Abstract
Background
Hybrid immunity is associated with more durable protection against coronavirus disease 2019 (COVID-19). We describe the antibody responses following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in vaccinated and unvaccinated individuals.
Methods
The 55 vaccine arm COVID-19 cases diagnosed during the blinded phase of the Coronavirus Efficacy trial were matched with 55 placebo arm COVID-19 cases. Pseudovirus neutralizing antibody (nAb) activity to the ancestral strain and binding antibody (bAb) responses to nucleocapsid and spike antigens (ancestral and variants of concern [VOCs]) were assessed on disease day 1 (DD1) and 28 days later (DD29).
Results
The primary analysis set was 46 vaccine cases and 49 placebo cases with COVID-19 at least 57 days post–first dose. For vaccine group cases, there was a 1.88-fold rise in ancestral antispike bAbs 1 month post–disease onset, although 47% had no increase. The vaccine-to-placebo geometric mean ratios for DD29 antispike and antinucleocapsid bAbs were 6.9 and 0.04, respectively. DD29 mean bAb levels were higher for vaccine vs placebo cases for all VOCs. DD1 nasal viral load positively correlated with bAb levels in the vaccine group.
Conclusions
Following COVID-19, vaccinated participants had higher levels and greater breadth of antispike bAbs and higher nAb titers than unvaccinated participants. These were largely attributable to the primary immunization series.
Keywords: COVID-19, antibodies, boost, immunogenicity, prime, seroprevalence, vaccines
Following disease, the previously vaccinated have a more potent and broader response to spike compared to unvaccinated disease cases. Roughly half of the vaccinated cases show minimal immune response to disease.
The immunogenicity and efficacy of the primary regimens of coronavirus disease 2019 (COVID-19) vaccines have been well characterized, with multiple vaccine platforms inducing high levels of antibodies (Abs) and protection [1–3]. Multiple lines of evidence have supported Ab response to vaccination as a correlate of protective efficacy against COVID-19, enabling authorization of modified vaccines based on Ab responses [4–6]. Recent studies characterized the Ab response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection postvaccination demonstrating a potent and broad serologic response [7–13]. Most studies evaluating immune response post–breakthrough infection enrolled individuals during convalescence with a variable time from symptom onset to sample collection. Our blinded placebo-controlled study specified sampling at symptom onset and 28 days later, in addition to prevaccination, postvaccination, and before study unblinding, which occurred upon regulatory authorization of the vaccine. This uniformity of sampling and prospective follow-up allows for a standardized evaluation of Ab kinetics before and throughout the disease process. In this analysis, we evaluated neutralization Ab titers and binding Ab levels to ancestral and variant spike (S) and nucleocapsid (N) antigens in participants who acquired COVID-19 during the blinded period of the mRNA-1273 phase 3 efficacy trial, and who were previously immunized with mRNA-1273 vaccine or placebo.
METHODS
Study Design and Population
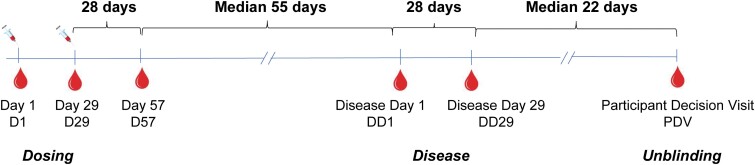
Between July 27 and October 23, 2020, the Coronavirus Efficacy (COVE) trial (NCT04470427; ClinicalTrials.gov) enrolled 30 420 adults ≥18 years of age at appreciable risk of SARS-CoV-2 infection and/or high risk of severe COVID-19 in the United States. Participants were randomized 1:1 to two 100-μg doses of mRNA-1273 vaccine or saline placebo, given at day 1 (baseline) and day 29. Participant characteristics, study procedures, and primary efficacy results are described elsewhere [2, 14]. COVID-19 diagnosis was based on symptom-prompted polymerase chain reaction testing [2]. Blood was collected for serology in all participants prevaccination on day 1, day 29, day 57, and at the participant decision visit (PDV), when participants were informed about their blinded randomization assignment. For participants diagnosed with COVID-19, blood was also collected on the first day of symptoms (disease day 1 [DD1]) and 28 days later (DD29). During the blinded phase of COVE, a total of 55 vaccinated and 744 placebo participants in the per-protocol set were diagnosed with COVID-19 at least 14 days post–dose 2. The 55 vaccinated COVID-19 cases were matched on age and sex with 55 placebo group COVID-19 cases and constitute the analysis set for this report. Figure 1 provides a schematic representation of the sampling time points for the 110 COVID-19 cases. The analysis is based on data through the completion of the blinded phase of the study with a data cutoff date of March 26, 2021, and focuses on post-D57 diagnoses.
Figure 1.
Schematic representation of the sampling time points for the 110 COVID-19 cases in relation to the dosing, disease, and unblinding dates. Abbreviation: COVID-19, coronavirus disease 2019.
Patient Consent
The central institutional review board approved the protocol and the consent forms. All participants provided written informed consent before enrollment in the COVE trial.
Laboratory Assays
On DD1, DD29, and PDV, serum Ab levels were measured using 3 different assays: (1) a Meso Scale Discovery (MSD) 4-plex assay [4], which measures binding immunoglobulin G (IgG) to the ancestral SARS-CoV-2 S (S-2P version), receptor binding domain (RBD), and N antigens with a readout of binding Ab units (BAU)/mL; for S-2P the upper limit of quantitation was determined to be 10 155.95 BAU/mL; (2) an MSD 10-plex assay [15] (Panel 25 IgG Kit), which measures binding IgG to S antigen (S-2P version) from 10 SARS-CoV-2 strains (ancestral, Alpha, Beta, Delta, 5 versions of Omicron [BA.1, BA.2, BA.3, BA.1 + R346K, BA.1 + L452R], and France-IHU [B.1.640.2]) with a readout of area under the titration curve (AUC); and (3) the Monogram PhenoSense pseudovirus D614G neutralization assay [16] with readouts for number of virus particles at 50% (80%) inhibition (ID50 or ID80) titers in international units (IU) per mL. From the previous immune correlates analysis [4], we utilized binding antibodies (bAbs) and neutralizing antibodies (nAbs) from 38 vaccinated COVID-19 cases and 32 placebo group COVID-19 cases on D1, D29, and D57.
Viral Sequence Analysis
The amino acid sequences for the 10 S antigens used in the 10-plex assay were multiply aligned with the mRNA-1273 vaccine insert sequence using MAFFT [17]. We calculated the Hamming distances between each of the 10-plex S sequences and the mRNA-1273 vaccine insert sequence.
Statistical Analysis
Geometric mean Ab levels, geometric mean ratios, and confidence intervals were calculated on the log10 scale and then back-transformed to original units. Linear regression was used to assess the associations between viral load, time interval between D57 and DD1, baseline demographics, and antibody levels. The concordance correlation coefficient (CCC) was used to assess agreement between the predicted and actual Ab on DD1. Magnitude-breadth (MB) curves [18] were used quantify Ab response across the VOCs and compared using the Wilcoxon rank-sum test. Two-sided P values <.05 were considered statistically significant. There was no adjustment for multiplicity. Analyses were done in R, version 4.1.3.
RESULTS
Fifty-four of the 55 vaccinated COVID-19 cases and 54 of the placebo group COVID-19 cases had Ab levels measured. A total of 46 of the 54 vaccine arm cases and 49 of the 54 placebo arm cases were diagnosed with COVID-19 after day 57, and these individuals are the focus of the tables and figures below. Supplementary Table 1 shows the number of participants with Ab data at each time point by assay. COVID-19 cases that occurred between days 43 and 57 are reported in Supplementary Figure 1. The baseline and clinical characteristics of the 108 COVID-19 cases (54 vaccine arm, 54 placebo arm) were well balanced and reflective of the overall COVE study population (Supplementary Table 2) [2].
Kinetics of 4-Plex Binding Ab Level
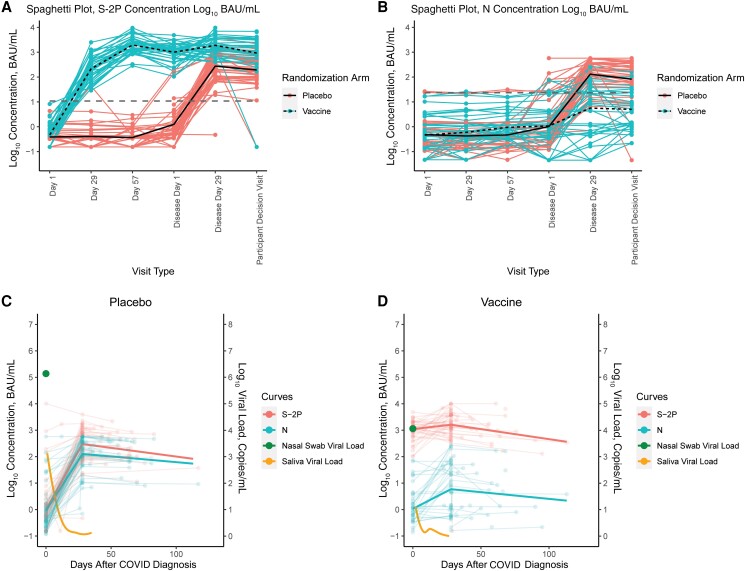
A robust bAb anti-S response to the 2 doses of vaccine was followed by an approximate halving of the bAb levels over a median of 55 days between D57 and DD1 (DD1/D57 geometric mean ratio [GMR], 0.52; 95% CI, 0.36–0.74) (Figure 2A). The bAb level on DD1 was similar to a prediction based on the decay from the D57 value, where decay was estimated from a separate longitudinal immunogenicity study of vaccinated and uninfected individuals (CCC, 0.72) (Supplementary Figure 2) [19]. Following a diagnosis of COVID-19, there was a modest boost in anti-S GM level 28 days after symptom onset (DD29/DD1 GMR, 1.88; 95% CI, 1.28–2.78), followed by an approximate halving of the anti-S GM level over a median of 21 days between DD29 and PDV (PDV/DD29 GMR, 0.49; 95% CI, 0.24–0.98) (Table 1). Interestingly, 18 of the 38 (47%) vaccinated COVID-19 cases with complete data showed no increase in S-2P anti-S bAb levels between DD1 and DD29. In contrast, 47 of 49 (96%) of the placebo group COVID-19 cases with complete data showed an increase in anti-S bAb to a mean level of 275 (95% CI, 177–428) binding bAb units (BAU)/mL at 29 days postinfection, similar to the level seen in the vaccine arm after 1 dose of mRNA-1273 of 214 (95% CI, 152–300) BAU/mL. The anti-S bAb levels declined by about a third between DD29 and PDV (PDV/DD29 GMR, 0.70; 95% CI, 0.35–1.41) in the placebo group. At DD29, the vaccine group anti-S bAb levels were 6.85-fold greater (95% CI, 4.03–11.67) than the placebo group, and 4.81-fold greater (95% CI, 2.10–10.99) at PDV. The DD29 anti-S bAb level following infection was less variable in the vaccine group than in the placebo group (standard deviation log10 bAb level, 0.40 for the vaccine group vs 0.69 for the placebo group).
Figure 2.
Four-plex S-2P and N bAb responses over the 6 different visits. A, Four-plex S-2P bAb concentrations in (coral, solid line) placebo arm COVID-19 cases and (turquoise, dashed line) vaccine arm COVID-19 cases by visit day. B, Anti-N bAb concentrations in (coral, solid line) placebo arm cases and (turquoise, dashed line) vaccine arm cases by visit day. C, Mean anti-S-2P and N bAb concentrations for (left panel) placebo arm and (right panel) vaccine arm COVID-19 cases at disease day 1, disease day 29, and the participant decision visit, along with the mean viral load at disease day 1 and over the 28 days of convalescence. Dashed gray lines in (A) and (B) are the positivity cutoff. Abbreviations: bAb, binding antibody; COVID-19, coronavirus disease 2019.
Table 1.
Geometric Mean of the 4-plex S-2P (A) and N (B) Binding Antibody Levels (Means and 95% CIs) Over the 6 Different Visits
| A | |||||
|---|---|---|---|---|---|
| Visit Type | GM Placebo (95% CI) | Change Ratio Placebo (95% CI) | GM Vaccine (95% CI) | Change Ratio Vaccine (95% CI) | GM Ratio (95% CI) |
| D1 | 0.39 (0.29–0.52) |
NA (NA–NA) |
0.47 (0.34–0.64) |
NA (NA–NA) |
1.21 (0.77–1.88) |
| D29 | 0.41 (0.27–0.62) |
1.05 (0.62–1.79) |
213.69 (152.2–300.01) |
458.38 (287.03–732) |
526.05 (302.35–915.27) |
| D57 | 0.37 (0.27–0.5) |
0.9 (0.53–1.54) |
1941.4 (1515.05–2487.74) |
9.09 (5.92–13.94) |
5292.59 (3539.67–7913.58) |
| DD1 | 1.25 (0.67–2.33) |
3.39 (1.67–6.9) |
1001.77 (772.71–1298.73) |
0.52 (0.36–0.74) |
804.44 (403.51–1603.73) |
| DD29 | 275.19 (176.83–428.27) |
220.99 (101.65–480.44) |
1886.1 (1424.23–2497.75) |
1.88 (1.28–2.78) |
6.85 (4.03–11.67) |
| PDV | 192.13 (113.08–326.44) |
0.7 (0.35–1.41) |
924.11 (499.35–1710.19) |
0.49 (0.24–0.98) |
4.81 (2.1–10.99) |
| B | |||||
| D1 | 0.48 (0.3–0.77) |
NA (NA–NA) |
0.46 (0.27–0.78) |
NA (NA–NA) |
0.96 (0.47–1.97) |
| D29 | 0.43 (0.27–0.68) |
0.89 (0.45–1.75) |
0.6 (0.37–0.97) |
1.29 (0.62–2.68) |
1.4 (0.7–2.8) |
| D57 | 0.47 (0.29–0.76) |
1.1 (0.55–2.19) |
0.95 (0.65–1.38) |
1.58 (0.84–2.97) |
2.02 (1.08–3.78) |
| DD1 | 1.02 (0.58–1.79) |
2.18 (1.03–4.62) |
1.08 (0.61–1.91) |
1.14 (0.57–2.29) |
1.06 (0.47–2.38) |
| DD29 | 130.83 (85.71–199.69) |
128.28 (62.99–261.24) |
5.65 (2.57–12.39) |
5.24 (1.95–14.09) |
0.04 (0.02–0.11) |
| PDV | 81.98 (44.75–150.19) |
0.63 (0.3–1.33) |
4.99 (2.64–9.44) |
0.88 (0.32–2.47) |
0.06 (0.02–0.15) |
Abbreviations: DD, disease day; GM, geometric mean; PDV, participant decision visit.
We next examined anti-N bAb levels. Both study groups had low anti-N bAb levels between D1 and DD1 (Figure 2B). At DD29, there was an anti-N bAb response to infection in the placebo group COVID-19 cases that was largely absent in the vaccine group; the anti-N bAb levels were 130.8 BAU/mL and 5.7 BAU/mL, respectively, for a ratio of 0.04 (95% CI, 0.02–0.11). The anti-N bAbs were detectable in 83.7% of placebo group COVID-19 cases on DD29 and in 79.5% at PDV. In contrast, only 15 of the 41 vaccinated COVID-19 cases (37%) were anti-N seropositive on DD29, and 10 of 35 (29%) were seropositive at PDV. The DD29 anti-N bAb level following infection was more variable in the vaccine group (standard deviation in log10 bAb level, 1.11 for the vaccine group vs 0.66 for the placebo group).
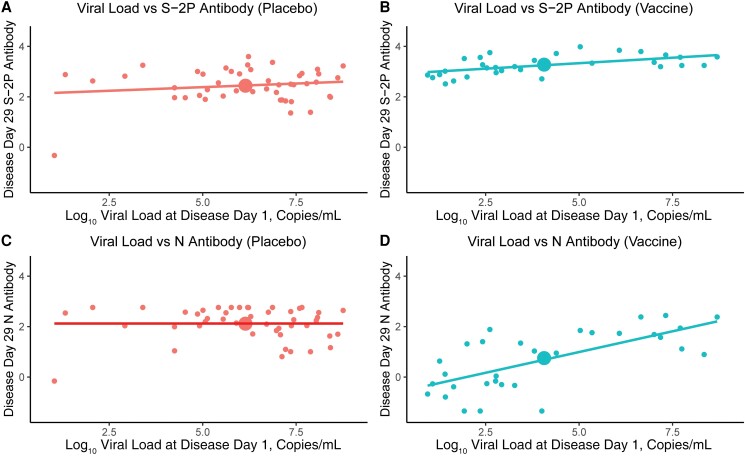
Viral load levels and kinetics were markedly different between arms. The mean nasal viral loads at DD1 were 6.04 log10 copies/mL (placebo group) vs 4.06 log10 copies/mL (vaccine group); the mean saliva viral load AUCs were 22.13 log10 copies/mL (placebo group) vs 5.61 log10 copies/mL (vaccine group) (Figure 2C, Figure 3). There was a positive correlation between DD1 nasal viral load and anti-S and anti-N bAb levels at DD29 (P = .001 and .00002, respectively) in the vaccine group. However, the magnitude of the association differed between the 2 antigens: a 1-log increment in DD1 viral load was associated with a 0.09 (95% CI, 0.04–0.14)-log10 BAU/mL increase in DD29 anti-S bAb levels, compared with a 0.33 (95% CI, 0.19–0.46)-log10 BAU/mL increase in DD29 anti-N bAb levels. We found no correlation between viral load and DD29 anti-S and anti-N bAb levels (P = .29 and 1 for S-2P and N, respectively) in the placebo group. Longer intervals between dose 2 (D29) and infection (DD1) were associated with higher viral loads (P = .02) and higher anti-S (P = .01) and anti-N (P = .003) bAb levels on DD29 in the vaccine group. However, in a multivariate analysis, the effect of interval since the second dose of vaccination was nonsignificant when controlling for viral load, indicating that viral load is the dominant correlate of the Ab level (Supplementary Table 3). Among vaccinees, the response to vaccination as measured by day 57 anti-S bAb level was not a significant predictor of the DD29 anti-N bAb level (P = .12). For both arms, body mass index, race/ethnicity, and age were not associated with either anti-S or anti-N bAb level at DD29. On DD29, female sex was associated with lower anti-S and anti-N bAb levels in the placebo arm and with anti-N bAb levels in the vaccine arm (all P < .05).
Figure 3.
Four-plex binding antibody concentration on disease day 29 for (A, B) S-2P and (C, D) N in the (B, D) vaccine and (A, C) placebo groups by disease day 1 viral load. The large dot in each panel denotes the mean response.
Binding Ab Levels to Variants of Concern
Across the 10 S antigens, there was a modest increase in bAb response after COVID-19 diagnosis in the vaccine group, a larger increase following COVID-19 diagnosis in the placebo group, and a uniformly higher mean anti-S bAb level at DD29 for the vaccine group COVID-19 cases (Supplementary Figure 3, Supplementary Table 4). bAb decay between DD29 and PDV was similar in the 2 arms. At DD29, all the vaccinated COVID-19 cases had geometric mean bAb levels >3 log10 AUC for all 10 antigens, while only 79.8% of the placebo group cases did (Supplementary Figure 4). Overall, the vaccinated COVID-19 cases had substantially greater postinfection bAb breadth (P < .001).
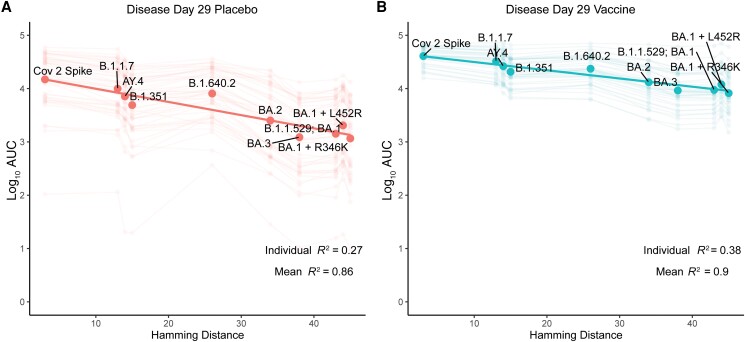
The variability in bAb levels to the VOCs was largely explained by the Hamming distance between the vaccine strain and the VOCs (Figure 4). In each group, there was a strong linear relationship between the DD29 anti-S bAb level and Hamming distance, with 90% and 86% of the variability in vaccine and placebo group mean bAb levels across antigens explained by Hamming distances, respectively. For the ancestral strain (Hamming distance of 0), the bAb level was 4.65 log10 AUC for vaccinated COVID-19 cases compared with 4.24 log10 AUC for placebo group COVID-19 cases. At a Hamming distance of 50, the analogous levels were 3.88 and 3.01, respectively, indicating relatively greater loss of bAb level for placebo cases for variants far distant from the ancestral strain (Figure 4).
Figure 4.
Relationship between the Hamming distance for spike from SARS-CoV-2 variants causing COVID-19 cases (vs spike from the mRNA-1273 vaccine SARS-CoV-2 strain) in (A) the placebo or (B) vaccine arm and binding antibody level (assessed by the 10-plex assay, see Supplementary Figure 4) at disease day 29. Each large dot is a mean value over all participants, each faded dot/faded line is from an individual participant. Abbreviations: AUC, area under the receiver operating curve; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
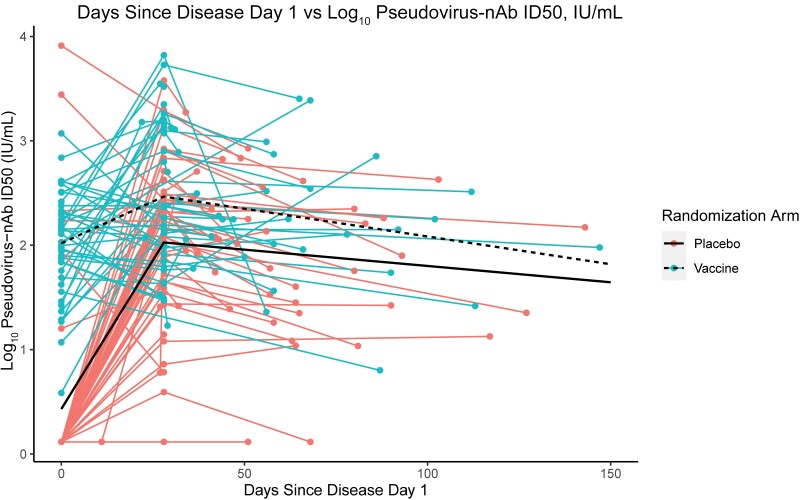
Neutralizing Antibody Response
The nAb kinetics on DD1, DD29, and PDV against the ancestral strain were similar to the anti-S bAb kinetics (Table 2, Figure 5). Between DD1 and DD29, nAb titers increased in 89.8% of placebo group COVID-19 cases and in 51.3% of vaccinated COVID-19 cases. On DD29, the nAb titer was 2.73-fold higher (95% CI, 1.53–4.88) than on DD1 for vaccinated COVID-19 cases. The rate of decline in nAbs between DD29 and PDV was similar in the 2 arms (vaccine: GMR PDV/DD29, 0.65; 95% CI, 0.33–1.30; placebo: GMR PDV/DD29, 0.54; 95% CI, 0.27–1.06). For the vaccine group, the predicted and actual nAb titers on DD1 were similar with no evidence of an anamnestic response (CCC, 0.66) (Supplementary Figure 2).
Table 2.
Postinfection Neutralizing Antibody Responses, Provided as Summary Statistics of the Monogram Phenosense D614G Pseudovirus Neutralization Assay in International Units/mL at DD1, DD29, and PDV, by Randomization Arm
| Visit | GMT Placebo | Change Ratio Placebo | GMT Vaccine | Change Ratio Vaccine | GMT Ratio |
|---|---|---|---|---|---|
| DD1 | 2.79 (1.58–4.91) |
NA (NA–NA) |
103.73 (72.98–147.43) |
NA (NA–NA) |
37.24 (18.91–73.34) |
| DD29 | 121.73 (76.85–192.8) |
43.71 (20.86–91.59) |
283.11 (180.07–445.1) | 2.73 (1.53–4.88) |
2.33 (1.21–4.47) |
| PDV | 65.66 (40.3–106.98) |
0.54 (0.27–1.06) |
184.45 (111.07–306.3) |
0.65 (0.33–1.3) |
2.81 (1.38–5.74) |
Abbreviations: DD, disease day; GM, geometric mean; PDV, participant decision visit.
Figure 5.
Pseudovirus neutralizing antibody ID50 titers for the (coral) placebo and (turquoise) vaccine arms by day post–disease onset.
DISCUSSION
The kinetics of the antibody response to symptomatic SARS-CoV-2 infection differ markedly by prior mRNA-1273 vaccination. The S-specific bAb level was 6.9-fold larger 28 days post–symptom onset in vaccinated vs placebo COVID-19 cases. Vaccinees had a greater breadth of S-specific antibody response against 10 VOCs, especially for VOCs with greater Hamming distance from the ancestral strain. The DD29 anti-N bAb levels were much lower and more variable in the vaccine group. The decay in DD29 antibody levels a median of 22 days later was similar for both arms. Overall, 47% of the vaccine cases showed no increase in anti-S bAbs, and 60% remained N-seronegative following infection. This modest immune response to infection in vaccine disease cases is potentially explained by the powerful effects of vaccination observed on viral replication upon infection (Table 2) [20].
SARS-CoV-2 infection can be considered as an uncontrolled first or third immunizing event for the placebo and vaccine arms, respectively. In the placebo arm, “dose” or viral load in the upper airways at onset of symptoms was 100-fold higher than in the vaccine arm [20] and resulted in similar antibody responses throughout the range of viral load. Interestingly, the anti-S response to infection was similar to 1 dose of mRNA. In contrast, the vaccine arm had 100-fold less nasal viral load “dose” at onset of symptoms, and anti-N antibody levels correlated with viral load. The interval from second dose to third immunizing event was independently associated with a higher anti-N bAb level in vaccinated COVID-19 cases. This association was not statistically significant in a multivariate analysis that controlled for viral load, suggesting that it is the viral replication rather than interval from second dose to third immunizing event that impacts N-response. In other studies, with mRNA vaccine boosting following infection, shorter intervals from infection to boost diminished B-cell responses [21]. A booster dose of 50 µg of mRNA-1273 at 6–8 months post–primary immunization series resulted in nAb titers at day 28 postboost that were 1.7-fold higher than those at 28 days post–second dose [22]. We observed a 2.7-fold increase in nAb titers 28 days postinfection compared with DD1 in the vaccine group.
In previous work [23], we reported that using the qualitative Roche Elecsys Anti-SARS-Cov immunoassay, the anti-N seroprevalence at the unblinding visit (PDV, a median of 53 days postdisease) was 40% for 52 vaccinated COVID-19 disease cases and 93% for 648 placebo group COVID-19 disease cases, for COVID-19 cases accrued during the blinded period of COVE. This analysis demonstrates that the absence of N Abs at the PDV was not attributable to transient or low-level Abs, but rather to a low N seroconversion rate of 37% 28 days post–symptom onset. This reinforces that vaccination status should be considered when interpreting seroprevalence and seropositivity data based solely on anti-N antibody testing.
Our analyses showed a strong and intriguing linear relationship between bAb level and Hamming distances for multiple variants of concern, with lower mean antibody levels for more distant VOCs and 86% and 90% of variation explained for the placebo and vaccine cases. A similar strong result has been demonstrated between Hamming distance and vaccine efficacy on clinical disease using a meta-analysis of multiple clinical studies [24]. Taken together, these data support the utility of Hamming distance as a tool to make predictions of antibody cross-reactivity against future variants. Hamming distance plots may complement antigenic cartography [25, 26] as a means of visualizing the antibody response to antigenically divergent virus.
Using data from a separate longitudinal immunogenicity study of mRNA-1273, we estimated the kinetics of anti-S and neutralizing antibody decay following D57. This model allowed an independent prediction of the antibody level on disease day 1 in the absence of infection. The predicted values were similar to the actual antibody levels on DD1, indicating that symptom onset occurred largely before the development of an anamnestic response, supporting the use of methods to assess immune correlates of risk (CoR) that rely on antibody measured at the time of detection of disease [27–29].
Our study has several limitations. The COVE trial was conducted in the United States, blinded follow-up ended on March 26, 2021, and virtually all sequenced strains in our analysis were ancestral [20], precluding analysis of kinetics by infecting strain. Our analysis set was restricted to baseline SARS-CoV-2-negative participants who received placebo or 2 doses of mRNA-1273. The force of infection was variable and related to risk and host factors, which included age, risk status, sex, and, for vaccinees, primary antibody response, though not time since vaccination. Nonetheless, the infected group should be representative of those who naturally were infected and diagnosed with COVID-19. Disease occurred a median of 55 days post–full immunization (day 57), and the kinetics of the antibody levels may be different for later disease cases. Our study focused on COVID-19 cases. Sampling was not conducted for asymptomatic cases, and there was only 1 case of severe disease in each arm, thus precluding a comparison of kinetics by severity of infection. Finally, because our focus was on quantifying and exploring multiple aspects of the antibody response to COVID-19, there was no adjustment for multiplicity.
In summary, the kinetics of the anti-N and anti-S responses to symptomatic SARS-CoV-2 infection differed markedly between vaccinated and unvaccinated persons. The powerful effects of vaccination on prevention of disease also manifested during COVID-19, with a relatively immunologically silent response for approximately half of the cases in the early period postvaccination. Nonetheless, the previously vaccinated persons had more potent and broader anti-S antibody levels than the unvaccinated disease cases. Additional studies of disease caused by variants of interest and concern, of more complex infection/vaccination histories, and of wider intervals from last antigen exposure to disease are needed.
Supplementary Material
Acknowledgments
We thank Lindsay Carpp for assistance with technical editing.
Financial support . This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (UM1 AI68635 to H.E.J. and 3UM1AI148575-01S2 to H.M.E.S.) and by the Administration for Strategic Preparedness and Response, Biomedical Advanced Research and Development Authority Contract No. 75A50122C00008 with Labcorp—Monogram Biosciences.
Contributor Information
Dean Follmann, Biostatistics Research Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Holly E Janes, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Center, Seattle, Washington, USA.
Eric Chu, Biostatistics Research Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Lakshmi Jayashankar, Biomedical Advanced Research and Development Authority, Washington, DC, USA.
Christos J Petropoulos, LabCorp-Monogram Biosciences, South San Francisco, California, USA.
Leonid Serebryannyy, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Robin Carroll, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Naz Jean-Baptiste, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Sandeep Narpala, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Bob C Lin, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Adrian McDermott, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Richard M Novak, Section of Infectious Diseases, University of Illinois at Chicago, Chicago, Illinois, USA.
Daniel S Graciaa, Hope Clinic, Emory Vaccine Center, Division of Infectious Diseases, Emory University School of Medicine, Decatur, Georgia, USA.
Stephanie Rolsma, Vanderbilt University Medical Center, Vanderbilt University, Nashville, Tennessee, USA.
Craig A Magaret, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Center, Seattle, Washington, USA.
Nicole Doria-Rose, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Lawrence Corey, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Center, Seattle, Washington, USA; Department of Laboratory Medicine and Pathology, University of Washington, Seattle, Washington, USA.
Kathleen M Neuzil, Center for Vaccine Development and Global Health, University of Maryland School of Medicine, Baltimore, Maryland, USA.
Rolando Pajon, Moderna, Inc., Cambridge, Massachusetts, USA.
Jacqueline M Miller, Moderna, Inc., Cambridge, Massachusetts, USA.
Ruben O Donis, Biomedical Advanced Research and Development Authority, Washington, DC, USA.
Richard A Koup, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Lindsey R Baden, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Hana M El Sahly, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, Texas, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Polack FP, Thomas SJ, Kitchin N, et al. . Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baden LR, El Sahly HM, Essink B, et al. . Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021; 384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palacios R, Batista AP, Albuquerque CSN, et al. . Efficacy and safety of a COVID-19 inactivated vaccine in healthcare professionals in Brazil: the PROFISCOV study. SSRN 3822780 [Preprint]. April 11,2021. Access date September 15, 2022. Available at: Available at: https://ssrn.com/abstract=3822780.
- 4. Gilbert PB, Montefiori DC, McDermott AB, et al. . Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022; 375:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khoury DS, Cromer D, Reynaldi A, et al. . Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 2021; 27:1205–11. [DOI] [PubMed] [Google Scholar]
- 6. US Department of Health and Human Services, Food and Drug Administration, and Center for Biologics Evaluation and Research . Emergency Use Authorization for vaccines to prevent COVID-19: guidance for industry. Appendix 2. 2022. Available at: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/emergency-use-authorization-vaccines-prevent-covid-19. Accessed September 8, 2022.
- 7. World Health Organization . Interim statement on hybrid immunity and increasing population seroprevalence rates. 2022. Available at: https://www.who.int/news/item/01-06-2022-interim-statement-on-hybrid-immunity-and-increasing-population-seroprevalence-rates. Accessed September 8, 2022.
- 8. Edara VV, Norwood C, Floyd K, et al. . Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe 2021; 29:516–21 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen X, Chen Z, Azman AS, et al. . Neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants induced by natural infection or vaccination: a systematic review and pooled analysis. Clin Infect Dis 2022; 74:734–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manali M, Bissett LA, Amat JAR, et al. . SARS-CoV-2 evolution and patient immunological history shape the breadth and potency of antibody-mediated immunity. J Infect Dis 2022; 227:40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walls AC, Sprouse KR, Bowen JE, et al. . SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell 2022; 185:872–80 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wratil PR, Stern M, Priller A, et al. . Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat Med 2022; 28:496–503. [DOI] [PubMed] [Google Scholar]
- 13. Evans JP, Zeng C, Carlin C, et al. . Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci Transl Med 2022; 14:eabn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sahly HM E, Baden LR, Essink B, et al. . Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N Engl J Med 2021; 385:1774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kardava L, Rachmaninoff N, Lau WW, et al. . Early human B cell signatures of the primary antibody response to mRNA vaccination. Proc Natl Acad Sci U S A 2022; 119:e2204607119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y, Borisov O, Kee JJ, et al. . Calibration of two validated SARS-CoV-2 pseudovirus neutralization assays for COVID-19 vaccine evaluation. Sci Rep 2021; 11:23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002; 30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y, Gilbert PB, Montefiori DC, Self SG. Simultaneous evaluation of the magnitude and breadth of a left and right censored multivariate response, with application to HIV vaccine development. Stat Biopharm Res 2009; 1:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doria-Rose N, Suthar MS, Makowski M, et al. . Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for COVID-19. N Engl J Med 2021; 384:2259–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pajon R, Paila YD, Girard B, et al. . Initial analysis of viral dynamics and circulating viral variants during the mRNA-1273 phase 3 COVE trial. Nat Med 2022; 28:823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buckner CM, Kardava L, El Merhebi O, et al. . Interval between prior SARS-CoV-2 infection and booster vaccination impacts magnitude and quality of antibody and B cell responses. Cell 2022; 185:4333–46.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chu L, Vrbicky K, Montefiori D, et al. . Immune response to SARS-CoV-2 after a booster of mRNA-1273: an open-label phase 2 trial. Nat Med 2022; 28:1042–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Follmann D, Janes HE, Buhule OD, et al. . Antinucleocapsid antibodies after SARS-CoV-2 infection in the blinded phase of the randomized, placebo-controlled mRNA-1273 COVID-19 vaccine efficacy clinical trial. Ann Intern Med 2022; 175:1258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cao L, Lou J, Chan SY, et al. . Rapid evaluation of COVID-19 vaccine effectiveness against symptomatic infection with SARS-CoV-2 variants by analysis of genetic distance. Nat Med 2022; 28:1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fonville JM, Wilks SH, James SL, et al. . Antibody landscapes after influenza virus infection or vaccination. Science 2014; 346:996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Straten K, Guerra D, van Gils MJ, et al. . Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of omicron. Immunity 2022; 55:1725–31 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis 2012; 54:1615–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Follmann DA, Dodd L. Immune correlates analysis using vaccinees from test negative designs. Biostatistics 2022; 23:507–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergwerk M, Gonen T, Lustig Y, et al. . COVID-19 breakthrough infections in vaccinated health care workers. N Engl J Med 2021; 385:1474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.