Abstract
The Drosophila neuralized gene shows genetic interactions with Notch, Enhancer of split, and other neurogenic genes and is thought to be involved in cell fate specification in the central nervous system and the mesoderm. In addition, a human homologue of the Drosophila neuralized gene has been described as a potential tumor suppressor gene in malignant astrocytomas. We have isolated a murine homologue of the Drosophila and human Neuralized genes and, in an effort to understand its physiological function, derived mice with a targeted deletion of this gene. Surprisingly, mice homozygous for the introduced mutation do not show aberrant cell fate specifications in the central nervous system or in the developing mesoderm. This is in contrast to mice with targeted deletions in other vertebrate homologues of neurogenic genes such as Notch, Delta, and Cbf-1. Male Neuralized null mice, however, are sterile due to a defect in axoneme organization in the spermatozoa that leads to highly compromised tail movement and sperm immotility. In addition, female Neuralized null animals are defective in the final stages of mammary gland maturation during pregnancy.
The Drosophila neuralized gene has been identified in screens for embryonic lethal mutations with defective lateral specification (5, 24). Hypomorphic neuralized mutations cause neuronal hyperplasia, which is characteristic of neurogenic genes, suggesting that neuralized is involved in cell fate specifications in the neurectoderm. Genetic interaction analysis suggests that neuralized appears to act upstream of Notch and E(spl) since increased Notch or E(spl) expression can partially rescue the neuralized phenotype (9). The Drosophila neuralized gene encodes a C3HC4 ring finger protein and is expressed throughout the ectoderm during neuronal cell fate specification, consistent with its proposed role in this process (6, 33). As with other neurogenic mutants, mesodermal cell fates also appear to be defective in Drosophila carrying a neuralized mutation (4, 27). neuralized mutant files produce an excess number of cells expressing the MyoD homologue, nautilus, at the expense of surrounding mesodermal cells, indicating aberrant cell fate specifications in this tissue (8).
A human homologue of the Drosophila neuralized gene has been identified in a region of chromosome 10q24-25 which shows frequent alterations in malignant astrocytomas (28). Like the Drosophila gene, the human Neuralized gene encodes a C3HC4 ring finger-containing protein of 574 amino acids. Interestingly, while the expression of human Neuralized is high in normal human brain tissue, expression is very low or absent in astrocytomas and multiple glioma cell lines. It has been postulated that human Neuralized, like the Drosophila gene, is involved in cell fate specifications or maintenance in the central nervous system and that loss of Neuralized expression is an important step in the development of malignant transformation in the central nervous system (CNS).
The Notch signaling pathway has been implicated in cell fate decisions in a variety of developmental contexts in Drosophila, Caenorhabditis elegans, and vertebrates (2, 16, 44). Notch encodes a transmembrane receptor that binds to the membrane-bound ligands Delta and Serrate. On ligand binding, Notch is cleaved by a Presenilin-dependent mechanism (13, 40) and an intracellular portion of the molecule translocates, together with the Suppressor of Hairless [Su(H)] gene product, to the nucleus, where the protein complex acts as a transcriptional activator (22, 39). As in Drosophila, Notch signaling in vertebrates is involved in specifying cell fates in a variety of developmental processes. Targeted disruption of Notch expression or disruption of its downstream targets in mice leads to embryonic death by about midgestation. Notch null embryos have a variety of developmental defects including severe disruption of development of the CNS and somites (7, 10, 41). Constitutive activation of Notch signaling by expression of hypermorphic Notch alleles or downstream targets of Notch interferes with cell fate specifications during neurogenesis and myogenesis in vitro as well as in vivo (30, 31, 37, 43). In addition to its well-established role in cell fate specification during development, recent evidence suggests that Notch signaling might be involved in the maintenance and homeostasis of differentiated cells. Neurite outgrowth of cortical neurons appears to be regulated by Notch signaling, indicating that Notch might be involved in maintenance or plasticity in the CNS (36).
In this study, we report the isolation and characterization of a murine homologue of the Drosophila neuralized gene. Our analysis indicates that expression of Neuralized is not essential for development and survival in vertebrates since Neuralized knockout mice are fully viable. Neuralized null mice, however, have defects in mammary gland development leading to deficient lactation. In addition, defects in the axonemes of spermatozoa isolated from Neuralized null mice result in immotile spermatozoa and male sterility. The axonemal and spermatid abnormalities seen in Neuralized null mice in part mimic the defects seen in many human spermatogenic disorders (32, 45, 46).
MATERIALS AND METHODS
Isolation of the murine Neuralized gene.
Degenerate PCR primers were designed based on the alignment of Drosophila melanogaster and D. virilis neuralized sequence. Primers were designed to the amino acid sequence AITFS and FWAKA, respectively, and PCR was performed using a mouse skeletal muscle cDNA library (Clontech) as template. The resulting 200-bp mouse Neuralized fragment was used to screen mouse skeletal muscle and brain cDNA libraries (Stratagene) by standard procedures (3). To obtain a full-length cDNA clone, 3′ rapid amplification of cDNA ends was performed using murine brain and skeletal muscle libraries (Marathon-ready cDNA; Clontech) and Advantage cDNA polymerase mix (Clontech). The gene-specific primer used in this reaction was 5′-GCT GTC CTT CGG GGT CAC CAC GTG TGA GGC-3′.
Northern blot analysis.
Total cytoplasmic RNA was isolated from murine tissues using RNA STAT (Tel-Test Inc.). For the expression analysis in adult mouse tissues, 2 μg of poly(A) RNA was loaded on a 1% agarose gel containing formaldehyde, transferred onto GeneScreen Plus membranes (NEN), and hybridized to a mouse Neuralized cDNA fragment using standard procedures. For the expression analysis in Neuralized null animals, 10 μg of total cytoplasmic RNA isolated from brain and skeletal muscle of Neuralized null adult mice and wild-type littermate controls was used. An 820-bp Neuralized cDNA fragment corresponding to amino acids 1 to 209 was used as a 5′ probe, a 1-kbp Neuralized cDNA fragment corresponding to amino acids 210 to 557 was used as a 3′ probe. A murine glyceraldehyde 3-phosphate dehydrogenase full-length cDNA probe was used for the loading control. For the expression analysis in human tissues, a human RNA master blot (Clontech) was hybridized with a full-length human Neuralized cDNA probe.
Expression constructs.
The full-length human Neuralized coding sequence was PCR amplified from a cloned human Neuralized cDNA using nrzF1 (GAA GCT TCC GAA GAT GGG GGG ACA GAT CAC CCG G) and nrzR1 (CGG TGG ATC CCG GGA GCT GCG GTA GGT CTT GAT GAT). The PCR product was cloned into pCMV-EGFP using HindIII and BamHI restriction sites creating an in-frame fusion between human Neuralized and enhanced green fluorescent protein. This construct was used to monitor subcellular localization in various cell lines by fluorescence microscopy.
The retroviral Neuralized vector pLIA-Neuralized was constructed by cloning the full-length human Neuralized cDNA coding sequence into the SmaI site of pLIA (generously donated by C. Cepko, Harvard Medical School). This construct places the human Neuralized coding sequence under the transcriptional control of the Moloney murine leukemia virus long terminal repeat promoter and contains an internal ribosome entry site (IRES)-alkaline phosphatase cassette that allows staining of infected cells by established methods (3).
The constitutive active allele of Notch contained the intracellular domain of Notch 1 under the control of the cytomegalovirus CMV promoter. Functional activity of the construct used was assessed by cotransfection with a Notch signaling reporter construct containing four Cbf-1 binding repeats and a minimal simian virus 40 promoter driving luciferase transcription. Notch signaling activity was assessed after 24 h using a luciferase assay (Boehringer Mannheim).
Cell lines and tissue culture.
PC-12 (mouse pheochromocytoma) cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) containing 10% fetal calf serum, 3 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. PC-12 cells were differentiated on polylysine-treated tissue culture dishes by adding 50 ng of nerve growth factor (NGF) (2.5 S NGF; Promega) per ml For infection of PC-12 cells with retrovirus, the cells were allowed to reach 60 to 70% confluency. They were fed prior to infection, and Polybrene was added to a final concentration of 80 μg/ml. C2C12 cells (mouse myoblast cells; American Type Culture Collection) were cultured in DMEM containing 20% fetal calf serum, 3 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. C2C12 cells were differentiated into myocytes by cultivating cells DMEM containing 10% horse serum, 3 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml.
Retroviral stocks were prepared in Phoenix cells by a procedure adapted from that of Cepko and Pear (3).
Targeted disruption of mouse Neuralized in ES cells and generation of Neuralized null mice.
A murine genomic fragment was isolated by screening a mouse 129/SvEv genomic BAC library (Genome Systems) using the 200-bp partial cDNA fragment described above. From the isolated BAC clone containing murine Neuralized sequence, two NotI fragments of 15 kbp (G) and 6 kbp (K) were subcloned into pBluescript. A NotI-XbaI 1.4-kbp Neuralized genomic fragment isolated from K was inserted into pOSdupdel (a kind gift from Oliver Smithies) digested with NotI and XbaI. The resulting plasmid was digested with PmlI and an 8.8-kbp NotI genomic fragment isolated from G was inserted. An IRES-green fluorescent protein (GFP) cassette was constructed by digesting pIRES-neo (Gibco BRL) with XbaI and SmaI, which removes the neo cassette. Into this vector, a GFP cDNA fragment isolated by PCR from the vector pLantern GFP (Gibco BRL) was cloned to yield pIRES-GFP. pIRES-GFP was digested with SalI and XhoI, and the IRES-GFP cassette was cloned into the targeting vector at the XhoI site, yielding pNRZKO-1.
TC1 embryonic stem cells (ES cells) derived from 129/SvEv mice (11) were electroporated with NotI-linearized pNRZKO-1 and selected with G418 and 1-(2-deoxy-2-fluoro-1-β-d-abino-furanosyl)-5-iodouracil as described previously (12). Genomic DNA from G418- and FIAU-resistant clones was isolated as described previously (12) and screened for targeting by digestion with BamHI followed by Southern blot analysis. The blots were hybridized with an 800-bp BamHI-EcoRV genomic fragment located 5′ to the genomic region used for targeting vector construction and isolated from a BAC subclone. Two positive ES cell clones were microinjected into C57BL/J6 blastocysts, which were transferred into pseudopregnant Swiss Webster foster mothers (Taconic). High-grade chimeras judged by agouti coat color of the offspring were mated to 129/SvEv mice (Taconic), and germ line transmission was confirmed by Southern blot analysis. Heterozygous offspring from this F1 cross were intercrossed to derive the mouse colony.
PCR genotyping was performed on genomic DNA using primers nrzF (5′-GAC AGC GAG CTG GTG CTG CCC GAC TG-3′), nrzR (5′-GAA GAT GGT TTC GGC CAC GCG CAC AGG CCG-3′), and nrzIRES (5′-GGA CGC GGC CAC CCT CAA AGG CAT C-3′). The wild-type allele (product nrzF-nrzR) was expected to give a PCR product of 367 bp, and the mutant allele (product nrzF-nrzIRES) was expected to give a PCR product of 190 bp.
In situ hybridizations.
Embryos were dissected from pregnant wild-type animals (FVB, Taconic) at various time points of pregnancy (8.5, 9.5, 10.5, and 12.5 days postcoitum (p.c.) and fixed overnight in 4% paraformaldehyde at 4°C. After incubation overnight in methanol, the embryos were rehydrated in a series of methanol–Tris-buffered saline with Tween 20 (PBT), bleached with 6% hydrogen peroxide, treated with 10 μg of proteinase K (Boehringer Mannheim) per ml for 15 min at room temperature, and washed with 2 mg of glycine per ml in PBT for 10 min at room temperature. The embryos were then postfixed with 4% paraformaldehyde–0.2% glutaraldehyde in PBT for 10 min and prehybridized in 50% formamide–5× SSC (pH 4.5) (1× SSC in 0.15 NaCl plus 0.015 sodium citrate)–1% sodium dodecyl sulfate (SDS)–50 μg of yeast RNA (Boehringer Mannheim) per ml–50 μg of heparin per ml for at least 1 h at 70°C.
Mouse Neuralized riboprobes (a 1,326-bp partial mouse Neuralized cDNA fragment; 1 μg per reaction) were digoxigenin DIG-labeled using T7 and T3 RNA polymerases (DIG RNA labeling kit; Boehringer Mannheim) and purified using ethanol precipitation. The embryos were then hybridized in 50% formamide–5× SSC (pH 4.5)–1% SDS–50 μg of yeast RNA per ml–50 μg of heparin per ml overnight at 70°C.
After hybridization, the embryos were washed in 50% formamide–5×SSC (pH 4.5)–1% SDS and blocked in 10% sheep serum–TBST, and the transcript was detected using an anti-DIG antibody (Boehringer Mannheim) and nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) staining.
Histology.
For histological analysis, tissues were fixed in Omnifix (Zymed), dehydrated in a graded alcohol series, and embedded in paraffin. Sections 4 to 6 μm thick were stained with hematoxylin and eosin using standard procedures.
For analysis of brain and pituitary, tissues were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) and embedded in paraffin. Sections 4 to 5 μm thick were stained with hematoxylin and eosin as well as by the Gordon-Sweet silver method to demonstrate the reticulin fiber network. Immunocytochemical stains to localize adenohypophysial hormones were performed using the avidin-biotin-peroxidase complex method. Primary antibodies against the following antigens were used at the specific dilutions: adrenocorticotropin (ACTH), 1:15; growth hormone, 1:2,500; prolactin, 1:2,500; β-thyroid-stimulating hormone, 1:3,000; β-follicle-stimulating hormone (β-FSH) 1:6,000; and luteinizing hormone (LH) 1:2,500. All antibodies were donated by the National Hormone and Pituitary Program (NHPP), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Child Health and Human Development (Bethesda, Md.) except for the ACTH antibody which was purchased from Dako Corp. (Carpinteria, Calif.). Primary antibodies were incubated at 4°C for 24 h prior to detection.
For skeletal staining, animals were euthanized with CO2 and the skin was removed. The animals were then eviscerated, fixed in 95% ethanol, and stained with Alizarin red S and Alcian blue.
Fertility and analysis of sperm motility.
Male Neuralized null animals aged between 12 weeks and 9 months were housed with 129/SvEv (Taconic), NIH Black Swiss (Taconic), or Neuralized heterozygous females, and the females were analyzed for the presence of vaginal plugs each morning. Sperm was isolated by flushing the epididymidis with Tyrode's solution (0.15 M NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 1 mM NaHCO3, 5 mM glucose). Sperm motility was assessed by light microscopy in Tyrode's solution after incubation at 30°C for at least 30 min to allow dissociation of spermatozoa.
Electron microscopy.
Testes and epididymides were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.2 M sodium cacodylate [pH 7.6]) at 4°C for ca. 12 h. The fixed tissues were dehydrated through a graded series of alcohols and finally embedded in Spurr's low-viscosity embedding medium. Thick (1-μm) and ultrathin (60 to 80-nm) sections were cut on an LKB MarkIII ultramicrotome. Thick sections were stained with 1% toluidine blue for routine light microscopic examination. Contrast was enhanced in ultrathin sections by sequential staining with a saturated uranyl acetate solution in 50% ethanol–25% methanol for 10 min followed by incubation in lead citrate. Ultrathin sections were examined under a Zeiss 10 electron microscope. Sperm abnormalities were quantified by analyzing sections of the cauda epididydimis at a magnification of ×10,000. The frequency of abnormalities was obtained by counting the incidence of flagellar structural defects in 100 cross-sections of the flagellum for both Neuralized knockout and wild-type mice.
Nucleotide sequence accession number.
The complete mouse neuralized cDNA sequence was deposited in GenBank under accession number AF401228.
RESULTS
Isolation of a murine homologue of the Drosophila neuralized gene.
We have identified a murine homologue of the Drosophila and human Neuralized genes with a cDNA sequence of 3.63 kbp and an open reading frame of 1,674 bp. The gene encodes a protein of 557 amino acids and a predicted molecular mass of 59.9 kDa (Fig. 1A). The sequence identity between the mouse and human Neuralized proteins is 93%, and the sequence identity between the mouse and Drosophila Neuralized proteins is 34% (28, 33). The murine Neuralized gene isolated here is syntenic to the human Neuralized gene isolated previously (28) and thus is very likely to be the mouse ortholog. The only detectable protein motif is a C3HC4-type ring finger at the carboxy terminus of the protein. In addition to the ring finger, Neuralized proteins contain two regions of approximately 160 amino acids, each of which appears to be composed of tandem repeats although the sequence identity between the two repeats in each protein is limited (Fig. 1A). These regions have previously been termed neuralized homology repeats (NHR), and their function has not yet been established. Sequence similarity searches in the database using the BLAST algorithm revealed significant sequence homology in the ring finger domain between Neuralized proteins and proteins of the IAP (inhibitor of apoptosis) family (Fig. 1B). No sequence similarity between Neuralized and IAP proteins was detected outside the ring finger domain, suggesting that they belong to distinct protein families.
FIG. 1.
(A) Alignment between Neuralized proteins from Drosophila, mouse, and human. NHR-1 and NHR-2 are marked by a dashed line above the amino acid sequence, and identical residues are boxed. The Drosophila neuralized gene encodes a protein of 754 amino acids, while the human and mouse gene encode proteins of 574 and 557 amino acids, respectively. Neuralized proteins from all three organisms show significant sequence homology in the two NHR domains and in the ring finger (solid line). (B) Alignment of the Neuralized ring finger domain with ring finger domains of the IAP protein family. Sequences from human (h-IAP 1 and h-IAP 2), mouse (m-IAP 1 and m-IAP 2), and rat (r-IAP 2) are shown.
Expression analysis of various murine tissues by Northern blot hybridization revealed a transcript of approximately 4,400 bp in adult brain and a slightly smaller transcript in skeletal muscle (Fig. 2A). Interestingly, no Neuralized transcript was detected in heart muscle, indicating a high degree of tissue specificity in myocytes. The size of the transcript is consistent with the cDNA sequence described above. The reason for the size difference in transcripts from brain and skeletal muscle is so far unknown. However, 3′ rapid amplification of cDNA ends from skeletal muscle and brain libraries yielded identical clones, suggesting that the presumed alternative splicing event occurs at the 5′ end of the transcript. Using a dot blot analysis with poly(A) RNA from human tissues and a human Neuralized cDNA probe, we also detected transcript in testis, pituitary gland, pancreas, and bone marrow (data not shown). This analysis of human tissues also confirmed the expression of Neuralized in all regions of the brain, including fetal brain and adult skeletal muscle.
FIG. 2.

Expression pattern and subcellular localization of mouse Neuralized. (A) Poly(A) RNA Northern blot analysis of mouse tissues with a mouse Neuralized cDNA probe detects a transcript of approximately 4 kbp in adult brain and a slightly smaller transcript in skeletal muscle. (B) In situ hybridization on day 10.5 p.c. mouse embryos using a mouse Neuralized probe shows expression of mouse Neuralized in the somites. (C) Dorsal view of the same embryo as shown in panel B at higher magnification. (D) Littermates were hybridized to a mouse myogenin probe as a positive control. Mouse Neuralized is expressed in the dermomyotomes of the somites in a pattern highly similar to myogenin. (E) Subcellular localization of a human Neuralized-GFP fusion protein in differentiating C2C12 cells. C2C12 myoblasts were transfected with a CMV-Neuralized-GFP construct and induced to differentiate by serum starvation. An equivalent subcellular localization was observed in a variety of different human and murine cell lines.
Using whole-mount in situ hybridizations on mouse embryos at various stages of development, we were able to detect Neuralized transcript in the somites from day 9.5 p.c. (Fig. 2B and C shows an embryo on day 10.5 p.c). The expression pattern of Neuralized is very similar to the expression found for the myotome marker myogenin (Fig. 2D). myogenin is expressed in the myotome from day 8.5 p.c. and is maintained in skeletal muscle throughout embryonic development (35). Control hybridizations using a sense Neuralized probe did not show staining above background throughout the embryo's torso. This whole-mount analysis did not reveal Neuralized expression in the developing brain due to high background in this tissue.
Neuralized is not localized to the nucleus.
We expressed a full-length human Neuralized cDNA-GFP fusion construct under the control of the CMV promoter in a variety of human and mouse cell lines including HeLa, C2C12, COS, and NIH 3T3. In all cell lines, the human Neuralized-GFP fusion was excluded from the nucleus, with perinuclear, Golgi-like staining detectable in many cells (Fig. 2E). Our results are consistent with those of Yeh et al. (48), who reported a function of Drosophila Neuralized outside the nucleus. Cotransfection with constructs encoding constitutively active forms of Notch 1 did not change this cellular localization (data not shown). However, these constitutive alleles of Notch 1 did activate Cbf-1-dependent reporter gene transcription in a Notch reporter assay (data not shown), indicating that the Notch constructs are functionally active. These data suggest that cellular localization of Neuralized is not regulated by Notch signaling.
Targeted disruption of Neuralized in ES cells and generation of Neuralized null mice.
A targeting strategy for the mouse Neuralized locus was designed that inserts PGK-neo and IRES-GFP cassettes into the exon encoding NHR2 (Fig. 3A). The IRES-GFP cassette includes transcriptional termination sequences 3′ to the GFP coding sequence. The insertion of the targeting cassette is therefore expected to disrupt the expression of all portions of the gene encoding the NHR2 domain and the ring finger, thus eliminating the majority of the coding sequence from the transcript.
FIG. 3.
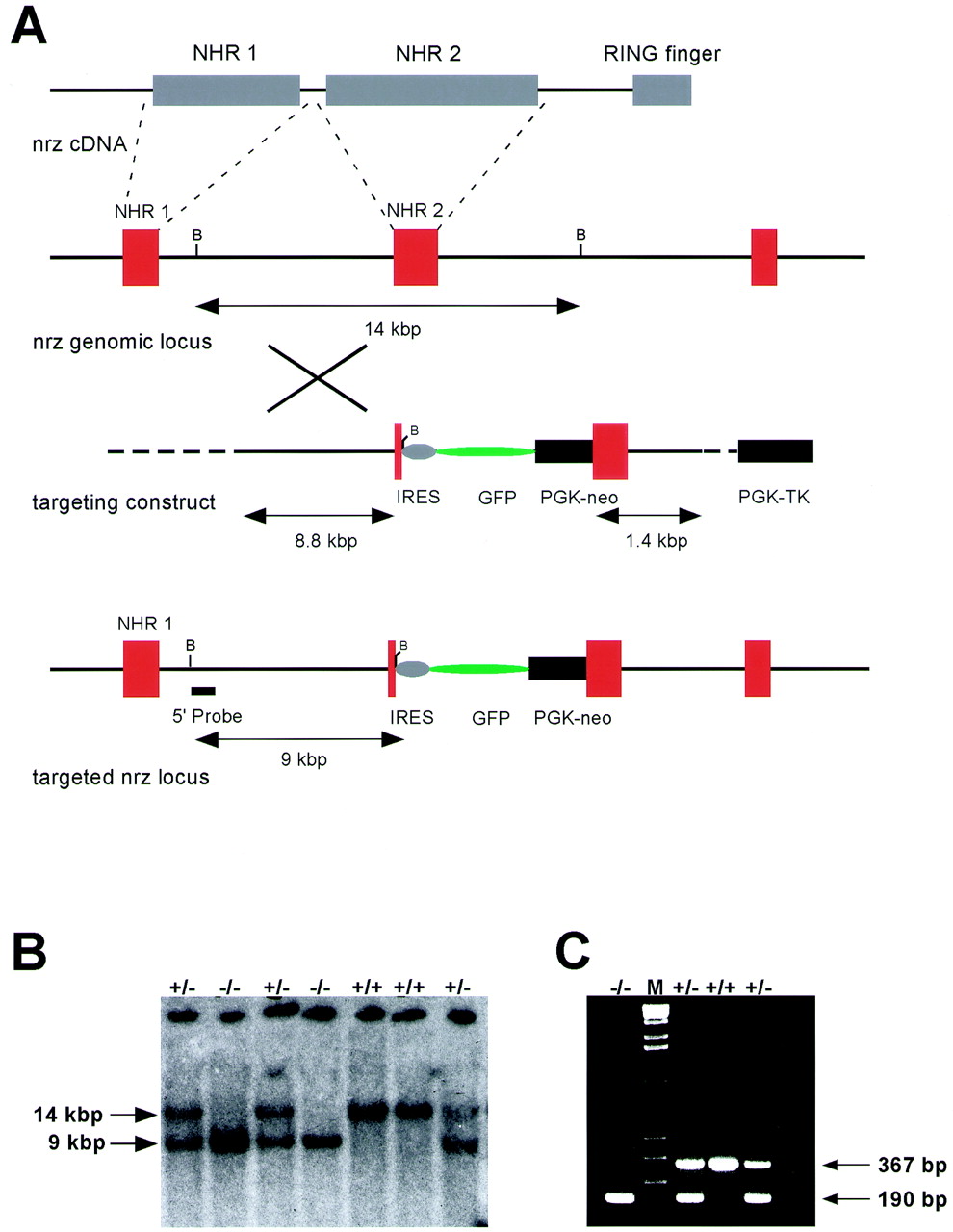
(A) Targeting strategy for disruption of the mouse Neuralized locus. A targeting cassette containing an IRES-GFP construct and a PGK-neo cassette for positive selection of transfected clones was inserted into the exon encoding the NHR-2 region of the protein. A thymidine kinase cassette (PGK-TK) was used for negative selection with FIAU. (B) Genotyping was performed by Southern blot analysis using a 5′ external probe and genomic DNA digested with BamHI. The wild-type allele yields a 14-kbp fragment, and the mutant allele yields a 9-kbp band. The Southern blot shows a typical result from tail DNA of offspring from a heterozygous intercross and the appearance of all three expected genotypes. (C) PCR genotyping assay on tail DNA from offspring of a heterozygous intercross using a three-primer setup. The wild-type allele yields a PCR product of 367 bp, the mutant allele yields a product of 190 bp, and all three genotypes are readily detectable.
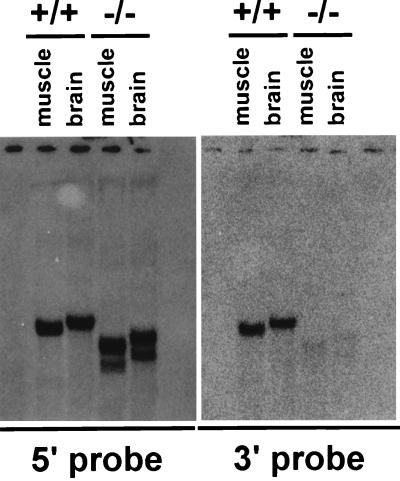
To confirm that we generated a true targeted deletion of the murine Neuralized gene, we performed Northern blot RNA analysis on wild-type controls and knockout animals. Using a cDNA probe containing sequence 5′ to the integration site of the IRES-GFP-PGK-neo cassette, we were able to detect two aberrantly migrating transcripts in the mutant animals compared to wild-type control RNA derived from skeletal muscle and brain (Fig. 4). The sizes of these transcripts are consistent with the generation of fusion transcripts containing the 5′ end of the murine Neuralized gene and the IRES-GFP cassette. Using a cDNA probe containing sequence 3′ to the integration site of the IRES-GFP-PGK-neo cassette, we failed to detect any Neuralized transcripts in the mutant animals, indicating that the transcripts produced in mutant animals lack sequence encoding the NHR 2 domain as well as the ring finger. This deletion is expected to result in a null allele.
FIG. 4.
Loss of full-length Neuralized transcript in Neuralized knockout mice. Total RNA isolated from adult brain or skeletal muscle was analyzed using a cDNA probe containing sequence 5′ (left panel) or 3′ (right panel) to the integration site of the targeting cassette (see Materials and Methods). Northern hybridization with the 5′ probe yields transcripts of the expected size in wild-type tissues and aberrantly migrating transcripts in the tissues derived from Neuralized null animals. The right panel shows normal transcripts in the wild-type controls but loss of transcription of sequence 3′ to the integration site of the targeting vector in Neuralized null animals.
In multiple heterozygous intercrosses from F1 or later generations for a total of 444 animals, we observed live Neuralized null animals at the expected Mendelian frequency of approximately 1:4 (the ratio of Neuralized null to Neuralized heterozygous to wild type is 1.2:2.0:1.1). This indicates full viability of animals carrying a targeted deletion of the Neuralized gene. The oldest animals in our colony have now reached ages of over 1 year, showing that murine Neuralized expression is not required for full development and survival. These data are consistent with observations on two mouse lines derived from two independently targeted ES cell clones.
Neuralized null mice do not show aberrant cell fate specifications in the CNS and somites.
Notch null mice die during embryogenesis with severe defects in neurogenesis. On histological analysis of the CNS, we did not detect any differences between brains derived from Neuralized knockout mice and wild-type controls (data not shown). All major brain structures and cell types were present, and the cortex showed correct layering.
Notch signaling has been implicated in cell fate specifications during somitogenesis, and targeted deletion of Notch or genes involved in Notch signaling leads to aberrant somite development (7, 23). Since Neuralized is expressed in developing somites, we investigated whether loss of Neuralized expression interferes with skeletal or myocyte differentiation. On histological analysis, we found that skeletal muscle from Neuralized null animals showed clearly developed myotubes and we failed to detect any differences between skeletal muscle from Neuralized null animals and wild-type controls (data not shown). Likewise, the skeletons of Neuralized null animals appeared to be identical to skeletons derived from wild-type controls after staining with Alizarin red S and Acian blue (data not shown). These observations indicate that loss of Neuralized expression does not interfere with cell fate specifications during somitogenesis.
Ectopic expression of Neuralized does not interfere with differentiation of PC-12 or C2C12 cells in vitro.
The PC-12 cell line has been extensively studied in vitro as a model for neuronal differentiation and neurotropin signaling. Stimulation of PC-12 cells with NGF induces neuronal differentiation, with differentiated cells expressing neuronal markers and forming long neurite extensions from the cell bodies. Activation of Notch signaling blocks the differentiation of neuronal precursor cells during development of the CNS in vivo and the Notch target Hes-1 modulates the differentiation of PC-12 cells in vitro (38). To assess whether ectopic expression of Neuralized was able to block neuronal differentiation of PC-12 cells in a similar manner, PC-12 cells were infected with Neuralized-expressing IRES alkaline phosphatase virus and differentiated by stimulation with NGF. After treatment of infected cells with NGF, neurite extensions were clearly detectable in virus-infected cells after staining for the viral marker alkaline phosphatase and no difference compared to cells infected with the empty virus (pLIA) or uninfected cells could be detected (Fig. 5), indicating that ectopic expression of Neuralized does not interfere with neuronal differentiation in PC12 cells. In addition, Neuralized-expressing cells still required NGF for neuronal differentiation (data not shown). These observations are consistent with our observed lack of defects in neuronal differentiation in Neuralized null mice, suggesting that, in contrast to Drosophila, Neuralized is not necessary for the regulation of neuronal differentiation in vertebrates.
FIG. 5.
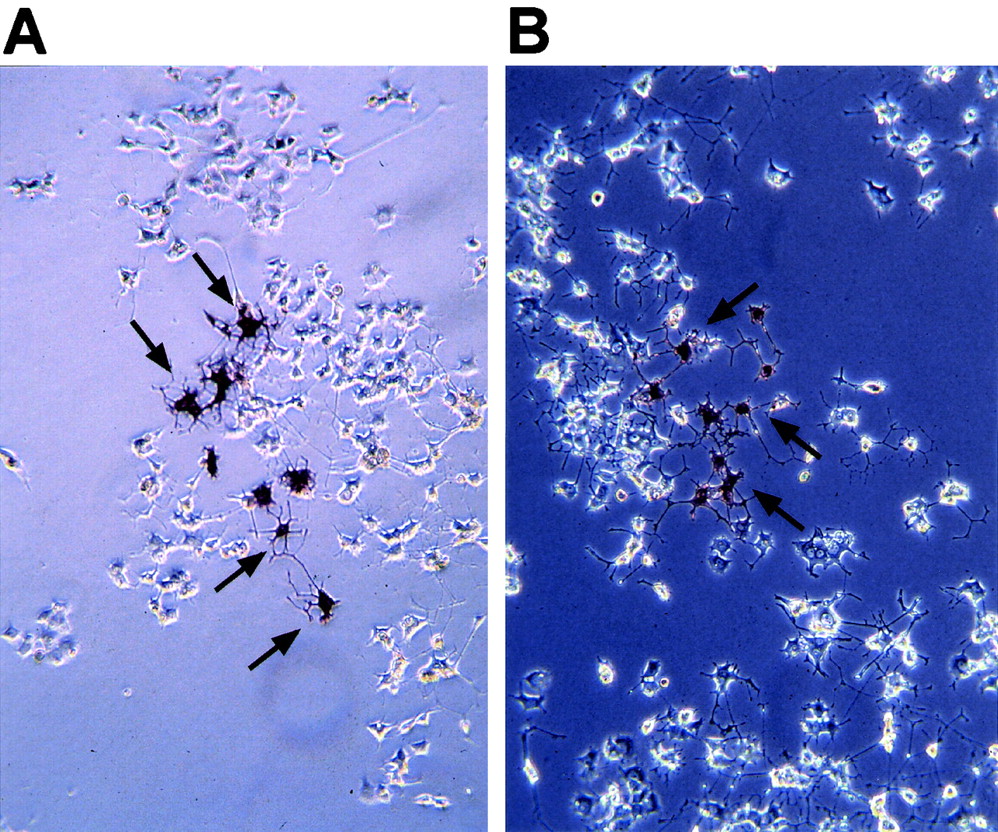
Ectopic expression of human Neuralized in PC-12 cells does not interfere with neuronal differentiation through NGF in vitro. The viral expression contruct expresses a bicistronic Neuralized alkaline phosphatase fusion transcript that allows translation of two independent proteins through an IRES sequence inserted between the two genes. Some of the infected, alkaline phosphatase-positive cells are marked by arrows in both panels. (A) PC-12 cells were infected with pLIA control virus, differentiated 24 h after infection by treatment with NGF for 4 days, and stained for alkaline phosphatase. Extensive neurite outgrowths are easily detectable in cell clones infected with pLIA. (B) PC-12 cells infected with pLIA-hNeuralized are indistinguishable from control-infected cells.
A similar experiment was performed to assess the effect of Neuralized expression on myocyte differentiation in vitro. The C2C12 cell line has been studied as an in vitro model for this process, and, in a manner highly similar to the situation found in PC-12 cells, ectopic expression of activated alleles of Notch interferes with the differentiation of C2C12 cells into fused myocytes (30, 37). Ectopic expression of Neuralized in C2C12 cells did not affect myocyte differentiation, consistent with the correct formation of myocytes observed in Neuralized null animals (Fig. 2E). This result, together with results obtained with Neuralized null mice, suggests that Neuralized is not necessary for regulating myocyte differentiation in vertebrates. However, ectopic expression of murine Neuralized does induce apoptosis in various cell lines (data not shown).
Male Neuralized null animals are sterile and have defective spermatozoa.
Male Neuralized null animals failed to fertilize females in matings with either 129/SvEv wild-type mice, NIH Black Swiss wild-type mice, or Neuralized heterozygous littermates. Of 10 male homozygote Neuralized animals aged between 12 weeks and 9 months, none gave rise to a pregnancy in matings with NIH BI/SW females, while matings of Neuralized heterozygous males or wild-type control males yielded pregnancies and litters of normal sizes.
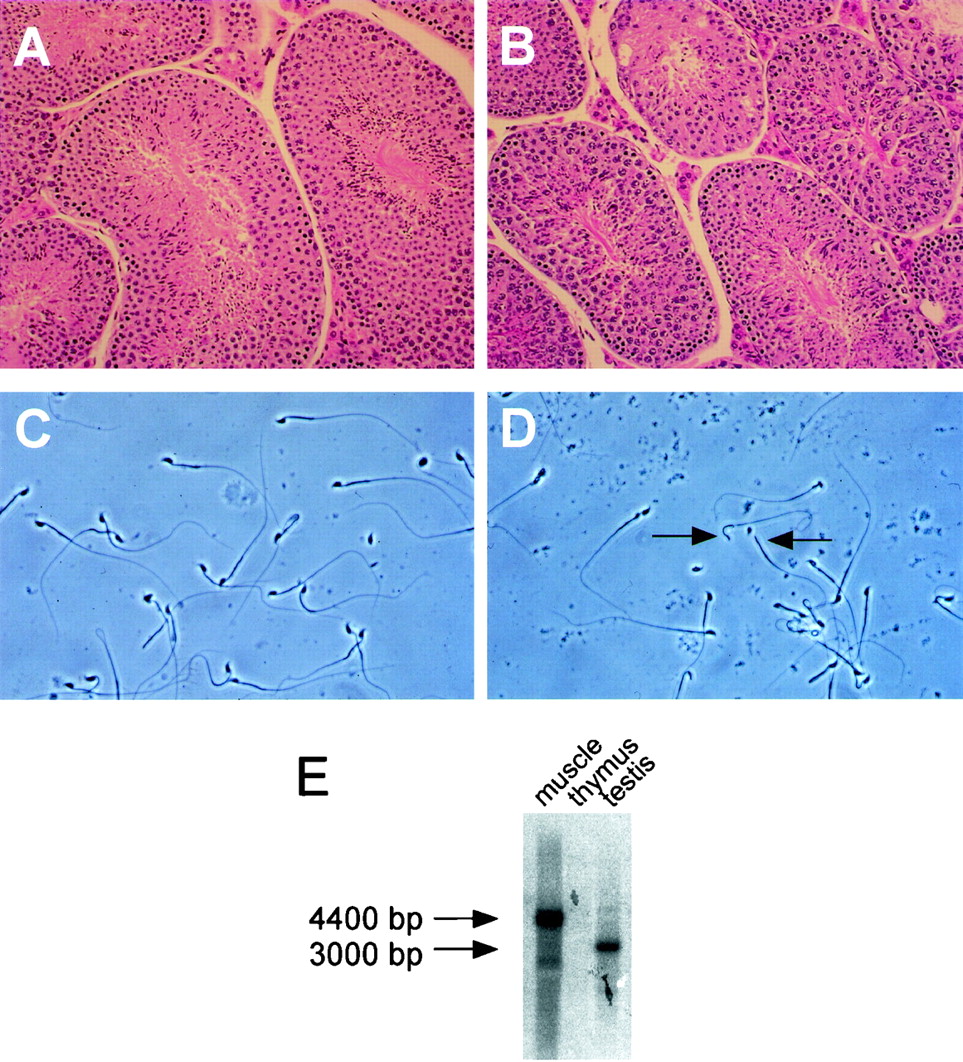
Surprisingly, standard histological analysis of male reproductive organs failed to reveal significant differences between Neuralized null animals and wild-type controls. When analyzed by histological staining, testes derived from Neuralized null animals showed proper spermatogenesis and the appearance of mature sperm in the seminiferous tubules (Fig. 6A and B). In addition, no defect in the histology of the prostate could be detected (data not shown).
FIG. 6.
Histological analysis of testis and spermatozoa isolated from Neuralized null animals. (A) Wild-type testis. There is normal morphology and active spermatogenesis in the seminiferous tubules. (B) Testis isolated from Neuralized null animals. No difference from wild-type tissues could be detected at this level of resolution. (C) Spermatozoa isolated from wild-type control animals. Spermatozoa were isolated from the epididydimus and showed high mobility. (D) Spermatozoa isolated from Neuralized null animals. Most spermatozoa lacked tail motility. In addition, defective spermatozoa were detected frequently in Neuralized null samples (arrows). (E) A Neuralized transcript of approximately 3,000 bp is detected in wild-type mouse testis by poly(A) RNA Northern blot hybridization (lane 3). The transcript is significantly smaller than the transcript found in skeletal muscle (lane 1). As shown in Fig. 1, the Neuralized transcript is absent in the thymus (lane 2).
However, a striking difference between Neuralized wild-type and null animals was detected when sperm motility and morphology of epididymal spermatozoa were analyzed. Spermatozoa isolated from wild-type or Neuralized heterozygous animals showed a homogenous population of intact sperm with high mobility. In contrast, spermatozoa isolated from Neuralized null animals showed only marginal motility, with a large majority of spermatozoa in a sample showing no detectable motility under standard light microscopic analysis. In addition, we frequently observed spermatozoa with missing head structures (Fig. 6D, arrows)
Electron microscopy analysis of cauda epididymal spermatozoa revealed the presence of structural defects of the spermatozoa involving the flagellum. Cross sections through different regions of the flagellum showed that the middle piece appeared consistently normal, being composed of the appropriate 9+2 configuration of doublets (Fig. 7A). Morphological aberrations occurring in the flagellum were most commonly seen in the principal piece and primarily involved the structural integrity of the axoneme (Fig. 7B to D). These defects were readily observed in the cross-sectional profiles of the axoneme and were also noticeably displayed by longitudinal sections through the flagellum. The most prevalent structural abnormality detected involved missing axonemal doublets that varied from either loss of one doublet (usually no. 7) to loss of half the axonemal complex (usually no. 4 to 7). Other, less common morphological defects included translocation of doublets outside of the axonemal complex, total disorganization of the axoneme, and dislocation of dense fibers. In addition, for some spermatozoa, regions of the fibrous sheath were found to be malformed or entirely missing, and this was often accompanied by disruption of the axonemal complex. A large number of spermatozoa were found to have developed double or sometimes triple axonemes that usually were defective. Moreover, these axonemal aberrations not only were now confined to the principal piece but also occurred in the midpiece of the flagellum. Abnormal axonemes were detected in approximately 30% of flagellar cross sections through the principal piece. This does not suggest, however, that normal axonemes prevail in the remaining 70% of the cross sections. Longitudinal sections through the flagellum clearly showed that structural defects of the axonemal complex were localized within the principal piece (Fig. 7F and G). Thus, depending on where the flagellum was sectioned, the axonemal profile could appear normal even if the principal piece was structurally compromised elsewhere along the length of the flagellum. Taken together, these aberrations are expected to significantly compromise the integrity of the flagellum and thus the motility of the spermatozoa.
FIG. 7.
Electron microscopy analysis of spermatozoa and testis from Neuralized null animals. (A) Cross section of a flagellar midpiece showing normal 9+2 arrangement of axoneme doublets. (B) Cross sections of the principal piece of spermatozoa from Neuralized null animals, showing that although some cross sections appear to be normal with the expected 9+2 configuration (no. 1 to 3), several other cross sections clearly show defects in axonemal organization (no. 4 to 6). (C) Cross section showing a common flagellar defect, i.e., deletion of half the axonemal complex. (D) Cross section of principal piece showing a flagellum where a portion of the fibrous sheath is absent as well as a highly disorganized axonemal complex. (E) Abnormal biflagellate spermatozoa identified in samples from Neuralized null animals. (F and G) Abnormal axonemal complexes occur along the length of the flagellum (arrowheads in panel F), as revealed by various longitudinal and grazing planes of sections. (H) Testicular spermatozoa of Neuralized null animals. Although cross-sections of the principal piece present a normal profile of the axoneme (arrows), it is apparent from grazing planes of sections that in localized regions of the flagellum, the axonemal complex is disorganized (arrowheads). Bars, 0.2 μm.
We also performed an ultrastructural analysis of the testes of Neuralized null mice to establish whether abnormal flagellar development could be detected in germ cells during spermiogenesis. In the lumen of seminiferous tubules, cross sections through the middle and principal piece of testicular spermatozoa mostly showed a normal organization of the axoneme. Longitudinal and grazing planes of sections through the flagellum, however, provided evidence that regions of the axonemal complex were in fact clearly disorganized (Fig. 7H). Fine-structure analysis of spermatids during late stages of maturation showed that although flagellar development appeared morphologically normal in some spermatids, other spermatids showed distinct aberrations in the organization and structure of the burgeoning neck piece.
Neuralized null females fail to lactate and successfully nurse their pups and have defective mammary gland development during lactation.
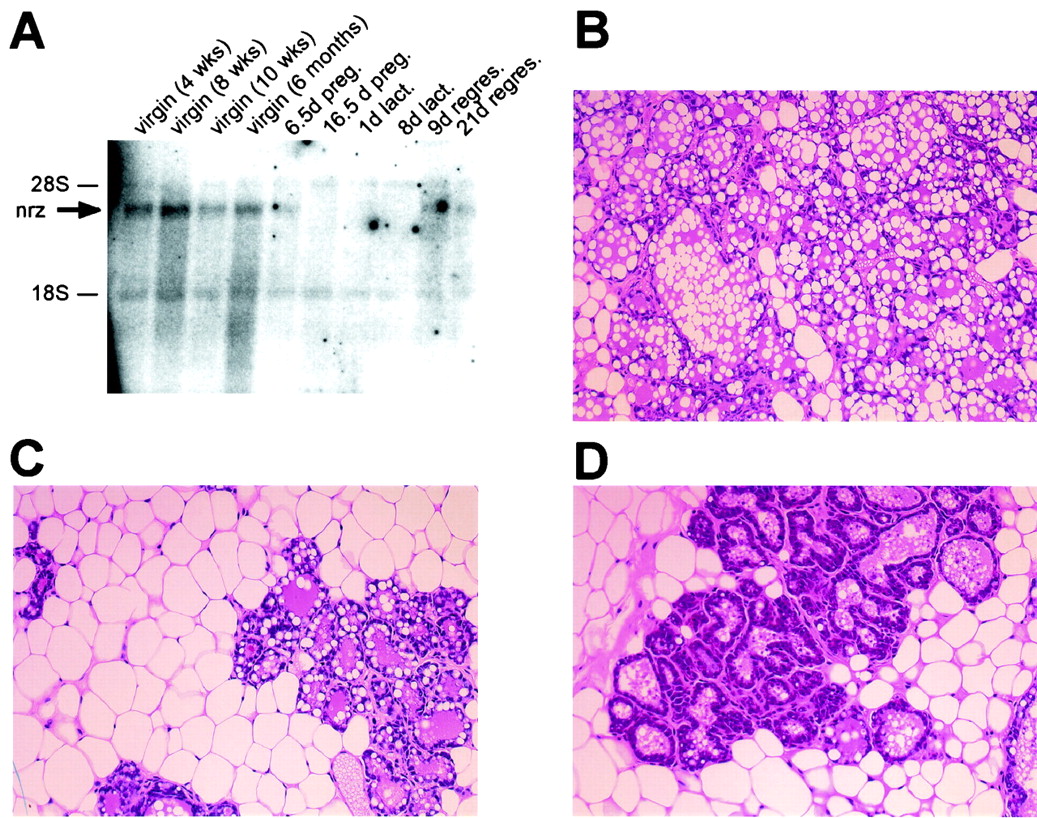
In analyzing the fertility of Neuralized female mice, we observed that pups born from Neuralized null females in matings with either wild-type control or Neuralized heterozygous males failed to survive beyond day 3 after birth. In addition, litters from Neuralized null females were often scattered throughout the cage and we failed to detect milk in the stomachs of the pups, whereas pups in control litters were clearly nursing and had milk in their stomachs. This suggests that Neuralized female mice were fully fertile and able to support pregnancies to full term but had defects in lactation or maternal behavior, leading to defective nursing. Since pups of all genotypes are able to actively nurse and survive until adulthood when born from heterozygous or wild-type mothers, this observed nursing defect was clearly maternal. We therefore analyzed the mammary glands of Neuralized null females on day 1 of lactation (L1). Mammary glands from Neuralized null females were clearly defective on L1, with significantly less alveolar structures penetrating the mammary fat pad compared to the glands from wild-type control animals at the same stage of lactation (Fig. 8B and C). In addition, some samples from Neuralized null females showed defective lipid production in the ducts (Fig. 8D). This result shows that the failure to properly nurse their pups and the observed death early after birth is clearly related to a severely underdeveloped mammary gland at lactation in Neuralized female animals.
FIG. 8.
Analysis of mammary glands during lactation in Neuralized null females and wild-type controls. Mammary glands were harvested on the first postpartum day (L1) and analyzed by standard histological hematoxylin and eosin staining. (A) Expression of Neuralized in mammary glands during pregnancy (preg.), lactation (lact.), and regression (regres.). (B) Wild-type control mammary gland on day L1. The mammary fat pad is filled with alveoli, and lipid droplets are easily detected in the ducts. (C) Mammary gland from a Neuralized null female on L1. The mammary gland is significantly underdeveloped, with few alveoli in the fat pad. (D) Mammary gland from a Neuralized null female on L1. Alveoli appear to be more developed than in panel C; however, lipid vacuoles are not detectable in the ducts.
The defective maturation of mammary glands during pregnancy in Neuralized null animals is consistent with expression of Neuralized in this tissue. Using Northern blot hybridization of mRNA derived from mammary glands at various stages of maturation and development, we could detect Neuralized transcripts in virgin mammary glands as well as during the earlier stages of pregnancy (Fig. 8A). We failed to detect Neuralized transcripts at very late stages of pregnancy and during lactation. However, this is a frequently observed phenomenon since mammary glands at these stages produce vast amounts of milk proteins, resulting in lactation-related transcripts outcompeting any other transcripts that might be present.
Neuralized heterozygous and null animals do not develop malignancies at a significantly higher frequency than wild-type controls.
Human Neuralized is localized on chromosome 10q24–25, a region showing frequent deletion in malignant astrocytomas, leading to the hypothesis that the Neuralized gene is involved in the formation of malignant tumors of the CNS in humans (28). In addition, loss of Neuralized transcript has been found in astrocytoma tissue derived from human patients and in glioblastoma cell lines, suggesting that loss of Neuralized transcription might be associated with malignant transformation (28). Therefore, we analyzed Neuralized null animals for the development of tumors, particularly in the CNS. The oldest animals in our colony are now older than 1 year. Neuralized null animals did not appear to become moribund at a rate significantly higher than their wild-type littermates, and we failed to detect CNS malignancies at a rate above background. Of 15 Neuralized null animals analyzed by histological examination, 1 showed a pituitary adenoma that invaded upwards from the sella turcica into the base of the brain. This tumor was characterized as a gonadotroph adenoma with nuclear immunoreactivity for steroidogenic factor 1 and cytoplasmic staining for FSH and LH. Since this is a tumor occasionally found in older wild-type animals (34), we are unable to definitively conclude that Neuralized null animals show a predisposition to tumor development in the CNS. The proposed link between loss of Neuralized expression and tumor formation in humans is therefore not yet supported by our mouse model.
DISCUSSION
In this study, we report the isolation and characterization of a murine homologue of the Drosophila neuralized gene. The gene isolated is syntenic to the human neuralized gene isolated recently and encodes a protein which is almost identical to human Neuralized.
Sequence similarity searches in the database using the BLAST algorithm revealed significant sequence homology in the ring finger domain between Neuralized proteins and proteins of the IAP family. IAP proteins were initially identified in baculoviruses, and the related viral and mammalian proteins all contain RING finger domains at their carboxy terminus (14). Expression of IAP proteins inhibits the induction of apoptosis by various stimuli in vitro (25, 42). Interestingly, ectopic expression of Neuralized appears to induce rapid cell death in a variety of different cell lines in vitro, suggesting a role of vertebrate Neuralized in apoptosis (K. Fitzgerald, unpublished observations). However, the sequence homology between Neuralized and IAP proteins is limited to the ring finger, suggesting that these two groups of proteins are distinct. Recent evidence indicates that ring finger-containing proteins can mediate ubiquitin-conjugating enzyme-mediated ubiquitination of receptor protein tyrosine kinases, leading to termination of signaling through protein degradation (20, 21, 26). Interestingly, IAP proteins have now also been shown to catalyze their own ubiquitination in response to apoptotic stimuli, an activity that requires the presence of the ring finger domain (47). Further experiments will be needed to establish if Neuralized has ubiquitin protein ligase activity.
Targeted deletion of the murine Neuralized gene reveals that expression of murine Neuralized is not essential for development and survival of the animal. In contrast, Notch null mice die around midgestation with severe defects in development, as do mice with null mutations in most other components of the Notch signaling pathway (10, 17, 18, 41). In addition, we failed to detect any aberrant cell fate specifications in Neuralized null animals during neurogenesis and somitogenesis, two processes where Notch signaling has been shown to be involved. This evidence suggests that the murine Neuralized isolated in this study is not an essential component of the Notch signaling cascade, at least during most developmental processes. However, two other possibilities must be considered. (i) Neuralized function is essential for Notch signaling, as suggested from genetic evidence in Drosophila, but other vertebrate Neuralized homologues or unrelated proteins compensate for loss of the Neuralized gene isolated here. A search of sequence databases using the BLAST algorithm did not reveal sequences showing significant homology to the gene described in this report. However, since the mouse genome sequence is not complete, this possibility cannot be ruled out. (ii) The Neuralized allele produced by targeting the exon encoding NHR-2 is a hypomorph, but not a complete null. Our targeting strategy removes sequence encoding most of the protein including domains highly conserved between Drosophila and vertebrates and the ring finger, which is thought to be a crucial component of a functional Neuralized protein. Since we have shown here that our targeting does result in the expected changes in Neuralized transcript, we think that this possibility is highly unlikely.
The human Neuralized homologue was isolated from a region at chromosome 10q24–25 that shows frequent alterations in malignant astrocytomas. In addition, loss of Neuralized transcription has been described in human astrocytoma tissue and glioma cell lines (28). The postulated link between loss of Neuralized transcription and neoplastic transformation in the CNS in humans is particularly interesting since Notch signaling is involved in cell fate specification during development of the CNS but has not, at least so far, been linked to the development of CNS tumors. Thus far, our Neuralized null mice have failed to develop any gliablastomas or astrocytomas. This observation questions the proposed link between loss of Neuralized transcription and malignant transformation in the CNS.
Neuralized null females failed to nurse their pups and support a litter. We show here that this is caused by defective lobular development of the mammary gland during pregnancy, leading to an insufficiently developed mammary gland at the end of pregnancy. However, we did not detect any clear differences between virgin Neuralized null animals and wild-type controls at the end of sexual maturation or during early stages of pregnancy. These observations suggest a role of Neuralized in later stages of mammary gland maturation during pregnancy. Whether the effect of Neuralized on mammary gland differentiation is cell autonomous or is caused by a defective hormonal environment during pregnancy in Neuralized null animals awaits the results of transplantation experiments. Interestingly, transgenic mice expressing constitutive active alleles of the Notch receptor also fail to lactate, with lobular development being retarded during late stages of pregnancy (15, 19). However, unlike these transgenic mice, Neuralized null animals do not develop mammary gland tumors.
While male Neuralized null mice were sterile and appeared to have normal reproductive organs when analyzed by standard histological techniques, spermatozoa isolated from these mice were immobile or displayed only residual motility. Electron microscopy clearly revealed structural abnormalities in the flagella of epididymal spermatozoa from Neuralized null animals. The most common defect observed was in the axoneme, which displayed missing microtubular doublets. The defects ranged from loss of one doublet to the deletion of up to half of the axonemal complex. These defects in the structural integrity of the axoneme would directly compromise the motility of the affected spermatozoa, leading to the observed infertility of male Neuralized null mice. Normal 9+2 doublet structures of the axoneme consist mainly of αβ-tubulin polymers as well as microtubule-associated proteins. However, the molecular events required for proper assembly of axonemal microtubule structures have not yet been identified. Indeed, this is the first evidence that Neuralized functions in this process. Interestingly, results from C. elegans indicate that the presenilin family member spe-4 is involved in tubulin localization during spermatogenesis (1). Since presenilins are thought to be regulators of Notch and amyloid precursor protein processing, this result provides a link between signal transduction events and microtubule assembly during spermatogenesis.
The severity of the observed axonemal defects identified in Neuralized null animals suggests that the structural alterations occur during spermiogenesis. This observation is confirmed by the fact that similar flagellar defects were detected in testicular spermatozoa in the lumen of the seminiferous tubules. Analysis of spermatid maturation in testes of Neuralized null mice showed that the structure of both the proximal and distal centrioles appeared normal. In addition, alignment and orientation of the centrioles with proper migration to the posterior pole of the nucleus appeared to be normal during initial development of the flagellum. It is therefore likely that the observed defects in axonemal structure occurred during subsequent growth and formation of the nascent flagellum. A striking feature of the morphology of spermatozoa from Neuralized null mice was the fact that axonemal abnormalities occurred as localized defects along the length of the axoneme. Longitudinal sections of flagella observed under the electron microscope displayed regions containing normal axonemal structures followed by a region where the axonemal complex was clearly disrupted. This observation suggests that proper construction of the 9+2 microtubular structure during spermiogenesis involves the presence of local regulatory factors along the length of the flagellum.
Defects in spermatogenesis account for more than 50% of human male infertility (29). The axonemal and spermatid abnormalities seen in Neuralized null mice in part mimic the defects seen in many human spermatogenic disorders (32, 45, 46). Thus, Neuralized null mice may thus be a valid model to study human infertility syndromes and to gain a better understanding of male infertility.
ACKNOWLEDGMENTS
We thank H. Nakamura for providing the full-length human Neuralized cDNA clone, Charles Murtaugh and Andrew Lassar for providing the constitutive active Notch allele, Diana Hayward for providing the CBF1-luciferase reporter constructs, and Rachel Neve for providing the PC-12 cell line. We also thank Jan Pinkas for help with the analysis of the mammary gland phenotype, Frank Kuo for advice on the initial analysis of the pituitary glands, Kelvin So (Toronto) for histological analysis of the pituitaries, Richard V. Pierce for advice in the early stages of spermatozoa analysis, and members of the Leder laboratory and the HMS Department of Genetics for helpful comments and suggestions throughout the project.
ADDENDUM IN PROOF
While this paper was in proof, Ruan et al. reported the isolation of a mouse Neuralized homolog and described the phenotype of the knockout mouse (Y. Ruan, L. Tecott, M. M. Jiang, L. Y. Jan, and Y. N. Jan, Proc. Natl. Acad. Sci. USA 98:9907–9912, 2001). The gene described in this report is identical to the gene described in our study with the exception of the amino-terminal 28 amino acids. We postulated in our study the existence of two splice variants of the mouse Neuralized gene, and comparison of the two sequences clearly suggests that this is the case. Two differences between our observations and those of Ruan et al. are noteworthy. First, Ruan et al. failed to detect any expression of mouse Neuralized in adult skeletal muscle and did not show convincingly expression in the developing somites. We believe that this is due to the fact that Ruan et al. used a probe from the very 5′ end of the cDNA in these experiments, thus presumably detecting only one splice variant, while our probe is expected to detect both splice variants. Second, Ruan et al. report normal reproductive behavior in both male and female mice. This is clearly distinct from our observations, and the cause for this discrepancy is currently unknown. However, strain differences or differences in the targeting strategy could account for these different observations. Further analysis is required to clarify these issues.
REFERENCES
- 1.Arduengo P M, Appleberry O K, Chuang P, L'Hernault S W. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111:3645–3654. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 4.Bate M, Rushton E, Frasch M. A dual requirement for neurogenic genes in Drosophila myogenesis. Dev Suppl. 1993;1993:149–161. [PubMed] [Google Scholar]
- 5.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 6.Boulianne G L, de la Concha A, Campos-Ortega J A, Jan L Y, Jan Y N. The Drosophila neurogenic gene neuralized encodes a novel protein and is expressed in precursors of larval and adult neurons. EMBO J. 1991;10:2975–2983. doi: 10.1002/j.1460-2075.1991.tb07848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlon R A, Reaume A G, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 8.Corbin V, Michelson A M, Abmayr S M, Neel V, Alcamo E, Maniatis T, Young M W. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991;67:311–323. doi: 10.1016/0092-8674(91)90183-y. [DOI] [PubMed] [Google Scholar]
- 9.de la Concha A, Dietrich U, Weigel D, Compos-Ortega J A. Functional interactions of neurogenic genes of Drosophila melanogaster. Genetics. 1988;118:499–508. doi: 10.1093/genetics/118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 11.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 12.Deng C X, Wynshaw-Boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 14.Deveraux Q L, Reed J C. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 15.Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith G H. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56:1775–1785. [PubMed] [Google Scholar]
- 16.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 17.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman J R, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 18.Ishibashi M, Ang S L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 19.Jhappan C, Gallahan D, Stahle C, Chu E, Smith G H, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 20.Joazeiro C A, Weissman A M. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 21.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 22.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusumi K, Sun E S, Kerrebrock A W, Bronson R T, Chi D C, Bulotsky M S, Spencer J B, Birren B W, Frankel W N, Lander E S. The mouse pudgy mutation disrupts Delta homologue DII3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann R, Jimenez F, Dietrich U, Campos-Ortega J A. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Roux's Arch Dev Biol. 1983;192:62–74. doi: 10.1007/BF00848482. [DOI] [PubMed] [Google Scholar]
- 25.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J E, MacKenzie A, Korneluk R G. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 26.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Bermudo M D, Carmena A, Jimenez F. Neurogenic genes control gene expression at the transcriptional level in early neurogenesis and in mesectoderm specification. Development. 1995;121:219–224. doi: 10.1242/dev.121.1.219. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura H, Yoshida M, Tsuiki H, Ito K, Ueno M, Nakao M, Oka K, Tada M, Kochi M, Kuratsu J, Ushio Y, Saya H. Identification of a human homolog of the Drosophila neuralized gene within the 10q25.1 malignant astrocytoma deletion region. Oncogene. 1998;16:1009–1019. doi: 10.1038/sj.onc.1201618. [DOI] [PubMed] [Google Scholar]
- 29.Namiki M. Genetic aspects of male infertility. World J Surg. 2000;24:1176–1179. doi: 10.1007/s002680010198. [DOI] [PubMed] [Google Scholar]
- 30.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 31.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 32.Palmblad J, Mossberg B, Afzelius B A. Ultrastructural, cellular, and clinical features of the immotile-cilia syndrome. Annu Rev Med. 1984;35:481–492. doi: 10.1146/annurev.me.35.020184.002405. [DOI] [PubMed] [Google Scholar]
- 33.Price B D, Chang Z, Smith R, Bockheim S, Laughon A. The Drosophila neuralized gene encodes a C3HC4 zinc finger. EMBO J. 1993;12:2411–2418. doi: 10.1002/j.1460-2075.1993.tb05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano T, Kovacs K, Stefaneanu L, Asa S L, Snyder D L. Spontaneous pituitary gonadotroph nodules in aging male Lobund-Wistar rats. Lab Investig. 1989;61:343–349. [PubMed] [Google Scholar]
- 35.Sassoon D, Lyons G, Wright W E, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- 36.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 37.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 38.Strom A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11:3168–3181. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 40.Struhl G, Greenwald I. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc Natl Acad Sci USA. 2001;98:229–234. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 42.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Sdrulla A D, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres B A. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 44.Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 45.Williamson R A, Koehler J K, Smith W D, Stenchever M A. Ultrastructural sperm tail defects associated with sperm immotility. Fertil Steril. 1984;41:103–107. doi: 10.1016/s0015-0282(16)47549-7. [DOI] [PubMed] [Google Scholar]
- 46.Wilton L J, Temple-Smith P D, de Kretser D M. Quantitative ultrastructural analysis of sperm tails reveals flagellar defects associated with persistent asthenozoospermia. Hum Reprod. 1992;7:510–516. doi: 10.1093/oxfordjournals.humrep.a137681. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Fang S, Jensen J P, Weissman A M, Ashwell J D. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 48.Yeh E, Zhou L, Rudzik N, Boulianne G L. Neuralized functions cell autonomously to regulate drosophila sense organ development. EMBO J. 2000;19:4827–4837. doi: 10.1093/emboj/19.17.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]