Abstract
The human RNA polymerase II transcription factor B-TFIID consists of TATA-binding protein (TBP) and the TBP-associated factor (TAF) TAFII170 and can rapidly redistribute over promoter DNA. Here we report the identification of human TBP-binding regions in human TAFII170. We have defined the TBP interaction domain of TAFII170 within three amino-terminal regions: residues 2 to 137, 290 to 381, and 380 to 460. Each region contains a pair of Huntington-elongation-A subunit-Tor repeats and exhibits species-specific interactions with TBP family members. Remarkably, the altered-specificity TBP mutant (TBPAS) containing a triple mutation in the concave surface is defective for binding the TAFII170 amino-terminal region of residues 1 to 504. Furthermore, within this region the TAFII170 residues 290 to 381 can inhibit the interaction between Drosophila TAFII230 (residues 2 to 81) and TBP through competition for the concave surface of TBP. Biochemical analyses of TBP binding to the TATA box indicated that TAFII170 region 290-381 inhibits TBP-DNA complex formation. Importantly, the TBPAS mutant is less sensitive to TAFII170 inhibition. Collectively, our results support a mechanism in which TAFII170 induces high-mobility DNA binding by TBP through reversible interactions with its concave DNA binding surface.
Initiation of mRNA synthesis by RNA polymerase (pol) II requires the basal transcription factors TFIID, TFIIB, TFIIE, TFIIF, and TFIIH. Recognition of the TATA box within core pol II promoters by TFIID initiates the assembly of a functional pol II preinitiation complex. TFIID is a multisubunit complex consisting of TATA-binding protein (TBP) and at least 10 TBP-associated factors (TAFIIs) ranging in size from 15 to 250 kDa. TAFIIs play several roles in transcriptional regulation, serving as coactivators, promoter selectivity factors, or enzymes that modify surrounding transcriptional proteins (3). The TBP subunit recognizes the TATA box forming the TBP-TATA complex (24). Structural studies of the TBP-TATA complex indicate that the β sheets of TBP form a concave undersurface which contacts the minor groove of the TATA box (28, 29).
Of relevance to this study is that the largest TAFII subunit of TFIID (dTAFII230 in Drosophila, hTAFII250 in humans, and yTAFII145/130 in yeast), can inhibit TBP binding to the TATA box (11, 31, 33). dTAFII230 achieves this by interacting with the concave DNA binding surface of TBP (see references cited above). Residues 2 to 81 (called TAND I) undergo a disorder-to-order transition upon interaction with TBP. The induced structure of TAND I mimics the structure of the TATA box when bound to TBP (34). A second N-terminal inhibitory domain (TAND II; residues 82 to 156) blocks the TBP-TFIIA interaction by competitively binding to helix 2 (H2) on the convex surface of TBP (10). The orthologous TAFII subunits from yeast and humans contain regions which are functionally equivalent to TAND I and TAND II despite a low degree of primary sequence similarity (33).
TAFII-containing complexes distinct from TFIID have been described. Examples include the yeast SAGA and human PCAF and TFTC complexes (3). In addition to these, it has been reported previously that the majority of TBP in mammalian cell extracts is present in the B-TFIID complex (49). In this distinct complex TBP is associated with a single TAFII, TAFII170 (49, 50). B-TFIID can support TATA-dependent transcription in reconstituted basal transcription experiments as efficiently as TBP or TFIID (49). Notably the complex displays several biochemical properties that make it distinct from traditional TFIID. Firstly, pol II transcription with B-TFIID is not responsive to activators (49). Secondly, B-TFIID exhibits dATPase activity that can be attributed to the TAFII170 subunit (50). Finally, B-TFIID binds unstably to the TATA box, which may explain its inability to commit a template for transcription (49).
hTAFII170 has also been cloned as TAF172 (16; referred to in this paper as TAFII170). hTAFII170 is the human ortholog of yMot1 (16, 50), and both are members of the SNF2 family of DNA-dependent ATPases (42). MOT1 was originally identified by genetic screens for enhanced basal transcription in yeast (20). Independent studies showed Mot1 protein to exist in a complex with TBP distinct from yTFIID (43). Mot1 protein was identified as an ATP-dependent inhibitor activity in yeast nuclear extracts that removed TBP from the TATA box using the energy of ATP hydrolysis (6). Interestingly, amounts of Mot1 stoichiometric to TBP weakly stimulate TBP-dependent transcription, whereas a molar excess of Mot1 leads to a repression of transcription (17, 36, 39). Studies with Mot1 have shown that the release of TBP from DNA does not appear to involve an ATP-dependent DNA tracking over distances as short as 40 bp (2, 7). Furthermore, Mot1-catalyzed disruption of TBP-DNA complexes does not appear to involve DNA strand separation, DNA bending, or twisting of the helix (2, 7). Rather, it has been proposed that Mot1 displaces TBP from DNA by translocation through the TATA box in an ATP-dependent manner (2, 7). Deletion analysis shows that this function requires a 17-bp double-stranded DNA “handle” upstream of the TATA box, indicating that Mot1 contacts upstream DNA (19). Several observations indicate that Mot1 interacts with H2 on the convex surface of yTBP (1, 5, 6, 15), which led to the hypothesis that Mot1 can also distort TBP conformation in a “power stroke” action leading to TBP-TATA dissociation (2, 8). However, to date there have been no experimental data to support this. Besides Mot1 being a repressor of transcription, there is also a link between Mot1 and activation of transcription. Mot1 may activate transcription by redistributing a limiting pool of TBP between DNA sites in vitro and in vivo (17, 36, 39). Notably, TAFII170, like Mot1, uses the energy of ATP hydrolysis to dissociate TBP from DNA (16).
An important issue is the observation that in contrast to TBP or TFIID, B-TFIID does not form highly stable complexes with the TATA box in the absence of ATP (49). We therefore set out to establish how the hTAFII170/hTBP interaction relates mechanistically to the TATA box-binding properties of B-TFIID. We first sought to establish the hTBP interaction domain of hTAFII170 and show that three regions within the amino-terminal third of hTAFII170 are involved in the interaction. We found that these regions are critical for the binding of hTAFII170 to hTBP and exhibit species-specific interactions with TBP family members. Strikingly, the concave DNA binding surface of hTBP is crucial for this interaction. This led to the finding that a small domain of hTAFII170 within its amino-terminal part could inhibit TBP-TATA complex formation. Our results support a model in which insertion of this hTAFII170 domain into the DNA binding cavity of hTBP inhibits its interaction with the TATA box and explains the DNA binding properties of B-TFIID. Reversibility of this process would allow for rapid redistribution and increased mobility of hTBP on DNA.
MATERIALS AND METHODS
Plasmids.
pJGhTBP was created by cloning a SalI (blunt)-XhoI fragment from pRSVhIID into the EcoRI (blunt) and XhoI sites of pJG4-5. pJGTBPAS was generated by cloning a DraIII-ClaI fragment from pSRaMSVtkneohTBPm3e (14) into the DraIII and ClaI sites of pJGTBP. To create the pEG202-hTAFII170 derivatives NdeI-AocI (1-504), BamHI-AocI (136-504), BspHI (1133-1849), and XmaI (blunt)-BspHI (505-1133), fragments from hTAFII170 cDNA were cloned into pEG202. The remaining pEG-hTAFII170 plasmids were created from pEG-hTAFII170 (1-504) by using a double-stranded, nested-deletion kit (Pharmacia Biotech). pAC-TAF (2-460) was created by cloning a PCR fragment generated from hTAFII170 cDNA into the XhoI and NotI (blunt) sites of pACYC. All other pAC-hTAFII170 constructs were created by cloning PCR fragments amplified from pAC-TAF (2-460) into the SpeI and XhoI sites of pAC-TAF (2-460). To generate glutathione S-transferase (GST)-Flag-TAF fusions, NdeI-SpeI fragments from pAC-hTAFII170 derivatives were cloned into the NdeI and SpeI sites of modified pGEX-2T. To prepare His-tagged dTAFII230 (2-81), dTAFII230 (82-156), and dTAFII230 (2-156), corresponding DNA from pGEX-dTAFII230 (2-156) (a kind gift from Y. Nakatani) was PCR amplified as SalI-BamHI fragments and subcloned into pET15b (Novagen). pGST-TFIIS was created by cloning an NdeI-BamHI fragment from pET22-TFIIS (a kind gift from C. Kane) into the NdeI and BamHI sites of pGEX-2T. For expression of full-length hTAFII170 in mammalian cells, pMT2-hTAFII170 was created by cloning a NaeI-StuI fragment from pET-hTAFII170 into the SmaI site of pMT2-SM. pSG-ehTBP and pSG-ehTBPm3e (kind gifts from H. Stunnenberg) and pSG-ehTBPK243E were used for expression of full-length hTBP in mammalian cells (see Fig. 5B). pSG-ehTBPK243E was created by oligonucleotide-mediated, site-directed mutagenesis using pSG-ehTBP as the template.
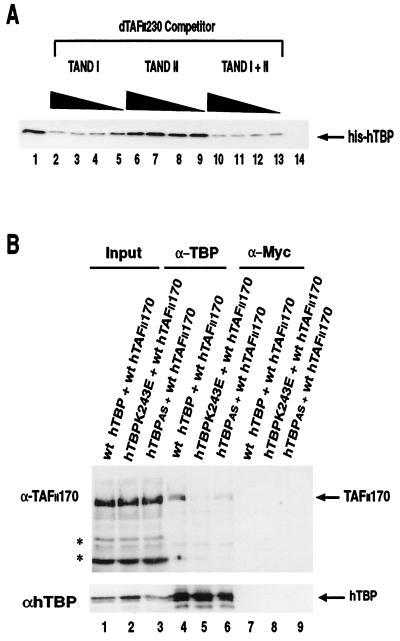
FIG. 5.
Interaction of full-length hTAFII170 with the convex and concave surfaces of hTBP. (A) The amino termini of dTAFII230 (TAND I) and hTAFII170 bind competitively to hTBP. The bacterial lysate of GST-Flag-hTAFII170 residues 290 to 381 was incubated with His-hTBP (2 pmol) in the absence (lane 1) or presence of batch-purified lysates of His-dTAFII230 (TAND I, residues 2 to 81; lanes 2 to 5), His-dTAFII230 (TAND II, residues 82 to 156; lanes 6 to 9), and His-dTAFII230 (TAND I and II, residues 2 to 156; lanes 10 to 13) and was assayed for hTBP binding as described in Materials and Methods. As a control for background binding GST-TFIIS was incubated with His-hTBP (2 pmol) in the absence of the His-dTAFII230 TANDs (lane 14). For His-dTAFII230 TAND I and His-dTAFII230 TAND II, the titration series of 180, 60, 20, and 6.6 pmol of batch-purified His-dTAFII230 lysates was used. For His-dTAFII230 TAND I and II, the titration series of 60, 20, 6.6, and 2.2 pmol of batch-purified His-dTAFII230 lysates was used. (B) Coimmunoprecipitation of exogenously expressed full-length hTAFII170 and the hTBPK243E and hTBPAS mutants. Immunoprecipitation from COS-7 cell lysates was performed with α-hTBP (lanes 4 to 6) or, as a control, α-c-Myc (lanes 7 to 9) as described in Materials and Methods. The input lane represents 20% of the cell extract used in the immunoprecipitations (lanes 1 to 3). wt, wild type. Asterisks indicate nonspecific bands.
Yeast two-hybrid analysis.
Yeast strain EGY48 was transformed with the hTAFII170 derivatives harbored in the bait vector pEG202, either pJG-hTBP or pJG-hTBPAS accompanied by pSH18-34 (Invitrogen) harboring the lacZ reporter in accordance with the standard lithium acetate method (9). Transformants were selected on His/Trp/Ura-deficient plates and patched onto His/Trp/Ura/Leu-deficient plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and galactose. Interactions were scored by measurement of growth of blue colonies and quantitative determination of β-galactosidase activity as described previously (38).
Protein expression.
Escherichia coli strain BL21(DE3) containing pGEX-hTBP, pGEX-TFIIS, pGEX-Flag-hTAFII170 derivatives, pET15b-dTAFII230 derivatives, or pAC-hTAFII170 derivatives was cultured at 37°C in 50 ml of Luria-Bertani medium containing ampicillin (100 μg/ml) or, in the case of pAC-hTAFII170 derivatives, chloramphenicol (25 μg/ml). At an optical density at 600 nm of 0.5 to 0.6, the cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.4 mM). After 3 h cells were harvested and the cell pellet was resuspended in 4 ml of lysis buffer (50 mM Tris-HCl, pH 8.0, 20% sucrose, 1 mM EDTA, 0.3 M KCl, 0.01% Triton X-100, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.2 mM sodium metabisulfite, 1 μg of aprotinin/ml, and 1 μg of leupeptin/ml). The cells were incubated on ice with 200 μg of lysozyme/ml and lysed by freeze-thawing and sonication, and the bacterial lysate was cleared by ultracentrifugation.
Purification of GST-Flag-hTAFII170, His-dTAFII230 derivatives, and hTBP proteins.
GST-Flag-TAFII170-containing lysates were prepared on a 2-liter scale as described above and loaded onto a glutathione agarose column. The column was washed with lysis buffer (20 mM HEPES-KOH, pH 8.05, 20% glycerol, 300 mM KCl, 0.01% Triton X-100, 1 mM EDTA, and protease inhibitors) and eluted with lysis buffer containing 15 mM glutathione. Peak fractions were pooled and dialyzed against A100 buffer (20 mM HEPES-KOH, pH 7.9, 20% glycerol, 100 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, and 0.5 mM PMSF). The amount of eluted protein was determined by Bio-Rad protein assays (Bio-Rad) using bovine gamma globulin as a standard and Coomassie blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels. Proteins were negative for RNase activity as analyzed by incubation with radiolabeled RNA. His-dTAFII230 derivatives were batch purified by mixing clarified lysates containing 20 mM imidazole with Ni-nitrilotriacetic acid agarose (Qiagen) for 2 h at 4°C with gentle shaking. The slurry was washed two times in wash buffer (50 mM Tris-HCl, pH 8.0, 20% sucrose, 1 mM EDTA, 0.3 M KCl, 0.01% Triton X-100, 20 mM imidazole, 0.5 mM PMSF, 0.2 mM sodium metabisulfite, 1 μg of aprotinin/ml, and 1 μg of leupeptin/ml), and the His-dTAFII230 proteins were eluted in wash buffer containing 300 mM imidazole for 30 min at 4°C with gentle shaking. His-hTBP was expressed in E. coli and purified as described previously (26). For Fig. 6C, recombinant hTBP and altered- or relaxed-specificity hTBP (hTBPAS) were expressed as GST fusions in E. coli and purified as described for the GST-Flag-TAFII170 derivatives, with the following modifications: after glutathione agarose chromatography, peak hTBP fractions, as judged by Coomassie staining of protein gels, were pooled and the GST tag was removed by cleaving the pooled hTBP preparations with thrombin (1 IU/0.8 mg of protein) for 4 h at 37°C. Thrombin was removed by batch purifying the hTBP preparations with benzamidine Sepharose 6B (Sigma). The preparations were dialyzed against A100 buffer and subsequently batch purified with glutathione agarose to remove GST and uncleaved hTBP fusion protein. The preparations were then applied to a 5-ml HiTrap heparin cartridge (Pharmacia Biotech). The column was developed by a linear gradient from 0.1 to 1.0 M KCl in buffer A over 10 column volumes. hTBP fractions with the highest specific activity, as judged by gel shift analysis, were used for subsequent experiments.
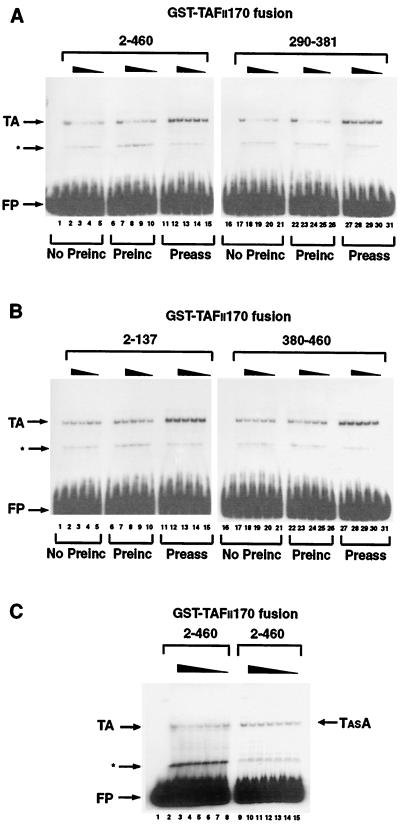
FIG. 6.
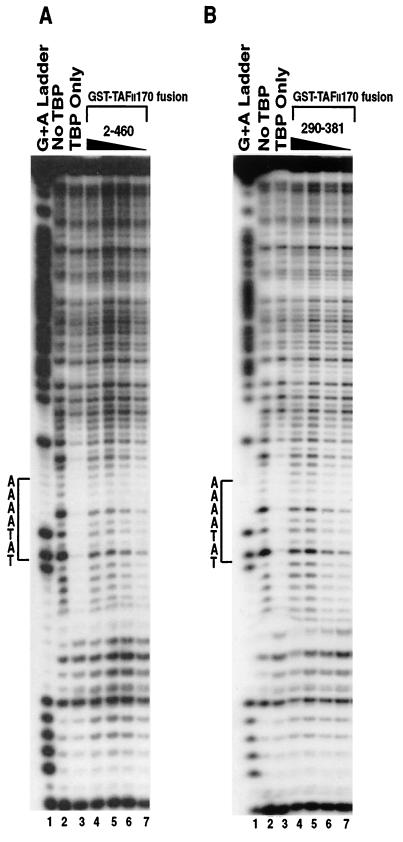
The amino terminus of hTAFII170 negatively regulates the TATA box-binding activity of hTBP. (A) Gel mobility shifts were performed with an AdMLP probe (−55 to +33) incubated with no protein (lane 16); with His-hTBP (0.2 pmol) and hTFIIA (lane 1, 6, 11, 17, 22, and 27); with hTBP, hTFIIA, and GST-Flag-hTAFII170 residues 2 to 460 (lanes 2 to 5, 7 to 10, and 12 to 15); or residues 290 to 381 (lanes 18 to 21, 23 to 26, and 28 to 31). (B) Gel mobility shifts were performed as described for panel A with no protein (lane 16); His-hTBP and hTFIIA (lane 1, 6, 11, 17, 22, and 27); with hTBP, hTFIIA, and GST-Flag-hTAFII170 residues 2 to 137 (lanes 2 to 5, 7 to 10, and 12 to 15); or residues 380 to 460 (lanes 18 to 21, 23 to 26, and 28 to 31). (C) Gel mobility shifts were performed as described for panel A with no protein (lane 1); hTBP and hTFIIA (lane 2); hTBP, hTFIIA, and GST-Flag-hTAFII170 residues 2 to 460 (lanes 3 to 8); hTBPAS and hTFIIA (lane 9); or hTBPAS, hTFIIA, and GST-Flag-hTAFII170 residues 2 to 460 (lanes 10 to 15). For panels A and B, an equivalent amount of each GST-Flag-hTAFII170 protein was used (25.0, 12.5, 6.25, and 3.12 pmol for each titration series). For panel C, 17.0, 13.0, 9.0, 7.0, 5.0, and 3.0 pmol of GST-Flag-hTAFII170 2-460 protein was used in the titration series. No preinc, GST-Flag-hTAFII170 was included simultaneously in reactions with TBP and hTFIIA (lanes 2 to 5 and 18 to 21); Preinc, GST-Flag-hTAFII170 was preincubated with TBP on ice for 30 min prior to the addition of probe and hTFIIA (lanes 7 to 10 and 23 to 26); Preass, TBP and TFIIA were allowed to form a complex on the AdMLP before the addition of GST-Flag-hTAFII170 (lanes 12 to 15 and 28 to 31). TA indicates the hTBP-hTFIIA-AdMLP complex. TASA indicates the hTBPAS-hTFIIA-AdMLP complex. The asterisks indicate a nonspecific interaction due to contaminating proteins present in the hTFIIA fractions. FP indicates free probe.
GST fusion protein interactions.
To analyze interactions between hTAFII170 and hTBP or TBP family members, bacterial lysates containing equivalent levels of Flag-hTAFII170 derivatives were incubated with bacterial lysate containing equivalent levels of GST-TBPs or GST-TFIIS and 100 μl of prewashed glutathione beads in 500 μl of binding buffer (50 mM Tris-HCl, pH 8.0, 1 mM EDTA, 20% glycerol, 300 mM KCl [unless otherwise indicated], 0.01% Triton X-100, and protease inhibitors). Reactions were performed at 4°C for 3 h with rotation. Complexes were washed in binding buffer eluted in SDS sample buffer, and Flag-tagged hTAFII170 proteins were analyzed by Western blotting with αFlag M2 (Kodak). For analysis of hTAFII170 and His-hTBP interactions, bacterial lysates containing equivalent levels of GST-Flag-hTAFII170 derivatives or GST-TFIIS were incubated with His-hTBP (460 ng) as described above. His-hTBP was analyzed by Western blotting with 20C7 (α-TBP). To examine interactions between GST-Flag-hTAFII170, His-hTBP, and His-dTAFII230 derivatives, bacterial lysates containing equivalent levels of GST-Flag-hTAFII170 derivatives were incubated with His-hTBP (72 ng) and 180, 60, 20, and 6.6 pmol of batch-purified His-dTAFII230 TAND I or II lysate or 60, 20, 6.6, and 2.2 pmol of batch-purified His-dTAFII230 TAND I and II lysate as described above. His-hTBP was analyzed by Western blotting with 20C7 (α-TBP).
Coimmunoprecipitation analysis.
COS-7 cells at approximately 70% confluency were washed in phosphate-buffered saline; resuspended with 5 μg of pSG-ehTBP, pSG-ehTBPm3e, or pSG-ehTBPK243E and 15 μg of pMT2-hTAFII170; and electroporated with a 1.2-kV pulse at 25 μF (gene pulser; Bio-Rad). After transfection (48 h) cells were lysed in ELB buffer (50 mM HEPES-KOH, pH 7.9, 150 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.1% NP-40, 1 mM β-mercaptoethanol, and protease inhibitors). For coimmunoprecipitation, COS-7 cell lysates were incubated in ELB buffer with 10 μg of α-TBP (1F8) or 10 μg of α-c-Myc (9E10) with rotation at 4°C. After 4 h, prewashed protein G beads were added to recover immunoprecipitates and were washed in ELB buffer and hTAFII170 proteins were analyzed by Western blotting with 5115 (α-TAFII170) (50).
Protein-DNA interaction assays.
Gel mobility shifts were performed with 5,000 cpm (approximately 0.1 ng) of 32P-labeled adenovirus major late promoter (AdMLP) probe (−53 to +33), 0.2 pmol of His-hTBP or GST-cleaved TBPs, 160 ng of partially purified hTFIIA, and purified GST-Flag-hTAFII170 derivatives for 30 min at 30°C in a buffer containing 18.5 mM HEPES-KOH, pH 7.9, 18.5% glycerol, 5 mM MgCl2, 0.5 mM EDTA, 50 μg of poly(dG-dC)/ml, 0.2 mg of bovine serum albumin/ml, and 60 to 80 mM KCl. Reactions were resolved on a 4% Tris-glycine gel. DNase I footprinting was performed with 12,500 cpm (approximately 0.5 ng) of 32P-labeled AdMLP probe (−17 to +33), purified GST-Flag-hTAFII170 derivatives, and 6.0 pmol of His-hTBP under standard gel shift conditions. DNase I (1.5 U) was added and the reactions were incubated for 1 min. DNase I was inactivated by addition of an equal volume of stop mix (2% SDS, 200 mM EDTA). DNA was recovered by ethyl alcohol precipitation and analyzed on a 6% urea–Tris-borate-EDTA gel.
In vitro transcription.
Transcription reactions using 25 ng of linearized template pAdML(C2AT)Δ400 were performed as previously described (49). Reactions contained bacterially derived His-hTBP (0.24 pmol), TFIIB (60 ng), TFIIE (150 ng), purified GST-Flag-hTAFII170 derivatives and baculovirus-expressed TFIIF (40 ng), purified HeLa cell-derived TFIIH (0.5 μl), and calf thymus RNA pol II (0.4 μl). Purification of transcription factors was performed as previously described (26). RNA was recovered as previously described (49). Recovery control RNA was synthesized as described above from linearized pAdML(C2AT)Δ200.
RESULTS
The amino terminus of hTAFII170 interacts with hTBP.
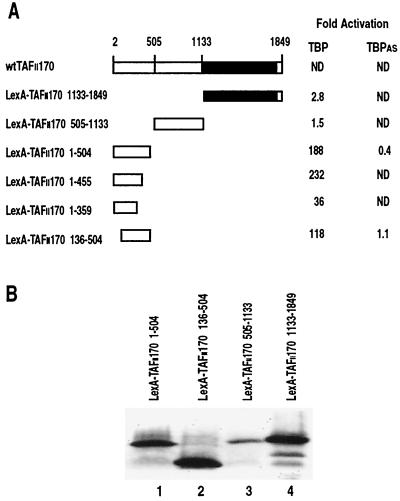
Determining interaction surfaces within the B-TFIID complex is hampered because B-TFIID cannot be efficiently reconstituted from its isolated components (16, 50). Therefore, we exploited the yeast two-hybrid system to reconstitute hTBP and hTAFII170 interactions in vivo. Full-length hTAFII170 is not expressed efficiently as a LexA fusion in yeast. Therefore, three LexA DNA binding domain fusion constructs spanning hTAFII170 (residues 1 to 504, 505 to 1133, and 1133 to 1849) were constructed and tested for interaction with hTBP fused to the B42 activation domain (Fig. 1A). The 1-504 construct shows a strong interaction with hTBP in contrast to the 505-1133 and 1133-1849 constructs (Fig. 1A). Region 1-455 is sufficient for hTBP interaction, but continued deletions result in hTAFII170 fusions that activate transcription in the absence of exogenous B42-hTBP (Fig. 1A and data not shown). Nevertheless, with the 1-359 construct, an interaction with hTBP was still seen above this background (Fig. 1A). Deletion of the first 135 residues results in a twofold decrease in hTBP interaction (Fig. 1A). Western blot analysis confirmed that all LexA-hTAFII170 deletion mutants that failed to interact with hTBP were expressed (Fig. 1B, lanes 1 to 4) and resided in the nucleus as indicated by the JK101 repression test (data not shown). Thus, the yeast two-hybrid analysis of the hTAFII170-hTBP interaction indicates that amino-terminal residues 1 to 504 of hTAFII170 are important for the interaction with hTBP. This agrees with deletion studies of Mot1 which showed that the first 1,200 amino-terminal residues are involved in contacting DNA-bound yTBP (8) while the first 800 amino-terminal residues can contact free yTBP (1).
FIG. 1.
Interaction of hTAFII170 with hTBP and hTBPAS in the yeast two-hybrid system. (A) Schematic representation of the various LexA-hTAFII170 deletion mutants and relative β-galactosidase activities. Yeast strain EGY48 was transformed with the indicated LexA-hTAFII170 expression plasmids and B42-hTBP or B42-hTBPAS expression plasmids together with a lacZ reporter gene containing LexA operators. β-Galactosidase activities were expressed as fold activation relative to the β-galactosidase activity of yeast cells expressing the LexA fusion and the unfused B42 activation domain only. Quantifications were performed in triplicate. ND, not determined. (B) Expression of LexA-hTAFII170 derivatives in yeast. Yeast samples containing equal amounts of cells were prepared from cultures harboring the LexA-hTAFII170 expression plasmids (lanes 1 to 4) and were analyzed using LexA antibodies.
Three regions in the amino terminus of TAFII170 can independently bind hTBP.
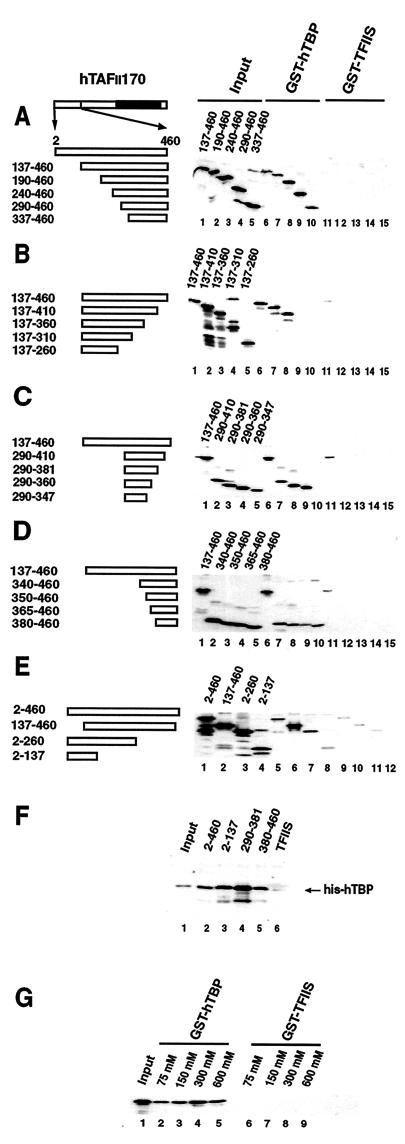
To avoid self-activation problems in the yeast two-hybrid system, we developed a direct protein-protein binding assay to further delineate the hTBP interaction domain of hTAFII170. A series of amino- and carboxy-terminal deletion mutants of Flag-tagged hTAFII170 residues 2 to 460 were analyzed for their ability to interact with GST-hTBP (Fig. 2A to E). As specificity controls, binding to a GST fusion of the active part of the elongation factor hTFIIS (Fig. 2A to E) or to GST alone (data not shown) was analyzed. Consistent with the yeast two-hybrid data, region 137-460 is specifically retained by GST-hTBP (Fig. 2A to C, lane 6; D, lane 7; and E, lane 6). Analysis with amino-terminal truncations revealed that residues up to 336 could be removed without affecting the interaction with hTBP (Fig. 2A, lanes 6 to 10). Similarly, carboxy-terminal truncations demonstrated that removal of residues to 361 had no effect (Fig. 2B, lanes 6 to 8). However, deletions extending to residues 311 abolished the interaction with hTBP (Fig. 2B, lanes 9 and 10). Thus, these analyses define a region of 23 residues between 337 and 360 important for the interaction with hTBP. Consistently, a more extensive carboxy-terminal deletion removing residues to 347 abolished hTBP-binding activity (Fig. 2C, lane 10).
FIG. 2.
Mapping domains in the amino terminus of hTAFII170 required for interaction with hTBP. (A to E) The schematics represent the hTAFII170 amino- and carboxy-terminal derivatives. Bacterial lysates containing equivalent amounts of the Flag-tagged-hTAFII170 derivatives were analyzed for binding to GST-hTBP (A to D, lanes 6 to 10; E, lanes 5 to 8) or, as a control, to GST-TFIIS (A to D, lanes 11 to 15; E, lanes 9 to 12) as described in Materials and Methods. The input lanes represent 10% of the hTAFII170 proteins used (A to D, lanes 1 to 5; E, lanes 1 to 4). (F) Bacterial lysates containing the GST-Flag-hTAFII170 regions (lanes 2 to 5) or, as a control, GST-TFIIS (lane 6) were tested for binding to His-hTBP (460 ng) as described in Materials and Methods. The input lane represents 10% of the hTBP used in the binding reactions (lane 1). (G) Salt sensitivity of the interaction between the amino terminus of hTAFII170 and hTBP. Bacterial lysates containing Flag-tagged-hTAFII170 2-460 were analyzed for binding to GST-hTBP (lanes 2 to 5) or, as a control, GST-TFIIS (lanes 6 to 9) in the presence of the indicated concentrations of KCl. The input lane represents 10% of the hTAFII170 protein used.
Amino-terminal truncations extending to residue 379 revealed binding of hTBP above background (Fig. 2D, lane 10). Thus, hTAFII170 appears to have a second region spanning residues 380 to 460 that is capable of independently interacting with hTBP. In addition, the yeast two-hybrid analysis suggested that the extreme amino terminus of hTAFII170 may also contain a hTBP-binding domain (Fig. 1A, compare LexA–hTAFII170 1-504 and LexA–hTAFII170 136-504). We tested this directly, and this revealed that the first 137 residues are able to interact with hTBP weakly but consistently (Fig. 2E, lane 8).
The in vitro GST-hTBP binding indicates that the amino-terminal third of hTAFII170 harbors three regions capable of binding hTBP. To confirm that these regions can independently interact, GST-Flag-hTAFII170 residues 2 to 460, 2 to 137, 290 to 381, and 380 to 460 were tested for their ability to retain hTBP. Each of these regions can specifically bind hTBP (Fig. 2F, lanes 2 to 6). Notably, region 290-381 binds hTBP most efficiently with approximately 40% of hTBP retained (Fig. 2F, compare lanes 1 and 4). These combined results indicate that three regions within the amino-terminal part of hTAFII170 can interact with hTBP independently.
The protein-protein binding assays used above were performed with a 300 mM concentration of potassium chloride. The B-TFIID complex is stable under these conditions, and the initial identification of B-TFIID relied on gel filtration chromatography performed with 300 mM potassium chloride (49). In fact, immunopurified hTBP/hTAFII170 complexes are stable with up to 1 M potassium chloride (16). However, a recent study by Adamkewicz and colleagues indicated that Mot1 binding to yTBP is compromised by increasing the ionic strength of the reaction (1). Therefore, we tested the effect of ionic strength on the efficiency of the hTAFII170 (2-460)/hTBP interaction. This interaction was not affected by increasing the salt concentration to 600 mM (Fig. 2G, lanes 2 to 5). In fact, with 75 mM potassium chloride, the interaction seemed somewhat reduced (Fig. 2G, compare lanes 2 and 4). Analysis of the individual regions 2-137, 290-381, and 380-460 showed the same pattern (data not shown). This indicates that electrostatic interactions are not the primary determinants for hTAFII170/hTBP interaction.
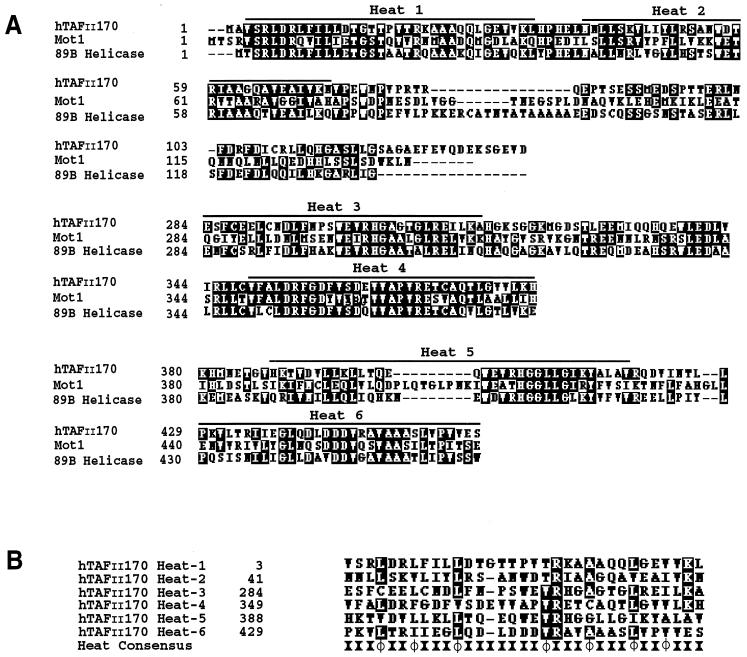
Comparison of the hTAFII170 regions extending from residues 2 to 137, 290 to 381, and 380 to 460 with those of TAFII170 from Saccharomyces cerevisiae and Drosophila melanogaster reveals significant similarity (Fig. 3A). Outside these domains there is little similarity. Furthermore, these regions are well conserved in the apparent TAFII170 orthologs in Arabidopsis thaliana, Schizosaccharomyces pombe, and Caenorhabditis elegans (data not shown). An iterative search-and-alignment method for repeated protein motifs indicated that these regions of hTAFII170 and Mot1 share clusters of tandemly arrayed Huntington elongation-A subunit-TOR (HEAT) repeats (40). HEAT repeats have been identified in a variety of proteins of distinct cellular function and can be defined as degenerate sequences rich in hydrophobic residues that are arranged into a pair of amphipathic α-helices (22, 40). Strikingly, the predicted HEAT repeats of hTAFII170 overlap with its TBP-binding regions, with each containing a pair of HEAT repeats (Fig. 3A and B). Outside these regions, hTAFII170 orthologs display a lower degree of similarity (16, 50). Collectively, this suggests that the TBP-binding regions of hTAFII170 have been conserved during evolution and that the HEAT repeat motifs mediate TBP interaction.
FIG. 3.
Sequence analysis of the hTBP binding regions of hTAFII170. (A) Alignment of hTAFII170 amino-terminal residues 2 to 137, 284 to 381, and 380 to 460 with yeast and Drosophila TAFII170 sequences. White letters on a black background indicate amino acid identity. The black bars indicate positions of the putative HEAT repeats (40). Alignments were performed using the CLUSTAL W algorithm (48). The sequence for Mot1 and Drosophila 89B helicase was obtained using hTAFII170 amino-terminal residues 1 to 460 as a query sequence in searches of the yeast and Drosophila databases with the BLAST Network service. (B) Alignment of the putative hTAFII170 HEAT repeat motifs within regions 2 to 137, 284 to 381, and 380 to 460 with the HEAT/ARM repeat consensus (4, 40). The HEAT/ARM repeat consensus is derived from sequence alignments of repeat-containing eukaryotic proteins identified in database searches and consists of highly conserved hydrophobic residues (φ) at positions 10, 13, 17, 24, 28, 32, and 35 (4, 40). Alignments were performed using the CLUSTAL W algorithm (48).
The hTAFII170 amino-terminal regions show species-specific interactions with TBP family members.
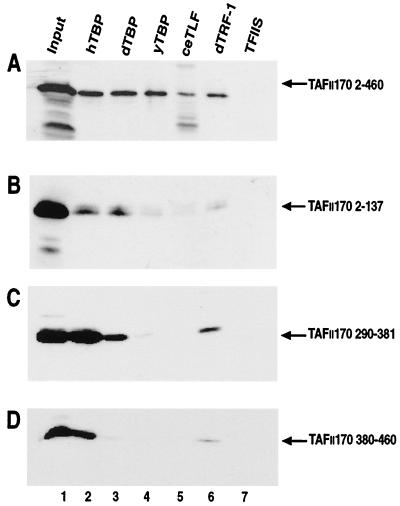
The core region of hTBP is highly conserved throughout evolution (25). In addition, recent studies have revealed the existence of the close TBP homologs TBP-like factor (TLF) and TBP-related factor (TRF) and have demonstrated their importance for the regulation of transcription (18). This prompted us to examine if the hTAFII170 amino terminus could interact with various TBP family members. GST fusions of dTBP, yTBP, and C. elegans TLF (ceTLF) and dTRF-1 were tested for their ability to retain the hTAFII170 amino terminus. The hTAFII170 amino terminus (residues 2 to 460) was specifically retained by yTBP and dTBP with comparable efficiency to that of hTBP (Fig. 4A; lanes 2 to 4). In contrast, poor binding to ceTLF was observed (Fig. 4A, lane 5). The hTAFII170 amino terminus also interacted with dTRF-1 but with a slightly lower efficiency than did hTBP (Fig. 4A, lane 6).
FIG. 4.
The hTAFII170 amino-terminal regions show species-specific interactions with TBP family members. Bacterial lysates containing GST-TBP family members (lanes 2 to 6) or, as a control, GST-TFIIS (lane 7) were assayed for binding to the amino-terminal Flag-hTAFII170 derivatives (A to D) as described in Materials and Methods. The input lanes represent 10% of the hTAFII170 proteins used (lane 1).
We tested whether the hTAFII170 regions 2-137, 290-381, and 380-460 were required for binding between the hTAFII170 amino terminus and TBP family members. As expected the regions 2-137, 290-381, and 380-460 could be specifically retained by hTBP (Fig. 4B to D, lanes 2). Region 2-137 and 290-381 but not 380-460 bound to dTBP (Fig. 4B to D, lanes 3). Surprisingly, each bound poorly, if at all, to yTBP (Fig. 4B to D, lanes 4). ceTLF bound poorly to all regions (Fig. 4B to D, lanes 5). In contrast, region 290-381 bound dTRF-1, but regions 380-460 and 2-137 exhibited poor binding (Fig. 4B to D, lanes 6). Western blot analysis and Coomassie staining confirmed that all GST fusion proteins were present in the reactions at comparable levels (data not shown). These results show that the amino-terminal regions of hTAFII170 exhibit species-specific interactions with various TBP family members.
The amino terminus of hTAFII170 interacts with concave DNA binding surface of hTBP.
Biochemical and genetic evidence suggests that Mot1 interacts with specific residues on H2 at the convex surface of yTBP overlapping the TFIIA-binding site (1, 5, 6, 15). To investigate the hTBP determinants for binding hTAFII170, we planned to test in the yeast two-hybrid assay hTBP convex surface mutants previously examined for loss of specific biochemical activities (14). These mutants were generated in the background of TBPAS. This TBP molecule contains a triple mutation in its concave surface (I292F, V301T, L303V) resulting in a relaxed specificity for TATA binding (45). As a control in the two-hybrid system we tested the interaction between hTBPAS and the amino terminus of hTAFII170 (Fig. 1A). To our surprise, hTBPAS failed to interact with hTAFII170 regions 1-504 and 136-504 (Fig. 1A). Western blot analysis indicated that wild-type hTBP and hTBPAS were expressed to similar levels (data not shown). This suggests that residues in the concave DNA binding surface of hTBP are important for interaction with the TAFII170 amino terminus, so we decided to test this possibility further.
TAND I (residues 2 to 81) of dTAFII230 blocks TBP-TATA box binding by direct occupancy of the concave DNA binding face of TBP, whereas TAND II competes with TFIIA for binding to H2 on the convex surface of TBP (31, 33, 34, 41). To elaborate the interaction between the amino terminus of hTAFII170 and the concave surface of hTBP, we tested whether TAND I of dTAFII230 and the hTAFII170 regions compete in an hTBP-binding assay. Histidine-tagged TAND I and TAND I and II together, but not TAND II alone, inhibited the interaction between hTBP and GST-Flag-hTAFII170 region 290-381 in a dose-dependent manner (Fig. 5A, lanes 2 to 13). In conclusion, results of the yeast two-hybrid and GST pulldown assays indicate that the amino terminus of hTAFII170 contains a region which can bind to the concave DNA binding surface of hTBP and influences its DNA binding properties.
In light of the observed interactions of Mot1 and hTAFII170 with the convex surface of TBP, this was unexpected. Therefore, we addressed this issue by analyzing in our in vitro GST pulldown assay the binding of hTBP convex mutants to three identified TBP interaction regions of hTAFII170. In this assay we failed to observe any effect of convex mutations (data not shown). Possibly, other regions of hTAFII170 are required for convex interactions. Alternatively, convex contacts escaped detection because a nonlinear domain is involved. To test this we used a coimmunoprecipitation assay, employing full-length hTAFII170. We constructed plasmids expressing either the K243E mutant or the hTBPAS mutant of hTBP. Mutation K243E lies in H2 on the convex surface of hTBP and corresponds to yTBP (K145E), which was shown to be defective for interaction with Mot1 (15). Immunoprecipitates from cotransfected cell lysates were analyzed for the presence of hTAFII170 and hTBP (Fig. 5B). Expression of wild-type hTBP resulted in coprecipitation of hTAFII170 (Fig. 5B, lane 4). In contrast, the convex K243E mutant inefficiently coimmunoprecipitated hTAFII170 (Fig. 5B, lane 5). As expected from our previous analyses, the concave hTBPAS was severely compromised in its interaction with hTAFII170 (Fig. 5B, lane 6). The coimmunoprecipitation results suggest that, in the context of full-length hTAFII170, both the convex and concave surfaces of hTBP are used.
The amino-terminal regions of hTAFII 170 inhibit hTBP-TATA box interaction.
To address whether the interaction between the amino-terminal part of hTAFII170 and the concave surface of hTBP influences the hTBP-TATA interaction, gel mobility shift analyses with hTBP, TFIIA, and a DNA fragment of the AdMLP TATA box were performed in the presence of the hTAFII170 amino-terminal regions. A complex corresponding to hTBP-TFIIA-TATA, TA, could be observed in the absence of hTAFII170 (Fig. 6A and B, lanes 1, 6, 11, 17, 22, and 27). Addition of purified GST-Flag-hTAFII170 region 2-460 or 290-381 simultaneously with the probe, hTBP, and TFIIA inhibited the TA complex in a dose-dependent manner (Fig. 6A, lanes 2 to 5 and 18 to 21). Consistent with its strong hTBP-binding activity, region 290-381 efficiently inhibited the TA complex (Fig. 6A, lanes 18 to 21). However, the TA complex was less efficiently inhibited by region 380-460 (Fig. 6B, lanes 18 to 21) and was unaffected by region 2-137 (Fig. 6B, lanes 2 to 5), both of which bind hTBP weakly. The inhibition was specific since the GST-Flag-hTAFII170 fusion proteins had no effect on protein-DNA complexes formed by the E-box protein, Max (data not shown). Inhibitory effects were also observed if the hTAFII170 amino-terminal regions were preincubated with hTBP prior to the addition of TFIIA and probe (Fig. 6A and B, lanes 7 to 10 and 23 to 26). Notably, preassembly of the TA complex rendered it largely refractory to the inhibitory effect of hTAFII170 regions 2-460, 290-381, and 380-460 (Fig. 6A and B, lanes 12 to 15 and 28 to 31). This is expected because TBP-TATA complexes are relatively stable and, thus, the concave DNA binding surface is not available after TA complex formation. Similarly, hTBP-TFIIA-TFIIB-TATA complex formation was also inhibited by the hTAFII170 regions 2-460 and 290-381 (data not shown). Together these results indicate that the amino-terminal regions 2-460, 290-381, and, to a lesser extent, 380-460 are capable of inhibiting the TBP-TATA interaction.
Our interaction analysis in yeast and mammalian cells indicated that mutations at the concave surface of hTBPAS abolish hTAFII170 interaction (Fig. 1A). This predicts that DNA complexes formed with this TBP mutant should be less sensitive to the hTAFII170 amino terminus. To test this we expressed and purified recombinant hTBPAS protein. Gel shift analysis indicated that hTBPAS forms a TA complex with an efficiency equal to that of wild-type hTBP (Fig. 6C, lanes 2 and 9). As expected, titration of hTAFII170 region 2-460 into the DNA binding reactions with wild-type hTBP inhibited the TA complex (Fig. 6C, lanes 3 to 8). In contrast, titration of hTAFII170 region 2-460 into the DNA binding reactions with hTBPAS failed to significantly inhibit the TA complex (Fig. 6C, lanes 10 to 15). This finding is consistent with the conclusion that the hTAFII170 amino terminus contacts the concave surface of TBP.
Because Mot1 has been proposed to use a binding site on the convex surface of yTBP overlapping with the TFIIA-binding site (1, 5, 6, 15) the possibility remained that the hTAFII170 derivatives inhibited the TA complex by binding competitively with TFIIA to hTBP. Under the gel mobility shift conditions used, the hTBP-TATA complex was not stable enough to be detected (data not shown). To clarify TFIIA involvement, the effects of the hTAFII170 amino-terminal regions on hTBP-TATA box complex formation in the absence of TFIIA were examined by DNase I footprinting experiments. The analysis was first performed with hTBP and a DNA probe that included the AdMLP TATA box in the presence of purified GST-Flag-hTAFII170 region 2-460 (Fig. 7A). In the presence of saturating amounts of hTBP, DNA was fully protected over the TATA box, indicating efficient binding of hTBP (Fig. 7A, lane 3). Addition of hTAFII170 region 2-460 caused the hTBP footprint to disappear in a dose-dependent manner, indicating that TBP was displaced from the TATA box (Fig. 7A, lanes 4 to 7). Extended analysis with region 290-381 also revealed displacement of hTBP (Fig. 7B, lanes 4 to 7). By contrast, hTAFII170 regions 2-137 and 380-460 failed to significantly inhibit hTBP binding to the TATA box (data not shown). Together with the gel mobility shift data (Fig. 6), these results show that subregion 290-381 of the amino-terminal region 2-460 is responsible for inhibiting the hTBP-TATA interaction by blocking the concave DNA binding surface of hTBP.
FIG. 7.
The amino terminus of hTAFII170 inhibits the TATA box-binding activity of hTBP. (A) DNase I footprinting was performed with a AdMLP probe (−17 to +33) and 6.0 pmol of His-hTBP in the presence of GST-Flag-hTAFII170 residues 2 to 460 (lanes 4 to 7; 60, 30, 15, and 7.5 pmol, respectively). Lane 1, chemical degradation of the probe on A + G's; lane 2, DNase I digestion of the probe; lane 3, His-hTBP binding to the probe in the absence of hTAFII170 2-460. Protected regions over the TATA box are indicated by brackets. (B) DNase I footprinting was performed as described for panel A in the presence of GST-Flag-hTAFII170 residues 290 to 381 (lanes 4 to 7; 44, 22, 11, and 5.5 pmol, respectively). Lane 1, chemical degradation of the probe on A + G's; lane 2, DNase I digestion of the probe; lane 3, His-hTBP binding to the probe in the absence of hTAFII170 290-381. Protected regions over the TATA box are indicated by brackets.
The amino-terminal regions of hTAFII170 inhibit RNA pol II transcription in vitro.
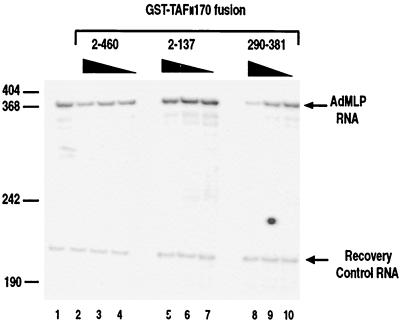
The full-length yMot1 (D1408N) mutant and yMot1 amino-terminal fragments whose ATPase activity has been abolished act as dominant negatives by forming stable dead-end complexes with yTBP (1, 8). The ability of the isolated 290-381 region of hTAFII170 to inhibit hTBP-TATA interaction by blocking the concave surface of hTBP suggests that these regions could also act in a dominant-negative fashion to inhibit in vitro transcription by binding and sequestering hTBP. To test this idea, hTBP binding regions 2-460 and 290-381 were added to a reconstituted pol II transcription system dependent upon hTBP. In vitro transcription reactions were reconstituted with bacterially derived hTBP, TFIIB, and TFIIE; baculovirus-expressed TFIIF; purified HeLa cell-derived TFIIH; and immunoaffinity-purified calf thymus pol II (see Materials and Methods). Transcription was analyzed from a linearized AdMLP core promoter (−53 to +10) that drives transcription of a 380-bp guanosine (G)-less cassette. Addition of equivalent amounts of purified GST-Flag-hTAFII170 region 2-460 or 290-381 to the reactions decreased transcription from the AdMLP (Fig. 8, lanes 2 to 4 and 8 to 10). Quantitation of transcript levels corrected by using a labeled internal standard RNA indicated that the regions 2-460 and 290-381 repressed pol II transcription by approximately 1.6 and 3.0-fold, respectively, at the highest amount used (Fig. 8, lanes 2 and 8). The stronger inhibition seen with the 290-381 region correlates well with its ability to bind hTBP more strongly, inhibit the dTAFII230/hTBP interaction, and inhibit hTBP-TATA complex formation, compared to region 2-460. Notably, the effect of these regions in transcription is less pronounced than that observed in the gel mobility shift and DNase I footprint analyses (Fig. 6 and 7). Possibly basal transcription factors present in the transcription reactions can partially relieve the hTAFII170-mediated inhibition (see Discussion). As expected, the weak TBP-binding region 2-137 had little or no effect (Fig. 8, lanes 5 to 7). These results indicate that pol II transcription can be directly repressed by region 290-381 of hTAFII170. This is consistent with the view that this region is capable of inhibiting the hTBP-TATA interaction by directly blocking the DNA binding surface of hTBP.
FIG. 8.
The amino terminus of hTAFII170 inhibits pol II transcription. In vitro transcription reactions were performed as described in Materials and Methods in the absence (lane 1) or presence of GST-Flag-hTAFII170 residues 2 to 460 (lanes 2 to 4), 2 to 137 (lanes 5 to 7), or 290 to 381 (lanes 8 to 10). An equivalent amount of each GST-Flag-hTAFII170 protein was used (46, 28, and 14 pmol for each titration series). Positions of comigrated DNA size marker fragments are indicated on the left.
DISCUSSION
The basal transcription factor B-TFIID supports pol II transcription as efficiently as TBP or TFIID in reconstituted basal transcription assays (49). However, in contrast to TBP or TFIID, B-TFIID displays dynamic binding to the TATA box. In an effort to understand the apparent high on and off rates of B-TFIID for DNA binding, we analyzed the interaction regions of its hTBP and hTAFII170 constituents. Our analysis indicates that the concave surface of TBP responsible for DNA binding is an important determinant for hTAFII170 action.
Our biochemical analyses mapped hTBP-binding domains in hTAFII170 to three regions within its amino-terminal third: residues 2 to 137, 290 to 381, and 380 to 460. The stable interaction of these regions with hTBP at increasing salt concentrations (300 to 600 mM) suggests that the interaction of the amino terminus of hTAFII170 with hTBP is sustained mainly by hydrophobic interactions. These regions are also important in the context of full-length hTAFII170, as we were unable to coimmunoprecipitate hTBP and full-length hTAFII170 from cell extracts when these regions were removed (data not shown). Yeast two-hybrid analysis indicated that the hTAFII170 ATPase domain alone is unable to interact detectably with hTBP. In addition we were also unable to coimmunoprecipitate hTBP and the isolated hTAFII170 ATPase domain from cellular extracts (data not shown). Consistent with this, the Mot1 ATPase domain cannot bind yTBP in the absence or presence of DNA (1, 2, 8). Together our results fit with the suggestion that the ATPase domain of hTAFII170-related proteins like Mot1 is targeted to its TBP substrate via the amino terminus (6, 8).
The importance of the hTAFII170 amino-terminal regions in contacting hTBP is consistent with previous findings that the first 800 residues of Mot1 are responsible for binding yTBP (1, 8). Furthermore, in the hTAFII170 ortholog, Drosophila 89B helicase (21), the first 825 residues can bind to dTBP (1). The corresponding TBP-binding regions in hTAFII170, Mot1, and 89B helicase also share common, highly related blocks (Fig. 3A). These observations indicate that the role of the amino terminus of hTAFII170 orthologs in TBP binding has been conserved during evolution. Furthermore, recent computational analysis indicated that these regions of hTAFII170 and Mot1 share clusters of tandemly arrayed HEAT repeats (4, 40). hTAFII170 contains six HEAT repeats within the first 460 residues. A pair of predicted HEAT repeats of hTAFII170 overlap with each of its TBP-binding regions (Fig. 3A and B). Together this suggests that the HEAT repeat protein motifs mediate TBP interaction in hTAFII170 orthologs.
Our results show that the amino-terminal regions of hTAFII170 exhibit species-specific interactions with TBP family members. While hTAFII170 region 2-460 interacted with yTBP remarkably well, when individually assayed the TBP-interacting regions showed poor binding. This may relate to specific differences between yTBP and hTBP. The binding of hTAFII170 region 290-381 to dTRF-1 suggests a functional link between 89B helicase (21) and dTRF-1 (47). Consistent with this, residues 1 to 825 of 89B helicase can bind dTRF-1 (1). This latter protein plays a role in directing transcription by RNA pol III (47). In this context it is interesting that hTAFII170 has been linked to RNA pol III transcription in vitro (46). However, other biochemical and genetic analyses have failed to observe an involvement of B-TFIID and Mot1 in pol III transcription (6, 37).
The most striking finding of our analyses is that an amino-terminal region of hTAFII170 can target the DNA binding concave surface of hTBP. First, we have not only shown that hTBPAS does not bind efficiently to the amino terminus of hTAFII170 or to full-length hTAFII170 in vivo but also that the region 290-381 of hTAFII170 can compete with dTAFII230 (TAND I) for binding the concave face of hTBP. Second, our biochemical evidence clearly shows that both regions 2-460 and 290-381 can inhibit hTBP-TATA box interaction. Notably, the effect of the hTAFII170 regions 2-460 and 290-381 in TATA-dependent in vitro transcription was less pronounced, in spite of the strong effect that they had on TBP-TATA complex formation in gel mobility shift and DNase I footprinting analyses. It is possible that stabilization of the TBP-TATA interaction by basal transcription factors such as TFIIA, TFIIB, and TFIIE (27, 51) partially counteracts the repressing activity of this hTAFII170 region. Alternatively, the effects of TFIIA, TFIIB, or TFIIE may be attributable to a block of hTAFII170 interaction with hTBP. Thirdly, the hTBPAS-TATA box interaction is less sensitive to inhibition by the hTAFII170 amino terminus. Together these results provide a rationale for the previous observation that B-TFIID cannot bind stably to the TATA box (49). Region 290-381 of hTAFII170 can compete with the TATA box for interaction with the concave DNA binding surface of hTBP. This is supported by the recent observations of Darst and coworkers (19) that preincubation of Mot1 with yTBP off DNA inhibits subsequent DNA binding by yTBP.
Previous mutational analyses indicate that amino acid residues of the yTBP H2 convex surface are important for the interaction between yTBP and Mot1 (1, 15). Furthermore, TFIIA has been shown to bind to H2 of the convex surface of yTBP competitively with Mot1 (5, 6, 15). Our observation that a single mutation at K243 of hTBP severely disrupts binding of full-length hTAFII170 in vivo suggests that hTAFII170 also interacts with the convex surface of hTBP. Together, with our observation that hTAFII170 binds the concave surface of hTBP, these results suggest that hTAFII170 can interact with multiple surfaces of hTBP. The behavior of hTAFII170 regions 2-137 and 380-460 was distinct from that of 2-460 and 290-381. Both bound to hTBP but had relatively minor, if any effects, on dTAFII230/TBP or TBP-TATA interactions (Fig. 7 and 8; data not shown). This distinct behavior may reflect an unstable interaction with the hTBP concave surface. Alternatively, these regions may contact the convex surface of hTBP. In that case one would expect higher-order complexes with these regions in gel shift analyses. Although this was not observed, TFIIA present to stabilize hTBP-TATA binding in these analyses may have competed with hTAFII170 regions 2-137 and 380-460. Therefore, in this context we cannot conclude whether these regions bind the hTBP convex surface.
It has been proposed that displacement of TBP from DNA by Mot1 involves an ATP-driven power stroke which relieves a conformationally strained Mot1 to pull the TBP-DNA complex apart (8). This was suggested to involve conformational changes in TBP (2). Our findings imply a different mechanism for the regulation of the TBP-TATA interaction by hTAFII170/Mot1 and how this relates to the TATA box-binding properties of B-TFIID. In its simplest form we propose a model for B-TFIID function in which B-TFIID can convert from an active to a blocked conformation and vice versa. In the blocked conformation, access of TBP to promoter DNA would be precluded by the presence of region 290-381 of hTAFII170 in the DNA binding cavity of TBP. This would explain the inability of B-TFIID to stably interact with TATA box DNA (49). In the active conformation, region 290-381 is absent from the concave surface and TBP is free to interact with promoter DNA. The ability of B-TFIID to support TATA-dependent transcription (49) could reflect this conformation. In this respect it is important to determine whether hTAFII170/Mot1 is still associated with TBP when the latter is bound to active promoters. Alternatively, hTAFII170/Mot1 may dissociate when TBP is assembled into an active preinitiation complex. An essential feature of our proposed model would be conversion between the active and blocked states. It is likely that the ATPase function of hTAFII170/Mot1 is involved in this conversion. As mentioned, dissociation of preformed TBP-TATA complexes by hTAFII170/Mot1 requires ATP hydrolysis (5, 6, 16). Future work using mutagenesis and single-molecule studies is required to more directly test these possibilities.
hTAFII170 shows the same pattern of TBP-TATA complex inhibition as dTAFII230 and yTAFII145/130 in gel mobility shift, DNase I footprint, and in vitro transcription analyses (31, 33). Furthermore, TBPAS residues affecting hTAFII170 binding contact dTAFII230 (34). These results support the concept that occupancy of the TBP concave surface may be a general mechanism for TAFII regulation of TBP-DNA binding. Most TBP is associated with other proteins in the cell (44, 49). Given the present results, it seems that several TBP/TAF complexes have a built-in module that regulates TBP access to DNA to prevent spurious TATA-DNA interactions. Negative regulation of TBP-binding activity may therefore be a general conserved property of TBP/TAF complexes. This appears relevant given that TBP has a relatively high affinity for nonpromoter DNA (23). Additionally, the weak consensus for the TATA element results in numerous nonpromoter binding sites for TBP within the genome. Consistent with this view, TAFI48 of SL1 can prevent binding of TBP to the TATA box element of pol I promoters (12). Furthermore, the association of TBP with the SAGA subunits Spt3 and Spt8 regulates TBP-DNA binding (13). Reversibility of this concave inhibition would be a key feature to allow TBP to bind DNA. It has been proposed that transcriptional activators like c-Jun, VP16, GAL4, and EBNA2 may regulate dTAFII230/hTAFII250 inhibition of TBP binding to DNA (30, 32, 35, 41). Might hTAFII170 inhibition of TBP be similarly modulated? While hTAFII170 does not respond to activators tested so far (49), it may respond to other activators or components of the general transcription apparatus releasing its inhibition of TBP. Our present results provide a framework for these studies.
ACKNOWLEDGMENTS
We thank A. Berk for the pSRaMSVtkneoTBPAS convex mutants; H. Stunnenberg for pSG-ehTBP and pSG-ehTBPm3e; C. Kane for pET22-TFIIS, Y. Nakatani for pGST-dTAFII230 (2-81), pGST-dTAFII230 (82-156), and pGST-dTAFII230 (2-156); M. Rabenstein for pGST-dTRF-2; R. Romier for pACYC; L. Tora for pGST-ceTLF; and N. Zak for pGST-dTBP. We are grateful to N. Zak for communication of unpublished results. We acknowledge the contributions of F. Holstege in providing TFIIH. We thank F. Kavelaars for technical assistance and members of our laboratory for discussions and critical reading of this manuscript.
This work was supported by a grant to H.T.M.T. from Human Sciences Frontier Program Organization (HSFPO) and The Netherlands Organization for Scientific Research-Medical Sciences (NWO-MW).
REFERENCES
- 1.Adamkewicz J I, Hansen K E, Prud'homme W A, Davis J L, Thorner J. High-affinity interaction of yeast transcriptional regulator, Mot1, with TATA box-binding protein (TBP) J Biol Chem. 2001;276:11883–11894. doi: 10.1074/jbc.M010665200. [DOI] [PubMed] [Google Scholar]
- 2.Adamkewicz J I, Mueller C G F, Hansen K E, Prud'homme W A, Thorner J. Purification and enzymatic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J Biol Chem. 2000;275:21158–21168. doi: 10.1074/jbc.M002639200. [DOI] [PubMed] [Google Scholar]
- 3.Albright S R, Tjian R. TAF's revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 4.Andrade M A, Petosa C, O'Donoghue S I, Muller C W, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 5.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 6.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 7.Auble D T, Steggerda S M. Testing for DNA tracking by Mot1, a SNF2/SWI2 protein family member. Mol Cell Biol. 1999;19:412–423. doi: 10.1128/mcb.19.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auble D T, Wang D, Post K W, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member Mot1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 10.Bagby S, Mal T K, Liu D, Raddatz E, Nakatani Y, Ikura M. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 2000;468:149–154. doi: 10.1016/s0014-5793(00)01213-8. [DOI] [PubMed] [Google Scholar]
- 11.Bai Y, Perez G M, Beecham J M, Weil P A. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckmann H, Chen J-L, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 13.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 15.Cang Y, Auble D T, Prelich G. A new regulatory domain on the TATA-binding protein. EMBO J. 1999;18:6662–6671. doi: 10.1093/emboj/18.23.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chicca J J, II, Auble D T, Pugh B F. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol Cell Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dantonel J-C, Wurtz J-M, Poch O, Moras D, Tora L. The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci. 1999;285:335–339. doi: 10.1016/s0968-0004(99)01436-x. [DOI] [PubMed] [Google Scholar]
- 19.Darst R P, Wang D, Auble D T. MOT1-catalyzed TBP-DNA disruption: uncoupling DNA conformational change and role of upstream DNA. EMBO J. 2001;20:2028–2040. doi: 10.1093/emboj/20.8.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman-Levi R, Miller C, Bogoch J, Zak N B. Expanding the mot1 subfamily: 89B helicase encodes a new Drosophila melanogaster SNF2-related protein which binds to multiple site on polytene chromosomes. Nucleic Acids Res. 1996;24:3121–3128. doi: 10.1093/nar/24.16.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves M R, Hanlon N, Turowski P, Hemmings B A, Barford D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell. 1999;96:99–110. doi: 10.1016/s0092-8674(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 23.Hahn S, Buratowski S, Sharp P A, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 26.Holstege F C P, van der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 27.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of the yeast factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi A, Miyake T, Ohyama Y, Kawaichi M, Kokubo T. Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J Biol Chem. 2000;276:395–405. doi: 10.1074/jbc.M008208200. [DOI] [PubMed] [Google Scholar]
- 31.Kokubo T, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc Natl Acad Sci USA. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotani T, Banno K-I, Ikura M, Hinnebusch A G, Nakatani Y, Kawaichi M, Kokubo T. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 2000;97:7178–7183. doi: 10.1073/pnas.120074297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotani T, Miyake T, Tsukihashi Y, Hinnebusch A G, Nakatani Y, Kawaichi M, Kokubo T. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J Biol Chem. 1998;273:32254–32264. doi: 10.1074/jbc.273.48.32254. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Solution structure of a TBP-TAFII230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 35.Lively T N, Ferguson H A, Galasinski S K, Seto A G, Goodrich J A. c-Jun binds the N terminus of human TAFII250 to derepress RNA polymerase II transcription in vitro. J Biol Chem. 2001;276:25582–25588. doi: 10.1074/jbc.M100278200. [DOI] [PubMed] [Google Scholar]
- 36.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyers R E, Sharp P A. TATA-binding protein and associated factors in polymerase II and polymerase III transcription. Mol Cell Biol. 1993;13:7953–7960. doi: 10.1128/mcb.13.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 39.Muldrow T A, Campbell A M, Weil P A, Auble D T. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol. 1999;19:2835–2845. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuwald A F, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishikawa J-I, Kokubo T, Horikoshi M, Roeder R G, Nakatani Y. Drosophila TAFII230 and the transcriptional activator VP16 bind competitively to the TATA box-binding domain of the TATA box-binding protein. Proc Natl Acad Sci USA. 1997;94:85–90. doi: 10.1073/pnas.94.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 43.Poon D, Campbell A M, Bai Y, Weil P A. Yeast TAF170 is encoded by MOT1 and exists in a TATA box-binding (TBP)-TBP associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 44.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 45.Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 46.Taggart A K P, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 47.Takada S, Lis J T, Zhou S, Tjian R. A TRF:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmers H T M, Sharp P A. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 1991;5:1946–1956. doi: 10.1101/gad.5.11.1946. [DOI] [PubMed] [Google Scholar]
- 50.van der Knaap J A, Borst J W, Gentz R, van der Vliet P C, Timmers H T M. Cloning of the cDNA for the TAFII170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc Natl Acad Sci USA. 1997;94:11827–11832. doi: 10.1073/pnas.94.22.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokomori K, Verrijzer C P, Tjian R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci USA. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]