Abstract
Phosphoinositides are membrane-localized phospholipids that regulate a plethora of essential cellular processes. These lipid signaling molecules are critical for cell homeostasis and therefore their levels are strictly regulated by the coordinated action of several families of lipid kinases and phosphatases. In this review, we provide a focused perspective on the phosphatidylinositol phosphate kinase (PIPK) family and the three subfamilies that compose it: Type I PIPKs or phosphatidylinositol-4-phosphate 5-kinases (PI4P5Ks), Type II PIPKs or phosphatidylinositol-5-phosphate 4-kinases (PI5P4Ks), and Type III PIPKs or phosphatidylinositol-3-phosphate 5-kinases (PIKfyve). Each subfamily is responsible for catalyzing a hydroxyl phosphorylation on specific phosphoinositide species to generate a double phosphorylated lipid, therefore regulating the levels of both substrate and product. Here, we summarize our current knowledge about the functions and regulation of each PIPK subfamily. Further, we highlight the roles of these kinases in various in vivo genetic models and give an overview of their involvement in multiple pathological conditions. The phosphoinositide field has been long focused on targeting PI3K signaling, but growing evidence suggests that it is time to draw attention to the other phosphoinositide kinases. The discovery of the involvement of PIPKs in the pathogenesis of multiple diseases has prompted substantial efforts to turn these enzymes into pharmacological targets. An increasingly refined knowledge of the biology of PIPKs in a variety of in vitro and in vivo models will facilitate the development of effective approaches for therapeutic intervention with the potential to translate into meaningful clinical benefits for patients suffering from cancer, immunological and infectious diseases, and neurodegenerative disorders.
Keywords: Phosphoinositides, Phosphatidylinositol phosphate kinase, PI4P5K (PIP5K), PI5P4K (PIP4K), PIKfyve, Mechanisms of disease
1. Introduction
Phosphoinositides are low-abundant lipid signaling molecules that govern key cellular processes including membrane trafficking, cytoskeletal rearrangement, and cell signaling to regulate cell growth, metabolism, survival, and proliferation [1, 2]. Phosphoinositide levels are tightly regulated by several kinases, phosphatases, and lipases, and dysregulation of these enzymes is linked to multiple diseases, including immune and developmental disorders, inflammatory diseases, and cancer [2].
All phosphoinositides derive from the addition or removal of phosphate groups to the inositol ring of phosphatidylinositol (PI) [1]. PI contains two distinct modules: a polar inositol head bound to glycerol and two non-polar fatty acid tails. The inositol ring can be phosphorylated at positions 3, 4, and 5 to generate seven phosphoinositide species: single phosphorylated PI(3)P, PI(4)P, and PI(5)P; double phosphorylated PI(3,4)P2, PI(3,5)P2, and PI(4,5)P2; and triple phosphorylated PI(3,4,5)P3 [3]. Dynamic interconversion of all these phosphoinositide species is regulated by specific lipid kinases and phosphatases to maintain the structural and functional integrity of the cell and to allow efficient adjustments to the changing cellular requirements.
The PI phosphate kinase (PIPK) family uses PI monophosphates to generate respective PI bisphosphate (PIP2) products [2]. The PIPK family is divided into three major subfamilies: Type I PIPKs or phosphatidylinositol-4-phosphate 5-kinases (PI4P5Ks / PIP5Ks), Type II PIPKs or phosphatidylinositol-5-phosphate 4-kinases (PI5P4Ks / PIP4Ks), and Type III PIPKs or phosphatidylinositol-3-phosphate 5-kinase (PIKfyve / PIPKIII). Each subfamily phosphorylates specific hydroxyl groups of PI monophosphates but has different substrate specificity. Type I PIPKs phosphorylate PI(4)P to PI(4,5)P2, Type II PIPKs catalyze the phosphorylation of PI(5)P to PI(4,5)P2, and Type III PIPKs primarily generate PI(3,5)P2 from PI(3)P but can also phosphorylate PI to produce PI(5)P [2, 4, 5] (Figure 1). Even though Type I and Type II PIP kinases generate the same lipid product, their functions are very different [6]. All members have a highly conserved catalytic kinase domain but have sequence variability in the activation loop region at the C-terminus, which is responsible for substrate specificity [7]. This review aims to give an overview of the knowledge we have today on the three PIPK subfamilies.
Figure 1. The Phosphatidylinositol phosphate kinase (PIPK) family.
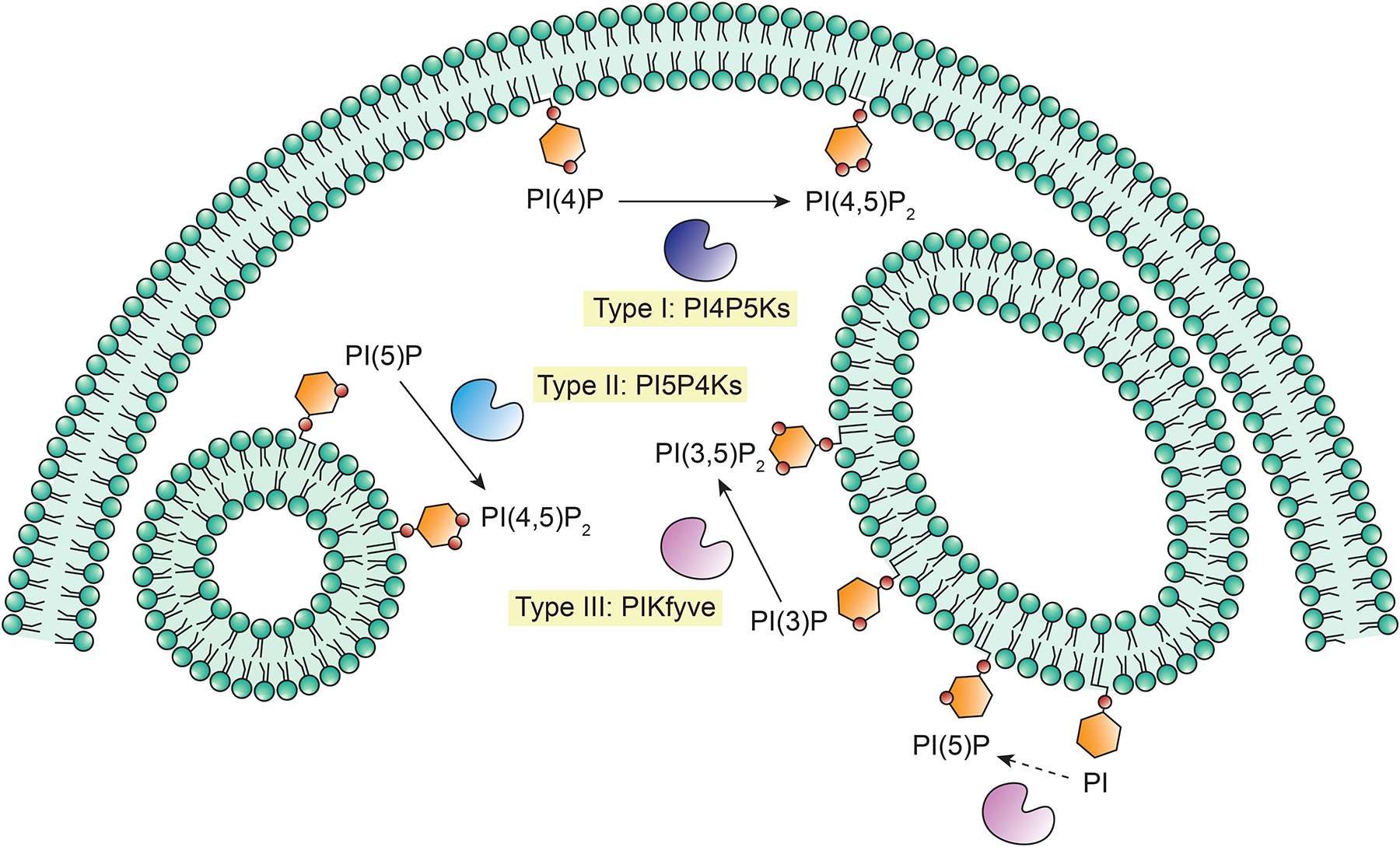
Phosphoinositides are generated by the addition or removal of phosphate groups to the 3, 4, or 5 positions of the hydroxyl groups of phosphatidylinositol’s inositol ring. Their levels are strictly regulated by several kinases and phosphatases. Among these enzymes, the PIPK family is responsible for the conversion of singly phosphorylated phosphatidylinositol (PI) to phosphatidylinositol bisphosphates (PIP2). The PIPK family contains three subfamilies: Type I, Type II, and Type III, with significant sequence homology but distinct substrate specificity. Type I PIPKs phosphorylate PI(4)P to PI(4,5)P2 mainly at the plasma membrane and are named Phosphatidylinositol-4-phosphate 5-kinases (PI4P5Ks). Type II PIPKs are called Phosphatidylinositol 5-phosphate 4-kinases (PI5P4Ks) and also generate PI(4,5)P2, but they do it by phosphorylating PI(5)P at intracellular locations. There is only one Type III PIPK, named phosphatidylinositol-3-phosphate 5-kinase or PIKfyve. This kinase mainly phosphorylates PI(3)P at endosomal compartments to generate PI(3,5)P2 but can also generate PI(5)P by phosphorylation of PI.
1.1. Type I: Phosphatidylinositol-4-phosphate 5- kinases (PI4P5Ks)
1.1.1. Substrate and product
PI4P5Ks are responsible for generating PI(4,5)P2 by phosphorylating PI(4)P at the 5th position of the inositol ring. Phosphorylation of PI(4)P by Type I PIPKs is the major route for PI(4,5)P2 production as opposed to the phosphorylation of PI(5)P by Type II PIPKs. Evidence for this comes from pulse-labeling studies with [32P] orthophosphate, which showed a higher labeling rate at the 5th position of the inositol ring as compared to the 4th position [8, 9]. Further reasoning comes from studies demonstrating that PI(5)P levels within cells are extremely low (~2% of total PI monophosphates), about 10-fold less than PI(4)P, making Type I-dependent activity the major synthetic pathway for PI(4,5)P2 [10]. It is the localization of Type I kinases as well as the activation loop sequence which dictates the substrate specificity, since specific amino acid changes in the activation loop of PI4P5Ks to the corresponding amino acids in PI5P4Ks alters the substrate specificity from PI(4)P to PI(5)P [6, 11]. PI4P5Ks can additionally phosphorylate PI(3,4)P2 at the 5th position to generate PI(3,4,5)P3 in vivo [12]. This ability has also been demonstrated for Its3, the Schizosaccharomyces pombe homolog of PI4P5K [13]. The physiological significance of the other products possibly generated by Type I kinases is mostly unknown.
1.1.2. PI4P5Ks location and structure
There are three isoforms of Type I PIPKs (designated as PI4P5Kα, PI4P5Kβ, and PI4P5Kγ, which are encoded by the genes PIP5K1A, PIP5K1B, and PIP5K1C, respectively; however, historically the nomenclature for murine and human α and β isoforms are reversed) with multiple alternative splice variants [14, 15]. The structural details for Type I PIPKs have been discussed in detail in several reviews [16]. The isoforms are widely expressed and have been shown to have distinct localizations as well as functions to ensure the generation of optimal, localized, and functionally distinctive pools of PI(4,5)P2 [15, 17, 18]. The isoforms are thus subjected to extensive feedback regulations. PI4P5Ks are located primarily on the plasma membrane but have also been shown to be present in the nucleus, perinuclear region on intracellular organelles such as endosomes and Golgi [17, 19] where they generate the multifunctional lipid PI(4,5)P2 [1, 20].
1.1.3. PI4P5Ks functions
The product of PI4P5Ks, PI(4,5)P2, is central to cellular functioning since it controls fundamental processes such as vesicular transport, membrane dynamics, actin cytoskeleton remodeling, cell cycle regulation [21–23] and therefore impaired homeostasis results in neurological diseases, metabolic disorders, ciliopathies and cancer [20, 24]. PI(4,5)P2 also acts as a substrate for Phospholipase C (PLC) signaling and for PI3K to generate PI(3,4,5)P3 [25]. Both of these pathways trigger major signaling alterations in cells, with critical implications in carcinogenesis. Further, pathogens are capable of hijacking PI(4,5)P2 to allow entry as well as replication [26]. These details have been described in several reviews [15, 27, 28].
1.1.4. Regulation of PI4P5Ks
There have also been extensive studies on understanding the regulation of Type I kinases [15, 29], where it is mostly evident that these kinases are regulated by the Rho family of GTPases [30] such as Rho [31], Rac1 [32] and Cdc42 as well as ARF GTPases [33]. Arf6 and Rho have been proposed to regulate PI4P5Ks to maintain high levels of PI(4,5)P2 at the cleavage furrow for efficient cytokinesis in mammalian cells [22]. Further, phosphatidic acid, generated by Phospholipase D has been shown to stimulate Type I PIP kinase isoforms [34, 35]. Several studies have also indicated a phosphorylation-mediated alteration in the activity of PI4P5Ks. These details are described in detail in several reviews [15, 36].
Interestingly, recent studies have highlighted a novel negative regulation of the Type I kinases through the formation of a complex with the Type II kinases [37, 38] to maintain PI(4,5)P2 levels because even excess of the lipid can be detrimental to cellular survival.
1.1.5. PI4P5Ks and pathology
Given the role of Type I PIP kinases in cytoskeletal functions and in the generation of plasma membrane PI(4,5)P2, which is a substrate for PI3K-based signaling, it is not surprising that PI4P5Ks have relevance in human diseases.
Cancer:
Higher expression of PI4P5Kα in both prostate and triple-negative breast cancers correlates with poor patient outcome [39, 40] and knockdown of PI4P5Kα in prostate and breast cancer cells inhibited tumor growth through reduced AKT signaling. Further, a positive correlation between PI4P5Kα and AR expression was observed in prostate cancer biopsy samples and compared with primary tumors, metastatic lesions were found to express significantly higher levels of PIP5K1A and AR mRNA. In both studies, treatment of the cancer cells with a PI4P5Kα inhibitor, ISA-2011B, had the same effect on signaling and tumorigenesis [39, 40]. PI4P5Kα has recently been shown to interact directly with p53 in the nucleus [41] and with oncogenic KRAS [42], providing a rationale for a potentially stronger and personalized therapeutic benefit of PI4P5K inhibitors in cancers with KRAS or TP53 mutations. Indeed, downregulation of PI4P5Kα reduced the viability of KRAS-mutant PDAC cell lines. PI4P5Kα downregulation by protein degradation by NEDD4, a ubiquitin ligase, resulted in impairment of proliferation of breast cancer cells through PI3K-Akt pathway [43]. Ubiquitination-mediated regulation of protein expression has also been demonstrated for PI4P5Kγ, where Smurf1 was shown to directly interact with and degrade PI4P5Kγ, with implications in lung cancer, providing a rationale for the development of PROTAC-based PI4P5K inhibitors. PI4P5Kγ knockdown decreased the proliferation of lung cancer cells and also repressed their ability to form tumors and the signaling for this was driven through the Wnt/β-catenin pathway [44]. PI4P5Kγ has been implicated in breast cancer [45] where PI4P5Kγ along with talin [46] regulates epithelial to mesenchymal transition and can promote malignancy again through the Wnt/β-catenin pathway. Further, PI4P5Kγ has also been implicated in the invasiveness of colorectal cancer due to its role in focal adhesion assembly and disassembly as well as in the facilitation of Warburg effect [47] and pharmacological inhibition with the inhibitor UNC3230 targeting PI4P5Kγ also disrupted glycolysis and tumorigenesis. Besides ubiquitination, phosphorylation modifications of PI4P5Kγ by EGFR [45, 48] and CDK5 [49] also influence tumor formation and metastasis.
Immunological diseases:
PI(4,5)P2 metabolism is being increasingly linked to immunological diseases due to its role in cytoskeletal dynamics including the organization of the immunological synapse, and due to it being a precursor for second messengers and a substrate for PI3K, and the downstream effects of both these pathways in the cellular immune response [50–52]. Each of the three isoforms of PI4P5K is expressed in the T lymphocytes [36, 53, 54]. PI4P5Kα has been demonstrated to be the predominant isoform to be recruited by CD28 [53] and it regulates both CD28 costimulatory as well as CD28 autonomous signals and use of PI4P5Kα inhibitor, ISA-2011B, significantly impairs both the signals, resulting in disruption of TCR-stimulated Ca2+ influx, NF-AT transcriptional activity, gene expression of cytokines and NF-κB activation [55]. NF-κB activation was also disrupted with PI4P5Kα knockdown in ganglioside-stimulated astrocytes indicating a potential role of PI4P5K in the immune response under pathologic neurological conditions also [56]. Metabolic disorders such as diabetes are also inflammatory conditions and ISA-2011B had promising anti-inflammatory effects in type 1 diabetes patients suggesting PI4P5Kα could be a promising anti-inflammatory target [55].
Host-pathogen Interaction:
The α and γ isoforms of PI4P5K, and its product PI(4,5)P2 are crucial for the HIV cycle, specifically the entry phase and for the targeting of Pr55Gag at the plasma membrane for HIV assembly [57]. The involvement of Type I PIP kinases in controlling the pathogen’s cycle comes from their ability to generate plasma membrane PI(4,5)P2 [26] and to alter cytoskeletal dynamics.
1.1.6. Genetic models of targeting PI4P5Ks and their phenotypes
Budding yeast expresses only one type I PIP kinase, Mss4, without which cells are not viable and inactivation of Mss4 results in altered cellular morphology and phenocopies defects seen in mutants of actin-binding proteins [58, 59]. Plants also express multiple copies of a protein showing similarity to PI4P5K [60]. However, there are distinct structural differences between the yeast and plant homologs when compared to the vertebrates signifying organism-specific variations in cellular functions. Fission yeast also expresses a single homolog of PI4P5K, Its3, and similar to its budding yeast homolog Mss4, mutations result in actin-associated aberrations in cellular structural integrity [61, 62]. Functionally, Its3 has been shown to also generate phosphatidylinositol 3,4,5-trisphosphate, and considering the absence of a class-I PI3K in fission yeast, the production of PI(3,4,5)P3 by PI4P5K signifies the early evolution for PI(3,4,5)P3 synthesis.
Caenorhabditis elegans expresses a single homolog of the Type I PIP kinase referred to as ppk-1. Downregulation of ppk-1 in C.elegans resulted in impairment of ovulation and sterility due to defects in cytoskeletal organization in the somatic gonad [63]. The C. elegans model also helped in demonstrating the involvement of ppk-1 in the nervous system and specifically in the formation of growth cones suggesting the importance of optimal PI(4,5)P2 levels in preventing neurodegeneration [64]. The functional significance of Type I PIP kinase in fertility, as well as brain function through effects on cytoskeletal organization, is evident also from studies in mouse models of PI4P5Ks [65, 66]. There are two genes in the Drosophila genome that encode PI4P5K, sktl [67] and dPIP5K [68]. sktl is required for cellular and organismal viability as well as germline development and has been demonstrated to be involved in cytoskeletal-associated processes such as vesicle trafficking, apical polarity [69], and wound healing [70]. dPIP5K is required for regulating PI(4,5)P2 dynamics in photoreceptors cell for phototransduction in Drosophila [68].
Among the mouse models for PI4P5Ks, the most striking phenotype was observed in mice lacking Pip5k1c, which die soon after birth due to neuronal defects [71], whereas mice lacking Pip5k1a or Pip5k1b (based on the nomenclature of human PI4P5Ks) continued to adulthood, only with certain specific cell type phenotypes [65]. In fact, a single allele of Pip5k1c, in the absence of both Pip4k1a and Pip4k2b, resulted in survival to adulthood [65]. On the other hand, the inactivation of PI4P5Kγ by the gene-trap method resulted in embryonic lethality with neural tube closure defects consistent with results showing that PI4P5Kγ isoform is the major contributor of PI(4,5)P2 in the brain. Inhibition of PI4P5Kγ using in utero electroporation also revealed its role in the neuronal migration [72]. The relevance in brain function is also seen in humans since disruption of the kinase activity of PI4P5Kγ by homozygous (D253N) mutation results in perinatal lethality associated with spinal defects, referred to as Lethal congenital contractural syndrome (LCCS) [73]. In a recent study, inhibition of Pip5k1c in the mesenchymal stem cells was seen to induce osteopenia in adult mice [74] (Figure 2).
Figure 2. Selected genetic models used as in vivo tools for PIPK research.
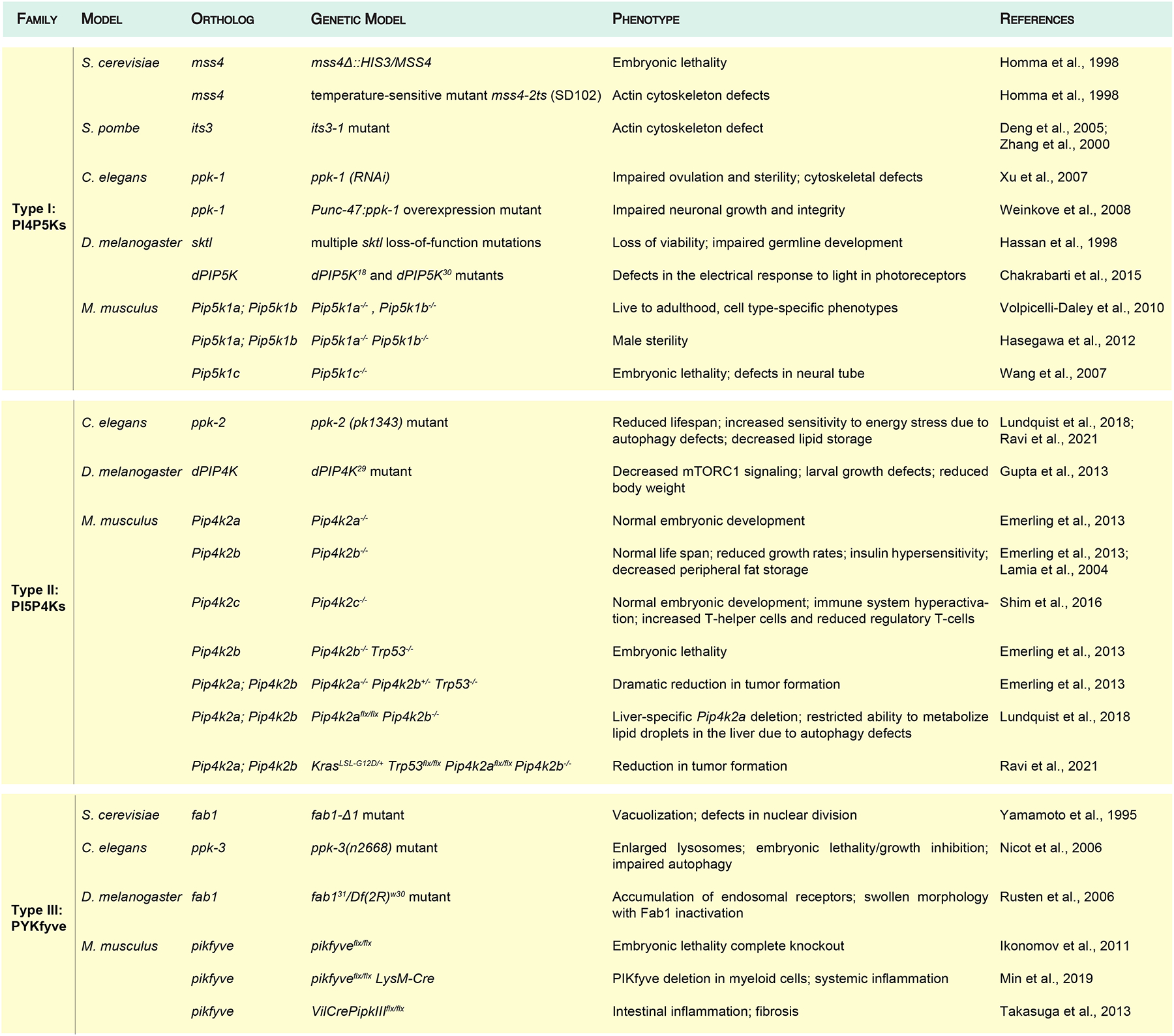
Summary of the main phenotypes of representative Type I, Type II and Type III PIPKs in vivo models used to study the role of these enzymes in development and disease.
1.1.7. Therapeutic targeting of PI4P5Ks
There has been development of inhibitors to target PI4P5Ks and the details of the inhibitors have been described in the recent review [75]. Briefly, as discussed above ISA-2011B [39] and UNC3230 [76] are compounds that have been shown to target PI4P5Kα and PI4P5Kγ respectively, with studies showing inhibitory effects on the proliferation of multiple cancer models [39, 40, 47]. ISA-2011B demonstrated similar potency to the chemotherapeutic docetaxel at stimulating tumor regression, however, the combination therapy significantly reduced the toxic effects of docetaxel [39], indicating a potential for possible combination therapies with existing drugs.
Moreover, a combination therapy of enzalutamide with ISA-2011B has also been proposed for prostate cancer to overcome resistance difficulties faced with antiandrogen therapies [77] and another combination therapy of tamoxifen and ISA-2011B resulted in increased tumor regression in castration-resistant prostate cancer [78]. Recently, a pan-isoform PI4P5K selective inhibitor was discovered using a high throughput screen of the AstraZeneca collection and this study also provides the opportunity to continue the development of potent and selective inhibitors since there have always been issues of having off-target effects on other lipid kinases with drugs targeting PI4P5Ks [79]. Besides the need for these inhibitors to be more specific, more studies are required to determine the pharmacokinetics and toxicity profiles in vivo so that they could make a quick transition to the clinics. The above studies, along with the potential to effectively target KRAS-mutant cancers, highlight that PI4P5Ks could be a promising therapeutic target for multiple cancer types and potentially other disease conditions (Figure 2) and thus requires increased attention from drug discovery and development point of view.
1.2. Type II: Phosphatidylinositol 5-phosphate 4- kinases (PI5P4Ks)
1.2.1. Substrate and product
PI5P4Ks generate PI(4,5)P2 by phosphorylating the 4-position of PI(5)P [10]. The majority of PI(4,5)P2 is generated by type I PIPK by phosphorylation of PI(4)P, which is far more abundant than PI(5)P. Thus, it is generally assumed that the main function of PI5P4Ks is to reduce the amount of cellular PI(5)P or to generate a minor pool of PI(4,5)P2 at specific cellular locations [80–82].
1.2.2. PI5P4Ks location and structure
This family of kinases includes three different isoforms α, β, and γ, encoded by the genes PIP4K2A, PIP4K2B, and PIP4K2C, respectively. PI5P4Kα is the most active of the three, being considerably more active than PI5P4Kβ and especially than PI5P4Kγ, which has little catalytic activity [82, 83]. PI5P4Ks are largely located in intracellular membranes. PI5P4Kα localizes to autophagosomes, lysosomes, and peroxisomes; PI5P4Kβ is predominantly located in the nucleus but can also be found in autophagosomes, and PI5P4Kγ is present in autophagosomes, Golgi, and endomembrane compartments [84–88]. The three PI5P4K isoforms have been reported to homodimerize or heterodimerize with each other, which can influence their localization [84, 89, 90]. PI5P4Ks consist of a dimerization domain and a lipid kinase domain with variable N- and C-terminal lobes that mediate specific functions for each isoform [16]. PI5P4Kα and PI5P4Kβ are more homologous to each other than either are to PI5P4Kγ. Although the three PI5P4K isoforms share significant sequence similarity, there are certain structural differences that account for their distinct enzymatic activities, such as their ATP affinity [90]. Another major difference is the glycinerich loop, which is supposed to interact with the substrate [83].
1.2.3. PI5P4Ks functions
PI5P4Ks play a role in a variety of essential cellular processes by facilitating the recruitment and allosteric activation of proteins at specific intracellular locations, modulating integral membrane protein signaling, and regulating lipid transport. It is worth noting that besides PI phosphorylation, PI5P4Ks have been reported to perform catalytic-independent functions [37, 91–94].
PI5P4Ks regulate insulin signaling and autophagy and are key for surviving metabolic stress by modulating PI3K/Akt/mTORC pathways [37, 85, 91, 95–99]. It has been shown that the Mesencephalic Astrocyte-derived Neurotrophic Factor (MANF) increases the localization of PI5P4Kβ in the endoplasmic reticulum to regulate insulin signaling [100]. Additionally, evidence has demonstrated that loss of PI5P4Ks results in an increase in PI(4,5)P2 levels, which serve as a substrate for insulin-stimulated production of PI(3,4,5)P3 by PI3K [37, 91]. Silencing of the three PI5P4K isoforms and the subsequent changes in PI(5)P levels result in an increase in autophagosome formation [87]. Consistently, loss of PI5P4Kα and PI5P4Kβ limits the ability to metabolize lipid droplets and leads to an accumulation of autophagic vesicles. This autophagy defect results in a reduction of nutrient supply, which impairs mTORC1 activation and triggers the activation of the transcription factor EB (TFEB), responsible for the expression of lysosomal and autophagic genes [85]. Recent data has shown that PI5P4Kα can interact with the long noncoding RNA lncSAMD11–1:1 in human endometrial stromal cells, which results in the inhibition of Akt phosphorylation and stabilization of the nuclear localization of FoxO1 to promote endometrial decidualization and successful pregnancy [101]. PI5P4Ks also regulate oxidative stress. PI5P4Kα overexpression reduces the expression of NRF2 target genes by H2O2, decreases the rate of reactive oxygen species (ROS) accumulation, and reduces resistance to oxidative stress [102]. Interestingly, it has been shown that induction of oxidative stress mitigates PI5P4Kα and PI5P4Kβ activity [80, 103].
Type II PIP kinases can bind to both ATP and GTP. In particular, the PI5P4Kβ isoform has a strong preference for GTP over ATP and its activity changes as GTP levels change, working as an intracellular GTP sensor [104]. It has been demonstrated that the ability of PI5P4Kβ to couple changes in GTP into changes in PI(5)P levels influences tumor growth. The preference of PI5P4Kβ for GTP over ATP was evolutionarily acquired by the generation of the guanine efficient association (GEA) motif, which uses its main chain atoms for adenine recognition and the side chain atoms for guanine recognition [105].
Cellular trafficking is also regulated by these lipid kinases. The activity of these enzymes is required for the crosstalk between peroxisomes and mitochondria. Loss of PI5P4Kα and PI5P4Kβ leads to changes in the peroxisomal PI(4,5)P2 pool, which impairs the trafficking of very long fatty acids to peroxisomes and results in mitochondrial dysfunction [106]. PI5P4Kα regulates intracellular cholesterol transport from lysosomes to peroxisomes by maintaining PI(4,5)P2 homeostasis on peroxisomes [107]. Moreover, PI5P4Kγ regulates the Notch pathway by controlling the transit of the receptor through recycling endosomes [108]. In Drosophila, the PI5P4Ks homolog dPIP4K regulates the trafficking of secretory granule proteins [109]. Similarly, dPIP4K regulates clathrin-mediated endocytosis by controlling the localization of rhodopsin 1 [94].
Evidence supports a role for PI5P4Ks in immune modulation. The lack of PI5P4Kγ in mice results in high levels of proinflammatory cytokines, an increase in T-helper cells, and a decrease in regulatory T-cells [110]. This enhanced immune response results from mTORC1 signaling hyperactivation upon Pip4k2c deletion and can be reduced after rapamycin treatment. Interestingly, various studies have described PIP4K2C single nucleotide polymorphisms (SNPs) associated with different autoimmune diseases [111, 112]. In addition, a recent study has shown that PI5P4Kβ and PI5P4Kγ regulate the immunosuppressive activity of regulatory T-cells by regulating PI3K, mTORC1, and MAPK signaling pathways and controlling the expression of the transcriptional regulator FOXP3 [113].
PI(5)P in the nucleus, regulated by PI5P4Ks, interacts with chromatin-associated proteins and regulates gene transcription [82]. For instance, PI(5)P can induce a conformational change in UHRF1 that mediates binding to H3K9me3, and can interact with ING2, which results in the repression of ING2 target genes [114, 115]. Consistently, overexpression of PI5P4Kβ results in the release of ING2 from chromatin [116]. Furthermore, PI5P4Kβ influences TAF3 association with H3K4me3 and gene expression during myoblast differentiation [117].
1.2.4. Regulation of PI5P4Ks
Although delineating the upstream regulation of PI5P4Ks requires further investigation, some studies suggest that these enzymes are negatively regulated by phosphorylation. Protein kinase D (PKD) phosphorylates PI5P4Kα at Thr376, located within the activation loop, and reduces its enzymatic activity [118]. Under stress conditions, PI5P4Kβ activity can be inhibited through direct phosphorylation at Ser326 by the p38 stress-activated protein kinase [80]. mTORC1 phosphorylates PI5P4Kγ at Ser324 and Ser328 depending on nutrient availability, which results in a decrease in the kinase functions. The protein kinase CK2 phosphorylates PI5P4Kα at Ser304, and mutation of this residue to aspartate to mimic phosphorylation leads to a redistribution from the cytoplasm to the plasma membrane [119]. Besides phosphorylation, the activity of PI5P4Ks might be regulated by interaction with other proteins. For instance, the interaction of PI5P4Kα and PI5P4Kβ with the proline isomerase Pin1 reduces the activity of the kinases in vitro [102]. Moreover, PI5P4Kβ can interact with the ubiquitin ligase complex Cul3-SPOP, which mediates its ubiquitylation [120].
1.2.5. PI5P4Ks and pathology
PIs are crucial signaling molecules involved in a variety of essential cellular processes and therefore it is imperative to maintain a tight balance between the enzymes that regulate the cellular pool of PIs. Alterations in PI5P4Ks have been linked to several pathological processes including cancer, psychiatric disorders, and infectious diseases.
Cancer:
The expression of the three PI5P4Ks is altered in different cancer types such as breast cancer, leukemias, soft tissue sarcomas, and glioblastomas [121]. PI5P4Kα and PI5P4Kβ are overexpressed in breast tumors and their depletion results in impaired tumor growth, selectively in p53-deficient tumors [122]. Depletion of the kinases is linked to increased AKT phosphorylation, oxygen consumption and ROS, and reduced glucose metabolism. Therefore, under oxidative stress (i.e., p53 deficiency), PI5P4Ks are required for cancer cell survival. Along these lines, downregulation of PI5P4Kα and PI5P4Kβ has also been shown to impair cell proliferation in multiple triple-negative breast cancer models in vitro [106]. A different study showed that both high and low PI5P4Kβ expression compared with intermediate expression levels correlate with poor patient survival [123]. Moreover, high throughput proteomics of breast cancer interstitial fluid identified PI5P4Kβ among a panel of 10 biomarkers for stratification of breast cancer subtypes [124]. PI5P4Ks are also involved in the pathogenesis of hematological malignancies including acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL). PI5P4Kα is essential for the proliferation and survival of AML cells but not for primary normal hematopoietic stem and progenitor cells [125]. Similarly, PI5P4Kα is overexpressed in multiple types of leukemia and its expression is significantly associated with unfavorable clinical outcomes [126]. High PI5P4Kα and PI5P4Kγ expression levels are associated with unfavorable cytogenetic risk and correlate with worse survival outcomes in AML patients [127]. Additionally, genome-wide association studies (GWAS) in multiple patient cohorts have identified PIP4K2A SNPs associated with ALL susceptibility through inducing PI5P4Kα overexpression [126, 128–131]. Although most studies point towards oncogenic roles for PI5P4Ks, in vivo RNAi screening of glioblastoma patient-derived xenografts identified PI5P4Kα as a putative tumor suppressor [92]. The authors report that in PTEN-deficient glioblastomas, PI5P4Kα inhibits PI3K signaling by inducing the degradation of the p85 regulatory subunit of PI3K.
Neurodegenerative / Psychiatric disorders:
Both PI5P4Kα and PI5P4Kβ are present in axon terminals and dendritic spines, suggesting a role in synaptic vesicle trafficking [132]. Silencing or inhibition of PI5P4Kγ reduces the levels of mutant huntingtin protein in fibroblasts, clears aggregates in neurons, and ameliorates neuronal dysfunction and degeneration in Drosophila [133]. Multiple GWAS studies have revealed that genetic variants of PIP4K2A are associated with an increased predisposition to schizophrenia [134–137] and with a poor antipsychotic response [138]. In addition, genetic variants of PIP4K2A have been associated with antidepressant treatment response, time to recurrence of depressive and manic/mixed episodes, and depression severity among patients with depression and bipolar disorder [139].
Infectious diseases:
PI metabolism plays a key role in the life cycles of several infectious agents by favoring viral replication and assembly [140]. Furthermore, it has been shown that PI5P4Kα is imported from red blood cells into Plasmodium falciparum, Plasmodium berghei, and Toxoplasma gondii, where it associates with specific parasite RNAs [93, 141]. The RNA binding activity of PI5P4Kα is conserved across species from Drosophila and C. elegans to humans, suggesting that this kinase is also important for posttranscriptional gene regulation [93].
1.2.6. Genetic models of targeting PI5P4Ks and their phenotypes
PPK-2 (PI5P4K ortholog) deficient C. elegans have reduced lifespan and a decrease in lipid storage, show autophagy defects, and are more sensitive to metabolic stress [85]. Deletion of PIP4K in Drosophila leads to a decrease in mTORC1 signaling, lower body weight, and a shortening of larval development [96]. In mice, homozygous germline deletion of Pip4k2a results in normal embryonic development [122]. Pip4k2b knockout mice are viable but show a mild reduction in growth rates, decreased fat content, and are hypersensitive to insulin, which protects them from obesity, insulin resistance, and type 2 diabetes when exposed to a high-fat diet [142]. Mice lacking Pip4k2c are normal regarding growth and viability, but they show hyperactivation of the immune system. These mice have increased mTORC1 activation in multiple tissues, which results in a higher expression of proinflammatory cytokines, increased immune cell infiltrates, increased T-helper cell populations, and decreased regulatory T-cell populations [110]. Strikingly, germline deletion of Pip4k2b in mice lacking Trp53 results in early embryonic lethality, with pups dying within 10–12h after birth [122]. It is important to mention that deletion of Pip4k2a in Pip4k2b+/− Trp53−/− mice lead to viable mice with a dramatic reduction in tumor formation compared to littermates Trp53−/− and wild-type for the kinases, uncovering a critical role for PI5P4Ks in maintaining glucose metabolism and ROS homeostasis in the context of p53 loss. Further, deletion of the kinases in a sarcoma mouse model (KrasLSL-G12D/+ Trp53flx/flx Pip4k2aflx/flx Pip4k2b−/−) also impaired tumor formation [106]. Mice with liver-specific deletion of Pip4k2a and germline deletion of Pip4k2b show a deficiency in the ability to metabolize lipid droplets in the liver upon fasting, indicating an autophagy defect [85] (Figure 2).
1.2.7. Therapeutic targeting of PI5P4Ks
Multiple studies have demonstrated the potential of PI5P4Ks to become effective therapeutic targets for the treatment of metabolic and psychiatric disorders, infectious diseases, and cancer. The isoform-specific and multi-isoform PI5P4K inhibitors developed to date have been thoroughly discussed in [75].
Briefly, isoform-specific inhibitors include the ATP-competitive inhibitor of PI5P4Kα I-OMe Tyrphostin AG-538 [143], the pyrimidine-2,4-diamine SAR088m which specifically inhibits PI5P4Kβ [144], and the specific inhibitors for PI5P4Kγ NCT-504 and NIH-12848 [133, 145]. Efforts to improve the physicochemical properties of NIH-12848 have led to the development of “compound 40” [146]. More recently, two potent and highly selective PI5P4Kα inhibitors, named BAY-091 and BAY-297 were identified using high throughput screening and subsequent structure-based optimization. However, although cellular target engagement was demonstrated, inhibition of PI5P4Kα with these compounds did not inhibit proliferation in p53-deficient cancer cells [147].
Inhibitors that target more than one PI5P4K isoform include a131, which selectively kills cancer cells but not normal cells [148]. THZ-P1–2 is also a potent and reasonably selective pan-PI5P4K covalent inhibitor that showed modest anti-proliferative activity and compromised mitochondrial homeostasis and autophagy in a panel of leukemia cell lines [149, 150]. Optimization of this molecule lead to the development of a compound with improved selectivity and potent biochemical and cellular activity [151]. CC260 is a highly potent and selective noncovalent dual inhibitor for both PI5P4Kα and PI5P4Kβ which incorporates a hydrophobic side chain that occupies a pocket unique to the lipid kinase family [152]. Inhibition of the kinases with this compound leads to disruption of cellular energy homeostasis, AMPK activation, and mTORC1 inhibition. This induced energy stress was selectively toxic for p53-null tumor cells. However, this compound showed some off-target activity against PI3K-δ.
1.3. Type 3: phosphatidylinositol-3-phosphate 5-kinase / PIKfyve
1.3.1. Substrate and product
Type III PIPK is comprised of a single member: phosphatidylinositol-3-phosphate 5-kinase type III or PIKfyve [153]. PIKfyve primarily functions in biosynthesis of PI(3,5)P2 from the substrate PI(3)P via phosphorylation of the 5th position of the PI(3)P inositol ring, however it has also been demonstrated to generate PI(5)P through direct and indirect processes [4]. The alteration of lipid combination from PI(3)P to PI(3,5)P2 is thought to drive vesicle maturation, regulating cellular endocytosis (reviewed in:[154]).
1.3.2. PIKfyve location and structure
In the cell, PIKfyve is found throughout the endosomal compartments, and colocalizes with early and late endosomal markers, including EEA1, along with endo-lysosomal markers and the retromer complex [155]. PIKfyve functions as a multidomain protein complex with the scaffolding protein VAC14 and lipid phosphatase Fig4, catalyzing the reverse reaction of PI(3,5)P2 to PI(3)P [153], for which structure and individual reactions have been thoroughly discussed [154, 156]. Interestingly, while interactions contribute to both PIKfyve autophosphorylation and lipid kinase activity, members of this complex have been shown to contribute to the vacuolization defect phenotype associated with PIKfyve inhibition [157]. The PIKfyve protein itself is a large 240kDa protein and contains three generally recognized domains including the FYVE, GroEL-like chaperonin, and lipid kinase domains [158]. Several isoforms of PIKfyve lacking the lipid kinase domain have been suggested by deep sequencing analyses but have not been experimentally verified [159]. The FYVE zinc finger domain, named for its association with the proteins Fab1, YOTB, Vac1, and EEA1, exhibits high specificity to PI(3)P lipids and is associated with endosomal trafficking [160]. The kinase domain of PIKfyve has been suggested to exhibit dual specificity as both a lipid kinase and protein kinase [161], and highlighted in a recent review by Hayakawa et. al.[160], the structure of which has yet to be solved despite shared homology with PI kinases. Combined, the lack of mechanistic understanding and characterization of PIKfyve and its proteincomplex highlights a need for further research in this area.
1.3.3. PIKfyve functions
At the cellular level, PIKfyve is associated with numerous cell functions including signaling, membrane homeostasis, and migration through its regulatory function in vesicle trafficking. PIKfyve is best known as a regulator of cellular membrane homeostasis and vesicular membrane trafficking, including autophagy, macropinocytosis, and receptor-mediated endocytosis [4]. This endocytic association is supported by evidence suggesting that PIKfyve regulates early endosomes in simian-derived fibroblasts [162] and phagosome maturation in macrophages [163]. Numerous studies using genetic and pharmacological inhibitors have also identified PIKfyve to be essential for lysosome function and acidification in neurons, fibroblasts, and cancer cells, further complicating its role in vesicle compartments [164–167]. Early studies reported that PIKfyve/Fab1p possessed protein kinase and autophosphorylation activities in addition to its lipid kinase activities [161]. These observations support evidence suggesting that protein kinase activity [168] and signaling effects [169] by PIKfyve are involved in vesicle trafficking, though these aspects have not been thoroughly investigated. Despite recent advancements, full understanding of the pleiotropic functions of PIKfyve remains to be explored despite its fundamental role in cell biology and clinical relevance.
1.3.4. Regulation of PIKfyve
PIKfyve has been found to be positively regulated by the class III PI3 lipid kinase (PIKC3 or Vps34), which produces the lipid PI(3)P, but can also be activated by cell stress responses via phosphorylation, increasing intracellular PI(3,5)P2 pools [155, 170]. These regulatory processes may in part explain the mechanism in which PIKfyve can regulate dynamic cellular processes such as macropinocytosis, autophagy, phagocytosis, and lysosomal function [165, 166].
1.3.5. PIKfyve and pathology
Recent translational interest of this lipid kinase has been spurred by the identification of PIKFyve as the target of the small molecule inhibitors apilimod and YM201636, implicating PIKFyve in several disease phenotypes. Alternatively, Loss-of-function PIKfyve mutations have been associated with development of Fleck Corneal Dystrophy, which is characterized by small white flecks, consisting of vacuolized keratocytes that are found in the corneal stroma of patients [171], and congenital cataracts [172].
Infectious diseases:
To date, PIKfyve, through pharmacological inhibition, has been identified as a potential antiviral against SARS-CoV-2, Ebola, and Malburg viruses, among others [173–178]. Multiple publications have suggested that PIKfyve inhibition viral entry into endocytic vesicles and cellular cytoplasm [173, 176]. Complementary evidence has suggested that viral infection is prevented by blocking endolysosome fusion upon PIKfyve inhibition [175].
Neurodegenerative disorders:
PIKfyve has also been demonstrated to be a potential therapeutic target in the tauopathies, such as Alzheimer’s disease, and neurodegenerative disorders reviewed in: [178–180]. Evidence from studies in taupathies suggests that PIKfyve inhibition impairs the trafficking of proteins into lysosomes [181].
Cancer:
Targeting PIKfyve exhibits therapeutic efficacy in both preclinical and clinical studies in a variety of cancer types, including non-Hodgkin’s B cell lymphoma, small cell lung cancer, and liver cancer, among others (reviewed in [182]. Studies using pharmacological PIKfyve inhibitors have associated this lipid kinase with transcription factor EB (TFEB) [183] and epidermal growth factor receptor (EGFR) [184] signaling involved in tumor progression. Expectedly, PIKfyve has also been shown to be essential in endocytic pathways which contribute to tumor progression and aggressiveness including autophagy [167] and macropinocytosis [166].
The role of PIKfyve in these pathologies has been extensively highlighted and will not be discussed in depth in this review.
1.3.6. Genetic models of targeting PIKfyve and their phenotypes
Type III PIPK function was originally identified in the ortholog formation of aploid and binucleate cells protein (Fab1p) in S. cerevisiae where loss-of-function mutations resulted in the development of cytoplasmic vacuoles and defects in nuclear division [185]. PIKfyve/Fab1p was also found to be functionally conserved across eukaryotes and is thought to be the only enzyme to produce intracellular PI(3,5)P2 [156]. In mammals, the PIKfyve protein was originally known as p235 which was shown to regulate actin fiber length in platelets [186] prior to lipid kinase identification by domain homology and verification of PI(3,5)P2 synthesis [4]. Parallel studies of PIKfyve orthologs including the phosphatidylinositol phosphate kinase-3 (PPK-3) in C. elegans [187] and fab1 in D. melanogaster [188] have contributed to the mechanistic understanding of PIKfyve function and its regulation at the cellular and physiological level. Given the variety of orthologs, often referenced interchangeably, PIKfyve is referred to as multiple aliases including: 1-phosphatidylinositol 3-phosphate 5-kinase, Fab1, or type III PIPK.
Mounting evidence using in vivo models strongly supports the integral role of PIKfyve expression at the organismal level despite incomplete understanding of its cellular mechanisms. PIKfyve expression has also been shown to be essential for embryonic survival and development in genetic knockout studies of D. melanogaster and mouse models [187–189]. Interestingly, heterozygous expression was shown to be sufficient for normal physiological function in mouse models, suggesting compensatory processes [189]. In the nervous system, PIKfyve and PI(3,5)P2 synthesis have been shown to be necessary for normal function through regulation of synaptic signaling [190]. Additionally, secondary systemic effects from tissue-specific knockout studies in the intestine and macrophage populations suggest that PIKfyve expression is not only important to tissues but also physiological homeostasis [191, 192] (Figure 2).
1.3.7. Therapeutic targeting of PIKfyve
Pharmacological inhibition of PIKfyve occurs through targeting of the kinase domain [169] and is generally observed to induce vesicle trafficking and endosomal defects, leading to the formation of characteristic cytoplasmic vacuoles. This phenomenon is observed in tauopathies, where PIKfyve inhibition by YM201636 reduced the aggregation of tau protein, a pathologic signature of multiple neurodegenerative diseases, where lysosomal trafficking is prevented [181]. Similarly, it is the inhibition of lysosome function by apilimod treatment that prevents viral infection of SARS-CoV-2 and ebola virus [173], and induces anti-cancer effects in Ras-driven cells and B-cell lymphoma [166] [193]. In other examples, PIKfyve targeting results in the inhibition and dysregulation of autophagy, in both prostate and liver cancers [167, 184]. Several of these preclinical studies have progressed to clinical trials of apilimod with varied effectiveness along with numerous confounding factors including low plasma concentration [173, 194]. Novel inhibitory compounds targeting PIKfyve are currently being developed and tested to overcome limited clinical effectiveness. Despite these results, PIKfyve is still considered to have significant therapeutic potential but its utility as a target will need to overcome the challenge of completely understanding the mechanism of action for the various compounds being developed.
2. Concluding remarks
Research on phosphoinositide signaling has underscored how indispensable these minor lipids are for the regulation of a myriad of fundamental cellular processes [2]. However, more cellular and physiological studies are required to further understand the phosphoinositide signaling pathways and importantly the controlling enzymes themselves. The importance of spatial and temporal organization of phosphoinositide levels proves beyond a doubt that there is a need for techniques capable of achieving a subcellular resolution to follow changes in live cells. Some tools such as the use of the widely used PH domain of PLCδ to image PI(4,5)P2 are used for specifically detecting phosphoinositides, but these techniques have limitations [195]. Super-resolution imaging would allow visualization of these low-abundant lipids with great precision to better interrogate phosphoinositide metabolism. A greater understanding of the spatial and temporal regulation of phosphoinositide signaling promises to determine how alterations in phosphoinositide pools at specific cellular and organelle membranes translate to human disease.
It has become clear that PIPKs represent potential and druggable targets for the treatment of these diseases and the development of multiple molecules to inhibit PIPKs proves that substantial efforts are currently being made to turn these proteins into druggable targets [75]. These efforts are most likely to have a significant translational value, however, there is still a need to further improve the pharmacological properties of PIPK inhibitors. The current development of potent and specific small molecule inhibitors to target PIPKs gives hope for future therapeutic options for the treatment of multiple diseases. It is important to consider that the emerging proteolysis targeting chimeric (PROTAC) technology offers the possibility to therapeutically target proteins that have proved difficult to modulate using small molecules [196]. These heterobifunctional compounds induce the degradation of a protein of interest by linking it to an E3 ligase, which induces ubiquitination and subsequent proteasomal degradation [197]. Given that PROTACs could target both catalytic and non-catalytic functions of the kinases, these molecules could certainly be more advantageous than other compounds to pharmacologically target PIPKs and are definitely worth exploring.
The three different PIPK families are implicated in a variety of mostly overlapping diseases (Figure 3) and despite the prospects of each of the PIP kinase family members to be a promising therapeutic option on their own, there is also a need to study them in combination. Targeting multiple PIPKs families and isoforms may be necessary to eliminate the constant metabolic rewiring that cancer cells undergo to survive various stresses and could represent a significant step forward in cancer research and possibly help to resolve the issues of drug resistance and relapse. The PIP kinase combinatorial targeting approach could also extend to other diseases and might prove to be more effective. The existing knowledge on the PIP kinases establishes beyond doubt that this family of enzymes demands further scientific as well as therapeutic attention.
Figure 3. The PIPK family is involved in various human diseases.
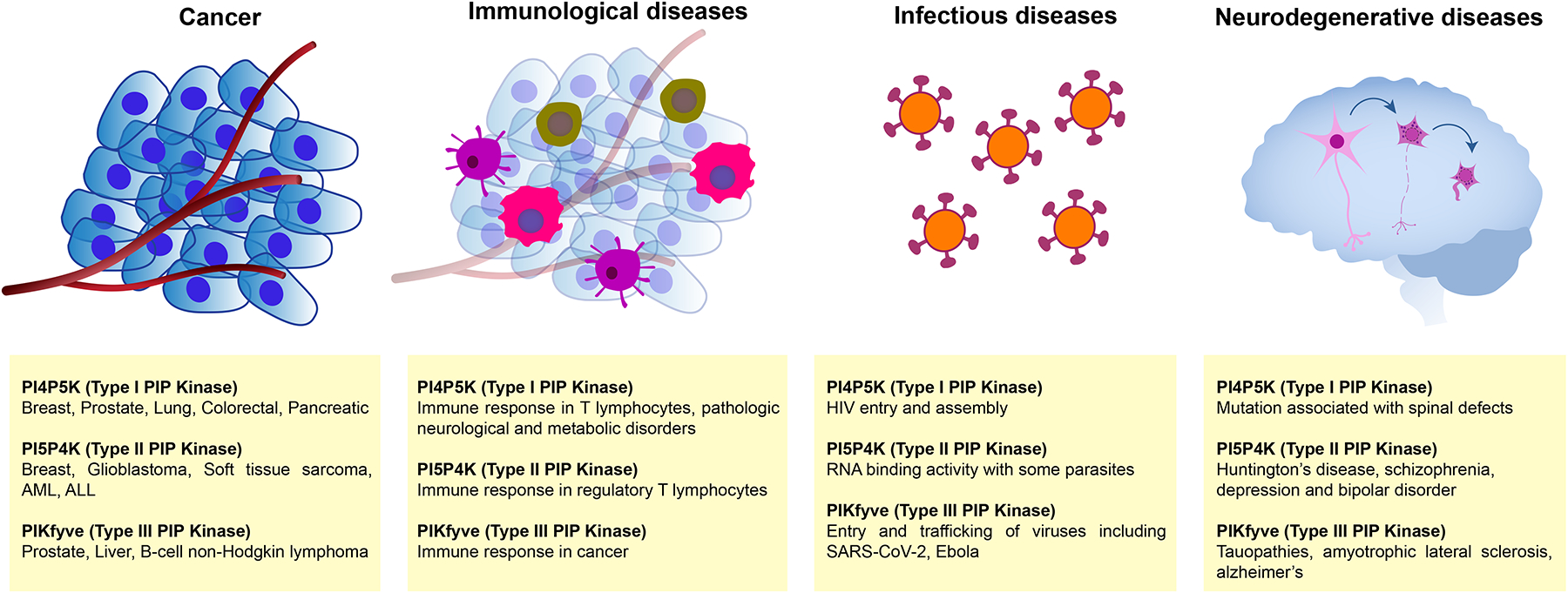
Schematic overview of the human pathologies that have been associated with alterations in PIPKs and have the potential of benefiting from therapies that are targeted towards these enzymes. Members of Type I, Type II and Type III PIPKs subfamilies have been linked to multiple types of cancer, immunological, infectious and neurodegenerative disorders.
Acknowledgments
BME receives research funding from NCI (R01 CA237536, CBC under Contract No. 75N91019D00024, Task Order No. 75N91020F00003), NIH (R01 GM143583) and ACS (RSG-20-064-01-TBE, TLC-21-156-01).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Balla T, Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev, 2013. 93(3): p. 1019–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond GRV and Burke JE, Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr Opin Cell Biol, 2020. 63: p. 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posor Y, Jang W, and Haucke V, Phosphoinositides as membrane organizers. Nat Rev Mol Cell Biol, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sbrissa D, Ikonomov OC, and Shisheva A, PIKfyve, a Mammalian Ortholog of Yeast Fab1p Lipid Kinase, Synthesizes 5-Phosphoinositides: EFFECT OF INSULIN *. Journal of Biological Chemistry, 1999. 274(31): p. 21589–21597. [DOI] [PubMed] [Google Scholar]
- 5.Posor Y, Jang W, and Haucke V, Phosphoinositides as membrane organizers. Nature Reviews Molecular Cell Biology, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunz J, et al. , The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol Cell, 2000. 5(1): p. 1–11. [DOI] [PubMed] [Google Scholar]
- 7.Liu A, et al. , The activation loop of PIP5K functions as a membrane sensor essential for lipid substrate processing. Sci Adv, 2016. 2(11): p. e1600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens LR, Hughes KT, and Irvine RF, Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature, 1991. 351(6321): p. 33–9. [DOI] [PubMed] [Google Scholar]
- 9.Whiteford CC, Brearley CA, and Ulug ET, Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J, 1997. 323 (Pt 3): p. 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rameh LE, et al. , A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature, 1997. 390(6656): p. 192–6. [DOI] [PubMed] [Google Scholar]
- 11.Kunz J, et al. , Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J Biol Chem, 2002. 277(7): p. 5611–9. [DOI] [PubMed] [Google Scholar]
- 12.Halstead JR, et al. , A novel pathway of cellular phosphatidylinositol(3,4,5)-trisphosphate synthesis is regulated by oxidative stress. Curr Biol, 2001. 11(6): p. 386–95. [DOI] [PubMed] [Google Scholar]
- 13.Mitra P, et al. , A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J Cell Biol, 2004. 166(2): p. 205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishihara H, et al. , Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J Biol Chem, 1996. 271(39): p. 23611–4. [DOI] [PubMed] [Google Scholar]
- 15.van den Bout I and Divecha N, PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci, 2009. 122(Pt 21): p. 3837–50. [DOI] [PubMed] [Google Scholar]
- 16.Burke JE, Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol Cell, 2018. 71(5): p. 653–673. [DOI] [PubMed] [Google Scholar]
- 17.Doughman RL, Firestone AJ, and Anderson RA, Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J Membr Biol, 2003. 194(2): p. 77–89. [DOI] [PubMed] [Google Scholar]
- 18.Choi S, et al. , PIP kinases define PI4,5P(2)signaling specificity by association with effectors. Biochim Biophys Acta, 2015. 1851(6): p. 711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boronenkov IV, et al. , Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell, 1998. 9(12): p. 3547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandal K, Review of PIP2 in Cellular Signaling, Functions and Diseases. Int J Mol Sci, 2020. 21(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibasaki Y, et al. , Massive actin polymerization induced by phosphatidylinositol-4-phosphate 5-kinase in vivo. J Biol Chem, 1997. 272(12): p. 7578–81. [DOI] [PubMed] [Google Scholar]
- 22.Brill JA, Wong R, and Wilde A, Phosphoinositide function in cytokinesis. Curr Biol, 2011. 21(22): p. R930–4. [DOI] [PubMed] [Google Scholar]
- 23.Kwiatkowska K, One lipid, multiple functions: how various pools of PI(4,5)P2 are created in the plasma membrane. Cellular and Molecular Life Sciences, 2010. 67(23): p. 3927–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, et al. , Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res, 2010. 12(1): p. R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katan M and Cockcroft S, Phosphatidylinositol(4,5)bisphosphate: diverse functions at the plasma membrane. Essays Biochem, 2020. 64(3): p. 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha-Perugini V, Gordon-Alonso M, and Sanchez-Madrid F, PIP2: choreographer of actin-adaptor proteins in the HIV-1 dance. Trends Microbiol, 2014. 22(7): p. 379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanaho Y, Kobayashi-Nakano A, and Yokozeki T, The phosphoinositide kinase PIP5K that produces the versatile signaling phospholipid PI4,5P(2). Biol Pharm Bull, 2007. 30(9): p. 1605–9. [DOI] [PubMed] [Google Scholar]
- 28.Wills RC and Hammond GRV, PI(4,5)P2: signaling the plasma membrane. Biochemical Journal, 2022. 479(21): p. 2311–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oude Weernink PA, Schmidt M, and Jakobs KH, Regulation and cellular roles of phosphoinositide 5-kinases. Eur J Pharmacol, 2004. 500(1–3): p. 87–99. [DOI] [PubMed] [Google Scholar]
- 30.Weernink PA, et al. , Activation of type I phosphatidylinositol 4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J Biol Chem, 2004. 279(9): p. 7840–9. [DOI] [PubMed] [Google Scholar]
- 31.Chong LD, et al. , The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell, 1994. 79(3): p. 507–13. [DOI] [PubMed] [Google Scholar]
- 32.Tolias KF, et al. , Type Ialpha phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr Biol, 2000. 10(3): p. 153–6. [DOI] [PubMed] [Google Scholar]
- 33.Honda A, et al. , Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell, 1999. 99(5): p. 521–32. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins GH, Fisette PL, and Anderson RA, Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem, 1994. 269(15): p. 11547–54. [PubMed] [Google Scholar]
- 35.Jones DR, Sanjuan MA, and Merida I, Type Ialpha phosphatidylinositol 4-phosphate 5-kinase is a putative target for increased intracellular phosphatidic acid. FEBS Lett, 2000. 476(3): p. 160–5. [DOI] [PubMed] [Google Scholar]
- 36.Porciello N, et al. , Phosphatidylinositol 4-Phosphate 5-Kinases in the Regulation of T Cell Activation. Front Immunol, 2016. 7: p. 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang DG, et al. , PIP4Ks Suppress Insulin Signaling through a Catalytic-Independent Mechanism. Cell Rep, 2019. 27(7): p. 1991–2001.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wills RC, et al. , A Novel Homeostatic Mechanism Tunes PI(4,5)P2-dependent Signaling at the Plasma Membrane. bioRxiv, 2022: p. 2022.06.30.498262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semenas J, et al. , The role of PI3K/AKT-related PIP5K1alpha and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc Natl Acad Sci U S A, 2014. 111(35): p. E3689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarwar M, et al. , The role of PIP5K1alpha/pAKT and targeted inhibition of growth of subtypes of breast cancer using PIP5K1alpha inhibitor. Oncogene, 2019. 38(3): p. 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi S, et al. , A nuclear phosphoinositide kinase complex regulates p53. Nat Cell Biol, 2019. 21(4): p. 462–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adhikari H and Counter CM, Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nat Commun, 2018. 9(1): p. 3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran MH, et al. , NEDD4-induced degradative ubiquitination of phosphatidylinositol 4-phosphate 5-kinase alpha and its implication in breast cancer cell proliferation. J Cell Mol Med, 2018. 22(9): p. 4117–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. , Smurf1 regulates lung cancer cell growth and migration through interaction with and ubiquitination of PIPKIgamma. Oncogene, 2017. 36(41): p. 5668–5680. [DOI] [PubMed] [Google Scholar]
- 45.Chen C, et al. , Targeting type Igamma phosphatidylinositol phosphate kinase inhibits breast cancer metastasis. Oncogene, 2015. 34(35): p. 4635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SY, et al. , Regulation of the interaction between PIPKI gamma and talin by proline-directed protein kinases. J Cell Biol, 2005. 168(5): p. 789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng W, et al. , Type Igamma phosphatidylinositol phosphate kinase promotes tumor growth by facilitating Warburg effect in colorectal cancer. EBioMedicine, 2019. 44: p. 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, et al. , EGFR-induced phosphorylation of type Igamma phosphatidylinositol phosphate kinase promotes pancreatic cancer progression. Oncotarget, 2017. 8(26): p. 42621–42637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L, et al. , Cdk5-mediated phosphorylation regulates phosphatidylinositol 4-phosphate 5-kinase type I gamma 90 activity and cell invasion. FASEB J, 2019. 33(1): p. 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fruman DA and Bismuth G, Fine tuning the immune response with PI3K. Immunol Rev, 2009. 228(1): p. 253–72. [DOI] [PubMed] [Google Scholar]
- 51.Finlay DK, Regulation of glucose metabolism in T cells: new insight into the role of Phosphoinositide 3-kinases. Front Immunol, 2012. 3: p. 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kane LP, et al. , Induction of NF-κB by the Akt/PKB kinase. Current Biology, 1999. 9(11): p. 601–S1. [DOI] [PubMed] [Google Scholar]
- 53.Muscolini M, et al. , Phosphatidylinositol 4-phosphate 5-kinase alpha activation critically contributes to CD28-dependent signaling responses. J Immunol, 2013. 190(10): p. 5279–86. [DOI] [PubMed] [Google Scholar]
- 54.Kallikourdis M, et al. , Phosphatidylinositol 4-Phosphate 5-Kinase beta Controls Recruitment of Lipid Rafts into the Immunological Synapse. J Immunol, 2016. 196(4): p. 1955–63. [DOI] [PubMed] [Google Scholar]
- 55.Kunkl M, et al. , ISA-2011B, a Phosphatidylinositol 4-Phosphate 5-Kinase alpha Inhibitor, Impairs CD28-Dependent Costimulatory and Pro-inflammatory Signals in Human T Lymphocytes. Front Immunol, 2017. 8: p. 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SY, et al. , Phosphatidylinositol 4-phosphate 5-kinase alpha is induced in ganglioside-stimulated brain astrocytes and contributes to inflammatory responses. Exp Mol Med, 2010. 42(9): p. 662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzales B, et al. , Type I Phosphatidylinositol-4-Phosphate 5-Kinases alpha and gamma Play a Key Role in Targeting HIV-1 Pr55(Gag) to the Plasma Membrane. J Virol, 2020. 94(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Homma K, et al. , Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem, 1998. 273(25): p. 15779–86. [DOI] [PubMed] [Google Scholar]
- 59.Desrivieres S, et al. , MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem, 1998. 273(25): p. 15787–93. [DOI] [PubMed] [Google Scholar]
- 60.Mikami K, et al. , A gene encoding phosphatidylinositol-4-phosphate 5-kinase is induced by water stress and abscisic acid in Arabidopsis thaliana. Plant J, 1998. 15(4): p. 563–8. [DOI] [PubMed] [Google Scholar]
- 61.Deng L, et al. , Phosphatidylinositol-4-phosphate 5-kinase regulates fission yeast cell integrity through a phospholipase C-mediated protein kinase C-independent pathway. J Biol Chem, 2005. 280(30): p. 27561–8. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, et al. , Phosphatidylinositol 4-phosphate 5-kinase Its3 and calcineurin Ppb1 coordinately regulate cytokinesis in fission yeast. J Biol Chem, 2000. 275(45): p. 35600–6. [DOI] [PubMed] [Google Scholar]
- 63.Xu X, et al. , Role of phosphatidylinositol-4-phosphate 5’ kinase (ppk-1) in ovulation of Caenorhabditis elegans. Exp Cell Res, 2007. 313(11): p. 2465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinkove D, et al. , Overexpression of PPK-1, the Caenorhabditis elegans Type I PIP kinase, inhibits growth cone collapse in the developing nervous system and causes axonal degeneration in adults. Dev Biol, 2008. 313(1): p. 384–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volpicelli-Daley LA, et al. , Phosphatidylinositol-4-phosphate 5-kinases and phosphatidylinositol 4,5-bisphosphate synthesis in the brain. J Biol Chem, 2010. 285(37): p. 28708–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hasegawa H, et al. , Phosphatidylinositol 4-phosphate 5-kinase is indispensable for mouse spermatogenesis. Biol Reprod, 2012. 86(5): p. 136, 1–12. [DOI] [PubMed] [Google Scholar]
- 67.Hassan BA, et al. , skittles, a Drosophila phosphatidylinositol 4-phosphate 5-kinase, is required for cell viability, germline development and bristle morphology, but not for neurotransmitter release. Genetics, 1998. 150(4): p. 1527–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chakrabarti P, et al. , A dPIP5K dependent pool of phosphatidylinositol 4,5 bisphosphate (PIP2) is required for G-protein coupled signal transduction in Drosophila photoreceptors. PLoS Genet, 2015. 11(1): p. e1004948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Claret S, et al. , PI(4,5)P2 produced by the PI4P5K SKTL controls apical size by tethering PAR-3 in Drosophila epithelial cells. Curr Biol, 2014. 24(10): p. 1071–9. [DOI] [PubMed] [Google Scholar]
- 70.Park SH, Lee CW, and Choe KM, Interplay between integrins and PI4P5K Sktl is crucial for cell polarization and reepithelialisation during Drosophila wound healing. Sci Rep, 2019. 9(1): p. 16331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, et al. , PIP5KIγ is required for cardiovascular and neuronal development. Proceedings of the National Academy of Sciences, 2007. 104(28): p. 11748–11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hara Y, et al. , Type I phosphatidylinositol 4-phosphate 5-kinase gamma is required for neuronal migration in the mouse developing cerebral cortex. Eur J Neurosci, 2013. 38(5): p. 2659–71. 73. [DOI] [PubMed] [Google Scholar]
- 73.Narkis G, et al. , Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI gamma of the phophatidylinsitol pathway. Am J Hum Genet, 2007. 81(3): p. 530–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan Q, et al. , Loss of phosphatidylinositol-4-phosphate 5-kinase type-1 gamma (Pip5k1c) in mesenchymal stem cells leads to osteopenia by impairing bone remodeling. J Biol Chem, 2022. 298(3): p. 101639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burke JE, et al. , Beyond PI3Ks: targeting phosphoinositide kinases in disease. Nature Reviews Drug Discovery, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wright BD, et al. , Development of a High-Throughput Screening Assay to Identify Inhibitors of the Lipid Kinase PIP5K1C. J Biomol Screen, 2015. 20(5): p. 655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarwar M, et al. , Targeted suppression of AR-V7 using PIP5K1alpha inhibitor overcomes enzalutamide resistance in prostate cancer cells. Oncotarget, 2016. 7(39): p. 63065–63081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Semenas J, et al. , Targeted inhibition of ERalpha signaling and PIP5K1alpha/Akt pathways in castration-resistant prostate cancer. Mol Oncol, 2021. 15(4): p. 968–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andrews DM, et al. , Identification and optimization of a novel series of selective PIP5K inhibitors. Bioorg Med Chem, 2022. 54: p. 116557. [DOI] [PubMed] [Google Scholar]
- 80.Jones DR, et al. , Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell, 2006. 23(5): p. 685–95. [DOI] [PubMed] [Google Scholar]
- 81.Wilcox A and Hinchliffe KA, Regulation of extranuclear PtdIns5P production by phosphatidylinositol phosphate 4-kinase 2alpha. FEBS Lett, 2008. 582(9): p. 1391–4. [DOI] [PubMed] [Google Scholar]
- 82.Bulley SJ, et al. , Exploring phosphatidylinositol 5-phosphate 4-kinase function. Adv Biol Regul, 2015. 57: p. 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clarke JH and Irvine RF, The activity, evolution and association of phosphatidylinositol 5-phosphate 4-kinases. Adv Biol Regul, 2012. 52(1): p. 40–5. [DOI] [PubMed] [Google Scholar]
- 84.Bultsma Y, Keune WJ, and Divecha N, PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J, 2010. 430(2): p. 223–35. [DOI] [PubMed] [Google Scholar]
- 85.Lundquist MR, et al. , Phosphatidylinositol-5-Phosphate 4-Kinases Regulate Cellular Lipid Metabolism By Facilitating Autophagy. Mol Cell, 2018. 70(3): p. 531–544 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu A, et al. , PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P(2) homeostasis. J Lipid Res, 2018. 59(3): p. 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vicinanza M, et al. , PI(5)P regulates autophagosome biogenesis. Mol Cell, 2015. 57(2): p. 219–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Clarke JH, Emson PC, and Irvine RF, Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am J Physiol Renal Physiol, 2008. 295(5): p. F1422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang M, et al. , Genomic tagging reveals a random association of endogenous PtdIns5P 4-kinases IIalpha and IIbeta and a partial nuclear localization of the IIalpha isoform. Biochem J, 2010. 430(2): p. 215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clarke JH and Irvine RF, Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem J, 2013. 454(1): p. 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sharma S, et al. , Phosphatidylinositol 5 Phosphate 4-Kinase Regulates Plasma-Membrane PIP(3) Turnover and Insulin Signaling. Cell Rep, 2019. 27(7): p. 1979–1990.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shin YJ, et al. , PIP4K2A as a negative regulator of PI3K in PTEN-deficient glioblastoma. J Exp Med, 2019. 216(5): p. 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Behari J, et al. , Conserved RNA Binding Activity of Phosphatidyl Inositol 5-Phosphate 4-Kinase (PIP4K2A). Front Mol Biosci, 2021. 8: p. 631281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamalesh K, et al. , Phosphatidylinositol 5-phosphate 4-kinase regulates early endosomal dynamics during clathrin-mediated endocytosis. J Cell Sci, 2017. 130(13): p. 2119–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carricaburu V, et al. , The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc Natl Acad Sci U S A, 2003. 100(17): p. 9867–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gupta A, et al. , Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc Natl Acad Sci U S A, 2013. 110(15): p. 5963–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mackey AM, et al. , PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci Signal, 2014. 7(350): p. ra104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bulley SJ, et al. , In B cells, phosphatidylinositol 5-phosphate 4-kinase-alpha synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc Natl Acad Sci U S A, 2016. 113(38): p. 10571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palamiuc L, Ravi A, and Emerling BM, Phosphoinositides in autophagy: current roles and future insights. FEBS J, 2020. 287(2): p. 222–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang S, et al. , MANF regulates hypothalamic control of food intake and body weight. Nat Commun, 2017. 8(1): p. 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang M, et al. , A novel lncRNA lncSAMD11–1: 1 interacts with PIP4K2A to promote endometrial decidualization by stabilizing FoxO1 nuclear localization. Int J Biochem Cell Biol, 2022. 151: p. 106280. [DOI] [PubMed] [Google Scholar]
- 102.Keune WJ, et al. , Regulation of phosphatidylinositol-5-phosphate signaling by Pin1 determines sensitivity to oxidative stress. Sci Signal, 2012. 5(252): p. ra86. [DOI] [PubMed] [Google Scholar]
- 103.Jones DR, et al. , PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. Faseb j, 2013. 27(4): p. 1644–56. [DOI] [PubMed] [Google Scholar]
- 104.Sumita K, et al. , The Lipid Kinase PI5P4Kβ Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol Cell, 2016. 61(2): p. 187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takeuchi K, et al. , The GTP responsiveness of PI5P4Kβ evolved from a compromised trade-off between activity and specificity. Structure, 2022. 30(6): p. 886–899.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ravi A, et al. , PI5P4Ks drive metabolic homeostasis through peroxisome-mitochondria interplay. Dev Cell, 2021. 56(11): p. 1661–1676.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu A, et al. , PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P2 homeostasis. J Lipid Res, 2018. 59(3): p. 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zheng L and Conner SD, PI5P4Kγ functions in DTX1-mediated Notch signaling. Proc Natl Acad Sci U S A, 2018. 115(9): p. E1983–e1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burgess J, et al. , Type II phosphatidylinositol 4-kinase regulates trafficking of secretory granule proteins in Drosophila. Development, 2012. 139(16): p. 3040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shim H, et al. , Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc Natl Acad Sci U S A, 2016. 113(27): p. 7596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raychaudhuri S, et al. , Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nature Genetics, 2008. 40(10): p. 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fung EYMG, et al. , Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes & Immunity, 2009. 10(2): p. 188–191. [DOI] [PubMed] [Google Scholar]
- 113.Poli A, et al. , PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc Natl Acad Sci U S A, 2021. 118(31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gelato KA, et al. , Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol Cell, 2014. 54(6): p. 905–19. [DOI] [PubMed] [Google Scholar]
- 115.Bua DJ, et al. , Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci Rep, 2013. 3: p. 2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gozani O, et al. , The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell, 2003. 114(1): p. 99–111. [DOI] [PubMed] [Google Scholar]
- 117.Stijf-Bultsma Y, et al. , The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol Cell, 2015. 58(3): p. 453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hinchliffe KA and Irvine RF, Regulation of type II PIP kinase by PKD phosphorylation. Cell Signal, 2006. 18(11): p. 1906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hinchliffe KA, et al. , Regulation of type IIalpha phosphatidylinositol phosphate kinase localisation by the protein kinase CK2. Curr Biol, 1999. 9(17): p. 983–6. [DOI] [PubMed] [Google Scholar]
- 120.Bunce MW, Boronenkov IV, and Anderson RA, Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem, 2008. 283(13): p. 8678–86. [DOI] [PubMed] [Google Scholar]
- 121.Arora GK, Palamiuc L, and Emerling BM, Expanding role of PI5P4Ks in cancer: A promising druggable target. FEBS Lett, 2022. 596(1): p. 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Emerling BM, et al. , Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell, 2013. 155(4): p. 844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keune W-J, et al. , Low PIP4K2B Expression in Human Breast Tumors Correlates with Reduced Patient Survival: A Role for PIP4K2B in the Regulation of E-Cadherin Expression. Cancer Research, 2013. 73(23): p. 6913–6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Terkelsen T, et al. , High-throughput proteomics of breast cancer interstitial fluid: identification of tumor subtype-specific serologically relevant biomarkers. Mol Oncol, 2021. 15(2): p. 429–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jude JG, et al. , A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene, 2015. 34(10): p. 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang S, et al. , Regulatory Network and Prognostic Effect Investigation of PIP4K2A in Leukemia and Solid Cancers. Front Genet, 2018. 9: p. 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lima K, et al. , PIP4K2A and PIP4K2C transcript levels are associated with cytogenetic risk and survival outcomes in acute myeloid leukemia. Cancer Genet, 2019. 233–234: p. 56–66. [DOI] [PubMed] [Google Scholar]
- 128.Walsh KM, et al. , Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood, 2013. 121(23): p. 4808–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liao F, et al. , Association Between PIP4K2A Polymorphisms and Acute Lymphoblastic Leukemia Susceptibility. Medicine (Baltimore), 2016. 95(18): p. e3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Migliorini G, et al. , Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood, 2013. 122(19): p. 3298–307. [DOI] [PubMed] [Google Scholar]
- 131.Xu H, et al. , Novel susceptibility variants at 10p12.31–12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst, 2013. 105(10): p. 733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Noch EK, et al. , Distribution and localization of phosphatidylinositol 5-phosphate, 4-kinase alpha and beta in the brain. J Comp Neurol, 2021. 529(2): p. 434–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Al-Ramahi I, et al. , Inhibition of PIP4Kγ ameliorates the pathological effects of mutant huntingtin protein. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.He Z, et al. , The PIP5K2A gene and schizophrenia in the Chinese population — A case-control study. Schizophrenia Research, 2007. 94(1): p. 359–365. [DOI] [PubMed] [Google Scholar]
- 135.Réthelyi JM, et al. , Association study of NRG1, DTNBP1, RGS4, G72/G30, and PIP5K2A with schizophrenia and symptom severity in a Hungarian sample. Am J Med Genet B Neuropsychiatr Genet, 2010. 153b(3): p. 792–801. [DOI] [PubMed] [Google Scholar]
- 136.Schwab SG, et al. , Evidence for association of DNA sequence variants in the phosphatidylinositol-4-phosphate 5-kinase IIalpha gene (PIP5K2A) with schizophrenia. Mol Psychiatry, 2006. 11(9): p. 837–46. [DOI] [PubMed] [Google Scholar]
- 137.Thiselton DL, et al. , Association analysis of the PIP4K2A gene on chromosome 10p12 and schizophrenia in the Irish study of high density schizophrenia families (ISHDSF) and the Irish case-control study of schizophrenia (ICCSS). Am J Med Genet B Neuropsychiatr Genet, 2010. 153b(1): p. 323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaur H, et al. , Genetic variations of PIP4K2A confer vulnerability to poor antipsychotic response in severely ill schizophrenia patients. PLoS One, 2014. 9(7): p. e102556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Levchenko A, et al. , NRG1, PIP4K2A, and HTR2C as Potential Candidate Biomarker Genes for Several Clinical Subphenotypes of Depression and Bipolar Disorder. Front Genet, 2020. 11: p. 936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Beziau A, Brand D, and Piver E, The Role of Phosphatidylinositol Phosphate Kinases during Viral Infection. Viruses, 2020. 12(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vindu A, Dandewad V, and Seshadri V, Identification of human Phosphatidyl Inositol 5-Phosphate 4-Kinase as an RNA binding protein that is imported into Plasmodium falciparum. Biochem Biophys Res Commun, 2018. 498(3): p. 529–536. [DOI] [PubMed] [Google Scholar]
- 142.Lamia KA, et al. , Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol Cell Biol, 2004. 24(11): p. 5080–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Davis MI, et al. , A Homogeneous, High-Throughput Assay for Phosphatidylinositol 5-Phosphate 4-Kinase with a Novel, Rapid Substrate Preparation. PLOS ONE, 2013. 8(1): p. e54127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Voss MD, et al. , Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem Biophys Res Commun, 2014. 449(3): p. 327–31. [DOI] [PubMed] [Google Scholar]
- 145.Clarke JH, et al. , The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem J, 2015. 466(2): p. 359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Boffey HK, et al. , Development of Selective Phosphatidylinositol 5-Phosphate 4-Kinase γ Inhibitors with a Non-ATP-competitive, Allosteric Binding Mode. Journal of Medicinal Chemistry, 2022. 65(4): p. 3359–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wortmann L, et al. , Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A. Journal of Medicinal Chemistry, 2021. 64(21): p. 15883–15911. [DOI] [PubMed] [Google Scholar]
- 148.Kitagawa M, et al. , Dual blockade of the lipid kinase PIP4Ks and mitotic pathways leads to cancer-selective lethality. Nat Commun, 2017. 8(1): p. 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sivakumaren SC, et al. , Targeting the PI5P4K Lipid Kinase Family in Cancer Using Covalent Inhibitors. Cell Chemical Biology, 2020. 27(5): p. 525–537.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lima K, et al. , The PIP4K2 inhibitor THZ-P1–2 exhibits antileukemia activity by disruption of mitochondrial homeostasis and autophagy. Blood Cancer Journal, 2022. 12(11): p. 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Manz TD, et al. , Structure–Activity Relationship Study of Covalent Pan-phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors. ACS Medicinal Chemistry Letters, 2020. 11(3): p. 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen S, et al. , Pharmacological inhibition of PI5P4Kα/β disrupts cell energy metabolism and selectively kills p53-null tumor cells. Proc Natl Acad Sci U S A, 2021. 118(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Akiba Y, et al. , Localization of mRNAs for phosphatidylinositol phosphate kinases in the mouse brain during development. Gene Expression Patterns, 2002. 1(2): p. 123–133. [DOI] [PubMed] [Google Scholar]
- 154.Hasegawa J, Strunk BS, and Weisman LS, PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct Funct, 2017. 42(1): p. 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Giridharan SSP, et al. , Lipid kinases VPS34 and PIKfyve coordinate a phosphoinositide cascade to regulate retriever-mediated recycling on endosomes. eLife, 2022. 11: p. e69709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lees JA, et al. , Insights into Lysosomal PI(3,5)P2 Homeostasis from a Structural-Biochemical Analysis of the PIKfyve Lipid Kinase Complex. Mol Cell, 2020. 80(4): p. 736–743 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ikonomov OC, et al. , PIKfyve-ArPIKfyve-Sac3 Core Complex. Journal of Biological Chemistry, 2009. 284(51): p. 35794–35806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Efe JA, Botelho RJ, and Emr SD, The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Current Opinion in Cell Biology, 2005. 17(4): p. 402–408. [DOI] [PubMed] [Google Scholar]
- 159.Kronenberg ZN, et al. , High-resolution comparative analysis of great ape genomes. Science, 2018. 360(6393): p. eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hayakawa A, et al. , Structural Basis for Endosomal Targeting by FYVE Domains *. Journal of Biological Chemistry, 2004. 279(7): p. 5958–5966. [DOI] [PubMed] [Google Scholar]
- 161.Sbrissa D, Ikonomov OC, and Shisheva A, PIKfyve Lipid Kinase Is a Protein Kinase: Downregulation of 5’-Phosphoinositide Product Formation by Autophosphorylation. Biochemistry, 2000. 39(51): p. 15980–15989. [DOI] [PubMed] [Google Scholar]
- 162.Ikonomov OC, Sbrissa D, and Shisheva A, Localized PtdIns 3,5-P2 synthesis to regulate early endosome dynamics and fusion. American Journal of Physiology-Cell Physiology, 2006. 291(2): p. C393–C404. [DOI] [PubMed] [Google Scholar]
- 163.Kim GHE, et al. , PIKfyve Inhibition Interferes with Phagosome and Endosome Maturation in Macrophages. Traffic, 2014. 15(10): p. 1143–1163. [DOI] [PubMed] [Google Scholar]
- 164.Tsuruta F, et al. , PIKfyve regulates CaV1.2 degradation and prevents excitotoxic cell death. Journal of Cell Biology, 2009. 187(2): p. 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Choy CH, et al. , Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. Journal of Cell Science, 2018. 131(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krishna S, et al. , PIKfyve Regulates Vacuole Maturation and Nutrient Recovery following Engulfment. Dev Cell, 2016. 38(5): p. 536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Qiao Y, et al. , Autophagy inhibition by targeting PIKfyve potentiates response to immune checkpoint blockade in prostate cancer. Nature Cancer, 2021. 2(9): p. 978–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ikonomov OC, et al. , Active PIKfyve Associates with and Promotes the Membrane Attachment of the Late Endosome-to-trans-Golgi Network Transport Factor Rab9 Effector p40*. Journal of Biological Chemistry, 2003. 278(51): p. 50863–50871. [DOI] [PubMed] [Google Scholar]
- 169.Cai X, et al. , PIKfyve, a Class III PI Kinase, Is the Target of the Small Molecular IL-12/IL-23 Inhibitor Apilimod and a Player in Toll-like Receptor Signaling. Chemistry & Biology, 2013. 20(7): p. 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hill EV, et al. , Regulation of PIKfyve phosphorylation by insulin and osmotic stress. Biochemical and Biophysical Research Communications, 2010. 397(4): p. 650–655. [DOI] [PubMed] [Google Scholar]
- 171.Li S, et al. , Mutations in PIP5K3 Are Associated with François-Neetens Mouchetée Fleck Corneal Dystrophy. The American Journal of Human Genetics, 2005. 77(1): p. 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mei S, et al. , Disruption of PIKFYVE causes congenital cataract in human and zebrafish. eLife, 2022. 11: p. e71256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kang Y-L, et al. , Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proceedings of the National Academy of Sciences, 2020. 117(34): p. 20803–20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Su J, et al. , PIKfyve inhibitors against SARS-CoV-2 and its variants including Omicron. Signal Transduction and Targeted Therapy, 2022. 7(1): p. 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nelson EA, et al. , The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLOS Neglected Tropical Diseases, 2017. 11(4): p. e0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Riva L, et al. , Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature, 2020. 586(7827): p. 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baranov MV, Bianchi F, and van den Bogaart G, The PIKfyve Inhibitor Apilimod: A Double-Edged Sword against COVID-19. Cells, 2021. 10(1): p. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Huang PT, Einav S, and Asquith CRM, PIKfyve: a lipid kinase target for COVID-19, cancer and neurodegenerative disorders. Nat Rev Drug Discov, 2021. 20(10): p. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rivero-Ríos P and Weisman LS, Roles of PIKfyve in multiple cellular pathways. Current Opinion in Cell Biology, 2022. 76: p. 102086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shisheva A, PIKfyve and its Lipid Products in Health and in Sickness, in Phosphoinositides and Disease, Falasca M, Editor. 2012, Springer Netherlands: Dordrecht. p. 127–162. [DOI] [PubMed] [Google Scholar]
- 181.Soares AC, et al. , PIKfyve activity is required for lysosomal trafficking of tau aggregates and tau seeding. Journal of Biological Chemistry, 2021. 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ikonomov OC, Sbrissa D, and Shisheva A, Small molecule PIKfyve inhibitors as cancer therapeutics: Translational promises and limitations. Toxicology and Applied Pharmacology, 2019. 383: p. 114771. [DOI] [PubMed] [Google Scholar]
- 183.Gayle S, et al. , B-cell non-Hodgkin lymphoma: Selective vulnerability to PIKFYVE inhibition. Autophagy, 2017. 13(6): p. 1082–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hou JZ, et al. , Inhibition of PIKfyve using YM201636 suppresses the growth of liver cancer via the induction of autophagy. Oncol Rep, 2019. 41(3): p. 1971–1979. [DOI] [PubMed] [Google Scholar]
- 185.Yamamoto A, et al. , Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Molecular Biology of the Cell, 1995. 6(5): p. 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Collier NC and Wang K, Human platelet P235: A high M r protein which restricts the length of actin filaments. FEBS Letters, 1982. 143(2): p. 205–210. [DOI] [PubMed] [Google Scholar]
- 187.Nicot A-S, et al. , The Phosphoinositide Kinase PIKfyve/Fab1p Regulates Terminal Lysosome Maturation in Caenorhabditis elegans. Molecular Biology of the Cell, 2006. 17(7): p. 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rusten TE, et al. , Fab1 Phosphatidylinositol 3-Phosphate 5-Kinase Controls Trafficking but Not Silencing of Endocytosed Receptors. Molecular Biology of the Cell, 2006. 17(9): p. 3989–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ikonomov OC, et al. , The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J Biol Chem, 2011. 286(15): p. 13404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.McCartney AJ, et al. , Activity-dependent PI(3,5)P2 synthesis controls AMPA receptor trafficking during synaptic depression. Proc Natl Acad Sci U S A, 2014. 111(45): p. E4896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Min SH, et al. , PIKfyve Deficiency in Myeloid Cells Impairs Lysosomal Homeostasis in Macrophages and Promotes Systemic Inflammation in Mice. Molecular and Cellular Biology, 2019. 39(21): p. e00158–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Takasuga S, et al. , Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proceedings of the National Academy of Sciences, 2013. 110(5): p. 1726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gayle S, et al. , Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood, 2017. 129(13): p. 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Magid Diefenbach CS, et al. , Results of a completed phase I study of LAM-002 (apilimod dimesylate), a first-in-class phosphatidylinositol-3-phosphate 5 kinase (PIKfyve) inhibitor, administered as monotherapy or with rituximab or atezolizumab to patients with previously treated follicular lymphoma or other B-cell cancers. Journal of Clinical Oncology, 2020. 38(15_suppl): p. 8017–8017. [Google Scholar]
- 195.Idevall-Hagren O and De Camilli P, Detection and manipulation of phosphoinositides. Biochim Biophys Acta, 2015. 1851(6): p. 736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Békés M, Langley DR, and Crews CM, PROTAC targeted protein degraders: the past is prologue. Nature Reviews Drug Discovery, 2022. 21(3): p. 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nalawansha DA and Crews CM, PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem Biol, 2020. 27(8): p. 998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]


