, Barrett SP, Waters HJ, Song S, Chavez A, Abdallah KO, et al. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev. 2013;27(12):1406–1420. doi: 10.1101/gad.218776.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poptsova MS, Il’Icheva IA, Nechipurenko DY, Panchenko LA, Khodikov MV, Oparina NY, Polozov RV, Nechipurenko YD, Grokhovsky SL. Non-random DNA fragmentation in next-generation sequencing. Sci Rep. 2014;4:1–6. doi: 10.1038/srep04532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Herzog M, Selivanova A, Wang S, Zhu J, Klein-Lavi S, Gordon M, Meirman R, Millan-Zambrano G, Ayestaran I, et al. Genome architecture and stability in the S. cerevisiae knockout collection. Nature. 2019;573(7774):416–420. doi: 10.1038/s41586-019-1549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Kumar N, Roginskaya V, Fouquerel E, Opresko PL, Shiva S, Watkins SC, Kolodieznyi D, Bruchez MP, Van Houten B. Chemoptogenetic damage to mitochondria causes rapid telomere dysfunction. Proc Natl Acad Sci U S A. 2019;116(37):18435–18444. doi: 10.1073/pnas.1910574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin JD, Jacome Burbano MS, Peng H, Croce O, Thomas JL, Laberthonniere C, Renault V, Lototska L, Pousse M, Tessier F, et al. Mitochondrial function in skeletal myofibers is controlled by a TRF2-SIRT3 axis over lifetime. Aging Cell. 2020;19(3):1–16. doi: 10.1111/acel.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M, Hall B. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109(4):437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe’er D, Ruppin E, et al. Environmental stresses disrupt telomere length homeostasis. PLoS Genet. 2013;9(9):e1003721. doi: 10.1371/journal.pgen.1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Miller LR, Kreisberg R, Vazquez L, Wan Y, Aitchison JD. Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing. Mol Syst Biol. 2011;7(1):455. doi: 10.1038/msb.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Pillus L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol. 1996;135(3):571–583. doi: 10.1083/jcb.135.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattini L, Tellini N, Mozzachiodi S, D’Angiolo M, Loeillet S, Nicolas A, Liti G. Accurate tracking of the mutational landscape of diploid hybrid genomes. Mol Biol Evol. 2019;36(12):2861–2877. doi: 10.1093/molbev/msz177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub MA, Conomos MP, Keener R, Iyer KR, Joshua S, Yanek LR, Lane J, Miller-fleming TW, Brody JA, Mchugh CP, et al. Novel genetic determinants of telomere length from a trans-ethnic analysis of 109.122 whole genome sequences in TOPMed. bioRxiv 749010. 10.1101/749010, 7 December 2020, preprint: not peer reviewed. [DOI]
- Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13(5):535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(12):8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar L, Harari Y, Toren A, Kupiec M. Tor complex 1 controls telomere length by affecting the level of Ku. Curr Biol. 2011;21(24):2115–2120. doi: 10.1016/j.cub.2011.11.024. [DOI] [PubMed] [Google Scholar]
- Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, Kupiec M. A genome-wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Res. 2009;37(12):3840–3849. doi: 10.1093/nar/gkp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239(94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191(4):1073–1105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FCP, Jennings EG, Causton HC, Shore D, Grustein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402(6760):418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Ye J, Renault VM, Jamet K, Gilson E. Transcriptional outcome of telomere signalling. Nat Rev Genet. 2014;15(7):491–503. doi: 10.1038/nrg3743. [DOI] [PubMed] [Google Scholar]
- Young AJ. The role of telomeres in the mechanisms and evolution of life-history trade-offs and ageing. Philos Trans R Soc B Biol Sci. 2018;373(1741):20160452. doi: 10.1098/rstb.2016.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Cosentino Lagomarsino M, et al. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet. 2017;49(6):913–924. doi: 10.1038/ng.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansooorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci U S A. 1997;94(14):7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The simulated reads of the designed genomes generated in this study are available at http://mdangiolo.ircan.org/sim_reads/. The phenotype data and other resources are available at: https://github.com/mdangiolo89/Different-telomere-length-patterns-between-domestic-and-wild-S.-cerevisiae-isolates.
Supplemental material available at GENETICS online.