Abstract
Purpose:
Osteoarthritis (OA) is a joint disease that is clinically diagnosed using components of history, physical exam, and radiographic evidence of joint space narrowing. Currently, there are no laboratory findings that are specific to a diagnosis of OA. The purpose of this systematic review is to evaluate the state of current studies of metabolomic biomarkers that can aid in the diagnosis and treatment of OA.
Methods:
Articles were gathered from PubMed and Web of Science using the search terms “osteoarthritis” and “biomarkers” and “metabolomics”. Last search of databases took place December 3rd, 2022. Duplicates were manually screened, along with any other results that were not original journal articles. Only original reports involving populations with diagnosed primary or secondary OA (human participants) or surgically induced OA (animal participants) and a healthy control group for comparison were considered for inclusion. Metabolites and metabolic pathways reported in included articles were then manually extracted and evaluated for importance based on reported a priori p-values and/or area under the receiver-operator curve (AUC).
Results:
Of the 161 results that were returned in the database searches, 43 unique articles met the inclusion criteria. Articles were categorized based on body fluid analyzed: 6 studies on urine samples, 13 studies on plasma samples,11 studies on synovial fluid (SF) samples, 11 studies on serum samples, 1 study on both synovial fluid and serum, and 1 study that involved both plasma and synovial fluid. To synthesize results, individual metabolites, as well as metabolic pathways that involve frequently reported metabolites, are presented for each study. Indications as to whether metabolite levels were increased or decreased are also included if this data was included in the original articles.
Conclusions:
These studies clearly show that there are a wide range of metabolic pathways perturbed in OA. For this period, there was no consensus on a single metabolite, or panel of metabolites, that would be clinically useful in early diagnosis of OA or distinguishing OA from a healthy control. However, many common metabolic pathways were identified in the studies, including TCA cycle, fatty acid metabolism, amino acid metabolism (notably BCAA metabolism and tryptophan metabolism via kynurenine pathway), nucleotide metabolism, urea cycle, cartilage matrix components, and phospholipid metabolism. Future research is needed to define effective clinical biomarkers of osteoarthritis from metabolomic and other data.
Keywords: osteoarthritis, biomarkers, metabolomics, metabolism
Introduction
Osteoarthritis (OA) is the most prevalent form of arthritis and has profound detrimental impacts on the patient population burdened by this disease. Approximately 54 million adults in the US have been diagnosed with some form of arthritis, with about 33 million of these being diagnosed with OA.1 Factors such as age, obesity, and injury, among many others, are involved in the development of OA; as the US population ages and rates of obesity are on the rise, the prevalence of OA is expected to increase as the total number of arthritis cases, which includes OA diagnoses, is predicted to rise to 78 million US adults by the year 2040.1 As OA negatively affects activity levels and quality of life for patients, it can be more difficult to control chronic conditions that OA patients may be concurrently facing, such as diabetes, hypertension, or mental health disorders. OA is also associated with an increased lifetime risk of developing heart disease by 50% as well as an increased all-cause mortality of 55%.1,2 As there are no treatments or therapies currently FDA approved and available that can reverse the progression of OA, it is important to make the diagnosis early so that changes can be made to slow the progression and limit the associated negative impacts on quality of life and overall health of patients. Metabolomic profiling is an emerging technology that provides systemic insight into disease processes by characterizing large numbers of biochemical reactants and products, termed metabolite features. As such, there is great potential for using metabolomics to support improved and early diagnosis of OA. Development of metabolomic biomarkers of OA has further potential to identify novel targets for both therapeutic intervention and monitoring disease progression.
OA has historically been considered a degenerative disease, and its development and progression originally were attributed to “wear and tear”.3 However, in recent decades our understanding of OA development has evolved to include chronic low-grade inflammation in addition to maladaptive signaling that activate a cascade of catabolic processes across multiple tissues in the joint.4 Ultimately in OA the homeostatic balance between maintenance and degradation is upset resulting in deterioration of bone and cartilage.4 However the metabolic links between disrupted homeostasis and the pathogenesis of OA are not completely understood, and are an emerging topic of active research utilizing metabolomic analysis.3–5 Metabolomics is a growing research field that characterizes large numbers of small-molecules using advanced technology. Metabolomic profiling provides insight into the myriad biochemical reactions that occur in cells and tissues, and pathologically-altered metabolites have the potential to serve as molecular biomarkers of disease processes. The objective of this review is to build upon prior knowledge6 and address the current state of metabolomic biomarkers for OA.
The scope of this review includes metabolomic findings by H1-NMR, various mass spectroscopy techniques, and immunosorbent assays related to primary and secondary OA. By performing a systematic review involving individual metabolites associated with OA and metabolic pathways that are perturbed in OA disease pathogenesis, this review will summarize both insight into OA provided by metabolomics and candidate biomarkers for future studies. The insight provided here may support and guide further research to enable earlier diagnosis of OA using metabolite biomarkers.
Methods
The aim of this study was to perform a systematic review of the current state of published metabolomic attempts to discover biomarkers of osteoarthritis.7 A search of PubMed and Web of Science was performed and included the search terms: “metabolomics” and “osteoarthritis” and “biomarkers”. A table depicting the search terms for each database is given below in Table 1. Included articles were published up until December 3, 2022, with no specific start date filter. To ensure a broad range of studies, there were no filters used for species or language, although all results were in the English language. Initial results were then manually reviewed by PVP to detect duplications. After duplicate articles were removed, the remaining articles were independently reviewed by PVP to remove any results that were not original refereed journal articles (e.g. conference proceedings) or did not meet the inclusion and exclusion criteria stated below. Reasons for excluding articles were noted and discussed between PVP and RKJ until a consensus was reached. The articles remaining to be included were then grouped based on body fluid utilized in the study: 13 studies of plasma, 6 studies of urine, 11 studies of synovial fluid (SF), 11 studies of serum, one study of both SF and plasma, and one study of both SF and serum.
Table 1.
Search Strategy including search terms used for identifying articles in each database.
| Database Searched | Search Terms Used | Results |
|---|---|---|
| PubMed | “osteoarthritis” AND “biomarkers” AND “metabolomics” | 81 |
| Web of Science (advanced search) | TS = (“osteoarthritis” AND “biomarkers” AND “metabolomics”) | 80 |
Inclusion and Exclusion Criteria
Only original journal articles were included in this review; reviews, abstracts, and brief reports were screened out. Articles were considered for inclusion only if the study population included a group with diagnosed primary or secondary OA (for studies of human participants) or OA induced through surgery or other accepted protocol (for non-human subjects) and a healthy control to allow for comparison. Publications were excluded if they (a) evaluated metabolites specific to therapeutic interventions but did not identify baseline metabolomic profiles or (b) did not identify potential biomarkers of OA with statistical significance or potential diagnostic importance. For inclusion, articles also needed to include data describing the metabolomic baseline of healthy controls, or the metabolomics involved in osteoarthritic joints. Additional exclusion criteria included (a) evaluation of animal OA with a distinct pathogenesis differing from that of humans (i.e., OA in equine athletes arising specifically due to carpal osteochondral fragmentation). Articles also needed to include both measures of statistical significance for metabolites and predictive diagnostic value, including p-values or area under the receiver-operator curve (AUC). For this review, for metabolite features or pathways to be deemed important they needed to reach either an a priori significance threshold of p < 0.05 or AUC > 0.8.
Data Abstraction
After abstracts were screened using the inclusion and exclusion criteria mentioned, papers considered for inclusion were read in their entirety. Key metabolite features and metabolic pathways were manually extracted based on p-values below the a priori threshold of 0.05 or AUCs greater than 0.80. Additional data that was extracted from the articles included the name of first author, year published, body fluid sampled, method of metabolite analysis, and number of subjects in the groups studied. Metabolites that were identified as important (either by statistical significance or AUC) were then categorized according to the particular body fluid that was studied, and metabolites were further grouped based on metabolic pathway involved and biochemical classification. Key metabolic pathways were also categorized according to the body fluid sampled. Data extraction was performed independently by PVP and then reviewed with RKJ.
Result Synthesis
Eligibility for inclusion in data synthesis was determined based on if important a priori or AUC values were included in the studies. Data from each study was manually extracted and transferred into Microsoft Excel. The data was then further synthesized by grouping key metabolites based on biochemical class metabolic pathway for each fluid type sampled. This allowed for evaluation of metabolic pathways that were perturbed and offers insight into pathogenesis of osteoarthritis as a disease. No methods were needed for data conversion or handling of missing summary statistics, as each study included a priori and/or AUC values for metabolites presented.
Tables were used to present the extracted metabolite and metabolic pathway. For each body fluid studied, a table was created; important metabolite and metabolic pathway data extracted was then populated in a row in the table corresponding to the fluid evaluated in the study. Grouping of results based on the fluid that was studied allowed for identification of replicated metabolites, as well as common metabolic pathways identified.
Bias Evaluation
Bias was analyzed at the study and fluid sample level. All disclaimers and statements of competing interest were evaluated. Patient recruitment/selection processes were evaluated; information, or lack thereof, on concurrent health conditions of patients were taken into consideration; targeted versus untargeted metabolomic analysis strategies were noted; finally, methods of acquiring fluid samples (e.g. clinical patient samples vs cadaver samples) were all taken into consideration. All results reported should be evaluated with the consideration that there are comorbidities that can confound systemic metabolomic attempts to identify biomarkers specific to OA in unknown ways, as the pathophysiologic link between OA and many health conditions have not been discovered or fully investigated.
Results
Using the search terms described above, 161 total articles were gathered from PubMed and Web of Science. After applying inclusion and exclusion criteria, 43 articles remained for this review. 62 excluded papers were duplicates between the search results of the two databases. 30 excluded results were not original articles and instead consisted of brief reports, reviews, abstracts, editorial material, and a textbook chapter. One article was unable to be accessed in full. The 68 remaining articles were then analyzed to determine inclusion: 25 of these were excluded that were either not relevant to OA metabolomics or possible biomarkers, intended to evaluate therapeutic effects of herbal medicine for treatment of OA, evaluated metabolomics to predict responses to total joint replacements or correlate OA severity without a healthy control, or analyzed metabolomics of OA in a species and disease model that was specific to the species and therefore not generalizable to OA in humans. The remaining 43 articles were included in the review as they offered insight into metabolomics of OA versus healthy controls (41 articles) or provided data on baseline healthy serum (1 article) and plasma (1 article) amino acid levels for reference (Figure 1).
Figure 1:
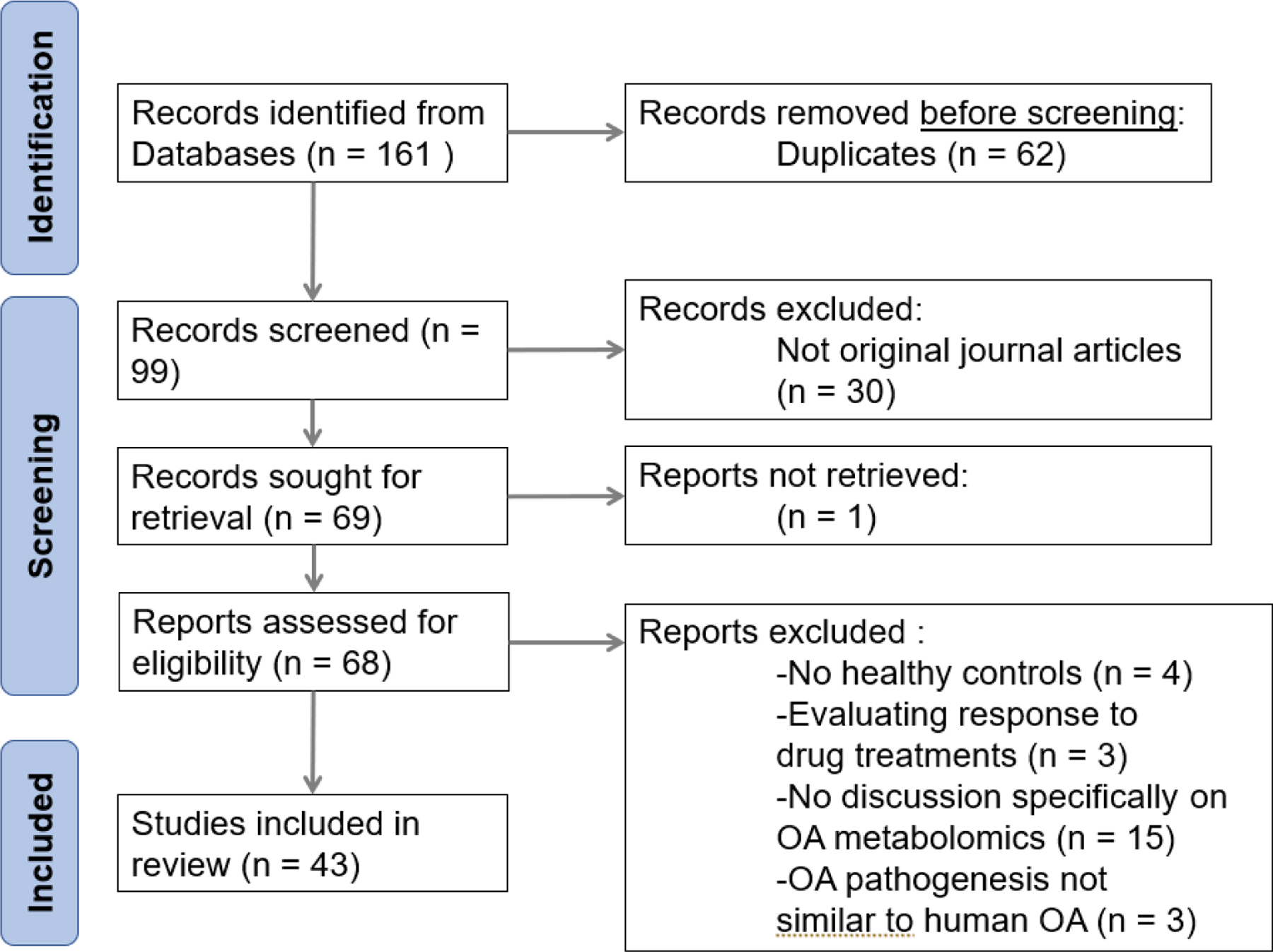
Flow diagram of literature search performed on December 3, 2022, displaying the study selection process.
Study Characteristics
Upon initial review of the articles, general characteristics of each study were extracted (Table 2). These data included the article author, specific body fluid that was used for metabolomic profiling, the model species, and the sample sizes of the various groups. This allowed for further categorization of metabolomic results into the fluids that were analyzed in the studies: plasma, urine, synovial fluid, and serum.
Table 2.
Study characteristics
| Article Author | Fluid Sampled | Model Species | Fluid Analysis Method | Sample size(s) |
|---|---|---|---|---|
| Rockel et al., 2018 8 | Plasma | Human | LC-MS/MS | OA=152, HC=194 |
| Steventon et al., 2012 9 | Plasma | Human | RP-HPLC with fluorescence detection | HC=100 |
| Smilde et al., 2005 10 | Urine | Guinea Pig | H1 NMR | OA=12, HC=6 |
| Carlson et al., 2018 11 | Synovial Fluid | Human | LC-MS | OA=5, HC=5 |
| Loeser et al., 2016 12 | Urine | Human | H1 NMR, ELISA, automated immunoanalyzer | OA1=22, OA2=22 |
| Carlson et al., 2019 13 | Synovial Fluid | Human | LC-MS | OA GI-II=55, OA GIII-IV=17, HC=7 |
| Thompson et al., 2011 14 | Serum | Human | FI-ESI-MS | OA=40 |
| Costello et al., 2020 15 | Plasma | Human | UPLC-MS, FI-ESI-MS | OA=704 |
| Hu et al., 2020 16 | Synovial Fluid | Rabbit | UPLC-MS | OA=12, HC=12 |
| Kang et al., 2015 17 | Synovial Fluid | Human | UPLC-QTOF-MS | OA=10 |
| Jiang et al., 2018 18 | Urine | Rat | GC-TOF-MS | OA=8, HC=8 |
| Maher et al., 2012 19 | Serum | Ovine | H1 NMR | Oam=24, HC=12 |
| Datta et al., 2017 20 | Plasma | Mouse | RP-LC-MS-MS | n=undefined |
| Adams et al., 2014 21 | Synovial Fluid | Human | LC-MS, GC-MS, ELISA | OA=20, HC=20 |
| Huang et al., 2020 22 | Plasma | Human | GC-QTOF-MS | OA=12, HC=20 |
| Senol et al., 2019 23 | Serum | Human | LC-QTOF-MS-MS | OKOA=14, NOKOA=14, HC=15 |
| Pousinis et al., 2020 24 | Plasma | Mouse | UHPLC-HR-MS | OA=8, HC=7 |
| Zhang et al., 2016 25 | Plasma | Human | TQ-UPLC-MS | DC-TKR=64, DC-NTKR=45, RC-TKR=72, RC-NTKR=76, LC-TKR=36, LC-NTKR=122 |
| Tootsi et al., 2018 26 | Serum | Human | UPLC-MS | OA=70, HC=82 |
| Mickiewicz et al., 2015 27 | Synovial Fluid | Human | H1 NMR, GC-MS | OA=55, HC=13 |
| Mickiewicz et al., 2015 28 | Synovial Fluid | Ovine | H1 NMR | OA=9, HC=9 |
| Abdelrazig et al., 2021 29 | Urine | Human | LC-HR-MS | OA=74, HC=68 |
| Liu et al., 2021 30 | Plasma | Human | UPLC-MS, FI-ESI-MS | n=610 |
| Yin et al., 2017 31 | Urine | Rat | GC-MS | OA=8, HC=8 |
| Zhang et al., 2016 32 | Plasma | Human | TQ-UPLC-MS | DC-OA=64, DC-HC=45, RC-OA=72, RC-HC=76 |
| Zhang et al., 2016 33 | Synovial Fluid, Plasma | Human | TQ-UPLC-MS | DC-OA=54, DC-HC=30, RC-OA=72, RC-HC=71 |
| Maerz et al., 2018 34 | Serum | Rat | H1 NMR, LC-MS-MS | OA=18, HC=18 |
| Hu et al., 2016 35 | Plasma | Human | TQ-UPLC-MS | DC-OA=64, DC-HC=45, RC-OA=72, RC-HC=76 |
| Chen et al., 2018 36 | Serum | Human | TQ-UPLC-MS | OA=32, HC=35 |
| Zhai et al., 2021 37 | Plasma | Human | TQ-UPLC-MS | Oaup=57, Oanup=206, Oabp=177, Oanbp=116 |
| Zhai et al., 2010 38 | Serum | Human | LC-MS-MS | DC-OA=123, DC-HC=299, RC-OA=76, RC-HC=100 |
| Zhang et al., 2015 39 | Serum | Human | UPLC-MS | OA=40, HC=20 |
| Kosinska et al., 2014 40 | Synovial Fluid | Human | FI-ESI-MS-MS, LC-MS-MS | eOA=17, lOA=13, HC=9 |
| Hügle et al., 2012 41 | Synovial Fluid | Human | H1 NMR | OA=15 |
| He et al., 2021 42 | Plasma | Human | GC-MS | OA=12, HC=12 |
| Li et al., 2009 43 | Urine | Human | GC-MS | OA=37, HC=37 |
| Zhao et al., 2021 44 | Serum | Rat | LC-MS | OA=6, HC=6 |
| Wallace et al., 2022 45 | Synovial Fluid, Serum | Mouse | LC-MS | OA=8, HC=4 |
| Hahn et al., 2021 46 | Synovial Fluid | Mouse | LC-MS | OA=9, HC=9 |
| Abshirini et al., 2021 47 | Plasma | Rats | LC-MS | OA=13, HC=11 |
| Xiao et al., 2022 48 | Serum | Rabbit | LC-MS-MS | OA=6, HC=3 |
| Pertusa et al., 2022 49 | Serum | Human | H1-NMR | OA=23, HC =53 |
| Nieminen et al., 2022 50 | Synovial Fluid | Human | UHPLC-QTOF-MS | OA=10, HC=5 |
General characteristics of each study include first author and date of publication, fluid sampled, model species, method of fluid analysis for metabolomics, and sample sizes. LC=Liquid chromatography, MS=Mass spectrometry, RP=Reversed phase, HPLC=High-performance liquid chromatography, H1-NMR=Proton Nuclear Magnetic Resonance, ELISA=Enzyme-linked immunosorbent assay, FI-ESI=Flow-injection electrospray ionization, QTOF=quadrupole time-of-flight, GC=Gas chromatography, TOF=Time-of-flight, TQ=Triple quadrupole, HR=High resolution, UPLC=Ultra performance liquid chromatography, OA=Osteoarthritis subjects, HC=Healthy control for comparison, OA1=Evidence of OA progression on radiographic findings 18 months from baseline, OA2=No evidence of OA progression on radiographic findings 18 months from baseline, OA GI-II=OA grades I-II using Outerbridge scale scoring, OA GIII-IV=OA grades III-IV using Outerbridge scale scoring, Oam=Models with OA induced by meniscal destabilization (MD, 12) or anterior cruciate ligament transection (ACLT, 12), OKOA=Knee OA and obesity (BMI≥30), NOKOA=Knee OA without obesity (BMI<30), TKR=Total knee replacement, NTKR=No total knee replacement performed, DC=Discovery cohort subjects, RC=Replication cohort subjects, LC=Longitudinal study cohort subjects, Oaup=OA patients with radiographic evidence of unilateral knee OA progression, Oanup=OA patients without radiographic evidence of unilateral knee OA progression, Oabp=OA patients with radiographic evidence of bilateral knee OA progression, Oanbp=OA patients without evidence of bilateral knee OA progression, eOA=OA with Outerbridge classification scale score ≤ 2, lOA=OA with Outerbridge classification scale score >2.
Risk of Bias in Studies
Bias was difficult to assess. In human studies, patient ethnicities were only reported in 3 of 29 articles.12,37,49 Potentially confounding concurrent health conditions were usually not determined or reported. Since obesity accompanies ~90% of knee OA cases, hypertension accompanies about 40% of knee OA cases, and diabetes accompanies about 16% of knee OA cases2 this may impact patient metabolomic profiles. For selection of human control groups, patients were usually selected on the basis that they lacked a diagnosis of OA by a family medicine provider and/or lacked radiographic evidence of OA. However subclinical or asymptomatic forms of OA may be present in these HC groups, and this could potentially confound metabolomic comparisons. For human cases where body fluids were taken from live patients, controls were also matched for age, sex, and BMI to limit demographic variation between groups.
In animal models where OA was surgically induced, controls underwent sham surgeries in attempts to ensure that differences in metabolite profiles were a reflection of OA and not surgical complications (e.g. incision-related inflammation). The final source of bias occurs at the study level and involves the use of either targeted or untargeted metabolomics: both have limitations for analyzing the comprehensive body of metabolites present in a body fluid due to the large numbers of potential metabolites.
Synthesis of Results
For analysis, studies were grouped by body fluid; metabolites reported with statistical significance or AUC>0.8 were extracted along with identified metabolic pathways, and are presented below.
Urine
Six studies evaluated metabolites found in urine. Important metabolites are summarized in Table 3 below; three of these studies used humans as subjects,12,29,43 two of the studies used Sprague-Dawley rats,18,31 and one study used the Hartley outbread guinea pig model that spontaneously develops progressive knee OA starting at ~10 months of age10. No single metabolite was identified to be important across all six of the urine metabolite studies. Increased alanine concentrations in OA versus HC were identified in two studies with p-values <0.01.18,31 Histidine was identified to be significantly altered in three studies;12,31,43 two studies reported a statistically significant decrease in histidine concentrations between OA and HC with p-values <0.05.31,43 Histidine levels were 1.3x higher (p-value 0.016) in OA patients with BMI ≥ 27kg/m2 who had radiographic progression of knee OA at 18 months versus OA patients with BMI ≥ 27kg/m2 who did not have radiographic progression of knee OA at 18 months12 (Radiographic progression was defined as decrease in minimum joint space width on radiograph ≥0.7mm compared to baseline radiographs 18 months prior). Glutamine was found to be significantly decreased in three studies with p-value <0.05.18,31,43
Table 3:
Important Metabolites Found in Urine
| Author | Sample Model | Metabolites/Metabolic Pathways of Importance |
|---|---|---|
| Smilde et al., 2005 10 | Guinea Pig | Acetate(↑), Lactate(↑), Glycine(↑), Creatinine(↑) in OA model versus control |
| Loeser et al., 2016 12 | Human | Trigonelline(↓) at both baseline and 18 months in patients with radiographic OA progression versus OA patients without radiographic progression, Histidine(↑) at 18 months in OA patients with radiographic progression versus OA patients without radiographic progression. |
| Jiang et al., 2018 18 | Rat | Alanine(↑), Alpha-Ketoglutarate(↑), Homogentisic acid(↑) in OA versus HC Glutamine(↓), aminomalonic acid(↓), shikimic acid(↓) in OA versus HC. |
| Abdelrazig et al., 2021 29 | Human | TCA cycle metabolites(↑), Glutathione synthesis metabolites(↓), Metabolites of Tryptophan, Lysine, Serine, and Glycine metabolism(↓) in OA versus HC. |
| Yin et al., 2017 31 | Rat | 5-hydroxy indole acetic acid(↑), Alanine(↑), TCA cycle metabolites (↓), tryptophan(↓),Glutamine(↓) in OA versus HC. |
| Li et al., 2010 43 | Human | TCA cycle metabolites(↑), Histamine(↑), Histidine(↓),Glutamine(↓) in OA versus HC. |
Key: (↑) indicates an increase in concentration in OA compared to controls, (↓) indicates a decrease in concentration in OA compared to controls.
The remaining importantly altered metabolites detected in the studies of urine mapped to many metabolic pathways. These include amino acid metabolism (phenylalanine, tryptophan, alanine, histidine, lysine, serine, threonine, glycine, glutamine, asparagine), lipid metabolism, nucleotide metabolism, the TCA cycle, ATP storage and utilization, and oxidative stress.
Plasma
13 studies evaluated OA metabolites found in plasma (Table 4). Ten studies evaluated plasma metabolites of OA in humans8,9,15,22,25,30,32,33,35,37,42, two studies evaluated plasma metabolites of OA in mice20,24, and one study evaluated plasma metabolites of OA in rats47. One study evaluated baseline plasma amino acid levels and was included for reference of absolute concentrations of plasma amino acids.9 Ten of the 13 plasma metabolite studies found lysophosphatidylcholine (LysoPC) and phosphatidylcholine (PC) species to be statistically significantly altered in OA8,15,20,24,25,30,32,33,35,37. Among these ten studies that reported LysoPC and PC metabolism to be altered in OA, Rockel et al found LysoPC and PC signatures predictive of total knee replacement only in males over the age of 50 years old compared to HC,8 while Liu et al found LysoPC and PC levels to be statistically significant only between OA with multi-joint pain versus OA with single joint pain but not OA versus HC.30
Table 4:
Important Metabolites Found in Plasma
| Author | Sample Model | Metabolites/Metabolic Pathways of Importance |
|---|---|---|
| Rockel et al., 2018 8 | Human | Lysophosphatidylcholine and phosphatidylcholine species were found to be altered in OA versus healthy, strongest signature in males >50 years of age. |
| Steventon et al., 2012 9 | Human | This study did not report metabolites, and instead was included as a reference as it reported normal plasma amino acid levels. |
| Costello et al., 2020 15 | Human | Glutamine, BCAAs, PcaaC38:0, and PcaaC40:6 found to overlap most between OA patients without function or pain response after undergoing total knee replacement. |
| Datta et al., 2017 20 | Mouse | Lysophosphatidylcholine and phosphatidylcholine levels remained elevated 3 months after returning to a normal chow diet from a high fat diet in OA mice model |
| Huang et al., 2020 22 | Human | Succinic acid(↓), Xanthurenic acid(↓), L-tryptophan(↓) in OA versus HC. |
| Pousinis et al., 2020 24 | Mouse | Cholesteryl ester species(↑), phosphatidylcholine species(↑), Sphingomyelin d34:1(↑) in OA versus HC. |
| Zhang et al., 2016 25 | Human | Ratio of total lysophosphatidylcholines to total phosphatidylcholines was on average 0.05μM higher in OA versus HC. Increased BCAA to histidine ratio seen in OA versus HC. |
| Liu et al., 2021 30 | Human | Lysophosphatidylcholine and phosphatidylcholine metabolism was altered in OA patients with multi-joint pain versus OA patients with pain in one joint, however not significant when comparing OA versus HC. |
| Zhang et al., 2016 32 | Human | Arginine(↓) Lysophosphatidylcholines species(↓), Phosphatidylcholine species(↓), Proline(↑) in OA versus HC. |
| Zhang et al., 2016 33 | Human | Altered phosphatidylcholine metabolism with phosphatidylcholine species(↓) was shared between OA without diabetes and OA with diabetes and also found independently in OA and diabetes, although the link between these diseases and pathways is unknown. Separation between OA with MetS and OA with type II diabetes mellitus versus OA without MetS appeared to be due to type II diabetes as opposed to hypertension, dyslipidemia, or obesity in MetS. |
| Hu et al., 2016 35 | Human | In differential correlation network, Ac-ornithine, Arginine, Alanine, and ornithine were found to be significant and made up the core of the network. Glycerophospholipids (lysophosphatidylcholine and phosphatidylcholine) species prominent in between the core and periphery of the network. |
| Zhai et al., 2021 37 | Human | Phenylalanine was significantly associated with both unilateral and bilateral knee OA progression from baseline to 30-month follow-up, noted to be a better predictor in women. Arginine, Leucine, Isoleucine, and Carnitine species also associated with unilateral progression. Serine, Tryptophan, Histidine, Lysine, Ornithine, Asparagine, and Phosphatidylcholine species were associated with bilateral progression. |
| He et al., 2021 42 | Human | Cholesterol(↑), Stearic acid(↑), L-Lactic acid(↑), Alpha-Tocopherol(↑), Oxalic Acid(↑) in OA versus HC. |
|
Abshirini et al.,
2021 47 |
Rats | 3-Hydroxybutyric Acid(↓), Tryptophan(↓), Cholic Acid(↑) in OA versus HC. |
Key: (↑) indicates an increase in concentration in OA compared to controls, (↓) indicates a decrease in concentration in OA compared to controls.
Although some articles evaluated LysoPC and PC levels in OA overall, others found these two classes of molecules to be altered in various conditions and models. In addition to findings of altered plasma LysoPC and PC levels broadly in OA, LysoPC and PC levels were found to be altered in other settings; for example, altered LysoPC and PC levels compared to health control were seen in the context of metabolic changes in OA with high fat diets20, differential correlation networks for patient pain and function responses to total joint replacements15, progression of OA in either unilateral and/or bilateral knee OA37, or in OA associated with metabolic syndrome (MetS) or components of MetS33. While LysoPC and PC species were mentioned in these ten plasma metabolomics articles, there was not a clear consensus as to whether LysoPC and PC species were increased or decreased in OA plasma compared to HC.
Amino acid levels were also altered in many studies. However, as with LysoPC and PCs, increases or decreases were not always specified. Glutamine, isoleucine, proline, tryptophan, arginine, alanine, phenylalanine, serine, histidine, lysine, and asparagine were all reported to be significantly altered with p-values <0.05.15,22,25,32,35,37,47
The remaining altered metabolites of importance corresponded largely to altered fatty acid metabolism, TCA cycle, and steroid biosynthesis.
Synovial Fluid
13 studies analyzed SF. Of these, nine used a human model11,13,17,21,27,33,40,41,50, two used a mouse model45,46, one used a New Zealand White rabbit model16, and one study used an ovine model28 (Table 5). In the synovial fluid studies, many metabolites mapped to metabolic pathways involving fatty acids, phospholipids, amino acids, vitamins, TCA cycle, collagen degradation and biosynthesis, glycosaminoglycan and keratin sulfate degradation, urea cycle, and oxidative stress.
Table 5:
Important Metabolites Found in Synovial Fluid
| Authors | Sample Model | Metabolites/Metabolic Pathways of Importance |
|---|---|---|
| Carlson et al., 2018 11 | Human | Phospholipid species, Fatty Acid metabolites, Acylcarnitine species, Ursolic acid and Sulfonic acid derivatives were associated with differentiating OA versus HC. |
| Carlson et al., 2019 13 | Human | Altered extracellular matrix component metabolism (glucosamine and galactosamine biosynthesis, ascorbate metabolism, keratin sulfate metabolism, N-glycan metabolism), amino acid metabolism, lipid metabolism (glycosphingolipid, glycerophospholipid, and carnitine/FA metabolism), leukotriene metabolism, central energy (glycolysis, gluconeogenesis, TCA cycle), oxidative stress (vitamin E and glutathione) and vitamin C, E, B1, B3, B6, and B9 metabolism in OA versus HC. |
| Hu et al., 2020 16 | Rabbit | Polyamines (acetylspermidine species), Phospholipid species, Tryptophan and Tryptophan metabolites (including kynurenic acid), Acylcarnitine Species, Phenylalanine and Phenylalanine metabolites, Lysine, Arginine, and Histidine were associated with differentiating OA versus HC. |
| Kang et al., 2015 17 | Human | Tryptophan metabolism, arachidonic acid metabolism, acylcarnitine metabolism, phospholipid metabolism, phenylalanine and tyrosine metabolism altered in OA. |
| Adams et al., 2014 21 | Human | IL-1Ra(↑), IL-6(↑), IL-8(↑), IL-10(↑), IL-15(↑), MCP-1(↑), Tryptophan(↑), Kynurenine(↑), pro-hydroxy-proline(↑), FA species(↑), Glutamate(>7x↑), Glutathione metabolites(↑), Isoleucine(↑) in post-traumatic ankle arthritis versus HC. |
| Mickiewicz et al., 2015 27 | Human | Citrate(↑), Malate(↓), Acylcarnitine species(↓), Creatine(↓) in OA versus HC. |
| Mickiewicz et al., 2015 28 | Ovine | Serine(↓), Asparagine(↓), Hydroxyproline(↓), Uridine(↓), Isobutyrate(↑), and Glucose(↑) reported as signs of early degenerative OA changes after surgical ACL-injury induced OA versus sham surgery HC. |
| Zhang et al., 2016 33 | Human | Altered phosphatidylcholine metabolism with phosphatidylcholine species(↓) was shared between OA without diabetes and OA with diabetes and also found independently in OA and diabetes, although the link between these diseases and pathways is unknown. Separation between OA with MetS and OA with type II diabetes mellitus versus OA without MetS appeared to be due to type II diabetes as opposed to hypertension, dyslipidemia, or obesity in MetS. |
| Kosinska et al., 2014 40 | Human | Sphingomyelin, Ceramide, Hexosylceramides, Phosphatidic Acid, Lysophosphatidic Acid, Phosphatidylglycerol, and Lysylphosphatidylglycerol species all increased in OA versus HC, and also increased between lOA compared to eOA. |
| Hügle et al., 2012 41 | Human | Hippuric acid, Phenylalanine, Tyrosine, D-Glucose, Valine, TCA cycle metabolites, Carnitine species, Glutamine, Lactate, Alanine, and Lysine signatures present on H1 NMR of OA. |
| Wallace et al., 2022 45 | Mouse | Day 1 Following Injury-Induced OA: Asparagine Metabolism(↑), Tryptophan Metabolism(↓), Tyrosine and Phenylalanine Degradation(↓), tRNA Charging Pathways(↓) in OA versus HC. Day 8 Following Injury-Induced OA: Asparagine Metabolism(↓), Tryptophan Metabolism(↓), Tyrosine and Phenylalanine Degradation(↑), tRNA Charging Pathways(↑) in OA versus HC. |
| Hahn et al., 2021 46 | Mouse | All Upregulated: Amino Acid Metabolism (Beta-Alanine, BCAA, Tryptophan, Lysine), Fatty Acid Metabolism (Propanoate, Butanoate), TCA Cycle and Energy (Pantothenate and CoA Biosynthesis, Ketone Body Metabolism), Terpenoid Backbone Synthesis, and Aminoacyl-tRNA Biosynthesis in Diet-Induced OA versus HC. |
| Nieminen et al., 2022 50 | Human | Phosphatidylethanolamine (22:6, 16:0)(↑), LysoPC16:0(↑), Palmitoleamide(↓), Oleamide(↓), Oleoyl Ethylamide(↓), Linoleamide(↓), 7-Keto-8-Aminopelargonic Acid(↑), Deoxyguanosine(↑), Gluconic Acid(↑) in OA versus HC. |
Key: (↑) indicates an increase in concentration in OA compared to controls, (↓) indicates a decrease in concentration in OA compared to controls.
Phospholipid species were not only frequently found to be significantly altered between OA and controls,11,13,16,17,33,40,50 but phospholipid levels also continued to increase when comparing early OA disease (Outerbridge score ≤ 2) to late OA (Outerbridge score > 2) and the HC population.40
Metabolites indicating altered energy metabolism were also commonly identified in SF of OA compared to HC. These included FA metabolites, TCA cycle metabolites, gluconeogenic amino acids, and sugars in the synovial fluid samples.11,13,16,17,21,27,28,41,45,46 Multiple carnitine species were perturbed; one study indicated decreased levels of two acylcarnitine species in OA synovial fluid.27 Two of the other studies mentioned altered acylcarnitine species as OA predictors (e.g. AUC >0.90), although it was not clear if the particular acylcarnitine species increased or decreased in OA compared to HC.11,16 Altered acylcarnitine levels in the SF of OA compared to HC may indicate altered energy production, as carnitine and carnitine-acylcarnitine translocase allow acyl chains to move across the inner mitochondrial membrane and into the mitochondrial matrix where beta-oxidation occurs.51
The prominence of altered metabolites that are involved in mitochondrial energy production and metabolism, as well as evidence of oxidative stress, may indicate mitochondrial dysfunction in the osteoarthritic joint.
Serum
Twelve studies involving serum were included in this review (Table 6): six articles analyzed human serum OA metabolites23,26,36,38,39,49, two article studied serum in rats34,44, one article examined serum in an ovine model19, one article examined serum from New Zealand rabbits48, one article studied serum from a mouse model45, and one article evaluated daily variation of acylcarnitines and amino acids in OA patients without comparison to a HC group14. Five of the articles reported altered levels of branched-chain amino acids between OA and HC groups.19,23,36,38,48 Two studies reported increases in valine, isoleucine, and leucine.36,38 One study reported increased valine levels in OA serum.23 The fourth article reported decreases in BCAAs in an ovine model of induced OA via surgical ACL transection.19 Maher et al. suggest that their observed decrease in BCAAs may be specific to the ovine species, as literature reporting increased BCAAs present in human OA serum was reported prior to their publication38 and has been replicated in other studies,23,36 and therefore should be investigated further.19 The final study to mention BCAAs reported that both biosynthesis and degradation of BCAAs was upregulated, along with findings of elevated levels of L-(+)-Valine.48
Table 6:
Important Metabolites Found in Serum
| Author | Sample Model | Metabolites/Metabolic Pathways of Importance |
|---|---|---|
| Thompson et al., 2011 14 | Human | This study did not report metabolites, and instead was included as a reference as it reported daily variations of serum amino acid and acylcarnitine levels. |
| Maher et al., 2012 19 | Ovine | BCAAs(↓), 3-Methylhistidine(↑), Glycine(↑), Creatinine(↑), Creatine(↑) at 4 weeks and 12 weeks in post-ACL Transection OA model versus HC. |
| Senol et al., 2019 23 | Human | Phospholipid species(↑), Indoleacetic acid(↑), Urea(↑), Valine(↑), Alanine(↑), FA metabolites(↑) increased in OA versus Control. |
| Tootsi et al., 2018 26 | Human | Total Acylcarnitine species to Carnitine species ratio was decreased in OA versus HC. Carnitine Palmitoyltransferase 1 activity was decreased in OA versus HC. |
| Maerz et al., 2018 34 | Rat | Acylcarnitine species increased at 72 hours, 4 weeks, and 10 weeks post-ACL rupture OA model versus HC. Most significantly affected pathways in OA versus HC identified as cyanoamino acid metabolism, methane metabolism, sphingolipid metabolism, histidine metabolism, glutathione metabolism, tryptophan metabolism, beta-alanine metabolism, fructose and mannose metabolism, and amino and nucleotide sugar metabolism. |
| Chen et al., 2018 36 | Human | BCAAs(↑), Tryptophan(↑), Alanine(↑), 4-Hydroxy-L-Proline(↑), Creatine(↑), Arginine(↑), Lysine(↑), Tyrosine(↑), Glutamine (↓), Phenylalanine(↓), Serine(↓), Proline(↓), Gamma-Aminobutyric Acid(↓), Creatinine(↓), Taurine(↓), Asparagine(↓), Acetyl-Carnitine(↓), Citrulline(↓) in OA versus HC. |
| Zhai et al., 2010 38 | Human | Increased BCAA to Histidine ratio in OA versus HC, with ratio of Valine to Histidine and Isoleucine to Histidine having strongest statistical significance. |
| Zhang et al., 2015 39 | Human | CRP(↑), Homocysteine(↑), Tryptophan(↑), Glycine(↓), Histidine(↓) in OA versus HC. |
| Zhao et al., 2021 44 | Rat | L-Tryptophan(↑), Gamma-Aminobutyric Acid(↑), Carbamic Acid(↑), L-Carnitine(↑), Stearic Acid(↑), L-Arginine(↓) in OA versus HC. |
| Wallace et al., 2022 45 | Mouse | Day 1 Following Injury-Induced OA: Arginine Synthesis(↑), Proline Degradation(↑), Nicotine Degradation(↑), Phospholipase Pathway(↓) in OA versus HC. Day 8 Following Injury-Induced OA: Tyrosine Degradation(↑), |
| Phospholipase Pathway(↑), Nicotine Degradation(↓) in OA versus HC. | ||
| Xiao et al., 2022 48 | Rabbit | ɛ, ɛ, ɛ Trimethyllysine(↓), Ascorbic acid(↑), Otonecine(↑), and Tranexamic Acid(↓) in OA versus HC. BCAA Biosynthesis and Degradation (Leucine-Valine, L-(+)-Valine)(↑), HIF-1 Pathway(↑), Antioxidant Metabolism (Glutathione and Ascorbic Acid)(↑), Phenylalanine Metabolism(↑), Pantothenate and CoA Biosynthesis(↑), Tyrosine Metabolism(↓) in OA versus HC. |
| Pertusa et al., 2022 49 | Human | Amino Acid Metabolism (Alanine, Glycine, Phenylalanine, and Tyrosine), Lactate, Acetate, Phosphocholine, Bacterial Co-Metabolism (2-Aminobutyrate, 4-Aminobutyrate, N(CH3)3, Dimethylamine) significantly altered in OA versus HC following adjustment for age, BMI, and bone mineral density. |
Key: (↑) indicates an increase in concentration in OA compared to controls, (↓) indicates a decrease in concentration in OA compared to controls.
Acylcarnitine species were serum metabolites associated with OA. One study found a lower ratio of total acylcarnitine to carnitine with lower levels of 8 out of 14 acylcarnitines species measured in OA versus HC.26 They also found lower levels of carnitine palmitoyltransferase 1 activity.26 CPT-1 is present on the outer mitochondrial membrane and is the rate limiting enzyme for long-chain fatty acid beta-oxidation the mitochondrial matrix. Decreased activity of this enzyme may drive mitochondrial dysfunction in OA. Another study noted a decrease in acetyl-carnitine in OA compared to HC36, while another noted a significant increase in L-Carnitine levels in OA versus HC44. The fourth serum study to mention acylcarnitine species noted significant increases in seven acylcarnitine species (p = 0.008–0.041) over 10 weeks after ACL rupture-induced post-traumatic OA.34
Other statistically significant serum metabolites mapped to phospholipid metabolism, amino acid metabolism (tryptophan, arginine, glutamate, proline, serine, alanine, glycine, histidine, tyrosine), TCA cycle, nucleotide metabolism, energy metabolism, and the urea cycle.
Discussion
Metabolomics is a growing field of research that can offer insight into disease pathophysiology and develop metabolic biomarkers that support early diagnosis and intervention, as well as both monitoring of disease progression and evaluation of treatment efficacy. OA is a disease with emerging metabolic features, and there are currently no treatment options that stop or reverse disease progression.52 Therefore, identification and diagnosis of OA early in the disease course is critical to prevent extensive cartilage destruction and allow modifiable risk factors to be addressed to slow disease progression.
The studies included in this review analyzed the metabolomics of OA in urine, serum, plasma, and synovial fluid. The studies were done with humans, rats, mice, sheep, guinea pigs, and rabbits. Various analytical methods including mass spectrometry were performed to identify and compare metabolites between OA and comparison groups in each biological fluid. Metabolites that resulted from analysis of body fluids were either reported as being altered, while other studies further clarified whether the measured concentrations of reported metabolites were increased or decreased in comparison to control groups. In studies that reported increased or decreased concentrations, the data compiled by the authors shows that approximately 168 metabolites were increased in OA versus control out of 239 metabolites. This proportion was more or less consistent across each individual fluid sample.
While metabolomic profiling has great potential for the development of diagnostic biomarkers for OA, the field is at an early stage. Typically studies report intensity levels of hundreds to thousands of metabolites for tens to hundreds of patients. As such multiple testing corrections (e.g. false-discovery rates) are required to minimize spurious discoveries. Few studies in this review report performance evaluation of candidate metabolite biomarkers, and for biomarker development rigorous analytical quantification including precision and accuracy are needed. Furthermore, validation across multiple clinical cohorts will also be required. Metabolite abundance can differ between serum and plasma 53, and it is not clear which blood preparation is best for detecting metabolomic biomarkers for OA. Depending on the candidate biomarker, metabolite abundance may be affected by diet or other factors 54, and robust performance evaluation is needed to ensure the utility of metabolomic biomarkers for OA. In this context, it remains to be determined if metabolomic biomarkers can identify patients with symptomatic OA.
In Maher et al19, the findings of decreased BCAA concentrations in ovine OA subjects is contrary to reported increases in BCAA concentrations in OA in other studies.23,36,38,48 Maher et al. suggest that their observation of decreased BCAA concentrations in OA may result from interspecies differences in disease metabolic processes. This highlights the need for further studies of OA metabolomics and pathophysiology, as well as further evaluation of animal models of OA. Identifying both similarities and differences between animal and human OA is important to ensure that metabolomic findings are relatable across species. Biomarker candidates identified in animal models must be evaluated in human clinical populations to ensure their utility.
The heterogeneity of OA is reflected in the reported metabolites and pathways detailed in these studies. From these studies, there was no consensus on either a single metabolite or a panel of metabolites that could serve as predictive biomarkers for early diagnosis of OA. This reflects the complexity of the disease and the multi-tissue pathophysiology that drive the progression of OA.
Although there was variation in the specific metabolites reported in these studies, common metabolic themes arose among many of the reports. Metabolites associated with mitochondrial dysfunction were detected in several studies. These include alterations in fatty acid metabolism, acylcarnitine and carnitine metabolites, and TCA cycle metabolites. Additionally several studies found metabolites indicating the presence of reactive oxygen species, gluconeogenic amino acid metabolites, and metabolites of antioxidants. There is potential for future studies to expand upon these results by focusing on metabolites associated with mitochondrial dysfunction.
Common themes seen across many studies include metabolism of phospholipids and articular cartilage matrix components, as well as metabolism of many amino acids, notably BCAAs. Altered levels of phospholipid metabolites including lysophosphatidylcholine and phosphatidylcholine species, sphingomyelins, ceramides, hexosylceramides, phosphatidylglycerols were noted. Levels of key precursors to articular cartilage matrix components including proline, hydroxyproline, and pro-hydroxy-proline, as well as glutamate (and molecules of glutamate metabolism) which is used by mammals to synthesize proline, were noted in many studies.. Altered amino acid metabolism was a common finding among many of the articles, with BCAA to histidine ratio being reported as a possible diagnostic marker of OA. Although the changes in the levels of BCAAs and histidine in the studies were not unanimously agreed upon across the studies, it warrants further investigation and research as two studies claimed that BCAA to histidine ratio could be an important marker of OA. Due to their frequent recurrence across studies, metabolites of phospholipid and articular cartilage matrix components, as well amino acid metabolism, could be topics of further investigation in future studies.
Tryptophan Metabolism
Tryptophan can be synthesized and degraded through multiple pathways including the serotonin and melatonin pathway, the kynurenine pathway, and the indole pathway. Tryptophan metabolism and metabolite levels are altered in many diseases, ranging from chronic inflammatory diseases, autoimmune disorders, and neurodegenerative diseases.21 21 articles included in this review found tryptophan levels to be significantly altered in OA, and 5 articles suggested that the kynurenine pathway may be involved in OA pathogenesis in some way. Kynurenine is produced from tryptophan by indolamine-2,3-oxidase, and this kynurenine production is enhanced after treatment by either interferon gamma or tumor necrosis factor-alpha.55,56 Metabolites further downstream in the kynurenine pathway can lead to increased production of reactive oxygen species, as well as modulation of the inflammatory T-cell response to induce tolerance to mild inflammatory attack.55 Further study of the tryptophan-kynurenine axis in OA may yield important information for better understanding OA pathogenesis.
Limitations
43 articles met the inclusion criteria for this review. However, there exists literature on the topic of metabolomics of OA that did not meet our inclusion criteria, or possibly were not displayed in the results of the database searches due to keyword or database limitations, and therefore were not included in this review. As such this review focused on identifying basic metabolite features altered in OA, analyzing how metabolomic profiles change during OA disease progression, and finding relationships between OA and other health conditions like metabolic syndrome, orthopaedic trauma, and cardiovascular disease. The reviewed articles did not always report data on these other health conditions which limits the generalizability of these findings. Additionally, Zhang et al., 2016, reported metabolite ratios that were only significant in advanced knee OA but not in early-stage OA.25 This suggests that metabolite biomarkers of OA may be stage-specific which suggests their potential to detect progression of OA.
Among all articles in this review, more than 350 metabolites were reported to be altered between OA and control subjects. These metabolites achieved varying degrees of significance with p-values smaller than 0.01 to barely distinguishable statistical differences between OA and HC. While some of these findings were replicated across many studies,, such as LysoPC and PC species8,11,15,20,23–25,32,33,35,37,50 and the amino acids glycine,10,19,29,39,49 alanine,18,23,31,34–36,41,46,49 and histidine,12,16,25,34,37–39,43 and many others, a large majority of individual metabolites were not. However, because metabolic pathways involve detection of subsets of their individual metabolites, comparing reported metabolic pathways was more feasible than detailing individual metabolites in many cases. Thus, it is possible that future studies may identify additional biomarkers based on the pathways reported herein.
As plasma, urine, and serum are all fluids that reflect systemic metabolism, future studies should identify and control for confounding factors, such as concurrent medical conditions, patient demographics, and dietary habits. These key covariates may drive metabolomic changes that require additional statistical modeling for developing biomarkers of OA. Clinical sampling of SF for metabolomics involves inherent risks as it is an invasive procedure. This challenge renders obtaining HC samples challenging. For future studies of OA synovial fluid, it would be beneficial to obtain healthy age- and sex-matched SF samples to allow for direct comparison to OA SF. Because SF directly contacts multiple tissues of the diseased joint, in contrast to systemic fluids, it contains great potential for developing biomarkers of OA and OA progression.
Conclusions
OA is a heterogenous disease with a wide range of altered metabolic pathways and processes that result in disease pathogenesis and progression. These studies demonstrate this heterogeneity through the vast array of metabolites and pathways reported. Through this review, metabolites indicated abnormalities in fatty acid metabolism and the TCA cycle suggesting mitochondrial dysfunction, altered energy metabolism, amino acid metabolism, nucleotide metabolism, phospholipid metabolism, urea cycle, and metabolism of articular cartilage matrix components.
Many reports find differences in lysophosphatidylcholine/phosphatidylcholine metabolism, tryptophan and kynurenine metabolism, and branched-chain amino acid metabolism in OA. These metabolites and pathways have potential both for developing prognostic biomarkers of OA and for improved understanding of OA pathogenesis and progression. Sub-groups and endotypes of OA were also proposed in several reports. Future studies might characterize metabolomic differences between phenotypes and endotypes of OA. The identification of distinct phenotypes or endotypes may support development of novel therapeutic targets and help address modifiable risk factors involved in OA development and progression.
While metabolomic profiling is relatively new to the OA field, future primary and replication studies have good potential to provide reliable, sensitive, and specific panels of biomarkers to allow for early diagnosis of OA.
Acknowledgements
The authors are grateful for funding support from NIH (NIAMS 5R01AR073964) and NSF (1554708 and 2140127).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Osteoarthritis Action Alliance. OA Prevalence and Burden Osteoarthritis Action Alliance. https://oaaction.unc.edu/oa-module/oa-prevalence-and-burden/. Published 2021. Accessed 8/23/2021, 2021. [Google Scholar]
- 2.Arthritis Foundation. Arthritis by the Numbers 2021.
- 3.Arthritis Foundation. Osteoarthritis https://www.arthritis.org/diseases/osteoarthritis. Published 2021. Accessed 8/24/2021, 2021.
- 4.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol 2011;23(5):471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams SB Jr., Setton LA, Nettles DL. The role of metabolomics in osteoarthritis research. J Am Acad Orthop Surg 2013;21(1):63–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaggard MKJ, Boulangé CL, Akhbari P, et al. A systematic review of the small molecule studies of osteoarthritis using nuclear magnetic resonance and mass spectroscopy. Osteoarthritis Cartilage 2019;27(4):560–570. [DOI] [PubMed] [Google Scholar]
- 7.Harris JD, Quatman CE, Manring MM, Siston RA, Flanigan DC. How to write a systematic review. Am J Sports Med 2014;42(11):2761–2768. [DOI] [PubMed] [Google Scholar]
- 8.Rockel JS, Zhang W, Shestopaloff K, et al. A classification modeling approach for determining metabolite signatures in osteoarthritis. PLoS One 2018;13(6):e0199618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steventon GB, Mitchell SC, Angulo S, Barbas C. An investigation into possible xenobiotic-endobiotic inter-relationships involving the amino acid analogue drug, S-carboxymethyl-L-cysteine and plasma amino acids in humans. Amino Acids 2012;42(5):1967–1973. [DOI] [PubMed] [Google Scholar]
- 10.Smilde AK, Jansen JJ, Hoefsloot HC, Lamers RJ, van der Greef J, Timmerman ME. ANOVA-simultaneous component analysis (ASCA): a new tool for analyzing designed metabolomics data. Bioinformatics 2005;21(13):3043–3048. [DOI] [PubMed] [Google Scholar]
- 11.Carlson AK, Rawle RA, Adams E, Greenwood MC, Bothner B, June RK. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochemical and Biophysical Research Communications 2018;499(2):182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loeser RF, Pathmasiri W, Sumner SJ, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage 2016;24(8):1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson AK, Rawle RA, Wallace CW, et al. Characterization of synovial fluid metabolomic phenotypes of cartilage morphological changes associated with osteoarthritis. Osteoarthritis and Cartilage 2019;27(8):1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson DK, Sloane R, Bain JR, et al. Daily variation of serum acylcarnitines and amino acids. Metabolomics 2012;8(4):556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costello CA, Hu T, Liu M, et al. Differential correlation network analysis identified novel metabolomics signatures for non-responders to total joint replacement in primary osteoarthritis patients. Metabolomics 2020;16(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu YW, Wu Q, Qiao Y, et al. Disturbances in Metabolic Pathways and the Identification of a Potential Biomarker Panel for Early Cartilage Degeneration in a Rabbit Anterior Cruciate Ligament Transection Model. Cartilage 2020. [DOI] [PMC free article] [PubMed]
- 17.Kang KY, Lee SH, Jung SM, Park SH, Jung BH, Ju JH. Downregulation of Tryptophan-related Metabolomic Profile in Rheumatoid Arthritis Synovial Fluid. J Rheumatol 2015;42(11):2003–2011. [DOI] [PubMed] [Google Scholar]
- 18.Jiang H, Liu J, Qin XJ, et al. Gas chromatography-time of flight/mass spectrometry-based metabonomics of changes in the urinary metabolic profile in osteoarthritic rats. Exp Ther Med 2018;15(3):2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher AD, Coles C, White J, et al. H-1 NMR Spectroscopy of Serum Reveals Unique Metabolic Fingerprints Associated with Subtypes of Surgically Induced Osteoarthritis in Sheep. Journal of Proteome Research 2012;11(8):4261–4268. [DOI] [PubMed] [Google Scholar]
- 20.Datta P, Zhang Y, Parousis A, et al. High-fat diet-induced acceleration of osteoarthritis is associated with a distinct and sustained plasma metabolite signature. Sci Rep 2017;7(1):8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams SB, Nettles DL, Jones LC, Miller SD, Guyton GP, Schon LC. Inflammatory Cytokines and Cellular Metabolites as Synovial Fluid Biomarkers of Posttraumatic Ankle Arthritis. Foot & Ankle International 2014;35(12):1241–1249. [DOI] [PubMed] [Google Scholar]
- 22.Huang ZS, He ZR, Kong Y, Liu ZQ, Gong LZ. Insight into osteoarthritis through integrative analysis of metabolomics and transcriptomics. Clinica Chimica Acta 2020;510:323–329. [DOI] [PubMed] [Google Scholar]
- 23.Senol O, Gundogdu G, Gundogdu K, Miloglu FD. Investigation of the relationships between knee osteoarthritis and obesity via untargeted metabolomics analysis. Clinical Rheumatology 2019;38(5):1351–1360. [DOI] [PubMed] [Google Scholar]
- 24.Pousinis P, Gowler PRW, Burston JJ, Ortori CA, Chapman V, Barrett DA. Lipidomic identification of plasma lipids associated with pain behaviour and pathology in a mouse model of osteoarthritis. Metabolomics 2020;16(3):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Sun G, Aitken D, et al. Lysophosphatidylcholines to phosphatidylcholines ratio predicts advanced knee osteoarthritis. Rheumatology (Oxford) 2016;55(9):1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tootsi K, Kals J, Zilmer M, Paapstel K, Ottas A, Märtson A. Medium- and long-chain acylcarnitines are associated with osteoarthritis severity and arterial stiffness in end-stage osteoarthritis patients: a case-control study. Int J Rheum Dis 2018;21(6):1211– 218. [DOI] [PubMed] [Google Scholar]
- 27.Mickiewicz B, Kelly JJ, Ludwig TE, et al. Metabolic analysis of knee synovial fluid as a potential diagnostic approach for osteoarthritis. Journal of Orthopaedic Research 2015;33(11):1631–1638. [DOI] [PubMed] [Google Scholar]
- 28.Mickiewicz B, Heard BJ, Chau JK, et al. Metabolic Profiling of Synovial Fluid in a Unilateral Ovine Model of Anterior Cruciate Ligament Reconstruction of the Knee Suggests Biomarkers for Early Osteoarthritis. Journal of Orthopaedic Research 2015;33(1):71–77. [DOI] [PubMed] [Google Scholar]
- 29.Abdelrazig S, Ortori CA, Doherty M, Valdes AM, Chapman V, Barrett DA. Metabolic signatures of osteoarthritis in urine using liquid chromatography-high resolution tandem mass spectrometry. Metabolomics 2021;17(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M, Xie Z, Costello CA, et al. Metabolomic analysis coupled with extreme phenotype sampling identified that lysophosphatidylcholines are associated with multisite musculoskeletal pain. Pain 2021;162(2):600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin H, Wang L, Li QL, Zhang JH, Zhang L, Wang X. Metabolomic Analysis of Biochemical Changes in Urine of Osteoarthritis Rat and Interventional Effects of Bushen-Huoxue Herb Couple. Chinese Herbal Medicines 2017;9(4):369–375. [Google Scholar]
- 32.Zhang W, Sun G, Likhodii S, et al. Metabolomic analysis of human plasma reveals that arginine is depleted in knee osteoarthritis patients. Osteoarthritis Cartilage 2016;24(5):827–834. [DOI] [PubMed] [Google Scholar]
- 33.Zhang WD, Sun G, Likhodii S, et al. Metabolomic analysis of human synovial fluid and plasma reveals that phosphatidylcholine metabolism is associated with both osteoarthritis and diabetes mellitus. Metabolomics 2016;12(2). [Google Scholar]
- 34.Maerz T, Sherman E, Newton M, et al. Metabolomic serum profiling after ACL injury in rats: A pilot study implicating inflammation and immune dysregulation in post-traumatic osteoarthritis. J Orthop Res 2018;36(7):1969–1979. [DOI] [PubMed] [Google Scholar]
- 35.Hu T, Zhang W, Fan Z, et al. METABOLOMICS DIFFERENTIAL CORRELATION NETWORK ANALYSIS OF OSTEOARTHRITIS. Pac Symp Biocomput 2016;21:120–131. [PubMed] [Google Scholar]
- 36.Chen R, Han S, Liu XF, et al. Perturbations in amino acids and metabolic pathways in osteoarthritis patients determined by targeted metabolomics analysis. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences 2018;1085:54–62. [DOI] [PubMed] [Google Scholar]
- 37.Zhai GJ, Sun XB, Randell EW, et al. Phenylalanine Is a Novel Marker for Radiographic Knee Osteoarthritis Progression: The MOST Study. Journal of Rheumatology 2021;48(1):123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai G, Wang-Sattler R, Hart DJ, et al. Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Annals of the Rheumatic Diseases 2010;69(6):1227–1231. [DOI] [PubMed] [Google Scholar]
- 39.Zhang QM, Li H, Zhang ZD, Yang F, Chen JY. Serum Metabolites as Potential Biomarkers for Diagnosis of Knee Osteoarthritis. Disease Markers 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosinska MK, Liebisch G, Lochnit G, et al. Sphingolipids in human synovial fluid--a lipidomic study. PLoS One 2014;9(3):e91769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hügle T, Kovacs H, Heijnen IA, et al. Synovial fluid metabolomics in different forms of arthritis assessed by nuclear magnetic resonance spectroscopy. Clin Exp Rheumatol 2012;30(2):240–245. [PubMed] [Google Scholar]
- 42.He ZR, Liu ZQ, Gong LZ. Systematic evaluation of sample preparation strategy for GC-MS-based plasma metabolomics and its application in osteoarthritis. Analytical Biochemistry 2021;621. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Yang SB, Qiu YP, et al. Urinary metabolomics as a potentially novel diagnostic and stratification tool for knee osteoarthritis. Metabolomics 2010;6(1):109–118. [Google Scholar]
- 44.Zhao J, Liu M, Shi T, et al. Analysis of Serum Metabolomics in Rats with Osteoarthritis by Mass Spectrometry. Molecules 2021;26(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace CW, Hislop B, Hahn AK, Erdogan AE, Brahmachary PP, June RK. Correlations between metabolites in the synovial fluid and serum: A mouse injury study. J Orthop Res 2022;40(12):2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn AK, Batushansky A, Rawle RA, Prado Lopes EB, June RK, Griffin TM. Effects of long-term exercise and a high-fat diet on synovial fluid metabolomics and joint structural phenotypes in mice: an integrated network analysis. Osteoarthritis Cartilage 2021;29(11):1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abshirini M, Cabrera D, Fraser K, et al. Mass Spectrometry-Based Metabolomic and Lipidomic Analysis of the Effect of High Fat/High Sugar Diet and Greenshell. Metabolites 2021;11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Z, Zhang Z, Huang S, Lon JR, Xie S. Metabolic Profiling of Serum for Osteoarthritis Biomarkers. Dis Markers 2022;2022:1800812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pertusa C, Mifsut D, Morales JM, et al. Metabolomic Analysis of Severe Osteoarthritis in a Spanish Population of Women Compared to Healthy and Osteoporotic Subjects. Metabolites 2022;12(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nieminen P, Hämäläinen W, Savinainen J, et al. Metabolomics of Synovial Fluid and Infrapatellar Fat Pad in Patients with Osteoarthritis or Rheumatoid Arthritis. Inflammation 2022;45(3):1101–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talley JT. Biochemistry, Fatty Acid Oxidation 2022. doi:NBK556002. Accessed 5/11/22. [PubMed]
- 52.Mayo Clinic. Osteoarthritis https://www.mayoclinic.org/diseases-conditions/osteoarthritis/symptoms-causes/syc-20351925. Published 2021. Accessed 8/31/2021, 2021.
- 53.Liu X, Hoene M, Wang X, et al. Serum or plasma, what is the difference? Investigations to facilitate the sample material selection decision making process for metabolomics studies and beyond. Anal Chim Acta 2018;1037:293–300. [DOI] [PubMed] [Google Scholar]
- 54.Bar N, Korem T, Weissbrod O, et al. A reference map of potential determinants for the human serum metabolome. Nature 2020;588(7836):135–140. [DOI] [PubMed] [Google Scholar]
- 55.Mándi Y, Vécsei L. The kynurenine system and immunoregulation. J Neural Transm (Vienna) 2012;119(2):197–209. [DOI] [PubMed] [Google Scholar]
- 56.Malone DG, Dolan PW, Brown RR, et al. Interferon gamma induced production of indoleamine 2,3 dioxygenase in cultured human synovial cells. J Rheumatol 1994;21(6):1011–1019. [PubMed] [Google Scholar]


