Abstract
Incorporating genetics into risk-stratification for treatment of childhood B-progenitor acute lymphoblastic leukaemia (B-ALL) has contributed significantly to improved survival. In about 30% B-ALL (B-other-ALL) without well-established chromosomal changes, new genetic subtypes have recently emerged, yet their true prognostic relevance largely remains unclear. We integrated next generation sequencing (NGS): whole genome sequencing (WGS) (n = 157) and bespoke targeted NGS (t-NGS) (n = 175) (overlap n = 36), with existing genetic annotation in a representative cohort of 351 B-other-ALL patients from the childhood ALL trail, UKALL2003. PAX5alt was most frequently observed (n = 91), whereas PAX5 P80R mutations (n = 11) defined a distinct PAX5 subtype. DUX4-r subtype (n = 80) was defined by DUX4 rearrangements and/or ERG deletions. These patients had a low relapse rate and excellent survival. ETV6::RUNX1-like subtype (n = 21) was characterised by multiple abnormalities of ETV6 and IKZF1, with no reported relapses or deaths, indicating their excellent prognosis in this trial. An inferior outcome for patients with ABL-class fusions (n = 25) was confirmed. Integration of NGS into genomic profiling of B-other-ALL within a single childhood ALL trial, UKALL2003, has shown the added clinical value of NGS-based approaches, through improved accuracy in detection and classification into the range of risk stratifying genetic subtypes, while validating their prognostic significance.
Subject terms: Cancer genomics, Risk factors
Introduction
In paediatric B-progenitor acute lymphoblastic leukaemia (B-ALL), genetic aberrations are important prognostic markers. A number of well-established abnormalities define specific subtypes, which are used to inform treatment [1]. Among approximately 30% of B-ALL patients (B-other-ALL) lacking these subtype-defining abnormalities, distinct genetic entities have emerged [2–10]. For example, DUX4-rearranged (DUX4-r) and patients with ABL-class fusions have been shown to have good and poor outcomes, respectively [4, 10–21], while the clinical relevance of other subtypes, including alterations of PAX5 (PAX5alt) and ETV6::RUNX1-like, remain unclear. Although these subtypes display characteristic gene expression signatures, their underlying genetic profiles are heterogeneous. For example, PAX5alt is associated with a wide spectrum of PAX5 abnormalities, including deletions, mutations and fusions with multiple partner genes [2].
We have demonstrated that whole genome sequencing (WGS) provides an excellent method for classifying B-ALL patients into clinically relevant genetic subtypes [22]. Here, we combine results from cytogenetics, fluorescence in situ hybridisation (FISH) and Multiplex Ligation-dependent Probe Amplification (MLPA) with both WGS and targeted next generation sequencing (t-NGS) of a large cohort of B-other-ALL from a single highly successful UK childhood ALL clinical trial, UKALL2003. Using this integrated approach, we have accurately classified these patients into 15 distinct genetic subtypes, described the spectrum of underlying abnormalities, and clarified their frequency and clinical significance.
Methods
Patient cohort
Patients were diagnosed with B-ALL and treated on the UKALL2003 trial (NCT00222612) (age 1–24 years) [23, 24]. The Scottish Multi-Centre Research Ethics Committee approved the trial and written informed consent was obtained in accordance with the Declaration of Helsinki. Among the total trial recruitment (n = 3204), 741 patients were classified as B-other-ALL, which excluded Down Syndrome individuals (n = 65), patients not fully tested (n = 38) and cases with normal karyotypes and ≥4 additional RUNX1 signals by FISH (n = 75), as they were likely to be undetected high hyperdiploidy or iAMP21-ALL (Supplementary Fig. 1). Samples were available for genetic testing on a representative cohort of 351/741 B-other-ALL patients. Initially patients were assigned to three or four drug induction based on NCI risk status. High-risk patients (slow early response or high-risk genetics) were assigned to augmented post-induction therapy (regimen C), while the remaining patients were randomised to either treatment reduction (minimal residual disease (MRD) low-risk) or intensification (MRD high-risk).
Targeted NGS
Whole-genome amplification (WGA) of 30 ng genomic DNA was performed using the Repli-G Mini kit (Qiagen). RNA baits were designed to capture the whole gene sequence of 23 genes and exonic regions of 35 genes, using SureDesign and the SureSelect XT2 platform (Agilent Technologies, Santa Clara, USA), covering 97% (average) of the target regions, ranging from 69% (IKZF1) to 100% (Supplementary Table 1). DNA was fragmented into 800–1000 bp fragments by sonication using the M220 Focused Ultrasonicator (Covaris, USA) or Bioruptor Pico (Diagenode, USA). Sequencing libraries were prepared using the custom-designed SureSelect XT2 kit (Agilent Technologies, USA) according to manufacturer’s protocol, with the following modifications for enrichment of larger DNA fragments: 1) 1 and 2 min annealing and elongation stage, respectively, during the pre- and post-hybridisation PCR and 2) the ratio of AMPure XP beads (Beckman Coulter, USA) to sample volume was reduced to 0.7:1. Individual samples were barcoded for pooling at equal volumes prior to sequencing. The libraries were sequenced on a HiSeq2500 or NextSeq (Illumina, UK) using 125–150 bp paired-end chemistry. Samples were sequenced to a mean coverage of 300-fold.
Raw Fastq reads were processed using the Genome Analysis Toolkit (GATK) [25]. Reads were aligned to hg19/GRCh37 and duplicates removed using BWA-MEM [26] and Picard [27]. Structural variants (SVs) were manually interrogated from deduplicated bam files, using Integrated Genomics Viewer (IGV) (Broad Institute, USA) [28], with the minimum and maximum insert size set to 50 bp and 5000 bp, respectively. Single nucleotide variations (SNVs) and indels were called using GATK HaplotypeCaller (version 3.8) [29]. Default settings were used but ploidy was increased to eight for the detection of subclonal variants and Base Quality Score Recalibration (BQSR) was not applied due to the small size of the targeted region. Hard filtering was performed and variants that passed filtering with an allele depth (AD) of 10 were annotated using Variant Effect Predictor (VEP) (version 102.0) [30], adding information from SIFT (version 5.2.2) [31] and Polyphen (version 2.2.2) [32].
Whole genome sequencing
WGS was performed on matched diagnostic and remission DNA samples, as previously described [22].
Fluorescence in-situ hybridisation
FISH results were available for rearrangements associated with B-other-ALL from our previously published studies, including: ABL-class genes: ABL1, ABL2, PDGFRB/CSF1R; JAK-STAT pathway genes: CRLF2, JAK2; other newly defined subtypes: ZNF384, MEF2D, and NUTM1; [12] as well as IGH and associated partner genes [33]. Additionally, FISH was performed to identify rearrangements of ETV6, PAX5, and IKZF1, using commercial or home-grown break-apart FISH probes [34] (Cytocell, UK; Leica Microsystems, UK).
Multiplex ligation-dependent probe amplification
MLPA results using the SALSA P335-ALL-IKZF1 and P327-iAMP21-ERG MLPA kits (MRC Holland, the Netherlands) were also available from our previous studies [12, 35, 36].
Single nucleotide polymorphism arrays
SNP array data were available for 148 patients from this cohort, using SNP6.0 (Affymetrix, Santa Clara, CA) or Infinium CytoSNP-850K (Ilumina Inc., San Diego, CA). SNP arrays were analysed using Nexus Copy Number 10 (Bio-discovery, El Segundo, CA), as previously reported [37].
Statistical analyses
Event-free survival (EFS) was defined as time-to-relapse, second tumour or death, censoring at date of last contact. Relapse rate (RR) was defined as time-to-relapse for those achieving complete remission, censoring at date of death in remission or last contact. Overall survival (OS) was defined as time-to-death, censoring at date of last contact. The median follow-up time for the whole cohort was 10.98 years (IQR 3.83 years). Kaplan-Meier methods were used to estimate survival rates and univariate Cox regression models were used to determine hazard ratios. Other comparisons were performed using χ2 or Fisher’s exact tests, as appropriate. All p-values were two-sided and values <0.05 were considered statistically significant. All analyses were performed using Intercooled Stata (Stata Statistical Software Release 16; StataCorp, USA).
Results
Classification of the B-other-ALL cohort
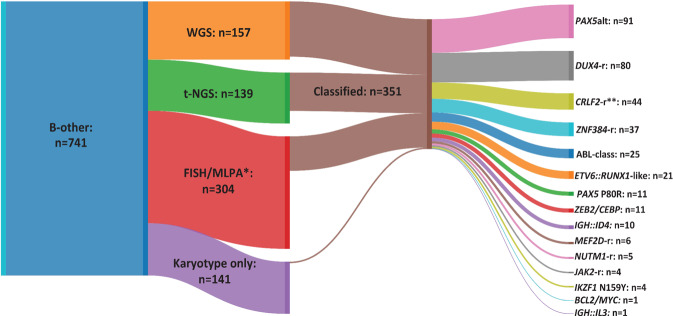
Data from WGS (n = 157) and t-NGS (n = 175) (36 patients tested by both techniques) were integrated with cytogenetics, FISH and MLPA to classify 351 patients into one of 15 distinct subtypes (Fig. 1, Table 1, Supplementary Tables 2–7). Among those patients tested by WGS, 94% (n = 147/157) were classified compared to 77% (n = 107/139) tested by t-NGS only. Samples for additional testing, including FISH, MLPA and NGS, were unavailable for some patients (n = 141). These remained unclassified, except for four patients who presented with a subtype-defining chromosomal abnormality by cytogenetic analysis: PAX5alt with dic(9;20)(p11~13;q11) (n = 3) and t(6;14)(p22;q32)/IGH::ID4 (n = 1). Additionally, 304 patients, not tested by NGS, were screened using FISH and/or MLPA, with 93 classified as previously described [12] (Fig. 1).
Fig. 1. Classification of B-other-ALL cohort according to technique.
Sankey plot showing the number of patients tested by each technique and those classified into genomic subtypes of B-other-ALL, as defined in Table 1. *Cases tested by FISH and/or MLPA, not all cases by all kits and probes. **Excludes 9 patients with other subtype defining abnormalities. WGS Whole genome sequencing, t-NGS Targeted next generation sequencing, FISH Fluorescence in situ hybridisation, MLPA Multiplex Ligation-dependent Probe Amplification.
Table 1.
Classification of B-other-ALL by standard and NGS techniques.
| Genomic Subtype | Abnormality | Standard techniques | NGS |
|---|---|---|---|
| ABL-class | ABL1 fusion | ABL1, ABL2, PDGFRB or CSF1R rearrangement by FISH | ABL1, ABL2, PDGFRB or CSF1R fusion |
| ABL2 fusion | |||
| CSF1R fusion | |||
| PDGFRB fusion | |||
| ETV6::RUNX1-like | ETV6 rearrangement | ETV6 rearrangement by FISH | ETV6 fusion |
| IKZF1 rearrangement | IKZF1 rearrangement by FISH | IKZF1 fusion and/or deletion | |
| Other ETV6::RUNX1-like | Not applicable | ETV6 biallelic inactivation in patients that lack other defining features | |
| IGH::ID4 | t(6;14)(p22;q32) by karyotype and/or IGH::ID4 positive by FISH | IGH::ID4 | |
| CRLF2-r | IGH::CRLF2 | IGH::CRLF2 positive by FISH | IGH::CRLF2 |
| P2RY8::CRLF2 | P2RY8::CRLF2 by FISH and/or PAR1 deletion by MLPA or SNP array | P2RY8::CRLF2 | |
| JAK2-r | JAK2 rearrangement by FISH | JAK2 fusion | |
| ZNF384-r | ZNF384 rearrangement by FISH | ZNF384 fusion | |
| MEF2D-r | MEF2D rearrangement by FISH | MEF2D fusion by WGS* | |
| NUTM1-r | NUTM1 rearrangement by FISH | NUTM1 fusion | |
| PAX5alt | PAX5 rearrangement | PAX5 rearrangement by FISH | PAX5 fusion |
| PAX5-ITD | Internal Tandem Duplication (Amplification) of PAX5 exons 2–5 by MLPA or SNP array | PAX5-ITD by NGS | |
| PAX5 mutation | Not applicable | Clonal PAX5 mutation (VAF = > 35%) not P80R that lack other defining features | |
| dic(9;20) | dic(9;20) by karyotype or loss of 9p and 20p by SNP array | dic(9;20) i.e. loss of 9p and 20p and/or PAX5::NOL4L | |
| dic(9;12) | dic(9;12) by karyotype or loss of 9p and 12p with retention of 5’PAX5 and 3’ETV6 by SNP array | dic(9;12) i.e. loss of 9p and 12p and PAX5::ETV6 | |
| Other PAX5alt | Not applicable | Biallelic inactivation of PAX5 or PAX5 loss [CN = 1] in cases with biallelic CDKN2A/B loss [CN = 0], and MTAP CNV/SV [CN = 0/1] that lack other defining features | |
| IKZF1 N159Y | Not applicable | IKZF1 N159Y mutation and/or IKZF-ITD (Internal Tandem Duplication) | |
| PAX5 P80R | Not applicable | PAX5 P80R mutation | |
| BCL2/MYC | IGH::BCL2 and/or IGH::MYC positive by FISH | Gene rearrangement involving BCL2, BCL6 or MYC | |
| DUX4-r | DUX4-r | Not applicable | DUX4 rearrangement by WGS* |
| ERG-d | Intragenic ERG deletion by MLPA or SNP array | Intragenic deletion, mutation or other rearrangement of ERG | |
| ZEB2/CEBP | IGH::CEBP family gene positive by FISH | CEBP family gene rearrangement and/or ZEB2 H1038R mutation | |
| IGH::IL3 | t(5;14)(q31;q32) by karyotype and/or IGH::IL3 positive by FISH | IGH::IL3 | |
Standard-of-care techniques include cytogenetics, FISH, MLPA and SNP array. NGS includes WGS and t-NGS. Abnormal FISH signal patterns classed as balanced rearrangements: 1R1G1F, or unbalanced: 1R0G1F or 0R1G1F, with evidence of fusion from karyotype, partner gene FISH, SNP array or RT-PCR, as previously published [12]. MEF2D::CSF1R and ETV6::ABL1 are classified as ABL-class fusions, PAX5::JAK2 and ETV6::JAK2 are classified as JAK2-r, PAX5::ETV6 are classified as PAX5alt according to previously published data [2]. All other rearrangements of ETV6 are assigned to the ETV6::RUNX1-like subtype. PAX5 mutations and CN abnormalities of PAX5, CDKN2A/B and MTAP are classified as PAX5alt, only in the absence of other subtype defining abnormalities. *DUX4 and MEF2D were not included in the t-NGS kit. CN copy number, SV structural variant, CNV copy number variant.
The most common subtypes were PAX5alt (n = 91), DUX4-r (n = 80), CRLF2-r (n = 53), ZNF384-r (n = 37), ABL-class (n = 25) and ETV6::RUNX1-like (n = 21). Less common were: CEBP/ZEB2 (n = 12), PAX5 P80R (n = 11), IGH::ID4 (n = 10), MEF2D-r (n = 6), NUTM1-r (n = 5), IKZF1 N159Y (n = 4), JAK2 fusions (n = 4), IGH::IL3 (n = 1) and BCL2/MYC (n = 1). In the majority of cases, the subtype-defining abnormalities were mutually exclusive, except for nine patients with P2RY8::CRLF2 coexisting with PAX5alt [dic(9;20) (n = 5) and PAX5-ITD (n = 2)], TCF3::ZNF384 (n = 1) and ETV6::RUNX1-like (ETV6::IKZF1, n = 1). One patient (22355) harboured both IGH::DUX4 and IGH::CEBPD.
Comparison of techniques
There was high concordance between WGS and t-NGS results, with the same subtype-defining abnormality identified in 28/32 (88%) cases (Supplementary Table 8). Four cases tested by both techniques remained unclassified, although WGS identified novel abnormalities, including three in the MAP kinase pathway, as previously reported [22]. Although the t-NGS panel did not include DUX4, t-NGS identified ERG abnormalities in 8/11 DUX4-r cases analysed by WGS and t-NGS, including deletions (n = 7) and inversions (n = 1).
Both WGS and t-NGS detected subtype-defining mutations in PAX5 (P80R and others) (n = 22), IKZF1 (N159Y) (n = 4), and ZEB2 (H1038R) (n = 4). They also identified secondary mutations in a wide range of genes, although none was associated with a particular subtype and numbers were small (Supplementary Table 9).
WGS detected a higher number of focal copy number abnormalities (CNA) than MLPA, notably of ERG deletions, as we have previously reported [22]. Similarly, t-NGS identified ERG deletions in three patients that had been called normal by MLPA. These deletions were either focal, with evidence of a single probe deletion only by MLPA, which is insufficient to call a deletion (n = 2), or sub-clonal, where the MLPA ratio for the deleted probes was 0.76–0.9 but not below the required 0.75 cut-off level to call a deletion (n = 1) [35, 38].
FISH and MLPA detected the highest number of CRLF2 rearrangements, in 100% agreement with WGS. Detection of CRLF2-r by t-NGS was inconsistent, identifying only 2/4 IGH::CRLF2 and 2/14 P2RY8::CRLF2 cases observed by FISH and/or MLPA. This was due to incomplete coverage across the PAR1 region, with only CRLF2 being included in the t-NGS panel design, with 82% coverage.
There was high concordance between our existing FISH data and both WGS and t-NGS for other genes (98–100%, Supplementary Table 10) [12, 33], with only four additional cases identified by NGS, where FISH had shown a normal result. These were MEF2D::CSF1R, SMARCA2::ZNF384, CUX1::NUTM1 and IGH::CEBPA. It is likely that these fusions resulted from complex rearrangements, as seen for MEF2D::CSF1R (9850), with multiple rearrangements, including deletions and inversions involving the MEF2D gene identified by WGS. Alternatively, the breakpoints were outside the region covered by the FISH probes, as demonstrated in the patient with the CUX1::NUTM1 fusion (20750). The translocation, t(7;15)(q22;q14), was present in the karyotype but NUTM1 was not seen to be rearranged by FISH.
FISH alone could not fully characterise rearrangements of PAX5 and ETV6. Among PAX5alt and ETV6::RUNX1-like patients, different FISH signal patterns were observed, including whole and partial deletions targeting PAX5 and ETV6, as well as balanced rearrangements (Supplementary Tables 2 & 3). Many of these cases were further characterised by t-NGS, although two cases of PAX5alt, with dic(9;20) by cytogenetics (10061 and 22861), were not called by t-NGS. Despite the presence of large abnormalities involving 9p, the breakpoints were outside the regions of PAX5, MTAP and CDKN2A/B covered by the t-NGS panel and were therefore undetected.
PAX5alt
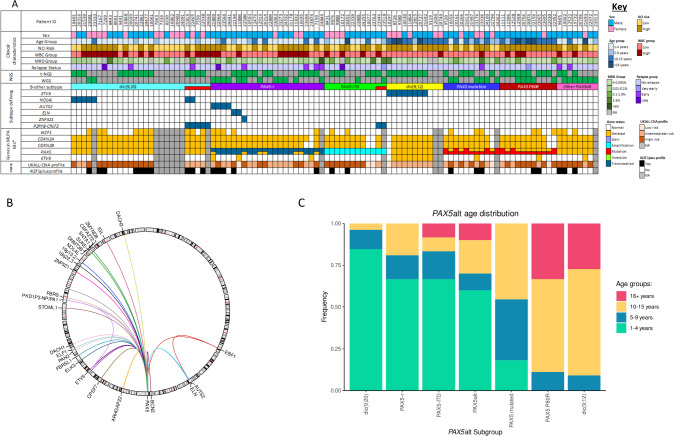
PAX5alt was the most frequently observed subtype (n = 91), including patients with dic(9;20)(p11~13;q11) (n = 27, 30%), dic(9;12)(p13;p13) (n = 11, 12%), PAX5 rearrangements (PAX5-r) (n = 22, 24%), PAX5 mutations (n = 11, 12%) and PAX5-ITD (n = 12,13%) (Fig. 2A). A further eight patients (9%) had a specific genomic profile of PAX5 loss, CDKN2A/B biallelic loss and MTAP abnormalities, with absence of other subtype-defining genetic abnormalities, which was associated with a PAX5alt gene expression profile in our WGS study [22]. Overall, the PAX5alt subtype had an increased frequency of CDKN2A/B (94 v 35%, p < 0.001) and PAX5 deletions (74 v 20%, p < 0.001) compared to other subtypes (Supplementary Fig. 2). Patients with PAX5 P80R mutations (n = 11) were classified as a distinct subtype due to their reported differential gene expression signature [2].
Fig. 2. Genomic and clinical features of the PAX5alt subtype.
A Oncoplot showing the distribution of clinical features and genetic abnormalities within the PAX5alt subtype and associated copy number profile risk status (UKALL-CNA [36] and IKZF1plus [40]). Coexistence of CRLF2-r is indicated in red in the B-other-ALL subtype row. Copy number profile status was unavailable for patients lacking Multiplex Ligation-dependent Probe Amplification (MLPA) data. *The SALSA P335-ALL-IKZF1 and P327-iAMP21-ERG MLPA kits were used to determine gene copy number. Relapses were defined as follows: very early, < 18 months from diagnosis; early, within 6 months of end of treatment; late >6 months after end of treatment. B Circos plot illustrating the range of PAX5 translocation partner genes in PAX5-r. C Stacked bar plot showing the distribution of PAX5alt abnormalities amongst different age groups. NCI National Cancer Institute, WBC White blood cell count, MRD Minimal residual disease, t-NGS Targeted next-generation sequencing, WGS Whole genome sequencing, PAX5-r PAX5 rearranged, PAX5-ITD PAX5 internal tandem duplication, CNA Copy number alteration.
Among patients with dic(9;20), three were identified from chromosomal analysis alone. In the remaining cases, PAX5 involvement was confirmed by a variety of techniques, with 23 patients showing whole (n = 9) or partial gene deletions (n = 14). One dic(9;20) patient (10401) showed normal copy number for PAX5, however t-NGS identified a PAX5::NOL4L fusion. In total, NGS identified PAX5::NOL4L fusions in seven dic(9;20) patients. Other recurrent fusions of PAX5 were observed with ETV6 (n = 8), AUTS2 (n = 3), ELN (n = 3) and ZNF521 (n = 2), while other fusions were detected in single cases (n = 22) (Fig. 2B).
There was an association between different PAX5 abnormalities and age: dic(9;20) was more commonly observed in children aged 1–4 (p < 0.001), whilst both dic(9;12) and PAX5 P80R were seen in older children, aged 10–15 years (p = 0.005) (Fig. 2C).
ETV6::RUNX1-like
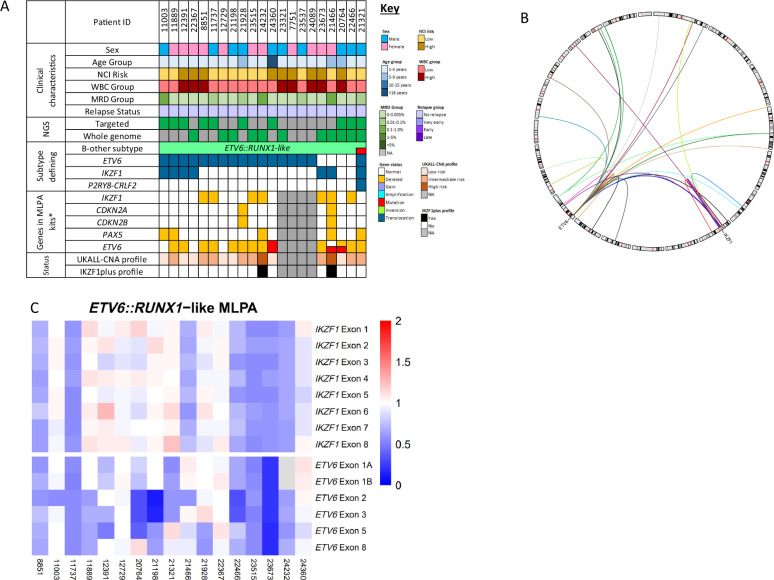
ETV6::RUNX1-like patients (n = 21) were characterised by multiple abnormalities of ETV6, including rearrangements with other genes (n = 17) and/or deletions (n = 12). The only recurrent ETV6 partner gene was IKZF1 (n = 2), although IKZF1 was rearranged with other genes (n = 7) and/or deleted (n = 7) (Fig. 3A–C).
Fig. 3. Genomic and clinical features of the ETV6::RUNX1-like subtype.
A Oncoplot showing the distribution of clinical features and genetic abnormalities within the ETV6::RUNX1-like subtype and associated copy number profile risk status (UKALL-CNA [36] and IKZF1plus [40]). Coexistence of CRLF2-r is indicated in red in the B-other subtype row. Copy number profile status was unavailable for patients lacking Multiplex Ligation-dependent Probe Amplification (MLPA) data. *The SALSA P335-ALL-IKZF1 and P327-iAMP21-ERG MLPA kits were used to determine gene copy number. Relapses were defined as follows: very early, < 18 months from diagnosis; early, within 6 months of end of treatment; late > 6 months after end of treatment. B Circos plot illustrating the constellation of ETV6 translocation partner genes which characterise ETV6::RUNX1-like. Only the recurrent translocation partners are labelled with gene names. C Heatmap of MLPA ratios for copy number detection of ETV6 and IKZF1 across the ETV6::RUNX1-like subtype.
ETV6 is known to rearrange with multiple genes in other B-ALL subtypes. In this study, we observed PAX5::ETV6 (n = 8), ETV6::ABL1 (n = 2) and ETV6::JAK2 (n = 1) fusions. These cases were excluded from the ETV6::RUNX1-like subtype, as previous studies have shown that these fusions do not drive the distinctive gene expression signature associated with this subtype [2, 4, 9].
Other subtypes
Within the DUX4-r subtype (n = 80), WGS identified DUX4 rearrangements in 61 patients (see accompanying article [22]), while ERG deletions were identified in a further 19 cases tested by t-NGS and/or MLPA (Supplementary Fig. 4).
Rearrangements of CRLF2 were observed in 53 patients, with P2RY8 (n = 33) or IGH (n = 20) partners, including the nine cases mentioned above, where the fusion co-existed alongside other subtype-defining abnormalities.
ABL-class fusions were observed in 25 patients, including PDGFRB (n = 15), ABL1 (n = 5), CSF1R (n = 4) and ABL2 (n = 1) (Supplementary Fig. 5A). Partner genes were identified in 22 cases, including a novel gene fusion, UBTF::CSF1R (Supplementary Fig. 5B).
ZNF384 fusions were found in 37 patients, involving nine different partner genes, including SPI1, which has not previously been reported in B-ALL (Supplementary Fig. 6).
New genomic subtypes and outcome
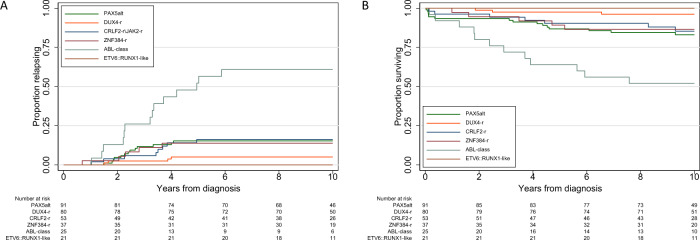
The 10-year RR and OS for the 741 patients with B-other-ALL in this study were 13% (95% CI 11–16) and 87% (84–89), respectively (Table 2, Fig. 4). There was no difference in outcome between patients assigned to a B-other-ALL subtype and those with incomplete or inconclusive testing (p-values: RR = 0.6, EFS = 0.6, OS = 0.7). Patients with DUX4-r had a lower RR (5%) and improved OS (96%) compared with other subtypes (hazard ratio (HR) for relapse = 0.28 (95% CI 0.10–0.79), p = 0.016; HR for death = 0.22 (0.07–0.72), p = 0.012). In addition, there were no relapses or deaths reported among ETV6::RUNX1-like patients. Patients with ABL-class fusions were associated with an inferior outcome compared to other subtypes (HR for relapse = 7.10 (3.79–13.27), p < 0.001; HR for death = 5.35 (2.77–10.36), p < 0.001). PAX5alt, CRLF2-r, and ZNF384-r patients had outcomes similar to B-other-ALL overall. Investigation of different abnormalities within the PAX5alt subtype revealed variation in RR, but none were significant (p-values all > 0.4).
Table 2.
10 year survival rates for 741 patients with B-other-ALL treated on UKALL2003 stratified by genomic subtype.
| Survival rates at 10 years | ||||
|---|---|---|---|---|
| Genomic Subtype | Cases (%) | Relapse | Event | Overall |
| Total B-other-ALL cohort | 741 (100) | 13% (11–16) | 82% (79–85) | 87% (84–89) |
| Unclassifieda | 390 (53) | 13% (10–17) | 82% (78–86) | 87% (83–90) |
| Classifiedb | 351 (47) | 14% (11–18) | 82% (77–85) | 86% (83–90) |
| PAX5altc | 91 (26) | 15% (9–25) | 74% (64–82) | 83% (73–89) |
| PAX5alt | ||||
| PAX5-ITD | 12 (13) | 18% (5–55) | 75% (64–82) | 92% (54–99) |
| PAX5 mutation | 11 (12) | 10% (1–53) | 73% (37–90) | 91% (51–99) |
| PAX5 fusion | 22 (24) | 24% (11–48) | 73% (49–87) | 77% (54–90) |
| dic(9;12) | 11 (12) | No relapses | 70% (32–89) | 80% (39–95) |
| dic(9;20) | 27 (30) | 12% (4–33) | 78% (57–89) | 84% (64–94) |
| Other | 8 (9) | 25% (7–69) | 75% (31–93) | 75% (31–93) |
| DUX4-rd | 80 (23) | 5% (2–13) | 95% (87–98) | 96% (89–99) |
| CRLF2-rc,e | 53 (14) | 16% (8–30) | 77% (63–86) | 85% (71–92) |
| ZNF384-rd | 37 (9) | 14% (6–30) | 81% (63–90) | 86% (71–94) |
| ABL-class | 25 (7) | 61% (42–80) | 36% (18–54) | 52% (31–69) |
| ETV6::RUNX1-likee | 21 (5) | No relapses | No Events | No Deaths |
| ZEB2/CEBPd | 12 (3) | 17% (4–52) | 83% (48–96) | 83% (48–96) |
| PAX5 P80R | 11 (3) | 10% (1–53) | 82% (45–95) | 82% (45–95) |
| IGH::ID4 | 10 (3) | No relapses | 90% (47–99) | 90% (47–99) |
| MEF2D-r | 6 (2) | No relapses | No Events | No Deaths |
| NUTM1-r | 5(1) | No relapses | No Events | No Deaths |
| IKZF1 N159Y | 4 (1) | No relapses | No Events | No Deaths |
| JAK2 fusion | 4 (1) | 25% (4–87) | 75% (13–96) | 75% (13–96) |
| IGH::IL3 | 1 (<1) | No relapses | No Events | No Deaths |
| BCL2/MYC | 1 (<1) | No relapses | No Events | No Deaths |
The italic values represent the different subsets of the PAX5Alt subtype underneath the values for the entire PAX5Alt subtype.
aCases where testing was either incomplete or inconclusive.
bCases with abnormalities detected by FISH, SNP array, MLPA, t-NGS or WGS which could be used to unequivocally assign them to one or more of the genomic subtypes listed.
cSeven cases had abnormalities consistent with both CRLF2-r and PAX5alt subtypes and were included in both subtypes for survival analysis.
dOne case had both IGH::DUX4 and ZEB2/CEPB and was included in both subtypes for survival analysis.
eOne case had both TCF3::ZNF384 and P2RY8::CRLF2 fusion and one case had P2RY8::CRLF2 and ETV6::RUNX1-like; both were included in both subtypes for survival analysis.
Fig. 4. Relapse rate and overall survival of B-other-ALL subtypes.
Kaplan-Meier survival curves showing the relapse rate (A) and overall survival (B) for patients treated on UKALL2003 classified into the six most prevalent genomic subtypes of B-other-ALL.
New genomic subtypes and additional risk factors
CNA identified from the P335 MLPA kit varied according to subtype, with several associations reaching statistical significance (Table 3). Previous studies have reported that specific copy number profiles have prognostic relevance. We investigated the distribution of the copy number profiles, UKALL-CNA [36, 39] or IKZF1plus [40]. among B-other-ALL (Table 3). Patients with PAX5alt and CRLF2-r had an increased frequency of the poor-risk IKZF1plus profile and were more likely to be classified as UKALL-CNA intermediate/poor-risk (IR/PR) compared to patients in other subtypes. ZNF384-r and DUX4-r cases were more likely to have a UKALL-CNA good risk (GR) profile. Unsurprisingly, patients with DUX4-r were less likely to have an IKZF1plus profile, given the association with ERG deletions.
Table 3.
Distribution of key copy number alterations by genomic subtype.
| PAX5alt | DUX4-r | CRLF2-r/JAK2-r | ZNF384-r | ABL-class | ETV6::RUNX1-like | |
|---|---|---|---|---|---|---|
| N = 91 | N = 80 | N = 53 | N = 37 | N = 25 | N = 21 | |
| IKZF1 deletion | ||||||
| No | 54 (70%) | 55 (71%) | 16 (46%) | 28 (93%) † | 9 (60%) | 10 (59%) |
| Yes | 23 (30%) * | 22 (29%) | 19 (54%) ‡ | 2 (7%) | 6 (40%) | 7 (41%) |
| PAX5 deletion | ||||||
| No | 20 (26%) | 70 (91%) ‡ | 20 (57%) | 29 (97%) ‡ | 10 (67%) | 12 (71%) |
| Yes | 57 (74%) ‡ | 7 (9%) | 15 (43%) | 1 (3%) | 5 (33%) | 5 (29%) |
| CDKN2A/B deletion | ||||||
| No | 5 (6%) | 55 (71%) ‡ | 19 (54%) | 24 (80%) † | 11 (73%) | 15 (88) † |
| Yes | 72 (94%) ‡ | 22 (29%) | 16 (46%) | 6 (20%) | 4 (27%) | 2 (12%) |
| BTG1 deletion | ||||||
| No | 75 (97%) | 76 (99%) | 28 (80%) | 28 (93%) | 12 (80%) | 14 (82%) |
| Yes | 2 (3%) | 1 (1%) | 7 (20%) ‡ | 2 (7%) | 3 (20%) | 3 (18%) |
| ETV6 deletion | ||||||
| No | 64 (83%) | 71 (92%) † | 29 (83%) | 20 (67%) | 13 (87%) | 5 (29%) |
| Yes | 13 (17%) | 6 (8%) | 6 (17%) | 10 (33%) | 2 (13%) | 12 (71%) ‡ |
| EBF1 deletion | ||||||
| No | 77 (100%) | 77 (100%) | 31 (89%) | 29 (97%) | 15 (100%) | 17 (100%) |
| Yes | 0 (0%) | 0 (0%) | 4 (11%) ‡ | 1 (3%) | 0 (0%) | 0 (0%) |
| RB1 deletion | ||||||
| No | 74 (96%) | 77 (100%)† | 31 (89%) | 25 (83%) | 14 (93%) | 16 (94%) |
| Yes | 3 (4%) | 0 (0%) | 4 (11%) | 5 (17%) | 1 (7%) | 1 (6%) |
| IKZF1plus | ||||||
| No | 54 (70%) | 76 (99%) ‡ | 20 (57%) | 29 (97%) | 11 (73%) | 15 (88%) |
| Yes | 23 (30%) *‡ | 1 (1%) | 15 (43%) ‡ | 1 (3%) | 4 (27%) | 2 (12%) |
| UKALL CNA profile | ||||||
| GR | 5 (6%) | 40 (52%) ‡ | 0 (0%) | 20 (67%) ‡ | 7 (47%) | 7 (41%) |
| IR/PR | 72 (94%) ‡ | 37 (48%) | 35 (100%) ‡ | 10 (33%) | 8 (53%) | 10 (59%) |
The UKALL-CNA or IKZF1plus profiles are based on the genes included within the P335-IKZF1 MLPA kit. Briefly, the UKALL-CNA profile classifies patients as good risk (CNA-GR), if they have no deletions among the genes tested for, isolated deletions of ETV6, PAX5, BTG1 or ETV6 with a single additional deletion of BTG1, PAX5, or CDKN2A/B. All other profile combinations are classified as intermediate/poor-risk (CNA IR/PR) [36]. The IKZF1plus profile defines patients with an IKZF1 deletion and at least one additional deletion of PAX5, CDKN2A/B, or PAR1, in the absence of an ERG deletion, as poor-risk [40].
‡significant increase compared to other genomic subtype, p < 0.001.
†significant increase compared to other genomic subtype, p < 0.01, p-values generated by Chi-squared testing.
*All 23 patients in the PAX5alt subtype who had an IKZF1 deletion had the IKZF1plus profile.
Where numbers permitted, we assessed whether the presence of deletions or copy number profiles modulated the outcome of patients within subtypes (Supplementary Table 11). Survival rates for patients with PAX5alt and CRLF2-r, who also had an IKZF1 deletion or IKZF1plus profile, appeared inferior, although log rank tests revealed borderline p values, suggesting that they were not the main drivers of poor outcome. In contrast, at end of induction (EOI), high levels of MRD were strongly associated with increased RR and lower EFS within the PAX5alt subtype. The UKALL-CNA profile was too tightly correlated with many subtypes to be assessable but was linked to outcome in ZNF384-r cases. Further analysis of PAX5alt and CRLF2-r revealed no difference in outcome within each subtype, according to NCI risk group.
As other studies [3, 41, 42], we observed that the prognostic impact of DUX4-r was equivalent to ETV6::RUNX1 and high hyperdiploidy (Supplementary Fig. 7A–C), despite its association with high-risk baseline features (male sex, older age, higher white-cell-count), resulting in twice as many patients categorised as NCI high-risk (Supplementary Table 12). Although all DUX4-r patients achieved complete remission on protocol therapy, 11/80 (14%) were slow early responders and 41/75 (55%) were MRD high-risk. There was no difference in the proportion of MRD high-risk patients by NCI risk group [20/39 (51%) v 21/36 (58%), p = 0.5]. In our cohort, only four DUX4-r patients relapsed (Supplementary Fig. 7D). Although these relapse patients were MRD high-risk, the difference was not significant (4/41 v 0/34, p = 0.06), while among 21 cases with MRD > 0.1%, only two relapsed. Notably, only 1/21 DUX4-r patients with an IKZF1 deletion relapsed compared with 3/54 without an IKZF1 deletion (p = 0.9). In contrast to some studies [43], the presence of an ERG abnormality was not linked to prognosis: within the WGS cohort, 2/45 v 1/16 patients with/without an ERG abnormality relapsed (p = 0.8). There was no evidence that RR varied by treatment regimen (on A/B/C, 1/29, 2/27, and 1/24, respectively, relapsed, p = 0.8). Among MRD high-risk patients, the relapse rate among those treated on regimen A/B was 3/18, not significantly higher than those treated on regimen C (1/23, p = 0.2). The long-term outcome of DUX4-r patients was excellent, with 72/77 (94%) surviving >7 years.
Discussion
In this study, we have comprehensively refined the classification of B-other-ALL by integrating NGS-based techniques with those that we have previously reported [12, 33]. We have demonstrated the value of incorporating both WGS and t-NGS for improved identification of a range of abnormalities associated with emerging subtypes, particularly, for detection of subtype-defining mutations.
While our previous approaches were highly successful in classification of B-other-ALL, sequencing-based methods provided valuable additional information in many cases. The increased sensitivity of NGS identified the full range of secondary and co-operating abnormalities, for example, ERG abnormalities in DUX4-r patients. Notably, NGS identified fusion partners, whereas FISH detected only the rearrangement of the relevant “hub” gene, such as ZNF384 or PDGFRB. These data may be important in future collaborative studies, to discern clinical associations for specific fusion genes within subtypes. For example, a recent international collaboration collected data from 218 patients with ZNF384-r and showed EP300::ZNF384 to be associated with a lower risk of relapse compared to other ZNF384 fusions [44].
NGS approaches were particularly informative in defining the genomic abnormalities characteristic of two subtypes, PAX5alt and ETV6::RUNX1-like, previously identified from gene expression profiling. Both subtypes are associated with a variety of abnormalities, which differ between patients and occasionally overlap with other subtypes, rendering them difficult to define by standard-of-care techniques. Building on our recent WGS study [22], here we have demonstrated that NGS can reliably detect these subtypes prospectively within a diagnostic setting without the need for expression profiling.
Neither DUX4 nor MEF2D were included in the t-NGS kit, as they were unknown at the time of design, thus highlighting the importance of flexibility when choosing tools for diagnostic testing. We were able to screen for MEF2D rearrangements by FISH [12], however, accurate DUX4-r identification was only possible using WGS with a bespoke analysis pipeline [22]. It remains to be determined whether standard PCR testing or t-NGS with a similar customised pipeline can reliably identify DUX4-r. In our recent WGS study, the occurrence of an ERG abnormality was pathognomonic of the DUX4-r subtype, although only present in 68% of cases [22], thus reliance on ERG deletion detection as a surrogate marker of the DUX4-r subtype would miss > 30% cases.
Due to the relatively small numbers of previously published cases, the true prognostic impact of these subtypes remains unresolved. The excellent long-term survival for DUX4-r patients in this study has extended the observations made by others, reporting high 5-year survival rates [3, 41, 42]. There is growing evidence that DUX4-r patients have low RR; possibly linked to the increased therapy that they receive based on EOI MRD [41, 42]. Compared to patients with ETV6::RUNX1 and high hyperdiploidy, DUX4-r patients were more often NCI and MRD high-risk, so more likely to be treated on more intensive treatment regimens (Supplementary Table 12). This phenomenon was mentioned in previous DUX4-r/ERG deletion studies [3, 41, 43, 45, 46], raising the question as to whether their excellent outcome was due to intensified treatment or that DUX4-r is an intrinsically chemo-sensitive good-risk subtype. Here we have shown no evidence that relapse is linked to therapy. Moreover, due to their long-term excellent outcome [47, 48], it is reasonable to consider these patients as cured.
It is now widely recognised that patients with ABL class-fusions not treated with tyrosine kinase inhibitors have a very poor prognosis [21, 49], as further reinforced here. No relapses or deaths were observed among 21 patients with ETV6::RUNX1-like-ALL after a median follow-up of 10 years. This excellent outcome differs from 22 and 13% 5-year cumulative incidence of relapse reported for 18 ETV6::RUNX1-like-ALL cases treated on Total Therapy 16 (n = 9) [42] or MS2003/2010 (n = 9) [41], respectively. Although here we primarily used DNA-based techniques to identify the genomic abnormalities associated with this subtype, we confirmed an ETV6::RUNX1-like gene expression signature in six patients by WTS [22]. Only four of the 21 ETV6::RUNX1-like patients received intensive therapy, suggesting that, when treated on UKALL2003, they have an excellent outcome. The outcome of patients with PAX5alt, CRLF2-r or ZNF384-r was very similar to B-other-ALL overall, broadly consistent with other paediatric ALL trial publications [41, 42, 50]. The MS2003/2010 study reported an adverse effect of IKZF1 deletions within the PAX5alt group [41]. Although our results were consistent with their observation, it was eclipsed by the negative effect of MRD. We identified too few patients with PAX5 P80R, IGH::ID4, ZEB2/CEBP, MEF2D-r, NUTM1-r or IKZF1 N159Y to reliably assess outcome. Given their rarity, international collaborative studies are needed to determine their true risk status.
It is evident that accurate classification of B-other-ALL is crucial to the success of future trials, thus access to a range of approaches for their detection is important. Other studies have applied WTS and subsequent cluster analysis to retrospectively classify B-other-ALL [41, 42, 51]. Here, we have chosen DNA-based approaches, for detection of the defining genetic abnormalities. As our associated study demonstrated high concordance between WGS and WTS, we are confident that our genomic approaches, specifically WGS, can accurately and prospectively classify B-other-ALL [22].
Our methodology has a number of advantages: while WTS requires a large reference cohort, these DNA-based techniques can be performed on individual or small numbers of cases. Both WGS and WTS are costly, requiring sophisticated bioinformatics pipelines for analysis, which will be prohibitive for many low and middle-income countries. As this study has demonstrated a high level of concordance between WGS and both t-NGS and standard techniques, although developed countries may adopt WGS as the predominant diagnostic test for ALL in future, laboratories with limited resources may choose standard techniques to screen only for those abnormalities linked to treatment implications. For example, in UK trials, we have previously shown that FISH testing for ABL-class fusions in patients with refractory ALL is highly effective for identification of the majority of patients [21, 52]. Choice of diagnostic testing will also be driven by the preferences of different centres and be dependent on individual trial requirements.
In conclusion, we have successfully classified 351 patients with B-other-ALL into key genomic subtypes, using both NGS and standard techniques; thereby providing screening options to suit all resource levels and trial protocols. As this study was based on a single clinical trial, we were able to provide robust and clinically useful prognostic information on six recently reported genomic subtypes.
Supplementary information
Acknowledgements
The authors thank member laboratories of the UK Cancer Cytogenetic Group for providing cytogenetic data and material. Primary childhood leukaemia samples used in this study were provided by the Blood Cancer UK Childhood Leukaemia Cell Bank. This work was funded by the Blood Cancer UK programme grant - 15036.
Author contributions
Conception and Design: SLR, CS, AVM, CJH. Generation, collection and assembly of data: CS, SLR, REC, EW, ZH, ZK, JM, AC. Hinchliffe, RB, EB, MB. Data analysis and interpretation: SLR, EB, CS, AE, REC, EW, ZH, JM, JFP, MTR, DRB, LJR, AV, CJH, AVM. Financial support: CJH, AVM. Administrative support: CJH, AVM. Provision of study materials or patients: AV, AVM. Manuscript writing: CS, REC, AVM, CJH. Final approval of manuscript: All authors.
Data availability
DNA and RNA Sequencing data have been deposited in the European Genome-phenome Archive (EGA) under the Accession Code EGAS00001006863. Alternatively these data will be made available upon request from Dr. Sarra Ryan (sarra.ryan@newcastle.ac.uk) or Prof Christine Harrison (christine.harrison@newcastle.ac.uk).
Competing interests
MTR, DRB, JFP, and ZK are employees of Illumina, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Claire Schwab, Ruth E. Cranston and Sarra L. Ryan.
These authors jointly supervised this work: Anthony V. Moorman, Christine J. Harrison.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01799-4.
References
- 1.Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: Results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–38. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 2.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci USA. 2018;115:E11711–20. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwab C, Harrison CJ. Advances in B-cell precursor acute lymphoblastic leukemia genomics. Hemasphere. 2018;2:e53. doi: 10.1097/HS9.0000000000000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102:118–29. doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. doi: 10.1038/ncomms13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohki K, Kiyokawa N, Saito Y, Hirabayashi S, Nakabayashi K, Ichikawa H, et al. Clinical and molecular characteristics of MEF2D fusion-positive B-cell precursor acute lymphoblastic leukemia in childhood, including a novel translocation resulting in MEF2D-HNRNPH1 gene fusion. Haematologica. 2019;104:128–37. doi: 10.3324/haematol.2017.186320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaliova M, Kotrova M, Bresolin S, Stuchly J, Stary J, Hrusak O, et al. ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype. Genes Chromosomes Cancer. 2017;56:608–16. doi: 10.1002/gcc.22464. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48:569–74. doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- 11.Roberts KG, Pei D, Campana D, Payne-Turner D, Li Y, Cheng C, et al. Outcomes of children with BCR-ABL1-like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol. 2014;32:3012–20. doi: 10.1200/JCO.2014.55.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwab CJ, Murdy D, Butler E, Enshaei A, Winterman E, Cranston RE, et al. Genetic characterisation of childhood B-other-acute lymphoblastic leukaemia in UK patients by fluorescence in situ hybridisation and Multiplex Ligation-dependent Probe Amplification. Br J Haematol. 2022;196:753–63. doi: 10.1111/bjh.17869. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48:1481–9. doi: 10.1038/ng.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reshmi SC, Harvey RC, Roberts KG, Stonerock E, Smith A, Jenkins H, et al. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children’s Oncology Group. Blood. 2017;129:3352–61. doi: 10.1182/blood-2016-12-758979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boer JM, Steeghs EM, Marchante JR, Boeree A, Beaudoin JJ, Beverloo HB, et al. Tyrosine kinase fusion genes in pediatric BCR-ABL1-like acute lymphoblastic leukemia. Oncotarget. 2017;8:4618–28. doi: 10.18632/oncotarget.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Veer A, Waanders E, Pieters R, Willemse ME, Van Reijmersdal SV, Russell LJ, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122:2622–9. doi: 10.1182/blood-2012-10-462358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwab C, Ryan SL, Chilton L, Elliott A, Murray J, Richardson S, et al. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): genetic profile and clinical implications. Blood. 2016;127:2214–8. doi: 10.1182/blood-2015-09-670166. [DOI] [PubMed] [Google Scholar]
- 20.Schwab C, Roberts K, Boer JM, Gohring G, Steinemann D, Vora A, et al. SSBP2-CSF1R is a recurrent fusion in B-lineage acute lymphoblastic leukemia with diverse genetic presentation and variable outcome. Blood. 2021;137:1835–8. doi: 10.1182/blood.2020008536. [DOI] [PubMed] [Google Scholar]
- 21.Moorman AV, Schwab C, Winterman E, Hancock J, Castleton A, Cummins M, et al. Adjuvant tyrosine kinase inhibitor therapy improves outcome for children and adolescents with acute lymphoblastic leukaemia who have an ABL-class fusion. Br J Haematol. 2020;191:844–51. doi: 10.1111/bjh.17093. [DOI] [PubMed] [Google Scholar]
- 22.Ryan SLPJ, Kingsbury Z, Schwab CJ, James T, Polonen P, Butler E, et al. Whole genome sequencing provides comprehensive diagnostic genetic testing in childhood B-cell acute lymphoblastic leukaemia (B-ALL). Leukemia 2022. [DOI] [PMC free article] [PubMed]
- 23.Vora A, Goulden N, Mitchell C, Hancock J, Hough R, Rowntree C, et al. Augmented post-remission therapy for a minimal residual disease-defined high-risk subgroup of children and young people with clinical standard-risk and intermediate-risk acute lymphoblastic leukaemia (UKALL 2003): A randomised controlled trial. Lancet Oncol. 2014;15:809–18. doi: 10.1016/S1470-2045(14)70243-8. [DOI] [PubMed] [Google Scholar]
- 24.Vora A, Goulden N, Wade R, Mitchell C, Hancock J, Hough R, et al. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): A randomised controlled trial. Lancet Oncol. 2013;14:199–209. doi: 10.1016/S1470-2045(12)70600-9. [DOI] [PubMed] [Google Scholar]
- 25.Van der Auwera GA, O’Connor BD, Safari aORMC. Genomics in the cloud: using Docker, GATK, and WDL in Terra, First edition. edn. Sebastopol, CA: O’Reilly Media; 2020. [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picard Toolkit. Broad Institute, GitHub Repository. https://broadinstitute.github.io/picard/; Broad Institute. 2019
- 28.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2018;201178.
- 30.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell LJ, Enshaei A, Jones L, Erhorn A, Masic D, Bentley H, et al. IGH@ translocations are prevalent in teenagers and young adults with acute lymphoblastic leukemia and are associated with a poor outcome. J Clin Oncol. 2014;32:1453–62. doi: 10.1200/JCO.2013.51.3242. [DOI] [PubMed] [Google Scholar]
- 34.Konn ZJ, Wright SL, Barber KE, Harrison CJ. Fluorescence In situ hybridization (FISH) as a tool for the detection of significant chromosomal abnormalities in childhood leukaemia. Methods Mol Biol. 2009;538:29–55. doi: 10.1007/978-1-59745-418-6_3. [DOI] [PubMed] [Google Scholar]
- 35.Schwab CJ, Jones LR, Morrison H, Ryan SL, Yigittop H, Schouten JP, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:1104–13. doi: 10.1002/gcc.20818. [DOI] [PubMed] [Google Scholar]
- 36.Moorman AV, Enshaei A, Schwab C, Wade R, Chilton L, Elliott A, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–44. doi: 10.1182/blood-2014-03-562918. [DOI] [PubMed] [Google Scholar]
- 37.Creasey T, Enshaei A, Nebral K, Schwab C, Watts K, Cuthbert G, et al. Single nucleotide polymorphism array-based signature of low hypodiploidy in acute lymphoblastic leukemia. Genes, chromosomes cancer. 2021;60:604–15. doi: 10.1002/gcc.22956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98:1081–8. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamadeh L, Enshaei A, Schwab C, Alonso CN, Attarbaschi A, Barbany G, et al. Validation of the United Kingdom copy-number alteration classifier in 3239 children with B-cell precursor ALL. Blood Adv. 2019;3:148–57. doi: 10.1182/bloodadvances.2018025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanulla M, Dagdan E, Zaliova M, Moricke A, Palmi C, Cazzaniga G, et al. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol. 2018;36:1240–9. doi: 10.1200/JCO.2017.74.3617. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Lee SHR, Chin WHN, Lu Y, Jiang N, Lim EH, et al. Distinct clinical characteristics of DUX4- and PAX5-altered childhood B-lymphoblastic leukemia. Blood Adv. 2021;5:5226–38. doi: 10.1182/bloodadvances.2021004895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeha S, Choi J, Roberts KG, Pei D, Coustan-Smith E, Inaba H, et al. Clinical significance of novel subtypes of acute lymphoblastic leukemia in the context of minimal residual disease–directed therapy. Blood Cancer Discov. 2021;2:326–37. doi: 10.1158/2643-3230.BCD-20-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaliova M, Potuckova E, Hovorkova L, Musilova A, Winkowska L, Fiser K, et al. ERG deletions in childhood acute lymphoblastic leukemia with DUX4 rearrangements are mostly polyclonal, prognostically relevant and their detection rate strongly depends on screening method sensitivity. Haematologica. 2019;104:1407–16. doi: 10.3324/haematol.2018.204487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirabayashi S, Butler ER, Ohki K, Kiyokawa N, Bergmann AK, Moricke A, et al. Clinical characteristics and outcomes of B-ALL with ZNF384 rearrangements: A retrospective analysis by the Ponte di Legno Childhood ALL Working Group. Leukemia. 2021;35:3272–7. doi: 10.1038/s41375-021-01199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clappier E, Auclerc MF, Rapion J, Bakkus M, Caye A, Khemiri A, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28:70–7. doi: 10.1038/leu.2013.277. [DOI] [PubMed] [Google Scholar]
- 46.Zaliova M, Zimmermannova O, Dorge P, Eckert C, Moricke A, Zimmermann M, et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia. 2014;28:182–5. doi: 10.1038/leu.2013.282. [DOI] [PubMed] [Google Scholar]
- 47.Pui CH, Pei D, Campana D, Cheng C, Sandlund JT, Bowman WP, et al. A revised definition for cure of childhood acute lymphoblastic leukemia. Leukemia. 2014;28:2336–43. doi: 10.1038/leu.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorman AV, Antony G, Wade R, Butler ER, Enshaei A, Harrison CJ, et al. Time to cure for childhood and young adult acute lymphoblastic leukemia is independent of early risk factors: Long-term follow-up of the UKALL2003 Trial. J Clin Oncol. 2022; JCO2200245. [DOI] [PubMed]
- 49.den Boer ML, Cario G, Moorman AV, Boer JM, de Groot-Kruseman HA, Fiocco M, et al. Outcomes of paediatric patients with B-cell acute lymphocytic leukaemia with ABL-class fusion in the pre-tyrosine-kinase inhibitor era: A multicentre, retrospective, cohort study. Lancet Haematol. 2021;8:e55–66. doi: 10.1016/S2352-3026(20)30353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Attarbaschi A, Morak M, Cario G, Cazzaniga G, Ensor HM, te Kronnie T, et al. Treatment outcome of CRLF2-rearranged childhood acute lymphoblastic leukaemia: A comparative analysis of the AIEOP-BFM and UK NCRI-CCLG study groups. Br J Haematol. 2012;158:772–7. doi: 10.1111/j.1365-2141.2012.09221.x. [DOI] [PubMed] [Google Scholar]
- 51.Brady SW, Roberts KG, Gu Z, Shi L, Pounds S, Pei D, et al. The genomic landscape of pediatric acute lymphoblastic leukemia. Nat Genet. 2022;54:1376–89. doi: 10.1038/s41588-022-01159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Connor D, Moorman AV, Wade R, Hancock J, Tan RM, Bartram J, et al. Use of minimal residual disease assessment to redefine induction failure in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2017;35:660–7. doi: 10.1200/JCO.2016.69.6278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA and RNA Sequencing data have been deposited in the European Genome-phenome Archive (EGA) under the Accession Code EGAS00001006863. Alternatively these data will be made available upon request from Dr. Sarra Ryan (sarra.ryan@newcastle.ac.uk) or Prof Christine Harrison (christine.harrison@newcastle.ac.uk).