Abstract
Species phylogenetic trees represent the evolutionary processes of organisms, and they are fundamental in evolutionary research. Therefore, new methods have been developed to obtain more reliable species phylogenetic trees. A highly reliable method is the construction of an ortholog data set based on sequence information of genes, which is then used to infer the species phylogenetic tree. However, although methods for constructing an ortholog data set for species phylogenetic analysis have been developed, they cannot remove some paralogs, which is necessary for reliable species phylogenetic inference. To address the limitations of current methods, we developed OrthoPhy, a program that excludes paralogs and constructs highly accurate ortholog data sets using taxonomic information dividing analyzed species into monophyletic groups. OrthoPhy can remove paralogs, detecting inconsistencies between taxonomic information and phylogenetic trees of candidate ortholog groups clustered by sequence similarity. Performance tests using evolutionary simulated sequences and real sequences of 40 bacteria revealed that the precision of ortholog inference by OrthoPhy is higher than that of existing programs. Additionally, the phylogenetic analysis of species was more accurate when performed using ortholog data sets constructed by OrthoPhy than that performed using data sets constructed by existing programs. Furthermore, we performed a benchmark test of the Quest for Orthologs using real sequence data and found that the concordance rate between the phylogenetic trees of orthologs inferred by OrthoPhy and those of species was higher than the rates obtained by other ortholog inference programs. Therefore, ortholog data sets constructed using OrthoPhy enabled a more accurate phylogenetic analysis of species than those constructed using the existing programs, and OrthoPhy can be used for the phylogenetic analysis of species even for distantly related species that have experienced many evolutionary events.
Keywords: bioinformatics, molecular evolution, ortholog, paralog, species phylogenetic inference
Significance.
For phylogenetic analyses, one of the most important tools is a program that constructs ortholog data sets. Among the many ortholog data set constructing programs currently available, OrthoPhy, which was specifically designed for phylogenetic analysis, shows the best performance in this regard, and ortholog data sets constructed by OrthoPhy are expected to enable accurate species phylogenetic inference for even distantly related species (for which accurate phylogenetic analysis is typically difficult, given that the underlying evolutionary events have accumulated to vast extents). Therefore, OrthoPhy has potential to become the standard tool for phylogenetic analysis in the future and will therefore have considerable impact on the field of phylogenetic analysis.
Introduction
Phylogenetic trees are important in evolutionary research because they clarify the evolutionary process of various species and provide a basis to clarify evolutionary events, such as the acquisition, deletion, and horizontal transfer of genes. Therefore, various methods have been developed to construct more reliable phylogenetic trees (Munjal et al. 2019). In the past, phylogenetic relationships of species were often inferred based on their morphological similarities, but performing such analysis was difficult because traits needed to be selected according to the target species (Sterner and Lidgard 2018). However, with the establishment of gene-sequencing technology, molecular phylogenetic analysis was developed, which infers the phylogenetic relationship between species by comparing the sequence of genes commonly present in the target species (Woese et al. 1990; Wainright et al. 1993) and is a more reliable and robust method for phylogenetic analysis. Therefore, it is necessary to detect genes that facilitate the determination of species phylogeny.
Homologs are genes derived from a common ancestor, and orthologs and paralogs are homologs derived from speciation and gene duplication, respectively. Homologs derived from horizontal gene transfer are called xenologs. In addition to speciation, paralogs and xenologs go through gene duplication or horizontal gene transfer; thus, their phylogenetic relationships do not reflect those of the species (Koonin 2005). Therefore, they should not be used for phylogenetic analysis of species since they would reflect incorrect phylogenetic relationships. When molecular phylogenetic analysis was first developed, it was common to use polymerase chain reaction (PCR) to amplify sequences that exist universally in species, such as ribosomal RNAs, and to use these sequences to infer phylogenetic relationships among species. This method uses high sequence similarity among genes amplified by the same primers to determine orthologs (Lang and Orgogozo 2011). However, because orthologs, paralogs, and xenologs are all homologs, their sequence similarity is high, and it is difficult to detect orthologs using PCR. Whole genomes of many species have been sequenced and are available in several databases (Pagani et al. 2012); thus, homolog data can be easily obtained using homology search programs such as basic local alignment search tool (BLAST; Altschul et al. 1990). However, when multiple homologs are detected in the genome of a species, it is difficult to determine orthologs. Therefore, it is crucial to develop methods to accurately identify orthologs using the accumulated genomic data.
For accurate phylogenetic analysis of species, in addition to developing methods for inferring orthologs, methods for inferring phylogenetic trees from gene sequences are also important. The methods commonly used in early molecular phylogenetic analysis presented disadvantages, such as the inability to accurately infer phylogenetic relationships of genes following complex evolutionary models and vulnerability to long-branch attraction (Felsenstein 1978; Philippe et al. 2005; Spencer et al. 2005). Therefore, the maximum likelihood (Felsenstein 1981) and Bayesian methods (Rannala and Yang 1996) were developed and are now commonly used as more accurate methods (Hall 2005). However, phylogenetic trees that include paralogs or xenologs do not reflect the phylogenetic relationships among species; thus, even if a tree is inferred accurately, the correct phylogenetic relationships among species cannot be determined. In other words, the accuracy of a phylogenetic analysis depends not only on the accuracy of the phylogenetic tree inference method but also on the accuracy of ortholog inference. Therefore, with whole-genome sequencing of many species and the availability of sequence information in databases, methods based on multiple orthologs have been developed to improve the accuracy of phylogenetic analysis of species. Typical examples of inference of a species phylogenetic tree (hereafter, species tree) include inference using concatenated data of ortholog sequences of each species and inference by integrating the topological information of each phylogenetic tree of orthologs. These methods are considered more robust (Gadagkar et al. 2005) and require an ortholog data set that is based on the complete genetic information of species for phylogenetic analysis.
Tree-based and graph-based methods are mainly used to construct ortholog data sets (Tekaia 2016). The tree-based method infers phylogenetic trees of homologs, obtained using programs such as BLAST, and compares them with species trees to identify orthologs. Tree-based methods, such as Hieranoid (Kaduk and Sonnhammer 2017), OrthoStrapper (Storm and Sonnhammer 2002), LOFT (van der Heijden et al. 2007), and TreeFam (Li et al. 2006), cannot be used to construct ortholog data sets for inferring species trees because they themselves require a species tree for ortholog inference.
The graph-based method is used to infer orthologs based on sequence similarity and does not require a species tree; thus, it can be used for phylogenetic analysis of species. Examples include COG (Tatusov et al. 2000), which builds orthologous clusters by connecting groups of three mutually orthologous sequences as the minimum unit; OrthoMCL (Li et al. 2003), which uses Markov clustering as the sequence clustering method; HaMStR (Ebersberger et al. 2009) and Orthograph (Petersen et al. 2017), which construct ortholog data sets based on transcript information; SonicParanoid (Cosentino and Iwasaki 2019) and SwiftOrtho (Hu and Friedberg 2019), which are programs that drastically reduce computational costs and enable rapid ortholog inference for large-scale analysis; OrthoFinder (Emms and Kelly 2019), which uses a normalized similarity score to improve accuracy; and OMA (Altenhoff et al. 2019), which verifies candidate ortholog pairs based on evolutionary distance. Of these, OrthoFinder and OMA also have paralog removal processes. OrthoFinder infers the rooted phylogenetic tree of the species based on the information of paralogs in the gene phylogenetic trees (hereafter, gene tree) of each Orthogroup. In addition, it removes paralogs by comparing the species tree of each Orthogroup phylogenetic tree. OMA removes paralogs based on the evolutionary distance between orthologs, which is shorter than that between paralogs.
Existing methods for obtaining ortholog data sets are not designed for the phylogenetic analysis of species and tend to focus on detecting a large number of orthologs rather than avoiding false orthologs. This occurs because ortholog information is used not only for phylogenetic analysis of species but also for function prediction of unknown genes. In function prediction, data sets containing more orthologs are useful because genes with known functions enable the prediction of the functions of numerous genes with unknown functions. Therefore, the threshold for detecting orthologs tends to be loosely set to obtain more orthologs, although the risk of interfusion of paralogs increases when constructing data sets for function prediction. As the functions of orthologs and paralogs are relatively similar, the effect of interfusion of paralogs on the accuracy of function prediction is minor compared with the risk of increasing the number of genes whose functions cannot be predicted because of the omission of orthologs. Therefore, ortholog data sets generated using the existing methods are not suitable for the phylogenetic analysis of species because they may contain many paralogs (Horiike et al. 2016). In addition, the paralog removal methods of OMA and OrthoFinder assume that no deletion occurs in any part of the duplicated genes and that two or more copies of the gene remain in the genome. Therefore, they cannot remove “hidden paralogs,” which are erroneously detected as single-copy genes because of differential gene loss (Kristensen et al. 2011), making them unsuitable for the construction of data sets for the phylogenetic analysis of species.
Existing ortholog inference methods should be improved in terms of the quality of ortholog data for species tree inference. Therefore, we focused on the taxonomic information of the target species to solve this problem. For instance, let us assume that species A, B, and C belonging to the same lineage are analyzed. In the phylogenetic tree of the inferred orthologs, if any of these three species is included in a different lineage, paralogs may be included because the tree topology is inconsistent with the known taxonomic information. Therefore, it is possible to detect and exclude paralogs based on taxonomic information without using the species tree. Xenologs can be detected and excluded in the same way because they are inconsistent with the taxonomic information of the target species. Thus, using taxonomic information for ortholog inference, the number of possible species for analysis could be limited, which is not a problem in practice because relatively high-level taxonomic information, such as phylum, class, order, and family, is often known for the analyzed species (Hug et al. 2016). Therefore, in many analyses, it is possible to divide all or part of the analyzed species into taxa and use this information for ortholog inference. In other words, it is possible to use taxonomic information that represents broad phylogenetic relationships to construct ortholog data sets to infer species trees that represent detailed relationships among species.
To the best of our knowledge, Ortholog Finder (OF), developed by our group, is the first program to detect and remove hidden paralogs that cannot be removed by the existing graph-based methods (Horiike et al. 2016). However, it has some limitations; hence, we developed OrthoPhy, which is an improved version of OF to overcome these limitations. For example, OF compares taxonomic information with the gene trees of candidate ortholog groups constructed based on sequence similarity. However, the available taxonomic information is limited to taxonomic groups that dichotomized the analyzed species, such as in an out-group and in-group, in phylogenetic tree inference. Therefore, hidden paralogs caused by gene duplication after the divergence of these two groups cannot be detected, which affects the accuracy of ortholog inference. Furthermore, even if detailed taxonomic information on the analyzed species is made available, users cannot take full advantage of that taxonomic information because OF only works with information that classifies the analyzed species into two monophyletic groups. Moreover, large-scale analysis is unsustainable because of high computational costs, and the graphical user interface (GUI) tool does not allow for an easy automatic run or comparison of performance with other programs.
In contrast, in OrthoPhy, to maximize the use of known taxonomic information, we enabled the use of taxonomic information that divides the analyzed species into three or more monophyletic groups, thereby removing more hidden paralogs and leading to the construction of a highly accurate ortholog data set. In addition, the source code was redesigned, and the algorithm and external programs were modified to reduce calculation time. Moreover, a command-line interface was adopted instead of a graphical one to facilitate automatic execution and comparison with other programs. To evaluate the performance of OrthoPhy, we conducted a test based on the sequences generated by the evolutionary simulation program and a test based on real sequence data from 40 gram-positive bacteria. We compared the accuracy of ortholog inference of OrthoPhy and existing ortholog data set construction programs. Furthermore, we conducted a benchmark test provided by Quest for Orthologs (Altenhoff et al. 2020) using real sequence data. Notably, to enable accurate species tree inference, OrthoPhy is specialized to increase the precision of ortholog inference using taxonomic information (e.g., phyla, classes, and orders) about analyzed species. It is not supposed to be used for analysis such as the prediction of gene function and may not be suitable for such applications.
Results and Discussion
Performance Test Using Simulated Sequences
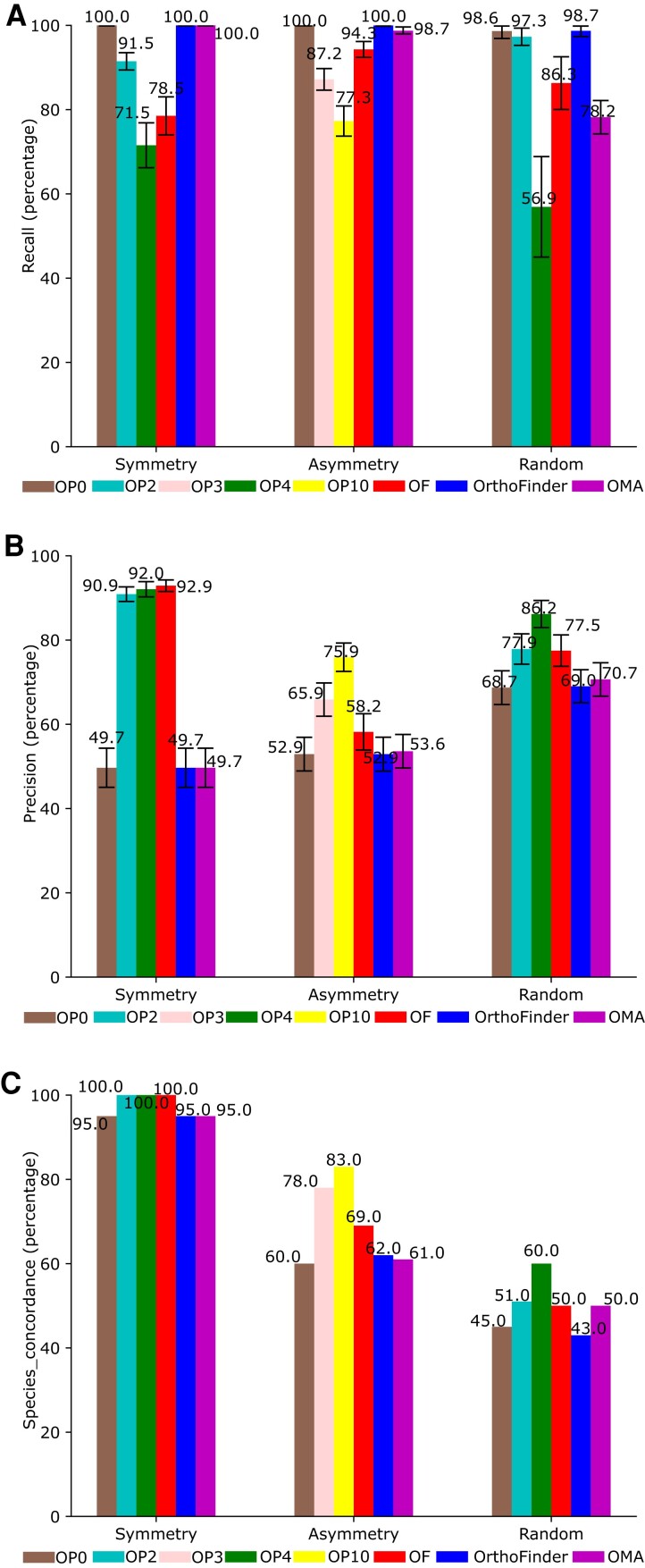
Based on the sequences generated using a symmetric phylogenetic tree model (complete binary tree), an asymmetric phylogenetic tree model (tree with topology such that a single operational taxonomic unit [OTU] diverged from each node), and random phylogenetic tree model (tree with random topology), ortholog data sets were constructed using OrthoPhy, OF, OrthoFinder, and OMA. All three phylogenetic tree models contained 32 OTUs (see Performance Test Using Simulation Sequences in Materials and Methods). For each data set, we evaluated “Recall,” “Precision,” and “Species_concordance” (concordance rate between the inferred species trees and the phylogenetic tree model; fig. 1).
Fig. 1.
Performance of ortholog inference for three model trees. Evaluation of (A) Recall, (B) Precision, and (C) Species_concordance (concordance rate between species phylogenetic trees and phylogenetic tree models) for three phylogenetic tree models: symmetry, asymmetry, and random. Brown: OrthoPhy without taxonomic information (OP0); light green: OrthoPhy with taxonomic information for two groups (OP2); light pink: OrthoPhy with taxonomic information for three groups (OP3); dark green: OrthoPhy with information for four groups (OP4); yellow: OrthoPhy with information for ten groups (OP10); red: OF (OF); blue: OrthoFinder; and purple: OMA.
When the symmetric phylogenetic tree was used as the phylogenetic tree model, the Recall of OrthoPhy with four taxonomic information that divided analyzed species into four monophyletic groups (OP4, see Symmetric Phylogenetic Tree in Materials and Methods) was the lowest at 71.5% (fig. 1A), whereas that of OrthoPhy with two taxonomic information that divided analyzed species into two monophyletic groups (OP2) at 91.5% was higher than that of OP4. This trend was also observed when other phylogenetic tree models were used. More detailed taxonomic information led to the removal of paralogs during ortholog inference because of information incensement for detecting paralogs. After paralog removal, if the number of sequences constituting an ortholog group was below the threshold (default: four), the ortholog group was removed, and the overall number of ortholog groups decreased. Consequently, the Recall of OP2, which uses less classification information, was higher than that of OP4. The difference in the Recall of OP2 and OF, which use the same taxonomic information, was attributed to their clustering method for ortholog inference. When the symmetric phylogenetic tree model was used, the Recall of OrthoPhy without taxonomic information (OP0), OrthoFinder, and OMA were all 100, and all orthologs in the input data set were obtained. However, this result does not indicate a high level of ortholog detection; this is because the hidden paralogs were included in the ortholog data set, as these programs cannot accurately detect hidden paralogs. Therefore, unlike OP2, OP4, and OF, no ortholog groups exist whose number of sequences was below the threshold and were removed from the data set due to paralog removal; thus, the quality of the ortholog data set was not high, but the Recall was extremely high. When an asymmetric phylogenetic tree model was used, OP0, OrthoFinder, and OMA showed high Recall values (100%, 100%, and 98.7%, respectively), similar to the results for symmetric phylogenetic trees; however, when a random phylogenetic tree model was used, the Recall values of OMA were the second-lowest after OP4 (78.2%) because OMA uses evolutionary distance as a criterion for paralog determination and removal. Therefore, sequences with long evolutionary distances were not considered orthologs and were removed.
When the symmetric phylogenetic tree was used as the model phylogenetic tree, the Precision was 92.0% (OP4), 90.9% (OP2), 49.7% (OP0), 92.9% (OF), 49.7% (OrthoFinder), and 49.7% (OMA; fig. 1C). Although OF had the highest value by a small margin, focusing only on the OrthoPhy results, Precision tended to be higher when more taxonomic information was given than when less was given. When the asymmetric phylogenetic tree model was used, Precision was 75.9% (OP10), 65.9% (OP3), 52.9% (OP0), 58.2% (OF), 52.9% (OrthoFinder), and 53.6% (OMA), and Precision for the random phylogenetic tree model was 86.2% (OP4), 77.9% (OP2), 68.7% (OP0), 77.5% (OF), 69.0% (OrthoFinder), and 70.7% (OMA). In all the model phylogenetic trees, Precision tended to be higher when more taxonomic information was given than when less information was given to OrthoPhy, and Precision tended to be similar for OP0, OrthoFinder, and OMA. This suggested that regardless of the phylogenetic tree model, increasing the amount of taxonomic information improved the Precision of ortholog inference, and thus, OrthoPhy can construct highly accurate ortholog data sets for various sets of species. In addition, the Precision was the highest when the symmetric phylogenetic tree model, which was less biased in branch lengths and patterns than the models for asymmetric or random phylogenetic trees, was used. Thus, OrthoPhy is particularly efficient at detecting and removing hidden paralogs in phylogenetic trees with simple topology such as the symmetric phylogenetic tree model, which is a complete binary tree. However, the results also showed that the improvement of Precision owing to the presence of more detailed taxonomic information used for ortholog inference is not large for the case with the symmetric phylogenetic tree model but is for cases with asymmetric and random phylogenetic tree models, which have complex topology.
When the symmetric phylogenetic tree model was used, the value for Species_concordance in OP2, OP4, and OF was 100.0. The value of OP0, OrthoFinder, and OMA was 95.0 (fig. 1C), which indicated that inferences of the species tree using OrthoPhy with taxonomic information and OF ortholog data sets were only slightly more accurate. In addition, as Species_concordance was 100% for cases where taxonomic information for two or four groups was provided, the improvement in the reliability of species phylogeny inference by increasing the amount of taxonomic information to construct the ortholog data set could not be verified. When the asymmetric phylogenetic tree model was used, Species_concordance generally showed the same trend as that of Precision. OP10 (83%), which is considered the most detailed taxonomic information, was the highest, followed by OP3 (78%) and OF (69%). These results suggest that the reliability of species phylogenetic inference based on an ortholog data set increased as the amount of taxonomic information increased. Species_concordance for OP0, OrthoFinder, and OMA were even lower at 60.0, 62.0, and 61.0, respectively. When a random phylogenetic tree model was used, Species_concordance values were generally lower than those obtained for other phylogenetic tree models. Species_concordance was the highest in OP4 (60.0%), followed by that in OP2 (51.0%). The OF and OMA values were both 50.0, the OP0 value was 45.0, and the OrthoFinder value was 43.0, the lowest among all programs. In all performance tests, the Species_concordance of OrthoPhy with more detailed taxonomic information was higher than that of OF, OrthoFinder and OMA, and OrthoPhy with less taxonomic information. Therefore, ortholog data sets constructed by OrthoPhy provide a more accurate inference of species trees using Astral than those constructed by the existing programs. Notably, in the future, when a new method for species tree inference is developed, the ortholog data set constructed by OrthoPhy will be available for using such method instead of Astral.
These results indicate that the performance of OrthoPhy without taxonomic information (OP0) is comparable with that of OrthoFinder and OMA. It is also clear that the use of taxonomic information enables the removal of hidden paralogs and the construction of more accurate ortholog data sets.
Note that OrthoFinder uses STAG (Emms and Kelly 2019), a built-in program, to infer species trees for ortholog inference. However, the accuracy of species trees inferred by STAG was lower than that inferred by Astral (Yin et al. 2019) under all conditions in the tests (supplementary fig. S1, Supplementary Material online). Therefore, when comparing the results of OrthoFinder with the results of other programs, we used the species trees inferred by Astral rather than STAG.
Astral-Pro (Zhang et al. 2020) is a program that can infer species trees even with ortholog data containing paralogs. We used Astral-Pro instead of Astral to infer the species tree using an ortholog data set constructed by OP0. However, the results showed that there was not much difference in the accuracy of species tree inference using Astral-Pro and using Astral (supplementary fig. S2, Supplementary Material online), possibly because Astral-Pro can handle ortholog data containing paralogs other than hidden paralogs but not ortholog data containing hidden paralogs. Therefore, we decided to use the simpler Astral in OrthoPhy.
Performance Test for 40 Gram-positive Bacteria
Ortholog data sets were constructed for 40 gram-positive bacteria (table 1) using each program (OrthoPhy given taxonomic information for five classes [Actinobacteria, Coriobacteriia, Bacilli, Clostridia, and Tissierellia] as OP5, OrthoPhy given taxonomic information for two phyla [Actinobacteria and Firmicutes] as OP2, OF, OrthoFinder, and OMA). Only orthologs in the ortholog data sets that satisfied the condition of each program were used for species tree inference as follows: The number of orthologs used to infer the species tree was 61 for OP5 (orthologs conserved in at least 70% of the analyzed species), 270 for OP2 (the condition of the orthologs used was the same as OP5), 262 for OF (orthologs conserved in at least 50% of the analyzed species), 120 for OrthoFinder (orthologs for which at least one sequence existed in all analyzed species), and 165 for OMA (the top 1% of the most conserved in the inferred OMA groups), respectively.
Table 1.
List of 40 Gram-positive Bacteria for Species Tree Inference
| Phylum | Class | Species |
|---|---|---|
| Actinobacteria | Actinobacteria | Bifidobacterium longum |
| Corynebacterium efficiens | ||
| Mycobacterium tuberculosis | ||
| Nocardia farcinica | ||
| Propionibacterium acnes | ||
| Streptomyces coelicolor | ||
| Thermobifida fusca | ||
| Tropheryma whipplei | ||
| Coriobacteriia | Atopobium parvulum | |
| Coriobacterium glomerans | ||
| Cryptobacterium curtum | ||
| Denitrobacterium detoxificans | ||
| Eggerthella lenta | ||
| Gordonibacter pamelaeae | ||
| Olsenella uli | ||
| Slackia heliotrinireducens | ||
| Firmicutes | Bacilli | Bacillus subtilis subsp. subtilis |
| Enterococcus faecalis | ||
| Geobacillus kaustophilus | ||
| Lactobacillus plantarum | ||
| Lactococcus lactis subsp. lactis | ||
| Oceanobacillus iheyensis | ||
| Staphylococcus epidermidis | ||
| Streptococcus thermophilus | ||
| Clostridia | Alkaliphilus metalliredigens | |
| Caldanaerobacter subterraneus subsp. tengcongensis | ||
| Carboxydothermus hydrogenoformans | ||
| Clostridium acetobutylicum | ||
| Desulfitobacterium hafniense | ||
| Moorella thermoacetica | ||
| Symbiobacterium thermophilum | ||
| Syntrophomonas wolfei subsp. wolfei | ||
| Tissierellia | Anaerococcus prevotii | |
| Caloranaerobacter azorensis | ||
| Finegoldia magna | ||
| Gottschalkia acidurici | ||
| Keratinibaculum paraultunense | ||
| Parvimonas micra | ||
| Peptoniphilus sp. ING2-D1G | ||
| Tissierella sp. JN-28 |
Note.—The first and second columns indicate the phylum and class into which the analyzed species are classified.
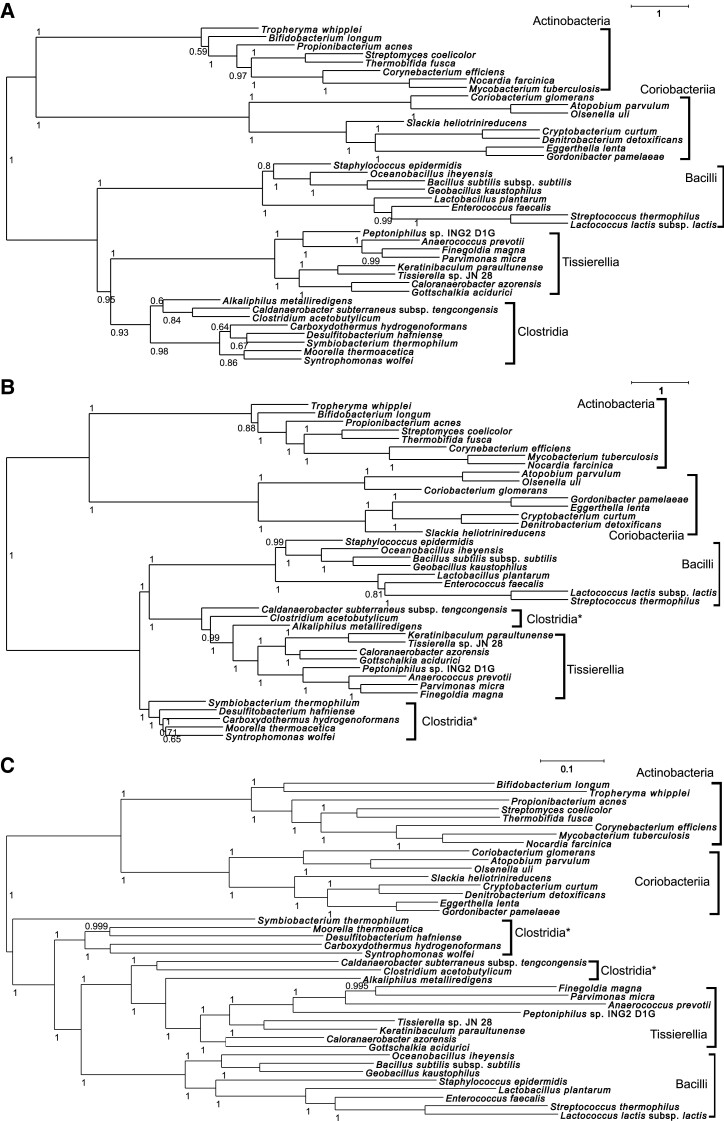
The species tree for the 40 gram-positive bacteria was inferred based on the ortholog data set constructed by each program (OP5, OP2, OF, OrthoFinder, and OMA; fig. 2). In the inferred species tree using the ortholog data set of OP5, the two phyla of Actinobacteria and Firmicutes were each monophyletic (fig. 2A). Moreover, the five classes of Actinobacteria, Coriobacteriia, Bacilli, Clostridia, and Tissierellia were also monophyletic. Thus, the species tree is supported by known taxonomy and is considered reliable. Similarly, the two phyla of Actinobacteria and Firmicutes were monophyletic in the species trees using the data set of OP2, OF, OrthoFinder, and OMA (fig. 2B–E, respectively), and the four classes of Actinobacteria, Coriobacteriia, Bacilli, and Tissierellia were also monophyletic. However, in these species trees, Clostiridia was polyphyletic, which was inconsistent with the taxonomic information. Since Clostridia is considered monophyletic, the position of some Clostridia species in the species tree might be inferred incorrectly in the species tree of figure 2B–E. The reason why the species trees obtained by OP2 and OF were not monophyletic for Clostridia was because only taxonomic information for two phyla was used for ortholog inference, and thus, the hidden paralogs caused by gene duplication and subsequent gene loss that occurred after the divergence of the two phyla were not detected, and the ortholog data set including these paralogs was used for species tree inference. Further, OrthoFinder and OMA could not remove hidden paralogs, which may have resulted in the inference of an incorrect tree. On the other hand, since OP5 can use taxonomic information for five classes for ortholog inference, it can detect and remove hidden paralogs that occurred after the divergence of the Firmicutes phylum and can infer the orthologs more accurately. Thus, a more reliable and accurate species tree could be obtained, in which all classes are monophyletic in the inferred species tree using an ortholog data set. This suggests that more accurate species trees can be obtained by constructing ortholog data sets using OrthoPhy with more detailed taxonomic information.
Fig. 2.
Phylogenetic tree for 40 gram‐positive bacteria. The phylogenetic tree includes two phyla (Actinobacteria and Firmicutes) and five classes (Actinobacteria, Bacilli, Clostridia, Coriobacteriia, and Tissierellia). An asterisk at the end of a class name indicates that the class is not monophyletic. (A) Species tree obtained using OrthoPhy with taxonomic information for five classes (branch lengths are in coalescent units, and all external branch lengths were set to 1; local posterior probability [Yin et al. 2019] is indicated in each internal branch). (B) Species tree obtained using OrthoPhy with taxonomic information for the two phyla (branch lengths in coalescent units, and all external branch lengths were set to 1; local posterior probability is indicated in each internal branch). (C) Species tree obtained using OF (local support values [Price et al. 2010] are indicated in each internal node). (D) Species tree obtained using OrthoFinder (support values are indicated in each internal node, summarizing the fraction of input trees that support its bipartition [Emms and Kelly 2019]). (E) Species tree obtained using OMA (branch lengths are in PAM units, and support values calculated by OMA are shown at each branch; Altenhoff et al. 2019).
Performance Test Using Quest for Orthologs Benchmark
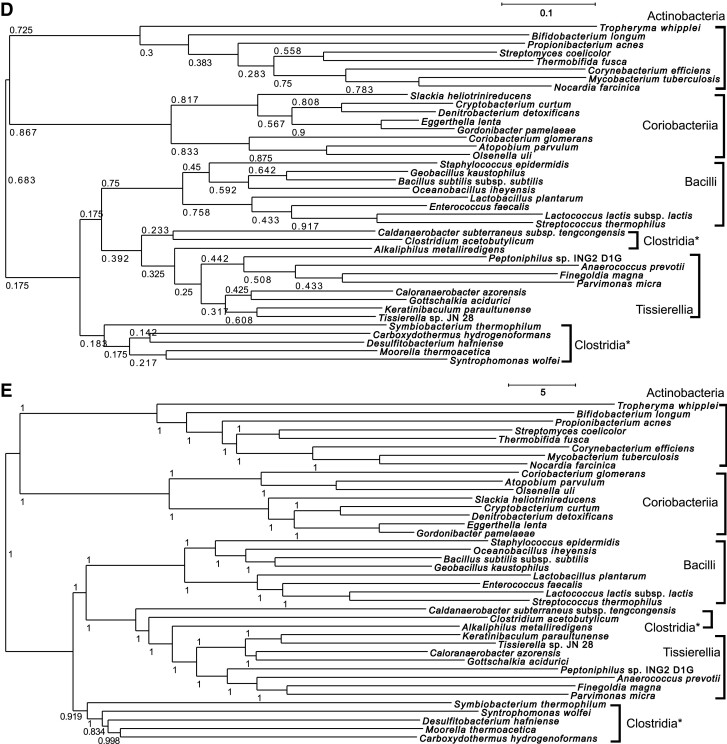
The orthologous data set constructed using OrthoPhy using the proteome data set “2018_4” provided by Quest for Orthologs (hereafter, QfO) was evaluated by a QfO benchmark test; the results are shown in figure 3. In this test, normalized Robinson–Foulds (RF) distances (Robinson and Foulds 1981) were calculated between the provided species trees by QfO and the ortholog phylogenetic trees inferred by OrthoPhy, followed by the Quest for Orthologs benchmark test method. The whole analyzed species tree is shown in the SwissTree database (https://swisstree.sib.swiss/cgi-bin/swisst?page=species_tree), and the phylogenetic range covered by each test depends on the test (see Performance Test Using Quest for Orthologs Benchmark in Materials and Methods).
Fig. 3.
Results of the benchmark test of Quest for Orthologs. The results of each program with the same benchmark test are indicated in the graph. The horizontal axis indicates the number of orthologous phylogenetic trees used in the test, and larger values indicate that more orthologs were inferred. The vertical axis shows the average Robinson–Foulds distance between the species phylogenetic tree and the ortholog phylogenetic trees. OrthoPhy is indicated by a yellow-green rhombus. OrthoPhy results of (A) “Generalized Species Tree Discordance Benchmark (LUCA)” with three-domain information; (B) “Generalized Species Tree Discordance Benchmark (Eukaryota)” with eukaryotes information; and (C) “Generalized Species Tree Discordance Benchmark (Fungi)” with fungi information.
For the ortholog data set constructed using OrthoPhy with information from three domains, the average RF distance between the phylogenetic trees of the inferred orthologs and phylogenetic trees containing species belonging to the domains Bacteria, Archaea, or Eukaryota (phylogenetic trees of LUCA provided by the QfO benchmark test) was 0.252 (standard deviation = 0.020), which was the smallest value obtained among all 22 analyzed programs (including the same program with different settings), as shown in figure 3A. This difference was large when compared with the second smallest RF distance of 0.285 (standard deviation = 0.016) for “Proteinortho_6.0.13_with-isoform” (Lechner et al. 2011). The average RF distance between the phylogenetic trees of orthologs inferred by OrthoPhy with “eukaryotes information,” which divided the analyzed eukaryotes species into five supergroups (see (2) of Performance Test Using Quest for Orthologs Benchmark in Materials and Methods), and the phylogenetic trees of eukaryotes was 0.117 (fig. 3B) and that between the phylogenetic trees of orthologs inferred by OrthoPhy with “fungi information,” which divided the analyzed fungi into three phyla (see (3) of “Performance Test Using Quest for Orthologs Benchmark” in Materials and Methods), and the phylogenetic trees of fungi was 0.188 (fig. 3C). These values were the smallest among all 22 tested programs, and they differed widely from the values obtained from programs with the second smallest RF distances (0.152 for “Proteinortho_6.0.13_with-isoform” in fig. 3B, and 0.223 for “OMA_Groups_2.0” in fig. 3C). Therefore, OrthoPhy is more suitable for phylogenetic analysis of species because the concordance rate between ortholog phylogenetic trees and species trees is higher than those obtained for phylogenetic trees constructed using existing programs. The number of ortholog phylogenetic trees used in the test was 1,132, 414, and 4,111 for OrthoPhy with “three-domain information” (fig. 3A), “eukaryotic information” (fig. 3B), and “fungi information” (fig. 3C), respectively, and they are lower than that of most of the 22 programs tested. However, in ortholog inference for the phylogenetic analysis of species, removing paralogs is more important than preventing missing orthologs. Furthermore, as the number of orthologs generally used for the phylogenetic analysis of species is not so large, the number of orthologs obtained by OrthoPhy is enough. The influence of the small number of orthologs inferred by OrthoPhy is insignificant compared with its advantage of high accuracy for species tree inference.
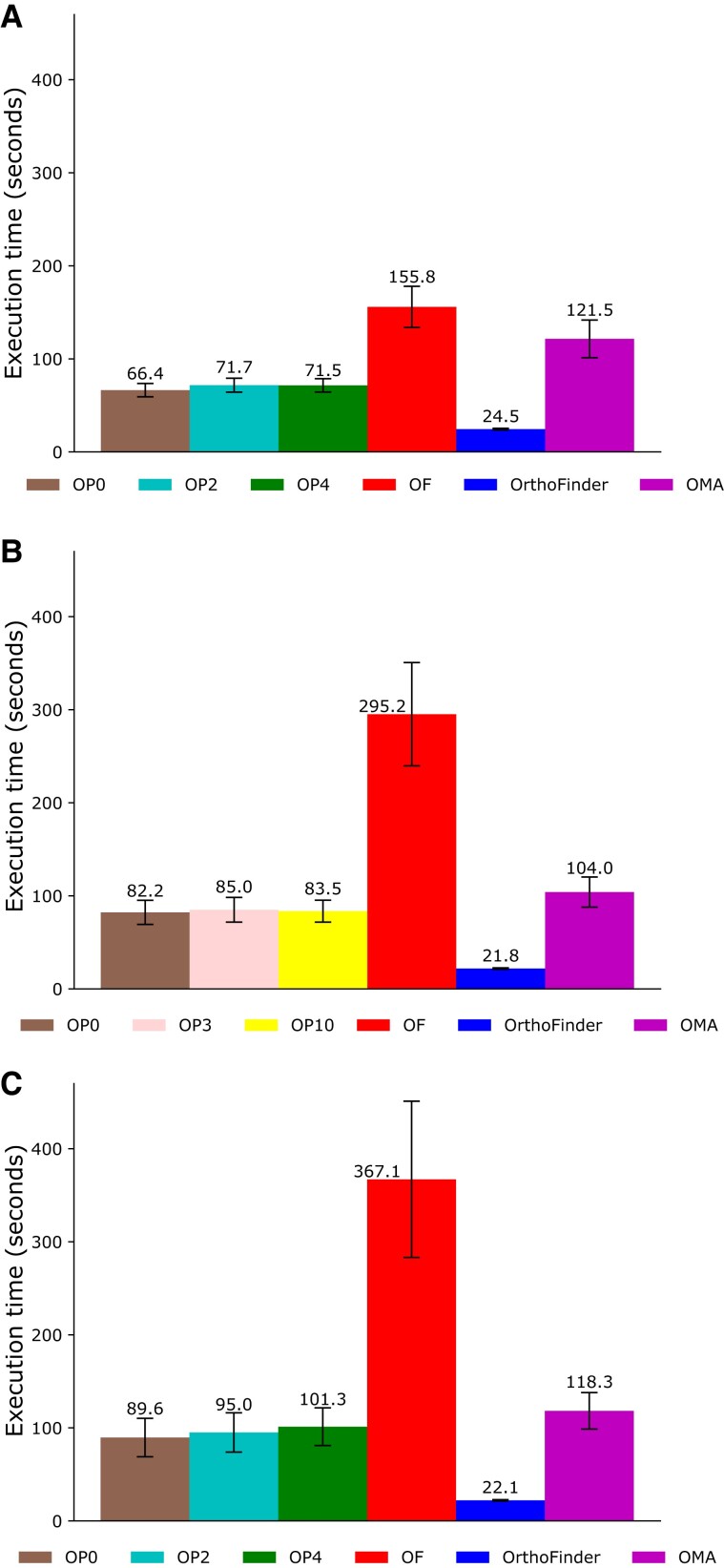
Comparison of Execution Time
In the performance test using simulated sequences, the time required to infer the orthologs (execution time) was measured for each program and the phylogenetic tree model (fig. 4). In the performance test using the symmetric phylogenetic tree model, the execution times of OP0, OP2, and OP4 were 66.4, 71.7, and 71.5 s, respectively, which were approximately half of the execution time of OF (155.8 s). This trend was more remarkable in the performance tests using other phylogenetic tree models, and the execution time of OrthoPhy was approximately one-third that of OF (fig. 4). Furthermore, the execution time of OrthoPhy remains almost the same regardless of the amount of taxonomic information, thereby suggesting that OrthoPhy can construct ortholog data sets faster than OF for a set of various species even when detailed taxonomic information is used. Although OrthoFinder presented the shortest execution time, it does not have the process to detect hidden paralogs or remove horizontal transfer genes. The execution time of OMA was shorter than that of OF but longer compared with OrthoPhy in all tests. These results indicate that OrthoPhy can be used to construct ortholog data sets within a reasonable time.
Fig. 4.
Execution time of ortholog inference by each program. The vertical axis represents the execution time required for ortholog inferences. Brown: OrthoPhy without taxonomic information (OP0); light green: OrthoPhy with taxonomic information for two groups (OP2); light pink: OrthoPhy with taxonomic information for three groups (OP3); dark green: OrthoPhy with taxonomic information for four groups (OP4); yellow: OrthoPhy with information for ten groups (OP10); red: Ortholog Finder (OF); blue: OrthoFinder; and purple: OMA. Tests using (A) symmetric, (B) asymmetric, and (C) random phylogenetic tree models.
Perspectives
One of the future challenges for OrthoPhy is enabling the use of hierarchical taxonomic information. For illustration, assume that each analyzed species is classified into one of two lineages, A and B, and further assume that lineage A comprises lineages a1 and a2, and lineage B comprises lineages b1 and b2. In this case, lineages A and B are monophyletic in the phylogenetic tree of the ortholog, as well as lineages a1, a2, b1, and b2. When taxonomic information for lineages a1, a2, b1, and b2 is input into OrthoPhy, the taxonomic information for lineages A and B cannot be provided at the same time because of different hierarchy. Therefore, if a1 and b1, and a2 and b2 are closely related, they are not excluded as paralogs, even though A and B are not monophyletic. Therefore, we plan to improve the algorithm to present group information with a hierarchical structure.
Conclusions
We developed a new program called OrthoPhy for constructing specialized ortholog data sets for the phylogenetic analysis of species. This program can construct ortholog data sets with a higher Precision than the existing programs by maximizing the use of taxonomic information. The results of the performance test using the sequences generated by the evolutionary simulation program suggest that OrthoPhy can infer orthologs more accurately by using more detailed taxonomic information than OF (developed in a previous study) and other existing programs. Additionally, species tree inference using ortholog data sets constructed by OrthoPhy was more accurate than those using data sets constructed by existing programs. In this analysis, the execution time of OrthoPhy was about half of that of OF (previous version of OrthoPhy), and it was sufficiently practical compared with that of other programs. We also constructed ortholog data sets and inferred species trees for 40 gram-positive bacteria as performance evaluation tests using real sequences. We obtained reliable species trees supported by taxonomic information only when OrthoPhy was run with detailed taxonomic information. When using OrthoPhy with less taxonomic information and other ortholog data set–constructing programs, inferred species trees were inconsistent with their taxonomy. This suggests that ortholog data sets constructed by OrthoPhy with more detailed taxonomic information can enable an improvement in the accuracy of species tree inference. Furthermore, we performed a benchmark test provided by QfO as a performance test and confirmed that the RF distance between phylogenetic trees of inferred orthologs and a species tree was the lowest for OrthoPhy of all other investigated programs, which suggests that, compared with existing programs, OrthoPhy can infer orthologs more accurately in an actual analysis for various species and the phylogenetic relationships of species inferred using ortholog data sets by OrthoPhy are more reliable.
In general, phylogenetic analysis of distantly related species is difficult, and the results are unreliable because of the long divergence time from the common ancestor and the many evolutionary events experienced. Therefore, OrthoPhy is suitable for elucidating highly accurate ortholog data sets even for distantly related phylogenetic analyses.
Materials and Methods
Overview of OrthoPhy
OrthoPhy constructed ortholog data sets in four steps using the gene or protein sequence data of the analyzed species in GenBank (gene) or FASTA (protein) format as follows: (1) prediction and removal of genes acquired by horizontal gene transfer, (2) inference of candidate ortholog groups, (3) inference of phylogenetic trees of candidate ortholog groups, and (4) detection and removal of paralogs from candidate ortholog groups using their phylogenetic trees and taxonomic information of the analyzed species. In addition, by default, the phylogenetic tree of species is inferred based on the constructed ortholog data set using Astral (Yin et al. 2019). The process of ortholog data set construction and phylogenetic inference of the species tree by OrthoPhy is described in the flowchart (supplementary fig. S3, Supplementary Material online). The details of each step are described below.
Prediction and Removal of Horizontally Transferred Genes
OrthoPhy used IslandPath-DIMOB (Bertelli and Brinkman 2018) to predict the regions acquired by horizontal transfer on the genome and then removed all genes encoded in the predicted regions. Because this process uses nucleotide sequence data, it can only be performed when the input sequence data are given in the GenBank format.
Inference of Candidate Ortholog Groups
A database for DIAMOND (Buchfink et al. 2021), a homology search program, was generated for each species using all sequences of that species. Using the databases, homology search between all sequences in all combinations was performed by DIAMOND. Subsequently, the bit scores (similarity scores) between all sequences were obtained. The calculated bit scores were then normalized using the OrthoFinder method (Emms and Kelly 2015) to remove sequence length bias. Based on the normalized bit scores, all reciprocal best-hit pairs and better-hit pairs (Emms and Kelly 2015) were detected. Finally, Markov clustering was performed on all reciprocal best-hit and better-hit pairs using the MCL program (Enright et al. 2002) to generate candidate ortholog groups.
Phylogenetic Tree Inference of Candidate Ortholog Groups
Each candidate ortholog group was aligned using the MAFFT software (Katoh and Standley 2013). The nonconserved region in each alignment sequence was removed using trimAl (Capella-Gutiérrez et al. 2009). The phylogenetic tree of each candidate ortholog group was inferred by FastTree (Price et al. 2010) using the obtained sequences. LG (Le and Gascuel 2008) was used as an amino acid substitution matrix.
Removal of Paralogs and Inference of Ortholog Groups
Using the phylogenetic trees of the inferred candidate ortholog groups and the taxonomic information of the analyzed species, paralogs were removed from each candidate ortholog group to generate groups consisting only of sequences that are orthologous to each other (ortholog groups). The paralog removal process can be divided into three phases: (i) removal of paralogs generated after the last speciation, (ii) removal of paralogs using overlapping species, and (iii) removal of paralogs using taxonomic information.
(i) Removal of Paralogs Generated After the Last Speciation.
If a clade contained only sequences of one species, the sequences were thought to be paralogs that were generated after the last speciation (supplementary fig. S4A, Supplementary Material online). OrthoPhy leaves only one sequence in the clade as an ortholog (supplementary fig. S4B, Supplementary Material online). The remaining sequence was the one with the shortest branch length in the clade.
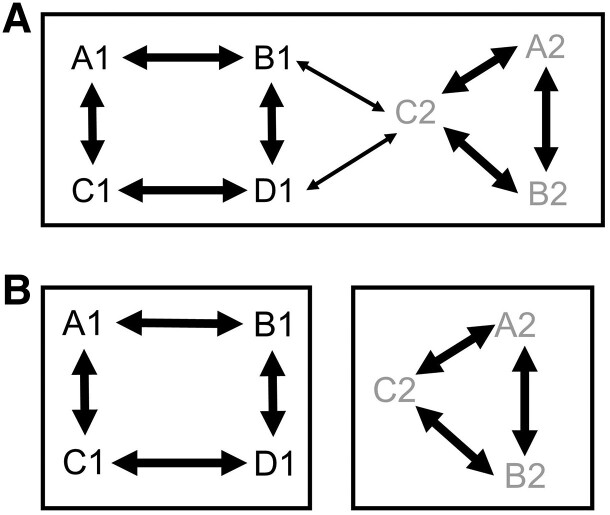
(ii) Removal of Paralogs Based on Overlapping Species.
If sequences of more than two species are included in both clades derived from a node, paralogs are thought to be included in the sequences (supplementary fig. S5A, Supplementary Material online). Therefore, for every two clades that diverge from each node of the phylogenetic tree of the inferred candidate ortholog group, OrthoPhy obtained information on the species contained in the clade and determined if the aforementioned conditions were met. If they did, the sequences in the two clades were considered paralogous to each other, and all sequences in one of the clades were removed (supplementary fig. S5B, Supplementary Material online). The sequences in the clade that branched off from the longer branch were preferentially removed.
(iii) Removal of Paralogs Using Taxonomic Information.
To illustrate the removal of paralogs using taxonomic information, we assumed that the analyzed species belong to either phylogeny X, phylogeny Y, or phylogeny Z. Herein, if a phylogenetic tree of the candidate ortholog group was inferred, the presence or absence of paralogs could be determined based on the species information in the clades located at both ends of a certain branch. For example, if one clade contains only sequences of the species belonging to lineage X and another one contains only sequences of the species belonging to lineages Y and Z, no phylogenetic overlap would occur between the clades. Therefore, no paralogs would be included. However, if both clades contained sequences of species belonging to lineages X, Y, and Z, the clades would have diverged owing to gene duplication before the divergence of the three lineages (supplementary fig. S6A, Supplementary Material online). In this case, the paralogs would be removed by retaining only the sequences in one clade (supplementary fig. S6B, Supplementary Material online). Therefore, in the phylogenetic tree of a candidate ortholog group, OrthoPhy obtained information about the lineage to which the species in the clades located at both ends of each branch belong and determined whether the aforementioned conditions are met. If they did, the sequences in the clades with fewer sequences were removed as paralogs.
Paralogs are detected in a bottom–up approach, tracing from each leaf of the phylogenetic tree to its parent nodes. If any of the conditions (i)–(iii) are met at each node, the paralogs are successively removed. These processes removed all paralogs from the candidate ortholog group, resulting in an ortholog group consisting only of sequences that are orthologous to each other.
Reanalysis With a Stepwise Increase in Inflation Value During Clustering
Some sequences removed as paralogs in the above process can still be partially usable as orthologs. Therefore, Markov clustering was performed for all removed sequences to generate candidate ortholog groups, and paralogs were removed from the obtained groups. Herein, by increasing the inflation value used for the Markov clustering, the generated groups included only sequences with high similarity, and sequences with low similarity were excluded, enabling the detection of orthologs of sequences that were removed as paralogs in the previous process (fig. 5). By default, an inflation value of 1.2 was set for the first Markov clustering, and the subsequent Markov clustering and paralog removal processes were performed by increasing the inflation value by 0.2 until it reached 2.0.
Fig. 5.
Markov clustering results based on different inflation values. Sequences connected by a two-way arrow indicate reciprocal best-hit pairs, and thick arrows indicate higher similarity. Letters and numbers represent species and sequences, respectively. Black and gray sequences are paralogous with each other, and sequences of the same color are orthologs. Candidate ortholog groups obtained for inflation values of (A) 1.2 and (B) 2.0. In (A), paralogs are included in the same group surrounded by a black border. In (B), paralogs are divided into two groups consisting only of orthologs.
Inference of a Species Tree Using an Ortholog Data Set
Ortholog groups in the inferred ortholog data set that satisfy the threshold of the number of species were obtained. By default, only ortholog groups that are conserved among more than half of the analyzed species were used. For each ortholog group, multiple alignments were performed using MAFFT, and nonconserved regions were removed by trimAl. The phylogenetic trees of each ortholog group were inferred using FastTree based on these sequences. Finally, the species tree was inferred by Astral using the phylogenetic trees of ortholog groups.
Performance Test Using Simulation Sequences
We generated protein sequence data using an evolutionary simulation program and constructed an ortholog data set using these sequences to evaluate the performance of OrthoPhy. Here, the ortholog data set consists only of ortholog groups. The following three tests were conducted, each with a different phylogenetic tree model used for sequence generation. The number of iterations for each analysis was set to 100.
Symmetric Phylogenetic Tree
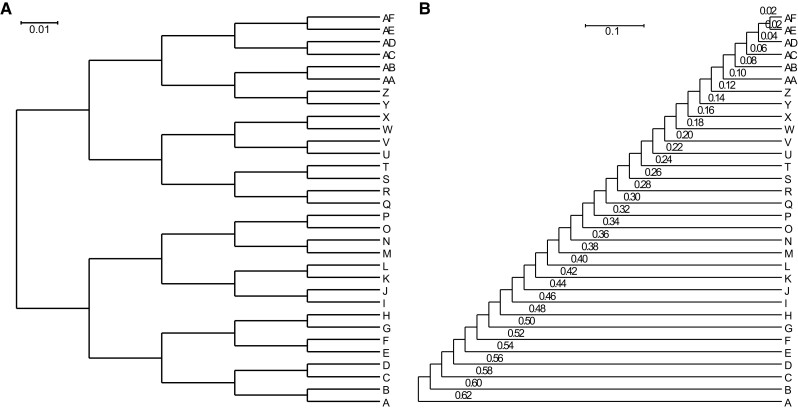
Fifty gene trees were generated by Zombi (Davín et al. 2020) using the symmetric phylogenetic tree model of 32 OTUs (fig. 6A). The paralogs were generated by gene duplicates with a certain probability during the generation of the gene trees. Next, the amino acid sequences of each homolog group were generated using INDELible (Fletcher and Yang 2009), with an LG substitution model based on each generated phylogenetic tree. The ancestral sequence length was set to a number that followed a gamma distribution, with a mean of 403 and a standard deviation of 299, which is the distribution of real protein sequence lengths (Tiessen et al. 2012). If more than one sequence of the same species was included in each generated homolog group, one was retained and the others were removed so that all included paralogs were considered hidden paralogs. Finally, the sequences of each generated homolog group were grouped by species, and the ortholog information was deleted.
Fig. 6.
Phylogenetic tree models. (A) Symmetric tree of 32 operational taxonomic units with the length of internal and external branches = 0.02. (B) Asymmetric tree of 32 operational taxonomic units with the length of internal branches = 0.02.
We constructed an ortholog data set in OrthoPhy based on the proteome data generated by the above process under the following conditions. Instead of the default settings, inflation was set in the range of 1.2–1.4. For species tree inference, we used orthologs that were conserved in at least 25% of species. We defined the following three conditions for the given taxonomic information: no taxonomic information (OP0, OrthoPhy without taxonomic information), species A-P and Q-AF were monophyletic (OP2, the number of groups is two), and species A-H, I-P, Q-X, and Y-AF were monophyletic (OP4, the number of groups is four). We also calculated the “Recall” (percentage of ortholog pairs included in the constructed ortholog data set compared with all ortholog pairs included in the input data set), “Precision” (percentage of correct ortholog pairs compared with all sequence pairs included in the constructed ortholog data set), and “Species_concordance” (percentage of species trees inferred from the ortholog data sets that matched the phylogenetic tree model). The same evaluation tests were also conducted for the existing programs, which can infer the ortholog group, OF and OMA. On the other hand, since OrthoFinder is the most popular program for constructing ortholog data sets but does not infer ortholog groups, we infer ortholog groups by the following method: First, orthogroup information was obtained by OrthoFinder. Next, in each orthogroup, a graph is constructed, with each sequence as a node and each orthologous node connected by edges at each node. The clique with the largest number of included sequences based on this graph is then extracted as the ortholog group. The ortholog data set was used to perform the same evaluation test as OrthoPhy.
Asymmetric Phylogenetic Tree
We used the asymmetric phylogenetic tree shown in figure 6B as a phylogenetic tree model to generate proteome data in the same manner as in Symmetric Phylogenetic Tree and ran OrthoPhy. We had two conditions of given taxonomic information. In the first condition, species A and B were independent lineages, and the other species (C-AF) were monophyletic (OP3, number of groups: 3). In the second condition, species A–I were independent lineages, and the other species (J–AF) were monophyletic (OP10, number of groups: 10). We also ran OrthoPhy without taxonomic information (OP0) and conducted the same tests for OF, OrthoFinder, and OMA. Finally, we evaluated each constructed ortholog data set and the inferred species tree in the same way as in Symmetric Phylogenetic Tree.
Random Phylogenetic Tree
When all lineages to which the analyzed species belong are known but the branching pattern within each lineage is unknown, ortholog inference by OrthoPhy and phylogenetic inference of the species using orthologs are considered particularly effective. We carried out the performance test according to the following procedure. A phylogenetic tree with four OTUs, which represents the branching pattern among the lineages to which the species to be analyzed belong, was randomly generated using Zombi (indicated in black in supplementary fig. S7, Supplementary Material online). For each leaf of the generated phylogenetic tree, a phylogenetic tree showing the branching pattern within the lineage was randomly generated using Zombi, and the generated phylogenetic trees were combined with the leaves (indicated in four colors in supplementary fig. S7, Supplementary Material online). The number of OTUs in the phylogenetic trees representing the branching within the lineage was randomized, but the minimum number of OTUs was set to 4. Then, the total number of OTUs in the phylogenetic trees after merging was set to 32. Based on the phylogenetic tree model created in the above process, proteome data were generated in the same manner as in Symmetric Phylogenetic Tree, and OrthoPhy was run. We investigated three conditions of given taxonomic information: no taxonomic information (OP0, OrthoPhy without taxonomic information), species in each of the two clades branching from the root are monophyletic (OP2, the number of groups is two), and species in each clade diverging from n1 and n2 are monophyletic when the nodes located one level downstream from the root are denoted as n1 and n2 (OP4, the number of groups is four). We conducted the same tests for OF, OrthoFinder, and OMA. Finally, we evaluated each constructed ortholog data set and the inferred phylogenetic tree of the species in the same way as in Symmetric Phylogenetic Tree.
Performance Test for 40 Gram-positive Bacteria
Using the genome data of 40 gram-positive bacteria (table 1), the ortholog data set was constructed, and a species tree was inferred by OrthoPhy. OrthoPhy was run with taxonomic information, that is, the analyzed species were classified into two phyla, and with taxonomic information, that is, they were classified into five classes, respectively. The process of prediction and removal of horizontal transferred genes was also performed, and orthologs that were conserved in at least 70% of the analyzed species were used for species tree inference. For comparison, ortholog data set construction and species tree inference were also performed in OF, OrthoFinder, and OMA using the same genome data. All programs were run with default settings except that the prediction of horizontal transferred genes was executed in OF.
Performance Test Using Quest for Orthologs Benchmark
To evaluate the performance of OrthoPhy using real sequence data, we conducted a benchmark test provided by QfO. Among the QfO tests, those classified as generalized species tree discordance tests were used to evaluate the ortholog data set based on the RF distance between the phylogenetic trees of inferred orthologs and the species tree (the phylogenetic range of the target species varies depending on the test). In other words, a lower score in the test represents a higher concordance rate between the phylogenetic tree constructed based on inferred orthologs and the species tree. Therefore, these tests can be used to evaluate the program to construct ortholog data sets for phylogenetic analysis of species. The proteome data set “2018_4” was used for tests, but variant data were not included. We ran OrthoPhy for three types of taxonomic information: (1) analyzed species were divided into three domains (bacteria, archaea, and eukaryotes) for a total of three groups (three-domain information); (2) eukaryotes were divided into five supergroups (Amoebozoa, Archaeplastida, Excavata, Opisthokonta, and SAR), archaea were divided into two groups (TACK and Euryarchaeota), and bacteria were classified into one group for a total of eight groups (eukaryotic information); and (3) other eukaryotes were divided into five supergroups, with fungi further divided into three phyla (Ascomycota, Basidiomycota, and Chytridiomycota), and archaea and bacteria were classified into one group each for a total of 10 groups (fungi information). The phylogenetic trees of orthologs inferred with the conditions in (3), (2), and (1) were compared with the phylogenetic trees of fungi and eukaryotes and the phylogenetic tree containing species from three domains, respectively, and they were evaluated based on the RF distance.
Supplementary Material
Acknowledgment
This research received no specific grant from any funding agency.
Contributor Information
Tomoaki Watanabe, United Graduate School of Agricultural Science, Gifu University, Gifu, Japan.
Akinori Kure, Graduate School of Integrated Science and Technology, Shizuoka University, Shizuoka, Japan.
Tokumasa Horiike, Department of Bioresource Sciences, Shizuoka University, Shizuoka, Japan.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
OrthoPhy, the program developed in the present study, is available on Github (https://github.com/tmakwtnb/OrthoPhy). The real sequence data used in the performance evaluation tests can be downloaded from Quest for Orthologs’ site (https://ftp.ebi.ac.uk/pub/databases/reference_proteomes/previous_releases/qfo_release-2018_04/QfO_release_2018_04.tar.gz).
Literature Cited
- Altenhoff AM, et al. . 2019. OMA Standalone: orthology inference among public and custom genomes and transcriptomes. Genome Res. 29:1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, et al. . 2020. The quest for orthologs benchmark service and consensus calls in 2020. Nucleic Acids Res. 48:W538–W545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Bertelli C, Brinkman FSL. 2018. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 34:2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost HG. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 18:366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino S, Iwasaki W. 2019. Sonicparanoid: fast, accurate and easy orthology inference. Bioinformatics 35:149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davín AA, Tricou T, Tannier E, de Vienne DM, Szöllosi GJ. 2020. Zombi: a phylogenetic simulator of trees, genomes and sequences that accounts for dead linages. Bioinformatics 36:1286–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A. 2009. HaMStR: profile hidden Markov model based search for orthologs in ESTs. BMC Evol Biol. 9:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. Orthofinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30:1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Biol. 27:401–410. [Google Scholar]
- Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 17:368–376. [DOI] [PubMed] [Google Scholar]
- Fletcher W, Yang Z. 2009. INDELible: a flexible simulator of biological sequence evolution. Mol Biol Evol. 26:1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Rosenberg MS, Kumar S. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zool B Mol Dev Evol. 304B:64–74. [DOI] [PubMed] [Google Scholar]
- Hall BG. 2005. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol. 22:792–802. [DOI] [PubMed] [Google Scholar]
- Horiike T, Minai R, Miyata D, Nakamura Y, Tateno Y. 2016. Ortholog-finder: a tool for constructing an ortholog data set. Genome Biol Evol. 8:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Friedberg I. 2019. Swiftortho: a fast, memory-efficient, multiple genome orthology classifier. GigaScience 8:giz118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, et al. . 2016. A new view of the tree of life. Nat Microbiol. 1:16048. [DOI] [PubMed] [Google Scholar]
- Kaduk M, Sonnhammer E. 2017. Improved orthology inference with Hieranoid 2. Bioinformatics 33:1154–1159. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2005. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 39:309–338. [DOI] [PubMed] [Google Scholar]
- Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. 2011. Computational methods for gene orthology inference. Brief Bioinform. 12:379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Orgogozo V. 2011. Identification of homologous gene sequences by PCR with degenerate primers. Methods Mol Biol. 772:245–256. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Lechner M, et al. . 2011. Proteinortho: detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. . 2006. Treefam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34:D572–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal G, Hanmandlu M, Srivastava S. 2019. Phylogenetics algorithms and applications. Adv Intell. 904:187–194. [Google Scholar]
- Pagani I, et al. . 2012. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 40:D571–D579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, et al. . 2017. Orthograph: a versatile tool for mapping coding nucleotide sequences to clusters of orthologous genes. BMC Bioinform. 18:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. 2005. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. Fasttree 2 - approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol. 43:304–311. [DOI] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math Biosci. 53:131–147. [Google Scholar]
- Spencer M, Susko E, Roger AJ. 2005. Likelihood, parsimony, and heterogeneous evolution. Mol Biol Evol. 22:1161–1164. [DOI] [PubMed] [Google Scholar]
- Sterner B, Lidgard S. 2018. Moving past the systematics wars. J Hist Biol. 51:31–67. [DOI] [PubMed] [Google Scholar]
- Storm CEV, Sonnhammer ELL. 2002. Automated ortholog inference from phylogenetic trees and calculation of orthology reliability. Bioinformatics 18:92–99. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F. 2016. Inferring orthologs: open questions and perspectives. Genom Insights. 9:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Pérez-Rodríguez P, Delaye-Arredondo L. 2012. Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes. 5:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden RTJM, Snel B, van Noort V, Huynen MA. 2007. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinform. 8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. 1993. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340–342. [DOI] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Zhang C, Mirarab S. 2019. ASTRAL-MP: scaling ASTRAL to very large datasets using randomization and parallelization. Bioinformatics 35:3961–3969. [DOI] [PubMed] [Google Scholar]
- Zhang C, Scornavacca C, Molloy EK, Mirarab S. 2020. ASTRAL-Pro: quartet-based species-tree inference despite paralogy. Mol Biol Evol. 37:3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
OrthoPhy, the program developed in the present study, is available on Github (https://github.com/tmakwtnb/OrthoPhy). The real sequence data used in the performance evaluation tests can be downloaded from Quest for Orthologs’ site (https://ftp.ebi.ac.uk/pub/databases/reference_proteomes/previous_releases/qfo_release-2018_04/QfO_release_2018_04.tar.gz).