Abstract
Purpose:
To examine variation in genetic testing between neonatal intensive care units (NICUs) across hospitals over time.
Methods:
We performed a multicenter large-scale retrospective cohort study using NICU discharge data from the Pediatric Hospital Information System Database between 2016 and 2021. We analyzed variation in the percentage of NICU patients who had any genetic testing between hospitals and over time. We used a multivariable multilevel logistic regression model to investigate the potential association of patient characteristics and genetic testing.
Results:
The final analysis included 207,228 neonates from 38 hospitals. Overall, 13% of patients had at least one genetic test sent, though this varied from 4% to 50% across hospitals. Over the study period, the proportion of patients tested increased, with the increase disproportionately borne by hospitals already testing high proportions of patients. On average, patients who received genetic testing had higher illness severity. Controlling for severity, however, only minimally reduced the degree of hospital-level variation in genetic testing.
Conclusion:
The percentage of NICU patients who undergo genetic testing varies among hospitals, and increasingly so over time. Variation is largely unexplained by differences in severity between hospitals. The degree of variation suggests that clearer guidelines for NICU genetic testing are warranted.
Keywords: Neonatology, genetic testing
BACKGROUND
The neonatal intensive care unit (NICU) has been a launch point for genetic testing.1,2 Many conditions warranting NICU admission have an underlying genetic etiology, and 9% of all level IV NICU patients received a genetic diagnosis in a recent study.3 Compared to patients with other types of disease, NICU patients with genetic diseases have longer, costlier admissions that more frequently end in death.3–6 Early identification of genetic disease promises to improve clinical care and inform decision making for this high acuity population.7 Yet, the complex and frequently uncertain information produced by modern genetic tests also has the potential to be misunderstood or misused by both clinicians and parents, particularly if not accompanied by sufficient support from clinical geneticists and genetic counselors.8–10
No guidelines exist to standardize genetic testing practices between NICUs. As such, hospitals are left to develop their own testing policies and practices. Genetic testing has evolved rapidly over the past two decades, but the absence of standardization means that progress may be unevenly distributed between hospitals.11 Insurance coverage of inpatient genetic testing varies between states, which likely increases variation in practice.12 Generally, extreme variation is considered a marker of poor quality care.15,16 The degree to which genetic testing practices vary between NICUs has not been studied.
Believing that describing genetic testing practices would be a first step toward assessing the need for standardization, we used data from NICU admissions at children’s hospitals in the Pediatric Hospital Information System (PHIS) Database to analyze variation in genetic testing among hospitals over time. PHIS compiles patient-level care data from 49 freestanding children’s hospitals including diagnostic codes, demographic and payer information, and billing data.
METHODS
We performed a retrospective multi-center large-scale cohort study using PHIS data from NICU discharges between January 2016 and December 2021. We excluded incomplete patient entries and hospitals with missing years of data for the study period.
Measurements
We extracted data on length of stay, admission year, gestational age, in-hospital mortality, biologic sex, race, ICD diagnosis codes, lab tests billed, binary flags signifying presence or absence of any complex chronic conditions (CCC)17 or congenital anomalies, All Patient Refined Diagnosis Related Groups (APR-DRG) severity levels, and whether a patient was inborn or transferred to the hospital. A medical geneticist (K.T.W) curated a list of 29 Current Procedural Terminology lab codes that denote genetic testing (Supplemental Table 1). We similarly refined a list of genetic diagnoses. We also extracted data from each hospital’s website on the number of clinical geneticists (MD or equivalent) working at the institution as of June 2022.
Statistical Analysis
The primary outcome was a binary indicator of any genetic testing during the hospital encounter. We used t-tests, chi-squared tests, and ANOVA tests as appropriate to compare patients that did and did not receive genetic testing. We calculated the percentage of patients who received genetic testing at each hospital for each year and across the study period. We performed linear regression analysis and graphed this percentage over time by hospital and looked separately at the trends for the five hospitals with the highest overall testing over the study period, heretofore “high testing hospitals,” and lowest overall testing percentages, or “low testing hospitals.” We also analyzed hospital-level variation in the mean number of genetic tests sent per patient.
We used a multivariable multilevel logistic regression model to investigate the potential association of patient or hospital characteristics with genetic testing. We included hospital as a random effect and then sought to assess variation attributable to the hospital after controlling for various patient characteristics. We ultimately fit a pair of models with differing fixed-effects – the first an intercept-only model, the second with patient-level fixed effects. We built the second model using forward stepwise procedures by adding patient- and hospital- level characteristic, including admission year, in-hospital mortality, CCC, congenital anomalies, severity level, gestational age, biologic sex, race, length of stay, and admission volume (annual hospital mean). We included characteristics that were statistically significant predictors (individually) at level α=.20 in a multivariable regression model. In the final multivariable model, we retained only those characteristics with P-values less than 0.05. We calculated the intra-class correlation coefficient (ICC) statistic for both models to measure the proportion of variation attributable to the random effects, in this case hospital. We obtained the Best Linear Unbiased Predictors (BLUPs) to estimate and graphically display the predicted effect each hospital had on the probability of testing. Using the final model, we made in-sample predictions of the probability of genetic testing and plotted the mean and standard deviation by hospital. We performed all data analysis using Stata version 16.1 (StataCorp, College Station, TX).
RESULTS
Our final analysis included 188,025 admissions from 38 hospitals (Supplemental Table 2). We excluded 95,878 entries representing 34% of all admissions due to missing data.
Of included admissions, 24,521 (13%) had genetic testing. Over the study period, the percentage of patients with testing increased (Figure 1; β=0.021, CI: 0.011–0.031, P<.001). The top five highest testing hospitals increased at a faster rate (β=0.063, CI: 0.024–0.10 P=.003) and the bottom five hospitals hospital increased at a slower rate (β=0.006, CI: 0.002–0.010, P=.008). Between hospitals, the percentage of patients with genetic testing ranged from 4% to 41% (Figure 2). Among patients who underwent genetic testing, the mean number of tests was 2.28 (SD 1.58), with an interhospital range of 1.14 (SD 0.49) to 3.50 (SD 1.46) (Supplemental Figure 1). Hospitals which tested a greater percentage of patients also sent more tests per patient (R2=0.75, P<.001). The most common tests sent were chromosomal microarray (16,668, 17%), tissue culture for chromosomal analysis (15, 056, 15%), chromosome analysis (13,342, 14%), sanger sequencing (10,588, 11%), and in situ hybridization (7,015, 7%). Over the study period microarrays and sanger sequencing accounted for an increasing proportion of testing. The most common genetic diagnoses among these patients are listed in the appendix (Supplemental Table 3).
Figure 1. Percent of patients who underwent genetic testing over time.
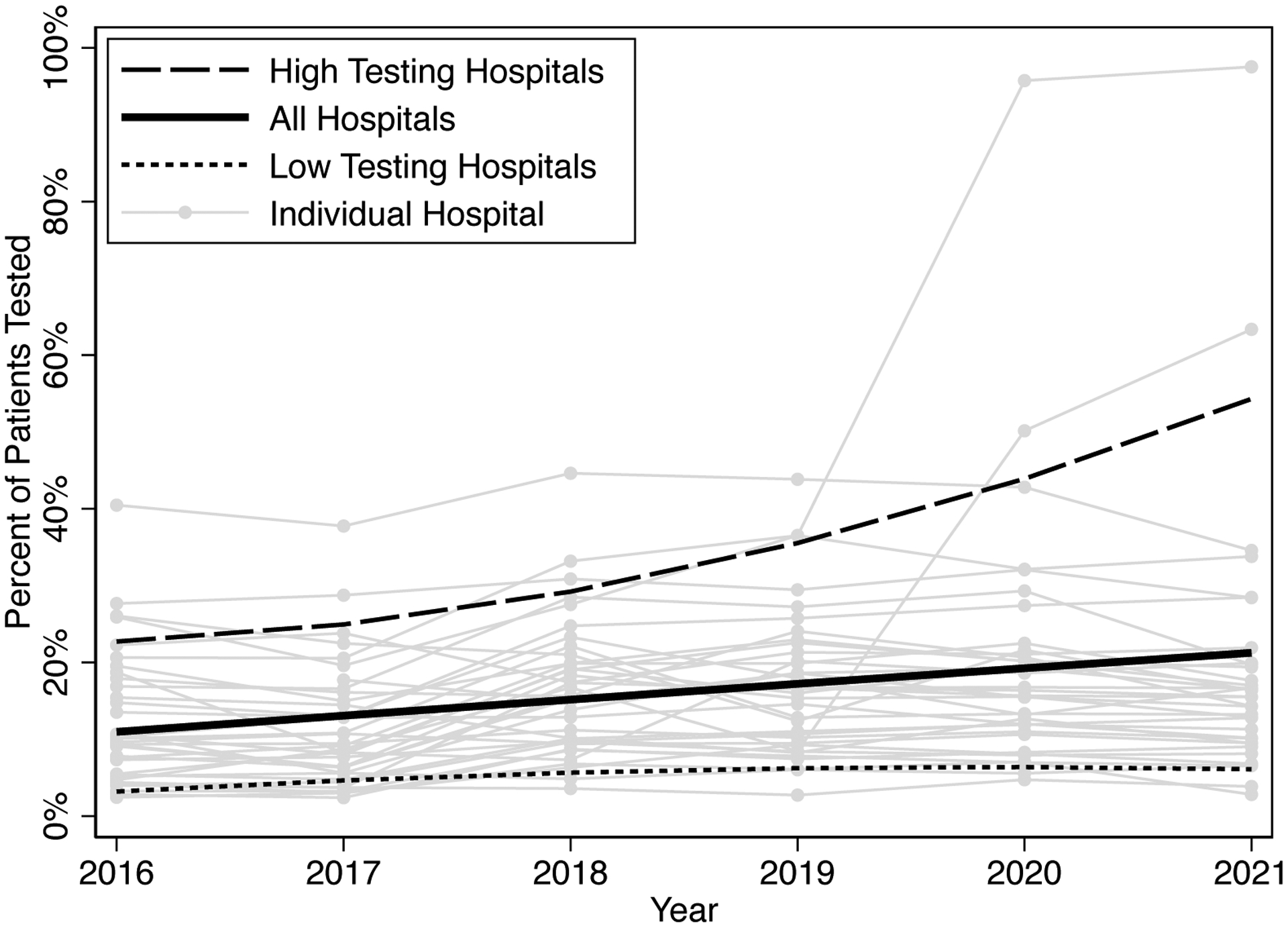
Fitted lines represent fractional polynomial lines for all hospitals, the top five highest testing hospitals, and bottom five lowest testing hospitals.
Figure 2. Unadjusted and adjusted genetic testing percentage by hospital.
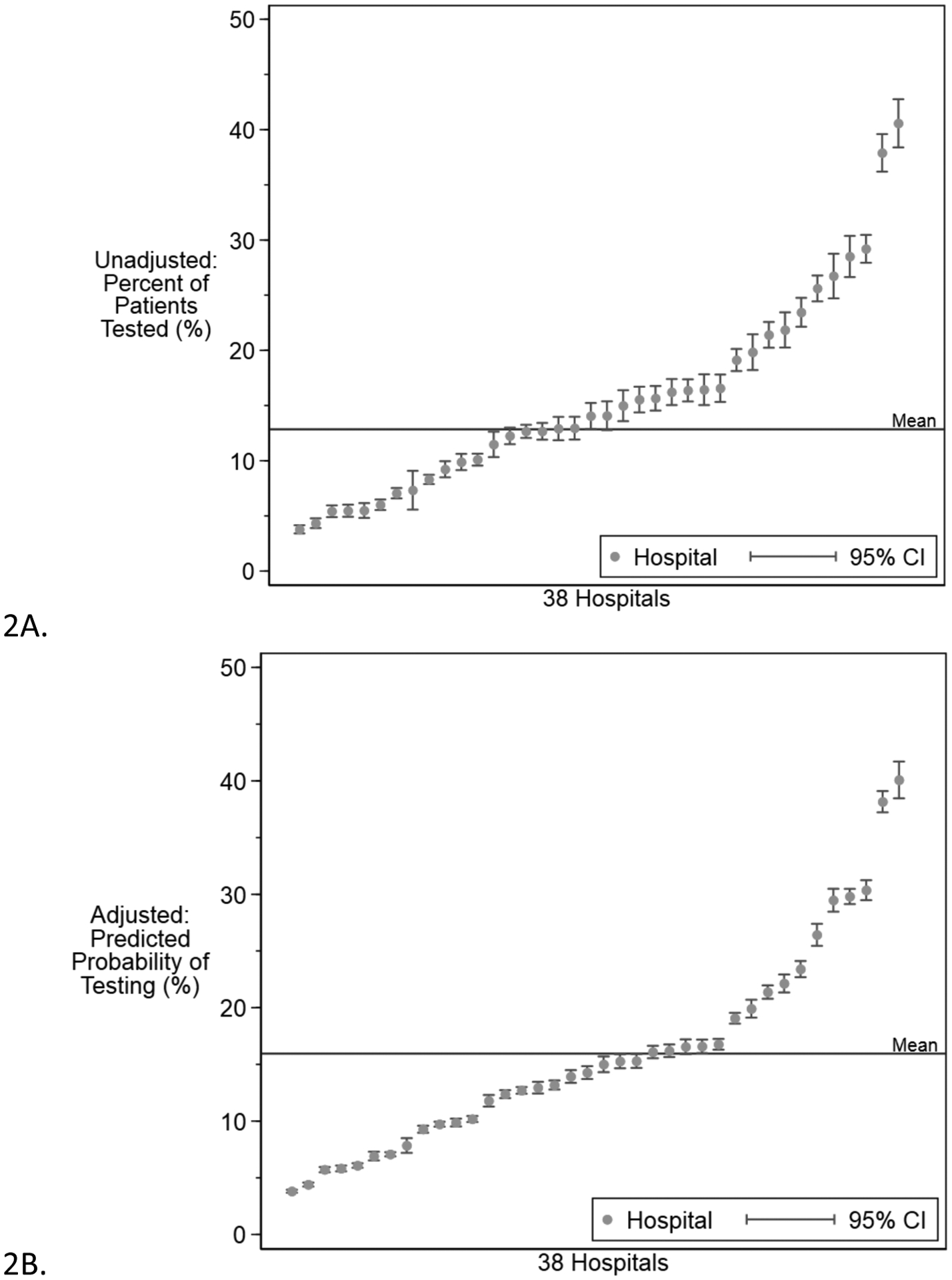
Each point represents a hospital and error bars indicate 95% confidence intervals. Panel 2A shows the actual testing variation. Panel 2B shows the variation in predicted testing rates calculated using best linear unbiased prediction based on the final multivariate regression model.
Compared to patients without genetic testing, patients with testing had higher mortality rate (Supplemental Table 4, 9.6% vs. 2.9%, P<.001), longer length of stay (7.0 weeks vs. 3.2 weeks, P<.001), were more likely to have a CCC (90.6% vs 48.0%, P<.001) and a congenital anomaly (29.4% vs 5.1%, P<.001), and were less likely to be of Black race (18.9% vs 21.4%, P<0.001). Genetic testing was increasingly likely as a patient’s APR-DRG severity level increased (P<.001). Patients admitted later in the study period were more likely to have a genetic test sent (R2=0.070, P<.001).
Consistent with these findings, increases in the following patient-level characteristics were associated with testing: year of admission, gestational age, in-hospital mortality, CCC and congenital anomaly flags, APR-DRG severity level, and length of stay (P<.001 for all). Patients who were Black were less likely and patients of race Other were slightly more likely to have a genetic test sent compared to White patients (P<0.001). Biologic sex was not associated with genetic testing (P=.058). We also individually considered the hospital-level characteristic of admission volume, which was significant (Supplemental Figure 2, P<.001). All significant patient-level factors retained significance while admission volume was no longer significant in the multivariate model. For continuous variables, we explored the relationship between deviance residuals and the patient-level fixed effect, which led us to log-transform length of stay resulting in a better fitting model. The final model included mortality, CCC, congenital anomalies, severity level, gestational age, length of stay (log-transformed), race, and year of admission (Supplemental Table 4). The random effects variance was significantly different from 0 in the intercept-only model (σ2=0.47; 95% CI 0.30 – 0.74) and patient fixed-effects model (σ2=0.35; 95% CI 0.22 – 0.56), justifying the choice of hospital as the random intercept. Compared to the intercept-only model, the multivariate model was associated with a 23% drop in the Akaike’s information criterion (136574 to 105587) and the Likelihood-ratio test comparing models was significant (P<0.001). The proportion of variation in genetic testing that was attributable to hospital as measured by the ICC was 12.3% (95% CI 8.2%–18.1%) in the unadjusted model and 9.3% (95% CI 6.1%–14.0%) in the adjusted model (Figure 2).
In several measures, high testing hospitals and low testing hospitals deviated in opposite directions from the remaining hospitals. High testing hospitals had a higher percentage of transferred patients (69% vs. 16%, P<.001), lower admission volume (Supplemental Figure 2, 644.41 vs. 1652.94, P<.001), and greater average number of clinical geneticists (7.14 vs. 3.93, P<.001).
DISCUSSION
This study of genetic testing in NICUs reveals large and increasing hospital-level variation that is not explained by patient-level characteristics. This suggests that if the same complex patient was born at different hospitals, their likelihood of receiving genetic testing could vary substantially.
We interpret these findings cautiously for three main reasons. First, coding practices may be inconsistent between hospitals.18 If hospitals vary in how they separate versus bundle billing codes, the total number reported per patient would be less precise. Given the standardization imposed by the PHIS reporting system, this is, however, unlikely to explain the degree of variation we observe. Second, the PHIS records likely exclude genetic testing that is done on a research basis. Research testing, though, is even more likely to differ across hospitals than clinical testing, so we expect this leads to an underestimation of variation. Finally, we are limited by the information in the database assessing whether genetic testing was appropriate. Even some represented variables may not fully capture difference in patient characteristics. For instance, the congenital anomaly flag may be enriched for particular diagnoses for which genetic testing is more or less appropriate at certain hospitals. However, since our model controlled for most factors that have been previously associated with likely genetic disease3,5,6, we believe the majority of the observed variation cannot be explained by differences in the patient characteristics between hospitals.
What might explain the observed variation in testing practices? The correlation between the percentage of testing sent and the mean number of tests sent per patient suggests that some hospitals have a generally more pro-testing culture than others. One reason for variation may be differences in reimbursement policies for inpatient genetic testing, varying ability to conduct genetic tests in house, or hospital administrations’ response to these policies. Also, varied prenatal testing practices and inter-state differences in the breadth of newborn screening may reasonably account for some variation. Hospitals may also be more likely to test because they have greater availability of genetic resources, which is supported by the higher number of geneticists at high-testing hospitals and the general observation that greater capacity in healthcare leads to greater utilization.19 High-testing hospitals seem to be referral centers, with a higher percentage of transferred patients and lower admission volume. Such centers may be culturally more likely to adopt new technology early. Patients may also be appropriately transferred to obtain genetic testing. The lower testing among Black patients raises the concerning possibility that bias may influence decisions about testing, though we cannot definitively exclude other explanations for this finding. Finally, the idiosyncrasy of medical cultural evolution in the absence of clear testing guidelines likely contributes to this variation. Further work is needed to explore the extent to which each of these factors contributes to variation.
This degree of variation for any reason would be troubling and the widening gap adds urgency to the problem. These findings imply that some patients are more likely than others to receive genetic testing based on where they are hospitalized. If testing is benefitting patients, denying patients access to testing based on location represents inequality.7 If testing poses risks, either to patients through misuse or discrimination or to society by creating an unjustified financial burden, the burden is unequally borne.8–10,14 As the types of genetic tests used become more complex, risks increase. A more complete understanding of the benefits and harms associated with genetic testing will facilitate consensus and generation of guidelines for genetic testing, which in turn can begin to reverse the trend of increasing variation. Future work should explore how inequalities in genetic testing may extend to or differ in other hospital and outpatient contexts.
Supplementary Material
Acknowledgments:
This study was supported by T32 HG009496 from the National Human Genome Research Institute (NHGRI, K.P.C.) and K23 HD102589 from the National Institute of Child Health and Human Development (M.H.W.). The work also benefitted from mentorship through the NHGRI Inter-Society Coordinating Committee for Practitioner Education in Genomics scholar program. The funder/sponsor had no role in the reported work. We would like to thank Scott Lorch, MD, MSCE, and Molly Passarella, MS, for their help in obtaining and managing the PHIS data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics Declaration: PHIS datasets are de-identified, so our study is not considered human subjects research by the Children’s Hospital of Philadelphia Institutional Review Board.
Data Availability:
All PHIS data are publicly available.
References
- 1.Powis Z, Hagman KDF, Speare V, et al. Exome sequencing in neonates: diagnostic rates, characteristics, and time to diagnosis. Genet Med. 2018;20(11):1468–1471. doi: 10.1038/gim.2018.11 [DOI] [PubMed] [Google Scholar]
- 2.Friedman JM, Bombard Y, Cornel MC, et al. Genome-wide sequencing in acutely ill infants: genomic medicine’s critical application? Genet Med. 2019;21(2):498–504. doi: 10.1038/s41436-018-0055-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swaggart KA, Swarr DT, Tolusso LK, He H, Dawson DB, Suhrie KR. Making a Genetic Diagnosis in a Level IV Neonatal Intensive Care Unit Population: Who, When, How, and at What Cost? J Pediatr. 2019;213:211–217.e4. doi: 10.1016/j.jpeds.2019.05.054 [DOI] [PubMed] [Google Scholar]
- 4.Gonzaludo N, Belmont JW, Gainullin VG, Taft RJ. Estimating the burden and economic impact of pediatric genetic disease. Genet Med. 2019;21(8):1781–1789. doi: 10.1038/s41436-018-0398-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa T, Scriver CR, Childs B, Opitz JM, Reynolds JF. The effect of Mendelian disease on human health: A measurement. Am J Med Genet. 1985;21(2):231–242. doi: 10.1002/ajmg.1320210205 [DOI] [PubMed] [Google Scholar]
- 6.Wojcik MH, Schwartz TS, Thiele KE, et al. Infant mortality: the contribution of genetic disorders. J Perinatol. 2019;39(12):1611–1619. doi: 10.1038/s41372-019-0451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NICUSeq Study Group Krantz ID, Medne L, et al. Effect of Whole-Genome Sequencing on the Clinical Management of Acutely Ill Infants With Suspected Genetic Disease: A Randomized Clinical Trial. JAMA Pediatr. Published online September 27, 2021. doi: 10.1001/jamapediatrics.2021.3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callahan KP, Flibotte J, Skraban C, et al. Influence of Genetic Information on Neonatologists’ Decisions: A Psychological Experiment. Pediatrics. Published online February 16, 2022:e2021052130. doi: 10.1542/peds.2021-052130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Char DS, Lee SSJ, Magnus D, Cho M. Anticipating uncertainty and irrevocable decisions: provider perspectives on implementing whole-genome sequencing in critically ill children with heart disease. Genet Med. 2018;20(11):1455–1461. doi: 10.1038/gim.2018.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janvier A, Barrington K, Lantos J. Next generation sequencing in neonatology: what does it mean for the next generation? Hum Genet. Published online March 29, 2022. doi: 10.1007/s00439-022-02438-9 [DOI] [PubMed] [Google Scholar]
- 11.Phillips KA, Deverka PA, Hooker GW, Douglas MP. Genetic Test Availability And Spending: Where Are We Now? Where Are We Going? Health Aff Proj Hope. 2018;37(5):710–716. doi: 10.1377/hlthaff.2017.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blue Shield of California Becomes First Health Plan in U.S. to Cover Cost of Rapid Whole Genome Sequencing for Critically Ill Children. RCIGM. Published March 13, 2020. Accessed June 22, 2022. https://radygenomics.org/2020/blue-shield-of-california-becomes-first-health-plan-in-u-s-to-cover-cost-of-rapid-whole-genome-sequencing-for-critically-ill-children/ [Google Scholar]
- 13.P Fishler K, Euteneuer JC, Brunelli L. Ethical Considerations for Equitable Access to Genomic Sequencing for Critically Ill Neonates in the United States. Int J Neonatal Screen. 2022;8(1):22. doi: 10.3390/ijns8010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan KP, Flibotte J, Skraban C, et al. How neonatologists use genetic testing: findings from a national survey. J Perinatol. Published online November 30, 2021:1–2. doi: 10.1038/s41372-021-01283-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason MJ, Moran JW. Understanding and controlling variation in public health. J Public Health Manag Pract JPHMP. 2012;18(1):74–78. doi: 10.1097/PHH.0b013e318233d5eb [DOI] [PubMed] [Google Scholar]
- 16.Fisher ES, Wennberg DE, Stukel TA, Gottlieb DJ, Lucas FL, Pinder ÉL. The Implications of Regional Variations in Medicare Spending. Part 2: Health Outcomes and Satisfaction with Care. Ann Intern Med. 2003;138(4):288–298. doi: 10.7326/0003-4819-138-4-200302180-00007 [DOI] [PubMed] [Google Scholar]
- 17.Feudtner C, Feinstein JA, Zhong W, Hall M, Dai D. Pediatric complex chronic conditions classification system version 2: updated for ICD-10 and complex medical technology dependence and transplantation. BMC Pediatr. 2014;14:199. doi: 10.1186/1471-2431-14-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch JA, Berse B, Dotson WD, Khoury MJ, Coomer N, Kautter J. Utilization of genetic tests: analysis of gene-specific billing in Medicare claims data. Genet Med Off J Am Coll Med Genet. 2017;19(8):890–899. doi: 10.1038/gim.2016.209 [DOI] [PubMed] [Google Scholar]
- 19.Delamater PL, Messina JP, Grady SC, WinklerPrins V, Shortridge AM. Do More Hospital Beds Lead to Higher Hospitalization Rates? A Spatial Examination of Roemer’s Law. PLOS ONE. 2013;8(2):e54900. doi: 10.1371/journal.pone.0054900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman DC, Ganduglia-Cazaban C, Franzini L, et al. Neonatal Intensive Care Variation in Medicaid-Insured Newborns: A Population-Based Study. J Pediatr. 2019;209:44–51.e2. doi: 10.1016/j.jpeds.2019.02.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All PHIS data are publicly available.


