Abstract
Interferon-gamma (IFNG) has long been regarded as the flag-bearer for the anti-cancer immunosurveillance mechanisms. However, relatively recent studies have suggested a dual role of IFNG, albeit there is no direct experimental evidence for its potential pro-tumor functions. Here we provide in vivo evidence that treatment of mouse melanoma cell lines with Ifng enhances their tumorigenicity and metastasis in lung colonization allograft assays performed in immunocompetent syngeneic host mice, but not in immunocompromised host mice. We also show that this enhancement is dependent on downstream signaling via Stat1 but not Stat3, suggesting an oncogenic function of Stat1 in melanoma. The experimental results suggest that melanoma cell-specific Ifng signaling modulates the tumor microenvironment and its pro-tumorigenic effects are partially dependent on the γδ T cells, as Ifng-enhanced tumorigenesis was inhibited in the TCR-δ knockout mice. Overall, these results show that Ifng signaling may have tumor-promoting effects in melanoma by modulating the immune cell composition of the tumor microenvironment.
Introduction
Cutaneous malignant melanoma is a complex, highly aggressive, and frequently chemoresistant cancer that continues to exhibit a positive rate of increase in the developed world 13, 40, 56. Numerous epidemiological studies have identified the solar ultraviolet radiation (UV/UVR) to be the major etiological risk factor for melanoma 14, 15, 35, with the highest risk associated with intermittent burning doses, especially during childhood 1, 3, 31, 52, 58. However, the underlying molecular mechanisms by which UVR (UVB and UVA wavebands) initiate melanomagenesis remain poorly understood. Although numerous studies have amassed strong evidence that UVB-induced signature DNA mutations play key roles in melanomagenesis, there is compelling evidence that non-mutational mechanisms, such as UVR-induced inflammation and immunosuppression, also contribute substantially to melanomagenesis 19, 32, 43, highlighting the importance of mechanisms other than direct DNA damage in UVR-induced initiation and/or progression of melanoma.
To uncover the molecular mechanisms underlying UVR-induced melanomagenesis, we previously investigated the genomic response of melanocytes to UVB and UVA radiation 59. We showed that answers to many of the questions regarding UVR-induced melanomagenesis lie not only in how UVR damages melanocyte DNA but also in how altered gene expression in the exposed melanocytes drives their interactions with the elements of the microenvironment to remodel damaged skin and allows UVR-damaged (mutated) melanocytes escape immunosurveillance-based destruction. We showed that UVB can directly upregulate melanocytic expression of ligands to the chemokine receptor CCR2, which recruits macrophages into the neonatal skin microenvironment. A subset of these macrophages produces interferon-gamma (IFNG) into the tumor microenvironment, which we postulated to be paradoxically pro-melanomagenic 59.
IFNG is known to play a central role in cancer immunosurveillance and immunoediting 21, 50. IFNG is also associated with anti-proliferative and anti-angiogenic, as well as anti-tumor immune responses against a variety of different cancers, including melanoma 5, 12, 21, 54, 57 For example, it has been reported that IFNG had significant growth inhibitory activity on four different human melanoma cell lines; albeit at concentrations that were up to 10,000-fold higher than physiologic levels 27. However, a potentially pro-tumorigenic role of IFNG has been postulated 60, 61. There is some indirect evidence that indicates that IFNG can have contrasting roles in tumorigenesis, i.e., it can exhibit both cytostatic/cytotoxic and anti-tumorigenic immunosurveillance functions as well as pro-tumorigenic immune-evasive effects in the tumor microenvironment in a context-dependent manner. Here we report in vivo evidence that activation of IFNG signaling directly in melanoma cells enhances their tumorigenicity and metastasis, which is partially dependent on the melanoma cell-driven modulation of the tumor immune microenvironment.
Results
Ifng treatment of mouse melanoma cells enhances lung colonization and metastasis
To understand the effects of IFNG signaling on melanoma cells, we studied mouse melanoma cell growth in vitro with and without treatment with 10 ng/ml of recombinant Ifng. The mouse melanoma cell lines tested showed differential response to Ifng. The B16 and its derivative cell line B16N showed significant reduction in proliferation (Extended Data Fig. 1a,b). Surprisingly, however, the proliferation of B2905, F5061, and YUMM1.1 cell lines was not affected by Ifng treatment, as no significant differences were seen in proliferation (Extended Data Fig. 1c-e). Similarly, while the soft agar colony formation of B16 and B16N cells was significantly inhibited by Ifng treatment (10 ng/ml) (Extended Data Fig. 2a,b), B2905 and F5061 cell lines did not show any difference in colony formation after Ifng treatment (Extended Data Fig. 2c,d).
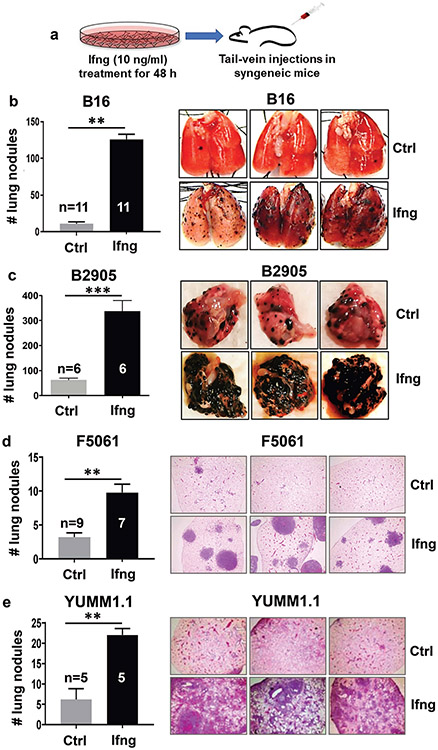
To assess the role of Ifng on melanoma tumorigenesis in mouse allograft model systems, we pretreated 5 mouse melanoma cell lines (B16, B16N, B2905, F5061, and YUMM1.1) in culture for 48 h with 10 ng/ml Ifng and implanted them into immune-competent syngeneic mice (C57BL/6 or FVB/N) via subcutaneous or tail-vein injections (Fig. 1a). This Ifng treatment regimen was selected because it produced optimal activation of the classic Ifng responsive genes Irf1 and Gbp4, which remained elevated 2 d following the removal of Ifng from the media but reduced to baseline level by 4 d (Extended Data Fig. 3). While no differences were detected in subcutaneous tumorigenesis between the mock-treated control and Ifng-treated groups (Extended Data Fig. 4), the tail-vein inoculation models showed a striking increase in the number of tumor nodules on the lung surfaces and/or by histopathological analyses of the lung tissues in the mice harboring Ifng-treated cells of all 5 melanoma cell lines (Fig. 1b-e, Fig. 2a, and Extended Data Fig. 5). The pigmented lung nodules of the B16, B16N, and B2905 cells could be observed both visually and histologically (Fig. 1b,c), whereas those of F5061 and YUMM1.1 cells were unpigmented and were only analyzable by histological analyses (Fig. 1d,e). To assess the effect of Ifng treatment on the metastatic potential of the B16N cells (a novel metastatic clone of B16) 34, we injected Ifng-treated and control B16N cells with ectopic expression of luciferase via tail vein in syngeneic C57BL/6 mice and monitored tumor growth by live animal bioluminescence imaging. We observed a significant increase in tumor growth in mice harboring Ifng-treated cells as compared to the controls, as measured by bioluminescence (Fig. 2b,c). Posthumous analyses also showed a statistically significant increase in extrapulmonary metastases in the ovary, bones, parametrium, and kidney tissues of the mice inoculated with Ifng-treated cells as compared with the mock-treated controls (Fig. 2d).
Figure 1.
Induction of Ifng signaling enhances melanoma tumorigenesis. a, An illustration of the experimental plan is shown. Mouse melanoma cell lines were treated with 10 ng/ml recombinant mouse Ifng (or mock treated, Ctrl) for 48 h, followed by inoculation in syngeneic mice via tail vein injections. b-e, Quantification of tumor nodules in lungs, either by counting surface nodules on gross lungs (for pigmented tumors) or in H&E-stained sections from paraffin-embedded lung tissues (for unpigmented tumors). Significantly increased tumor counts were observed in all mice inoculated with Ifng-treated cell lines. Lungs recovered 20-28 days after injection from mice bearing (b) B16 and (c) B2905 cells were paraformaldehyde-fixed, quantified, and photographed using a dissecting microscope. Pulmonary tumors appeared as black pigmented nodules on the surface of the lungs. For the unpigmented cell lines, (d) F5061 and (e) YUMM1.1, representative photomicrographs of 4x microscopic fields of view in H&E-stained sections are shown. 3 slides per mouse, 3 fields per slide, for a total of ≥45 fields were quantified per group. Data are presented as mean±SEM. n = number of mice used for each group. *P< 0.05, **P<0.01, ***P<0.001), Student’s t test.
Figure 2.
Ifng signaling enhances metastasis. Bioluminescent B16N metastatic melanoma cell line was treated (or mock treated, Ctrl) with Ifng (10 ng/ml) for 48 h, followed by tail vein injection into syngeneic C57BL/6 mice. Tumor growth was monitored by live animal bioluminescence imaging at days 5, 8, 12, 16 and 20. Lung surface nodules were counted at terminal autopsy on day 20. a, Quantification of lung surface tumor nodules. n = 5 mice each group. Representative lungs are shown. Data are presented as mean±SEM. **P<0.01, Student’s t test. b, Total tumor burden as measured by bioluminescence (photon flux) imaging in live mice. Metastasis by Ifng-treated B16N cells was significantly increased compared to the control cells. *P<0.05, **P<0.01, Student’s t test. c, Bioluminescence imaging on day 20 after cell inoculation. d, The frequency and distribution pattern of extrapulmonary metastases was determined in the indicated tissue samples collected at 20 d after tail vein injection by a detailed histopathological analysis. The frequency of extrapulmonary metastases was significantly greater (*p<0.05, chi -square test) in the mice inoculated with Ifng-treated B16N cells.
Stat1 but not Stat3 is the downstream effector of the pro-tumorigenic effects of Ifng signaling
The activation of the canonical Ifng signaling pathway leads to the phosphorylation of Jak1 and Jak2, followed by phosphorylation and homodimerization of the transcription factor Stat1, which translocates to the nucleus to activate its target genes. However, in some contexts, Ifng can also activate Stat3 45. We observed phosphorylation of both Stat1 and Stat3 upon Ifng treatment in mouse melanoma cell lines (Extended Data Fig. 6). To determine whether the Ifng-mediated tumorigenic effect on melanoma cells was routed through Stat1 or Stat3, we generated knockout (KO) clones for both Stat1 and Stat3 in the B16N cell line utilizing the CRISPR/Cas9 methodology. The knockout clones were verified by western blotting to confirm the absence of the respective proteins (Extended Data Fig. 6). The B16N-Stat1-KO and B16N-Stat3-KO cells were treated with 10 ng/ml Ifng or mock-treated as above and their proliferation and colony formation were assessed. The results showed that while the parental WT B16N cells and their Stat3-KO counterparts were significantly inhibited in proliferation by Ifng treatment, this effect was nullified in the Stat1-KO cells (Extended Data Fig. 7a). In the colony formation assay, Stat1-KO cells did not respond to Ifng treatment; however, colony formation was suppressed in Stat3-KO cells (Extended Data Fig. 7b,c).
To test the in vivo effects of Stat1-KO and Stat3-KO, cells were inoculated via tail vein in syngeneic mice. While the Ifng-treated B16N-Stat3-KO cells showed a statistically significant increase in lung tumorigenesis as compared to the controls (Fig. 3a), which was similar to the parental B16N cells, the tumorigenicity of the B16N-Stat1-KO cells was drastically and statistically significantly inhibited and there was no difference in lung colonization between Ifng-treated and control B16N-Stat1-KO cells (Fig. 3b). These results suggested that Stat1, but not Stat3, is the downstream mediator of the tumorigenic effects of Ifng signaling on melanoma cells.
Figure 3.
Ifng-mediated enhancement of lung tumorigenesis is through Stat1 but not Stat3. (a) Stat3-knockout (Stat3-KO) and (b) Stat1-KO B16N cells were treated with 10 ng/ml Ifng (or mock treated, Ctrl) and injected by tail vein into C57BL/6 mice. Lung surface tumor nodules were quantified at 20 d. n = 10 each group. n.s. = not significant; **P<0.01, Student’s t test. c, Ifng signaling in melanoma cells enhances tumorigenesis independently of the host systemic Ifng signaling, which is anti-tumorigenic. Untreated B16N cells were tail vein inoculated in either wild type C57BL/6 (n = 5) or C57BL/6-Ifng-knockout host mice (n = 9). The untreated cells exhibited significantly enhanced tumorigenesis in the Ifng-KO host mice. Ifng-treated B16N cells showed further enhancement of tumorigenesis in Ifng-KO host mice (n = 11). Data are presented as mean±SEM of lung surface tumors. *P<0.05, **P<0.01, Student’s t test.
To determine the comparative contribution of the systemic Ifng signaling of the host mice and the intracellular Ifng signaling in melanoma cells to the enhancement of tumorigenicity, we injected untreated parental B16N cells in wildtype C57BL/6 and Ifng-knockout (C57BL/6) host mice. There was a statistically significant increase in tumorigenicity of the cells in the Ifng-KO host mice, confirming the essential role of Ifng in the systemic anti-tumor immunosurveillance mechanisms. Remarkably, a further additive increase in the cells’ tumorigenicity was seen when the cells were treated with Ifng followed by injection into Ifng-KO host mice (Fig. 3c). These results suggest dual and opposite functions of Ifng signaling wherein it may play an anti-tumor role in the context of the systemic immunosurveillance but may have a pro-tumor effect on the melanoma cells.
Ifng-enhanced melanoma tumorigenicity is dependent on the immune system
To test whether the enhancement of tumorigenicity and metastatic capabilities of melanoma cells by Ifng is dependent on the presence of the immune system, we implanted B16N and B2905 cells, with or without treatment with Ifng, in the immunocompromised NOG mice via tail vein (Fig. 4a,b). There were no statistical differences in lung colonization between the treated and control groups, indicating that the Ifng-mediated enhancement of tumorigenesis required a functional immune system. Similar insignificant results were obtained when a human melanoma cell line A2058, with or without IFNG treatment (10 ng/ml for 48 h), was inoculated in the NOG mice via tail vein (Fig. 4c,d). To test whether the absence of the T cell compartment was responsible for the lack of Ifng-mediated enhancement of tumorigenesis in the immunocompromised context, we inoculated Ifng-treated and control B16N cells in the athymic Nude (Foxn1nu) mice via tail vein. Interestingly, we observed a greatly reduced number of lung tumor nodules but robust extrapulmonary metastatic spread, both of which were not statistically different between the Ifng-treated and control groups (Fig. 4e). These results implicated the modulation of the T cell-mediated immunity as an important player in the Ifng-mediated enhancement of melanoma tumorigenesis.
Figure 4.
Host immune response is necessary for the Ifng-mediated enhancement of melanoma lung colonization. Mouse melanoma cells (a) B16N and (b) B2905 were treated with 10 ng/ml of Ifng and inoculated into the immunodeficient NOG mice (n = 7 for each group) via tail vein. Quantification of lung surface tumor nodules is shown as mean±SEM. n.s. not significant; Student’s t test. c, A2058 human melanoma cells were treated (or mock treated, Ctrl) with human recombinant IFNG and injected into NOG mice through the tail vein and monitored by bioluminescence imaging (BLI). BLI images at day 0 immediately after tail vein injection and on 18 d are shown. d, Total tumor burden (photon flux) as measured by BLI. n.s. not significant, One-Way ANOVA with post-hoc analysis. e, Ifng-treated (n = 6) and Ctrl (n = 5) B16N cells were tail vein inoculated in the immunocompromised Nude mice. Lung surface tumor nodules were quantified at 18 d post inoculation. n.s. not significant; Student’s t test.
Ifng-treated melanoma cells modulate the tumor immune microenvironment
Since we found no difference in lung tumorigenesis of Ifng-treated versus untreated melanoma cells when inoculated in T cell-deficient hosts, we hypothesized that Ifng-treated melanoma cells modulate their tumor microenvironment where T cells play a crucial role. To compare the tumor microenvironment (TME) between untreated versus Ifng-treated melanoma lung tumor nodules, we microdissected the tumor nodules from the lungs and performed immune phenotyping by multi-color flow cytometry. Interestingly, the tumor nodules formed by the Ifng-treated melanoma cells and the tumors from the control melanoma cells did not show a statistically significant difference in the tumor-infiltrating CD45+ cells in the TME (Fig. 5a). We found no significant differences in the frequencies of tumor-infiltrating lymphocytes (TILs), CD4, CD8, and Treg cells between the two groups (Fig. 5b,d,e). However, further analyses revealed a statistically significant increase in CD4+CD8+ dual-positive (DP) T cells (Fig. 5d,i). Surprisingly, the frequency of tumor-associated neutrophils (TAN) was increased >2-fold in the Ifng-treated melanoma TME (Fig. 5c,i). Although the frequency of the infiltrating γδ T cell was similar, we observed a significant increase in the CD27-CCR6- γδ T cells with a concomitant decrease in the CD27+ γδ T cells in the Ifng-treated melanoma TME as compared to the control tumors (Fig. 5b,f,i,k). The CD27+ γδ T cell are well known to produce Ifng, whereas the CCR6+ γδ T cells produce IL-17 and are known to perform a pro-tumorigenic role 48. Therefore, we tested the cytokine production from γδ T cells, CD4, and CD8 T cells. As expected, γδ T cells from the Ifng-treated melanoma TME failed to produce Ifng (Fig. 5h,k), whereas they readily produced both TGFβ and IL-17 upon in vitro stimulation with PMA and Ionomycin (Fig. 5h,l). We also observed a trend of harboring a reduced frequency of Ifng-producing CD4+ cells in the TME in the Ifng-treated tumors (Fig. 5g). Altogether, our analyses suggested that Ifng-treated melanoma cells created a TME that was enriched in neutrophils and IL-17-producing and TGFβ-producing γδ T cells.
Figure 5.
Analyses of immune cell infiltration in the tumor microenvironment. Lung tumors were isolated by microdissection from Ifng-treated and untreated B2905 tumor bearing mice at 23 d post inoculation. Tumor infiltrating immune cell profiles were generated by flow cytometric analysis. Relative cell frequencies within the indicated gated immune cell subset are shown for total lymphocytes (a), B cells (b), αβ T cells (b, d, i), γδ T cells (b, f, h, k) and myeloid cells (c, j). Cytokine production of infiltrating T cells in response to PMA and Ionomycin stimulation was measured by intracellular staining (g, h, k, l). Data displayed in panels a-h represent mean ± SEM. Significance of differences was determined by One-Way ANOVA with post-hoc Bonferroni test, as marked by: *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
Ifng-enhanced metastasis of melanoma cells is partially dependent on γδ T cells
To further study whether the γδ T cells play a pivotal role in shaping a strong pro-tumorigenic microenvironment as opposed to the αβ T cells, we inoculated via tail-vein Ifng-treated and control B2905 melanoma cells in TCRδ−/− and TCRβ−/− mice. The TCRδ−/− mice lack the γδ T cell receptor expression in all adult lymphoid and epithelial cells and lack the γδ T cells 23. The TCRβ−/− mice lack the αβ T cells 36. In contrast to WT and TCRβ−/− host mice, we observed a statistically significant reduction in lung colonization and metastasis of Ifng-treated cells in the TCRδ−/− mice (Fig. 6a,b). In addition, the TCRδ−/− host mice exhibited a statistically significant reduction in the metastasis of the untreated control cells. A flow cytometry analysis of the TME revealed that lack of γδ T cells caused a substantial reduction in neutrophil infiltration in the TME of the tumors made by both Ifng-treated and untreated control cells (Fig. 6c,d). These results suggest a crucial role of γδ T cells in neutrophil infiltration. A deficiency of the γδ T cells enhanced the infiltration of B cells in the TME (Extended Data Fig. 8a-e), though we did not find any effect on the total T cells as CD4+, and CD8+ T cells in both groups (Extended Data Fig. 8b). Interestingly, we observed a marked decrease in the CD25-hi FoxP3+ Treg cells in the Ifng-treated TME in the TCRδ−/− mice, as compared to WT, though the frequency of total Treg in TME did not change significantly (Fig. 6f, g). Further analyses of the cytokine profile revealed that Ifng-producing CD4+ T cells were substantially increased in the Ifng-treated TME in the TCRδ−/− host mice, as compared to WT (Fig. 6h,i). We also observed a lack of γδ T cells-enhanced Ifng production by CD8+ TILs (Fig. 6j). Collectively, our data suggest that Ifng signaling in melanoma cells enhances lung colonization and tumorigenesis partially via driving the recruitment of IL-17 and TGFβ-producing γδ T cells to enhance neutrophil recruitment to create a pro-tumorigenic microenvironmental niche.
Figure 6.
Requirement for gamma-delta T cells in Ifng-enhanced melanoma lung tumorigenesis. Ifng-treated and untreated B2905 melanoma cells were inoculated into syngeneic wild type, TCRβ−/− and TCRγ−/− mice by tail vein inoculation. a, Mice were euthanized on 23 d and lung surface tumors were counted. b, The proportion of lungs affected by metastases was determined by histological analyses. Tumor infiltrating immune cell profiles were assessed by flow cytometry. Relative distributions of different populations of myeloid cells (c, d, e), and Tregs (f, g) in the tumor microenvironment were determined. IL-17 and Ifng production by the infiltrating T cells in response to PMA and Ionomycin stimulation was measured by intracellular staining (h, I, j). Data displayed in panels a, b, d, e, g, i and j represent mean±SEM. Significance of differences was determined by One-Way ANOVA with post-hoc Bonferroni test: *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
Discussion
The tumor microenvironment (TME) is a dynamic assortment of pro-tumor and anti-tumor molecular forces. The success or failure of the establishment and progression of a tumor is dependent on which forces outperform and overpower the others in the TME. Immunosuppressive cell networks and factors play a significant role in the failure of the anti-tumor immune responses and therapies 22. The interferon-gamma (IFNG) cytokine has long been regarded as a marquee orchestrator of the anti-tumor immunosurveillance mechanisms. A recent study suggested that the IFNG transcriptome response in melanoma cells served to amplify the magnitude of the antitumor T cell response, and the corresponding downstream IFNG signaling factors are the main drivers of the clinical responses to immune checkpoint inhibition 18. Paradoxically, we had previously identified a pro-melanomagenic role of Ifng in the context of UV irradiation in mice 59. However, it was unclear whether the potential pro-melanomagenic effects of Ifng were due to its systemic functions or via direct stimulation of melanoma cell-mediated events. Our results presented here provide evidence that intracellular Ifng signaling in melanoma cells promotes melanomagenesis and that this promotion is brought about partially via skewing of the tumor immune microenvironment towards a pro-tumor character.
The concentration of IFNG that we have used (10 ng/ml) is an important experimental variable to justify. We and others have extensively used this IFNG concentration for in vitro applications, as it is widely accepted to be within physiologically relevant range 34, 38, 53. While the normal serum IFNG concentration is in 1-10 pg/ml range, it reaches as high as 80 ng/ml under some viral infection and inflammatory conditions 2, 39. However, in the context of the tumor microenvironment (TME), it is not the serum IFNG concentration but rather the local tumor microenvironmental concentration of IFNG that we believe carries greater relevance and determines the IFNG-mediated pro-tumorigenic effects. In the TME, there are numerous types of immune cells, e.g., cytotoxic T cells, NK cells, NK-T cells, Treg, B cells, and even γδ T cells themselves, that produce and secrete copious amounts of IFNG in the immediate TME and can serve as constant and long-term sources of IFNG. Capture by the extracellular matrix can further increase local concentrations and form gradients of IFNG 17. We chose 10 ng/ml IFNG concentration because it produced the optimal activation of its classic downstream target gene IRF1, as previously published 37, and increasing the IFNG concentration higher did not further increase this activation. Therefore, although the local concentration of IFNG in the TME is difficult to measure and predict, we believe that 10 ng/ml is a reasonable in vitro experimental application.
The specific role of STAT1 in cancer remains ambiguous, as it is known to exhibit both tumor suppressor as well as oncogenic properties in a context-dependent manner 63. Several types of cancers, including melanoma, show loss of STAT1, and its tumor suppressor function is mainly attributed to its role in the activation of pro-apoptotic (such as BCL2 and BCL-xL) and cell cycle inhibitor genes (e.g. p27 and p21WAF1) as well as negative regulation of angiogenesis 20, 26, 62. Several lines of evidence also implicate STAT1 as a key regulator of anti-cancer immunosurveillance by its regulation of the major histocompatibility complex (MHC) Class I and other genes of the antigen presentation machinery 6, 28, 49. The most convincing case for the tumor suppressor gene designation for STAT1 is provided by the Stat1-knockout mice, which exhibit enhanced susceptibility to spontaneous and carcinogen-induced tumorigenesis 9, 29. On the other hand, there is also some evidence for the oncogenic role of STAT1, albeit relatively less convincing 63. One possible reason for this dichotomy may rest upon how the analysis of STAT1 expression has been performed in different types of cancers. For example, analysis of the whole tumor tissue fails to distinguish between the STAT1 expression in the tumor cells versus the immune cells. It is plausible to speculate that the expression of STAT1, or lack thereof, may have contrasting effects depending on the cellular context. Our results presented here provide experimental evidence that Stat1 may play an oncogenic role in a transplant model of murine melanoma cells. Further studies will be needed to delineate this oncogenic role.
The γδ T cells perform crucial roles in the anti-tumor immune responses, e.g., cytotoxicity, production of IFNG and TNFα, and inducing the maturation of dendritic cells (DC). A recent analysis of expression signatures from ~18,000 human tumors with overall survival outcomes across 39 malignancies identified tumor-infiltrating γδ T cells as the most significant favorable cancer wide prognostic signature 16. However, both positive and negative correlations have been found between clinical responses and tumor-infiltrating γδ T cells. The positive correlation between the tumor-infiltrating γδ T cells and the clinical survival of the patients was observed in necrotizing choroidal melanomas 4, ovarian cancer 47, and melanoma 10. Interestingly, in breast cancer, a potential pro-tumor function was reported 44, highlighting that the infiltrating γδ T cells were able to inhibit the function of several immune cell populations in vitro and were involved in suppression of anti-tumor responses. Consistent with these observations, the presence of γδ T cells was shown to positively correlate with advanced tumor stages and inversely correlated with patient survival. A positive correlation between disease progression and the number of tumor-infiltrating γδ T cells was also observed in a cohort of breast cancer patients 30. These findings strongly suggest that γδ T cells in the tumor microenvironment may play substantially disparate functions; hence positive or negative correlation with prognosis may depend on the specific γδ T cell subset present in the TME.
Interestingly, Singh et al. have previously shown that the loss of ELF5-regulated ubiquitin ligase FBXW7, which stabilizes the interferon-gamma receptor 1 (IFNGR1) and enhances the IFNG signaling, promoted the tumor growth and metastasis of triple-negative breast cancer 51. This enhancement of tumorigenesis was accompanied by accumulation of neutrophils in the tumor microenvironment and depletion of Ly6G+ neutrophils reduced the tumor growth and metastasis 51. In this study, we found that although the frequency of the γδ T cells remained approximately the same in the TME, the TME of the tumors made by the Ifng-treated cells harbored mainly CD27- γδ T cells, which have previously been shown to produce IL-17 rather than Ifng 48. We found that the Ifng-treated melanoma cells induced a tumor microenvironment that was enriched in pro-tumorigenic IL-17, TGFβ-producing γδ T cells, and accumulation of immunosuppressive neutrophils. We also demonstrated that the genetic ablation of γδ T cells not only reduced the tumor burden and metastasis but also substantially reduced the accumulation of neutrophils, thus providing evidence of the involvement of pro-tumorigenic γδ T cells in neutrophil accumulation and melanoma progression and metastasis.
Most interestingly, ablation of the γδ T cells could also restore anti-tumor immune response in the Ifng-treated melanoma microenvironment as evidenced by an increased frequency of Ifng-producing CD4+ and CD8+ T cells and striking down-modulation of CD25 and Foxp3 expression in the Treg akin to ‘non-Treg’ 55. It has been shown that blocking the γδ T cells with UC7–13D5 monoclonal antibody inhibited microbiota-promoted lung cancer development 24. Our data corroborate these results and provide rationale for the clinical application of this therapeutic strategy against metastatic melanoma, which frequently metastasizes to lungs.
An important question that arises is what the downstream effects of IFNG signaling are on the melanoma cells that then potentially chemoattract the γδ T cells to the TME. Our RNA-Seq analysis of gene expression in the B16 and YUMM1.1 cells, and validated in B16, YUMM1.1, and B2905 cell lines, identified CXCL9 chemokine, which is a known chemoattractant for γδ T cells 64, to be one of the highest upregulated genes in response to IFNG treatment (Extended Data Figs. 9 and 10, and Extended Data Spreadsheet). On the other hand, CCL5 and ICAM1, which are also known to be involved in the chemotaxis and activation of γδ T cells 7, 25, were not consistently activated by IFNG in all these cell lines (Extended Data Fig. 10). These data suggest that tumor microenvironmental IFNG signaling activates melanoma cells to actively participate in modulating the TME towards a tumor-promoting characteristic, likely by secreting immune modulating chemokines such as CXCL9.
While cell proliferation and colony formation of B16 and B16N cells were inhibited by Ifng treatment in vitro, the subcutaneous tumor growth in vivo did not show any difference. This is an intriguing phenomenon that we have found to be specific to the B16 and its derivative B16N cell line. Among all the other mouse melanocyte and melanoma cell lines we have tested, none has shown an effect of IFNG on proliferation. This B16-specific effect does not appear to be due to downstream phosphorylation of STAT1 or activation of IRF1, as both are activated normally in these cells (Extended Data Figs. 3 and 6). Interestingly, however, IFNG failed to inhibit proliferation of B16N-Stat1-KO cells, but proliferation of B16N-Stat3-KO cells continued to be inhibited by IFNG (Extended Data Fig. 7). These results suggest that it is dependent on a downstream target(s) of STAT1. Further research will be needed to delineate this phenomenon at the molecular level.
Our results suggest that IFNG is a potential driver of novel cellular/molecular inflammatory mechanisms that may promote the outgrowth of melanoma. Since the expression of the IFNG-induced genes seemed to normalize within 48-96 h after IFNG was removed from the culture media, it is plausible that the pro-tumorigenic effects of IFNG are likely dependent on the early cascade of events set forth immediately after the cells have been transplanted. For example, an early establishment of chemokine gradient that chemoattracts γδ T cells is likely to be sufficient to modulate the tissue microenvironment to establish a pro-tumorigenic niche and promote the outgrowth of the tumor. In the context of the TME, further infiltration of immune cells can be a constant source of IFNG and persistent activation of its downstream effectors to sustain a tumor-promoting niche. While it adds further complexity to the already intricate tumor microenvironmental interactions, it also offers an opportunity to understand the process of melanoma progression from a new perspective. Moreover, understanding the yin and yang of the IFNG signaling may have implications in the efficacy of cancer immunotherapy, e.g. antibody-mediated blockade of PD-1/PD-L1 and CTLA4 8, 11.
Materials and Methods
Cell lines
Five mouse melanoma cell lines were used in this study. B16 cell line was obtained from Dr. Glenn Merlino (NCI/NIH). B16N is a novel metastatic clone of B16, established at NCI B16 and B16N both are syngeneic to the C57BL/6 strain background. The B2905 (C57BL/6) and F5061 (FVB/N) cell lines were derived from spontaneous tumors induced by UV irradiation of the hepatocyte growth factor/scatter factor (HGF/SF) transgenic mice 41, 42. YUMM1.1 cell line was isolated from a BrafV600E; Pten−/−; Cdkn2a−/− mouse melanoma was obtained from Dr. Marcus Bosenberg 33. Human melanoma cell line A2058 was purchased from ATCC (CRL-11147). All cell culture media and supplements were purchase from Life Technology. All tumor cell lines were cultured at 37°C in DMEM supplemented with 10% FBS, l-alanyl-l-Glutamine (2 mmol/L), and Gentamycin (50 μg/mL) at 5% CO2. DMEM, FBS, and l-alanyl-l-Glutamine were purchased from Corning, Cellgro. All cell lines were authenticated by genotyping and/or RNA sequencing and were tested for mycoplasma contamination.
Generation of CRISPR/Cas9-mediated knockout cell lines
We designed two guide RNAs targeting different exons of Stat1 and Stat3 loci by online CRISPR Design Tool. The Cas9 expression construct pSpCas9(BB)-2A-GFP was purchased from Addgene (Plasmid ID 44758). Stat1 (NM_001205313.1) and Stat3 (NM_213659) were used to search gRNA using the online CRISPR Design Tool (http://tools.genome-engineering.org) 46. Two different gRNA targeting different exons were used for both Stat1 and Stat3. The sequence of gRNA_Stat1_#1: GGAAACTGTCATCGTACAGC. The sequence of gRNA_Stat1_#2: GGTCGCAAACGAGACATCAT. The sequence of gRNA_Stat3_#1: GCAGCTGGACACACGCTACC. The sequence of gRNA_Stat3_#2: TTCTTCACTAAGCCGCCAAT. Plasmid construction and molecular cloning were done by following the previously published protocol 46. B16N cells were transfected with each construct using Lipofectamine 3000 (Invitrogen) following the manufacturer’s protocol. Single GFP+ cells were sorted into each well of multiple 96-well plates by BD Influx Cell Sorter after 48 h post-transfection. Selected clones were screened for expression of either Stat1 or Stat3 by quantitative real-time PCR, western blotting, and Surveyor mutation detection assay.
Mice
The C57BL/6, FVB/N, athymic Nude, Ifng-knockout (Ifngtm1Ts), and B6-TCRdelta-knockout (Tcrdtm1Mom allele), B6-TCRbeta-knockout (Tcratm1Mom allele) mice were purchased from The Jackson Laboratory (Jax Stock #002120 and 002118, respectively). The NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (CIEA NOG) immunodeficient mice were purchased from Taconic Biosciences. Both female and male mice were used at 6-8 weeks of age, with individual experiments using mice of a single sex. All mice were housed in the AAALAC accredited animal facility of Temple University. All animal procedures were performed under the guidelines of the protocols approved by the Temple University Institutional Animal Care and Use Committee.
Antibodies
All fluorescently labeled antibodies used were obtained from commercial sources (eBioscience or Biolegend), including anti-Thy1, TCRβ, γδTCR, CD4, CD8α, CD11b, CD19, Ly6g (1A1), CD45.2, Foxp3, CD25, GITR, PD1, and CD27. CCR6 antibody was purchased from R&D Systems.
Interferon-gamma treatment of cells
Recombinant mouse Ifng and human IFNG proteins (with carrier) were purchased from Cell Signaling Technology (catalog #39127 and 8901). They were reconstituted with sterile water at a concentration of 0.1 mg/ml, then diluted into a final concentration of 10 ng/ml with fresh DMEM with 10% FBS. According to safety data sheets provided by the manufacturer, the bioactivity of h-IFNG was determined in a virus protection assay. The ED50 of each lot was between 0.3 and 1.2 ng/mL. The conversion of 10 ng/mL to biological activity was 8.33 to 33.33 U/mL. The cells were treated with 10 ng/ml Ifng/IFNG in culture at 50% confluency for 48 h followed by 3x washes with PBS just before inoculation in mice and other assays.
Tumor cell inoculation and tumor analysis
All animal studies were approved by the Institutional Animal Care and Use Committee at Temple University. Melanoma cells with or without GFP-luciferase reporter were cultured and counted using an automated cell counter (BioRad). Single cell suspension in 100 μL of 1×PBS were transplanted subcutaneously to mice or introduced by tail vein injection with a 1-ml syringe with 30 gauge ½ inch needle. For subcutaneous injection: For B16, B16N, and B2905 cells, 2.5×10 cells were injected; for F5061 and YUMM1.1 cell, 1×106 cells were injected in flanks. Tumor latency was defined as the period between injection of tumorigenic cells into mice and the appearance of tumors of ≥1 mm in diameter. The endpoint was when the largest tumor reached a diameter of 1.3 cm, which ranged from 3 to 4 weeks post inoculation for the different cell lines. Tumor growth was measured by caliper, and tumor volumes were calculated using the formula ½(L × W× H). For tail vein injection: For B16, B16N, and B2905 cells, 1.25×105 cells were inoculated through the tail vein. 5×105 cells were injected for F5061 and YUMM1.1 cells. All cell lines were injected in mice on the C57BL/6 strain background, except for the F5061, which were injected in FVB/N mice. Bioluminescent imaging (BLI) was performed in a Xenogen IVIS imaging system (Perkin Elmer) after intraperitoneal injection of luciferin (100 μl of 15 mg/ml solution per 10 g). The endpoint was the day of euthanasia as determined by >10% body weight loss, hind limb paralysis or fracture, immobility, or a total photon flux >1 × 108, a value that initial results indicated reliably predicts death in less than one week in this model. Lungs and other distant organs (such as liver, spleen, kidney, right femur for bone marrow collection, brain, and any observed potential metastatic tissues) were removed from mice at day 21 post-injection. Some samples were collected for flow cytometry, while others were perfused and fixed in 4% paraformaldehyde (Fisher Scientific) overnight at 4°C, rinsed, and transferred to 30% ethanol, and stored at 4°C until further analysis. Lung surface gross tumor nodules were counted under a dissecting microscope. Images of whole mouse lungs were captured using a Nikon SLR camera with AF 60mm 1:2.8 D lens (magnification 1×).
Preparation of single cell suspensions for flow cytometry:
Tumors were micro-dissected from mouse tissues at the end of each experiment, dissociated through a 50-μm filter, and washed with PBS. In some cases, immune cells were further enriched by layering cell suspension γδ on the Ficoll–Hypaque, followed by centrifugation for 15–30 min at 400 g. The buffy layer was isolated and washed twice with RPMI before staining and FACS analysis. To measure cytokine production, cells were stimulated with PMA/ionomycin in presence of Brefeldin A for 6 h before intracellular staining for FACS. analyses.
Intracellular Staining:
Cells were stained for surface markers, then fixed in 100 μl of Cytofix/Cytoperm solution for 30min at 4°C, washed two times in perm/wash solution, pelleted by centrifugation and resuspended in 100 μl of perm/wash solution with or without (FMO control) fluorochrome-conjugated antibody at room temperature, using the BD Permeabilization Solution obtained from Transcription Factor Phospho Buffer Set (Cat. No. 565575), according to manufacturer’s instructions.
Fluorescence-activated cell sorting
B16N melanoma cells were transfected with a Green Fluorescent Protein (GFP) and Firefly Luciferase lentivirus after single-cell growth selection process. GFP-positive cells were sorted using the BD FACSAria IIu or BD FACS Vantage (BD Biosciences) systems. FACS DIVA software was used during cell sorting and the FlowJo software for analysis. Cells were initially identified on forward scatter (FSC) vs. side scatter (SSC). Single cells were identified using FSC and SSC pause width. Cell doublets were excluded from the analysis. Untransfected cells were used as negative controls. Cells were sorted based on GFP expression and SSC-A. GFP-positive cells were identified using appropriate gates based on negative controls. Due to low sample cell number, reanalysis of sorted cells was not usually done, but representative post-sort analyses confirmed that presort purities of 0.74–0.75% were enriched to 98–99.5%.
Western blotting
Human melanocytes and melanoma cells were lysed in Pierce RIPA buffer (Thermo Scientific) containing 1 × Halt protease inhibitor cocktail (100×; Thermo Scientific) and 1x Halt phosphatase inhibitor cocktail (100×; Thermo Scientific), and the protein concentration was measured with the Bio-Rad Protein Assay following the manufacturer's protocol. The same amounts of protein extracts were subjected to polyacrylamide gel electrophoresis using the 4% to 20% Mini-Protean TGX gel system (Bio-Rad), transferred to PVDF (0.45 μm pore size; Millipore) membranes, and immunoblotted using antibodies that specifically recognize STAT1 (1:1,000; Cell Signaling Technology), pSTAT1 (Y701, 58D6, 1:1,000; Cell Signaling Technology), pSTAT1 (Y727, D3B7, 1:2,000; Cell Signaling Technology), STAT3 (124H6, 1:1,000; Cell Signaling Technology), pSTAT3 (Y705, D3A7, 1:2,000; Cell Signaling Technology), GAPDH-HRP (D16H11, 1:1,000; Cell Signaling Technology), IRF1 (D5E4, 1:1,000; Cell Signaling Technology). The secondary antibodies used for detection were horseradish peroxidase (HRP)–conjugated goat anti-mouse and goat anti-rabbit IgG (1:5,000; Thermo Scientific). The blots were incubated with Luminata Western HRP substrate (Millipore) for 5 minutes. Band intensities of Tiff images were quantified by using Image J software.
Quantification and statistical analyses
All sample sizes were determined based on preliminary studies and prior knowledge of expected variability within assays. The minimum sample size was 5 mice per group, which was calculated by the G Power (v. 3.1.9.7) software with the following input parameters: two tails, effect size d=3, α err prob=0.05, and power=0.95. Age-matched (6–8 weeks) mice were randomly assigned to control and experimental groups. Quantification of the lung nodule counts were performed blindly by the pathologist. Where blinding was not used, data were analyzed using automated image analysis software when possible. All statistical tests used were deemed appropriate and met the assumptions required; when parametric tests were used, normal distribution was assumed. Where necessary unequal variance was corrected for, or if no correction was used, variation was assumed equal based on prior knowledge of the experimental assay. All experiments were performed in triplicate, data are presented as mean ± SEM, and graphs were prepared with GraphPad Prism 7. To analyze the statistical difference between two groups, a two-tailed unpaired Student’s t-test was used. Comparisons involving multiple groups were assessed by one-way ANOVA with post-hoc Tukey analysis. P value < 0.05 was considered as statistically significant.
Cell proliferation assay
A total of 3,000 cells in 200 μl of DMEM plus 10% FBS was plated in 6 wells per cell type per condition in a 96 well plate. To measure cells input on day 0, an additional set of 6 wells per cell type was seeded and assayed 2 hours later. Plates were developed by discarding 100 ul from each well and adding 100 ul of solution containing DMEM with 10% FBS and WST-1 reagent (Roche Diagnostics) diluted 1:10. Plates were incubated for 1 h at 37°C and absorbance at 450 nm was measured using a plate reader.
In vitro Tumor Growth Assays
Neutralized rat tail collagen solution was prepared at 0.8 mg/ml in DMEM by adding appropriate amounts of 10× DMEM concentrate and 1 N NaOH. Next, melanoma cells were resuspended at 2.86 × 104 cells/ml in the collagen solution, and 350 μl of cell suspension was plated per well in 24 well plates (for a final cell number of 10,000 cells per well). After 20 min at 37 °C, wells containing cells, suspended in polymerized collagen, were overlaid with 3,000 tumor cells per well in 500 μl of DMEM with 10% FBS. Tumor cell growth was photographed, and images were analyzed by Image J software.
Soft-Agar Colony Formation Assay
A soft-agar colony formation assay was done using 6-well plates. Each well contained 2 mL of 0.8% agarose base layer in complete medium (DMEM with 10% fetal bovine serum and 1% antibiotics) as the bottom layer and 1 mL of 0.4% agarose in complete medium and 3,000 cells (untreated and Ifng treated cells) as the top layer. Cultures were maintained under standard conditions for 14-21 days. The colonies were stained with cell stain solution (Chemicon) overnight at 37°C and counted the following morning. The number of colonies was determined with a microscope at 100x magnification; a group of >20 cells were counted as a colony. For colony quantification 1.4 mL cell quantification solution (Chemicon) was added to each well and incubated for 4 h at 37°C. Absorbance was measured at 490 nm.
Quantitative Real-Time RT-PCR
Cells were lysed and RNA was purified by RNeasy Mini Kit with DNase I digestion (Qiagen) following the manufacturer’s protocol. Generation of cDNA was performed by GoTaq 2-step RT system (Qiagen). Real-time PCR reactions were measured by ABI StepOnePlus system using SYBR Green qPCR master mix (ThermoFisher). 18s rRNA was used as the reference control. The ΔΔCT method was used to calculate relative expression level.
RNA Sequencing
RNA was isolated from B16 and YUMM1.1 cells after 48 h treatment with 10 ng/ml recombinant Ifng in biological triplicates using RNeasy Mini Kit. On column DNase treatment was done. Strand-specific RNA libraries were generated from 1 μg of RNA using NEBNext Ultra II RNA library Prep Kit for Illumina following the manufacturer’s instructions. Libraries were sequenced 75 bases single-end on Illumina HiSeq 2500. Quality monitoring was done using FastQC. Cutadapt was used to remove adapter sequences. Sequence alignment to the mouse genome and gene counting was performed by STAR. Genes that had 0 reads across all samples were excluded from the analysis. Differential expression analysis was performed using DESeq2. Genes were considered upregulated with a fold change (FC) > 2 and downregulated with an FC < 0.5 and QFDR < 0.05, unless otherwise stated.
Extended Data
Extended Data Fig. 1.
Ifng treatment showed differential effects on cell proliferation in five indicated mouse melanoma cell lines. The proliferation of (a) B16 and (b) B16N cell lines was significantly inhibited. In contrast, (c) B2905, (d) F5061, and (e) YUMM1.1 cell lines were not affected. For proliferation, WST-1 assay was performed with mock- or Ifng treatment with 10 ng/ml conc. Melanoma cells in medium with or without Ifng were seeded in triplicates and absorbance was measured at indicated time points. All data are plotted as mean±SEM. *P<0.05; **P<0.01; n.s. not significant; Student’s t test.
Extended Data Fig. 2.
Ifng treatment showed differential effects on soft agar colony formation in four indicated melanoma cell lines. (a) B16 and (b) B16N cell lines were significantly inhibited in colony formation by Ifng. In contrast, (c) B2905 and (d) F5061 cell lines were not affected. The number of colonies per well were counted after indicated number of days of incubation. Representative microscopic images are shown. All experiments were performed in triplicates and plotted as mean±SEM. **P<0.01; n.s. not significant; Student’s t test.
Extended Data Fig. 3.
Graphs representing mRNA expression of a) Interferon Regulatory Factor 1 (Irf1) and b) Guanylate Binding Protein 4 (Gbp4) for B16 cells in response to Interferon gamma (Ifng). Cells were treated with Ifng (10ng/ml) for 48 hours and to investigate posttreatment stimulation of Irf1 and Gbp4, cells were allowed to grow further without Ifng for 4 days. Isolation of total RNA and cDNA synthesis was done at each time point. 2μg of total RNA was reverse transcribed into cDNA using Applied Biosystems cDNA synthesis kit (Thermo Fisher, USA) and mRNA expression was quantified using real time PCR (Quant studio 3, Thermo Fisher, USA) through ΔΔCT method using 18s rRNA as a normalizer. Results are representative of 3 independent experiments. ****P<0.0001, **P<0.01, *P<0.05.
Extended Data Fig 4.
Ifng treatment did not affect subcutaneous growth of melanoma cells. The mouse melanoma cells were mock- or Ifng-treated (10 ng/ml for 48 h) and subcutaneous injections were performed at back flank of 8-week-old syngeneic mice (C57BL/6 for B16, B16N, and B2905; FVB/N for F5061). For B16, B16N, and B2905 cells, 2.5×105 cells were injected; for F5061 and YUMM1.1 cell, 1×106 cells were injected in flanks. Once tumors became palpable, tumor volumes were measured up to 3x a week with digital calipers. The final volumes were measured at the time of termination when the largest tumor reached a tumor diameter of 1.3 cm, which ranged from 3 to 4 weeks post inoculation for the different cell lines. There was no significant difference in mean subcutaneous tumor volume between control and Ifng-treated (a) B16, (b) B2905, (c) B16N, and (d) F5061 cells. Data are presented as mean±SEM; n.s. not significant, Student’s t test.
Extended Data Fig. 5.
H&E stained FFPE sections for lung tissues from mice injected via tail-vein with (a) mock-treated control or (b) Ifng-treated B16 cells. Cells were treated with 10 ng/ml mouse recombinant Ifng in culture for 48 h, followed by 3x washes with PBS and injected into syngeneic C57BL/6 mice via tail vein (n=7 each). Tumors are seen as darker purple lesions. Only a few random lesions are marked by arrows, especially in (b).
Extended Data Fig. 6.
Western blot of pStat1 (Y701), Stat1, Irf1, pStat3 (Y705), Stat3, and Gapdh expressions in parental B16N cells, Stat1-KO and Stat3-KO clones (2 clones each, #1 and #2) in response to 10 ng/ml Ifng treatment for 24 h.
Extended Data Fig. 7.
The effect of Ifng on B16N cell proliferation and colony formation is dependent on Stat1 but not Stat3. (a) Ifng treatment inhibited the growth of Stat3 knockout (KO) and parental B16N cells, but not Stat1-KO cells. Cell proliferation assay (WST-1) showed an Ifng-induced reduction of cell growth on WT and Stat3-KO cells (both P<0.05 on day 2 onward), but not Stat1-KO cells. Six replicates per sample were measured and plotted as mean±SEM. Plates were read at 450nm for absorbance after 4 days culture. (b) Ifng had no effect on colony formation on Stat1-KO but had reduced colony formation on Stat3-KO cell lines. The number of colonies per well for Stat1-KO and Stat3-KO cells in soft agar assay with representative microscopic images are shown. Images were taken at day 27 (Magnification 4×). (c) Quantified colonies data are plotted as mean±SEM of three independent experiments. *P<0.05; n.s. not significant, Student’s t test.
Extended Data Fig. 8.
Requirement for gamma-delta T cells in Ifng-enhanced metastasis. Ifng treated and untreated B2905 melanoma cells were inoculated into syngeneic WT, TCRβ−/− and TCRγ−/− mice by tail vein injection. Mice were euthanized on day 23. (a) Tumor infiltrating immune cell profiles were assessed by flow cytometry. (b-d) Relative distributions of different populations of myeloid cells and Tregs in tumor microenvironment were determined. (e) IL-17 and Ifng production by infiltrating T cells in response to PMA and Ionomycin stimulation was measured by intracellular staining. Data displayed represent mean ± SEM. Significance of differences was determined by one-way ANOVA with post-hoc Bonferroni test, as indicated by asterisks: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
Extended Data Fig. 9.
RNA sequencing data for B16 and YUMM1.1 mouse melanoma cells in relation to Ifng treatment as compared to untreated cells. Cells were treated with Ifng (10 ng/ml for 48 h) followed by RNA isolation by Qiagen’s RNeasy Plus Mini kit (Qiagen, Germany). RNA libraries were generated using NEBNext Ultra II RNA library Prep Kit which were further sequenced on Illumina HiSeq 2500. Differential expression analysis was performed using DESeq2. a) Approximately 30.2% genes were found to be concordantly upregulated in both B16 and YUMM1.1 cell lines after Ifng treatment. b) Heatmap of top 100 common most significantly upregulated genes are presented, indicating high expression of Cxcl9, a chemokine known to be involved in chemotaxis of γδ T cells.
Extended Data Fig. 10.
Graphs representing mRNA expression of C-X-C motif chemokine ligand 9 (Cxcl9), Intercellular Adhesion Molecule 1 (Icam1) and Chemokine ligand 5 (Ccl5) for a) YUMM1.1, b) B16, and c) B2905 cells in response to Interferon gamma (Ifng). Cells were treated with Ifng (10 ng/ml) for 48 hours followed by total RNA isolation and cDNA synthesis. 2μg of total RNA was reverse transcribed into cDNA using Applied Biosystems cDNA synthesis kit (Thermo Fisher, USA) and mRNA expression was quantified using real time PCR (Quant studio 3, Thermo Fisher, USA) through ΔΔCT method using 18s rRNA as a normalizer. Results are representative of three independent experiments. ***P<0.001, **P<0.01, *P<0.05.
Acknowledgments
Funding:
USA National Institutes of Health, R01CA193711 (MRZ); R01CA236391 (DJK), R01AI068907 (DJK), P30CA006927 (FCCC Comprehensive Cancer Center Core Grant).
Footnotes
Authors have no conflicts of interest.
Data Availability
RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) and can be accessed through the accession number GSE214873 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214873).
References
- 1.Austin MT, Xing Y, Hayes-Jordan AA, Lally KP, Cormier JN. Melanoma incidence rises for children and adolescents: an epidemiologic review of pediatric melanoma in the United States. J Pediatr Surg 2013; 48: 2207–2213. [DOI] [PubMed] [Google Scholar]
- 2.Balkhy HH, Heinzel FP. Endotoxin fails to induce IFN-gamma in endotoxin-tolerant mice: deficiencies in both IL-12 heterodimer production and IL-12 responsiveness. J Immunol 1999; 162: 3633–3638. [PubMed] [Google Scholar]
- 3.Bennett DC. Ultraviolet wavebands and melanoma initiation. Pigment Cell Melanoma Res 2008; 21: 520–524. [DOI] [PubMed] [Google Scholar]
- 4.Bialasiewicz AA, Ma JX, Richard G. Alpha/beta- and gamma/delta TCR(+) lymphocyte infiltration in necrotising choroidal melanomas. Br J Ophthalmol 1999; 83: 1069–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown TJ, Lioubin MN, Marquardt H. Purification and characterization of cytostatic lymphokines produced by activated human T lymphocytes. Synergistic antiproliferative activity of transforming growth factor beta 1, interferon-gamma, and oncostatin M for human melanoma cells. J Immunol 1987; 139: 2977–2983. [PubMed] [Google Scholar]
- 6.Brucet M, Marques L, Sebastian C, Lloberas J, Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF-1. Genes Immun 2004; 5: 26–35. [DOI] [PubMed] [Google Scholar]
- 7.Byeseda SE, Burns AR, Dieffenbaugher S, Rumbaut RE, Smith CW, Li Z. ICAM-1 is necessary for epithelial recruitment of gammadelta T cells and efficient corneal wound healing. Am J Pathol 2009; 175: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callahan MK, Postow MA, Wolchok JD. Immunomodulatory therapy for melanoma: ipilimumab and beyond. Clin Dermatol 2013; 31: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM et al. STAT1-deficient mice spontaneously develop estrogen receptor alpha-positive luminal mammary carcinomas. Breast Cancer Res 2012; 14: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordova A, Toia F, La Mendola C, Orlando V, Meraviglia S, Rinaldi G et al. Characterization of human gammadelta T lymphocytes infiltrating primary malignant melanomas. PLoS One 2012; 7: e49878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol 2014; 11: 24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dummer R, Hassel JC, Fellenberg F, Eichmuller S, Maier T, Slos P et al. Adenovirus-mediated intralesional interferon-gamma gene transfer induces tumor regressions in cutaneous lymphomas. Blood 2004; 104: 1631–1638. [DOI] [PubMed] [Google Scholar]
- 13.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer 2005; 41: 45–60. [DOI] [PubMed] [Google Scholar]
- 14.Garibyan L, Fisher DE. How sunlight causes melanoma. Curr Oncol Rep 2010; 12: 319–326. [DOI] [PubMed] [Google Scholar]
- 15.Garland CF, Garland FC, Gorham ED. Epidemiologic evidence for different roles of ultraviolet A and B radiation in melanoma mortality rates. Annals of epidemiology 2003; 13: 395–404. [DOI] [PubMed] [Google Scholar]
- 16.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015; 21: 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon-Alonso M, Hirsch T, Wildmann C, van der Bruggen P. Galectin-3 captures interferon-gamma in the tumor matrix reducing chemokine gradient production and T-cell tumor infiltration. Nat Commun 2017; 8: 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M et al. Conserved Interferon-gamma Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell 2020; 38: 500–515 e503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat 2007; 28: 578–588. [DOI] [PubMed] [Google Scholar]
- 20.Hsu KS, Zhao X, Cheng X, Guan D, Mahabeleshwar GH, Liu Y et al. Dual regulation of Stat1 and Stat3 by the tumor suppressor protein PML contributes to interferon alpha-mediated inhibition of angiogenesis. J Biol Chem 2017; 292: 10048–10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev 2002; 13: 95–109. [DOI] [PubMed] [Google Scholar]
- 22.Ilkovitch D, Lopez DM. Immune modulation by melanoma-derived factors. Exp Dermatol 2008; 17: 977–985. [DOI] [PubMed] [Google Scholar]
- 23.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR et al. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell 1993; 72: 337–348. [DOI] [PubMed] [Google Scholar]
- 24.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019; 176: 998–1013 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabelitz D, Wesch D. Features and functions of gamma delta T lymphocytes: focus on chemokines and their receptors. Crit Rev Immunol 2003; 23: 339–370. [DOI] [PubMed] [Google Scholar]
- 26.Kachroo P, Lee MH, Zhang L, Baratelli F, Lee G, Srivastava MK et al. IL-27 inhibits epithelial-mesenchymal transition and angiogenic factor production in a STAT1-dominant pathway in human non-small cell lung cancer. J Exp Clin Cancer Res 2013; 32: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortylewski M, Komyod W, Kauffmann ME, Bosserhoff A, Heinrich PC, Behrmann I. Interferon-gamma-mediated growth regulation of melanoma cells: involvement of STAT1-dependent and STAT1-independent signals. J Invest Dermatol 2004; 122: 414–422. [DOI] [PubMed] [Google Scholar]
- 28.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol Immunother 2011; 60: 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesinski GB, Anghelina M, Zimmerer J, Bakalakos T, Badgwell B, Parihar R et al. The antitumor effects of IFN-alpha are abrogated in a STAT1-deficient mouse. J Clin Invest 2003; 112: 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma C, Zhang Q, Ye J, Wang F, Zhang Y, Wevers E et al. Tumor-infiltrating gammadelta T lymphocytes predict clinical outcome in human breast cancer. J Immunol 2012; 189: 5029–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maddodi N, Setaluri V. Role of UV in cutaneous melanoma. Photochem Photobiol 2008; 84: 528–536. [DOI] [PubMed] [Google Scholar]
- 32.Matsumura Y, Ananthaswamy HN. Molecular mechanisms of photocarcinogenesis. Front Biosci 2002; 7: d765–783. [DOI] [PubMed] [Google Scholar]
- 33.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res 2016; 29: 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo X, Kazmi HR, Preston-Alp S, Zhou B, Zaidi MR. Interferon-gamma Induces Melanogenesis Via Post-Translational Regulation of Tyrosinase. Pigment Cell Melanoma Res 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moan J, Porojnicu AC, Dahlback A. Ultraviolet radiation and malignant melanoma. Adv Exp Med Biol 2008; 624: 104–116. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 1992; 360: 225–231. [DOI] [PubMed] [Google Scholar]
- 37.Murtas D, Maric D, De Giorgi V, Reinboth J, Worschech A, Fetsch P et al. IRF-1 responsiveness to IFN-gamma predicts different cancer immune phenotypes. Br J Cancer 2013; 109: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Natarajan VT, Ganju P, Singh A, Vijayan V, Kirty K, Yadav S et al. IFN-gamma signaling maintains skin pigmentation homeostasis through regulation of melanosome maturation. Proc Natl Acad Sci U S A 2014; 111: 2301–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen KB, Biron CA. Synergism for cytokine-mediated disease during concurrent endotoxin and viral challenges: roles for NK and T cell IFN-gamma production. J Immunol 1999; 162: 5238–5246. [PubMed] [Google Scholar]
- 40.Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. Br J Dermatol 2014; 170: 11–19. [DOI] [PubMed] [Google Scholar]
- 41.Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL et al. Neonatal sunburn and melanoma in mice. Nature 2001; 413: 271–272. [DOI] [PubMed] [Google Scholar]
- 42.Noonan FP, Zaidi MR, Wolnicka-Glubisz A, Anver MR, Bahn J, Wielgus A et al. Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat Commun 2012; 3: 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norval M, McLoone P, Lesiak A, Narbutt J. The effect of chronic ultraviolet radiation on the human immune system. Photochem Photobiol 2008; 84: 19–28. [DOI] [PubMed] [Google Scholar]
- 44.Peng G, Wang HY, Peng W, Kiniwa Y, Seo KH, Wang RF. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity 2007; 27: 334–348. [DOI] [PubMed] [Google Scholar]
- 45.Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem 2004; 279: 41679–41685. [DOI] [PubMed] [Google Scholar]
- 46.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raspollini MR, Castiglione F, Rossi Degl'innocenti D, Amunni G, Villanucci A, Garbini F et al. Tumour-infiltrating gamma/delta T-lymphocytes are correlated with a brief disease-free interval in advanced ovarian serous carcinoma. Ann Oncol 2005; 16: 590–596. [DOI] [PubMed] [Google Scholar]
- 48.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ et al. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol 2009; 10: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez T, Mendez R, Del Campo A, Jimenez P, Aptsiauri N, Garrido F et al. Distinct mechanisms of loss of IFN-gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer 2007; 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331: 1565–1570. [DOI] [PubMed] [Google Scholar]
- 51.Singh S, Kumar S, Srivastava RK, Nandi A, Thacker G, Murali H et al. Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-gamma signalling. Nat Cell Biol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slade AD, Austin MT. Childhood melanoma: an increasingly important health problem in the USA. Curr Opin Pediatr 2014; 26: 356–361. [DOI] [PubMed] [Google Scholar]
- 53.Son J, Kim M, Jou I, Park KC, Kang HY. IFN-gamma inhibits basal and alpha-MSH-induced melanogenesis. Pigment Cell Melanoma Res 2014; 27: 201–208. [DOI] [PubMed] [Google Scholar]
- 54.Tamura K, Makino S, Araki Y, Imamura T, Seita M. Recombinant interferon beta and gamma in the treatment of adult T-cell leukemia. Cancer 1987; 59: 1059–1062. [DOI] [PubMed] [Google Scholar]
- 55.Togashi Y, Nishikawa H. Regulatory T Cells: Molecular and Cellular Basis for Immunoregulation. Curr Top Microbiol Immunol 2017; 410: 3–27. [DOI] [PubMed] [Google Scholar]
- 56.Tran TT, Schulman J, Fisher DE. UV and pigmentation: molecular mechanisms and social controversies. Pigment Cell Melanoma Res 2008; 21: 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wall L, Burke F, Smyth JF, Balkwill F. The anti-proliferative activity of interferon-gamma on ovarian cancer: in vitro and in vivo. Gynecol Oncol 2003; 88: S149–151. [DOI] [PubMed] [Google Scholar]
- 58.Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control 2001; 12: 69–82. [DOI] [PubMed] [Google Scholar]
- 59.Zaidi MR, Davis S, Noonan FP, Graff-Cherry C, Hawley TS, Walker RL et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature 2011; 469: 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res 2011; 17: 6118–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaidi MR. The Interferon-Gamma Paradox in Cancer. J Interferon Cytokine Res 2019; 39: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Molavi O, Su M, Lai R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer 2014; 14: 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med 2017; 24: 19–29. [PubMed] [Google Scholar]
- 64.Zhao Y, Niu C, Cui J. Gamma-delta (gammadelta) T cells: friend or foe in cancer development? J Transl Med 2018; 16: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) and can be accessed through the accession number GSE214873 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214873).