Abstract
Motivation
As the number of public data resources continues to proliferate, identifying relevant datasets across heterogenous repositories is becoming critical to answering scientific questions. To help researchers navigate this data landscape, we developed Dug: a semantic search tool for biomedical datasets utilizing evidence-based relationships from curated knowledge graphs to find relevant datasets and explain why those results are returned.
Results
Developed through the National Heart, Lung and Blood Institute’s (NHLBI) BioData Catalyst ecosystem, Dug has indexed more than 15 911 study variables from public datasets. On a manually curated search dataset, Dug’s total recall (total relevant results/total results) of 0.79 outperformed default Elasticsearch’s total recall of 0.76. When using synonyms or related concepts as search queries, Dug (0.36) far outperformed Elasticsearch (0.14) in terms of total recall with no significant loss in the precision of its top results.
Availability and implementation
Dug is freely available at https://github.com/helxplatform/dug. An example Dug deployment is also available for use at https://search.biodatacatalyst.renci.org/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The ability to interrogate large-scale data resources is becoming a central focus of many research efforts. The U.S. National Institutes of Health (NIH) and other public funding agencies have supported data generation at unprecedented scales through projects such as Trans-Omics for Precision Medicine (TOPMed) (University of Washington Department of Biostatistics, 2020), All of Us (The ‘All of Us’ Research Program, 2019) and Helping to End Addiction Long-Term (HEAL) (Collins et al., 2018). From these efforts, the ability to integrate data within and across disjoint and complex public data repositories is quickly replacing data scarcity as a primary bottleneck to research progress. While successful data integration efforts have resulted in novel diagnostics, therapies and prevention strategies, researchers often lack the necessary tools for navigating this complex data landscape (Powell, 2021).
There is a growing need for comprehensive search tools that identify datasets relevant to a researcher's particular scientific question. Despite recent NIH emphasis on making research data more Findable, Accessible, Interoperable and Re-Usable (‘FAIR’ data principles) (Wilkinson et al., 2016), the diversity of public data repositories has proven to be a formidable barrier to developing intelligent search strategies. To illustrate, consider that the NIH alone currently refers to data submission to more than 95 domain-specific repositories (NIH Data Sharing Resources, 2020). Often, more established repositories like the NIH database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) require studies to submit only free-text descriptions of experimental variables. The complexity of ‘heart attack’ versus ‘myocardial infarction’ exemplifies the challenges of identifying relevant datasets among the growing corpus of non-standardized biomedical datasets.
Emerging techniques for natural language processing (NLP) are enabling semantic search over biomedical datasets. We define semantic search as a search that considers the intent and context of the query as opposed to a purely lexical approach (Tran et al., 2007). Many existing search and annotation tools successfully employ methods for named entity recognition (NER) and disambiguation to annotate free text with synonyms or ontology terms (Bell et al., 2019; Canakoglu et al., 2019; Chapman et al., 2020; Chen et al., 2018; Huang et al., 2016; Laulederkind et al., 2012; Pang et al., 2015; Soto et al., 2019). An ontology defines a set of entities in a subject area as well as the relationships between them. By mapping free text to ontology terms, semantically related ideas can be explored by traversing related nodes on the knowledge graph structure defining the ontology.
Similar NLP techniques have been successfully employed by semantic-aware dataset search engines like Google Dataset Search (Brickley et al., 2019) and Datamed (Bell et al., 2019) to make data more discoverable. Broadly, both tools index study-level metadata describing high-level features of each dataset (e.g. study description and abstract) to increase findability. For Datamed, study metadata are augmented with Unified Medical Language System (UMLS) concept identifiers extracted by NLP annotation tools to increase findability. Google Datasets extend this concept by mapping study metadata elements to Google’s internal knowledge graph, used to augment metadata with additional relevant search terms. For a more complete review of dataset search tools, see Chapman et al. (2020).
Despite the utility of these tools, there remains a need for a biomedical dataset search engine that can empower true data re-use. A critical shortcoming of the search engines described above is a focus on study-level metadata to the exclusion of variable-level metadata (i.e. the features measured as part of this study); two or more studies can be combined and integrated with only a single conceptually overlapping variable, enabling a broader set of possible data integrations.
There also remains a need for a biomedical dataset search engine that combines context-awareness with domain specificity. The ability to show second- and third-order connections can be helpful for data discovery but requires an ontological knowledge graph describing how biological entities and phenomena are related. Despite the practical utility of Google’s proprietary knowledge graph for general search, the provenance, depth and quality of its biomedically relevant connections are not easily verifiable. There remains a need for a search tool capable of leveraging evidence-based biological connections to show researchers datasets useful for hypothesis generation or scientific support.
Therefore, we present Dug (https://github.com/helxplatform/dug): a semantic search tool for biomedical datasets that leverages NLP technologies and ontological knowledge graphs curated by biomedical subject matter experts to intelligently identify datasets relevant to a user query. Dug is designed specifically for data integration and re-use by allowing users to search over variable-level metadata and high-level study metadata. Dug is also the first biomedical search engine that can explain why it returns what it returns (Fig. 1). For example, if a search for ‘cancer’ were to return a dataset that measured ‘asbestos exposure’, Dug can show the user—often with links to supporting literature—that this variable was returned due to the causal linkage between asbestos exposure and certain types of cancer. Here, we discuss Dug's motivations, architecture, functionality and evaluation and demonstrate its successful deployment in the NHLBI’s BioData Catalyst Ecosystem (National Heart Lung and Blood Institute et al., 2020).
Fig. 1.
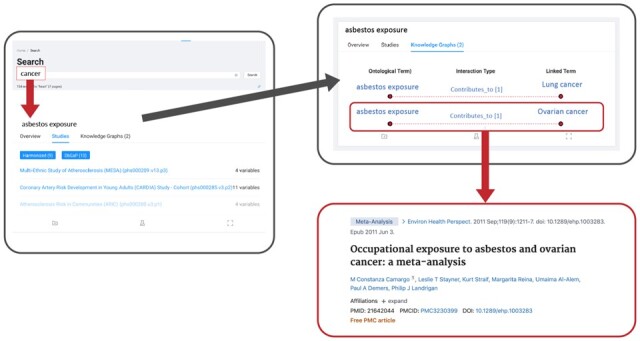
The Dug web portal leverages knowledge-graph connections with supporting links to PubMed literature to explain why certain results are relevant to a user’s query
2 Materials and methods
Dug consists of two components: A Dug API service that orchestrates metadata ingestion, indexing and search, and the Dug search web portal that displays results to end users (Fig. 2). The ingestion/indexing pipeline is designed to:
Fig. 2.
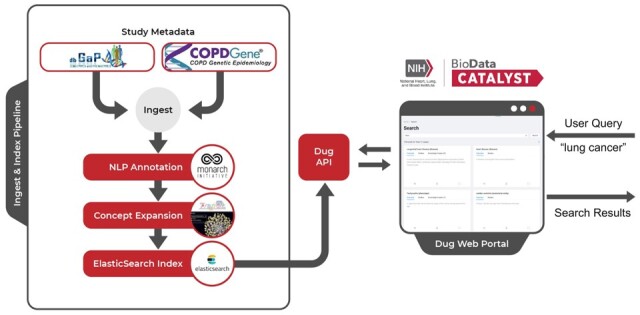
High-level Dug architecture. Dug makes study metadata searchable by parsing heterogenous metadata formats into a common format (ingest), annotating metadata using NLP tools to extract ontology identifiers from prose text (NLP annotation), searching for relevant connections in federated knowledge graphs using translator query language (TranQL) (concept expansion) and indexing this information into an Elasticsearch index. Dug’s web portal utilizes a flexible API to query and display search results back to end users
Parse heterogenous study metadata formats into a common Dug metadata format.
Automatically annotate free-text descriptions of study variables with a set of ontological identifiers.
Expand annotations with relevant terms returned from knowledge-graph queries.
Index each study variable, associated ontological concepts and set of knowledge-graph answers to an Elasticsearch (Kuć and Rogozinski, 2016) endpoint. Elasticsearch is a stack of flexible components that allow users to quickly index and search over large volumes of structured text data.
2.1 Data and ingestion pipelines
2.1.1 Data ingestion
Dug ingests and indexes study metadata (e.g. text descriptions of study variables) as opposed to actual study data. To accommodate the diversity of metadata formats available across public data repositories, our ingestion pipeline abstracts out retrieval modes (e.g. local file and network file) and data parsing formats (e.g. XML, JSON). Similar to the Data Tags Suite (DATS) metadata schema (Sansone et al., 2017), Dug parses diverse metadata formats into a common DugElement metadata model, which defines a standard set of metadata required for indexing. Dug can be flexibly adapted to ingest nearly any metadata format by extending its plug-in interface to parse input data into DugElement objects. The DugElement is designed to make study artifacts searchable through the Dug interface. Unlike many dataset search engines that collect only study-level metadata, Dug does not index any study-level metadata. A study/collection is made findable because its underlying artifacts are findable and can link back to a parent/containing study The primary fields of the DugElement describe basic attributes of the parent study (id, name and external link) as well as attributes of the element itself (id, name, description and external link). Supplementary materials include a detailed discussion of the DugElement metadata model.
2.1.2 Data annotation
Dug’s data annotation module extracts a set of biomedical ontology identifiers from ingested metadata elements using tools for NER (Fig. 3). Typically, we use the/nlp/annotate endpoint exposed by the Monarch Initiative’s (Mungall et al., 2017) Biolink API. The underlying Biolink model provides a high-level data model for representing biomedical knowledge (Reese et al., 2021) and can be used to integrate across domain-specific ontologies. Monarch’s API service accepts prose text as input and returns a set of ontological identifiers with additional information in JSON. We chose the Biolink API for NER because it natively interoperates with the Translator Query Language, which we use for concept expansion (TranQL, see below).
Fig. 3.
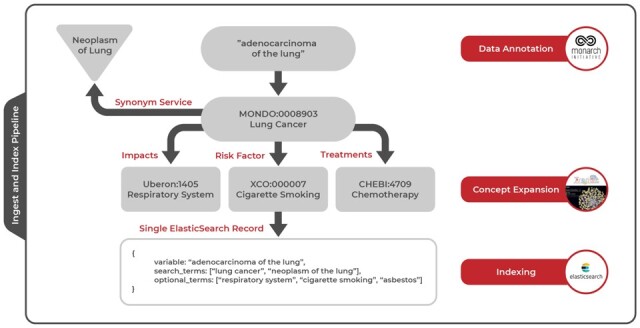
Detailed example of ingest and index pipeline. After ingesting a variable called ‘adenocarcinoma of the lung’ from study metadata, Dug uses NLP methods to annotate the variable with an ontology identifier for ‘Lung Cancer’ from the Mondo disease ontology. The resulting identifier is used to gather synonyms for lung cancer such as ‘Neoplasm of lung’ from an external API service. During concept expansion, Dug leverages TranQL to query knowledge graphs for other ontological concepts related to lung cancer through certain predicates; in the figure, we are looking for risk factors, treatments and anatomical entities impacted by lung cancer. During indexing, all terms discovered through annotation and concept expansion are combined with the original metadata into a single Elasticsearch record so that queries against any of these terms will yield the initial variable measuring ‘adenocarcinoma of the lung’
To accommodate NLP services and tools for biomedical NER, Dug also abstracts out the Annotation module. To extend Dug’s annotation interface, developers can create a child class specifying the new API endpoint and define a function converting successful API responses into an internal data structure called a DugIdentifier, which defines a minimal set of ontological information needed for downstream processing (e.g. id, name and Biolink type).
By converting free text to ontology identifiers, we can leverage semantic web services supporting this nomenclature to gather additional information about each identifier. Dug utilizes a normalization service (necessary to interoperate with Biolink knowledge graphs, discussed below) to transform identifiers to the preferred equivalents (https://github.com/TranslatorSRI/NodeNormalization). Additionally, Dug uses an ontology metadata service for fetching identifier names, descriptions, synonyms and Biolink types (https://onto.renci.org/apidocs/). These specific tools were chosen by default as they are, to our knowledge, the only tools capable of performing their respective functions.
2.1.3 Concept expansion
Dug’s ability to retrieve contextualized search results and explain these connections to end users is undergirded by a process we call concept expansion, which further annotates ontological identifiers by identifying relevant connections within ontological knowledge graphs (Fig. 3).
The data structure for concept expansion is a knowledge graph, with nodes representing entity types (e.g. disease, gene and chemical exposure) and edges providing predicates that describe the relationship between entities; Biolink predicates include ‘causes’, ‘is associated with’, and ‘is expressed in’. Biolink provides an ‘upper ontology’ that defines connections across domain-specific ontologies (e.g. Mondo, ChEBI and HP). For example, the ChEBI ‘asbestos’ identifier might be linked to the Mondo ‘lung cancer’ identifier via the Biolink ‘risk_factor’ predicate. Inclusion/exclusion of specific ontologies, and creation of links and predicates across ontologies, is curated by subject matter experts as part of the Biolink initiative.
To contextualize metadata within a knowledge graph, we leverage data integration approaches developed through the NCATS Biomedical Data Translator (Biomedical Data Translator Consortium, 2019). Chief among these are ROBOKOP (Bizon et al., 2019) and TranQL (Translator Query Language; https://tranql.renci.org) (Cox et al., 2020).
Dug leverages TranQL to gather an expanded set of ontological concepts related to those extracted via NLP annotation. Because entities in TranQL knowledge graphs may have hundreds of connections, Dug defines a set of TranQL query templates prior to indexing to establish which ontology identifiers and predicates should be considered relevant. End users will never interact with Dug’s query templates. Though technically configurable, Dug’s default query templates were selected through iterative feedback from beta testers to expose the most interesting connections. Below is an example of a query template used to retrieve chemical risk factors for a disease:
FIND Chemical_Entity −> Risk_Factors −> Disease WHERE Disease == {Query Ontology ID}
During concept expansion, Dug uses templates to substitute actual ontological identifiers from the previous NLP annotation step to retrieve a set of relevant terms for a specific variable. In Figure 4, Dug returns a list of chemical risk factors for lung cancer by substituting an ontology identifier for lung cancer (MONDO: 0008903) into the query template. The actual query becomes:
Fig. 4.
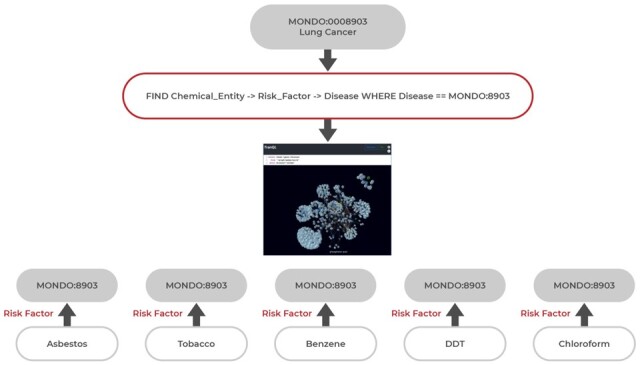
An example TranQL query for chemical risk factors of lung cancer. Each TranQL answer returned is a knowledge subgraph linking one chemical element node back to the query node
FIND Chemical_Entity −> Risk_Factors −> Disease WHERE Disease == MONDO:0008903
The ‘answers’ returned by TranQL queries are used to increase the search relevance of related concepts and to provide a basis for including explanations for links that led to the result (e.g. peer-reviewed literature supporting knowledge graph links). TranQL ‘answers’ are also knowledge graphs: a set of nodes (ontological identifiers), edges (predicates) and metadata including names, descriptions, synonyms of ontological identifiers and PubMed literature links supporting each edge (Fig. 4).
2.1.4 Data indexing
After annotation and concept expansion, the resulting data structure must be indexed. To maximize speed and flexibility, Dug’s back-end search architecture is implemented as a set of linked Elasticsearch indices. Elasticsearch provides the basic functionality to ‘save’ structured bits of text called ‘documents’ into databases called ‘indices’ that can be queried by downstream users. By default, Elasticsearch uses a ‘fuzzy’ term frequency-inverse document frequency (TF-IDF) scoring algorithm for text retrieval, a purely lexical search that allows some character mismatches and ranks the importance of results by the inverse frequency of each token in the complete set of documents.
Dug's semantic capabilities result from combining ingested study and variable metadata with terms harvested through annotation and concept expansion into a single Elasticsearch record (see Fig. 3). For each metadata variable, Dug’s indexer adds a ‘search_terms’ field to the original record containing the names and synonyms of each identifier added via data annotation. The indexer adds an ‘optional_terms’ field by traversing the names of knowledge graph nodes added during concept expansion. Dug’s results always include text matches from the original metadata, since Dug’s Elasticsearch query searches over the original metadata fields parsed during ingestion and the expanded fields added during annotation and concept expansion.
Dug quickly organizes search results by higher-level concepts through partitioning (i) ingested metadata records, (ii) core ontological concepts and (iii) expanded knowledge-graph answers respectively into three separate Elasticsearch indices. Any ontological identifier extracted is added to the concept index. By indexing study variables with ID pointers to concepts, Dug eliminates the need to calculate these groups dynamically and eliminates redundant text stored across study variables mapping to the same ontological concept. Each indexed knowledge-graph answer contains a JSON representation of the answer subgraph returned by TranQL and a pointer back to the ontological concept used in the original query.
2.2 Search engine
2.2.1 Search functionality
Dug’s search API exposes three search endpoints for querying each underlying Elasticsearch index.
/search_var—search for study variables matching a user’s query
/search_concepts—search for ontological concepts matching a user’s query
/search_kg—search for knowledge-graph answers matching user’s query and an ontological concept id
Dug’s search API uses the default Elasticsearch algorithm (cosine similarity based on vector space model using TF-IDF weighting) (Elasticsearch, 2021) to rank and retrieve indexed metadata records. The search field weighting scheme prioritizes exact matches from the originally ingested text first, followed by synonyms added through annotation, and lastly, related terms added through concept expansion.
The Dug search app is implemented in Python 3.9 (https://python.org) and is available via GitHub (https://github.com/helxplatform/dug). Supplementary materials include a detailed discussion of the Dug search API implementation.
2.2.2 Search user interface
Dug’s user interface (UI) is a stand-alone React JS-based web application designed to provide an intuitive interface for navigating large collections of data. Dug’s minimalist UI design is intended to reduce the burden on users and empower them to discover search terms they are interested in exploring. Dug provides a search box that prioritizes exact phrase matching (AND logic) over partial matching (OR logic) (Fig. 5). Other features include auto-generated tabbing of search results by data type.
Fig. 5.
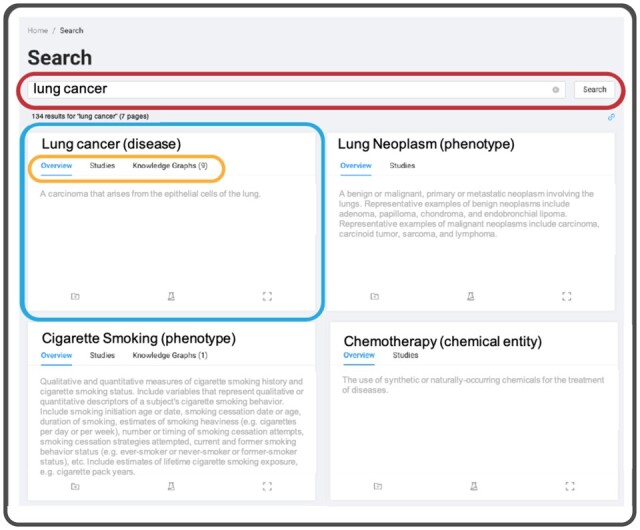
Dug UI. Dug’s UI aggregates search results for user queries (red) into higher-order ontological concepts (blue) based on NLP annotation. Links to knowledge graphs (orange) explain biological relationships between the query and each concept. Concepts may not contain knowledge-graph links if synonymous with the search query (e.g. ‘lung cancer’ versus ‘adenocarcinoma of the lung’) or if TranQL did not return answers during concept expansion (A color version of this figure appears in the online version of this article)
Dug’s UI organizes search results in two ways: by variable and by the concept. When organized by variable, Dug returns study variables sorted by relevance, with each result containing information about the variable returned (e.g. parent study). When organized by concept (Fig. 5), Dug aggregates results into higher-level ontological concepts. Dug can then be used as a preliminary harmonization step to create de novo groups of similar variables based on NLP annotations (Fig. 5). By providing both approaches, Dug’s concept-based search gives users an exploratory look at the data landscape, while its variable-based search allows users to investigate specific variables of interest.
The defining feature of Dug’s UI is its ability to explain why it returns certain results (Fig. 6). When Dug returns a result based on text added during concept expansion, the UI fetches and renders the corresponding knowledge-graph answer from the knowledge-graph index to explain the connection (if available, answers include links to supporting peer-reviewed literature).
Fig. 6.
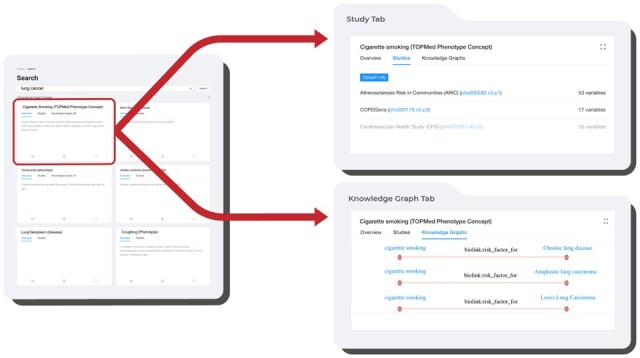
Dug UI. Dug results show datasets with variables relevant to a user’s query (e.g. ‘lung cancer’ returns ‘cigarette smoking’), and knowledge-graph disclosures for understanding why Dug considered those results relevant (e.g. cigarette smoking is a risk factor for lung cancer)
2.3 Deployment on BioData catalyst
We deployed Dug on the NHLBI’s BioData Catalyst ecosystem and indexed the TOPMed freeze 5b and 8 studies (excluding parent studies). These studies were chosen because they were the datasets available on the BioData Catalyst platform at the time of publication. Data dictionaries downloaded directly from dbGaP were ingested and indexed for each of the 76 datasets included in these freezes.
A total of 15 911 of these variables were manually harmonized into 65 higher-order groups called ‘phenotype concepts’ by data curation experts at the TOPMed Data Coordinating Center (DCC) (Stilp et al., 2021). The TOPMed harmonized phenotypes were created to enable interoperability between TOPMed datasets by manually combining semantically similar dbGaP variables under a single term.
2.4 Evaluation
We evaluated Dug’s performance against a purely lexical search strategy (default Elasticsearch; TF-IDF with ‘fuzzy matching’) to quantify the impact of Dug’s semantic awareness on recall and precision. We used the TOPMed phenotype concepts dataset as a framework for evaluation.
Variable descriptions were preprocessed prior to evaluation to characterize search performance more accurately. Steps included lowercasing descriptions, removing punctuation, removing the uninformative ‘Exam {integer}’ pattern, normalizing whitespace and de-duplicating to include only unique variable descriptions. We preprocessed the TOPMed Phenotype concept names by removing stop words (e.g. ‘the’, ‘in’, ‘of’) and expanding known abbreviations (e.g. ‘Resting arm systolic BP’ becomes ‘Resting arm systolic blood pressure’.).
For each of the 65 TOPMed phenotype concepts, we queried concept titles against Dug’s indexed collection of dbGaP variables to see how well Dug could recapitulate the manually curated set of variables. Nearly, 76% of the variables in each TOPMed phenotype concept contained a lexical match to the name of the concept we had used as a test query. To evaluate Dug’s performance on a less trivially simplistic dataset, we repeated the evaluation using synonyms of TOPMed phenotype concepts as test queries. We used synonyms from the Unified Medical Language System (UMLS) (Bodenreider, 2004) with no words in common with the original query; if no reasonable synonym could be chosen for a TOPMed phenotype, it was removed.
To quantify Dug’s performance, we used recall and precision. We report scores at the 10th result (P@10, R@10), at the 50th result (P@50, R@50) and at the nth result (P, R) for every search. While (P, R)@10 is a standard metric, we chose (P, R)@50 as the maximum number of results a person would reasonably scroll through, assuming 25 results per page (Jansen and Spink, 2005).
Within this framework, each variable belonging to a given TOPMed phenotype concept represents a condition positive (P) we would expect Dug to return. True positive (TP) results were variables returned by the TOPMed phenotype concept to which they belonged. Conversely, false positives (FP) were variables returned by any TOPMed phenotype concept to which they did not belong. Recall and precision are calculated at results returned (10, 50 or n) for each TOPMed phenotype concept query.
| (1) |
| (2) |
We evaluated Dug’s performance against a lexical search strategy as a baseline for how ‘findable’ test datasets were without Dug’s semantic enrichment. We indexed the same set of study metadata into Elasticsearch using Dug’s ingestion pipeline but did not add any semantic annotations. Search results were retrieved using the same default Elasticsearch scoring algorithm Dug uses. This was done to eliminate the variation between Dug and a baseline search due only to differences in the underlying lexical search strategies employed, allowing us to attribute differences in performance to Dug’s semantic annotation. There was no significance to using Elasticsearch beyond it providing a sophisticated lexical search strategy to make our evaluation results more directly interpretable.
A post-hoc analysis quantified the average semantic and lexical similarity between test queries and the top 50 results returned by Dug and Elasticsearch. Semantic similarity was measured using cosine similarity between test queries and search results after using the BioSentVec (Chen et al., 2019) model to transform queries and phrases into sentence embeddings. The sent2vec (v0.2.2) Python package was used to load the pretrained BioSentVec model (Pagliardini et al., 2018). Lexical similarity was measured using Levenshtein distance implemented in the python-Levenshtein Python package (v0.12.2).
To determine the statistical significance of differences in performance metrics between Dug and default Elasticsearch, we used nonparametric paired Wilcoxon signed-rank tests to test the hypothesis that location shift for each performance metric was non-zero. We chose this nonparametric test as it allows violations of normality in the sample distribution of differences. Wilcoxon tests were performed using the Python SciPy (v1.7.3) package using two-sided alternative hypotheses and a critical value of 0.05 (Virtanen et al., 2020).
3 Results
The initial Dug deployment on the NHLBI Biodata Catalyst ecosystem successfully indexed 15 991 study variables from 76 genomics datasets. Dug augmented these study variables with 573 ontological concepts and 11 752 knowledge-graph answers.
The results of our initial evaluation (Supplementary Fig. S1) showed Dug outperformed default Elasticsearch in terms of mean recall (0.79 versus 0.76, P < 0.001) but was not significantly different from Elasticsearch in terms of recall @ 10 and @ 50 (Supplementary Table S1). Additionally, Dug performed slightly but significantly worse than Elasticsearch in terms of precision @ 10 (P = 0.022) and @ 50 (P = 0.027). The biggest difference between Dug and a text-based method was Dug’s significantly lower total precision than Elasticsearch (0.07 versus 0.28, P < 0.001), though this was expected as Dug is designed to return a larger set of more exploratory results.
After repeating the evaluation with synonyms of original queries (e.g. ‘lung adenocarcinoma’ versus ‘lung cancer’), Dug vastly outperformed lexical search. Shown in Figure 7, Dug significantly outperformed default Elasticsearch in terms of mean recall (0.36 versus 0.14, P < 0.001), recall @ 10 (0.08 versus 0.04, P = 0.033) and recall @ 50 (0.18 versus 0.08, P = 0.007). We found no significant difference between Dug and Elasticsearch in precision @ 10 (P = 0.184) and precision @ 50 (P = 0.223); Dug’s total precision was significantly worse (P = 0.041), as to be expected with Dug’s exploratory results.
Fig. 7.
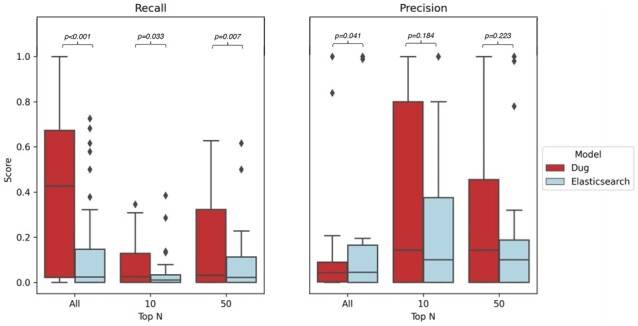
Dug versus lexical search performance using UMLS synonyms of TOPMed phenotype concepts to search for underlying variables. Left: Recall, R@10, R@50. Right: Precision, P@10, P@50. Diamonds represent outliers for each search method. Dug significantly outperforms lexical search (implemented in Elasticsearch) in terms of total recall, R@10 and R@50. Precision @10 and P@50 were not significantly different between Dug and Elasticsearch. Though Dug’s total precision was significantly lower, this was expected given Dug’s objective to uncover exploratory connections
We discount Dug’s lower total precision somewhat as an artifact of our evaluation rather than reflecting an excess of low-quality results. For practical reasons our test dataset narrowly defines ‘true’ search results to be only direct synonyms of the query. For example, our test dataset would not consider ‘smoking behavior’ relevant to a search for ‘lung cancer’ even though Dug would, and a user might. As a result, we would naturally expect Dug’s total precision to be lower than lexical search because Dug returns lexical results plus closely related but non-synonymous results. For this reason, Dug’s lower total precision might more accurately be characterized as reflecting a surplus of contextually relevant results.
Together, these results suggest Dug’s superior recall results from including semantically equivalent terms that do not appear lexically similar to the query (e.g. ‘heart attack’ versus ‘myocardial infarction’). To test this hypothesis, we quantified the average semantic and lexical distance between queries and results returned by Dug and Elasticsearch. Dug’s results are on average significantly (P = 0.021) more lexically divergent from the original query than Elasticsearch results, despite there being no significant difference (P = 0.878) in the semantic distances of search results returned by each (Fig. 8). Together, these results suggest Dug outperforms the most sophisticated lexical search strategies when the lexical diversity of the search space is high, as is typical for non-standardized biomedical datasets.
Fig. 8.
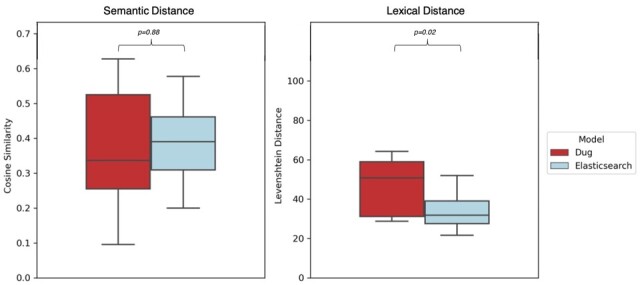
Semantic versus Lexical distance of Dug and Elasticsearch search results. Left: Semantic distance between queries and top 50 results returned by each method. Semantic distance for each query is defined as the mean cosine similarity of the top 50 results in the pre-trained BioSentVec model vector space. Right: Lexical distance between each query and its top 50 results (mean Levenshtein distance). Dug and Elasticsearch return semantically similar results on average (P = 0.88), yet Dug’s results are significantly more lexically divergent from the query (P = 0.02) with no loss in precision
4 Discussion
The exponential increase in publicly available datasets has created a need for comprehensive search tools that identify datasets relevant to a researcher's particular scientific question. To help researchers better navigate this new data landscape, we created Dug: a semantic search tool for biomedical datasets leveraging ontological knowledge graphs to intelligently suggest relevant connections derived from peer-reviewed research.
Our results demonstrate Dug’s ability to find relevant datasets regardless of the lexical expression of a query. When compared to a sophisticated lexical search strategy like Elasticsearch, Dug returns more relevant results with no significant difference in the precision of the top 10 or top 50 results. Like all search engines, Dug orders its result by relevance; as one looks deeper into the results, recall increases and precision decreases as Dug returns less directly related concepts. Dug also outperforms lexical search particularly when the search space is characterized by the type of high lexical diversity typical of biomedical data repositories.
Dug’s potential for data exploration and discovery is perhaps its greatest and least quantifiable strength. Dug’s ability to surface relevant datasets may be most useful when the user is not sure what they are looking for. For example, the relevance of ‘asbestos exposure’ variables to a search for ‘lung cancer’ datasets is contextual and subjective, making it difficult to quantify when and how second- and third-order connections might be useful. A key innovation of Dug is its ability to provide exploratory connections without sacrificing the precision of top results.
Dug’s modular design and stand-alone companion web portal can flexibly fit myriad use cases. For example, a centrally hosted version of Dug could index multiple data repositories to service a much larger user base. Additionally, smaller data coordinating centers like NIDDK central repository (Cuticchia et al., 2006; Rasooly et al., 2015) or large data ecosystem initiatives like NIH’s HEAL Data Ecosystem (U.S. Department of Health and Human Services) could use Dug to search across non-standardized data from diverse consortium members via a single portal without the need for significant manual curation.
Future work focuses on known limitations and responding to user feedback from the BioData Catalyst consortium. A principal concern is parallelizing the indexing process to index multiple datasets simultaneously and increase the throughput for larger datasets. Additionally, we are evaluating various strategies for improving ranking search results by relevance based on input from current users. We believe Dug provides a powerful, flexible tool for searching intuitively across complex data resources that are increasingly common in the biomedical data landscape.
Supplementary Material
Acknowledgements
The authors would like to thank Ben Heavner, Adrienne Stilp and Ingrid Borecki for their invaluable feedback during development. We also want to thank members of the BioData Catalyst Fellows program for feedback and testing.
Data availability
No new data were generated or analysed in support of this research.
Funding
This work was supported by the National Heart, Lung, and Blood Institute (NHLBI) [1-OT3-HL142479-01]; the National Center for Advancing Translational Sciences (NCATS) [1-OT3-TR002020-01]; and the Helping to End Addiction Long-Term (HEAL) Office [1-OT2-OD031940-01].
Conflict of Interest: none declared.
Contributor Information
Alexander M Waldrop, Center for Genomics, Bioinformatics, and Translational Research, RTI International, Research Triangle Park, NC 27709-2194, USA.
John B Cheadle, Research Computing Division, RTI International, Research Triangle Park, NC 27709-2194, USA.
Kira Bradford, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Alexander Preiss, Center for Data Science, RTI International, Research Triangle Park, NC 27709-2194, USA.
Robert Chew, Center for Data Science, RTI International, Research Triangle Park, NC 27709-2194, USA.
Jonathan R Holt, Center for Data Science, RTI International, Research Triangle Park, NC 27709-2194, USA.
Yaphet Kebede, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Nathan Braswell, Research Computing Division, RTI International, Research Triangle Park, NC 27709-2194, USA.
Matt Watson, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Virginia Hench, Center for Genomics, Bioinformatics, and Translational Research, RTI International, Research Triangle Park, NC 27709-2194, USA.
Andrew Crerar, Center for Genomics, Bioinformatics, and Translational Research, RTI International, Research Triangle Park, NC 27709-2194, USA.
Chris M Ball, Research Computing Division, RTI International, Research Triangle Park, NC 27709-2194, USA.
Carl Schreep, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
P J Linebaugh, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Hannah Hiles, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Rebecca Boyles, Research Computing Division, RTI International, Research Triangle Park, NC 27709-2194, USA.
Chris Bizon, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
Ashok Krishnamurthy, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA; Department of Computer Science, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7548, USA.
Steve Cox, Renaissance Computing Institute, University of Chapel Hill, North Carolina, Chapel Hill, NC 27599-7568, USA.
References
- Bell E. et al. (2019) Finding useful data across multiple biomedical data repositories using DataMed. Nat. Genet., 49, 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biomedical Data Translator Consortium (2019) The biomedical data translator program: conception, culture, and community. Clin. Transl. Sci, 12, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon C. et al. (2019) ROBOKOP KG and KGB: integrated knowledge graphs from federated sources. J. Chem. Inf. Model., 59, 4968–4973. [DOI] [PubMed] [Google Scholar]
- Bodenreider O. (2004) The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res., 32, D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley D. et al. (2019) Google dataset search: building a search engine for datasets in an open web ecosystem. In: The World Wide Web Conference, WWW ’19. Association for Computing Machinery, New York, NY, USA, pp. 1365–1375.
- Canakoglu A. et al. (2019) GenoSurf: metadata driven semantic search system for integrated genomic datasets. Database (Oxford), 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A. et al. (2020) Dataset search: a survey. VLDB J, 29, 251–272. [Google Scholar]
- Chen Q. et al. (2019) BioSentVec: creating sentence embeddings for biomedical texts. In: 2019 IEEE International Conference on Healthcare Informatics, Xi'an, China, ICHI 2019. pp. 0–4.
- Chen X. et al. (2018) DataMed – an open source discovery index for finding biomedical datasets. J. Am. Med. Informatics Assoc., 25, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F.S. et al. (2018) Helping to end addiction over the long-term: the research plan for the NIH HEAL initiative. JAMA, 320, 129–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox S. et al. (2020) Visualization environment for federated knowledge graphs: development of an interactive biomedical query language and web application interface. JMIR Med. Inform., 8, e17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuticchia A.J. et al. (2006) NIDDK data repository: a Central collection of clinical trial data. BMC Med. Inform. Decis. Mak., 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elasticsearch Theory Behind Relevance Scoring. https://www.elastic.co/guide/en/elasticsearch/guide/current/scoring-theory.html. (20 May 2021, date last accessed).
- Huang J. et al. (2016) OmniSearch: a semantic search system based on the ontology for MIcroRNA target (OMIT) for microRNA-target gene interaction data. J. Biomed. Semantics, 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen B.J., Spink A. (2005) Analysis of document viewing patterns of web search engine users. In: Scime A. (ed). Web Mining: Applications and Techniques, Idea Publishing Group, pp. 339–354. [Google Scholar]
- Kuć R., Rogozinski M. (2016) ElasticSearch server. Packt Publishing Ltd.
- Laulederkind S.J. et al. (2012) Ontology searching and browsing at the rat genome database. Database (Oxford), 2012, doi: 10.1093/database/bas016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C.J. et al. (2017) The monarch initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res., 45, D712–D722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services (2020). The NHLBI BioData Catalyst. Zenodo. https://doi.org/10.5281/zenodo.3822858
- NIH Data Sharing Resources (2020). https://www.nlm.nih.gov/NIHbmic/nih_data_sharing_repositories.html (30 June 2021, date last accessed).
- Pagliardini M. et al. (2018) Unsupervised learning of sentence embeddings using compositional n-gram features. In: NAACL 2018 - Conference of the North American Chapter of the Association for Computational Linguistics, New Orleans, LA, USA.
- Pang C. et al. (2015) BiobankConnect: software to rapidly connect data elements for pooled analysis across biobanks using ontological and lexical indexing. J. Am. Med. Inform. Assoc., 22, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. (2021) The broken promise that undermines human genome research. Nat. News. https://www.nature.com/articles/d41586-021-00331-5 (29 June 2021, date last accessed). [DOI] [PubMed] [Google Scholar]
- Rasooly R.S. et al. (2015) The national institute of diabetes and digestive and kidney diseases central repositories: a valuable resource for nephrology research. Clin. J. Am. Soc. Nephrol., 10, 710– LP – 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J.T. et al. (2021) KG-COVID-19: a framework to produce customized knowledge graphs for COVID-19 response. Patterns (NY), 2, 100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone S. et al. (2017) OPEN DATS, the data tag suite to enable discoverability of datasets. Sci Data, 4, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto A.J. et al. (2019) Thalia: semantic search engine for biomedical abstracts. Bioinformatics, 35, 1799–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilp A.M. et al. (2021) A system for phenotype harmonization in the NHLBI Trans-Omics for precision medicine (TOPMed) program. Am. J. Epidemiol., 190, 1977–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The “All of Us” Research Program. (2019) N. Engl. J. Med., 381, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T. et al. (2007) Ontology-based interpretation of keywords for semantic search. In: The Semantic Web. Springer, Berlin, Heidelberg, pp. 523–536. [Google Scholar]
- U.S. Department of Health and Human Services. What is the HEAL data ecosystem? Natl. Inst. Heal. [Google Scholar]
- University of Washington Department of Biostatistics (2020) NHLBI trans-omics for precision medicine WGS-About TOPMed. About TOPMed. https://topmed.nhlbi.nih.gov/ (20 June 2021, date last accessed).
- Virtanen P. et al. ; SciPy 1.0 Contributors. (2020) {SciPy} 1.0: fundamental algorithms for scientific computing in python. Nat. Methods, 17, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D. et al. (2016) The FAIR guiding principles for scientific data management and stewardship. Sci. Data, 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were generated or analysed in support of this research.


