Change history
10/17/2024
Editors Note: Readers are alerted that concerns have been raised about the data and statistical analysis reported in this article. A correction is forthcoming.
Abstract
Alzheimer’s disease (AD) is characterized by synaptic loss, which can result from dysfunctional microglial phagocytosis and complement activation. However, what signals drive aberrant microglia-mediated engulfment of synapses in AD is unclear. Here we report that secreted phosphoprotein 1 (SPP1/osteopontin) is upregulated predominantly by perivascular macrophages and, to a lesser extent, by perivascular fibroblasts. Perivascular SPP1 is required for microglia to engulf synapses and upregulate phagocytic markers including C1qa, Grn and Ctsb in presence of amyloid-β oligomers. Absence of Spp1 expression in AD mouse models results in prevention of synaptic loss. Furthermore, single-cell RNA sequencing and putative cell–cell interaction analyses reveal that perivascular SPP1 induces microglial phagocytic states in the hippocampus of a mouse model of AD. Altogether, we suggest a functional role for SPP1 in perivascular cells-to-microglia crosstalk, whereby SPP1 modulates microglia-mediated synaptic engulfment in mouse models of AD.
Subject terms: Neuroimmunology, Alzheimer's disease, Mechanisms of disease, Molecular neuroscience
Microglia mediate aberrant synapse engulfment in Alzheimer’s disease (AD), but the underlying mechanisms are poorly understood. Here the authors show a perivascular cells-to-microglia crosstalk that induces microglia phagocytic state resulting in synapse engulfment in two mouse models of AD.
Main
The integrity of neurons and synapses is critically dependent on brain-resident macrophages, which include microglia and perivascular macrophages (PVMs) that continuously monitor and clear phagocytic targets across the lifespan1,2. Microglia, tissue-resident macrophages of brain parenchyma, contribute to circuit refinement through engulfment of synapses and axon tracts during postnatal development, whereas PVMs associate with the perivascular space where they represent the first responders to toxic agents and pathogens that may cross the blood–brain barrier (BBB) into the brain parenchyma3–7. Microglia and PVMs both share similar yolk-sac origin and reside on the parenchymal side of the BBB, yet occupy distinct microenvironments and express unique cell-type-specific markers8–13. Because of the immediate juxtaposition of perivascular space with the brain parenchyma, we reasoned that microglia and PVMs could potentially influence each other to coordinate phagocytosis in response to central nervous system perturbations. For example, in Alzheimer’s disease (AD), many of the identified risk variants point toward defective phagocytic and endolysosomal pathways in microglia and PVMs; however, intercellular and intracellular mechanisms governing this impairment remain unclear14. In line, microglia mediate regional synapse loss in AD, but environmental cues that modulate potential microglial functional states are poorly understood15.
Here we uncovered secreted phosphoprotein 1 (SPP1/osteopontin), predominantly derived from PVM, as an extrinsic modulator of microglial phagocytosis. We observed that SPP1 is required for activation of complement-initiator C1q and synaptic engulfment by microglia in AD mouse models. SPP1 has been shown in multiple peripheral tissues to regulate phagocytosis by macrophages16–20. In AD patients, secreted SPP1 levels are increased in cerebrospinal fluid (CSF) and plasma; further, SPP1 is found on plaque-associated microglia and has been suggested as a conserved disease-associated marker among mice and human21–27. However, the role of SPP1 in the brain or its relevance in AD is unknown. Here we found a region-specific activation of SPP1 in the hippocampal perivascular space of AD mouse models as well as in AD patient tissues. Super- and ultra-structural examination revealed that SPP1 is expressed predominantly by PVMs, and to a lesser extent, by perivascular fibroblasts (PVFs) in the adult hippocampus. In mouse models of AD, we found that perivascular SPP1 is upregulated in a region-specific manner at onset of synaptic elimination by microglia. Genetic ablation of SPP1 ameliorated microglial phagocytic states in AD mouse models as well as C1q activation, leading to the prevention of microglia–synapse engulfment and synapse loss in these mice despite amyloid-β (Aβ) challenge. Using single-cell RNA sequencing (scRNA-seq) and computational ligand–target predictions (NicheNet28), we noted multiple autocrine and paracrine signaling pathways to be potentially modulated by perivascular SPP1, with many of these pathways converging on microglial phagocytic functional states. Altogether, our data suggest a functional crosstalk between PVMs and microglia and propose perivascular SPP1 as an extrinsic signal that modulates synapse phagocytosis by microglia.
Results
SPP1 upregulation at onset of microglia–synapse phagocytosis
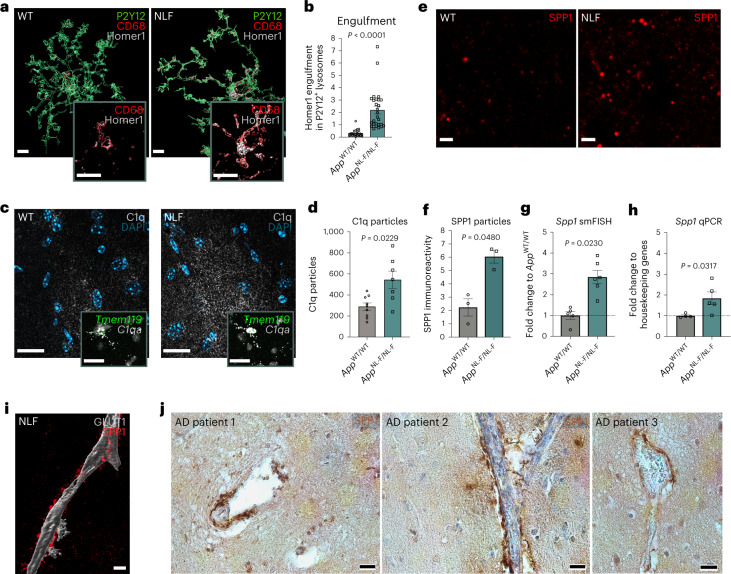
To address whether SPP1 is dysregulated at a time point when synapses are vulnerable to microglial engulfment in an AD-relevant context, we used the slow-progressing AppNL-F mouse model, where control by the endogenous App promoter allows for physiological cell-type specific and temporal regulation of Aβ production15,29,30. We first assessed phagocytic microglia–synapse interactions in the AppNL-F hippocampus at the age of 6 months, an age that precedes robust plaque deposition in the hippocampal parenchyma30. We quantified internalized level of Homer1-immunoreactive synaptic puncta within CD68+ P2Y12+ microglial lysosomes and observed an approximate sevenfold increase in synaptic engulfment by microglia in AppNL-F mice as compared to those of age- and sex-matched wild-type (WT) mice (Fig. 1a,b). We also observed upregulation of C1q, the initiating protein of the classical complement cascade that mediates microglial phagocytosis of synapses3,15,31 (Fig. 1c,d). C1qa expression was contained within Tmem119+ microglia as assessed by single-molecule fluorescent in situ hybridization (smFISH), confirming microglia as the main producers of C1q in the adult hippocampus (Fig. 1c, insert)15,32. We next assessed potential cross-regulation between synaptic engulfment and SPP1, a glycoprotein associated with phagocytosis in peripheral macrophages, in the 6-month AppNL-F hippocampus18. Using 3D-stimulated emission depletion (3D-τ-STED) super-resolution imaging, we found an approximately threefold increase of punctate SPP1 protein immunoreactivity in the CA1 hippocampus of AppNL-F mice as compared to age- and sex-matched WT controls (Fig. 1e,f). The SPP1 upregulation was region-specific, that is, in hippocampus but not cerebellum (Extended Data Fig. 1a,b). In line with increased SPP1 production, we found an approximate threefold increase of Spp1 mRNA expression levels in hippocampal CA1 sections of AppNL-F mice compared to WT mice by smFISH in intact tissue, which was further confirmed by qPCR analysis on brain homogenates (Fig. 1g,h). The specificity of SPP1 antibody and Spp1-targeting smFISH probes was validated by absence of signals in Spp1KO/KO mice (Extended Data Fig. 1c).
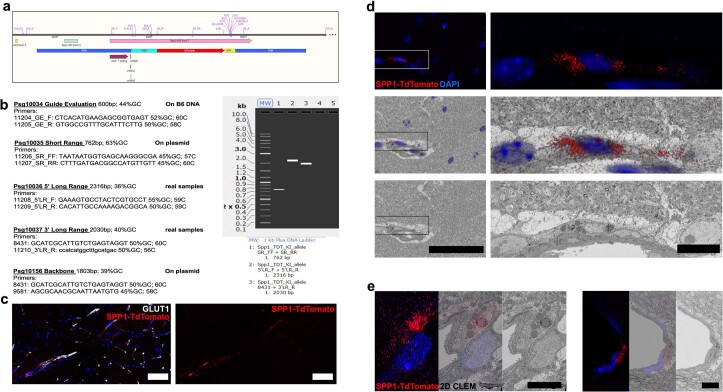
Fig. 1. SPP1 upregulation at onset of microglia–synapse phagocytosis.
a, Representative 3D reconstructed images showing Homer1 engulfment within CD68+ lysosomes of P2Y12+ microglia in 6-month AppWT (WT) versus AppNL-F CA1 hippocampal SLM. Scale bar represents 5 µm. b, Quantification of Homer1 engulfment in 6-month WT and AppNL-F P2Y12+ microglia. One datapoint represents one individual P2Y12+ microglia with a total of 24–27 ROIs pooled from n = 4 animals per genotype examined over two independent experiments. P values from two-tailed unpaired Student’s t-test. c, Representative confocal images of C1q protein expression in 6-month WT versus AppNL-F mice. Insert represents C1qa mRNA within Tmem119+ microglia. Scale bar represents 20 µm. Data are representative of two mice per genotype examined over at least five independent experiments. d, Quantification of C1q puncta in 6-month WT and AppNL-F CA1 hippocampus. One datapoint represents one ROI per mouse from n = 9 WT mice and n = 8 AppNL-F mice examined over two independent experiments. Average amount of cells per datapoint is 30–40. P values from two-tailed Mann–Whitney test. e,f, 3D-τ-STED imaging of secreted SPP1 in 6-month WT and AppNL-F SLM (e) and quantification of SPP1 total fluorescence particles (f). One datapoint represents one ROI per mouse, with total of n = 3 mice examined over one independent experiment. P values from two-tailed Mann–Whitney test. Scale bar represents 2 µm. g,h, Quantification of Spp1 expression within hippocampus of 6-month WT versus AppNL-F as measured by smFISH in hippocampus (g) or via qPCR on hippocampal homogenates (h). One datapoint represents one individual value per mouse, with total of n = 5–6 mice (per genotype) (g) or 4–5 mice (per genotype) (h) examined over two independent experiments. P values from two-tailed Mann–Whitney test. i, Representative 3D reconstruction of SPP1 adjacent to GLUT1+ vasculature in SLM of 6-month AppNL-F. Scale bar represents 10 µm. Image representative of two AppNL-F mice examined over four independent experiments. j, Representative images of SPP1 expression along vasculature in postmortem hippocampal brain slices of three AD patients. Images are representative of six AD patients (see also Supplementary Table 1). Scale bar represents 20 µm. Data are shown as mean ± s.e.m.
Extended Data Fig. 1. Pre-plaque vascular Aβ deposition in AppNL-F mice.
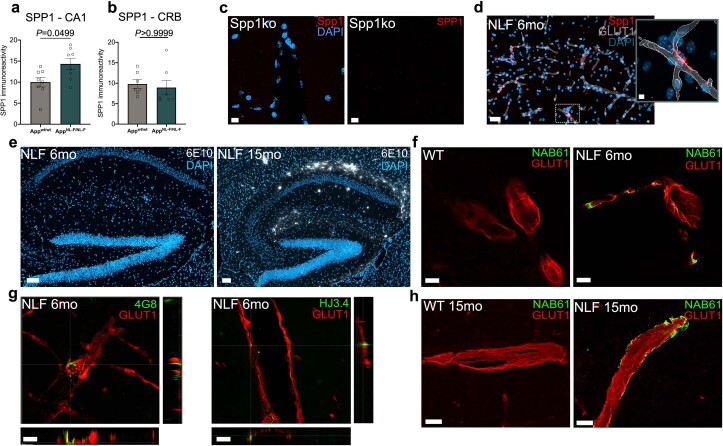
(a, b) Quantification of SPP1 immunoreactivity in CA1 hippocampus (a) versus cerebellum as control region (b), as measured by confocal imaging. 1 datapoint represents 1 ROI per mouse hippocampus, with total of 8 (CA1, a) and 7 (CRB, b) ROI from n = 3 mice per genotype examined over 2 independent experiments. P Values from Two-tailed Mann-Whitney test. Data are shown as Mean ± SEM. (c) Validation of SPP1 antibody (left) and anti-Spp1 probe for smFISH (right) in 3 mo Spp1KO/KO hippocampal tissue. Scale bar represents 10 µm. Data representative of n = 4 Spp1KO/KO mice examined over 1 independent experiment. (d) Representative confocal image of Spp1 mRNA expression along GLUT1+ vasculature in SLM of 6 mo AppNL-F mice as characterized by smFISH-IHC. Scale bar represents 20 µm. Insert: 3D reconstruction. Scale bar represents 5 µm. Data are representative of 3 AppNL-F mice examined over at least 4 independent experiments. (e) Representative confocal images showing 6E10 plaque staining in hippocampus of 6 mo versus 15 mo AppNL-F mice. Scale bar represents 100 µm. Data representative of n = 3 AppNL-F mice mice per time point, examined over 2 independent experiments. (f) Representative confocal images showing oAβ around GLUT1+ blood vessels as characterized by NAB61 immunoreactivity in 6 mo WT versus 6 mo AppNL-F SLM. Scale bar represents 10 µm. Data representative of n = 3 mice per genotype, examined over 2 independent experiments. (g) Representative confocal images showing vascular oAβ deposition using alternative antibodies 4G8 and HJ3.4 that recognize Aβ17-24 and Aβ1-13, respectively, in 6 mo AppNL-F SLM. Scale bar represents 10 µm. Data representative of n = 3 AppNL-F mice examined over 2 independent experiments. (h) Representative confocal images showing vascular oAβ (NAB61) in 15 mo WT versus AppNL-F SLM. Scale bar represents 20 µm.
Interestingly, Spp1 mRNA expression in the hippocampal CA1 was specifically enriched within the stratum lacunosum-moleculare (SLM) layer and Spp1+ cells displayed an elongated vascular-like pattern (Extended Data Fig. 1d). Costaining with pan-endothelial marker GLUT1 showed cellular zonation of Spp1 mRNA expression adjacent to vascular structures, whereas outside of the endothelial membrane, no Spp1 mRNA was detected (Extended Data Fig. 1d, inset). Using high-resolution confocal imaging and 3D surface rendering, we confirmed the close association of cytosolic SPP1 protein expression with GLUT1+ vasculature (Fig. 1i). Similar to the murine AppNL-F hippocampus, we also found a striking presence of SPP1 immunoreactivity along the vasculature in the hippocampus of postmortem AD patient tissues (Fig. 1j and Supplementary Table 1). Altogether, these data suggest that SPP1 expression is enriched along the vasculature in AppNL-F mice and AD patients. Further, we found negligible levels of Aβ oligomers (oAβ) or plaques at this early time point in the parenchyma of AppNL-F hippocampus (Extended Data Fig. 1e)30. In contrast, however, we observed oAβ deposition along the hippocampal vasculature at 6 months, assessed by the oAβ-specific NAB61+ immunostaining juxtaposed to GLUT1+ vasculature (Extended Data Fig. 1f)33. The latter was confirmed using two other anti-Aβ antibodies, the 4G8 and HJ3.4, which recognize Aβ17-24 and Aβ1-13, respectively (Extended Data Fig. 1g)34. The vascular oAβ deposition was more pronounced in 15-month AppNL-F mice, in contrast to age-matched WT controls in which no positive NAB61 staining was found (Extended Data Fig. 1h). These data suggest early, preplaque vascular Aβ deposition in the AppNL-F, coinciding with SPP1 activation in the perivascular space.
SPP1 is expressed by PVMs and fibroblasts
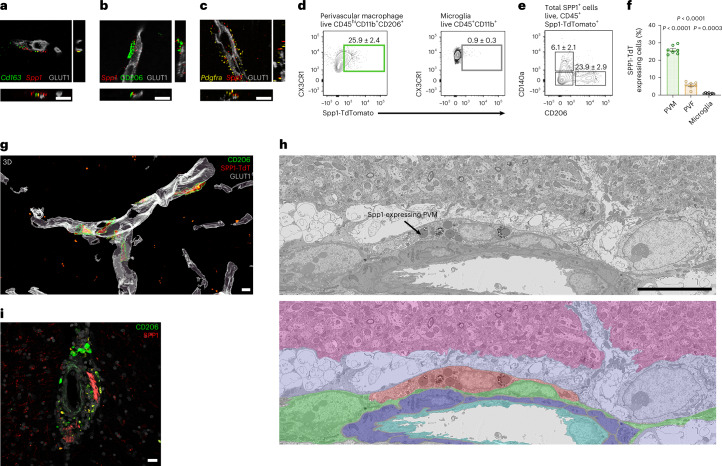
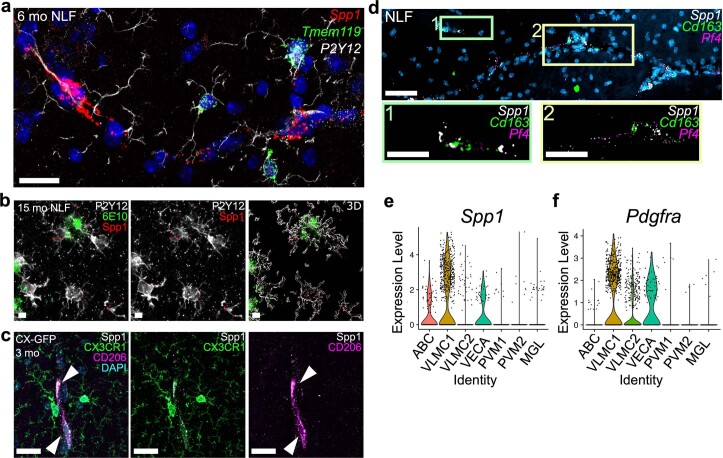
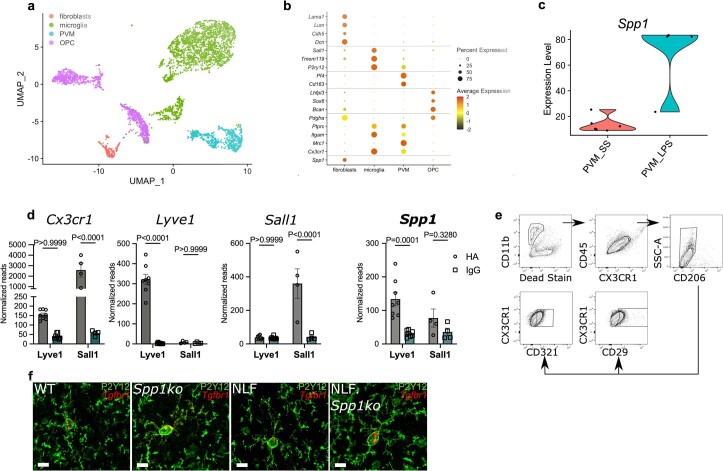
Spp1 expression in 6-month AppNL-F hippocampus was remarkably restricted to cells along the perivascular space (Fig. 1). Given the role of SPP1 as a marker of macrophage subsets in peripheral tissues, we assessed whether Spp1 is expressed by brain-resident macrophages associated with the vascular space, that is Cd163+ CD206+ PVM35. Spp1 mRNA expression predominantly colocalized with pan-PVM markers Cd163 and CD206 juxtaposed to GLUT1+ hippocampal vasculature (Fig. 2a,b). In contrast, we failed to observe Spp1 expression in Tmem119hi P2Y12+ microglia of 6-month AppNL-F animals (Extended Data Fig. 2a). Of note, Spp1 expression in microglia was detected only at later stages, that is, in 6E10+ plaque-rich 15-month AppNL-F hippocampus (Extended Data Fig. 2b), likely reflecting SPP1+ disease-associated microglia25,36. Further, immunophenotypic characterization revealed restricted expression of SPP1 within CD206+CX3CR1+ cells, and Spp1+ cells were found positive for PVM-specific platelet factor 4 (Pf4) and Cd163 (Extended Data Fig. 2c,d)11,13,37. Interestingly, we also found rare Spp1+ Cd163− Pf4− cells in the hippocampal perivascular space (Extended Data Fig. 2d). Spp1 mRNA has been suggested by scRNA-seq to be expressed by vascular leptomeningeal cells, also known as PVFs, which are located within parenchymal arteries (Extended Data Fig. 2e)37. Indeed, the second cluster of Spp1+ cells coexpressed the pan-PVF marker Pdgfra (encoding CD140a), suggesting PVFs as a second cellular source of SPP1 surrounding GLUT1+ vasculature in the hippocampus, albeit much less abundant compared to PVMs (Extended Data Fig. 2f,c).
Fig. 2. SPP1 is expressed by PVMs and fibroblasts.
a–c, Representative images of Spp1 mRNA expression juxtaposed to GLUT1+ vasculature, colocalizing with pan-PVM markers Cd163 (a), CD206 (b) and PVF (Pdgfra+) (c) in 6-month AppNL-F SLM as characterized by smFISH-IHC. Scale bar represents 10 µm. Data are representative of four AppNL-F mice examined over two independent experiments. d–f, Representative FACS plots to identify PVMs (CX3CR1+CD45+CD11b+CD206+ (d), microglia CX3CR1highCD45+CD11b+ (e) or gated on total TdT expressing cells isolated from Spp1TdT hippocampal homogenates and quantification (f)). One datapoint represents one cell type per mouse (PVM, PVF and microglia) pooled from n = 7 mice examined over two independent experiments (f). P values from one-way ANOVA, Kruskal–Wallis test. g, Three-dimensional reconstruction of CD206+ PVMs expressing SPP1-Td along GLUT1+ vessels in SLM from Spp1TdT mice. Scale bar represents 7 µm. Data are representative of three Spp1TdT mice examined over two independent experiments. h, Representative single serial section SEM backscatter electron image of a representative SPP1-TdT-positive PVM as identified by CLEM (Upper). SPP1-TdT-positive cell manually pseudocolored red, together with neuropil (pink), astrocytes (lilac), smooth muscle cells (purple), endothelial cells (cyan) and other perivascular cells (green), shown with reduced opacity over the electron microscopy data (lower). Accompanying confocal overlays and correlation images shown in Extended Data Fig. 3e,d array tomography data shown in Supplementary Video 1. Scale bar represents 10 µm. i, Representative image of perivascular SPP1 in AD postmortem hippocampal tissue, costained with CD206. Scale bar represents 25 µm. Data are representative of n = 6, six different patient tissues (Supplementary Table 1). Data are shown as mean ± s.e.m. TdT, TdTomato.
Extended Data Fig. 2. Spp1 is expressed in perivascular cells.
(a) Representative confocal image showing Spp1 and Tmem119 mRNA expression within P2Y12+ microglia of 6 mo AppNL-F SLM as characterized by smFISH-IHC. Scale bar represents 20 µm. Data representative of n = 8 AppNL-F mice examined over 3 independent experiments. (b) Representative confocal image showing Spp1 mRNA in P2Y12 microglia associated with 6E10+ plaques in 15 mo AppNL-F SLM. Scale bar represents 5 µm. Data representative of n = 3 AppNL-F mice examined over 2 independent experiments. (c) Representative confocal images of SPP1 protein expression in CD206+ PVMs (arrow head) and CX3CR1GFP myeloid cells of 3 mo Cx3cr1GFP/WT SLM86. Scale bar represents 20 µm. Data are representative of n = 2 Cx3cr1GFP/WT mice examined over at least 3 independent experiments. (d) Representative confocal image showing Spp1 mRNA expression occasionally colocalizing with pan-PVM markers Cd163 and Pf4 in 6 mo AppNL-F SLM as identified by smFISH. Scale bar represents 50 µm. Data representative of n = 6 AppNL-F mice examined over 2 independent experiments. (e, f) Violin plots of Spp1 (e) and Pdgfra (f) expression reanalyzed from Zeisel et al.37. VLMC, vascular leptomeningeal cells; ABC, Arachnoid barrier cells; VECA, Arterial vascular endothelial cells; PVM, Perivascular macrophage; MGL, Microglia.
To further assess in vivo SPP1 expression, we developed Spp1-IRES-TdTomato (TdT) (Spp1TdT) reporter mice, which carry an IRES-TdT cassette in Spp1 exon 7 retaining endogenous SPP1 expression (Extended Data Fig. 3a–c). Flow cytometric analysis of naïve animals revealed the expression of Spp1-TdT within 25.9% of pregated CD45hi CD206+ PVM, in contrast to CD11b+ CD45int CX3CR1hi microglia that were almost devoid of TdT expression (0.9% of which were TdT+) (Fig. 2d–f). Further, only approximately 6% of total live TdT+ cells were CD140a+ (gene product of Pdgfra), in contrast to 23.9% for CD206+, highlighting PVMs as a predominant cellular source of SPP1 (Fig. 2e,f). Using high-resolution confocal imaging, we found that the distribution of SPP1-TdT was comparable to the SPP1 immunoreactivity detected by IHC (Fig. 1i), that is, along GLUT1+ vasculature of the hippocampus (Extended Data Fig. 3d). Further, we saw similar cellular localization of SPP1-TdT within CD206+ PVMs in the Spp1TdT hippocampus (Fig. 2g). Finally, we used correlative light and electron microscopy (CLEM) to target and visualize the ultrastructure and environmental context of SPP1-TdT positive cells in the hippocampus (Fig. 2h, Extended Data Fig. 3e and Supplementary Video 1). CLEM identified SPP1-TdT expressing cells as lysosome-rich PVMs located within the basement membrane of the perivascular space (Fig. 2h and Extended Data Fig. 3e). CLEM and mRNA in situ localization identified PVMs as a source for SPP1 in mice, which translated to human tissue, where we found enrichment of SPP1 within the perivascular space of AD patients, occasionally overlaying with CD206+ cells (Fig. 2i). Collectively, our data suggest that SPP1 is predominantly expressed by CD206+Cd163+Pf4+ PVMs and Pdgfra+/CD140+ PVFs in the mouse hippocampus during onset of microglia–synapse engulfment. Similar to perivascular SPP1 upregulation in mice, we also found perivascular SPP1 expression in CA1 hippocampus of AD patient brains.
Extended Data Fig. 3. Spp1TdT reporter mouse development.
(a) Design of the knock-in allele is shown, with an IRES and TdTomato (TdT) reporter inserted at the stop codon in exon 7 in the mouse Spp1 locus. Top, the mouse locus with locations of primers used for validation of the inserted construct and later genotyping for the reporter construct is shown. Below, schematic displays the upstream (5’HA) and downstream (3’HA) homology arms in blue; IRES in aqua; TdT reporter gene in red; and bovine growth hormone poly(A) region in yellow. (b) PCR primers used to confirm incorporation of construct are listed, with primer names based on genomic locations as shown in B. Gel shows appropriate size bands for incorporation of plasmid construct (lane 1), for left arm integration event (lane 2) and right arm integration event (lane 3). Primer pairs for plasmid backbone alone are negative (lanes 4 and 5). (c) Representative confocal image showing SPP1-TdT expression along GLUT1+ vessels in hippocampus of 3 mo Spp1TdT reporter mice. (d) Left panels show low magnification overview of SPP1-TdT+ cell (red), and all nuclei (blue) in the hippocampus, as shown in Fig. 2h, scale bar 50 µm. Top, maximum intensity projection of confocal stack; middle, maximum intensity projection shown at reduced opacity over the correlated back scattered electron image of serial section 79 of the same region of the CLEM sample; bottom, back scattered electron image of serial section 79 of the same region of the CLEM sample alone. Box highlights region acquired across 200 serial sections. Right panels show boxed region at higher magnification with serial section 85. Scale bar represents 10 µm. Data representative of n = 4 Spp1TdT mice examined over 2 independent experiments. (e) Two additional CLEM examples of SPP1-TdT+ cells targeted and correlated with volume electron microscopy. Left panels show maximum intensity projection of confocal stack; middle panels, maximum intensity projection shown at reduced opacity over the correlated back scattered electron image of a central serial section of the same region of the CLEM sample; right panels, back scattered electron image of the same region of the CLEM sample alone. Scale bar represent 5 µm. Data representative of n = 4 Spp1TdT mice examined over 2 independent experiments.
SPP1 drives microglial engulfment of synapses in AD context
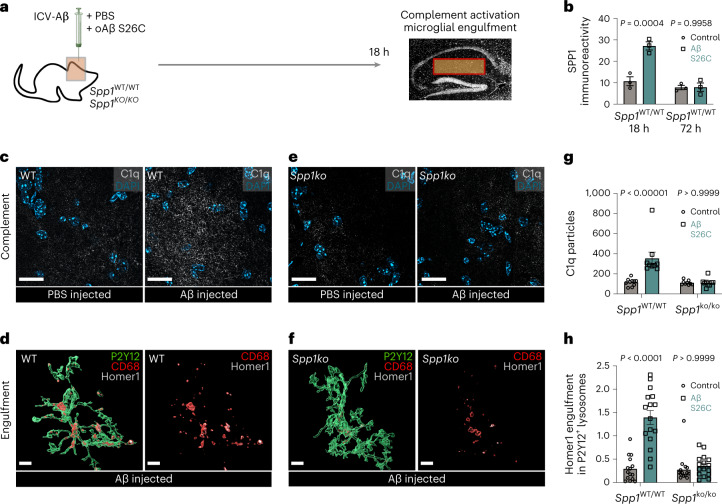
We next addressed the functional consequences of the SPP1 increase that spatiotemporally coincides with onset of microglia–synapse engulfment. To determine whether SPP1 has a role in microglial phagocytosis of synapses, we first performed intracerebroventricular (ICV) injections of oAβ in WT versus Spp1-deficient20 (Spp1KO/KO) mice. As we have previously shown, oAβ triggers microglia- and complement-mediated synapse engulfment15 (Fig. 3a). Concurrently, we observed a robust upregulation of SPP1 in the contralateral hippocampus of WT mice 18 h after ICV injection of oAβ (Fig. 3b). This upregulation coincided with microglial C1q activation and peak engulfment of Homer1-immunoreactive synapses by microglia in contralateral hippocampus of oAβ-injected WT mice (Fig. 3c,d,h). In contrast, oAβ failed to upregulate C1q or induce microglial synaptic engulfment in Spp1KO/KO mice, suggesting that SPP1 is necessary for microglia–synapse engulfment.
Fig. 3. Spp1 modulates complement activation and microglial synaptic engulfment upon acute oAβ challenge.
a, Scheme illustrating ICV injection of S26C oAβ versus PBS in WT versus Spp1KO/KO mice, 18 h before tissue collection and analysis. b, Quantification of SPP1 immunoreactivity within SLM of 3-month WT mice injected with oAβ versus PBS control, at either 18 h or 72 h post-ICV injection. One datapoint represents one ROI per mouse hippocampus, with total of n = 3 mice per genotype and time point, examined over one independent experiment. P values from two-way ANOVA. c, Representative images of C1q expression in SLM of 3-month PBS versus oAβ-injected WT mice. Scale bar represents 20 µm. d, Representative 3D reconstructed images showing Homer1 engulfment within CD68+ lysosomes of P2Y12+ microglia from WT mice injected with oAβ. Scale bar represents 5 µm. e, Representative images of C1q expression in Spp1KO/KO mice injected with PBS versus oAβ. f, Representative 3D reconstructed images showing Homer1 engulfment within CD68+ lysosomes of microglia from Spp1KO/KO mice injected with oAβ. Scale bar represents 5 µm. g, Quantification of C1q particles (puncta) in WT or Spp1KO/KO mice treated with either PBS or oAβ, as in c and e. One datapoint represents one ROI, with total of 10 (WT) and 9 (Spp1KO/KO) ROIs pooled from n = 3 mice per genotype, examined over two independent experiments. P values from two-way ANOVA. h, Quantification of Homer1 engulfment in WT or Spp1KO/KO P2Y12+ microglia, ICV treated with either PBS or oAβ, as in d and f. One datapoint represents one individual P2Y12+ microglia with a total of 15–16 cells pooled from n = 2 animals per genotype examined over two independent experiments. P values from two-way ANOVA. Data are shown as mean ± s.e.m.
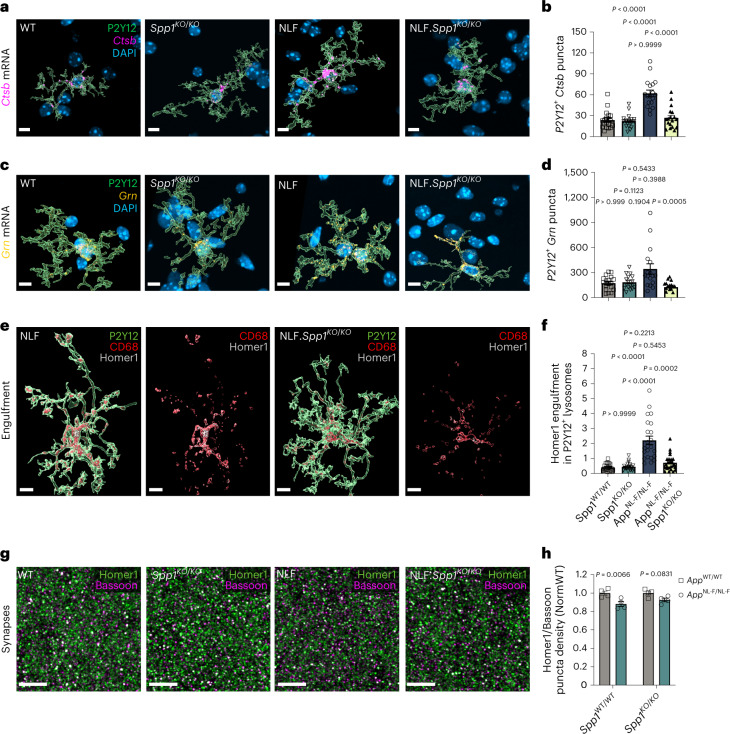
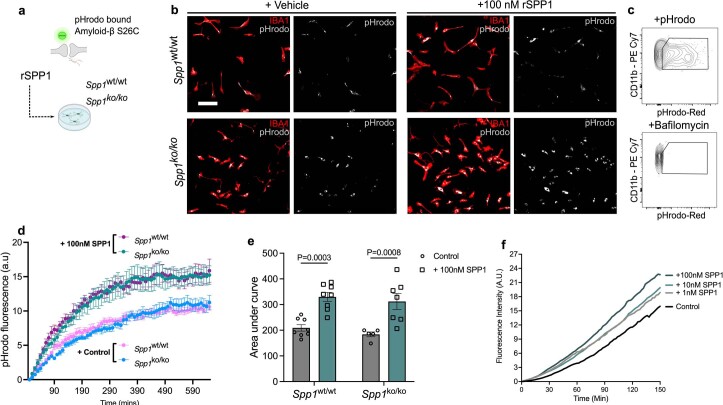
To test whether Spp1 deficiency altered phagocytic signature in microglia in 6-month AppNL-F mice, we crossed the Spp1KO/KO mice to AppNL-F mice (AppNL-F·Spp1KO/KO). Using smFISH-IHC and 3D reconstruction, we observed upregulation of Grn and Ctsb, encoding for progranulin and Cathepsin B, respectively, key components of the endolysosomal processing machinery in P2Y12+ microglia of AppNL-F animals (Fig. 4a–d)14. However, in the AppNL-F·Spp1KO/KO hippocampus, we found substantially decreased levels of Grn and Ctsb mRNA expression by P2Y12+ microglia (Fig. 4a–d). In fact, Grn and Ctsb expression levels were similar to WT and Spp1KO/KO mice. These results suggest that microglia fail to adopt phagocytic states in AppNL-F·Spp1KO/KO mice as compared to those of AppNL-F mice, despite oAβ production. Further, we observed a sharp reduction of engulfed synapses inside P2Y12+ microglia of AppNL-F·Spp1KO/KO mice (Fig. 4e,f). These data altogether suggest that the absence of SPP1 prevents microglial phagocytosis as well as synaptic engulfment in response to amyloidosis (Figs. 3–4). We next tested whether microglia in Spp1KO/KO mice display intrinsic defective phagocytic capacity. We assessed the ability of microglia to engulf synaptosomes in vitro using isolated primary microglia from postnatal WT versus Spp1KO/KO mice (Extended Data Fig. 4a). We found that engulfment of oAβ-bound synaptosomes was similarly performed by microglia isolated from either genotype (Extended Data Fig. 4b–e). This suggests that diminished microglia–synapse engulfment in Spp1KO/KO mice in vivo is not due to an inability of microglia to phagocytose or respond to oAβ, but likely results from defective extrinsic SPP1 signaling in the brain. Corroborating this hypothesis, the application of extracellular SPP1 was sufficient to promote microglia–synapse engulfment in Spp1KO/KO primary microglia, demonstrating similar efficiency in engulfing synaptosomes compared to their WT counterparts upon SPP1 pretreatment (Extended Data Fig. 4d–f).
Fig. 4. SPP1 drives microglial engulfment of synapses in AD context.
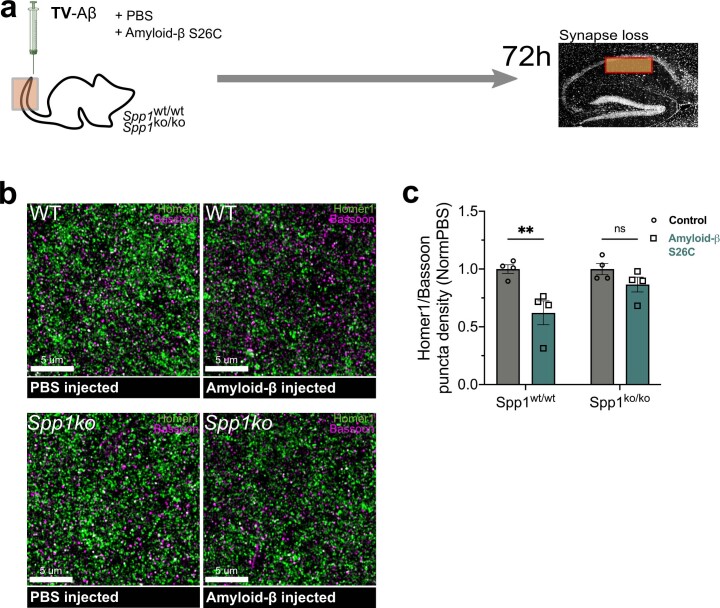
a–d, Representative image of 3D reconstructed P2Y12+ microglia expressing phagocytic markers Ctsb (a,b) and Grn (c,d) assessed by smFISH-IHC in 6-month WT, Spp1KO/KO, AppNL-F and AppNL-F·Spp1KO/KO SLM. Scale bar represents 7 µm. Quantification of Ctsb (b) and Grn (d) mRNA levels expression within P2Y12+ microglia. One datapoint represents one individual P2Y12+ microglia with a total of 18 microglia (b) and 18 (WT, Spp1KO/KO), 17 (AppNL-F) and 15 (AppNL-F·Spp1KO/KO) microglia (d) pooled from n = 3 mice per genotype examined over two independent experiments. P values from one-way ANOVA, Kruskal–Wallis test. e, Representative 3D reconstructed images showing Homer1 engulfment within CD68+ lysosomes of P2Y12+ microglia in 6-month AppNL-F versus AppNL-F·Spp1KO/KO SLM. Scale bar represents 7 µm. f, Quantification of Homer1 engulfment ratio in P2Y12+ microglia of WT versus Spp1KO/KO versus AppNL-F versus AppNL-F·Spp1KO/KO mice. One datapoint represents one individual P2Y12+ microglia with a total of 23 (WT, AppNL-F), 24 (Spp1KO/KO) and 25 (AppNL-F·Spp1KO/KO) microglia pooled from n = 3 mice per genotype examined over two independent experiments. P values from one-way ANOVA, Kruskal–Wallis. g, Representative super-resolution images of Homer1 and Bassoon puncta colocalization in 6-month WT, Spp1KO/KO, AppNL-F and AppNL-F·Spp1KO/KO SLM. Scale bar represents 5 µm. h, Quantification of Homer1/Bassoon colocalization density normalized to WT or Spp1KO/KO accordingly. One datapoint represents the average of one animal (3–5 ROIs per animal) with a total of n = 4 animals per genotype. P < 0.0066 (WT) and 0.0831 (Spp1KO/KO) from two-way ANOVA, Bonferroni’s multiple comparison test. Data are shown as mean ± s.e.m.
Extended Data Fig. 4. Primary microglia engulf synaptosomes through extrinsic SPP1.
(a) Schematic illustrating in vitro engulfment assay whereby primary microglia isolated from WT or Spp1KO/KO mice phagocytose S26C oAβ-treated synaptosomes that are tagged with pHrodo upon PBS or recombinant SPP1 treatment. (b) Representative images showing engulfment of pHrodo-synaptosomes by primary microglia isolated from WT mice (upper panel) or Spp1KO/KO mice (lower panel), treated with vehicle (left) or 100 nM recombinant SPP1 (right). Data are representative of primary microglial cultures prepared from n = 6−8 neonates per genotype, examined over 2 independent experiments. Scale bar represents 100 µm. (c) Representative FACS plot showing pHrodo signal in CD11b+ cells collected from primary microglial culture (upper). No pHrodo signal was observed after Bafilomycin treatment, confirming lysosomal acidification as the source of pHrodo signal (lower). Data are representative of 2 independent experiments. (d) pHrodo fluorescence (arbitary fluorescence units) over 600 min (3–5 min intervals, synaptosomes added at t = 0) in primary microglia isolated from WT mice or Spp1KO/KO mice, treated with control versus 100 nM recombinant SPP1. 1 datapoint represents 2–4 ROIs (approximately 40 microglia per ROI). Primary microglial cultures have been prepared from n = 6−8 neonates per genotype, examined over of 2 independent experiments. Data are shown as Mean ± SEM. (e) Area under curve quantification of pHrodo-synaptosome engulfment within primary WT versus Spp1KO/KO microglia (d), treated with control versus 100 nM recombinant SPP1. 1 datapoint represents area under the curve (AUC) at 3 h of 2–4 ROIs (approximately 40 microglia per ROI). Primary microglial cultures have been prepared from n = 6−8 neonates per genotype, examined over of 2 independent experiments. P Values from two way ANOVA, Bonferroni’s multiple comparison test. Data are shown as Mean ± SEM. (f) Graph showing fluorescence intensity of pHrodo within primary WT microglial lysosomes over time (min), either in the presence of control or 1, 10 and 100 nM recombinant SPP1. Representative of 2 independent experiments.
Finally, to determine the consequence of Spp1 deficiency on synapse numbers in 6 mo AppNL-F animals, we used super-resolution microscopy to analyze presynaptic and postsynaptic markers (Bassoon and Homer1, respectively) in the hippocampal CA1 stratum radiatum of 6 mo AppNL-F mice (Fig. 4g,h). While we observed synaptic loss in colocalized Bassoon and Homer1-immunoreactive synaptic puncta density in AppNL-F versus WT mice, Spp1 deficiency restored this synapse loss (Fig. 4h). Likewise, we observed that acute oAβ challenge failed to induce synapse loss in Spp1KO/KO mice as it did in WT mice, suggesting that SPP1 is required for oAβ-induced synapse loss (Extended Data Fig. 5). Altogether, these results suggest that SPP1, likely secreted from PVMs in vivo in response to oAβ vasculature deposition, acts as an extrinsic signal to promote microglial engulfment of synapses and synapse loss in 6-month AppNL-F animals.
Extended Data Fig. 5. SPP1 deficiency prevents synapse loss upon oAβ challenge.
(a) Scheme illustrating tail vein (TV) injection of 100ng S26C oAβ versus PBS in WT versus Spp1KO/KO mice, 72 h before tissue collection and analysis. (b) Representative super-resolution images of Homer1 and Bassoon puncta colocalization in 3 mo WT and Spp1KO/KO SR injected with oAβ versus PBS control, at 72 h post-TV injection. Scale bar represents 5 µm. (c) Quantification of Homer1/Bassoon colocalization density, represented as S26C oAβ-injected mice normalized to PBS injected mice. 1 datapoint represents the average of 1 animal (3–5 ROIs per animal, 40 cells per ROI) with a total of n = 4 mice per genotype. Data are from one experiment. P Values from two-way ANOVA, Bonferroni’s multiple comparison test. Data are shown as Mean ± SEM.
Spp1 regulates perivascular–microglial interaction networks
To obtain insight into how SPP1 may regulate interactions between perivascular cells, that is PVM and PVF, and parenchymal microglia in the AppNL-F hippocampus, we dissected hippocampi from 6-month WT, Spp1KO/KO, AppNL-F, and AppNL-F·Spp1KO/KO animals and sorted CD45highCD11intCD206+ PVMs, CD140a+ PVFs and CD45intCD11intCX3CR1high microglia for scRNA-seq analysis using the 10× Genomics platform. After quality control, we ran unsupervised clustering and annotated cell types based on expression of known marker genes (Extended Data Fig. 6a,b and Supplementary Table 2). Microglia expressed exclusive pan-markers such as Sall1, Tmem119 and P2ry12; PVMs showed enrichment in Mrc1, Pf4 and Cd163 (Extended Data Fig. 6b)11. Of note, the sorted CD140a+ cells also included oligodendrocyte precursors (OPCs), which were discerned according to expression of OPC-specific markers including Lhfpl3, Sox6 and Bcan (Extended Data Fig. 6b). Further, PVFs were positive for Cdh5, Lama1 and Dcn that distinguished them from OPC. Interestingly, we did not observe Spp1 mRNA expression levels in the sorted PVMs (Extended Data Fig. 2d). This conforms with previous scRNA-seq datasets from homogenized and isolated tissues35; however, translatome analysis of PVMs of brains of lipopolysaccharide (LPS) endotoxin challenged Cx3cr1ccre:Lyve1ncre:RiboTag mice have demonstrated the presence of Spp1 in Lyve1+ PVMs (Extended Data Fig. 6c)11,37. Indeed, comparative RiboTag analysis of challenged split-Cre animals that target PVMs and microglia, respectively11, confirmed prominent Spp1 expression in PVMs as compared to microglia (Extended Data Fig. 6d). The discrepancy of detected expression levels between scRNA-seq of isolated cells versus the RiboTag approach, which bypasses cell isolation and sorting38,39, and the highly sensitive smFISH methods suggest that Spp1 signature within PVMs is highly regulated and closely associated to changes in its microenvironment, as earlier demonstrated for many other microglial transcripts40. It further suggests that Spp1+ PVMs likely do not survive isolation and highlights the relevance of studying SPP1 in an intact spatial context.
Extended Data Fig. 6. scRNA-seq and translatome analysis of perivascular cells and microglia.
(a) UMAP plot of 4,238 cells, color-coded based on the inferred cell type. (b) DotPlot of gene expression for a series of known cell type markers for fibroblasts (Lama1, Lum, Cdh5, Dcn), microglia (Sall1, Tmem119, P2ry12), PVMs (Pf4, Cd163), and OPC (Lhfpl3, Sox6, Bcan). Expression of genes encoding for markers used for FACS of cells prior to scRNA-seq (Pdgfra, Ptprc, Itgam, Mrc1, Cx3cr1) and for Spp1 is also included. Radius of dot is proportional to the percentage of cells expressing the gene, color is the scaled gene expression level. (c) Violin plot of Spp1 expression reanalyzed from Jung-Seok Kim et al.11. (d) RiboTag-based translatome analysis of pooled oAβ and PBS-challenged brain homogenates of Cx3cr1ccre:Lyve1ncre (n = 8 mice) and Cx3cr1ccreSall1ncre:RiboTag-mice (n = 4 mice). Normalized reads of pan macrophage marker (Cx3cr1), PVM marker (Lyve1), microglia marker (Sall1) and Spp1. P Values from two-way ANOVA, Bonferroni’s multiple comparison test. Data are shown as Mean ± SEM. Data are from 1 independent experiment. (e) Gating strategy to identify CD321 (F11r) and CD29 (Itgb1) expression in microglia (CX3CR1highCD45+ CD206- CD11b+). (f) Representative image of P2Y12+ microglia expressing tgfbr1, assessed by smFISH-IHC in 6 mo AppNL-F and AppNL-F.Spp1KO/KO SLM. Scale bar represents 10 µm. Data representative of n = 3 mice per genotype examined over 1 independent experiment.
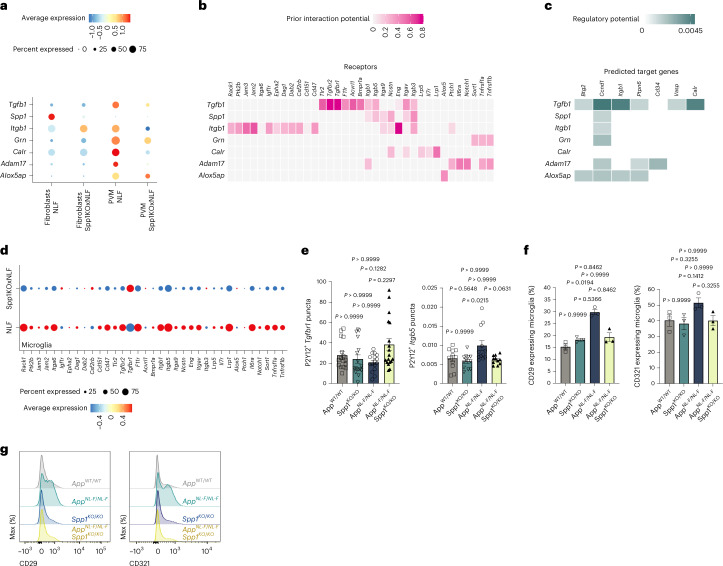
Next, we used NicheNet on the scRNA-seq datasets to predict ligand–target links between interacting cells28. Specifically, we applied NicheNet to investigate how intercellular communication between PVM/PVF (ligand) and microglia (target) is altered by Spp1 deficiency in 6-month AppNL-F mice (Fig. 5). We hypothesized SPP1 to function through paracrine and/or autocrine mechanisms, that is, modulating microglial function through direct or indirect signaling within the perivascular space. Among top predicted ligands affected in PVMs and PVFs by Spp1 deficiency in 6-month AppNL-F hippocampus is transforming growth factor-beta 1 (TGF-β1), a cytokine critical for microglial development and previously shown to be modulated by SPP1 within fibroblasts41,42 (Fig. 5a–c). Other ligands include A Disintegrin and Metallopeptidase Domain 17 (ADAM17) and calreticulin, the latter being reported as an ‘eat-me’ signal driving macrophage phagocytosis of apoptotic cells43. Further, expression of Itgb5, Itgb1 and Itgav, encoding integrin receptors and subunits of the SPP1 receptor, were affected in microglia of AppNL-F·Spp1KO/KO versus AppNL-F mice, as expected44. Other top affected receptors in microglia were Tgfbr1, Tgfbr2 and protein tyrosine phosphatase Ptpn6 (Fig. 5b,c)45,46. We further validated these results via smFISH-IHC; as suggested by NicheNet, Tgfbr1 and Itgb5 expression levels were dysregulated in microglia of AppNL-F·Spp1KO/KO versus AppNL-F mice (Fig. 5e and Extended Data Fig. 6e,f). In addition, we confirmed decreased expression of CD29 (Itgb1) and CD321 (F11r) in hippocampal microglia of AppNL-F·Spp1KO/KO compared to AppNL-F animals by flow cytometry (Fig. 5f,g and Extended Data Fig. 6f). Altogether, NicheNet revealed evidence for paracrine and autocrine crosstalk signals between the perivascular space and microglia in AppNL-F mice in the context of SPP1 signaling, with the majority of the top hits implicating potential alterations of microglial phagocytic states.
Fig. 5. Spp1 regulates perivascular–microglial interaction networks.
a, Expression level of selected ligands expressed by cell types known to express Spp1 (PVF and PVM), by cell type and genotype. Radius of dot is proportional to the percentage of cells expressing the gene; color is the scaled gene expression level. b, Predicted receptor genes for ligands represented in a, which show differential expression in microglia (receiver cells). Color represents the predicted interaction potential. c, Predicted target genes downstream of receptors identified in b, which show differential expression in microglia (receiver cells). Color represents the predicted regulatory potential. d, Expression of predicted receptor genes in microglia, by genotype. Radius of dot is proportional to the percentage of cells expressing the gene; color is the scaled gene expression level. e, Quantification of Tgfbr1 and Itgb5 mRNA levels expressed by P2Y12+ microglia assessed by smFISH-IHC in 6-month WT, Spp1KO/KO, AppNL-F and AppNL-F·Spp1KO/KO SLM. One datapoint represents one individual P2Y12+ microglia with a total of 11 (WT, AppNL-F·Spp1KO/KO) and 12 (Spp1KO/KO, AppNL-F) microglia (Itgb5) or 16 (WT, AppNL-F), 17 (Spp1KO/KO) and 18 (AppNL-F·Spp1KO/KO) microglia (Tgfbr1) pooled from n = 3 animals per genotype examined over one independent experiment. P values from one-way ANOVA, Kruskal–Wallis test with Dunn’s multiple comparisons test. f, Quantification of NicheNet hits CD29 (Itgb1) and CD321 (F11r) on microglia (CX3CR1highCD45+CD11b+ CD206-) isolated from hippocampal homogenates of 6-month WT, Spp1KO/KO, AppNL-F and AppNL-F·Spp1KO/KO animals. One datapoint represents one individual mouse (microglia) pooled from n = 3 mice from one experiment. P values from one-way ANOVA, Kruskal–Wallis test with Dunn’s multiple comparisons test. g, Flow cytometry profiles of the protein expression intensity of CD29 and CD321 on microglia of the four genotypes. Data are shown as mean ± s.e.m. See also Extended Data Fig. 6 and Supplementary Table 2.
Discussion
Microglia–synapse engulfment has been demonstrated to be relevant to synaptic loss and dysfunction in a wide variety of neurologic diseases across the lifespan, including neurodevelopmental and neuropsychiatric disorders, acute injury and virus-induced cognitive impairment, and multiple sclerosis and neurodegeneration15,29,47–56. Genetic studies in sporadic AD further highlight the importance of understanding microglial phagocytosis14. Therefore, our study demonstrating a functional role for SPP1 in microglia–synapse phagocytosis will likely have broad relevance to understanding neuroimmune mechanisms of synapse vulnerability in AD and other neurologic diseases involving synaptopathy.
Here we report a role for perivascular SPP1 as an upstream mediator of microglia–synapse engulfment in both genetic (AppNL-F) and acute oAβ challenge mouse models. SPP1 is a multifunctional glycoprotein that was originally identified as a proinflammatory cytokine secreted by T cells and later found to be expressed in distinct tissue-resident macrophages linked with active clearance of apoptotic cells, chemotaxis and macrophage migration18,57. In the brain, SPP1 expression appears to be highly regulated in a spatiotemporal and cell type-specific manner, depending on context, age and brain region. In the perinatal and prenatal brain, Spp1 is expressed by microglia and associates with axon tracts of the corpus callosum, in contrast to the adult brain where SPP1 expression is thought to be restricted to glutaminergic and GABAergic neurons in adult hindbrain11,37,58–60. Using various imaging tools including smFISH-IHC and CLEM in Spp1TdT mice, we demonstrate that SPP1 expression in the hippocampus of healthy adult WT animals is restricted to the perivascular space, that is, Pf4, CD163+ and CD206+ PVM and to a much lesser extent, the closely neighboring PVFs. The finding that PVMs act as a major source of SPP1 upon oAβ challenge is consistent with the enrichment of ribosome-attached Spp1 transcripts in Lyve1+ PVMs compared to Sall1+ microglia and is consistent with our earlier study that shows Spp1 upregulation in Lyve1+ PVMs upon LPS challenge in adult mice11. Further, we found PVMs as the predominant cellular origin for SPP1 activation in early App models coinciding with Aβ aggregation along the vasculature. Of note, a recent study demonstrated early Spp1 upregulation in spinal cord PVFs in a mouse model of amyotrophic lateral sclerosis, preceding microglial activation and onset of motor neuron loss61. This suggests that SPP1 activation might represent a conserved molecular response to perturbed perivascular homeostasis, potentially beyond Aβ pathology.
We further recapitulated the perivascular localization of SPP1 in postmortem human brains of AD patients. These results are in line with a recent single-nucleus transcriptomic study in vessel-associated structures of the human cortex that found SPP1 expression in perivascular myeloid cells62. Our study thus raises an intriguing question of what could possibly trigger SPP1 elevation within the perivascular space. In AD patients, SPP1 levels are elevated in CSF21–23, which flows via the perivascular space toward the subarachnoid space. The cellular origin or the functional relevance of SPP1 increase, however, has been unclear. PVMs are distributed along the perivascular space and may have a critical role as sentinels in amyloidosis (our data here and ref. 63). Clearance of Aβ across the BBB represents an important homeostatic function, the impairment of which has been linked to exacerbated vascular as well as parenchymal deposition of Aβ64,65. Blood vessels therefore may represent a frequent, early and vulnerable site for Aβ deposition. In support of this, vascular pathology is commonly observed in AD, afflicting approximately 80% of patients66. Our findings of oAβ deposition along the vasculature in early stages of amyloidosis in AppNL-F mice further support this view. Indeed, it was postulated that early failure of perivascular drainage could contribute to parenchymal Aβ deposition and associated neuronal toxicity67. Furthermore, SPP1 has been linked to the clearance of vascular Aβ deposits in mice68. Altogether, these and our studies highlight Aβ deposition along the vasculature as a possible trigger for SPP1 upregulation in PVMs and PVFs. Future studies allowing cell-type specific fate mapping and mutagenesis of SPP1 in hippocampal PVM and PVF subsets will be needed to provide insight into the origin and fate of SPP1-expressing perivascular subsets.
Our observation that SPP1 signaling promotes synaptic engulfment by microglia in the hippocampus of AD models suggests that SPP1 functions to regulate phagocytic cell states. Indeed, in the absence of Spp1, microglia fail to upregulate key phagocytic and AD-relevant genes such as progranulin and Ctsb in 6-month AppNL-F mice. Of note, phagocytosis induced by microglial progranulin as well as complement proteins including C3 has been considered protective against Aβ plaque load and neuronal loss in plaque-rich mouse models of AD. In line with our data, this suggests that the timing of phagocytic activity in different stages of disease might be critical to determine the protective versus detrimental nature of the outcome15,69,70. Another interesting question raised is how SPP1 modulates microglial phagocytosis in the presence of Aβ pathology. SPP1 functions as a secreted or intracellular isoform71. Based on our data, a direct effect of intracellular SPP1 on microglia function seems unlikely in our experimental setting. First, we found that microglia-intrinsic Spp1 signaling is not required for the engulfment of Aβ-treated synaptosomes, where primary microglia derived from WT and Spp1KO/KO mice exhibit similar engulfment capacities of synaptosomes in the presence of extracellular SPP1. This is consistent with work in primary gut macrophages from Spp1-deficient mice, whereby low doses of recombinant SPP1 restored phagocytic function toward opsonized bacteria, akin to what we observed for primary microglia and synaptosomes72. Second, we did not observe mRNA or protein SPP1 expression localized inside hippocampal microglia at the age of 6 months, yet the same cells fail to engulf synapses in AppNL-F·Spp1KO/KO mice or Spp1KO/KO mice challenged with synaptotoxic Aβ. Third, we found increased protein levels of SPP1-immunoreactive puncta within the SLM parenchyma using super-resolution imaging, suggesting that the observed punctate signals are mostly extracellular. Altogether, these results suggest that SPP1 is secreted by PVMs and PVFs and influences microglia phagocytosis.
Precise mechanisms of how microglial engulfment is initiated by SPP1 are to be investigated. SPP1 could opsonize neuronal material for example damaged synapses for microglial phagocytosis73. It could also engage directly with the canonical receptors, αvβ3, which are expressed on microglia37, to trigger downstream signaling pathways. Indeed, using NicheNet, we observed Itgb3 and Itgav, which encode αvβ3, to be downregulated in microglia of AppNL-F·Spp1KO/KO mice. Our NicheNet analysis further shows reduced expression levels of both Tgfb1 in PVMs and its receptor Tgfbr2 in microglia from AppNL-F·Spp1KO/KO mice, suggesting that SPP1 potentially coordinates perivascular–microglial crosstalk via autocrine TGF-β signaling. Indeed, SPP1 has been identified to promote autocrine TGF-β signaling in fibroblasts in a mouse model of muscular dystrophy, thereby promoting fibrosis by neighboring fibroblasts and macrophages41. Further, TGF-β is a major determinant of microglia maturation and homeostasis, and Tgfbr2 deficient microglia demonstrate dysregulated expression of transcripts related to phagosome formation and immune activation42,45. In contrast to decreased Tgfbr2, we observed increased Tgfbr1 signaling in microglia of AppNL-F·Spp1KO/KO versus AppNL-F animals. It is tempting to speculate that this paradoxical down- and upregulation reflects compensatory mechanisms to counteract microglia phagocytosis induced by SPP1. Finally, TGF-β1 has been shown to be increased in CSF and perivascular space of AD patients, where its levels positively correlate with Aβ deposition along blood vessels, and TGF-β1-overexpressing mice show enhanced microglial phagocytosis and plaque amelioration in a complement-dependent manner74,75. Further studies are needed to gain better insight into the role of TGF-β signaling in governing autocrine and paracrine PVM-microglial crosstalk in AD. Other predicted Spp1-specific pathways in microglia of AppNL-F mice include Calr, encoding for calreticulin, a multifunctional chaperone protein that has been described as an ‘eat-me’ signal43. Altogether, our results suggest multiple mechanisms by which microglial phagocytosis could be modulated by perivascular SPP1 signaling. Determining molecular mechanisms of how these pathways may contribute to synaptic engulfment will be important to better understand multicellular events and crosstalk between perivascular space and the brain parenchyma.
Altogether, our study nominates perivascular SPP1 as a modulator for microglial phagocytosis, impacting microglia–synapse interactions and synaptic homeostasis. In peripheral tissues, insight into stroma–macrophage interactions demonstrates intricate and functionally relevant immune crosstalk that underlies critical tissue remodeling and homeostasis76. Likewise, our results highlight a potential role for perivascular–microglia interactions in the brain, and the presence of an SPP1-producing perivascular niche may offer opportunities to specifically manipulate and block microglia-mediated synaptic engulfment.
Methods
Mice
All experiments were performed in accordance with the UK Animal (Scientific Procedures) Act, 1986 and following local ethical advice. Experimental procedures were approved by the UK Home Office and ethical approval was granted through consultation with veterinary staff at University College London (UCL).
For all experiments, C57BL/6J (WT) mice were obtained from Charles River UK. Spp1KO/KO (B6.129S6(Cg)-Spp1tm1Blh/J; stock 4936) and CX3CR-1GFP (B6.129P2(Cg)-Cx3cr1tm1Litt/J; stock 5582) mice were obtained from The Jackson Laboratory. AppNL-F/NL-F mice were obtained from F. A. Edwards (Department of Neuroscience, Physiology & Pharmacology, UCL, UK)30. Cx3cr1ccre:Sall1ncre (split-Cre) R26-LSL-tdTom:Rpl22HA (RiboTag) and Cx3cr1ccre:Lyve1ncreR26-LSL-tdTom:Rpl22HA homozygous animals were maintained in specific pathogen-free conditions and handled according to protocols approved by the Weizmann Institute Animal Care Committee as per international guidelines. All animals were housed under temperature-controlled (temperature, 23.1 °C; humidity, 30–60%) and pathogen-free conditions with 12 h light/12 h dark cycle with an ad libitum supply of food and water. Both male and female age-matched mice were used in this study: WT males and females, Spp1KO/KO males, CX3CR-1GFP males, Spp1tm1(tdTomato)Msasn males, AppNL-F/NL-F males and AppNL-F·Spp1KO/KO males.
Spp1 reporter mouse model development
All animal work was approved by the Jackson Laboratory Animal Care and Use Committee and adhered to the standards of the Guide for the Care and Use of Laboratory Animals set forth by the NIH. The Spp1tm1(tdTomato)Msasn mouse allele was generated using direct delivery of CRISPR–Cas9 reagents to mouse zygotes. An IRES-tdT construct was introduced in the mouse Spp1 gene (Ensembl Gene UID, ENSMUSG00000029304; Extended Data Fig. 3a). Analysis of genomic DNA sequence surrounding the target region, using the Benchling (www.benchling.com) guide RNA (gRNA) design tool, identified a gRNA sequence (AACAAGAAAAAGTGTTAGTG) with a suitable target endonuclease site at the stop codon of exon 7 of the mouse Spp1 locus. Streptococcus pyogenes Cas9 (SpCas9) V3 protein and gRNA were purchased as part of the Alt-R CRISPR–Cas9 system using the crRNA:tracrRNA duplex format as the gRNA species (IDT). Alt-R CRISPR–Cas9 crRNAs (1072532, IDT) were synthesized using the gRNA sequences specified in the DESIGN section and hybridized with the Alt-R tracrRNA (1072534, IDT) as per the manufacturer’s instructions. A plasmid construct with a 2 kB 5′ homology arm ending 62 bp past the Spp1 stop codon in exon 7, IRES, tdT coding sequence, bGH poly(A) signal and 1.5 kB 3′ homology arm was synthesized by Genscript (Extended Data Fig. 3b). To prepare the gene editing reagent for electroporation, SpCas9:gRNA ribonucleoprotein complexes were formed by incubating Alt-R–SpCas9 V3 (1081059, IDT) and gRNA duplexes for 20 min at room temperature in embryo tested Tris–EDTA (TE) buffer (pH 7.5). Fertilized mouse embryos were generated via natural mating and cultured as described previously77. C57BL/6J (Stock 000664, The Jackson Laboratory) donor female mice (3–4 weeks of age) were superovulated by administration of 5 IU of pregnant mare serum gonadotrophin via intraperitoneal (ip) injection (HOR-272, ProSpec) followed 47 h later by 5 IU (ip) human chorionic gonadotrophin (hCG) (HOR-250, ProSpec). Immediately postadministration of hCG, the female was mated 1:1 with a C57BL/6J stud male and 22 h later checked for the presence of a copulation plug. Female mice displaying a copulation plug were sacrificed, the oviducts excised and embryos collected. Electroporation was performed as described in ref. 77. In brief, zygotes were treated with the acidic Tyrode’s solution (T1788, Millapore-Sigma) for 10 s and washed extensively in prewarmed M2 media (M7167, Millapore-Sigma). Zygotes were then placed in 10 μl drops of Opti-MEM media (3198570, Thermo Fisher Scientific–Gibco). Ten microliters of gene editing reagent solution, including the SpCas9/gRNA and the ssDO, was mixed with the Opti-MEM drops with the embryos and deposited into a 1 mm electroporation cuvette (45-0124, Harvard Apparatus). Electroporation was performed in an ECM830 Square Wave Electroporation System (45-0661, BTX). The electroporation setting was a 1 ms pulse duration and two pulses with 100 ms pulse interval at 30 V. Following the electroporation, a prewarmed 100 μl aliquot of M2 media was deposited into the cuvette with a sterile plastic pipette to recover the embryos. The zygotes were removed from the cuvette and washed in prewarmed M2 media. Embryos were immediately transferred into B6Qsi5F1 pseudopregnant female mice, an F1 hybrid strain produced by breeding C57BL/6J female mice with the inbred Quackenbush Swiss line 5 mouse strain78.
Founders were first assayed by short-range PCR with primer SR-FF 5′-TAATAATGGTGAGCAAGGGCGA-3′ and reverse primer SR-RR 5′-CTTTGATGACGGCCATGTTGTT-3′ within the reporter construct (see schematic). Positive founders were then screened by PCR: across the 5′ homology arm with primer 5′LR_F 5′-GAAAGTGCCTACTCGTGCCT-3′ and reverse primer 5′LR_R: CACATTGCCAAAAGACGGCA-3′; across the 3′ homology arm with primers 8431 GCATCGCATTGTCTGAGTAGGT and 3′LR_R: ccatcatggctttgcatgac; and for the presence of plasmid backbone with primers 8431: GCATCGCATTGTCTGAGTAGGT and 9581: AGCGCAACGCAATTAATGTG. Sanger sequencing was performed across the homology arm junctions and portions of the knocked-in reporter gene (Extended Data Fig. 3c).
Founders were selected that were positive by short-range PCR assays, had appropriate sequence across the homology arm junctions, were negative for the plasmid backbone and had correct sequence of the inserted construct. These were bred to C57BL/6J.
Once the line was established, mice were genotyped using a Taqman qPCR protocol run on a real-time PCR instrument (Roche LightCycler480). Forward primers for the wild-type allele (AAACACAGTTCCTTACTTTGCAT producing a 94 bp product) and the mutant allele (AGGATTGGGAAGACAATAGCA in the bGH poly A producing an 84 bp product) were combined with a common reverse primer (CACTGAACTGAGAAATGAGCAGT) using an annealing temperature of 60 °C. The wild-type probe (5′ HEX fluorophore label) used was TGTTAGTGAGGGTTAAGCAGGAATA and the knock-in probe (5′ FAM fluorophore label) used was ATGCGGTGGGCTCTATGG, each utilizing a black hole quencher on their 3′ end. These were run with an EndPoint protocol, imaging negative (quenched) fluorescent values at the completion of the cycling protocol.
This new line of Spp1-IRES-TdT mice is available as B6J.Spp1tm1(tdTomato)Msasn/J (stock 33731) from The Jackson Laboratory.
ICV injections of S26C oAβ
S26C Aβ(1–40)2 dimers were purchased from Phoenix Pharmaceuticals15. Adult (2–3 months) mice were anesthetized with 4% isoflurane followed by maintenance at 1.5–2% isoflurane during surgery. Surgery was performed after head fixing in a stereotaxic frame (World Precision Instruments). Marcaine (0.025%) was applied locally, the skull was exposed by a single incision along the midline and a unilateral craniotomy was drilled with a 0.9-bit drill burr (Hager and Meisinger). Pulled long-shaft borosilicate pipettes (Drummond Scientific) were backfilled with mineral oil before loading with oAβ (1 ng µl−1) or sterile PBS vehicle. Four microliters total volume was injected into the right lateral ventricle (stereotaxic coordinates in millimeters from Paxinos and Franklin’s The Mouse Brain in Stereotaxic Coordinates, Fourth Edition; AP: −0.40, ML: 1.00, DV: −2.50) using a Nanofil 10 ml syringe (World Precision Instruments) connected to an UltraMicroPump-3 (World Precision Instruments) at a flow rate of 400 nl min−1. The pipette was left in place for 5 min after complete substance injection and slowly withdrawn to avoid backflow along the pipette track. The incision on the scalp was closed with Vetbond tissue adhesive (3 M). Subcutaneous carprofen (Carprieve, 5 mg g−1 body weight) and buprenorphine (Vetergesic, 0.1 mg g−1 body weight) diluted in 0.9% saline were administered perioperatively. Animals received carprofen (33.33 mg ml−1) in their drinking water until tissue collecting. The left hemisphere, contralateral to the injection site, was analyzed.
Immunohistochemistry
Deeply anesthetized mice were transcardially perfused with 25–30 ml of filtered PBS, followed by 20 ml of 4% ice-cold ultrapure PFA (Generon, 18814-20). Brains were removed from the skull and fixed in 4% PFA (Generon) for 24 h at 4 °C. For cryoprotection, brains were rinsed in PBS for at least 2 h to remove excess PFA and placed in 30% sucrose for 48 h before embedding in optimal cutting compound. Fresh cryosections were blocked with 5% BSA, 0.2% Triton X-100 and 5% Donkey serum in PBS at room temperature for 90 min before incubation with primary antibodies at 4 °C overnight. All primary antibodies were diluted in a blocking buffer. After 4X PBS washes for 15 min each, sections were incubated with secondary antibodies in blocking buffer for 2 h at room temperature and then washed again with PBS. All secondary antibody aliquots were centrifugated at 15,000g for 15 min before being diluted in blocking buffer. The sections were then incubated for 5 min in DAPI diluted in PBS before being mounted either in Prolong Gold Antifade Mounting Medium (Thermo Fisher Scientific, P36930) for confocal imaging or in Prolong Glass Antifade Mountant (P36982) for STED imaging.
For immunostaining of C1q and synapses, 30 µm free-floating tissue sections were washed in PBS, followed by pretreatment in 1% Triton X-100 for 20. Sections were then blocked in 20% NGS, 1% BSA and 0.3% Triton, in PBS for 2 h followed by standard primary antibody incubation overnight at 4 °C. Sections are washed in 0.3% Triton X-100 in PBS for 30 min followed by secondary incubation for 4 h at room temperature and wash for 30 min before mounting.
Postmortem human brain tissue
Brains were donated to the Queen Square Brain Bank (QSBB) for neurological disorders (UCL Queen Square Institute of Neurology). All tissue samples were donated with full, informed consent. Accompanying clinical and demographic data of all cases used in this study were stored electronically in compliance with the 1998 data protection act and are summarized in Supplementary Table 1. Ethical approval for the study was obtained from the NHS research ethics committee and in accordance with the human tissue authority’s code of practice and standards under license number 12,198, with an approved material transfer agreement. The cohort included pathologically diagnosed cases of AD (n = 6) and neurologically normal controls (n = 6). The level of AD pathology in all cases was assessed using current diagnostic consensus criteria79,80. The APOE genotype was also determined for each case as previously described81. Information regarding sex is included in Supplementary Table 1; however, sex was not taken into consideration when including patient samples. Findings did not apply to only one sex .
Immunohistochemistry on postmortem human brain
Slides with 8 µm mounted tissue sections from the frontal cortex were incubated at 60 °C overnight. Sections were deparaffinized in xylene and rehydrated in decreasing grades of alcohol. Slides were incubated in methanol/hydrogen peroxide (0.3%) solution for 10 min to block endogenous peroxidase activity. For heat-induced antigen retrieval, slides were then transferred to a boiling solution of 0.1 M citrate buffer (pH 6.0) and pressure cooked at maximum pressure for 10 min. Nonspecific binding was blocked by incubating slides in 10% nonfat milk for 30 min at room temperature. Sections were incubated in anti-SPP1 antibody for 1 h at room temperature. After three gentle 5 min washes in tris-buffered saline with tween (TBS-T); slides were incubated for 45 min in biotinylated goat anti-mouse IgG secondary antibody (Vector Laboratories, BA-9200, 1:200). Slides were washed and incubated in avidin–biotin complex (ABC; Vector Laboratories) for signal amplification. The slides were then washed for a final time and 3,3′-diaminobenzidine was used as the chromogen and counterstained in Mayer’s hematoxylin (BDH). Finally, slides were dehydrated in increasing grades of alcohol (70%, 90% and 100% industrial methylated spirits), cleared in xylene and mounted.
Double immunofluorescence staining was carried out on the frontal cortex. Sections were prepared as detailed above up to the incubation of the anti-SPP1 antibody, secondary biotinylated goat anti-mouse and ABC. Antibody binding was visualized using a TSA Cyanine 3 amplification kit (PerkinElmer), which was applied to sections for 20 min at room temperature. After TBS-T washing, sections were incubated with anti-CD206 antibody for 1 h at room temperature and species-appropriate Alexa Fluor 658 secondary antibodies (Invitrogen, 1:1,000) for 2 h at room temperature to visualize the antibody. Sections were washed a final three times in TBS-T and mounted using Vectashield antifade mounting medium (Vector Laboratories).
Image acquisition
Images were acquired using a Zeiss LSM800 confocal microscope (×40 objective, 1.3NA oil, ×20 objective 0.8-NA and 63×0.8-NA oil). Settings were kept constant for all sections in the same comparison group. Step size was determined using the optimal interval adjustment on the Zen blue software with a stack size of 10–14 µm for all on-slide IHC experiments and 6–10 µm for RNAScope experiments.
Secreted SPP1 fluorescence intensity analysis
For quantification of secreted SPP1, Triton X-100 was excluded from the blocking buffer to retain signals of secreted and membrane-bound protein immune reactivity. To quantify SPP1 protein expression, images were processed in Fiji ImageJ (NIH)82. An automated ImageJ macro was created to analyze the signal intensity of each slice in the z stack and select the plane with the highest signal intensity. Background was subtracted with a rolling ball radius of 10 pixels. Images were thresholded with consistent thresholding parameters and made into binary images based on intensity. As pixel intensity information has been translated into area, the particle analysis function was used to quantify the total immune-reactive area.
Antibodies
For immunostaining, all the antibodies used were rabbit anti-mouse C1q (Abcam, ab182451; Clone 4.8, 1/200), goat anti-mouse SPP1 (Bio-Techne, AF808; 1/50), rabbit anti-mouse IBA1 (Wako Chemicals, 019-19741; 1/500), rabbit anti-mouse GLUT1 (Merck Millipore, CBL242; 1/10,000), rat anti-mouse CD68 (Bio-Rad, MCA1957; Clone FA-11, 1/200), rabbit anti-mouse P2Y12 (Anaspec, AS-55043A; 1/500), rat anti-mouse CD206 (Bio-Rad, MCA2235; Clone MR5D3, 1/400), NAB61 (kindly provided by Virginia M-Y Lee, 1/500), HJ5.1 (kindly provided by John R. Cirrito, 1/500), chicken anti-mouse Homer1 (Synaptic System, 160 006; 1/500), mouse anti-mouse 6E10 (Biolegend, 803001; Clone 6E10, 1/200), mouse anti-mouse 4G8 (Biolegend, 800702; Clone 4G8, 1/200), rat anti-mouse CD140a (Thermo Fisher Scientific, 14-1401-82; Clone APA5, 1/50), rabbit anti-mouse Bassoon (Synaptic System, 141 003; 1/200) and Lyve1 (Abcam, ab14917; 1/500). Secondary antibodies used were a combination of Alexa Fluor 488, 594 and 647 (1/400, Jackson ImmunoResearch and Thermo Fisher Scientific) chosen from goat anti-rabbit, goat anti-rat, goat anti-mouse, donkey anti-goat, donkey anti-rat, donkey anti-mouse and donkey anti-rabbit secondaries. For flow cytometry, BUV395 CD45 (BD Biosciences, 564279; 1/400), Pe-Cy7 CD11b (BD Biosciences, 552850; 1/400), BV421 CX3CR1 (Biolegend, 149023; 1/400), PE CD140a (Miltenyi, 130-102-473; 1/50), APC CD206 (Biolegend, 141708; 1/400), FITC CD29 (Biolegend, 102205; 1/200) and BV711 CD321 (BD Biosciences, 745405; 1/200) were used.
In vivo microglial engulfment analysis
Engulfment analysis was performed as previously described15. Thirty micrometers of free-floating tissue sections were immunostained with IBA1, CD68 and Homer1. For each mouse, four to six regions of interest within CA1 SLM were acquired using a 63 × 1.4-NA objective on a Zeiss 800 microscope. Next, 60–80 z-stack planes were taken with 0.27-μm spacing and raw images were processed in Imaris (Bitplane) for analysis, after background subtraction of Homer1 channel. P2Y12+ cells and CD68 lysosomes were surface rendered with 0.25-μm and 0.1-μm smoothing, respectively. A mask was applied within P2Y12+ CD68+ reconstructed lysosomes for Homer1, and the percentage engulfment of Homer1 within lysosomes was calculated using the following formula: volume of engulfed material (Homer1 within CD68)/total microglial volume × 100.
Super-resolution imaging and synapse analysis
Super-resolution synapse images were acquired on a Zeiss LSM 880 microscope with Airyscan detector using a 63x, 1.4-NA oil immersion Plan-Apochromat objective (theoretical maximum resolution—140 nm lateral, 350 nm axial). A zoom factor of 1.8× and a 37 × 37 × 144 nm voxel pixel size. The Airyscan detector was aligned before imaging each new slide. Three regions of interest of 1.15 µm were acquired in the center of the hippocampal CA1 stratum radiatum for each brain section. Super-resolution synapse images were processed in Zen Black using 3D Airyscan Processing. Imaris software was used for presynaptic and postsynaptic puncta detection, using intensity centers to maximize the detection of immunoreactive spots. Presynaptic and postsynaptic spots were colocalized using a MATLAB colocalization script (Colocalize Spots XTension), using a colocalization distance of 0.25 μm between spot centers. Synapse density is shown as AppNL-F mice normalized to WT mice, and AppNL-F·Spp1KO/KO normalized to Spp1KO/KO mice.
smFISH (RNAscope) and smFISH combined with IHC (smFISH-IHC)
To detect single RNA molecules, the RNA probes for Spp1 (435191), C1q (441221), Bin1 (529541), Ctsb (561541), Grn (422861), Cd163 (406631), Pdgfra (480661), Pf4 (502391), Tmem119 (472901), Tgfbr1 (406201) and Itgb5 (404311) were purchased from advanced cell diagnostics. RNAscope was performed following the manufacturer’s protocol in RNAscopeFluorescent Multiplex Assay (320293). 15 μm frozen brain sections were collected on Superfrost Plus GOLD Slides (Thermo Fisher Scientific, K5800AMNZ72) and dried overnight at 40 °C. Briefly, slides were incubated in H2O2 for 4 min at room temperature and washed in RNAse-free water. Slides were placed in boiling target antigen retrieval for 4 min, dehydrated in 100% ethanol for 5 min and treated with Protease Plus for 15 min at room temperature before probe incubation. Further protocol as described by the manufacturer, and post-RNAscope immunostaining as standard immunohistochemistry procedure.
RNA isolation, reverse transcription and RT-qPCR
Mice were anesthetized and perfused with ice-cold PBS. Next, fresh cortex and hippocampus were dissected and homogenized with TissueLyser II (QIAGEN) using the mRNA easy mini kit (QIAGEN) as described by the manufacturer. Next, RNA purity and concentration were assessed by Nanodrop. mRNA was converted to cDNA using the qScript cDNA SuperMix reverse transcription kit as described by the manufacturer (95048, Quantabio). For RT-qPCR, 12 ng of cDNA was loaded in triplicates per gene in a total volume of 20 μl using the SYBR green PCR master mix as described by the manufacturer (4309155, Thermo Fisher Scientific). The reaction was run using a LightCycler 96 Instrument (Roche) with white 96-well plates (04729692001, Roche). Triplicate Ct values were averaged and data are shown as respective to the geomean of three housekeeping genes (Actb, Gapdh and Rpl32) using the Ct delta method (2–∆∆Ct). Primers purchased from IDT were used at a concentration of 200 nM. Actb, forward—CATTGCTGACAGGATGCAGAAGG, reverse—TGCTGGAAGGTGGACAGTGAGG; Gapdh, forward—CATCACTGCCACCCAGAAGACTG, reverse—ATGCCAGTGAGCTTCCCGTTCAG; Rpl32, forward—ATCAGGCACCAGTCAGACCGAT, reverse—GTTGCTCCCATAACCGATGTTGG. Spp1, forward—ATC TCACCATTCGGATGAGTCT, reverse—TGTAGGGACGATTGGAGTGAAA
Microglia isolation for flow cytometry and FACS sort
PBS-perfused brains were quickly isolated from the skull and the hippocampus was dissected on ice using chilled instruments. For scRNA sequencing, brains were perfused with inhibitor cocktail including Actinomycin D (5 μg ml−1) and triptolide (10 μM). Next, single-cell suspension was prepared using the Adult Brain Dissociation kit from Miltenyi Biotec (Bergisch Gladbach, Germany), according manufacturer’s instructions and ref. 25. Afterwards, cell suspension was filtered through a 70 μM cell strainer before mixing with debris removal solution. Cells were centrifuged (300g for 10 min), washed with ice-cold FACS buffer (PBS, 2% FBS, 0.78 mM EDTA) and incubated for 30 min at 4 °C with FACS buffer containing Fc block (BD Biosciences) and primary antibody mix. Flow cytometry data were analyzed using FACSDiva software 4.0 and FlowJo 10 software (Treestar).
Primary microglia isolation and culturing
For generation of microglial primary cultures, P0 WT or Spp1KO/KO mice were decapitated, the brain was dissected from the skull and meninges were removed in ice-cold HBSS with 5% FBS. Eight to ten mice were pooled per culture preparation. Tissue was homogenized first with 2 ml pipettes (15 strokes) in 15 ml falcon tube and subsequently transferred to a prewet 50 ml tube with 70 μM strainer. The 15 ml tube was washed with HBSS and then put through filter to ensure all tissues were collected. The supernatant was removed and the cell pellet was resuspended in ice-cold 35% isotonic percoll. The interface was carefully created with HBSS. The samples were centrifuged for 40 min at 4 °C at 2,800g with no break and with slow acceleration and deceleration. The myelin layer and supernatant layers were aspirated, and the cell pellet was washed in HBSS. Cells were centrifuged and resuspended in 1 ml microglial media (DMEM F12 (Gibco), 5% FBS (Gibco), 1% pen-strep (Gibco), 50 ng ml−1 CSF1 416-ML-010/CF (RnD Systems), 50 ng ml−1 TGFb1 7666-MB-005/CF (RnD Systems) and 100 ng ml−1 CX3CL1 472-FF-025/CF (RnD Systems)) for cell counting. Cells were used within 7–10 d of plating.
Isolation of synaptosomes and conjugation to pHrodo
Three to five 8-week-old animals were pooled per experiment. In brief, mice were intracardiac perfused with 10 ml ice-cold PBS. The hippocampi and cortices were dissected on ice. Tissue was weighed and homogenized in five volumes of sucrose homogenization buffer (5 mM HEPES pH 7.4, 320 mM sucrose and 1 mM EDTA) using a Dounce homogenizer with 15–20 strokes. The homogenate was centrifuged at 3,000g for 10 min at 4 °C and the supernatant was saved as total homogenate fraction (THF). The THF was centrifuged again at 14,000g for 12 min at 4 °C and supernatant was saved as cytosolic fraction. The pellet was carefully resuspended in 550 μl of Krebs–Ringer buffer (KRB: 10 mM HEPES, pH 7.4, 140 mM NaCl, 5 mM KCl, 5 mM glucose and 1 mM EDTA) and 450 µl of Percoll solution (for a final concentration of 45%). The solution was mixed by gently inverting the tube and an interface was slowly created with 400 µl of KRB. After centrifugation at 14,000g for 2 min at 4 °C, the synaptosomal fraction was recovered at the surface of the flotation gradient and carefully resuspended in 1 ml of KRB to wash. The functional synaptosomal preparation was centrifuged at 14,000g for 1 min at 4 °C. Fresh mouse synaptosomes were immediately divided into Eppendorfs at 2–2.5 mg of protein and resuspended in total 1 ml KRB in 50 nM of oAβ 40-S26C dimer or PBS as control and left overnight at 4 °C on nutator. Synaptosomes were then centrifuged at 14,000g for 1 min at 4 °C, supernatant was discarded and synaptosomes were washed in 1 ml PBS, after which they were centrifuged at 14,000g for 1 min at 4 °C to obtain oAβ-synaptosomes and control-synaptosomes. Briefly, 1 mg of synaptosomes were left at room temperature on nutator for 2 h in sodium bicarbonate 0.1 M with pHrodo Red, succinimidyl ester at a concentration of 1 mg ml−1. After conjugation, synaptosomes were centrifuged at 14,000g for 1 min and resuspended for use in the in vitro synaptosome engulfment assay.
In vitro synaptosome engulfment assay
Primary mouse microglia were treated with 1 µg of pHrodo-conjugated S26C oAβ-treated synaptosomes. Plates were then placed in a CD7 with the incubator at 37 °C and 5% CO2. Fluorescent (594 nm and 647 nm) and brightfield (oblique and phase) images were acquired at a ×20 objective (×0.5) at intervals of 3–5 min. A three-slice z stack was taken at a 1.5-μm interval to ensure that imaging was within focus throughout the imaging session; however, one plane was used for analysis. For analysis, the z-profile axis was plotted for respective pHrodos on ImageJ with respect to time. Fluorescence intensity at t = 0 was subtracted from subsequent time frames.
3D-τ-STED/STED-FLIM
For visualization of secreted SPP1 in mouse and human tissue, tissue sections were imaged with Leica STELLARIS 8 STED microscope using the ×100 objective (1.4 NA oil) (Leica Microsystems). Tissue sections were imaged at least 24 h after being coverslipped with mounting medium to avoid discrepancies in fluorescence lifetime within each section. STED microscope with Fluorescence Lifetime Imaging (STED-FLIM) was used to visualize secreted SPP1 in the extracellular space and fluorescence lifetime information was used to gate fluorescence signals. Alignment between STED laser and excitation laser was performed before each imaging session. A 775 nm STED laser was used to generate a doughnut beam to silence the peripheral fluorophores from Alexa Flour 647 photoexcitation to achieve subdiffractional resolution. For all STED images, pixel size was limited to at most 50 nm. STED laser intensity was set at 20%. All images were taken with a step size of 0.15 μm. Fluorescence signal was time-gated from −0.5 ms to 4.5 ms. For analysis, raw images were processed by thresholding, followed by binary image transformation to match ROI with intensity. Finally, fluorescence intensity was measured by the particle analysis function in ImageJ (NIH).
Volume correlative light and electron microscopy with array tomography
Mice were perfusion fixed with 4% PFA (EM grade) in ice-cold PBS as described above, the brain was dissected and left in 4% PFA in PBS overnight at 4 °C. The following day, coronal vibratome slices (100 μm) of the brain were collected and slices containing clear cross sections of the hippocampus were manually trimmed to minimally contain the hippocampus and ensure that the tissue piece was asymmetric. Low-resolution maps of the entire hippocampus tissue piece were taken using a confocal Zeiss LSM800 and ×10 lens with montaging for gross mapping and identification of regions of interest. High-resolution (63 × 1.4 NA) confocal stacks were taken of regions of interest. Slices were fixed further with 2% formaldehyde/1.5% glutaraldehyde in 0.1 M sodium cacodylate, before being processed for volume EM using a modified protocol based on the NCMIR protocol83. Briefly, tissue slices were incubated in 1% osmium tetroxide/1.5% potassium ferricyanide for 1 h at 4 °C, before being washed and left in 0.1 M sodium cacodylate in a fridge overnight. The following day, tissue was incubated in 1% thiocarbohydrazide for 15 min at 60 °C, 2% osmium tetroxide for 30 min, 1% uranyl acetate for 30 min and Walton’s lead aspartate for 30 min at 60 °C, with numerous distilled water washes between each step. The samples were consequently dehydrated through an ethanol series, embedded in Epon resin and baked overnight at 60 °C. Using the maps acquired by light microscopy, the block was trimmed down to the approximate region of interest and serial sections were collected on ITO-coated coverslips using an ultramicrotome (Leica) and diamond knife (Diatome). Coverslips were mounted on SEM stubs using carbon stickies and silver DAG and array tomography serial back scattered electron images (5 nm pixels) were acquired for each CLEM cell (n = 4) using Atlas 5 software and a Gemini 300 SEM (Zeiss) operating in high vacuum, at 4.5 kV with tandem decel operating at 3 kV. Serial images were registered using TrakEM2 (ref. 84) in Fiji (NIH), aligned with the confocal images in Photoshop (Adobe) using nuclei as unbiased fiducials and 2/3D reconstructed using Amira (Thermo Fisher Scientific).
scRNA-seq library preparation, expression and NicheNet analysis
For scRNA-seq, we pooled tissue from four mice for each genotype, to be able to sort rare cell populations. Indeed, from one such preparation, we could isolate 5,000 microglia but only limited PVMs (200) and PVFs (350) on average per genotype. For each cell type and each genotype, we prepared a single scRNA-seq library, and we validated the findings using orthogonal methods such as smFISH and flow cytometry.
Single-cell libraries were prepared using 10X Genomics Chromium Next GEM Single Cell 3′ kit v3.1, according to the manufacturer’s instructions. Cells were loaded on the chromium chip according to Supplementary Table 2; because sorted PVMs and fibroblasts were in low abundance, we loaded the whole sample, while for microglia cells we aimed for 5,000 cells per sample. Libraries were sequenced using an SP flowcell on an Illumina NovaSeq S6000 instrument, with sequencing parameters recommended by 10X Genomics, aiming for 40,000 reads per cell. Raw FASTQ files were preprocessed using 10X Genomics CellRanger (version 6.0) and the mm10 mouse reference genome. Filtered gene expression matrices obtained from CellRanger were imported in R (version 4.1.2) and analyzed using the Seurat (version 4.0.6) and NicheNet (version 1.0.0) packages28. Cells with more than 10% of reads aligning to mitochondrial transcripts were deemed to be low quality or damaged and removed from the dataset. Cells remaining after each filtering step are reported in Supplementary Table 2. Identification of high variable genes, PCA and clustering were performed using Seurat functions. Cluster annotation was performed considering the expression of a set of known cell type markers (Extended Data Fig. 6). Differential expression analysis between pairs of conditions was performed with Seurat’s FindMarkers function using the Wilcoxon Rank Sum test. Comparisons between selected cell populations were performed using NicheNet functions and standard workflow.
Ribosome immunoprecipitation (IP)
Brains were extracted from mice and homogenized in ice-cold homogenization buffer (50 mM Tris, pH 7.4, 100 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT, 1:100 protease inhibitor (Sigma-Aldrich), 200 units per ml RNasin (Promega) and 0.1 mg ml−1 cycloheximide (Sigma-Aldrich) in RNase-free DDW) 10% wt/vol with a Dounce homogenizer (Sigma-Aldrich) until the suspension was homogeneous. Cell debris was removed by transferring 1 ml of homogenate and centrifuging at 10,000g and 4 °C for 10 min. Next, 10 μl of supernatant was removed for ‘input’ analysis and 10 μg of anti-HA.7 antibody (H9658, Sigma-Aldrich) or 10 µg of mouse monoclonal IgG1 antibody (Merck, PP100) was added to the supernatant, followed by 4 h of incubation with slow rotation in a cold room at 4 °C. Afterward, Dynabeads Protein G (Thermo Fisher Scientific) were equilibrated to homogenization buffer by washing three times. Samples were then incubated for 4 h with antibody and afterward beads were added for overnight incubation at 4 °C. Afterward, samples were washed three times with high-salt buffer (50 mM Tris, pH 7.4, 300 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT, 1:200 protease inhibitor, 100 units per ml RNasin and 0.1 mg ml−1 cycloheximide in RNase-free DDW) for 5 min. At the end of the washes, beads were magnetized and excess buffer was removed, 100 µl Lysis Buffer was added to the beads and RNA was extracted with Dynabeads mRNA Direct purification kit (Thermo Fisher Scientific). RNA was eluted in 6 μl H2O and taken for RNA sequencing.
Bulk RNA sequencing for ribosome IP samples
mRNA was captured with Dynabeads oligo(dT) (Life Technologies) according to the manufacturer’s guidelines. Qubit fluorometer (Life Technologies) technology was used to calculate library concentration and mean molecule size was determined with a 2200 TapeStation instrument. Next, libraries were sequenced using Illumina NextSeq-500. Raw reads were mapped to the genome (NCBI37/mm10) using hisat (version 0.1.6). Only reads with unique mapping were considered for further analysis. Gene expression levels of genes of interest (Lyve1, Cx3cr1, Sall1 and Spp1) were calculated using the User-friendly Transcriptome Analysis Pipeline85. Next, DESeq2 R-package was used for normalization and differential expression analysis and pValue was adjusted by multiple gene testing <0.05.
Statistics
All statistical analysis was performed in Prism (GraphPad Software, Version 9.3.1). Two groups were compared using two-tailed unpaired Student’s t-test. For oAβ injections experiments, two-tailed paired Student’s t-test was used to normalize variances of injections on different days or in different oAβ batches. To compare more than two groups (WT, Spp1KO/KO, AppNL-F/NL-F, AppNL-F·Spp1KO/KO mice or PVMs, microglia and PVFs), one-way ANOVA with Kruskal–Wallis post hoc test was used. To compare multiple variables (WT and Spp1KO/KO injected with S26C oAβ), two-way ANOVA with Tukey’s post hoc test was used. Data in graphs are presented as mean ± s.e.m. Data distribution was assumed to be normal but this was not formally tested. Statistical methods were not used to predetermine study sizes but were based on similar experiments previously published. Experiments were blinded to the genotype of the animal as well as the treatment of the animal. Experiments involving human sections were blinded to the demographics of the patients. Analysis of flow cytometry data of Spp1TdT mice was performed in an unblinded fashion, for compensation purposes. No data were excluded for analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41593-023-01257-z.
Supplementary information
Supplementary Table 1—Case demographics for control and AD cases used in this study. Supplementary Table 2—Cell numbers of isolated and sequenced PVM, PVF and microglia of 6 mo wild-type, AppNL-F, Spp1KO/KO and AppNL-F·Spp1KO/KO mice after CellRanger filtering and QC filtering.
Supplementary Video 1—Video showing back scattered electron images of serial sections of SPP1-TdT PVM shown in Fig. 2h. The manually segmented nuclei of the SPP1-TdT positive cell is shown highlighted in blue, to mark the SPP1-TdT correlated cell. Video width is 60 µm.
Source data
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Acknowledgements
We thank M.L. Shinohara (Duke) for the valuable discussion and input. We thank S. West (Sainsbury Wellcome Centre, UCL), I.N. Chiong (King’s College London), E. Turkes (UK DRI at UCL) and P. Muza (UCL) for experimental help. We thank F. A. Edwards (UCL; PPL); T. Saido (RIKEN; AppNL-F mice), E. Ghiradello and P. Muckett (UK DRI at UCL; animal husbandry). We thank V. M. Lee (Department of Pathology and Laboratory Medicine, Institute on Aging and Center for Neurodegenerative Disease Research, University of Pennsylvania School of Medicine, Philadelphia, USA; NAB61 antibody), J. Cirrito (Department of Neurology, Washington University School of Medicine, St. Louis, USA; mHJ3.4antibody) and N. Cade (UK DRI at UCL for microscope imaging assistance). We thank J. Evans (FACS Core Facility at UCL, flow cytometry assistance and cell sort). We would like to acknowledge The Jackson Laboratory’s Genetic Engineering Technologies Scientific Service for mouse strain development. We thank our funders—UK Dementia Research Institute (UKDRI-1011) (which receives its funding from UK DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK), Bright Focus Foundation Grant (A2021032S) (S.H.) and UCL Neurogenetic Therapies Programme (NgTP07) funded by Sigrid Rausing Trust (S.H.). We acknowledge Wellcome Trust Sir Henry Wellcome Fellowship (221634/Z/20/Z) (S.D.S.), Wellcome Trust 4-year Neuroscience PhD studentship (219906/Z/19/Z) (G.C.) and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project-ID 259373024—TRR 167 (S.J.). The QSBB is supported by the Reta Lila Weston Institute of Neurological Studies and UCL Queen Square Institute of Neurology (C.E.T. and T.L.). We acknowledge core funding to the MRC Laboratory for Molecular Cell Biology at University College London (award code MC_U12266B) (J.J.B.). The LMCB volume EM facility was set up with funding from the Wellcome Trust (218278/Z/19/Z). The Spp1TdT reporter mouse model was developed with funding from an anonymous organization. The Jackson Laboratory gratefully acknowledges the contribution of Kim Martens and the Genetic Engineering Technologies Service at The Jackson Laboratory for expert assistance with the work described in this publication (M.S. and D.G.). The authors would like to acknowledge helpful discussions and feedback from the International Neuroimmune Consortium. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Extended data
Author contributions
Design of the study and conceptualization: S.D.S., S.H. and S.J. Experiments and data collection: S.D.S., J.Z.G., G.C., L.S.S.F., D.G., C.E.T., D.S., J.R-C., S-H.S., J-S.K., T.C., T.L., J.J.B., M.S. and C.S.F. Software and bioinformatic analysis: J.J.B. and C.S.F. Formal analysis of the data: S.D.S., J.Z.G., L.S.S.F., D.G., J.R-C., S-H.S., J-S.K., T.C., J.J.B. and C.S.F. Validation of the data: S.D.S., J.Z.G., G.C., L.S.S.F., C.E.T., D.S., T.C., T.L., J.J.B. and M.S. Responsible for resources: D.G., C.E.T., T.L., J.J.B., M.S. and C.S.F. Writing of the original and revised draft: S.D.S. and S.H. Supervision of research and funding acquisition: S.H.
Peer review
Peer review information
Nature Neuroscience thanks F. Chris Bennett, Robert Harris and Vivek Swarup for their contribution to the peer review of this work.
Data availability
The scRNA-seq data generated in this study are deposited at ArrayExpress (BioStudies, Annotare 2.0) under accession number E-MTAB-11918. The RiboTag-based RNA-seq data generated in this study are deposited at ArrayExpress (BioStudies, Annotare 2.0) under accession number E-MTAB-12515. Source data are provided within this paper. Raw data used in this study and requests for resources and reagents are available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41593-023-01257-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41593-023-01257-z.
References
- 1.Arandjelovic, S. & Ravichandran, K. S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol.16, 907–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kierdorf, K., Masuda, T., Jordão, M. J. C. & Prinz, M. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci.20, 547–562 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Schafer, D. P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron74, 691–705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cserép, C. et al. Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science367, 528–537 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Paolicelli, R. C. et al. Synaptic pruning by microglia is necessary for normal brain development. Science1456, 10–13 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Engelhardt, B., Vajkoczy, P. & Weller, R. O. The movers and shapers in immune privilege of the CNS. Nat. Immunol.18, 123–131 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Lapenna, A. M., De Palma, M. & Lewis, C. E. Perivascular macrophages in health and disease. Nat. Rev. Immunol.18, 689–702 (2018). [DOI] [PubMed]
- 8.Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utz, S. G. et al. Early fate defines microglia and non-parenchymal brain macrophage development. Cell181, 557–573 (2020). [DOI] [PubMed]
- 10.Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol.17, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J. S. et al. 2021 A binary Cre transgenic approach dissects microglia and CNS border-associated macrophages. Immunity54, 176–190 (2021). [DOI] [PubMed]
- 12.Masuda, T. et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature604, 740–748 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Van Hove, H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci.22, 1021–1035 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Podleśny-Drabiniok, A., Marcora, E. & Goate, A. M. Microglial phagocytosis: a disease-associated process emerging from Alzheimer’s disease genetics. Trends Neurosci.43, 965–979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirakawa, K. et al. IL (interleukin)-10-STAT3-galectin-3 axis is essential for osteopontin-producing reparative macrophage polarization after myocardial infarction. Circulation138, 2021–2035 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Remmerie, A. et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity53, 641–657 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rittling, S. R. Osteopontin in macrophage function. Expert Rev. Mol. Med.13, e15 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell178, 686–698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liaw, L. et al. Altered wound healing in mice lacking a functional osteopontin gene (Spp1). J. Clin. Invest.101, 1468–1478 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comi, C. et al. Osteopontin is increased in the cerebrospinal fluid of patients with Alzheimer’s disease and its levels correlate with cognitive decline. J. Alzheimers Dis.19, 1143–1148 (2010). [DOI] [PubMed]
- 22.Paterson, R. W. et al. A targeted proteomic multiplex CSF assay identifies increased malate dehydrogenase and other neurodegenerative biomarkers in individuals with Alzheimer’s disease pathology. Transl. Psychiatry6, e952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chai, Y. L. et al. Plasma osteopontin as a biomarker of Alzheimer’s disease and vascular cognitive impairment. Sci. Rep.11, 4010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med.26, 769–780 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sala Frigerio, C. et al. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep.27, 1293–1306(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deczkowska, A. et al. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell173, 1073–1081 (2018). [DOI] [PubMed]
- 27.Silvin, A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity55, 1448–1465 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Browaeys, R., Saelens, W. & Saeys, Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods17, 159–162 (2020). [DOI] [PubMed]
- 29.Gratuze, M. et al. Impact of TREM2R47Hvariant on tau pathology-induced gliosis and neurodegeneration. J. Clin. Invest.130, 4954–4968 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito, T. et al. Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci.17, 661–664 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Stevens, B. et al. The classical complement cascade mediates CNS synapse elimination. Cell131, 1164–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Fonseca, M. I. et al. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J. Neuroinflammation14, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, E. B. et al. Targeting amyloid-β peptide (Aβ) oligomers by passive immunization with a conformation-selective monoclonal antibody improves learning and memory in Aβ precursor protein (APP) transgenic mice. J. Biol. Chem.281, 4292–4299 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Koffie, R. M. et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl Acad. Sci. USA106, 4012–4017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, T., Guo, R. & Zhang, F. Brain perivascular macrophages: recent advances and implications in health and diseases. CNS Neurosci. Ther.25, 1318–1328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell10.1016/j.cell.2017.05.018 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell174, 999–1014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz, E. et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl Acad. Sci. USA106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haimon, Z. et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol.19, 636–644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science356, 1248–1259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramerova, I. et al. Spp1 (osteopontin) promotes TGFβ processing in fibroblasts of dystrophin-deficient muscles through matrix metalloproteinases. Hum. Mol. Genet.28, 3431–3442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci.17, 131–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krysko, D. V., Ravichandran, K. S. & Vandenabeele, P. Macrophages regulate the clearance of living cells by calreticulin. Nat. Commun.9, 4644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu, D. D., Lin, E. C. K., Kovach, N. L., Hoyer, J. R. & Smith, J. W. A biochemical characterization of the binding of osteopontin to integrins αvβ1 and αvβ5. J. Biol. Chem.270, 26232–26238 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Zöller, T. et al. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun.9, 4011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothman, S. M. et al. Human Alzheimer’s disease gene expression signatures and immune profile in APP mouse models: a discrete transcriptomic view of Aβ plaque pathology. J. Neuroinflammation15, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sellgren, C. M. et al. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat. Neurosci.22, 374–385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekar, A. et al. Schizophrenia risk from complex variation of complement component 4. Nature530, 177–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dejanovic, B. et al. Changes in the synaptic proteome in tauopathy and rescue of tau-induced synapse loss by C1q antibodies. Neuron100, 1322–1336 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Wu, T. et al. Complement C3 is activated in human AD brain and is required for neurodegeneration in mouse models of amyloidosis and tauopathy. Cell Rep.28, 2111–2123 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Lui, H. et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell165, 925–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasek, M. J. et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature534, 538–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holden, S. S. et al. Complement factor C1q mediates sleep spindle loss and epileptic spikes after mild brain injury. Science373, eabj2685 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Absinta, M. et al. A lymphocyte–microglia–astrocyte axis in chronic active multiple sclerosis. Nature597, 709–714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schirmer, L. et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature573, 75–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Werneburg, S. et al. Targeted complement inhibition at synapses prevents microglial synaptic engulfment and synapse loss in demyelinating disease. Immunity52, 167–182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber, G. F. & Cantor, H. The immunology of Eta-1/osteopontin. Cytokine Growth Factor Rev.7, 241–248 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron101, 207–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen, X., Qiu, Y., Wight, A., Kim, H. J. & Cantor, H. Definition of a mouse microglial subset that regulates neuronal development and proinflammatory responses in the brain. Proc. Natl Acad. Sci. USA119, e2116241119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes resource single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell. Immunity50, 253–271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Månberg, A. et al. Altered perivascular fibroblast activity precedes ALS disease onset. Nat. Med.27, 640–646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature603, 885–892 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawkes, C. A. et al. Perivascular drainage of solutes is impaired in the ageing mouse brain and in the presence of cerebral amyloid angiopathy. Acta Neuropathol.121, 431–443 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol.14, 133–150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nation, D. A. et al. Blood–brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med.25, 270–276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toledo, J. B. et al. Contribution of cerebrovascular disease in autopsy confirmed neurodegenerative disease cases in the National Alzheimer’s Coordinating Centre. Brain136, 2697–2706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardy, J. & Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci.12, 383–388 (1991). [DOI] [PubMed] [Google Scholar]
- 68.Rentsendorj, A. et al. A novel role for osteopontin in macrophage-mediated amyloid-β clearance in Alzheimer’s models. Brain. Behav. Immun.67, 163–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi, Q. et al. Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci. Transl. Med.9, 6295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Minami, S. S. et al. Progranulin protects against amyloid β 2 deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med.20, 1157–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shinohara, M. L., Kim, H. J., Kim, J. H., Garcia, V. A. & Cantor, H. Alternative translation of osteopontin generates intracellular and secreted isoforms that mediate distinct biological activities in dendritic cells. Proc. Natl Acad. Sci. USA105, 7235–7239 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toyonaga, T. et al. Osteopontin deficiency accelerates spontaneous colitis in mice with disrupted gut microbiota and macrophage phagocytic activity. PLoS ONE10, e0135552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin, Y. J. et al. Osteopontin: correlation with phagocytosis by brain macrophages in a rat model of stroke. Glia59, 413–423 (2011). [DOI] [PubMed]
- 74.Wyss-Coray, T. et al. TGF-β1 promotes microglial amyloid-β clearance and reduces plaque burden in transgenic mice. Nat. Med.7, 612–618 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Tesseur, I. et al. Deficiency in neuronal TGF-β signaling promotes neurodegeneration and Alzheimer’s pathology. J. Clin. Invest.116, 3060–3069 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guilliams, M., Thierry, G. R., Bonnardel, J. & Bajenoff, M. Establishment and maintenance of the macrophage niche. Immunity52, 434–451 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Qin, W. et al. Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics200, 423–430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holt, M., Nicholas, F. W., James, J. W., Moran, C. & Martin, I. C. A. Development of a highly fecund inbred strain of mice. Mamm. Genome15, 951–959 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Montine, T. J. et al. National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol.123, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiao, S., Qin, M. & Wang, H. Analysis of knee infrared image based on sample entropy algorithm. Proceedings of 2015 8th International Conference on Intelligent Networks and Intelligent Systems (ICINIS) 1–4 (IEEE, 2016).
- 81.Emi, M. et al. Genotyping and sequence analysis of apolipoprotein E isoforms. Genomics3, 373–379 (1988). [DOI] [PubMed] [Google Scholar]
- 82.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deerinck, T. J., Bushong, E. A., Thor, A. & Ellisman, M. H. NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial block face scanning electron microscopy. Microscopy1, 6–8 (2010). [Google Scholar]
- 84.Cardona, A. et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE1, e38011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kohen, R. et al. UTAP: user-friendly transcriptome analysis pipeline. BMC Bioinf.20, 154–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol.20, 4106–4114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1—Case demographics for control and AD cases used in this study. Supplementary Table 2—Cell numbers of isolated and sequenced PVM, PVF and microglia of 6 mo wild-type, AppNL-F, Spp1KO/KO and AppNL-F·Spp1KO/KO mice after CellRanger filtering and QC filtering.
Supplementary Video 1—Video showing back scattered electron images of serial sections of SPP1-TdT PVM shown in Fig. 2h. The manually segmented nuclei of the SPP1-TdT positive cell is shown highlighted in blue, to mark the SPP1-TdT correlated cell. Video width is 60 µm.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Raw data.
Data Availability Statement
The scRNA-seq data generated in this study are deposited at ArrayExpress (BioStudies, Annotare 2.0) under accession number E-MTAB-11918. The RiboTag-based RNA-seq data generated in this study are deposited at ArrayExpress (BioStudies, Annotare 2.0) under accession number E-MTAB-12515. Source data are provided within this paper. Raw data used in this study and requests for resources and reagents are available from the corresponding author upon request. Source data are provided with this paper.