Abstract
Summary
Here, we introduce SNIKT, a command-line tool for sequence-independent visual confirmation and input-assisted removal of adapter contamination in whole-genome shotgun or metagenomic shotgun long-read sequencing DNA or RNA data.
Availability and Implementation
SNIKT is implemented in R and is compatible with Unix-like platforms. The source code, along with documentation, is freely available under an MIT license at https://github.com/piyuranjan/SNIKT.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Implementing quality control steps on next-generation sequencing data improves the accuracy and efficiency of downstream analyses. For this reason, quality control is a highly recommended first step for all sequencing data, regardless of platform or sequencing chemistry. While platform-specific steps exist, removing sequencing adapter contamination is common to most quality control protocols for next-generation sequencing data (Breitwieser et al., 2019; Gu et al., 2019).
Sequencing adapters are stretches of oligonucleotides, added during library preparation to flank the nucleotide fragment, which facilitate barcoding and sequencing (Jain et al., 2016; Slatko et al., 2018). While a necessary sequencing step, these adapter sequences are a source of systemic contamination for any downstream analysis steps, which may impact assembly quality or metagenomic characterization. For this reason, Illumina data are often subjected to popular tools like fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), cutadapt (Martin, 2011) and Trimmomatic (Bolger et al., 2014), which perform quality assessment and remove adapters based on alignments with adapter sequences that are either user-specified or obtained from a local database. Porechop (https://github.com/rrwick/Porechop), Oxford Nanopore’s Guppy and NanoPlot (De Coster et al., 2018) are equivalent methods for quality control on Nanopore data. Yet a central limitation to all these approaches is the need for the user to pre-specify the adapter sequence. This is not always feasible, especially when using publicly available data with incomplete methodological reporting. To our knowledge, no existing tool provides the ability to visualize and simultaneously remove adapter contamination independent of the adapter sequence.
Using visually aligned nucleotide compositions to estimate sequence overrepresentation is potentially a faster alternative to identifying adapter contamination. By aligning reads at their 5′ and 3′ ends and visually identifying common positions that vary from the predicted (random) nucleotide distribution, users can readily determine the length and position of contaminating adapters, regardless of their specific sequence. This approach offers several potential advantages. Firstly, it can be performed without any prior knowledge of adapter sequences, as commonly occurs with publicly available data. Secondly, it is platform-agnostic, and works equally well on short-read (e.g. Illumina) and long-read (e.g. Oxford Nanopore) sequence data. Thirdly, unlike alignment-based methods, this approach is robust to platforms with higher error rates (e.g. Oxford Nanopore), which may introduce misalignments and decrease the efficiency and accuracy of alignment-based adapter removal. Once identified, the contaminating pattern can be easily removed by end-trimming the reads. While this visualization-based approach is partially implemented in fastqc, that specific implementation is optimized for Illumina reads, and performs suboptimally on long-read datasets (e.g. via Oxford Nanopore). Similarly, tools in the NanoPack suite can utilize visualization-based approach but they are inefficient when dealing with large long-read datasets.
Here, we present SNIKT, a tool that enables visualization and input-assisted trimming of adapter contamination in long-read data. Designed to work without knowledge of the adapter sequence used, SNIKT is suitable without modification for a wide range of existing and upcoming sequencing and barcoding kits. It uses seqtk (https://github.com/lh3/seqtk) for efficient manipulation of fastq data and summarizes essential quality control metrics in concise figures and reports, enhancing the reproducibility of the quality control process. SNIKT is resource efficient in comparison to other quality control methods and handles large next-generation sequencing (NGS) datasets with ease.
2 Description
SNIKT is available with the conda package manager for easy installation through the bioconda repository (Grüning et al., 2018). Alternatively, its source code can be directly pulled from the GitHub repository and executed as a program if all the dependencies are met. Its dependencies include the package seqtk, the R language (R Core Team, 2021), and the R libraries tidyverse (Wickham et al., 2019), gridExtra (Baptiste and Antonov, 2017), docopt (De Jonge, 2016) and lubridate (Grolemund and Wickham, 2011). SNIKT accepts a fastq file as input which ideally has not been processed by another adapter removing process.
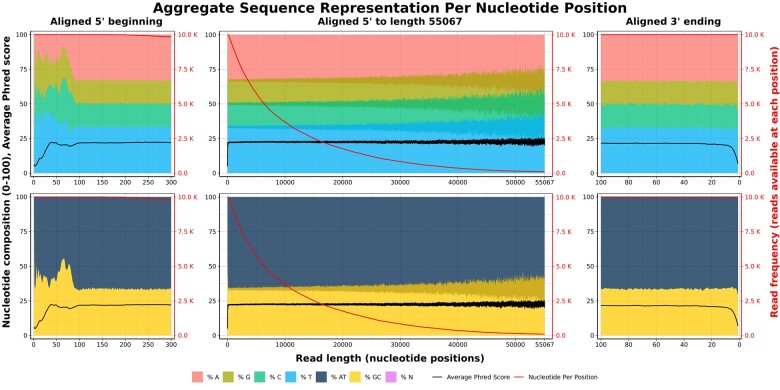
SNIKT works with the fastq file in two stages. In the first stage, it calculates forward and reverse compliment nucleotide compositions with seqtk. It then aligns sequences in a visualization that renders adapter contamination readily apparent (Fig. 1). Contamination can be seen as a sequence overrepresentation pattern (jagged lines) on the averaged compositions and Phred scores per base. Based on this visualization, users can choose the most suitable method of quality control. In the second stage, based on user input, SNIKT removes the contamination by end-trimming and filtering fastq reads by length. It then creates pre- and post-cleaning summary demonstrating the path that was taken along with the summary statistics of the resulting fastq data (Supplementary File S1). As products, SNIKT produces this report in HTML format along with the cleaned reads in fastq format.
Fig. 1.
SNIKT report identifying the presence and length of adapter contamination. Sequences (N = 412 984) are aligned at their 5′ and 3′ ends, and the relative composition of nucleotides at each position is visualized along with length statistics. In this instance, the nonrandom distribution of nucleotides extends from about 1–100 positions at the 5′ beginning. Lower quality bases can be observed from about 1–10 positions on the 3′ ending. This approach does not require prior knowledge of adapter sequences and overcomes the challenge of variable read length
SNIKT offers an interactive mode that guides users through these stages. It also offers to run these stages in batch jobs, facilitating running over remote servers. The execution of SNIKT is single-threaded due to its dependencies but the process can be parallelized over multiple fastq files using programs like GNU parallel (Tange, 2018).
SNIKT is more consistent in adapter and other technical artifact content removal than ONT’s Guppy suite that is the current alignment-based standard for adapter content removal for Nanopore long reads (Supplementary File S2). SNIKT is four times faster and finishes reporting using half the memory as NanoQC. SNIKT is 1.5 times faster than NanoFilt but uses more memory for trimming since it also generates reports with pre- and post-trimming read data. Taken together, SNIKT works more efficiently than a combined run of NanoQC and NanoFilt (Supplementary File S3).
3 Example and discussion
Figure 1 demonstrates the results of SNIKT implementation using a Nanopore-derived whole-genome shotgun dataset of Candida albicans CHN1 sequenced on an R9.4.1 MinION flow cell after a rapid library preparation with SQK-RAD004 kit (Garg et al., 2021). Reads were base-called with Guppy 3.2.9 and collected in a single fastq file containing 4.9 Gbp in 413 K reads. This read set is available on NCBI SRA under accession number SRR13441294. SNIKT was performed on this data set in batch processes on a laptop computer running Ubuntu 20 in Windows Subsystem Linux. Resource utilization was captured with the/usr/bin/time utility.
The first process (adapter visualization) finished in 1 min 5 s using peak memory of 815 MB and average CPU utilization of 107%, compiling a report with the first 10 000 reads that contained Figure 1. Sequence overrepresentation was readily apparent in nucleotide compositions and the Phred scores on the aligned 5′ and 3′ ends of the read-set. A marginal fraction of short-length reads was also observed with the read frequency over nucleotide positions acting as a proxy for a read length distribution. Based on this report, the second process (adapter removal) was executed with trim lengths of 110 and 20 nucleotides from the 5′ and the 3′ ends respectively and a filter length of 500 bases. The second process finished in about 4 min 25 s, using a peak memory of 760 MB and average CPU utilization of 130%. This process resulted in a report (Supplementary File S1) summarizing pre- and post-trimming statistics along with a cleaned fastq file containing 4.84 Gbp in 398 K reads with a minimum read length of 500 bases.
4 Conclusion
SNIKT is a command-line tool that efficiently and accurately identifies and removes adapter contamination in sequencing data. It works without knowledge of adapter sequence and is effective independent of sequencing platform, read length or error rate.
Funding
This work was supported by the National Institutes for Health (R01HL144599 and T32HL007749) and the Kahn Foundation.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Piyush Ranjan, Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI 48109, USA.
Christopher A Brown, Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI 48109, USA; Institute for Research on Innovation and Science, Institute for Social Research, University of Michigan, Ann Arbor, MI 48109, USA.
John R Erb-Downward, Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI 48109, USA.
Robert P Dickson, Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI 48109, USA; Department of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109, USA; Weil Institute for Critical Care Research & Innovation, Ann Arbor, MI 48109, USA.
References
- Baptiste A, Antonov A. (2017) Package ‘gridExtra’ Miscellaneous Functions for “Grid” Graphics. https://github.com/baptiste/gridextra. [Google Scholar]
- Bolger A.M. et al. (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitwieser F.P. et al. (2019) A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform., 20, 1125–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W. et al. (2018) NanoPack: visualizing and processing long-read sequencing data. Bioinformatics, 34, 2666–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge E. (2016) Docopt: Command-Line Interface Specification Language. R package version 0.4 5. https://github.com/docopt/docopt.R. [Google Scholar]
- Garg S. et al. (2021) High-quality genome reconstruction of Candida albicans CHN1 using nanopore and illumina sequencing and hybrid assembly. Microbiol. Resour. Announc., 10, e0029921. https://doi.org/10.1128/mra.00299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolemund G., Wickham H. (2011) Dates and times made easy with lubridate. J. Stat. Soft., 40, 1–25. [Google Scholar]
- Grüning B. et al. ; Bioconda Team. (2018) Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods, 15, 475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. et al. (2019) Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol., 14, 319–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M. et al. (2016) The oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol., 17, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J., 17, 10. [Google Scholar]
- R Core Team (2021) R: A language and environment for statistical ## computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Slatko B.E. et al. (2018) Overview of Next-Generation sequencing technologies. Curr. Protoc. Mol. Biol., 122, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. (2018) GNU Parallel 2018. https://doi.org/10.5281/ZENODO.1146014.
- Wickham H. et al. (2019) Welcome to the tidyverse. JOSS, 4, 1686. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.