Abstract
Background:
Polygenic risk scores (PRS) which summarize individuals’ genetic risk profile may enhance targeted colorectal cancer (CRC) screening. A critical step towards clinical implementation is rigorous external validations in large community-based cohorts. This study externally validated a PRS-enhanced CRC risk model comprising 140 known CRC loci to provide a comprehensive assessment on prediction performance.
Methods:
The model was developed using 20,338 individuals and externally validated in a community-based cohort (n=85,221). We validated predicted 5-year absolute CRC risk, including calibration using expected-to-observed case ratios (E/O) and calibration plots, and discriminatory accuracy using time-dependent AUC. The PRS-related improvement in AUC, sensitivity and specificity were assessed in individuals of age 45–74 years (screening-eligible age group) and 40–49 years with no endoscopy history (younger-age group).
Results:
In European-ancestral individuals, the predicted 5-year risk calibrated well (E/O=1.01 (95%CI 0.91–1.13)) and had high discriminatory accuracy (AUC=0.73 (95%CI 0.71–0.76)). Adding the PRS to a model with age, sex, family and endoscopy history improved the 5-year AUC by 0.06 (p-value<0.001) and 0.14 (p-value=0.05) in the screening-eligible age and younger-age groups, respectively. Using a risk-threshold of 5-year SEER CRC-incidence rate at age 50 years, adding the PRS had a similar sensitivity but improved the specificity by 11% (p-value<0.001) in the screening-eligible age group. In the younger-age group it improved the sensitivity by 27% (p-value=0.04) with similar specificity.
Conclusions:
The proposed PRS-enhanced model provides a well-calibrated 5-year CRC risk prediction and improves discriminatory accuracy in the external cohort.
Impact:
The proposed model has potential utility in risk-stratified CRC prevention.
Keywords: Colorectal cancer, Cancer risk assessment, Personalized cancer prevention, Discriminatory accuracy, Gene polymorphisms, Familial and hereditary cancers
Introduction
Colorectal cancer (CRC) is among the leading causes of cancer death.1 Despite decreasing CRC incidence overall, the incidence rate in individuals aged <50 years has been increasing over the last decades,2 leading to a recent recommendation by the US Preventive Services Task Force (USPSTF) to lower the age at screening initiation to 45 years for individuals at average risk.3 However, given the enormous burden of nearly 22 million additional people becoming eligible for screening and that CRC remains a rare event in younger individuals, targeted screening based on an individual’s risk factors has received much attention and may be an appealing alternative to a universal change of the screening age. 4–10 CRC risk prediction models summarize individuals’ CRC risk based on their risk profile and quantitatively position them on the risk spectrum. There is a growing interest in developing accurate and precise CRC risk prediction models to achieve risk stratification and targeted screening.
Polygenic risk scores (PRS), which uniquely summarize individuals’ genetic risk profile, have shown promising potential in CRC risk prediction.11–20 A PRS-enhanced CRC risk model is a model that adds genetic information via a PRS to other known CRC risk predictors, such as age and family history. Prior studies investigated the impact of PRS (comprising subsets of current known CRC loci) on CRC risk11,12,14 and assessed the contribution of these PRS to model discrimination, which is the ability to assign higher risk scores to cases than non-cases.11–13,15–17,19,20 However, due to the lack of external cohorts with sufficient events and available genetic information, earlier validation studies relied on either internal validation in a reserved subgroup or cross-validation, or external validation using UK Biobank16,17,20 which has been included in recent GWAS discoveries.21 Therefore, existing data are not adequate for externally validating a model incorporating the CRC loci discovered in these recent GWAS. Despite supportive findings from some of these studies, more real-world evidence on the validity of PRS-enhanced CRC risk models in large and diverse independent cohorts, particularly those reflecting sociodemographic diversity in consensus population, are warranted before the inclusion of PRS to CRC risk stratification can be considered at a population scale. In addition, limited consideration has been given to risk calibration, the closeness of the predicted and the observed risks. Without this crucial step in validation, it remains unclear if PRS-enhanced models reliably predict CRC risks observed in practice.
The Genetic Epidemiology Research on Adult Health and Aging cohort (GERA) established at Kaiser Permanente Northern California (KPNC) provides an opportunity, often rare in the era of large GWAS consortia, to externally validate PRS-enhanced CRC risk models as it has not been used in any CRC GWAS discoveries. KPNC, one of the large integrated health care systems serving 30–40% general population in northern California including Medicare and Medicaid patients, has a member cohort broadly representative of the regional consensus population’s demographics.22 The GERA, with its large sample size, community-based and sociodemographically diverse population, and detailed clinical and genetic information, is uniquely positioned for real-world community-based validation on PRS-enhanced CRC risk models.
In this study, we extended a PRS-enhanced model15 using individual-level demographic, clinical and genetic information, and externally validated the proposed model in the GERA, including a comprehensive assessment of absolute risk calibration and discriminatory accuracy. Our model includes an updated PRS comprising 140 CRC risk loci (including 77 additional loci from recent GWAS21,23,24 compared to the PRS validated in Jeon et al. 201815) in addition to age, sex, first-degree family history of CRC (hereafter termed as family history) and endoscopy (including colonoscopy and sigmoidoscopy) history. We conducted a time-dependent validation under a time-to-event framework accounting for competing risk of mortality, which is important as the onset of CRC occurs generally later in a person’s life. In addition, we evaluated the gain in prediction accuracy of including PRS in CRC risk prediction models in screening-eligible individuals with age 45–74 years, reflecting a recent USPSTF recommendation on CRC screening age,3 and individuals with age 40–49 years and no endoscopy history as the latter group may benefit from a risk-stratified screening-initiation strategy using the PRS-enhanced model.
Materials and Methods
Study Population: Model Building
We extended the risk prediction model using a subset of Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) and Colorectal Cancer Transdisciplinary Study (CORECT) (9,748 CRC cases and 10,590 controls of European ancestry) with detailed harmonized data on age, family history, endoscopy history, and genetic risk factors (Supplemental Materials in Jeon et al. (2018)15 and Supplemental Table S1) along with CRC-incidence rates in SEER data (https://seer.cancer.gov/, registry18, 2007–2015, RRID:SCR_006902).25
Study Population: Model Validation
External validation was carried out in the GERA (Supplemental Methods and Supplemental Table S2). Briefly, the cohort is comprised of study participants (survey respondents) from the Research Program on Genes, Environment, and Health (RPGEH) and California Men’s Health Study (CMHS), both nested within the member population of KPNC.26,27 In total, 110,266 consenting participants who provided saliva samples were selected for genotyping. All racial and ethnic minority participants who provided saliva samples (n=20,935, 19%) were selected for genotyping to maximize diversity and a random subset were selected from the approximately 140,000 available non-Hispanic White participants with biospecimens. 94,938 participants passed genotype quality control and had a valid PRS. The demographic characteristics of the GERA are generally representative of the RPGEH and CMHS survey respondents; compared with the KPNC member population, the GERA participants are on average older (average age at time of sample collection: 63 years), have a slightly greater proportion of non-Hispanic whites, and have higher levels of education and income. In the validation dataset, prevalent CRC cases were excluded from the analyses (n=865). Participants who entered the cohort at ages <40 years (n=6,622) and ≥85 years (n=2,230) were excluded since few CRC cases were diagnosed at ages <40 years and aggregated SEER CRC rates for ages ≥85 years hindered a reliable estimation of CRC risk in this age group. The number of participants after exclusion was 85,221. Due to the relatively small number of CRC cases in non-European ancestry groups, we restricted our primary analysis to European-ancestry participants and secondarily evaluated the calibration of relative risk of PRS in other racial and ethnic groups. A comparison on risk factors between the GERA and GECCO/CORECT was shown in Supplemental Table S3. All study participants provided written informed consent and the study was approved by the KPNC and Fred Hutchinson Cancer Research Center Institutional Review Board.
Developing Risk Prediction Models in GECCO and CORECT
We estimated sex-specific odds ratios (ORs) of PRS, family history and endoscopy history associated with CRC risk using logistic regression models, adjusting for study and age, using pooled GECCO and CORECT data. The PRS was calculated as a weighted sum of numbers of the effective alleles of the 140 known CRC loci (Supplemental Table S4 and S4a) with the marginal log-odds ratios estimated from >125,000 samples, predominantly of European ancestry, as weights.21 Baseline CRC hazard rates were derived using CRC-incidence rates in SEER18 (2007–2015). The absolute risk estimates for CRC were calculated based on estimated sex-specific baseline CRC hazard rates and ORs (Supplemental Table S5), while accounting for competing risks from death (Supplemental Methods).25,28
Projecting Risk in the GERA
The 5- and 10-year absolute CRC risks for each GERA participant were estimated from the model based on age, sex, family history, endoscopy history, and PRS of the 140 known loci (Supplemental Methods). To account for the diminishing protective impact of endoscopy over time, we classified patients who had received their last endoscopy greater than 10 years ago as no endoscopy. We chose 10 years based on the current recommended screening interval following a colonoscopy.
Statistical Analysis
We primarily focused on the predicted 5-year risk and secondarily on the 10-year risk given that both family history and endoscopy history in our model varied over time, and risk prediction every 5 years was likely more accurate. All analyses were conducted under survival analysis framework to account for varying follow-up duration across participants, with the observed time-to-event defined as the time from study entry to the earliest of the following events: CRC diagnosis, death, last follow-up, or 6-months after first colonoscopy post study entry (Supplemental Methods).
Calibration29
We compared model-based expected CRC cases (E) to observed CRC cases in the GERA (O) within t years (t=5 and 10) following study entry, where E was the sum of individuals’ absolute risk predictions at either the observed time-to-event or t-year, whichever comes first. The 95% confidence interval (CI) was calculated by using a normal approximation to Poisson distribution as . We assessed t-year E/O ratios overall and in clinically meaningful subgroups defined by age (age 40–49, 50–59 and ≥60), sex (men, women), endoscopy history within 10 years prior to study entry (yes, no), and family history (yes, no). We further evaluated the consistency between E and O across 10 risk-based deciles based on the predicted 5- and 10-year CRC risks using calibration plots (Supplemental Methods). O in the jth decile for the t-year risk was estimated by , where is the Kaplan-Meier estimator for CRC-free probability accounting for right-censoring and competing risk.30,31 As a secondary analysis, we evaluated how the relative risk (RR) of PRS calibrates in African American, Asian, European-ancestry, and Latinx groups by comparing the predicted and observed RR in 7 PRS-based strata in individual ancestral groups (Supplemental Methods). The stratum that included the sample medium of the PRS was set as the reference group in the RR calculation. The observed RR of a PRS stratum was estimated by fitting a Cox model with a 0–1 stratum indicator as the covariate using individuals included in this specific stratum and the reference stratum.
Discriminatory Accuracy
We evaluated the discriminatory accuracy of 5- and 10-year risk predictions by time-dependent AUC, accommodating time-varying CRC status and predictors during follow-up. We accounted for competing risks of death32 since 8.2% of participants experienced death without prior CRC diagnosis.
We compared the AUC of the proposed PRS-enhanced model with two reduced models that excluded PRS, Model 1 (age and first-degree family history; Supplemental Methods) as this is currently used to inform screening and Model 2 (age, first-degree family history, sex, and endoscopy history; Supplemental Methods) in all European-ancestry individuals and those aged 45–74 years eligible for screening (termed screening-eligible group hereafter). We also compared the AUC in individuals aged 40–49 years without endoscopy history (termed younger-age group hereafter) to explore whether the proposed PRS-enhanced model can better inform strategic screening initiation. Additionally, we compared the 5-year risk prediction of these models in terms of two clinically relevant measures, sensitivity and specificity of identifying a high 5-year risk group with a risk threshold of 0.29%, the SEER 5-year CRC incidence rate at age 50 when CRC screening initiation was recommended conventionally for people at average CRC risk in the US.
We obtained the 95% confidence intervals (CI) and p-values by bootstrapping with 500 resamples. A two-sided p-value ≤0.05 was considered statistically significant. All analyses were performed using R 3.633 and plots were generated using the R package ‘ggplot2’.34
Data availability
Access to GERA data used in this study may be obtained by application to the Kaiser Permanente Research Bank (KPRB) via ResearchBankAccess@kp.org. A subset of the GERA consented for public use can be found at NlH/dbGaP: phs000674. Genotype data from CORECT and GECCO are deposited at NIH/dbGaP: phs001415.v1.p1, phs001315.v1.p1, phs001078.v1.p1, phs001499.v1.p1, phs001903.v1.p1, phs001856.v1.p1, phs001045.v1.p1, and phs001499.v1.p1.
Results
Our primary analysis was restricted to GERA participants of European ancestry (n=66,282, 78%). Among them, 57.7% were women, 57.5% underwent endoscopy before study entry, and 9.9% had a positive family history. The mean age at study entry was 63.2 years (Supplemental Table S2). 30% of men were enrolled in 2002–2003, earlier than the rest of the GERA. However, no appreciable difference in CRC rate was observed between this subgroup and the rest of the GERA. Overall, men and women were comparable in age, family history, endoscopy history and PRS (Supplemental Table S2). The sex-specific odds ratio estimates of these risk factors are provided in Supplemental Table S5.
Calibration
The PRS-enhanced model accurately predicted the number of CRC cases in a 5-year window. The overall E/O ratio for 5-year risk was 1.01 (95% CI 0.91–1.13); the subgroup E/O ratios were close to 1 with 95% CIs including 1 (Table 1). The calibration for 10-year risk performed generally well (overall E/O ratio 1.00, 95% CI 0.92–1.09, Supplemental Table S6), except for an underestimation in individuals with a positive family history (E/O ratio 0.77, 95% CI 0.61–0.98), and an overestimation in the age subgroup of 50 to 59 years (E/O ratio 1.42, 95% CI 1.11–1.83). The risk calibration stratified by the risk-based deciles (Figure 1 left panel, Supplemental Figures S1–S5) demonstrated the same patterns of good calibration.
Table 1.
5-year E/O CRC case ratio in the GERA European-ancestry participants. We presented the comparison of expected (E) and observed (O) CRC cases based on the PRS-enhanced model (including age, first-degree CRC family history, sex, endoscopy history and polygenic risk score) in the 5 years follow-up via E/O ratio and the corresponding 95% confidence intervals in GERA participants of European ancestry. The number of expected cases was calculated based on age, first-degree family history, sex, endoscopy history and polygenic risk score (PRS); The number of observed cases was based on diagnosed cases in the cohort.
| Stratum | E | O | E/O ratio (95% CI) |
|---|---|---|---|
| Overall | 313.56 | 309 | 1.01 (0.91, 1.13) |
| Age at study entry, years | |||
| 40–49 | 13.96 | 11 | 1.27 (0.70, 2.29) |
| 50–59 | 48.40 | 37 | 1.31 (0.95, 1.81) |
| >=60 | 251.19 | 261 | 0.96 (0.85, 1.09) |
| First-degree CRC family history a | |||
| Negative | 279.24 | 270 | 1.03 (0.92, 1.17) |
| Positive | 34.32 | 39 | 0.88 (0.64, 1.20) |
| Sex | |||
| Women | 160.66 | 173 | 0.93 (0.80, 1.08) |
| Men | 152.90 | 136 | 1.12 (0.95, 1.33) |
| Endoscopy history b | |||
| No | 136.55 | 139 | 0.98 (0.83, 1.16) |
| Yes | 177.01 | 170 | 1.04 (0.90, 1.21) |
First-degree family history of ascertained from study survey questionnaires and electronic health records.
Endoscopy history in 10 years prior to study entry.
Figure 1.
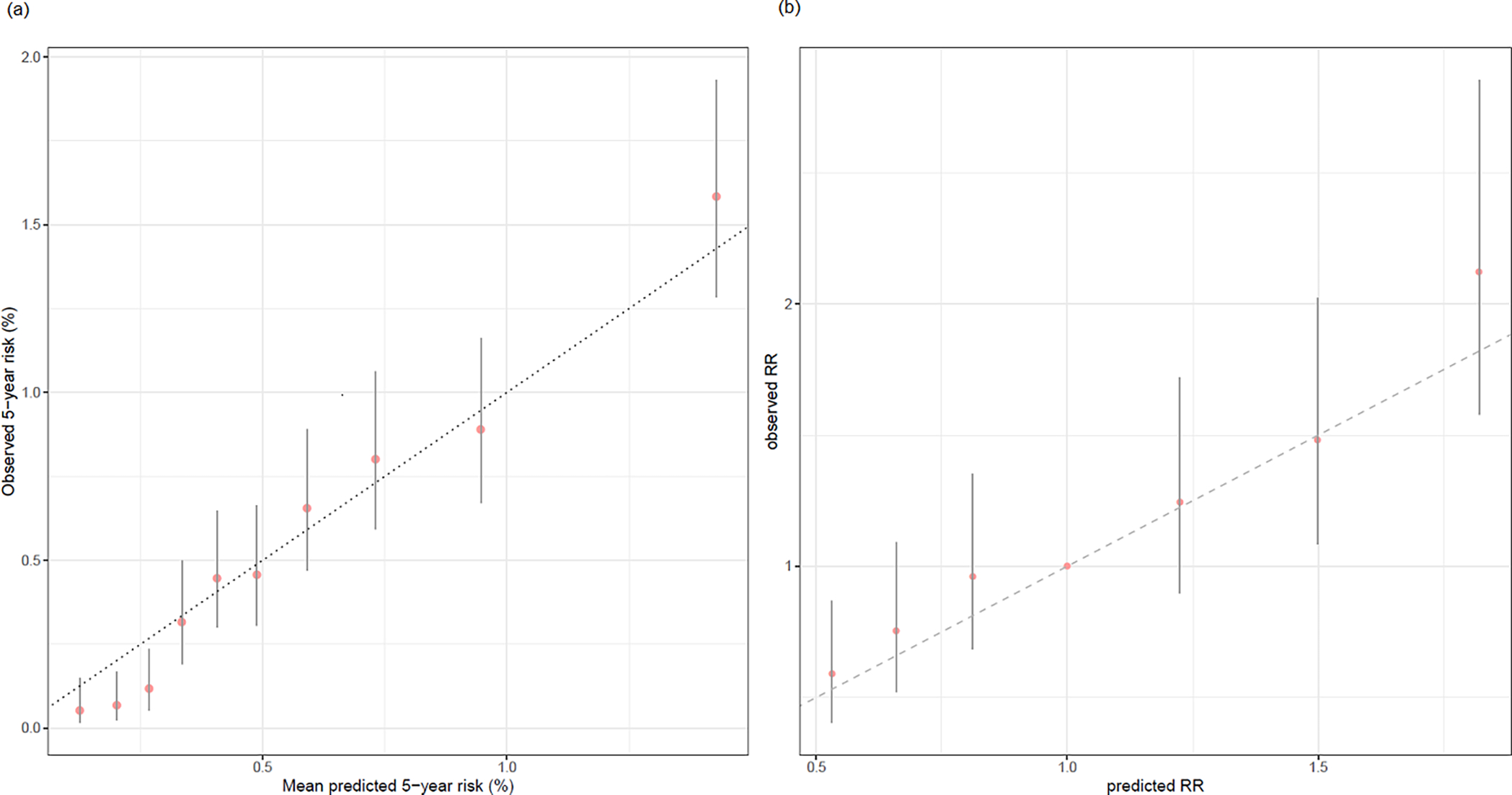
Calibration plots of 5-year absolute risk and relative risk of PRS with risk-based and PRS-based stratification. We demonstrated the calibration of 5-year absolute risk based on the PRS-enhanced model (1(a); left panel) and relative risk of PRS (1(b); right panel) stratified by deciles of predicted risk (left) and PRS (right) in GERA participants of European-ancestry. The x-axis is the average of predicted absolute (left) and relative (right) risk values by deciles. The y-axis is the average observed absolute (left) and relative (right) risks by deciles of predicted absolute risk (left) and PRS (right), along with their 95% CIs indicated by the vertical error bars.
The model-based relative risks (RR) of PRS calibrated well across the PRS range in the European-ancestry group (Figure 1 right panel). For the non-European ancestry groups, the model-based RR of PRS were generally lower than the observed RR (Supplemental Figure S6). However, the 95% confidence intervals of the observed RR were wide due to the limited non-European ancestry CRC cases and covered the model-based RR.
Discriminatory Accuracy
The overall 5-year AUC of the PRS-enhanced risk model was 0.73 (95%CI, 0.71–0.76, Table 2). The AUC did not vary by sex or endoscopy history. The AUC among individuals with a positive family history was 0.78 (95%CI 0.70–0.86), 5% (95%CI −3%–13%) higher than the AUC in those without (0.73, 95%CI 0.70–0.75). The AUC was 0.77 (95%CI 0.70–0.84) in individuals aged 40–49 years and 0.72 (95%CI 0.67–0.77) in 50–59 years, 10% (95%CI 2%–17%) and 5% (95%CI −1%–11%) higher than in those aged 60 years or older (0.67, 95%CI, 0.64–0.71), respectively. The 10-year AUCs generally followed a similar pattern (Supplemental Table S7).
Table 2.
5-year time-dependent AUC estimates of the PRS-enhanced model in the GERA European-ancestry participants. The time-dependent AUC estimates (95% confidence interval) of predicted 5-year absolute risk are based on the PRS-enhanced model including age, first-degree CRC family history, sex, endoscopy history and polygenic risk score.
| Stratum | AUC (95% CI) | Difference to the lowest AUC in each stratification (95% CI) |
|---|---|---|
| Overall | 0.73 (0.71, 0.76) | Not available |
| Age at study entry, years | ||
| 40–49 | 0.77 (0.70, 0.84) | 10% (2%, 17%) |
| 50–59 | 0.72 (0.67, 0.77) | 5% (−1%, 11%) |
| >=60 | 0.67 (0.64, 0.71) | Reference |
| First-degree CRC family History | ||
| Negative | 0.73 (0.70, 0.75) | Reference |
| Positive | 0.78 (0.70, 0.86) | 5% (−3%, 13%) |
| Sex | ||
| Women | 0.73 (0.70, 0.76) | Reference |
| Men | 0.74 (0.70, 0.79) | 1% (−4%, 6%) |
| Endoscopy history | ||
| No | 0.73 (0.69, 0.77) | Reference |
| Yes | 0.73 (0.70, 0.77) | 0% (−5%, 5%) |
The 5-year AUC of the PRS-enhanced model in the screening-eligible group was 0.70 (95%CI 0.67–0.74), which was 6% (p-value<0.001) higher than Model 2 (AUC 0.64; when including age and family history sex, endoscopy history; Table 3). An improvement was also observed in those aged 40–49 years without endoscopy history (0.76 vs. 0.62, p-value=0.04) and in all European-ancestry participants (0.73 vs. 0.71, p-value=0.04). The comparison of the PRS-enhanced model to Model 1 of age and family history showed very similar results. The 10-year AUC (Supplemental Table S8) also showed a similar pattern, except that the AUC improvements were not significant in the younger-age group.
Table 3.
Sequential model comparison using 5-year time-dependent AUC in the GERA European-ancestry participants. We compared time-dependent AUCs of 5-year absolute risks based on Model 1 (age and first-degree CRC family history) and Model 2 (age, first-degree CRC family history, sex, and endoscopy history) to the proposed PRS-enhanced model (age, first-degree CRC family history, sex, endoscopy history and polygenic risk score) in GERA European-ancestry participants.
| Model | AUC | P-valuea |
|---|---|---|
| All participants eligible for screening with ageb 45–74 years | ||
| Model 1 | 0.64 | <0.001 |
| Model 2 | 0.64 | <0.001 |
| PRS-enhanced model | 0.70 | Reference |
| All participants with ageb 40–49 years and having no endoscopy history | ||
| Model 1 | 0.60 | 0.04 |
| Model 2 | 0.62 | 0.05 |
| PRS-enhanced model | 0.76 | Reference |
| All participants | ||
| Model 1 | 0.71 | 0.04 |
| Model 2 | 0.71 | 0.01 |
| PRS enhanced model | 0.73 | Reference |
P-value for comparing the AUC estimates of a reduced model to the PRS-enhanced model.
Age at study entry.
Using a risk threshold of 0.29% to identify a high 5-year risk group in the screening-eligible group, no appreciable difference in sensitivity was observed across models, but specificity of the proposed PRS-enhanced model was 31.0% (95%CI: 30.6% – 31.4%), which was 15% (p-value<0.001) and 11% (p-value<0.001) higher than Model 1 and Model 2, respectively (Table 4). A similar pattern was observed in all European-ancestry individuals. In the younger-age group, using the PRS-enhanced model to define a high 5-year risk group demonstrated a sensitivity of 64% (95%CI 35%–92%), which was 55% (p-value<0.001) and 27% (p-value=0.04) higher than Models 1 and 2, respectively, and had a specificity of 80% (95%CI 79%–81%), which was 17% (p-value<0.001) lower than Model 1 and 1% (p-value=0.05) higher than Model 2.
Table 4.
Sequential model comparison using 5-year sensitivity/specificity in the GERA European-ancestry participants. We presented the comparison across Model 1 (age and first-degree CRC family history), Model 2 (age, first-degree CRC family history, sex, and endoscopy history) and the PRS-enhanced model (age, first-degree CRC family history, sex, endoscopy history and polygenic risk score) on sensitivity and specificity with a risk threshold of 0.29% among GERA participants of European ancestry.
| Measure estimatea (95% CI) | Difference to PRS-enhanced model (95% CIb) | P-valueb | |
|---|---|---|---|
| All participants eligible for screening with agec 45–74 years | |||
| Sensitivity (177 individuals) | |||
| Model 1 | 93.8% (85.2%, 96.9%) | 0% (−5.2%, 3.9%) | p>0.99 |
| Model 2 | 90.4% (85.1%, 94.3%) | −3.4% (−7.5%, 0.8%) | p=0.11 |
| PRS-enhanced model | 93.8% (85.2%, 96.9%) | Reference | Reference |
| Specificity (52,890 individuals) | |||
| Model 1 | 16.4% (16.1%, 16.7%) | −14.6% (−15.0%, −14.2%) | p<0.001 |
| Model 2 | 20.1% (19.8%, 20.4%) | −10.9% (−11.3%, −10.6%) | p<0.001 |
| PRS-enhanced model | 31.0% (30.6%, 31.4%) | Reference | Reference |
| All participants with agec 40–49 years and having no endoscopy history | |||
| Sensitivity (11 individuals) | |||
| Model 1 | 9% (0%, 26%) | −55% (−86%, −25%) | p<0.001 |
| Model 2 | 36% (8%, 65%) | −27% (−54%, 0%) | p=0.04 |
| PRS-enhanced model | 64% (35%, 92%) | Reference | Reference |
| Specificity (6889 individuals) | |||
| Model 1 | 97% (96%, 97%) | 17% (16%, 18%) | p<0.001 |
| Model 2 | 79% (78%, 80%) | −1% (−2%, 0%) | p=0.05 |
| PRS-enhanced model | 80% (79%, 81%) | Reference | Reference |
| All participants: age 40–84 years | |||
| Sensitivity (309 individuals) | |||
| Model 1 | 96.1% (94.0%, 98.3%) | 0% (−2.7%, 2.7%) | p>0.99 |
| Model 2 | 94.2% (91.6%, 96.8%) | −1.9% (−4.4%, 0.3%) | p=0.09 |
| PRS-enhanced model | 96.1% (94.0%, 98.3%) | Reference | Reference |
| Specificity (65973 individuals) | |||
| Model 1 | 17.1% (16.8%, 17.4%) | −11.7% (−12.0%, −11.4%) | p<0.001 |
| Model 2 | 20.0% (19.7%, 20.4%) | −8.8% (−9.0%, −8.4%) | p<0.001 |
| PRS-enhanced model | 28.8% (28.4%, 29.1%) | Reference | Reference |
We dichotomized 5-year risks derived from each model to high- (>0.29%) and low-risk (≤0.29%) categories. The risk threshold of 0.29% represents the 5-year incidence CRC rate for average-risk individuals at age 50 based on SEER18 CRC incidence rates (2007–2015).
The 95% confidence intervals (CI) of the differences and the corresponding p-values were obtained by bootstrap resampling with 500 resamples.
Age at study entry.
Discussion
The current analysis, using large training and validation datasets, substantially extends existing CRC risk prediction knowledge by incorporating an updated PRS and conducting a comprehensive external validation in a community-based cohort. The predicted 5- and 10-year CRC risks from the proposed PRS-enhanced risk model calibrated well overall in European-ancestry participants. The model accurately discriminated CRC cases and controls. In addition, the inclusion of the PRS in CRC risk models significantly improved the discriminatory accuracy of 5-year predicted risk in the screening-eligible group and the younger-age group. Using the average 5-year CRC risk at age 50 years as a threshold, adding the PRS leads to significant improvement in the specificity in the screening-eligible age group and the sensitivity in the younger-age group. The latter group with its low prevalence of CRC would likely benefit substantially from risk-stratified screening. Our study provides empirical support to warrant future studies on the evaluation of PRS-enhanced risk models focusing on younger population.
The PRS-enhanced model calibrated well overall and in subgroups defined by sex, family history, endoscopy history and age, with some overestimation of the 10-year risk in participants aged 50 to 59 years and underestimation among those with a family history of CRC. A possible explanation for the overestimation in those aged 50 to 59 years is that there were two spikes in the SEER CRC incidence rates around ages 50 and 65 years (Supplemental Figure S7), which were used to calculated baseline risk. These possibly result from increased screen-detected CRC as people tend to undergo screening at these ages, which include the historical initiation of screening at age 50 and the beginning of insurance coverage with Medicare at age 65. The underestimation of the 10-year risk among those with a family history is likely due to the underestimated effect of family history on CRC risk in GECCO/CORECT compared to that in the GERA. By fitting a Cox proportional hazards model on CRC with endoscopy history, family history and PRS in the GERA, we observed that positive family history is associated with 2.2- and 1.6-times higher CRC hazard ratios, in men and women, respectively, greater than the observed increased risk of 1.3 and 1.2 in men and women in GECCO/CORECT. Since it is difficult to reliably predict long-term (such as 10-year or lifetime) risk, one may consider updating risk prediction periodically using predicted 5-year risk based on one’s most recent risk profiles, which is more accurate35 and reflective of the clinical needs of a risk model36 when recommendation on screening/intervention for the near future is needed.
Prior studies have validated other CRC risk models incorporating genetic components.11–13,15–17,19,37–39 The AUCs of our model in external validation are comparable to or better than the best AUCs in these studies, especially considering that internal validation as used in prior studies tends to yield more optimistic results.40 Calibration of CRC absolute risk predicted by PRS-enhanced models has been given little emphasis in the literature and yet is crucial for determining if a model provides reliable predictions at an individual level. This study examined the absolute risk calibration of the proposed PRS-enhanced risk model among European-ancestry participants, overall and in several subgroups, and demonstrated that it provides accurate predictions. We also noted in an exploratory analysis comparing a risk model including endoscopy history versus not, that the endoscopy history does not show a strong predictive value in terms of AUC improvement (Supplemental Table S9) despite its significant association with CRC risk observed in the GERA [hazard rate 0.69 (p=0.004) in men and 0.73 (p=0.009) in women estimated from a Cox model] and an earlier study.41 This observation reflects a phenomenon that significant variables do not guarantee improvement in prediction, as discussed in Lo et al. (2015).42
The improved discrimination using the PRS-enhanced model in the younger-age group, although there are only 11 CRC cases in this group in our study, demonstrates the model’s potential in risk stratification and supports the warrant for a further evaluation in this cohort with a sufficient sample size and number of CRC cases to assess the model-based triage of screening initiation. The result also supports our prior finding of a stronger predictive value at younger ages for PRS comprising of 95 known loci.43 Recently the USPSTF recommended to lower the starting age for screening to 45 years,3 addressing rising incidence rates of early onset CRC over the last two decades.44 However, concerns remain with this new recommendation, including the lack of outcome data supporting earlier screening and whether existing screening resources can absorb the nearly 22 million newly screening-eligible adults. As such, this risk model could be a starting point for identifying young individuals of higher CRC risk for earlier screening.
Advances in PRS development have invigorated the interest in using PRS-enhanced models for risk stratification to facilitate targeted screening or intervention. The decreasing genome-wide genotyping cost has made it affordable to implement PRS broadly. However, barriers of implementation of PRS in healthcare remain and need further study, including PRS tailored for different ancestries, education of clinicians and public, and adaptation of health care systems to manage and utilize continuously improved PRS.45 For the first barrier, the PRS developed in cohorts predominately of European-ancestry individuals may not predict well in non-European ancestral groups, due to different linkage disequilibrium patterns resulting likely attenuated genetic associations.46–49 We anticipate an expansion of PRS development and validation to include more ancestrally diverse populations to ensure equity in targeted screening across all populations. The second barrier will require more promotion and education to enhance clinician and public awareness of utilizing genetic profile to inform complex disease risks. The last barrier is related to the continuous discovery of more CRC-associated loci and updates on scoring algorithms for the PRS. However, it does not require genome-wide genotyping on an individual multiple times since one’s genetic profile is time-invariant and genome-wide information can be stored digitally once it is collected.45 Prior to updating the PRS in healthcare implementation, a cost-benefit evaluation to weigh the cost of a system update to adapt a new scoring algorithm and the improvement in predictive performance of the updated PRS may be needed.
There are several strengths of our study. First, the proposed model includes an updated PRS comprising 140 known CRC loci. Since the publication of earlier studies on CRC risk models with PRS,11–16,19 a sizable number of CRC risk loci have been discovered in large-scale GWAS.21,23,24 Our work provides empirical evidence supporting that adding this updated PRS to a risk model improves risk stratification. Second, this is the first external validation for the updated PRS-enhanced CRC risk model. Assessing model performance in an independent validation cohort as shown in our study minimizes the impact of optimism which is usually a concern in internal validation.40 Third, the GERA provides a unique and rare opportunity with detailed demographics, clinical and genetic information to externally validate the proposed PRS-enhanced model in a community-based setting. The comprehensive assessment shown in this study, including absolute risk calibration and model discrimination, are more generalizable to census population compared to assessment obtained in typical research cohorts. Overall, our study supplies warranted evidence for moving the inclusion of PRS in CRC screening further toward clinical consideration.
Our study has some limitations. First, the GERA included participants from two large cohorts of KPNC members recruited in 2002–2003 and 2007–2008. As the baseline CRC hazard was derived using SEER18 2007–2015, the prediction of participants in the earlier cohort may be less accurate. However, there was no appreciable difference between the empirical CRC incidence rates in these two cohorts. Second, the GERA was drawn from a single US geographic region and may not reflect the overall US population. Additional validations are needed to comprehensively assess the model performance in the overall US population and more globally. Third, only 11 European-ancestry individuals in the younger-age group developed CRC within 5 years since study entry. Our findings of greater AUC improvement in this subgroup may not be robust and generalizable to other cohorts due to the limited number of CRC cases. Further study focusing on individuals of age 40–49 years is warranted to confirm these results. Fourth, our model did not include lifestyle and environmental risk factors such as smoking, dietary, and non-steroidal anti-inflammatory drugs use. These factors are difficult to measure precisely in clinical practice, as they are commonly obtained using questionnaires, hence prone to recall bias and measurement errors.12 Further research is needed before incorporating these factors in risk prediction models to ensure accuracy.
Conclusion
We externally validated that the PRS-enhanced CRC risk model was calibrated well and had high predictive performance among European-ancestral individuals in a large, community-based cohort. Our findings demonstrated improvement in risk discrimination and classification accuracy with the addition of the updated PRS in individuals eligible for screening and potentially in those aged <50 years. The proposed PRS-enhanced model may aid in developing targeted screening strategy to improve screening efficiency and further enhance CRC prevention.
Supplementary Material
Acknowledgement:
This work was primarily supported by the National Institutes of Health (NIH, grant number R01CA206279[MPI: U. Peters; D.A. Corley; R. B. Hayes], R01CA195789[PI: L. Hsu], R01CA189532[PI: L. Hsu], UM1CA222035[MPI: D.A. Corley; J. K. Lee], K07CA188142 [PI: L. C. Sakoda], R03CA215775 [PI: R. B. Hayes] and K07CA212057 [PI: J. K. Lee]). Scientific Computing Infrastructure at Fred Hutch supporting the analyses in this study was funded by NIH Office of Research Infrastructure Programs grant S10OD028685. Additionally, individual cohorts and studies in GECCO/CORECT/GERA were supported by other funding resources as listed in the supplement. The National Institutes of Health had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The authors greatly thank participants, interviewers, coordinators, data managing staff, and all researchers in the GERA, GECCO and CORECT. Y. Su thanks Kaiser Permanente Washington for supporting the writing of this manuscript.
Footnotes
Conflict of Interest Disclosure
The authors declared no conflicts of interest outside the grant funding listed in funding section (in the main manuscript and the supplementary document).
Ethics Declaration: The study was approved by the Fred Hutchinson Cancer Research Center and Kaiser Permanente Northern California Institutional Review Board. All study participants provided written informed consent as required by the IRBs. The data was de-identified.
References:
- 1.American Cancer Society. Cancer Facts & Figures for African Americans 2019–2021. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/cancer-facts-and-figures-for-african-americans/cancer-facts-and-figures-for-african-americans-2019-2021.pdf
- 2.Murphy CC, Singal AG, Baron JA, Sandler RS. Decrease in Incidence of Young-Onset Colorectal Cancer Before Recent Increase. Gastroenterology. 2018;155(6):1716–1719.e4. doi: 10.1053/j.gastro.2018.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Preventive Services Task Force. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2021;325(19):1965. doi: 10.1001/jama.2021.6238 [DOI] [PubMed] [Google Scholar]
- 4.Lieberman DA. Targeted colon cancer screening: a concept whose time has almost come. Am J Gastroenterol. 1992;87(9):1085–1093. [PubMed] [Google Scholar]
- 5.Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, et al. Estimation of Benefits, Burden, and Harms of Colorectal Cancer Screening Strategies: Modeling Study for the US Preventive Services Task Force. JAMA. 2016;315(23):2595–2609. doi: 10.1001/jama.2016.6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos FG. Colorectal cancer in young adults: A difficult challenge. World J Gastroenterol. 2017;23(28):5041–5044. doi: 10.3748/wjg.v23.i28.5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colorectal Cancer Screening: Recommendations for Physicians and Patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Am J Gastroenterol. 2017;112(7):1016–1030. doi: 10.1038/ajg.2017.174 [DOI] [PubMed] [Google Scholar]
- 8.Corley DA, Peek RM. When Should Guidelines Change? A Clarion Call for Evidence Regarding the Benefits and Risks of Screening for Colorectal Cancer at Earlier Ages. Gastroenterology. 2018;155(4):947–949. doi: 10.1053/j.gastro.2018.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68(4):250–281. doi: 10.3322/caac.21457 [DOI] [PubMed] [Google Scholar]
- 10.Weinberg BA, Marshall JL. Colon Cancer in Young Adults: Trends and Their Implications. Curr Oncol Rep. 2019;21(1):3. doi: 10.1007/s11912-019-0756-8 [DOI] [PubMed] [Google Scholar]
- 11.Dunlop MG, Tenesa A, Farrington SM, Ballereau S, Brewster DH, Koessler T, et al. Cumulative impact of common genetic variants and other risk factors on colorectal cancer risk in 42,103 individuals. Gut. 2013;62(6):871–881. doi: 10.1136/gutjnl-2011-300537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibáñez-Sanz G, Díez-Villanueva A, Alonso MH, Rodríguez-Moranta F, Pérez-Gómez B, Bustamante M, et al. Risk Model for Colorectal Cancer in Spanish Population Using Environmental and Genetic Factors: Results from the MCC-Spain study. Sci Rep. 2017;7:43263. doi: 10.1038/srep43263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu L, Jeon J, Brenner H, Gruber SB, Schoen RE, Berndt SI, et al. A model to determine colorectal cancer risk using common genetic susceptibility loci. Gastroenterology. 2015;148(7):1330–1339.e14. doi: 10.1053/j.gastro.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigl K, Chang-Claude J, Knebel P, Hsu L, Hoffmeister M, Brenner H. Strongly enhanced colorectal cancer risk stratification by combining family history and genetic risk score. Clin Epidemiol. 2018;10:143–152. doi: 10.2147/CLEP.S145636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon J, Du M, Schoen RE, Hoffmeister M, Newcomb PA, Berndt SI, et al. Determining Risk of Colorectal Cancer and Starting Age of Screening Based on Lifestyle, Environmental, and Genetic Factors. Gastroenterology. 2018;154(8):2152–2164.e19. doi: 10.1053/j.gastro.2018.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith T, Gunter MJ, Tzoulaki I, Muller DC. The added value of genetic information in colorectal cancer risk prediction models: development and evaluation in the UK Biobank prospective cohort study. Br J Cancer. 2018;119(8):1036–1039. doi: 10.1038/s41416-018-0282-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saunders GL, Kilian B, Thompson DJ, McGeoch LJ, Griffin SJ, Antoniou AC, et al. External Validation of Risk Prediction Models Incorporating Common Genetic Variants for Incident Colorectal Cancer Using UK Biobank. Cancer Prev Res. 2020;13(6):509–520. doi: 10.1158/1940-6207.CAPR-19-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGeoch L, Saunders CL, Griffin SJ, Emery JD, Walter FM, Thompson DJ, et al. Risk Prediction Models for Colorectal Cancer Incorporating Common Genetic Variants: A Systematic Review. Cancer Epidemiol Biomarkers Prev. 2019;28(10):1580–1593. doi: 10.1158/1055-9965.EPI-19-0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki M, Tanaka-Mizuno S, Kuchiba A, Yamaji T, Sawada N, Goto A, et al. Inclusion of a Genetic Risk Score into a Validated Risk Prediction Model for Colorectal Cancer in Japanese Men Improves Performance. Cancer Prev Res (Phila). 2017;10(9):535–541. doi: 10.1158/1940-6207.CAPR-17-0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kachuri L, Graff RE, Smith-Byrne K, Meyers TJ, Rashkin SR, Ziv E, et al. Pan-cancer analysis demonstrates that integrating polygenic risk scores with modifiable risk factors improves risk prediction. Nat Commun. 2020;11(1):6084. doi: 10.1038/s41467-020-19600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, et al. Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet. 2019;51(1):76–87. doi: 10.1038/s41588-018-0286-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon N, Lin T. The Kaiser Permanente Northern California Adult Member Health Survey. Perm J. 2016;20(4):15–225. doi: 10.7812/TPP/15-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The PRACTICAL consortium, Law PJ, Timofeeva M, Fernandez-Rozadilla C, Broderick P, Studd J, et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun. 2019;10(1):2154. doi: 10.1038/s41467-019-09775-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai Q, et al. Large-Scale Genome-Wide Association Study of East Asians Identifies Loci Associated With Risk for Colorectal Cancer. Gastroenterology. 2019;156(5):1455–1466. doi: 10.1053/j.gastro.2018.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–1886. doi: 10.1093/jnci/81.24.1879 [DOI] [PubMed] [Google Scholar]
- 26.Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1285–1295. doi: 10.1534/genetics.115.178616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1051–1060. doi: 10.1534/genetics.115.178905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freedman AN, Slattery ML, Ballard-Barbash R, Willis G, Cann BJ, Pee D, et al. Colorectal cancer risk prediction tool for white men and women without known susceptibility. J Clin Oncol. 2009;27(5):686–693. doi: 10.1200/JCO.2008.17.4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128–138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aalen O Nonparametric Estimation of Partial Transition Probabilities in Multiple Decrement Models. Ann Statist. 1978;6(3). doi: 10.1214/aos/1176344198 [DOI] [Google Scholar]
- 31.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data: Kalbfleisch/The Statistical. John Wiley & Sons, Inc.; 2002. doi: 10.1002/9781118032985 [DOI] [Google Scholar]
- 32.Saha P, Heagerty PJ. Time-dependent predictive accuracy in the presence of competing risks. Biometrics. 2010;66(4):999–1011. doi: 10.1111/j.1541-0420.2009.01375.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing. https://www.R-project.org/.
- 34.Wickham H Ggplot2. Springer; New York; 2009. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- 35.MacInnis RJ, Knight JA, Chung WK, Milne RL, Whittemore AS, Buchsbaum R, et al. Comparing 5-Year and Lifetime Risks of Breast Cancer using the Prospective Family Study Cohort. J Natl Cancer Inst. 2021;113(6):785–791. doi: 10.1093/jnci/djaa178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etzioni R, Shen Y, Shih YCT. Identifying Preferred Breast Cancer Risk Predictors: A Holistic Perspective. J Natl Cancer Inst. 2021;113(6):660–661. doi: 10.1093/jnci/djaa181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo J, Nam CM, Sull JW, Yun JE, Kim SY, Lee SJ, et al. Prediction of Colorectal Cancer Risk Using a Genetic Risk Score: The Korean Cancer Prevention Study-II (KCPS-II). Genomics Inform. 2012;10(3):175–183. doi: 10.5808/GI.2012.10.3.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang HM, Chang TH, Lin FM, Chao TH, Huang WC, Liang C, et al. A new method for post Genome-Wide Association Study (GWAS) analysis of colorectal cancer in Taiwan. Gene. 2013;518(1):107–113. doi: 10.1016/j.gene.2012.11.067 [DOI] [PubMed] [Google Scholar]
- 39.Yarnall JM, Crouch DJM, Lewis CM. Incorporating non-genetic risk factors and behavioural modifications into risk prediction models for colorectal cancer. Cancer Epidemiol. 2013;37(3):324–329. doi: 10.1016/j.canep.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 40.Collins GS, Reitsma JB, Altman DG, Moons K. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13(1):1. doi: 10.1186/s12916-014-0241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carr PR, Weigl K, Edelmann D, Jansen L, Chang-Claude J, Brenner H, et al. Estimation of Absolute Risk of Colorectal Cancer Based on Healthy Lifestyle, Genetic Risk, and Colonoscopy Status in a Population-Based Study. Gastroenterology. 2020;159(1):129–138.e9. doi: 10.1053/j.gastro.2020.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo A, Chernoff H, Zheng T, Lo SH. Why significant variables aren’t automatically good predictors. Proc Natl Acad Sci U S A. 2015;112(45):13892–13897. doi: 10.1073/pnas.1518285112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Archambault AN, Su YR, Jeon J, Thomas M, Lin Y, Conti DV, et al. Cumulative Burden of Colorectal Cancer–Associated Genetic Variants Is More Strongly Associated With Early-Onset vs Late-Onset Cancer. Gastroenterology. 2020;158(5):1274–1286.e12. doi: 10.1053/j.gastro.2019.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bailey CE, Hu CY, You YN, Bednarski BK, Rodriguez-Bigas MA, Skibber JM, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150(1):17–22. doi: 10.1001/jamasurg.2014.1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slunecka JL, van der Zee MD, Beck JJ, Johnson BN, Finnicum CT, Pool R, et al. Implementation and implications for polygenic risk scores in healthcare. Hum Genomics. 2021;15(1):46. doi: 10.1186/s40246-021-00339-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017;100(4):635–649. doi: 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, Manolio TA, et al. Prioritizing diversity in human genomics research. Nat Rev Genet. 2018;19(3):175–185. doi: 10.1038/nrg.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570(7762):514–518. doi: 10.1038/s41586-019-1310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to GERA data used in this study may be obtained by application to the Kaiser Permanente Research Bank (KPRB) via ResearchBankAccess@kp.org. A subset of the GERA consented for public use can be found at NlH/dbGaP: phs000674. Genotype data from CORECT and GECCO are deposited at NIH/dbGaP: phs001415.v1.p1, phs001315.v1.p1, phs001078.v1.p1, phs001499.v1.p1, phs001903.v1.p1, phs001856.v1.p1, phs001045.v1.p1, and phs001499.v1.p1.


