Abstract
In our previous study, black raspberry (BR) reduced the serum levels of trimethylamine-N-oxide and cholesterol in rats fed excessive choline with a high-fat diet (HFC). We hypothesized that gut microbiota could play a crucial role in the production of trimethylamine and microbial metabolites, and BR could influence gut microbial composition. This study aimed to elucidate the role of BR on changes in gut microbiota and microbial metabolites in the rats. The phylogenetic diversity of gut microbiota was reduced in the rats fed HFC, while that in the BR-fed group was restored. The BR supplementation enriched Bifidobacterium and reduced Clostridium cluster XIVa. In the BR-fed group, most cecal bile acids and hippuric acid increased, while serum lithocholic acid was reduced. The BR supplementation upregulated Cyp7a1 and downregulated Srebf2. These results suggest that BR extract may change gut bacterial community, modulate bile acids, and regulate gene expression toward reducing cholesterol.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-023-01267-4.
Keywords: Rubus occidentalis, Choline, TMAO, Gut microbiome, Bile acid, CYP7A1
Introduction
Dietary intake is a well-known factor affecting gut microbiota and metabolic phenotype (Cotillard et al., 2013; Meslier et al., 2020). Various studies have reported that not only macronutrients but also bioactive compounds can modulate the gut bacterial community by changing its richness and composition, which can affect host metabolism and health in chronic diseases (Cotillard et al., 2013). Therefore, researchers now consider the gut microbiota as a therapeutic target, thus trying to find some new natural and bioactive materials with a prebiotic-like activity from dietary interventions in both preclinical and clinical models (Luo et al., 2021).
Rubus occidentalis (black raspberry, BR) is relatively high in polyphenols and anthocyanins compared to other Rubus fruits (Jung et al., 2015; Moyer et al., 2002). The consumption of plant foods is known to be beneficial for cardiovascular health due to their antioxidant activity (Jeong et al., 2014). However, major bioactive polyphenols in BR, such as anthocyanins and ellagitannins, were reported to be relatively low in bioavailability (de Ferrars et al., 2014). Their intact form and metabolites are known to be rarely detected in peripheral organs and circulation system (de Ferrars et al., 2014). Therefore, polyphenols in berry fruits and gut microbiota are considered to be highly interacted, which can affect host health. Few studies have reported the modulating effect of whole berry or a fraction of BR on gut microbial composition so far (Gu et al., 2019; Lee et al., 2019).
Excessive choline intake has been linked to cardiovascular diseases (CVD) due to the production of trimethylamine-N-oxide (TMAO), a putative promoter of CVD (Romano et al., 2015; Wang et al., 2011). Some species of gut bacteria are capable of converting specific trimethylamine (TMA)-containing molecules, such as choline, l-carnitine, and betaine, into TMA through the action of TMA lyase (Romano et al., 2015; Wang et al., 2011; Zhu et al., 2016). It is thus of utmost importance to determine the dietary effect on gut microbial composition to potentially prevent or alleviate the development of CVD.
In addition to TMA, a precursor of TMAO, microbial metabolites such as secondary bile acids (BAs) and short-chain fatty acids (SCFAs) can also affect the pathogenesis of CVD (Ding et al., 2018; Poll et al., 2020; Wahlström et al., 2016). Primary BAs produced in hepatocytes are modified into secondary BAs by gut microbiota (Winston and Theriot, 2020). BAs regulate cholesterol metabolism and their own synthesis as hormones do via the gut-BAs-host axis (Winston and Theriot, 2020). When sufficient BAs bind to ileal farnesoid X receptor (FXR; Nr1h4), BA synthesis from cholesterol is repressed by inhibiting hepatic cholesterol 7α-hydroxylase (CYP7A1) (Winston and Theriot, 2020). However, the affinity of BAs toward FXR is varied depending on their structural characteristics (Winston and Theriot, 2020). Thus, BA profile and gut microbiota are highly associated with serum cholesterol levels (Huang et al., 2019; Winston and Theriot, 2020). SCFAs produced by gut microbiota, such as acetate, propionate, and butyrate, can also influence gene expression at peripheral organs and regulate metabolism and inflammatory response (Poll et al., 2020). Therefore, the connection between microbial metabolites and the pathogenesis of chronic diseases has recently become more understood.
Our previous study reported that high-choline intake increased serum cholesterol levels, one of the CVD risk factors, in rats fed a high-fat diet, and intake of BR extract could lower the levels of cecal TMA and serum TMAO as well as serum cholesterol (Lim et al., 2020). Based on the results of the previous study, we hypothesized that consistent intake of BR extract could improve the serum lipid profile and inflammatory biomarkers through prebiotic-like activity. It was because (1) major bioactive compounds in BR have been known to be hardly absorbed into the circulation system; and (2) the supplementation of BR could reduce the TMA level in the gut. Therefore, to comprehensively elucidate the role of BR in rats with elevated levels of serum TMAO and cholesterol, this study aimed to evaluate the effect of BR extract on gut microbiota, microbial metabolites, and cholesterol and BA metabolisms in rats fed excessive choline with a high-fat diet.
Materials and methods
Materials and reagents
Anti-β-actin and anti-CYP7A1 antibodies were purchased from Abcam (Cambridge, England); and anti-flavin monooxygenase 3 (FMO3) antibody was from Proteintech Group (Rosemont, IL, USA). Horseradish peroxidase-linked anti-rabbit immunoglobulin G was purchased from Cell Signaling Technology (Danvers, MA, USA). Enhanced chemiluminescence (ECL) solution was purchased from GenDEPOT (Katy, TX, USA). α-Muricholic acid (α-MCA), β-MCA, ω-MCA, tauro-α-MCA (Tα-MCA), tauro-β-MCA(Tβ-MCA), and glycine-β-MCA (Gβ-MCA) were purchased from Cayman Chemical (Ann Arbor, MI, USA). All the other standards were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Preparation of BR extract powder
BR fruits harvested in Gochang (Korea) were purchased and extracted to obtain the powder form of BR extract as previously described (Lim et al., 2020). Briefly, BR extract was filtered and lyophilized for further use after 1 h extraction with 80% (v/v) aqueous ethanol solution. The extracted BR powder mainly consisted of carbohydrates (approximately 70% of the powder) with bioactive compounds including phenolics and anthocyanins (Shaddel et al., 2018). The solvent was chosen based on the content of total phenolics (42.7 ± 6.9 mg gallic acid equivalent/g) and anthocyanins in the powder (Lim et al., 2020). Major anthocyanins detected in the powder were cyanidin-3-rutinoside (3.49 ± 0.13 μmol/g), cyanidin-3-xylosylrutinoside (1.39 ± 0.08 μmol C3R equivalent/g), and cyanidin-3-glucoside (1.03 ± 0.02 μmol/g) (Lim et al., 2020).
Animals and diets
Serum, liver, and cecal content were obtained from our previous animal experiment, and the experimental design was described in our previous publication (Lim et al., 2020). Briefly, after 1 week of acclimation, female Sprague–Dawley rats (initial body weight: 136.7 ± 7.4 g) were randomly divided into four groups (n = 10 each) and housed two per cage: CON, fed AIN-93G diet; HF, fed 45% high-fat diet; HFC, fed high-fat diet with water containing 1.5% (w/w) choline chloride (Jinan Pengbo Biotechnology Co., Ltd., Jinan, China); and HFCB, fed high-fat diet supplemented with 0.6% BR extract powder and water containing 1.5% (w/w) choline chloride. All the groups were fed experimental diets and water ad libitum for 8 weeks. Water was replaced every 2 days. All animal experiments were conducted according to the ARRIVE guidelines and all protocols were approved by the Institutional Animal Care and Use Committee of Seoul National University (Approval No.: SNU-171103-1-5). All the rats were fasted for 6 h and were euthanized by asphyxiation with CO2.
16S ribosomal RNA (rRNA) gene sequencing
Total bacterial genomic DNA (gDNA) was extracted from cecal content using QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction with some modifications. Briefly, about 100 mg of cecal content was added into 0.7 mL InhibitEX buffer containing sterilized 0.1 mm glass beads (Scientific Industries, Inc., Bohemia, NY, USA) and homogenized using TissueLyser (Qiagen) at 30 Hz for 5 min and another 5 min. The homogenated soup was then heated at 95 °C for 5 min. After the loading and washing steps, the gDNA was incubated with 50 μL ATE buffer for 5 min and eluted by centrifugation (16,000×g, 1 min).
The V3 and V4 hypervariable regions of the 16S rRNA gene were amplified with TaKaRa Ex Taq DNA Polymerase (Takara Bio, Otsu, Shiga, Japan) and universal primers (341F/805R) with overhang adapter attached, followed by AMPure XP bead (Beckman Coulter, Brea, CA, USA) cleanup. The amplicon was quantified by Qubit 3.0 System (Thermo Fisher Scientific, Hampton, NH, USA). The Miseq libraries were prepared using Illumina Nextera XT Index Kit (Illumina Inc.) according to the manufacturer’s instruction. Paired-end sequencing (2 × 300 bp) was performed on Illumina Miseq Platform (Illumina Inc., San Diego, CA, USA).
Bioinformatics analysis
Quantitative Insights Into Microbial Ecology 2 (QIIME2, v 2020.2) software (Bolyen et al., 2019) was used to analyze the raw data obtained from 16S rRNA gene sequencing. DADA2 plugin (Callahan et al., 2016) in QIIME2 was used to merge and filter the sequence data. Forward and reverse sequences were trimmed (--p-trim-left-f 17 and --p-trim-left-r 21, respectively), and the parameters for truncation was set at 280 bp and 220 bp (--p-trunc-len-f 280 and --p-trunc-len-r 220, respectively) to get rid of reads having low quality. For α- and β-diversity analyses, the feature table was rarefied not to exclude any individuals by subsampling randomly (15,883 reads). Taxonomic analysis was conducted by a pre-trained classifier based on Greengenes 13_8 99% operational taxonomic units (OTUs).
BA profile and hippuric acid concentration
To analyze the BA profile (in cecum and serum) and hippuric acid (in cecum), UPLC SYNAPT G2-Si Q-TOF mass spectrometer (Waters Co., Milford, MA, USA) was used. Each of the cecal content (about 40 mg) and serum samples (100 μL) was immersed in 80% (v/v) ice-cold methanol (800 μL and 400 μL, respectively), vortexed for 5 min and 1 min, respectively, and then centrifuged at 12,000×g for 5 min at 4 °C. The supernatant was filtered using a 0.22 μm syringe filter (Pall Co., Port Washington, NY, USA), and the filtrate was concentrated only for the cecal samples by centrifugation at 15,000×g for 25 min at 4 °C in a Vivaspin centrifugal concentrator (Vivaspin 500, MWCO 3000, VS0192; Sartorius Stedim Lab, Stonehouse, UK).
Sixteen BAs and hippuric acid (Table S1) were separated using an Acquity UPLC system (Waters Co.) equipped with an HSS T3 column (2.1 mm × 100 mm, 1.7 μm, Waters Co.) heated at 50 °C. The mobile phase consisted of two eluents: water with 0.1% (v/v) formic acid (A) and acetonitrile with 0.1% (v/v) formic acid (B). The flow rate was set at 0.4 mL/min, and the gradient was as follows: 0–2.5 min, 15% (B); 9 min, 95% (B); 9–13 min, 95% (B); 13.1 min, 95% (B); and 13.1–15 min 15% (B). BAs were detected by SYNAPT G2-Si Q-TOF mass spectrometer (Waters Co.) in negative ionization mode. Mass parameters were set as follows: Tof-MRM mode; capillary voltage, 2.5 kV; sampling cone voltage, 25 V; desolvation gas, 600 L/h; cone gas, 50 L/h; and desolvation temperature, 400 °C. The ion transitions used for quantitation are shown in Supplementary Material (Table S1). Data quantitation was performed using MassLynx software 4.1 (Waters Co.).
Cecal SCFA composition
SCFA concentrations in the cecum were determined by gas chromatography (GC) equipped with a Nukol fused silica capillary column (30 m × 0.25 mm, 0.25 μm) and a flame ionization detector. Cecal content (about 100 mg) was homogenized with 1 mL of distilled water. Sulfuric acid (20 μL) was then added into the homogenate and remained for 5 min at room temperature. The mixture was centrifuged at 13,000×g for 5 min, and 500 μL of the supernatant was mixed with the same volume of diethyl ether. The mixture was vortexed for 1 min and centrifuged for 13,000×g for 5 min. The organic phase was collected and transferred into a vial for GC analysis. GC analysis was performed as follows: Oven temperature was held at 170 °C, and the injector and detector temperatures were 225 °C. The injected volume was 2 μL, and the split ratio was 100:1.
Real-time quantitative polymerase chain reaction (qPCR) analysis
RNA extraction and cDNA synthesis were performed according to the method in our previous paper (Lim et al., 2020). qPCR was performed using Applied Biosystems StepOne Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with SYBR Green PCR Master Mix (Applied Biosystems) with 2 min denaturation at 95 °C followed by 40 cycles of 15 s at 95 °C and 60 s at 58 °C. All primer sequences used in this study are listed in Table S2. All the gene expressions were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression.
Western blot analysis
Liver tissue was added to a mixture of RIPA buffer and PIC #6 at a ratio of 100:1 and homogenized using TissueLyser (DE/85220, Qiagen). The homogenized liver samples were agitated at 4 °C for 1 h, followed by centrifugation (12,000×g, 4 °C, 30 min). The supernatants were mixed with 5× loading buffer (Biosesang, Seongnam, Korea) and heated at 100 °C for 15 min. The protein samples were loaded into Mini-protein TGX gel 10% (Bio-Rad Laboratories Inc., Hercules, CA, USA) and separated at 200 V for 45 min. The proteins were then transferred onto nitrocellulose membrane (Bio-Rad Laboratories Inc.) at 350 mA for 90 min. The blotted membranes were washed with Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBST) and then blocked with blocking buffer (TBST containing 5% skim milk) for 1 h. After blocking and washing, the membranes were incubated overnight with 1:1000 (anti-CYP7A1 and anti-β-actin) or 1:500 (anti-FMO3) dilution of primary antibodies. The membranes were washed three times with TBST for 5 min each and incubated with 1:1000 dilution of secondary antibody for 1 h. The membranes were then washed three times with TBST for 5 min each. Protein bands were detected using ECL solution and determined by densitometric analysis using Chemidoc XRS+ (Bio-Rad Laboratories Inc.).
Statistical analysis
All statistical analyses were conducted using SPSS program (version 26.0, SPSS, Chicago, IL, USA). One-way analysis of variance (ANOVA) with Duncan’s multiple range test (for SCFA composition, hippuric acid concentrations, and mRNA and protein expressions of genes; at p < 0.05) or Kruskal–Wallis test with Dunn’s test (for microbiome and BA composition) was used to compare statistical significance between the groups. Permutational multivariate analysis of variance (PERMANOVA) test was used to analyze significant differences in β-diversity analysis using QIIME2 command line (qiime diversity beta-group-significance) (Bolyen et al., 2019). Spearman correlation analysis was conducted to examine the association between either individual taxa or microbial metabolites, and biochemical data. Among the biochemical data for correlation analysis, TC, LDL-C, TMAO, and TMA levels were retrieved from the previous study (Lim et al., 2020).
Results and discussion
Changes in gut microbiome in the rat cecum
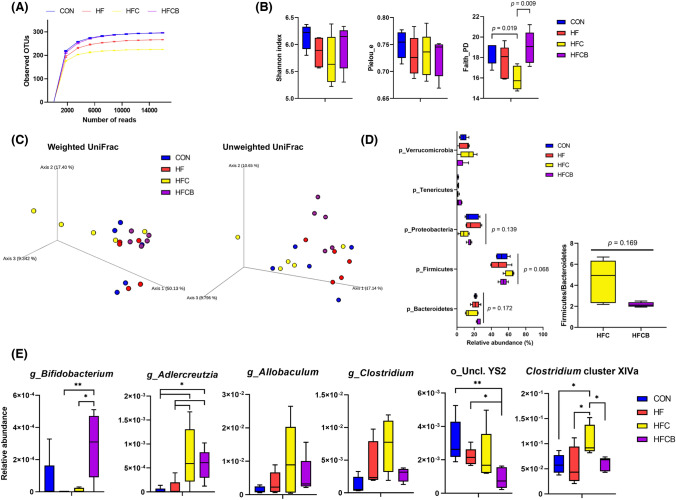
Polyphenols in BR were reported to have poor bioavailability and to be degraded in the gut despite several studies showing their cardio-protective effect (de Ferrars et al., 2014). Therefore, we hypothesized that they could act as prebiotics, remodeling the composition of the gut microbiota and further metabolite profile. α-Rarefaction (observed OTUs) curves at different sampling depths are shown in Fig. 1(A). The curves reached the plateau phase by 15,883 reads, indicating that sampling depths were sufficient to investigate following diversity analysis. α-Diversity (Shannon, Pielou’s evenness, and Faith’s phylogenetic diversity (PD)) was analyzed to investigate the richness and evenness of the gut microbiome in each rat (Fig. 1(B)). There were no significant differences in bacterial community diversity (Shannon) and evenness (Pielou’s evenness) among the groups; however, Faith’s PD index, phylogenetic distance between OTUs in each sample, was significantly lower in the HFC than in the CON (p = 0.019), which was reverted in the HFCB (p = 0.009). Depleted gut microbial diversity is generally associated with diseases (Wilmanski et al., 2019). Cho et al. (2017) reported that healthy men who can produce a high level of TMAO have a less diverse gut microbiome. In this study, it was revealed that BR extract could restore the decreased microbial diversity induced by high-choline intake.
Fig. 1.
Gut microbiome analysis in rats fed a high-fat diet with choline only or choline plus black raspberry extract (n = 5). (A) α-Rarefaction curves based on the number of observed operational taxonomic units (OTUs). (B) Box plots of α-diversity (Shannon, Pielou’s evenness (Pielou_e), and Faith’s phylogenetic diversity (Faith_PD)). (C) Principal coordinate analysis plots of β-diversity (weighted and unweighted UniFrac distances). (D) Taxonomic classification at the phylum levels of gut microbiota. (E) Relative abundance of 6 OTUs at the genus level of gut microbiota. CON (AIN-93G diet), HF (45% high-fat diet), HFC (HF + 1.5% choline water), and HFCB (HFC + 0.6% black raspberry extract)
Principal coordinate analyses (PCoA) were performed to assess the beta diversity of the gut microbiome between the groups (Fig. 1(C)). Based on the weighted and unweighted UniFrac distances, the HFC seemed to be clustered away from the other groups, which was confirmed by PERMANOVA tests (weighted UniFrac distance: pseudo-F = 1.65, p = 0.001; unweighted UniFrac distance: pseudo-F = 2.59, p = 0.011).
At the phylum level, the proportion in Firmicutes tended to increase in the HFC and decrease in the HFCB, although there was no significant difference among the 5 taxa (Fig. 1(D)). On the other hand, the proportion in Bacteroidetes showed the opposite tendency. The relative abundance of Proteobacteria tended to be lower in the HFC. The ratio of Firmicutes/Bacteroidetes tended to be higher in the HFC but lower in the HFCB.
At the genus level, the relative abundance of Adlercreutzia was higher in the HFC and HFCB than in the HF (Fig. 1(E)). The HFC showed higher tendencies in the relative abundances of Allobaculum and Clostridium (Clostridiaceae), while these shifts tended to be reversed by the BR supplementation (p = 0.130 and 0.144, respectively). There are several studies showing that the bacterial abundances of genera Adlercreutzia and Allobaculum increased in rats fed a high-choline diet (Lin et al., 2016). The BR-supplemented group was higher in Bifidobacterium than the HF (p < 0.01) and HFC (p < 0.05) but lower in unclassified YS2 (order) than the CON and HF.
Numerous studies have reported that intake of polyphenols or polyphenol-rich extracts has a beneficial effect on the growth of the Bifidobacterium, thereby ameliorating chronic diseases (Gowd et al., 2019). We also measured the abundances of bacterial genera with choline-TMA lyase (CutC) activity. The genera Clostridium (Clostridiaceae), Collinsella (Coriobacteriaceae), and Clostridium cluster XIVa including Clostridium, Eubacterium, Ruminococcus, Coprococcus, Dorea, Lachnospira, and Roseburia were reported to have TMA lyase activity or to be associated with blood TMAO level (Cho et al., 2017; Rath et al., 2020; Romano et al., 2015; Zhu et al., 2016). In this study, Clostridium cluster XIVa was enriched in the cecum of rats fed a high-choline diet, which was reversed by the BR supplementation (Fig. 1(E)). Unclassified YS2 was the only taxa not restored to the CON by the BR supplementation. It was reported that YS2 was the characteristic taxa of a high TMAO-producing strain (C57BL/6J) along with Prevotella compared to a low TMAO-producing one (NZW/LacJ) (Zhu et al., 2016), meaning that it could be likely to produce TMAO when unclassified YS2 is enriched in the gut. Therefore, it suggests that BR consumption might repress the accumulation of TMAO by reducing the relative abundance of YS2.
Therefore, these results suggest that the BR supplementation might lead to a reduction in the ability of the gut bacterial community to produce TMA from choline. In addition, it was reported that CutC was less abundant in herbivores than in omnivores and carnivores based on the result of a gene-targeted assay, suggesting that consumption of phytochemicals may reduce the TMA-producing ability of the gut bacterial community (Rath et al., 2020).
BA profile in cecum and serum
BAs can act as ligands for BA receptors, including FXR and TGR5 (G protein-coupled BA receptor 1) and regulate the expression of genes involved in their synthesis, transport, conjugation, and excretion (Ding et al., 2018). Primary BAs are synthesized in host hepatocytes and metabolized into secondary BAs by gut microbiota. Therefore, BA composition is highly associated with the gut microbial composition and can affect cholesterol and BA metabolisms (Winston and Theriot, 2020).
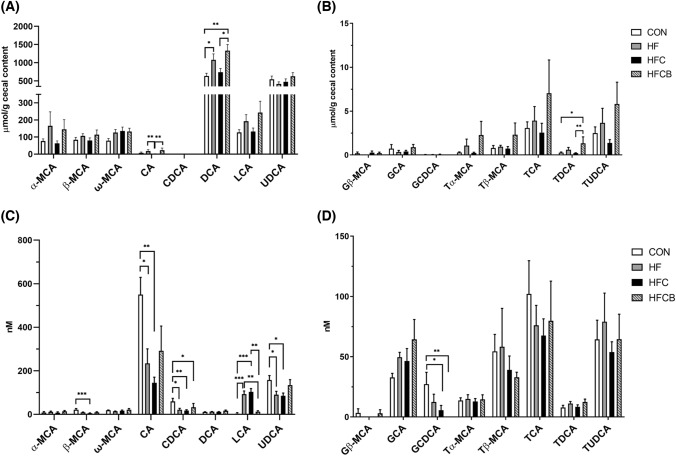
Most of the BAs showed relatively low concentrations in the cecum of the HFC compared to the HF and HFCB (Fig. 2(A), (B)). Especially, deoxycholic acid (DCA), one of the dominant secondary BAs, was lower in the HFC than in the HF and HFCB. Similar tendencies were observed in cholic acid (CA) and tauro-DCA (TDCA). This might be attributed to the increased activity of bile salt hydrolase in the gut microbiome, thereby resulting in increases in unconjugated secondary BAs.
Fig. 2.
Unconjugated (A) and conjugated (B) bile acids in the cecal content of rats fed a high-fat diet with choline only or choline plus black raspberry extract. Unconjugated (C) and conjugated (D) bile acids in the serum of the rats. All data represent the means and standard errors of the means (n = 9). Statistically significant differences between the groups were assessed by Kruskal–Wallis test and Dunn’s test (*p < 0.05, **p < 0.01, and ***p < 0.001). CON (AIN-93G diet), HF (45% high-fat diet), HFC (HF + 1.5% choline water), and HFCB (HFC + 0.6% black raspberry extract). MCA muricholic acid; CA cholic acid; CDCA chenodeoxycholic acid; DCA deoxycholic acid; LCA lithocholic acid; UDCA ursodeoxycholic acid; G glycine-conjugated; T taurine-conjugated
As shown in Fig. 2(C) and (D), primary BAs including CA, chenodeoxycholic acid (CDCA), and β-MCA significantly decreased in the serum of the HFC and were restored in that of the HFCB, but not significant between the two groups. On the other hand, interestingly, lithocholic acid (LCA), one of the major secondary BAs, was higher in the HF and HFC than in the CON and HFCB. The ratio of secondary BAs to primary BAs tended to increase in the serum of rats fed excessive choline. Likewise, it was reported that the relative percentage of secondary BAs increased in the serum of TMAO-fed Apoe knockout mice while that of primary BAs decreased (Ding et al., 2018).
Since BA receptors, including FXR, pregnane X receptor, and TGR5, were known to be differentially activated by specific individual BAs (Parks et al., 1999), changes in BA profile by BR extract might be a key factor regulating cholesterol levels in the host.
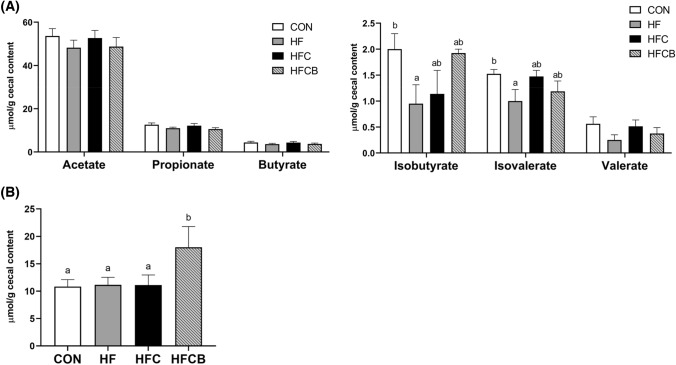
SCFA profile and hippuric acid concentration in cecum
Among six SCFAs (acetate, propionate, butyrate, isobutyrate, valerate, and isovalerate), the concentrations of major SCFAs, including acetate, propionate, and butyrate, in rat cecum were not significantly different among the treatments (Fig. 3(A)). Although a little change was observed in the branched SCFAs, there was no significant difference between the HFC and HFCB. Therefore, the cholesterol-lowering effect of BR extract may be due to changes in BA composition, not SCFA profile, which was similar to our previous results (Lim et al., 2022), since the supplementation of BR extract did not induce any significant difference in SCFA profile no matter which compound (choline as a precursor of TMA or TMAO itself) was used for cholesterol accumulation. The group fed BR extract showed a significant increase in the content of hippuric acid (Fig. 3(B)). In our previous study, hippuric acid also increased in the groups fed BR extract. We assume that hippuric acid might be one of the contributing metabolites that benefit the gut and cardiovascular health since it was reported to be correlated with the diversity of gut microbiota and metabolic syndrome (Pallister et al., 2017).
Fig. 3.
Short-chain fatty acid (A) and hippuric acid (B) concentrations in the cecal content of rats fed a high-fat diet with choline only or choline plus black raspberry extracts. All data represent the means and standard errors of the means (n = 9–10). Different small letters above bars indicate significant differences among the groups (p < 0.05 by one-way ANOVA and Duncan’s multiple range test). CON (AIN-93G diet), HF (45% high-fat diet), HFC (HF + 1.5% choline water), and HFCB (HFC + 0.6% black raspberry extract)
Expression of the genes involved in cholesterol and BA metabolisms
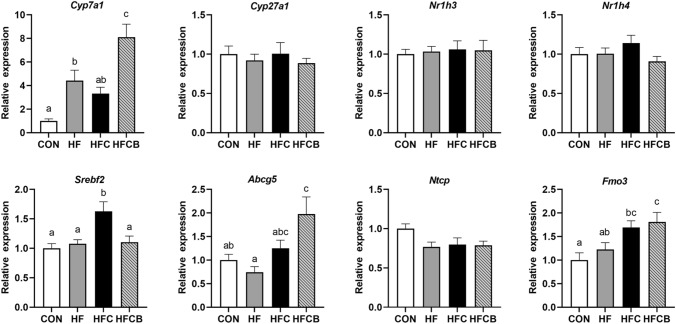
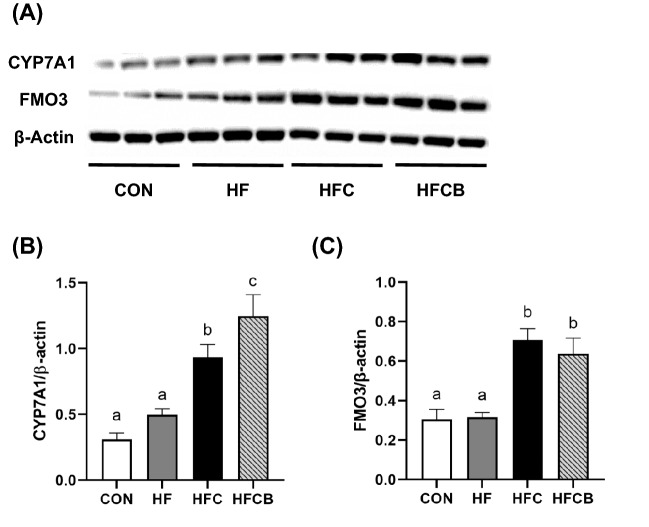
In our previous study, serum levels of total cholesterol and LDL-cholesterol were higher in the HFC than in the CON and HF but lower in the HFCB (p < 0.05) (Lim et al., 2020). Thus, to better understand the potential mechanism of BR extract in rats with elevated levels of cholesterol by administration of excessive choline, the mRNA expressions of the genes related to cholesterol and BA metabolisms in the liver tissues were explored. The mRNA (Fig. 4) and protein (Fig. 5(A), (B)) expressions of Cyp7a1, a rate-limiting enzyme of BA synthesis, were significantly (p < 0.05) higher in the HFCB than in the HFC. However, Cyp27a1 expression was not significantly (p > 0.05) different among the groups (Fig. 4). Among transcription factors that can activate transcription of genes involved in cholesterol and BA metabolisms, including liver X receptor-alpha (LXR-α), FXR, and sterol regulatory element-binding protein-2 (SREBP-2) (Nr1h3, Nr1h4, and Srebf2, respectively), only Srebf2 expression was significantly different among the groups (p < 0.05) and was the most upregulated in the HFC. Among cholesterol and BA transporters, the mRNA expression of Abcg5, a transporter facilitating the excretion of cholesterol into bile (Molusky et al., 2018), was the highest in the HFCB. The mRNA (Fig. 4) and protein (Fig. 5(A), (C)) expressions of FMO3, an enzyme generating the oxidized product TMAO from TMA, were upregulated in the HFC and HFCB, but not significantly (p > 0.05) different between the two groups regardless of the BR supplementation.
Fig. 4.
Hepatic mRNA expressions of the genes responsible for cholesterol and bile acid metabolisms in rats fed a high-fat diet with choline only or choline plus black raspberry extract. Relative gene expressions were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase. All data represent the means and standard errors of the means (n = 8–9). Different small letters above bars within the same genes indicate significant differences among the groups (p < 0.05 by one-way ANOVA and Duncan’s multiple range test). CON (AIN-93G diet), HF (45% high-fat diet), HFC (HF + 1.5% choline water), and HFCB (HFC + 0.6% black raspberry extract)
Fig. 5.
Protein expressions of cholesterol 7 α-hydroxylase (CYP7A1) (A, B) and flavin monooxygenase 3 (FMO3) (A, C) in the liver of rats fed a high-fat with choline only or choline plus black raspberry extract. All data represent the means and standard errors of the means (n = 8–9). Different small letters above bars indicate significant differences among the groups (p < 0.05; one-way ANOVA and Duncan’s multiple range test). CON (AIN-93G diet), HF (45% high-fat diet), HFC (HF + 1.5% choline water), and HFCB (HFC + 0.6% black raspberry extract)
Based on the results, it appeared that BR extract could reduce the elevated level of serum cholesterol via upregulation of Cyp7a1 and downregulation of Srebf2 expression. There are plenty of studies reporting that reverse cholesterol transport (RCT) is the major pathway to reduce CVD risk via the conversion of cholesterol into BAs (Chambers et al., 2019). CYP7A1 plays an important role in RCT since it can initiate BA synthesis from cholesterol (Chiang, 2004). Polyphenols, such as resveratrol, chlorogenic acid, and catechin, can modulate hepatic Cyp7a1 expression, enhancing RCT (Chambers et al., 2019). On the other hand, TMAO was reported to inhibit RCT and cholesterol removal from peripheral macrophages and to downregulate the hepatic mRNA expression of Cyp7a1 and Cyp27a1 (Koeth et al., 2013). SREBP-2 modulates the expressions of genes involved in the regulation of cellular cholesterol levels (Brown and Goldstein, 1997). Similar to our result, polyphenols from black chokeberry were reported to modulate Srebf2 expression in Caco-2 cells so that it can encourage cholesterol flux (Kim et al., 2013). The hepatic mRNA and protein expressions of FMO3, an enzyme converting TMAO from TMA in the liver (Koeth et al., 2013), was not significantly (p > 0.05) changed regardless of the BR supplementation, indicating that the TMAO-lowering effect of BR extract in our previous work (Lim et al., 2020) may be due to the less production of TMA in the intestine.
Correlation analysis
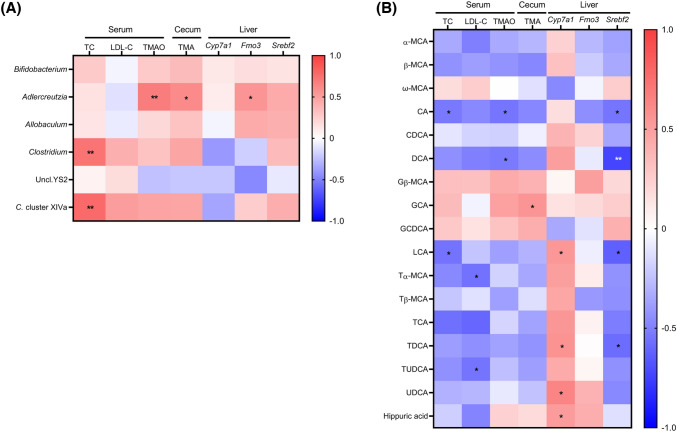
Clostridium (Clostridiaceae) and Clostridium cluster XIVa, known to have TMA lyase activity (Rath et al., 2020), were positively associated with serum TC level (Fig. 6(A)). The relative abundance of Adlercreutzia (Coriobacteriaceae) was positively associated with serum TMAO, cecal TMA, and hepatic Fmo3 expression. Microbial metabolites, including BAs and hippuric acid, except for ω-MCA and glycine-conjugated BAs, tended to be negatively associated with serum TC and LDL-C (Fig. 6(B)). Especially, serum TC level was negatively associated with cecal levels of CA, LCA, and TCA. Serum LDL-C level was negatively associated with taurine-conjugated BAs, including Tα-MCA, TCA, and TUDCA. Serum TMAO level was negatively associated with CA and DCA. Cecal TMA level was positively associated with GCA. Hepatic Cyp7a1 expression was positively associated with LCA, TCA, TDCA, UDCA, and hippuric acid, while hepatic Srebf2 expression was negatively associated with CA, DCA, LCA, and TDCA.
Fig. 6.
Heatmap generated by Spearman’s correlation analysis between gut microbiota and biochemical results (A) and between microbial metabolites and biochemical results (B) in the rats (n = 20). Red, positive correlation; white, no correlation; and blue, negative correlation. *p < 0.05; and **p < 0.01. Biochemical data, including total cholesterol (TC), LDL-cholesterol (LDL-C), trimethylamine-N-oxide (TMAO), and trimethylamine (TMA), were retrieved from the previous study (Lim et al., 2020). Uncl. Unclassified; Clostridium, Clostridiaceae Clostridium; MCA muricholic acid; CA cholic acid; CDCA chenodeoxycholic acid; DCA deoxycholic acid, LCA lithocholic acid, UDCA ursodeoxycholic acid; G glycine-conjugated; T taurine-conjugated
In our previous study, BR extract reduced cecal TMA and serum TMAO and cholesterols in rats fed excessive choline with a high-fat diet. We revealed that this might be because the BR extract increased the phylogenetic diversity of gut microbiota and changed the relative abundance of some specific genera, such as Bifidobacterium and Clostridium cluster XIVa. The altered gut microbiota might also lead to changing the profile of microbial secondary metabolites, mainly BAs. We also revealed that the genes involved in cholesterol metabolism, especially Cyp7a1 and Srebf2 were regulated by the BR extract in the rats fed a high-choline diet. The main limitation of this study is the problem of direct translation of the present results to human cases due to the difference in not only gut microbial composition but also BA metabolism between humans and rats (Nagpal et al., 2018). However, since the role of specific taxa in TMA production and the role of major microbial metabolites were similar between humans and rats, this study can provide insight into how BR rich in polyphenols can lower serum cholesterol levels in rats induced by high-choline administration in terms of changes in gut microbial composition and metabolite profile.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B03028407).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Taehwan Lim, Email: imtae86@snu.ac.kr.
Kiuk Lee, Email: leku@snu.ac.kr.
Ryun Hee Kim, Email: ryunheekim@snu.ac.kr.
Juhee Ryu, Email: issue221@snu.ac.kr.
Kwang Hyun Cha, Email: chakh79@kist.re.kr.
Sun Young Park, Email: sunyoung.park@snu.ac.kr.
Song Yi Koo, Email: ninesong2@kist.re.kr.
Keum Taek Hwang, Email: keum@snu.ac.kr.
References
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME2. Nature Biotechnology. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:31–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2 is an open-source software package that denoises and removes sequencing errors from Illumina amplicon. Nature Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers KF, Day PE, Aboufarrag HT, Kroon PA. Polyphenol effects on cholesterol metabolism via bile acid biosynthesis, CYP7A1: a review. Nutrients. 2019;11:2588. doi: 10.3390/nu11112588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JYL. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. Journal of Hepatology. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Cho CE, Taesuwan S, Malysheva OV, Bender E, Tulchinsky NF, Yan J, Sutter JL, Caudill MA. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: A randomized controlled trial. Molecular Nutrition & Food Research. 2017;61:1770016. doi: 10.1002/mnfr.201770016. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renault P, MicroObes consortium ANR, Doré J, Zucker JD, Clément K, Ehrlich SD. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- de Ferrars RM, Czank C, Zhang Q, Botting NP, Kroon PA, Cassidy A, Kay CD. The pharmacokinetics of anthocyanins and their metabolites in humans. British Journal of Pharmacology. 2014;171:3268–3282. doi: 10.1111/bph.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Chang M, Guo Y, Zhang L, Xue C, Yanagita T, Zhang T, Wang Y. Trimethylamine-N-oxide (TMAO)-induced atherosclerosis is associated with bile acid metabolism. Lipids in Health and Disease. 2018;17:286. doi: 10.1186/s12944-018-0939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowd V, Karim N, Shishir M, Xie L, Chen W. Dietary polyphenols to combat the metabolic diseases via altering gut microbiota. Trends in Food Science & Technology. 2019;93:81–93. doi: 10.1016/j.tifs.2019.09.005. [DOI] [Google Scholar]
- Gu J, Thomas-Ahner JM, Riedl KM, Bailey MT, Vodovotz Y, Schwartz SJ, Clinton SK. Dietary black raspberries impact the colonic microbiome and phytochemical metabolites in mice. Molecular Nutrition & Food Research. 2019;63:1800636. doi: 10.1002/mnfr.201800636. [DOI] [PubMed] [Google Scholar]
- Huang F, Zheng X, Ma X, Jiang R, Zhou W, Zhou S, Zhang Y, Lei S, Wang S, Kuang J, Han X, Wei M, You Y, Li M, Li Y, Liang D, Liu J, Chen T, Wei R, Rajani C, Shen C, Xie G, Bian Z, Li H, Zhao A, Jia W. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nature Communications. 2019;10:4971. doi: 10.1038/s41467-019-12896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HS, Hong SJ, Lee TB, Kwon JW, Jeong JT, Joo HJ, Park JH, Ahn CM, Yu CW, Lim DS. Effects of black raspberry on lipid profiles and vascular endothelial function in patients with metabolic syndrome. Phytotherapy Research. 2014;28:1492–1498. doi: 10.1002/ptr.5154. [DOI] [PubMed] [Google Scholar]
- Jung H, Lee HJ, Cho H, Hwang KT. Anti-inflammatory activities of Rubus fruit anthocyanins in inflamed human intestinal epithelial cells. Journal of Food Biochemistry. 2015;39:300–309. doi: 10.1111/jfbc.12130. [DOI] [Google Scholar]
- Kim B, Park Y, Wegner CJ, Bolling BW, Lee J. Polyphenol-rich black chokeberry (Aronica melanocarpa) extract regulates the expression of genes critical for intestinal cholesterol flux in Caco-2 cells. The Journal of Nutritional Biochemistry. 2013;24:1564–1570. doi: 10.1016/j.jnutbio.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kim J, An J, Lee S, Kong H, Song Y, Choi HR, Lee SJ, Chae KS, Kwon JW, Kim KJ. Amelioration of hyperglycemia by Rubus occidentalis (black raspberry) and increase in short-chain fatty acids producing bacteria. Journal of Functional Foods. 2019;54:433–439. doi: 10.1016/j.jff.2019.01.045. [DOI] [Google Scholar]
- Lim T, Ryu J, Lee K, Park SY, Hwang KT. Protective effects of black raspberry (Rubus occidentalis) extract on hypercholesterolemia and hepatic inflammation in rats fed high-fat and high-choline diets. Nutrients. 2020;12:2448. doi: 10.3390/nu12082448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim T, Lee K, Kim RH, Ryu J, Park SY, Cha KH, Koo SY, Hwang KT. Black raspberry extract can lower serum LDL cholesterols via modulation of gut microbial composition and serum bile acid profile in rats fed trimethylamine-N-oxide with a high-fat diet. Food Science and Biotechnology. 2022;31:1041–1051. doi: 10.1007/s10068-022-01079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, An Y, Hao F, Wang Y, Tang H. Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state. Scientific Reports. 2016;6:21618. doi: 10.1038/srep21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Lin X, Bordiga M, Brennan C, Xu B. Manipulating effects of fruits and vegetables on gut microbiota – a critical review. International Journal of Food Science and Technology. 2021;56:2055–2067. doi: 10.1111/ijfs.14927. [DOI] [Google Scholar]
- Meslier V, Laiola M, Roager HM, de Filippis F, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons N, Pasolli E, Rivellese A, Dragsted LO, Vitaglione P, Ehrlich SD, Ercolini D. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69:1258–1268. doi: 10.1136/gutjnl-2019-320438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molusky MM, Hsieh J, Lee SX, Ramakrishnan R, Tascau L, Haeusler RA, Accili D, Tall AR. Metformin and AMP Kinase activation increase expression of the sterol transporters ABCG5/8 (ATP-binding cassette transporter G5/G8) with potential antiatherogenic consequences. Atherosclerosis, Thrombosis, and Vascular Biology. 2018;38:1493–1503. doi: 10.1161/ATVBAHA.118.311212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer RA, Hummer KE, Finn CE, Frei B, Wrolstad RE. Anthocyanins, phenolics, and antioxidant capacity in diverse small fruits: Vaccinum, Rubus, and Ribes. Journal of Agricultural and Food Chemistry. 2002;50:519–525. doi: 10.1021/jf011062r. [DOI] [PubMed] [Google Scholar]
- Nagpal R, Wang S, Woods LCS, Seshie O, Chung ST, Shively CA, Register TC, Craft S, McClain DA, Yadav H. Comparative microbiome signatures and short-chain fatty acids in mouse, rat, non-human primate, and human feces. Frontiers in Microbiology. 2018;9:2897. doi: 10.3389/fmicb.2018.02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister T, Jackson MA, Martin TC, Zierer J, Jennings A, Mohney RP, MacGregor A, Steves CJ, Cassidy A, Spector TD, Menni C. Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Scientific Reports. 2017;7:13670. doi: 10.1038/s41598-017-13722-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Wilson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Poll BG, Cheema MU, Pluznick JL. Gut microbial metabolites and blood pressure regulation: focus on SCFAs and TMAO. Physiology. 2020;35:275–284. doi: 10.1152/physiol.00004.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath S, Rud T, Pieper DH, Vital M. Potential TMA-producing bacteria are ubiquitously found in mammalia. Frontiers in Microbiology. 2020;10:2966. doi: 10.3389/fmicb.2019.02966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6:e02481-14. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaddel R, Hesari J, Azadmard-Damirchi S, Hamishehkar H, Fathi-Achachlouei B, Huang Q. Double emulsion followed by complex coacervation as a promising method for protection of black raspberry anthocyanins. Food Hydrocolloids. 2018;77:803–816. doi: 10.1016/j.foodhyd.2017.11.024. [DOI] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall H, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metabolism. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung YM. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmanski T, Rappaport N, Earls JC, Magis AT, Manor O, Lovejoy J, Omenn GS, Hood L, Gibbons SM, Price ND. Blood metabolome predicts gut microbiome α-diversity in humans. Nature Biotechnology. 2019;37:1217–1228. doi: 10.1038/s41587-019-0233-9. [DOI] [PubMed] [Google Scholar]
- Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes. 2020;11:158–171. doi: 10.1080/19490976.2019.1674124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;24:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.