Abstract
The lactic acid bacteria, Lactococcus lactis subsp. lactis LM1185 was isolated from Hydrangea macrophylla. Strain LM1185 showed 50.5% of acid tolerance at pH 2.5 for 2 h and 30.4% of 0.3% (w/v) bile salt tolerance for 24 h. The antioxidant activity of this strain was measured at 99.4% of 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) radical scavenging activity. When RAW 264.7 macrophage cells were treated with strain LM1185, there was no observed cytotoxicity. This strain showed high nitric oxide production and mRNA expression levels of cytokines such as tumor necrosis factor-α and inducible nitric oxide synthase (iNOS). The nuclear factor-kB signaling pathway was activated by this strain resulting in the production of iNOS and cyclooxygenase-2 determined by western blotting. The present results indicated that L. lactis subsp. lactis LM1185 could be used as potential probiotics and may play a crucial role in the immunostimulatory effect on macrophages.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-022-01199-5.
Keywords: Hydrangea macrophylla, Lactic acid bacteria, Lactococcus lactis, Probiotics, Immunostimulatory activity
Introduction
Hydrangea (genus Hydrangea) is a popular and commonly cultivated shrub with a large, dome-shaped bloom native to Asia and America. Many species in the genus Hydrangea belong to the family Hydrangeaceae and have various bloom colors ranging from pink to white to blue (Khaing et al., 2016). The colors of the flowers depend on the cultivar and age and can also be influenced by chemical characteristics of the soil, such as pH and aluminum concentration (Khaing et al., 2016). Hydrangea can be differentiated into five representative types: bigleaf, smooth, paniculate, climbing, and oakleaf (Li et al., 2018). Seven hundred cultivars of Hydrangea macrophylla, the most popular hydrangea, have been described, but only two subspecies are commonly and commercially grown; H. macrophylla ssp. Macrophylla and H. macrophylla ssp. Serrata (Mmbaga et al., 2011). Traditionally, the components of hydrangea, the leaves, stems, and roots have been known for having therapeutic properties, regarding kidney and urinary diseases as well as exhibiting antiallergenic, antibacterial, and antioxidative properties (Bag et al., 2008; Geum et al., 2020).
Lactic acid bacteria (LAB) are bacteria that produce lactic acid by fermentation of carbohydrates. The main characteristics of LAB are that they are gram-positive, non-sporing, rod- or cocci-shaped, and anaerobes or facultative aerobes (Masood et al., 2011; Quinto et al., 2014). They are widespread microorganisms, mainly found in fermented foods as well as in the environment. LAB can be divided into two groups depending on its fermentation pathway. The homofermentative LAB ferments carbohydrates and produces lactic acid as the only and major end-product, while heterofermentative LAB produces more than one product such as CO2 and alcohol in addition to lactic acid (Zúñiga et al., 1993).
Probiotics are defined as ‘live microorganisms, which when administrated in adequate amounts confer a health benefit on the host’ (Masood et al., 2011). LAB can be used as ideal probiotics when the following criteria are satisfied: (1) acid and bile-tolerance, (2) adhesion to mucosal and epithelial surfaces, (3) antimicrobial activity against pathogenic bacteria, and (4) bile salt hydrolase activity (Abushelaibi et al., 2017). Interest in plant-origin LAB is increasing because they are known to be safer than animal-origin LAB from the viewpoint of antibiotic resistance and the environmental ecosystem (Mota-Gutierrez and Cocolin, 2021). They also have a variety of health-beneficial activities comparable to that of animal-derived lactic acid bacteria.
In this study, the LAB was isolated from the new variety of H. macrophylla, which was developed by Jeonnam Agricultural Research & Extension Services and named “White Ari.” The isolate was evaluated for potential probiotic utilization. Currently, the isolation of LAB from hydrangea has not been reported. This is the first study on the isolation of LAB from hydrangea and its immunostimulatory activity.
Materials and methods
Isolation of lactic acid bacteria
Hydrangea (three varieties: Morning Star, Pink Ari, and White Ari), twenty plant samples for each variety, was provided by Lactomason (Jinju, Korea), and its flower and stem were separated. Fifty grams of each sample were mixed with 200 mL of sterilized saline, respectively, and homogenized for 3 min using a stomacher (BNF KOREA Co., Ltd., Seoul, Korea). They were serially diluted with 0.88%(w/v) NaCl and cultured on Plate Count Agar with bromocresol (5 g peptone, 3 g beef extract, 1 g lactose, 0.025 g bromophenol purple, 15 g agar, per liter, pH 7.0, KisanBio, Seoul, Korea). The plates were incubated at 37 °C for 48 h in anaerobic conditions. Individual colonies, expected to be LAB, were chosen with sterile toothpicks and transferred to another Plate Count Agar with bromocresol (KisanBio). Finally, the colonies were sub-cultured to Plate Count Agar with bromocresol and incubated at 37 °C for 24 h. The pure isolate was cultured in de Man, Rogosa, and Sharpe (MRS) broth (BD BBL™, Franklin Lakes, NJ, USA) mixed equal volume with 40% glycerol, and stored at − 80 °C.
Gram staining, catalase test, and KOH test
Gram staining, catalase test, and KOH test of LAB were carried out according to the standard protocol described by Cappuccino and Sherman (1983).
API 50 CHL kit
The carbohydrate utilization of the strain was determined using an API 50 CH system (bioMérieux, Marcy l'Etoile, France). API 50 CH systems consisted of API 50 CHL medium, 49 strips which were used to examine the fermentation of carbohydrates, and one negative control strip. All steps were performed according to the manufacturer’s manual.
Identification of lactic acid bacteria
The isolated LAB were cultured in MRS broth for 16 h at 37 °C. Genomic DNA was isolated using AccuPrep® Genomic DNA Extraction Kit (Bioneer, Seoul, Korea). Polymerase chain reaction for the amplification of 16S rRNA gene was performed using AccuPower® PCR PreMix (Bioneer, Seoul, Korea) and isolated genomic DNA as a template. 27F (5′-AGA GTT TGA TCA TGG CTC AG-3′) and 1492R (5′-TAC GGT TAC CTT GTT ACG AC-3′) primers were used to amplify the 16 s rRNA gene using MyCycler™ Thermal Cycler (BIO-RAD, Hercules, CA, USA). Initial denaturation was conducted at 94 °C for 2 min, followed by 35 cycles including denaturation at 94 °C for 30 s, annealing at 50 °C for 1 min, and extension at 72 °C for 1 min. The final extension was performed at 72 °C for 7 min. The nucleotide sequence of 16 s rRNA gene was determined by Bionics Co., Ltd. (Seoul, Korea). The analysis of resulted nucleotide sequence of 16 s rRNA gene was performed using BLAST from NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
ClustalX v 2.1 (Larkin et al., 2007) was used to carry out an alignment on the sequences, and MEGA v 11.0.9 (Tamura et al., 2021) was used to construct a tree using the neighbor-joining method for the alignment file obtained from ClustalX. All programs were used with default parameters, except for the settings of MEGA. The number of the bootstrap test had changed to 1000 replications, and the Kimura 2-parameter model was selected as a substitution model.
Scanning electron microscopy
The bacterial sample was fixed for 2 h with 2.5% glutaraldehyde, followed by washing using 0.1 M phosphate buffer (pH 7.4) for 15 min. The secondary fixation was performed with 1% osmium tetroxide solution for 1 h. After fixing, the fixed specimen was washed with the same buffer in the same way as before. Dehydration was carried out for 15 min each in ethyl alcohol, which had gradient dilution of 50%, 60%, 70%, 90%, 95%, and 100%. For substitution, the specimen was reacted with a mixture of isoamyl acetate and 100% ethyl alcohol for 15 min, of which ratios were 1:2, 1:1, 2:1, and isoamyl acetate only. After the reaction, the specimen was dried with CO2 gas using Hitachi HCP-2 Critical point dryer (Hitachi, Tokyo, Japan). The specimen was mounted on carbon tape on the stub, and the platinum coating was carried out using Hitachi E-1030 (Hitachi). The coated stub was observed by Hitachi S-4700 (Hitachi) installed at the Industry-Academia Cooperation Center, Eulji University, Seongnam, Korea.
Antioxidant activity
The antioxidant activity of the strain was measured using 2,2-diphenyl-1-picrylhydrazyl (DPPH) and 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) assays. For the DPPH assay, 2,2-diphenyl-1-picrylhydrazyl was purchased from Sigma-Aldrich (St. Louis, USA). The overnight culture of the strain was centrifuged at 15,920×g for 5 min. Ten microliters of supernatant were mixed with 190 μL of 0.1 mM methanolic DPPH solution in a 96-well plate, resulting in a total volume of 200 μL. The reaction was carried out in the dark for 30 min. The absorbance of the resulted plating was measured at 517 nm to calculate the radical scavenging activity of the sample. With the following equation, the antioxidant activity could be calculated:
| 1 |
Distilled water was used as a negative control, and 2 mM ascorbic acid was used as a positive control. The standard curve of ascorbic acid from 0.0625 to 2 mM was drawn to use to determine the activity.
ABTS radical solution was prepared by mixing 7 mM of 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (Sigma-Aldrich) with 2.45 mM potassium persulfate (Sigma-Aldrich) (1:1, v/v). The solution resulted from the reaction in the dark for 16 h at 25 °C, was diluted with 0.1 M sodium phosphate buffer (pH 7.4) until its absorbance was reached at 0.7 at 734 nm. A total of 20 μL of the supernatant obtained from the culture broth cultured overnight at 37 °C was mixed with 980 μL of ABTS radical solution. The mixed sample was incubated at 37 °C for 5 min, followed by absorbance detected at 734 nm. ABTS radical scavenging activity was calculated by the equation below:
| 2 |
Same as the DPPH assay, distilled water, and 2 mM ascorbic acid were used as a negative and positive controls, respectively.
Acid-tolerance test
L. lactis subsp. lactis LM1185 was inoculated in MRS broth at 37 °C overnight. The seed culture was inoculated to another MRS broth and incubated at 37 °C for 16 h. The culture was centrifuged for 3 min at 15,920×g. The pellet was washed twice with sterile saline and resuspended in 100 mM glycine–HCl buffer (pH 2.5) followed by incubation at 37 °C for 2 h. The sample was spread on Petrifilm™ Lactic Acid Bacteria Count Plate (3 M™, St. Paul, MN, USA) and incubated at 37 °C overnight. The acid-tolerance (%) was calculated by the following equation:
| 3 |
The number of cells grown on the plate, which was spread immediately after inoculation, was used as a control.
Lacticaseibacillus rhamnosus GG (LGG) was provided by Lactomason (Jinju, Korea) and used as a positive control.
Bile-tolerance test
The strain LM1185 that was prepared in MRS broth at 37 °C overnight was inoculated in MRS broth and MRS broth containing 0.3% Difco™ Oxgall (BD BBL™) with the concentration of 1% (v/v). After incubation for 24 h at 37 °C, the sample was spread on Petrifilm™ Lactic Acid Bacteria Count Plate (3 M™) and also incubated at 37 °C overnight. The number of viable cells was counted, and bile-tolerance (%) was calculated using the equation below:
| 4 |
Cell lines and growth condition for the determination of immunostimulatory activity
RAW 264.7 macrophage cells (KCLB 40071) were obtained from the Korean Cell Line Bank (Seoul, Korea). Cells were cultured in high glucose Dulbecco’s Modified Eagle’s Medium (DEME; Gibco, Grand Island, NY, USA) containing 1% (v/v) penicillin–streptomycin (Gibco) and 10% (v/v) FBS (Gibco) at 37 °C with 5% CO2.
Preparation of lactic acid bacteria
L. lactis subsp. lactis LM1185 was cultured in MRS broth at 37 °C overnight. The culture broth was inoculated in MRS broth for 1% (v/v) concentration and incubated until its concentration reached 108 CFU/mL. The culture broth was centrifuged at 15,920×g for 1 min to obtain a concentration of 5 × 108 CFU/mL. The collected pellet was washed three times with Dulbecco’s Phosphate-Buffered Saline (DPBS; Welgene Inc., Gyeongsan, Korea). The washed pellet was resuspended in DMEM containing 1% penicillin–streptomycin and 10% FBS. The suspension was diluted to the required concentration and used.
Cell viability
Cell viability was determined using EZ-CYTOX (DoGenBio, Seoul, Korea). Briefly, RAW 264.7 cells were seeded in a 96-well plate with 5 × 104 cells/well and incubated at 37 °C in 5% CO2 for 20 h. L. lactis subsp. lactis LM1185 was prepared with the multiplicity of infection (MOI; number of lactic acid bacteria/number of macrophages) of 0.1, 1, 10, 100, and 500. After treatment of the sample, incubation was required for 24 h. Cells were washed three times with DPBS, and then DMEM and EZ-CYTOX were added at 200 μL and 20 μL, respectively. The reaction was ended after 20–30 min of incubation. The absorbance was measured at 450 nm with Epoch Microplate Spectrophotometer (Biotek, Santa Clara, CA, USA).
Nitric oxide assay
Cells were cultured in a 24-well plate with 5 × 105 cells/well. The cells were cultured at 37 °C in 5% CO2 for 20 h. The MOI of L. lactis subsp. lactis LM1185 was prepared with 50, 100, and 500. The macrophage cells treated with 1 μg/mL of lipopolysaccharides (LPS) were used as a positive control. After 24 h incubation, 100 μL of culture supernatant was transferred to a 96-well plate and Griess reagent, which mixed Griess A (0.1% N-1-naphtyl ethylene diamine, NEDHC) with Griess B (5% phosphoric acid containing 1% sulfanilamide) (1:1), was added. The mixture was incubated at 25 °C in the dark for 15 min, and the absorbance was measured at 540 nm using Epoch Microplate Spectrophotometer (Biotek). The absorbance was used to calculate the content of NO in the sample with standard curve obtained using sodium nitrite (Sigma-Aldrich) dissolved in DMEM in the range of 0–250 µM.
Cell culture for the quantitative analysis of immunostimulatory activity
RAW 264.7 cells (ATCC, Manassas, VA, USA), murine macrophage cell line, were cultured in Dulbecco’s Modified Eagle’s Medium (WELGENE Inc.), containing 10% fetal bovine serum (WELGENE Inc.) and 1% penicillin and streptomycin (WELGENE Inc.). The cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C. The cells were sub-cultured for 24 h prior to the addition of LPS and L. lactis subsp. lactis LM1185. Cells were then incubated with 10 ng/ml LPS or various concentrations of L. lactis subsp. lactis LM1185 (1 × 106, 1 × 107 cells/mL) for 24 h.
Real-time PCR (RT-qPCR)
To determine the expression of cytokines at the mRNA level, total RNA was extracted using MiniBEST Universal RNA Exraction Kit (Takara Bio Inc., Shiga, Japan). Next, cDNA was synthesized from total RNA (2 µg) using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA). qRT-PCR was performed using SYBR Green PCR master mix (Applied Biosystems) with a QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems). The oligonucleotide primers were as follows: TNF-α, forward 5′-ACAAGCCTGTAGCCCATGTT-3′ and reverse 5′-AAAGTAGACCTGCCCAGACT-3′; iNOS, forward 5′-CACCACCCTCCTTGTTCAAC-3′ and reverse 5′-CAATCCACAACTCGCTCCAA-3′; β-actin, forward 5′-ATTGCCGACAGGATGCAGAA-3′ and reverse 5′-AAGCATTTGCGGTGGACGAT-3′. The thermocycling conditions were as follows: 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s, followed by annealing and extension at 60 °C for 1 min. The fold changes of gene expression were calculated using the ΔΔCt method and normalized to that of β-actin in each sample.
Immunoblotting
The quantity of immune-related proteins was determined by automated capillary-based nanoimmunoassay using the Jess™ Simple Western system (ProteinSimple, Bio-Techne, Minneapolis, MN, USA) following the manufacturer’s standard method. Protein extracts (0.5 μg/μL) were mixed with 0.1 × sample buffer and fluorescent 5 × master mix to achieve a final concentration of 0.4 μg/μL in the presence of fluorescent molecular weight markers (ProteinSimple). Samples were denatured and equally loaded in 12–230 kDa Jess/Wes Separation Module. Primary antibodies were used for western blotting at a concentration of 40 μg/mL (1:25 dilution). Primary antibodies for western blotting included: rabbit-anti-α-tubulin (Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit-anti-Cox2 (Cell Signaling Technology, Inc.), rabbit-anti-NFκB p65 (Cell Signaling Technology, Inc.), rabbit-anti- pNFκB (Cell Signaling Technology, Inc.), and rabbit-anti-iNOS (GeneTex, Irvine, CA, USA). Next, HRP-conjugated anti-rabbit secondary antibody (ProteinSimple) was treated in Jess/Wes Separation Module, and HRP signal was visualized by using peroxyde/luminol-S (Bio-Techne). These chemiluminescent digital images in the capillary were evaluated automatically by Compass Simple Western software (version 4.1.0, Protein Simple) that calculated heights, area, and signal/noise ratio.
Antibiotics susceptibility test
The antibiotics susceptibility was examined by E-test. The colonies of the agar were inoculated in 2 mL 0.88%(w/v) NaCl and the turbidity was adjusted to match the turbidity of 1.0 McFarland standards (3 × 108 CFU/mL). The mixture was streaked on LSM agar plate composed of 90% Iso-Sensitest broth (Oxoid, Hampshire, UK) and 10% (w/v) MRS medium using a cotton swab and E-test strip (ETEST®, bioMérieux) was applied to the agar surface. The plates were incubated at 37 °C for 20 h. Minimum inhibitory concentration (MIC) values of the bacteria were determined by comparing the cut-off values of each antibiotic with the criteria recommended and given by European Food Safety Authority (ESFA) guidelines (EFSA, 2012).
Statistical analysis
Data were evaluated using GraphPad Prism software Version 5.0 (GraphPad Software, San Diego, CA, USA) and presented as box plots of at least three independent experiments. Statistical comparisons of vehicle controls versus treatment were performed with one-way ANOVA in conjunction with a Tukey’s multiple comparison test. Levels of significance are indicated as #p < 0.001, *p < 0.0001.
Results and discussion
Isolation and biochemical characteristics of lactic acid bacteria
To isolate LAB from hydrangea, three varieties of hydrangea including Morning Star, Pink Ari, and White Ari were used. When bacteria were isolated from the flower part and stem part of each plant, three LAB strains were isolated only in the flower part of White Ari, determined by Gram staining, 3% KOH test, and a catalase test. Among the isolated LAB, the strain LM1185 was selected as the strain with the highest immune activity through a preliminary experiment, and this strain was selected for this study.
Identification of lactic acid bacteria
The strain LM1185 was identified using the analysis of the nucleotide sequence of 16S rRNA gene using BLAST as well as a phylogenetic tree analysis. BLAST results indicated that strain LM1185 showed 100% identity with L. lactis strains. The prepared phylogenetic tree analysis confirmed that strain LM1185 belonged to L. lactis subsp. lactis as shown in Fig. S1. As a result, strain LM1185 was named L. lactis subsp. lactis LM1185. Lactococci are ovoid-shaped, present individually, in pairs, or in chains (Samaržija et al., 2001). The morphology of L. lactis subsp. lactis LM1185 is obtained by scanning electron microscopy as shown in Fig. S1. Lactococcus lactis subsp. lactis LM1185 has a size of ca. 1.0 µm in length and 0.5 µm in width as a form of paired ovoid. The carbohydrate utilization pattern of L. lactis subsp. lactis LM1185 was examined using API 50 CHL kit, and the result was shown in Table 1. Lactococcus lactis subsp. lactis LM1185 could utilize L-arabinose, D-mannitol, amygdalin, D-sucrose, starch, gentiobiose, and potassium gluconate, which could not be utilized by L. lactis subsp. lactis ATCC 19435, the type strain of L. lactis subsp. lactis.
Table 1.
Identification of LM1185 using API 50 CHL kit
| Tube | Active ingredients | ATCC 19435 | LM1185 | Tube | Active ingredients | ATCC 19435 | LM1185 |
|---|---|---|---|---|---|---|---|
| 0 | CONTROL | − | − | 25 | Esculin ferric citrate | + | + |
| 1 | Glycerol | − | − | 26 | Salicin | + | + |
| 2 | Erythritol | − | − | 27 | d-cellobiose | + | + |
| 3 | d-arabinose | − | − | 28 | d-maltose | + | + |
| 4 | l-arabinose | − | + | 29 | d-lactose (bovine origin) | + | + |
| 5 | d-ribose | + | + | 30 | d-melibiose | − | − |
| 6 | d-xylose | + | + | 31 | d-saccharose (sucrose) | − | + |
| 7 | l-xylose | − | − | 32 | d-trehalose | + | + |
| 8 | d-xylose | − | − | 33 | Inulin | − | − |
| 9 | Methyl-beta- d-xylopyranoside | − | − | 34 | d-melezitose | − | − |
| 10 | d-galactose | + | + | 35 | d-raffinose | − | - |
| 11 | d-glucose | + | + | 36 | Amidon (starch) | − | + |
| 12 | d-fructose | + | + | 37 | Glycogen | − | − |
| 13 | d-mannose | + | + | 38 | Xylitol | − | − |
| 14 | l-sorbose | − | − | 39 | Gentiobiose | − | + |
| 15 | l-rhamnose | − | − | 40 | d-turanose | − | − |
| 16 | Dulcitol | − | − | 41 | d-lyxose | − | − |
| 17 | Inositol | − | − | 42 | d-tagatose | − | − |
| 18 | d-mannitol | − | + | 43 | d-fucose | − | − |
| 19 | d-sorbitol | − | − | 44 | l-fucose | − | − |
| 20 | Methyl-alpha- d -mannopyranoside | − | − | 45 | d-arabitol | − | − |
| 21 | Methyl-alpha-d-glucopyranoside | − | − | 46 | l-arabitol | − | − |
| 22 | n-acetylglucosamine | + | + | 47 | Potassium gluconate | − | + |
| 23 | Amygdalin | − | + | 48 | Potassium 2-ketogluconate | − | − |
| 24 | Arbutin | + | + | 49 | Potassium 5-ketogluconate | − | − |
(+): turned to yellow (positive), expect for tube 25 which turned to black; (−): turned to violet
Lactococcus lactis subsp. lactis ATCC 19435 was used as a positive control
L. lactis subsp. lactis LM1185 was deposited in the Korean Culture Center of Microorganisms (Seoul, Korea) under the deposit number KCCM13146P.
Antioxidant activity
DPPH and ABTS radical scavenging activity were used to measure the antioxidant activity of L. lactis subsp. lactis LM1185. As shown in Table 2, Strain LM1185 showed high antioxidant activity, 58.3% and 99.4% of DPPH and ABTS radical scavenging activities, respectively. ABTS radical scavenging activity of L. lactis subsp. lactis LM1185 was higher than than of Lacticaseibacillus rhamnosus GG, although DPPH radical scavenging activity of L. lactis subsp. lactis LM1185 was lower than that of Lacticaseibacillus rhamnosus GG. Previous studies report that probiotics have a positive effect on oxidative stress. Reactive oxygen species (ROS), which includes free radicals and non-free radicals, could be generated by several factors, such as human metabolic reaction and exposure to oxidizing agents (Singh et al., 2022). An excessive increase in ROS results in an imbalance between generation and destruction of ROS and causes the bodily environment to be in a reducing state. As this reducing state is maintained, the peroxides and free radicals, which have toxicity causing cell damage or even death, were produced (Poljsak et al., 2013). To overcome these problems that lead to disease, studies of antioxidants were needed.
Table 2.
Antioxidant activity, acid- and bile-tolerance of Lactococcus lactis subsp. lactis LM1185
| Strain | Antioxidant activity (%) | Acid-tolerance (%) | Bile-tolerance (%) | |
|---|---|---|---|---|
| DPPH | ABTS | |||
| LM1185 | 58.3 ± 0.5 | 99.4 ± 0.0 | 50.5 ± 3.0 | 30.4 ± 2.2 |
| LGG | 80.4 ± 0.5 | 87.8 ± 0.5 | 58.0 ± 4.1 | 89.2 ± 1.3 |
Data are presented as mean ± SD of three independent experiments
Levilactobacillus brevis KU15153 and Enterococcus faecium MR-Pro3 showed 44.14% and 67.75% of DPPH antioxidant activities, respectively (Jang et al., 2019; Rezaei et al., 2020). Kunduhoglu and Hacioglu (2021) also reported Levilactobacillus brevis KT16-2 showed 71.0% and 54.1% DPPH and ABTS radical scavenging activities, respectively. Based on the studies on the antioxidant activity of LAB, the antioxidant activity of L. lactis subsp. lactis LM1185 was compatible with that of other LAB strains, and this high antioxidant activity could be used to improve the imbalance of ROS and reduce oxidative stress by utilizing as probiotics or applying them to food.
Acid/bile tolerance test
Acid and bile tolerances of L. lactis subsp. Lactis LM1185 were checked, and the result is shown in Table 2. Acid tolerance of stain LM1185 was similar to Lacticaseibacillus rhamnosus GG, a representative probiotic strain. However, the bile tolerance of stain LM1185 was lower than that of Lacticaseibacillus rhamnosus GG.
The acid and bile tolerances of LAB were indispensable characteristics of probiotics. This is because probiotics were consumed by the host, it was attacked by the physiological condition of the gastrointestinal tract, such as the acidic environment of the stomach and the secretion of bile acid in the duodenal section of the small intestine (Kimoto-Nira et al., 2015; Sahadeva et al., 2011). This tolerance could help the bacteria to enter the intestinal phase, and the survival probiotics could confer some health benefits, such as keeping the microbiome healthy (Sahadeva et al., 2011).
The mechanisms of bile on cell membrane were reported that membrane lipids were dissolved by bile, and the integral proteins were dissociated with the high concentration of bile (Begley et al., 2005). To solve this problem, Tween 80 and the change in the type of carbohydrates in the culture medium could be the key, but further studies are needed to determine correlation (Kimoto-Nira et al., 2015). In addition to these methods, several methods had been proposed, but microencapsulation was getting attention. Microencapsulation was widely used to enhance the viability of probiotics by protecting them from the critical environment (Vidyalakshmi et al., 2009). Hou et al. (2003) used sesame oil emulsion as the material of encapsulation, and it showed an increase of 4 log in the viability in acidic conditions as well as in the presence of bile.
Immunostimulatory effect of L. lactis subsp. lactis LM1185
To assess the cytotoxicity, the EZ-CYTOX method was used on RAW 264.7 macrophage cells treated with L. lactis subsp. lactis LM1185. As shown in Fig. 1A, there was no inhibition of cell growth caused by the treatment of L. lactis subsp. lactis LM1185 in all MOI, which showed there was no cytotoxicity of L. lactis subsp. lactis LM1185 on RAW 264.7 macrophage cells.
Fig. 1.
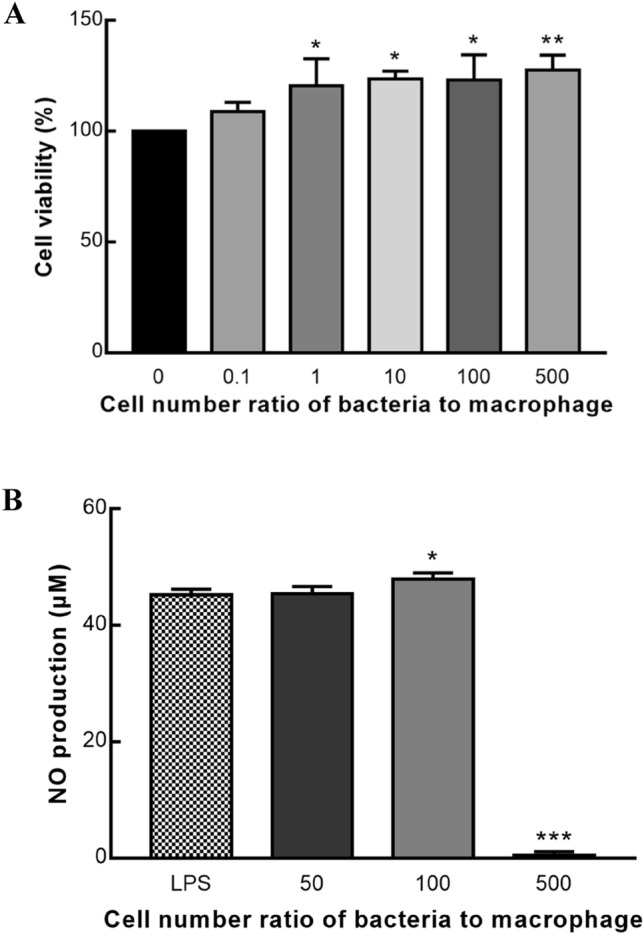
Cytotoxicity of Lactococcus lactis subsp. lactis LM1185 on RAW 264.7 cells and Effect of Lactococcus lactis subsp. lactis LM1185 on NO production in the RAW 264.7 cells. A Cytotoxicity of Lactococcus lactis subsp. lactis LM1185 on RAW 264.7 cells. Raw 264.7 cells (5 × 104 cells/well) were treated with different cell number of L. lactis subsp. lactis LM1185. The results showed that all MOI of L. lactis subsp. lactis LM1185 had no cytotoxicity on RAW 264.7 cells. Data are presented as mean ± SD of three independent experiments. Noted asterisks indicate statistical significance (*P < 0.05, **P < 0.01). B Effect of Lactococcus lactis subsp. lactis LM1185 on NO production in the RAW 264.7 cells. RAW 264.7 cells were cultured with 5 × 105 cells/well. RAW 264.7 cells treated with 1 μg/mL of LPS or L. lactis subsp. lactis LM1185. When RAW 264.7 cells treated with 5 × 107 cells of L. lactis subsp. lactis LM1185, the highest NO production was detected. Asterisks indicate statistical significance (*P < 0.05, *** P < 0.001)
When the human system is attacked by the pathogen, macrophages are activated and secrete various inflammatory mediators, such as NO and cytokines. NO is a short-lived radical synthesized by the oxidation of L-arginine induced by nitric oxide synthase (NOS) (Liu et al., 2006; Yang et al., 2009). There are three isoforms of NOS including neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS), which are named based on the dependence of Ca2+ calmodulin pathway (Kang et al., 2019; Liu et al., 2006). Among them, iNOS can synthesize a large amount of NO, which was known for its cytotoxicity to the infectious agents, pathogenic microorganisms, and tumor cells (Wang et al., 2018).
When LPS and strain LM1185 were used to induce the nitric oxide (NO) production of RAW 264.7 macrophages. The highest NO production of 47.9% was recorded when the macrophages were treated with MOI 100. The NO production at MOI 50 had no significant differences comparing LPS-treated cells (Fig. 1B).
Lipopolysaccharides are a component of the cell wall of gram-negative bacteria that cause an inflammatory response against invading microorganisms and pathogens by causing macrophages and monocytes to produce pro-inflammatory cytokines (Limtrakul et al., 2015). In the present study, the increasing production of NO by treatment of strain LM1185 was induced at MOI 100, and treatment of strain LM1185 could be considered to give a positive immune response to the macrophage cells and thereby have high immunostimulatory activity.
Analysis of immunostimulatory activity by real-time quantitative PCR (RT-qPCR)
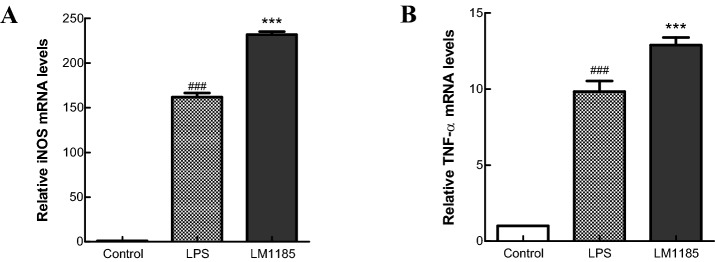
The mechanism of immunostimulatory activity of L. lactis subsp. lactis LM1185 was elucidated by analyzing the mRNA expression levels of tumor necrosis factor-alpha (TNF-α) and iNOS genes using RT-qPCR. Expression levels of TNF-α and iNOS genes in the cells treated with strain LM1185 were higher than in the cells treated with LPS alone (Fig. 2). TNF-α secreted by the activated macrophages and mast cells is a kind of proinflammatory cytokine (Delgado et al., 2003). iNOS associated with the synthesis of NO highly expressed by the activation of nuclear factor-kappa B (NF-κB), which is simulated by TNF- α, interferon-gamma (IFN-γ), interleukin-6 (IL-6), interleukin-1alpha (IL-1α), LPS, and bacterial and viral components (Soufli et al., 2016). This observation indicated that L. lactis subsp. lactis LM1185 had an important role as the immune-enhancing mediator in RAW 264.7 cells.
Fig. 2.
Effect of Lactococcus lactis subsp. lactis LM1185 on the mRNA expression of TNF-α and iNOS gene in the RAW 264.7 cells. RAW 264.7 cells (1 × 108 cells/well) were cultured for 24 h in medium with 10 ng/mL LPS (10 ng/mL) L. lactis subsp. lactis LM1185 (1 × 106 cells/mL). A TNF-α and B iNOS relative mRNA expression levels were evaluated by reverse transcription-quantitative polymerase chain reaction. Values are expressed as the mean ± SD of three independent experiments. A, B #P < 0.01 vs. control; *P < 0.001 vs. LPS
Analysis of immunostimulatory activity by immunoblotting
The activation of NF-κB/iNOS/COX-2 signaling pathway by treatment of L. lactis subsp. lactis LM1185 on macrophage cells was assessed by measuring the increase in protein expression levels via western blot analysis. RAW 264.7 cells were treated with 1 × 106 and 1 × 107 cells/mL of strain LM1185 or LPS for 24 h. As shown in Fig. 3, NF-κB activation and iNOS/COX-2 expression were both significantly increased by treatment of strain LM1185 in a concentration-dependent manner. Thus, L. lactis subsp. lactis LM1185 could promote the NF-κB signaling pathway with phosphorylation of NF-κB leading to produce pro-inflammatory mediator expressions such as iNOS and cyclooxygenase-2 (COX-2), which played a crucial role in the activation of innate immunity and inflammation (Fig. 4).
Fig. 3.
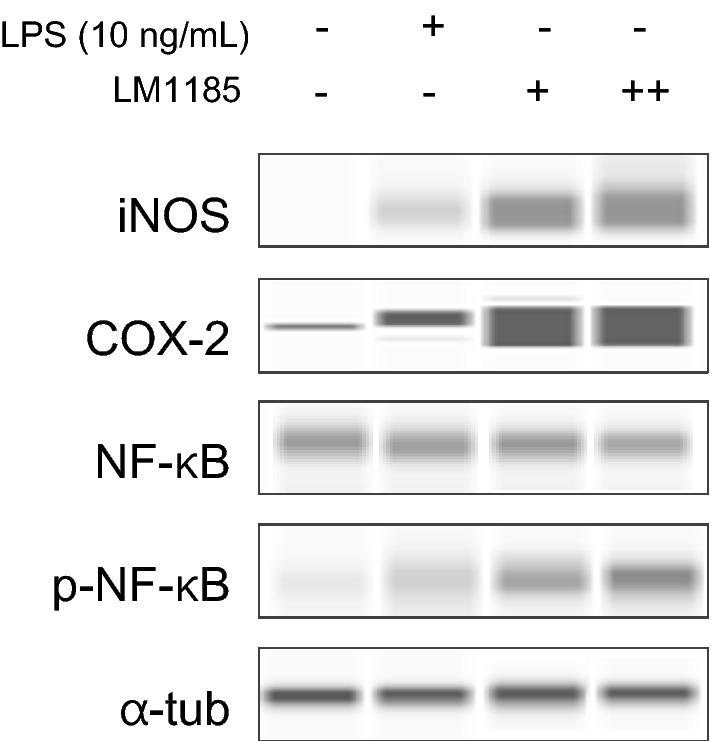
Concentration-dependent effect of Lactococcus lactis subsp. lactis LM1185 on pro-inflammatory factors (iNOS, COX-2) expression and NF-κB activation in the RAW 264.7 cells. RAW 264.7 cells (1 × 108 cell per well) were cultured for 24 h in medium with LPS (10 ng/mL) or L. lactis subsp. lactis LM1185 (+ ;1 × 106, ++;1 × 107 cells/mL). The expression of pro-inflammatory factors (iNOS, COX-2) and NFκB activation were measured using western blot analysis. The normalized iNOS, COX-2, and NFκB protein intensity with α-tubulin was acquired using Compass Simple Western software to determine the fold induction
Fig. 4.
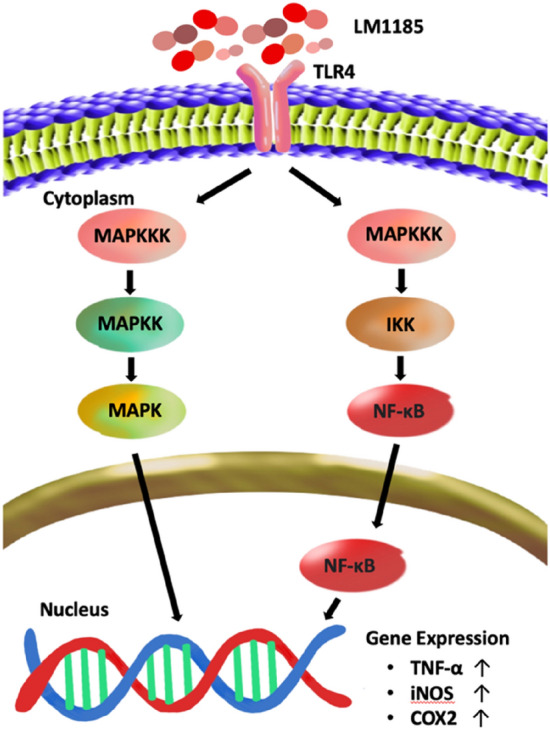
Diagram depicting the proposed immunostimulatory signaling mechanism of Lactococcus lactis subsp. lactis LM1185 in RAW 264.7 macrophages. LM1185, Lactococcus lactis subsp. lactis LM1185; TLR4, Toll-like receptor 4; MAPKKK, mitogen-activated protein kinase kinase kinase; MAPKK, mitogen-activated protein kinase kinase; MAPK, mitogen-activated protein kinase; IKK, IκB kinase; NF-κB, nuclear factor-kappa B; TNF-α, tumor necrosis factor-alpha; iNOS, inducible nitric oxide synthase; COX2, cyclooxygenase-2
NF-κB is a nuclear protein, first discovered by Sen and Baltimore (1986) as an expression regulator of the kappa light chain of immunoglobulin in the B-lymphocytes (Sen and Baltimore, 1986; Serasanambati and Chilakapati, 2016). In canonical NF-κB pathway, NF-κB dimer consisting of p50 and RelA, known as p65, is maintained in the cytoplasm in inactive form while interacting with inhibitor, IκB (especially IκBα) (Gilmore, 2006; Serasanambati and Chilakapati, 2016). When macrophages are activated by LPS, the toll-like receptor 4 (TLR4) signaling pathway that involves the IκB kinase (IKK) mediated-phosphorylation of NF-κB will be initiated resulting in the degradation of IκB inhibitor. The activated NF-κB binds to promoters to transcribe genes associated with inflammatory responses and inhibits apoptosis, which in turn induces the production of iNOS, COX-2, TNF-α, IL-1β, and IL-6 (Lee et al., 2016; Serasanambati and Chilakapati, 2016).
Antibiotics susceptibility
When the antibiotics susceptibility of L. lactis subsp. lactis LM1185 was checked for nine antibiotics using E-test, the MIC of all antibiotics was below the cut-off value, which meant the antibiotics susceptibility of L. paracasei EPS DA-BACS was acceptable (Table S1).
L. lactis strains are usually susceptible to vancomycin and erythromycin (Toomey et al. 2010). It is reported that Lactococcus sp. are frequently resistance to chloramphenicol, streptomycin, and tetracycline (Perreten et al., 1997). However, L. lactis subsp. lactis LM1185 had no resistance to all antibiotics examined in this study.
In conclusion, this study was conducted to isolate lactic acid bacteria from Hydrangea macrophylla var. White Ari and evaluate the probiotic properties. The isolated strain, L. lactis subsp. lactis LM1185 showed the possibility to be potential probiotics, with remarkable properties such as remarkable antioxidant and immunostimulatory activities. In RAW 264.7 macrophage cells, no cytotoxicity was observed, and increasing NO production at a specific concentration was measured. In the protein level, when RAW 264.7 cells were treated with L. lactis subsp. lactis LM1185, the mRNA expression of TNF-⍺ and iNOS was increased, and NF-κB signaling pathway was activated resulting in an increase of pro-inflammatory proteins such as iNOS and COX-2. Despite these positive results, L. lactis subsp. lactis LM1185 should be further studied because of its low tolerance to bile salt. LM1185 could survive in an acidic environment but showed low viability in the presence of bile. In addition, the effects of this strain were demonstrated using in vitro experiments, so further in vivo analysis will be required.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Jeonnam Agricultural Research & Extension Services (JNARES) for the collaboration and supply of Hydrangea (three varieties: Morning Star, Pink Ari, and White Ari).
Declarations
Competing interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yoonjeong Cho and Hyeon Tak Han have contributed equally to this work.
Contributor Information
Yoonjeong Cho, Email: whdbswjd505@gmail.com.
Hyeon Tak Han, Email: hthan@lactomason.com.
Tae-rahk Kim, Email: trkim@lactomason.com.
Minn Sohn, Email: ms@lactomason.com.
Young-Seo Park, Email: ypark@gachon.ac.kr.
References
- Abushelaibi A, Almahadin S, El-Tarabily K, Shah N, Ayyash M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT - Food Science and Technology. 2017;79:316–325. doi: 10.1016/j.lwt.2017.01.041. [DOI] [Google Scholar]
- Bag A, Bhattacharyya S, Chattopadhyay R. Medicinal plants and urinary tract infections: An update. Pharmacognosy Reviews. 2008;2:277–284. [Google Scholar]
- Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiology Reviews. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Delgado AV, McManus AT, Chambers JP. Production of Tumor Necrosis Factor-alpha, Interleukin 1-beta, Interleukin 2, and Interleukin 6 by rat leukocyte subpopulations after exposure to Substance P. Neuropeptides. 2003;37:355–361. doi: 10.1016/j.npep.2003.09.005. [DOI] [PubMed] [Google Scholar]
- EFSA Panel on Additives Products or Substances used in Animal Feed Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal. 2012;10:2740. [Google Scholar]
- Geum NG, Eo HJ, Kim HJ, Park GH, Son HJ, Jeong JB. Immune-enhancing activity of Hydrangea macrophylla subsp. serrata leaves through TLR4/ROS-dependent activation of JNK and NF-κB in RAW2647 cells and immunosuppressed mice. Journal of Functional Foods. 2020;73:104139. doi: 10.1016/j.jff.2020.104139. [DOI] [Google Scholar]
- Gilmore TD. Introduction to NF-κB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- Hou RCW, Lin MY, Wang MMC, Tzen JTC. Increase of Viability of Entrapped Cells of Lactobacillus delbrueckii ssp. bulgaricus in Artificial Sesame Oil Emulsions. Journal of Dairy Science. 2003;86(2):424–428. doi: 10.3168/jds.S0022-0302(03)73620-0. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Lee N-K, Paik H-D. Probiotic characterization of Lactobacillus brevis KU15153 showing antimicrobial and antioxidant effect isolated from kimchi. Food Science and Biotechnology. 2019;28:1521–1528. doi: 10.1007/s10068-019-00576-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C-H, Han SH, Kim J-S, Kim Y, Jeong Y, Park HM, Paek N-S. Inhibition of nitric oxide production, oxidative stress prevention, and probiotic activity of lactic acid bacteria isolated from the human vagina and fermented food. Microorganisms. 2019;7:109. doi: 10.3390/microorganisms7040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaing MT, Jung H-J, Han T-H. Trend of hydrangea cultivar development. Trends in Agriculture & Life Sciences. 2016;53:63–68. doi: 10.29335/tals.2016.53.63. [DOI] [Google Scholar]
- Kimoto-Nira H, Suzuki S, Suganuma H, Moriya N, Suzuki C. Growth characteristics of Lactobacillus brevis KB290 in the presence of bile. Anaerobe. 2015;35:96–101. doi: 10.1016/j.anaerobe.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Kunduhoglu Buket, Hacioglu Seda. Probiotic Potential and Gluten Hydrolysis Activity of Lactobacillus brevis KT16-2. Probiotics and Antimicrobial Proteins. 2021;13(3):720–733. doi: 10.1007/s12602-020-09723-x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee J, Ha SJ, Lee HJ, Kim MJ, Kim JH, Kim YT, Song K-M, Kim Y-J, Kim HK, Jung SK. Protective effect of Tremella fuciformis Berk extract on LPS-induced acute inflammation via inhibition of the NF-κB and MAPK pathways. Food & Function. 2016;7:3263–3272. doi: 10.1039/C6FO00540C. [DOI] [PubMed] [Google Scholar]
- Li Y, Mmbaga MT, Zhou B, Joshua J, Rotich E, Parikh L. Diseases of hydrangea. In: McGovern RJ, Elmer WH, editors. Handbook of florists’ crops diseases. Cham: Springer; 2018. pp. 987–1006. [Google Scholar]
- Limtrakul P, Yodkeeree S, Pitchakarn P, Punfa W. Suppression of inflammatory responses by black rice extract in RAW 264.7 macrophage cells via downregulation of NF-kB and AP-1 signaling pathways. Asian Pacific Journal of Cancer Prevention. 2015;16:4277–4283. doi: 10.7314/APJCP.2015.16.10.4277. [DOI] [PubMed] [Google Scholar]
- Liu K-L, Chen H-W, Wang R-Y, Lei Y-P, Sheen L-Y, Lii C-K. DATS reduces LPS-induced iNOS expression, NO production, oxidative stress, and NF-κB activation in RAW 264.7 macrophages. Journal of Agricultural and Food Chemistry. 2006;54:3472–3478. doi: 10.1021/jf060043k. [DOI] [PubMed] [Google Scholar]
- Masood MI, Qadir MI, Shirazi JH, Khan IU. Beneficial effects of lactic acid bacteria on human beings. Critical Reviews in Microbiology. 2011;37:91–98. doi: 10.3109/1040841X.2010.536522. [DOI] [PubMed] [Google Scholar]
- Mmbaga MT, Kim M-S, Mackasmiel L, Li Y. Evaluation of Hydrangea macrophylla for resistance to leaf-spot diseases. Journal of Phytopathology. 2011;160:88–97. doi: 10.1111/j.1439-0434.2011.01862.x. [DOI] [Google Scholar]
- Mota-Gutierrez J, Cocolin L. Current trends and applications of plant origin lactobacilli in the promotion of sustainable food systems. Trends in Food Science & Technology. 2021;114:198–211. doi: 10.1016/j.tifs.2021.05.030. [DOI] [Google Scholar]
- Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Antibiotic resistance spread in food. Nature. 1997;389:801–802. doi: 10.1038/39767. [DOI] [PubMed] [Google Scholar]
- Poljsak B, Šuput D, Milisav I. Achieving the balance between ROS and antioxidants: When to use the synthetic antioxidants. Oxidative Medicine and Cellular Longevity. 2013;2013:956792. doi: 10.1155/2013/956792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinto E, Jiménez P, Caro I, Tejero J, Mateo J, Girbés T. Probiotic lactic acid bacteria: A review. Food and Nutrition Sciences. 2014;5:1765–1775. doi: 10.4236/fns.2014.518190. [DOI] [Google Scholar]
- Rezaei M, Noori N, Shariatifar N, Gandomi H, Basti AA, Khaneghah AM. Isolation of lactic acid probiotic strains from Iranian camel milk: Technological and antioxidant properties. LWT. 2020;132:109823. doi: 10.1016/j.lwt.2020.109823. [DOI] [Google Scholar]
- Sahadeva R, Leong S, Chua K, Tan C, Chan H, Tong E, Wong S, Chan H. Survival of commercial probiotic strains to pH and bile. International Food Research Journal. 2011;18:1515–1522. [Google Scholar]
- Samaržija D, Antunac N, Havranek JL. Taxonomy, physiology and growth of Lactococcus lactis: A review. Mljekarstvo. 2001;51:35–48. [Google Scholar]
- Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-X. [DOI] [PubMed] [Google Scholar]
- Serasanambati M, Chilakapati SR. Function of nuclear factor kappa B (NF-kB) in human diseases-A review. South Indian Journal of Biological Sciences. 2016;2:368–387. doi: 10.22205/sijbs/2016/v2/i4/103443. [DOI] [Google Scholar]
- Singh V, Ahlawat S, Mohan H, Gill SS, Sharma KK. Balancing reactive oxygen species generation by rebooting gut microbiota. Journal of Applied Microbiology. 2022;132:4112–4129. doi: 10.1111/jam.15504. [DOI] [PubMed] [Google Scholar]
- Soufli I, Toumi R, Rafa H, Touil-Boukoffa C. Overview of cytokines and nitric oxide involvement in immuno-pathogenesis of inflammatory bowel diseases. World Journal of Gastrointestinal Pharmacology and Therapeutics. 2016;7:353–360. doi: 10.4292/wjgpt.v7.i3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidyalakshmi R, Bhakyaraj R, Subhasree RS. Encapsulation "the future of probiotics"-A review. Advances in Biological Research. 2009;3:96–103. [Google Scholar]
- Wang J, Wu T, Fang X, Min W, Yang Z. Characterization and immunomodulatory activity of an exopolysaccharide produced by Lactobacillus plantarum JLK0142 isolated from fermented dairy tofu. International Journal of Biological Macromolecules. 2018;115:985–993. doi: 10.1016/j.ijbiomac.2018.04.099. [DOI] [PubMed] [Google Scholar]
- Yang E-J, Yim E-Y, Song G, Kim G-O, Hyun C-G. Inhibition of nitric oxide production in lipopolysaccharide-activated RAW 264.7 macrophages by Jeju plant extracts. Interdisciplinary Toxicology. 2009;2:245–249. doi: 10.2478/v10102-009-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga M, Pardo I, Ferrer S. An improved medium for distinguishing between homofermentative and heterofermentative lactic acid bacteria. International Journal of Food Microbiology. 1993;18:37–42. doi: 10.1016/0168-1605(93)90005-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.