Abstract
Notch signaling commences with two ligand-mediated proteolysis events that release the Notch intracellular domain, NICD, from the plasma membrane. NICD then translocates into the nucleus and interacts with the DNA binding protein CSL to activate transcription. We found that NICD expression also potentiates activity of the transcription factor LEF-1. NICD stimulation of LEF-1 activity was context dependent and occurred on a subset of promoters distinct from those activated by β-catenin. Importantly, the effect of NICD does not appear to be mediated through canonical components of the Wnt signaling pathway or downstream components of the Notch pathway. In vitro assays show a weak association between the C-terminal transactivation domain of NICD and the high-mobility group domain of LEF-1, suggesting that the two proteins interact in vivo. Our data therefore describe a new nuclear target of Notch signaling and a new coactivator for LEF-1.
Notch signaling involves a series of precisely regulated events. Notch resides at the plasma membrane as a heterodimer due to proteolysis by a furin-like convertase at a site designated S1 (25). In response to ligand, Notch is cleaved at two additional sites, S2 and S3, by TACE and a γ-secretase-like activity, respectively (6; for a review, see references 20 and 27). Cleavage at S3 releases the Notch intracellular domain (NICD) from the membrane. NICD has two nuclear localization signals that target it to the nucleus where it interacts with the DNA binding factor CSL (CBF-1, Suppressor of Hairless, LAG-1; also known as RBP-J) (1, 18, 37, 38). In the absence of NICD, CSL acts as a transcriptional repressor. CSL can mediate repression in vitro by interacting with TFIID and TFIIA (29) and in vivo by interacting with corepressors and histone deacetylases (15, 19, 45). NICD and the corepressors bind to the same region of CSL; thus, entry of NICD into the nucleus leads to the displacement of the CSL-associated corepressors (15, 19, 45). Binding of NICD to CSL is mediated by the Notch RAM and Ankyrin domains (14), and transcriptional activation occurs as a consequence of the NICD transcription activation domain recruiting coactivators, such as PCAF and GCN-5 (21, 22).
The LEF-1 transcription factor was originally identified as a T-cell-specific factor that regulates the T-cell receptor α enhancer (40). While LEF-1 was once thought to play an architectural role in transcriptional activation, it is now clear that LEF-1 can act as a conventional transcription factor (10, 13). LEF-1 acts in conjunction with several other DNA binding proteins to activate the TCR-α enhancer (12) using a context-dependent activation domain (8, 11). LEF-1 can also interact with the coactivators β-catenin and ALY to induce gene expression (5, 7). The activities of β-catenin and ALY toward LEF-1 are highly context dependent and have not been found to overlap (16). While β-catenin can activate certain promoters containing multiple LEF-1 binding sites, ALY cannot. Conversely, ALY is able to stimulate activity of the TCR-α enhancer, while β-catenin has no effect.
Results presented here identify NICD as a coactivator for the LEF-1 transcription factor. The effects of Notch on LEF-1 activity are direct and not due to modulation of components of the Wnt signaling cascade or due to effects of Notch-mediated activation of CSL. Potentiation of LEF-1 activity by NICD defines new roles for both Notch and LEF-1 in the regulation of gene expression.
MATERIALS AND METHODS
Plasmids and transfections.
The Notch expression plasmids NICD, NICDΔR, NICDΔRA, and NICDΔTAD were generated by PCR from a full-length human Notch 1 cDNA. Amino acid positions included in each Notch fragment are 1760 to 2556 for NICD, 1859 to 2556 for NICDΔR, 2094 to 2556 for NICDΔRA, and 1760 to 2094 for NICDΔTAD. Each PCR product was cloned into pcDNA3.1(−) Myc-HisC (Invitrogen), in frame with the Myc-His tags, and sequenced to ensure correct cloning and sequence. NICD (or NICD fragments) was subcloned using traditional methods to create GAL4-NICDΔRA, GST-NICDΔTAD, GST-NICDΔRA, MIGR-NICD, and NICD-ER (details available upon request). The parental MIGR retroviral construct was provided by W. Pear (University of Pennsylvania), and the parental estrogen receptor (ER) fusion retroviral construct was given by M. McMahon (University of California, San Francisco). The LEF-1, Δ56LEF, β-catenin, and GST-ALY expression vectors, as well as the 7xLEF-luc and fos-luc reporter plasmids, were the generous gifts of R. Grosschedl (University of Munich). The LEF-OT and LEF-OF reporter constructs were gifts of B. Vogelstein (Johns Hopkins University). The Notch3 NICD, TCF-1, and CSL-VP16 (previously known as pCMX-VP16-RBP-J) expression vectors were gifts from U. Lendahl (Karolinska Institute), H. Clevers (University Hospital Utrecht Medical School), and T. Honjo (Kyoto University), respectively. Reporter constructs containing promoters from the Xtwn, Cyclin D1, and WISP-1 genes were gifts of L. Attisano (University of Toronto), A. Rustgi (University of Pennsylvania), and A. Levine (Rockefeller University), respectively.
All cell lines were maintained in Dulbecco's minimal Eagle's medium (Gibco-BRL) supplemented with 10% fetal bovine serum, Pen/Strep, and glutamine. To generate the MIGR, MIGR-NICD, ER, and NICD-ER transduced cells, NIH 3T3 cells were infected with ecotropic retrovirus and selected with 2 μg of puromycin per ml (for ER and NICD-ER) or by green fluorescent protein (GFP)-positive fluorescence-activated cell sorting (for MIGR and MIGR-NICD). Transfections of NIH 3T3 and Neuro-2A were carried out using CaPO4 DNA coprecipitation (Clontech) or Fugene (Boehringer-Mannheim), as per manufacturers' instructions. Jurkat cells were transfected by electroporation with the Gene Pulser II electroporator (Bio-Rad). Jurkat cells were transfected during logarithmic growth phase, using 2 × 106 cells, in 4 mM Gap cuvettes with settings of 0.250 kV and 975 μF. Transfections typically contained 100 ng of reporter and 1 ng of pRL-CMV (Promega) to assess relative transfection efficiencies. Unless otherwise noted, transfections also contained 500 ng of expression vector(s). All cells were harvested 42 to 48 h after transfection. Firefly and Renilla luciferases were assayed following the instructions provided with the Dual Luciferase Assay kit (Promega). All transfections are shown as the means ± standard errors of the means of at least three separate transfections.
GST interaction assays.
Glutathione-S-transferase (GST), GST-NICDΔTAD, GST-NICDΔRA, and GST-ALY were expressed in the BL21 strain of Escherichia coli (Stratagene). GST proteins were induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Promega), and the bacteria were allowed to grow an additional 4 to 5 h. Following induction, cells were lysed by freeze-thawing in phosphate-buffered saline and protease inhibitors. GST proteins were bound to glutathione resin (Pharmacia) and washed five times with phosphate-buffered saline–0.2% NP-40 (Sigma).
The LEF-1 deletion fragment proteins were generated in a manner similar to that described in reference 4. Briefly, two rounds of PCR amplification were used: the first, to generate the deletion fragments with a common 5′ end containing a consensus Kozak sequence and a 3′ stop codon; the second, to generate deletion fragments containing a 5′ T7 promoter. After the first amplification reaction the PCR products were gel purified, and after the second amplification reaction the samples were purified using the QIAquick PCR purification kit (Qiagen). The PCR products purified from the second round of PCR were then in vitro transcribed and translated with the TNT T7 coupled reticulocyte lysate system (Promega) in the presence of [S35]methionine.
GST interaction assays were performed with whole-cell extracts from transfected 293T cells or in vitro-synthesized proteins that were prebound with glutathione resin. GST fusion proteins were equilibrated in bead binding buffer (25 mM HEPES [pH 7.5], 150 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1% NP-40, 1% glycerol) and then incubated with the whole-cell extracts or in vitro-synthesized proteins at 4°C for 1 h. After incubation, glutathione bead-GST fusion protein complexes were collected by centrifugation and washed five times with bead binding buffer. The washed beads were then resuspended in sodium dodecyl sulfate loading buffer and boiled, and Western blotting was performed.
RESULTS AND DISCUSSION
NICD potentiates LEF-1 activity.
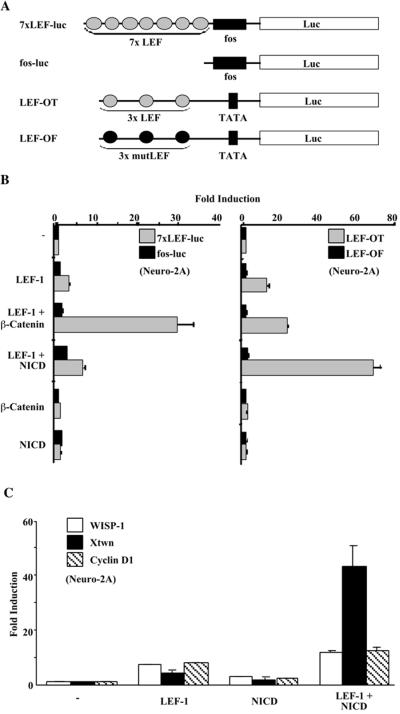
During experiments designed to investigate interactions between components of the Notch and Wnt signaling pathways, we noted the ability of Notch1 NICD to augment the activity of LEF-1 on certain promoters. The reporter 7xLEF-luc (Fig. 1A) harbors a promoter consisting of seven LEF-1 binding sites upstream of a minimal Fos promoter and is responsive to β-catenin acting through LEF-1 (16). When assayed in Neuro-2A cells, β-catenin alone did not activate 7xLEF-luc and LEF-1 activated the reporter only weakly. However, LEF-1 plus β-catenin had a marked effect, stimulating the reporter greater than ninefold (Fig. 1B, left panel). NICD had very little effect on the reporter either in the presence or in the absence of LEF-1. Very different results were obtained with a second LEF-1 responsive reporter, LEF-OT (Fig. 1B, right panel), whose promoter carries three LEF-1 binding sites upstream of the E1b TATA box. First, activity of LEF-OT was stimulated approximately 15-fold by LEF-1 alone and the addition of β-catenin gave rise to only a modest further increase in LEF-1 activity (less than twofold). Second, the addition of NICD stimulated reporter activity approximately 5-fold over that seen with LEF-1 alone (Fig. 1B) and up to 10-fold in other cell lines (data not shown). NICD had no effect if LEF-1 was not included in the transfection or if the reporter harbored mutant LEF-1 binding sites (LEF-OF). We conclude that NICD stimulation of LEF-OT is mediated through LEF-1.
FIG. 1.
Notch potentiates LEF-1 activity. (A) Schematic representations of reporters used to assay LEF-1 activity. Binding sites for LEF-1 are shown as gray (consensus) or black (mutant) ovals. (B) Effects of LEF-1, β-catenin, and NICD on 7xLEF-luc and LEF-OT in Neuro-2A cells. Relative luciferase values for reporters containing seven LEF-1 binding sites (7xLEF-luc; gray bars) or no LEF-1 binding sites (fos-luc; black bars) are shown in the left panel. Values for reporters having three consensus LEF-1 binding sites (LEF-OT; gray bars) or three mutant LEF-1 binding sites (LEF-OF; black bars) are shown in the right panel. Values for fold induction were determined relative to those obtained for each reporter in the absence of any expression plasmids. (C) Effects of LEF-1 and NICD on naturally occurring promoters. Transfections of Neuro-2A cells were carried out with reporters containing promoters from the WISP-1 (white bars), Cyclin D1 (striped bars), or Xtwn (black bars) genes. Expression plasmids that were cotransfected with each set of reporters are indicated. Values are given as fold induction relative to the reporter alone.
These findings are reminiscent of reports demonstrating promoter specificity for the LEF coactivators β-catenin and ALY (7, 16). We attempted to determine the nature of the NICD-LEF promoter specificity by generating hybrids of LEF-OF and 7xLEF-luc. In one instance we fused the LEF sites of LEF-OF to the core promoter in 7xLEF-luc (c-Fos), and in another, we fused the LEF sites of 7xLEF-luc to the core promoter of LEF-OF (a TATA box). Surprisingly, both hybrid reporters were activated by NICD in the presence of LEF-1 (data not shown). Hence, the NICD-LEF promoter specificity cannot be easily explained.
We also examined the responses of three naturally occurring promoters that carry LEF-1 binding sites (Fig. 1C). The WISP-1 promoter is an example of a promoter with LEF-1 binding sites that are not necessary for stimulation by β-catenin or Wnt signaling (43). Both LEF-1 alone and NICD alone activated the WISP-1 promoter to some degree, and LEF-1 plus Notch exerted a small additive effect. The LEF-1 sites in the Xenopus Twin (Xtwn) promoter are responsive to β-catenin and Wnt signaling, but are also active independently of β-catenin (23, 28). LEF-1 alone and NICD alone induced the Xtwn promoter fourfold and twofold, respectively. However, LEF-1 plus NICD gave a strong synergistic activation of the Xtwn promoter (43-fold). The Cyclin D1 promoter has also been identified as having LEF-1 binding sites; but unlike the WISP-1 promoter, these sites are highly responsive to Wnt signaling through LEF-1 and β-catenin (35, 39). As with the WISP-1 promoter, NICD did not potentiate LEF-1 activity on the Cyclin D1 promoter. We conclude that naturally occurring promoters, like the artificial promoters, fall into two groups: those that are stimulated by NICD through LEF-1 and those that are not.
Activation is limited to subsets of each protein family.
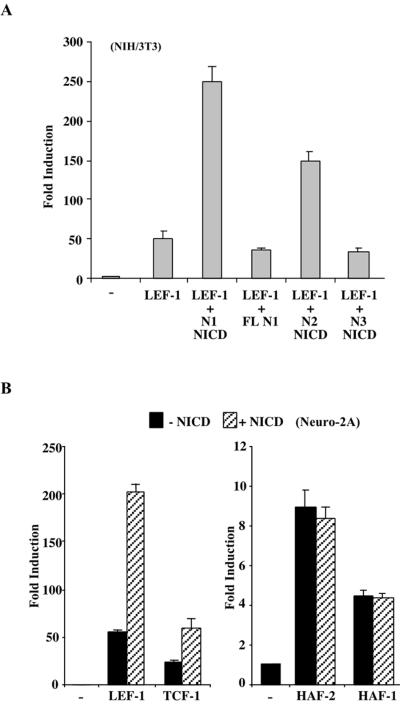
In vertebrates, multiple Notch genes exist; therefore, we sought to identify whether the potentiation of LEF-1 is specific to Notch1. Notch1 NICD stimulated LEF-1 activity approximately fivefold, while a full-length Notch1 receptor had no effect on LEF-1 activity (Fig. 2A). Notch2 and Notch3 were also tested for the ability to activate LEF-1. Notch2 NICD potentiated LEF-1 activity, although not as strongly as Notch1 NICD; however, Notch3 NICD was unable to stimulate LEF-1. The ability of Notch1 and Notch2, but not Notch3, to enhance LEF-1 activity is consistent with the idea that both Notch1 and Notch2 are transcriptional activators, while Notch3 is not (3, 22).
FIG. 2.
Specific Notch proteins activate a subset of HMG domain transcription factors. (A) Ability of Notch 1, 2, and 3 to potentiate LEF-1. LEF-1 activity was assayed in NIH 3T3 cells using the LEF-OT reporter in the presence of Notch1 (N1) NICD, full-length Notch1 (FL N1), Notch2 (N2) NICD, or Notch3 (N3) NICD. Values are given as fold induction relative to LEF-OT alone. (B) Effects of NICD on other HMG box transcription factors. Neuro-2A cells were transfected with LEF-OT and expression vectors for LEF-1, TCF-1, HAF-2, or HAF-1 in the presence (striped bars) or absence (black bars) of NICD. Values are give as fold induction as for panel A.
LEF-1 is a member of the high-mobility group (HMG) box family of DNA binding proteins. To determine if NICD potentiates the activity of other members of this family, we tested the response of additional HMG box transcription factors, including TCF-1, HAF-1, and HAF-2 (the last two are also referred to as Sox 17 and Sox 18, respectively [36]). Like LEF-1, TCF-1 was also stimulated by NICD, although not as strongly (Fig. 2B). Reasons for the variance in potentiation by NICD are not clear, as Western analysis shows no apparent difference in protein levels (data not shown). By contrast, HAF-1 and HAF-2 stimulated the activity of LEF-OT, but NICD had no additional effect. The effect of NICD is therefore restricted to a subset of HMG box proteins.
The effect of NICD on LEF-1 does not involve other components of the canonical Wnt or Notch signaling pathways.
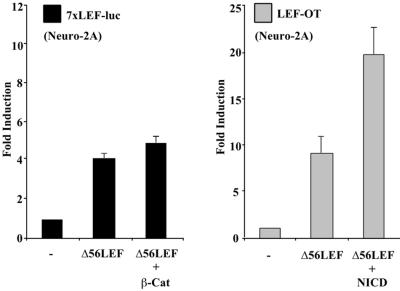
We considered the possibility that NICD may stimulate LEF-1 by modulating the components of the Wnt signaling pathway that lead to an increase in nuclear β-catenin. We felt that this was unlikely since NICD and β-catenin were most effective on distinct reporters (Fig 1A). However, to test directly if Notch potentiates LEF-1 through β-catenin, NICD was assayed in the presence of a LEF-1 deletion mutant, Δ56LEF, that lacks the β-catenin interaction domain. As expected, β-catenin was unable to stimulate 7xLEF-luc in the presence of Δ56LEF (Fig. 3, left panel). By contrast, Δ56LEF induced LEF-OT eightfold and NICD resulted in a further twofold stimulation (Fig. 3, right panel). While Western blot analysis comparing wild-type LEF-1 and Δ56LEF expressions indicated that Δ56LEF was present at lower levels, increasing the amount of transfected Δ56LEF was unable to match the degree of potentiation observed with wild-type LEF-1 and NICD (data not shown). Although this overall level of stimulation was below that obtained with wild-type LEF-1, we conclude that NICD does not augment LEF-1 activity through β-catenin.
FIG. 3.
NICD activation of LEF-1 is independent of β-catenin. NICD augments activity of Δ56LEF. Neuro-2A cells were transfected with 7xLEF-luc and expression plasmids for Δ56LEF (LEF-1 lacking the β-catenin interaction domain) and β-catenin as indicated (left panel, black bars). Cells were also transfected with LEF-OT and expression plasmids for Δ56LEF and NICD as indicated (right panel, gray bars). Results are given as fold induction relative to the reporters alone.
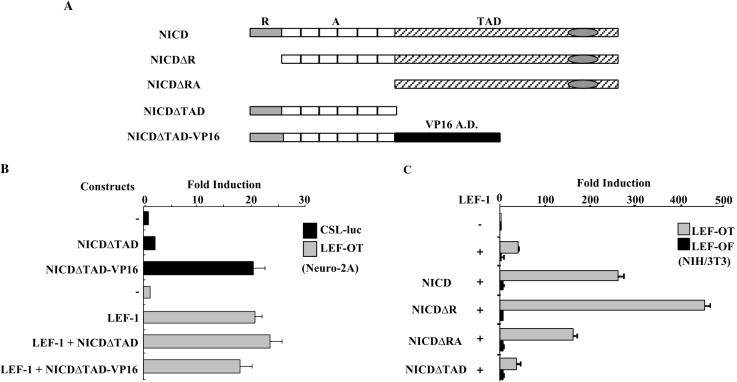
The effect of NICD is also not mediated by prototypical Notch target genes. CSL-VP16 is a fusion protein that activates Notch target genes in the absence of Notch signaling. Although CSL-VP16 was able to activate a CSL-dependent reporter, it had no effect on LEF-1 (data not shown). Additionally, potentiation of LEF-1 was not observed with a hybrid protein that carries the VP16 activation domain in place of the NICD activation domain (Fig. 4B). The latter result suggests that the effect of NICD specifically requires the Notch activation domain. NICD comprises three functional domains: the RAM domain (R), which mediates interaction with CSL; the Ankyrin repeats (A), which bind a number of proteins including CSL; and the C-terminal transcriptional activation domain (TAD). We generated a series of proteins that contain one or more of these domains and assessed their abilities to activate LEF-1 on the LEF-OT reporter (Fig. 4C). Both NICD and NICDΔR (lacking the RAM domain) potentiated LEF-1. The C terminus of NICD that encompasses the TAD but lacks both CSL-interaction domains (NICDΔRA) also activated LEF-1, while a fragment that contains the RAM and ankyrin domains (NICDΔTAD) did not. NICDΔRA and LEF-1 also gave synergistic activation of the Xtwn promoter, while NICDΔTAD-VP16 did not (data not shown). Although induction of LEF-1 by NICDΔRA was slightly less than that observed for NICD, this is likely due to lower protein levels (data not shown). These data show that the Notch TAD is necessary and sufficient for the observed effects on LEF-1 and argue further that the effects are not mediated indirectly through the induction of CSL-responsive genes.
FIG. 4.
NICD activates LEF-1 independently of CSL. (A) Schematic diagrams of the NICD deletion fragments are shown. R, RAM domain; A, Ankyrin repeats; TAD, C-terminal transcription activation domain; VP16 A.D., VP16 transcription activation domain. (B) NICDΔTAD-VP16 does not augment activity of LEF-1. Neuro-2A cells were transfected with a CSL-dependent reporter, CSL-luc (black bars), or LEF-OT (gray bars) and the expression vectors as indicated. NICDΔTAD-VP16 carries the VP16 TAD in place of the Notch TAD. Results are given as fold induction relative to the reporters alone. (C) The Notch TAD is sufficient for LEF-1 activation. NIH 3T3 cells were transfected with LEF-OT (gray bars) or LEF-OF (black bars) and the NICD fragments indicated. Results are presented as fold induction relative to the reporter alone.
NICD and LEF-1 interact physically.
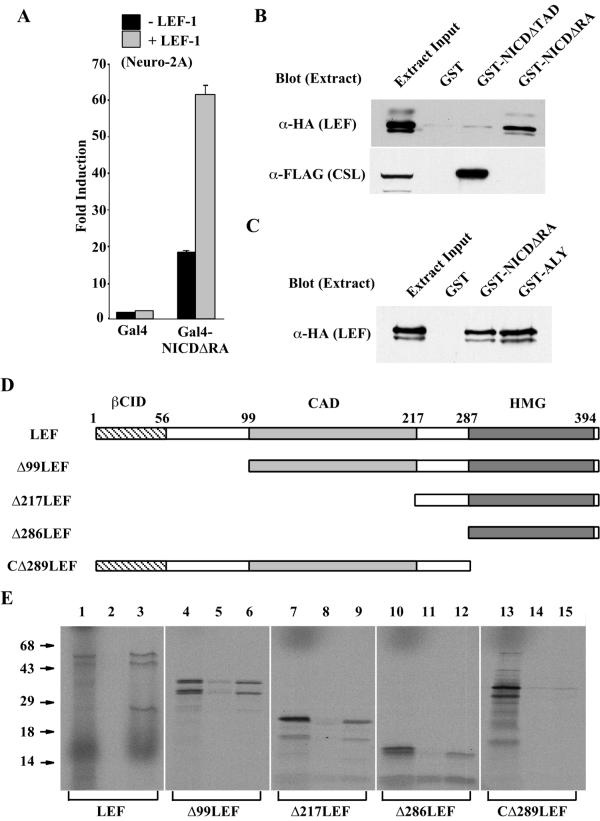
Next, we carried out experiments to investigate whether NICD and LEF-1 interact physically. First we used a modified mammalian two-hybrid assay to assess in vivo interactions. Specifically, a Gal4-NICDΔRA fusion protein was tested for transcriptional activity in the absence and presence of LEF-1 (Fig. 5A). Gal4-NICDΔRA alone was able to stimulate the Gal4 responsive reporter, confirming the presence of an activation domain within the C terminus of NICD (22). While LEF-1 had little effect on the Gal4 DNA binding domain alone or a Gal4-VP16 fusion, it enhanced the activity of Gal4-NICDΔRA approximately threefold to fourfold (Fig. 5A and data not shown). As anticipated by earlier results, Gal4-NICDΔRA was also activated by Δ56LEF (data not shown). The increase in transcriptional response is most likely due to the consequences of physically linking two activation domains, one provided by NICD and the other by LEF-1. This would occur when NICD interacts with DNA-bound LEF-1 (Fig. 1) or when LEF-1 interacts with DNA-bound NICD (Fig. 5A).
FIG. 5.
The Notch TAD physically interacts with the LEF-1 HMG domain. (A) LEF-1 activates Gal4-NICDΔRA. Neuro-2A cells were transfected with a Gal4 responsive reporter and either the DNA binding domain of Gal4 (Gal4) or a Gal4 fused to the TAD of Notch (Gal4-NICDΔRA) in the presence (gray bars) or absence (black bars) of LEF-1 as indicated. Results are shown as fold induction relative to the reporter plus the Gal4 DNA binding domain. (B) LEF-1 interacts with the Notch TAD in vitro. 293T cells were transfected with plasmids expressing HA-LEF-1 and FLAG-CSL, extracts were incubated with GST, GST-NICDΔTAD, or GST-NICDΔRA, and bound proteins were analyzed by Western analysis using anti-HA antibodies (top panel) or anti-FLAG antibodies (lower panel). Samples of untreated cell extracts were loaded in each of the far left lanes, corresponding to 1/350 of the input analyzed for HA-LEF-1 (top) and 1/5 of the input analyzed for FLAG-CSL (bottom). (C) LEF interacts with the Notch TAD and ALY with comparable affinities. 293T cells were transfected with HA-LEF, extracts were incubated with GST, GST-NICDΔRA or GST-ALY as indicated, and bound proteins were analyzed by Western analysis with an anti-HA antibody. Lane 1 contains untreated extract corresponding to 1/300 of the input. (D) Schematic diagram of the LEF-1 fragments used to map the interaction with NICD. Amino acid positions and several functionally defined domains are indicated. βCID, β-catenin interaction domain; CAD, context-dependent activation domain; HMG, HMG domain. (E) The LEF-1 HMG box mediates interactions with the Notch TAD. The indicated radiolabeled LEF-1 fragments were generated by in vitro transcription and translation (lanes 1, 4, 7, 10, and 13) and analyzed for binding to GST (lanes 2, 5, 8, 11, and 14) and GST-NICDΔRA (lanes 3, 6. 9, 12, and 15). Untreated samples represent 1/100 of the input used for each binding analysis. Positions of molecular mass standards (in kilodaltons) are shown at the left.
To measure interactions in vitro, GST-NICDΔRA (containing the Notch TAD) and GST-NICDΔTAD (lacking the TAD) fusion proteins were generated and tested for their abilities to interact with LEF-1. Initially, whole-cell extracts were used to investigate whether NICD and LEF-1 could interact in the context of a large array of cellular proteins. Cells transfected with hemagglutinin (HA)-tagged LEF-1 or FLAG-tagged CSL (as a positive control) were used in these assays (Fig. 5B). LEF-1 was retained by GST-NICDΔRA, but not by GST alone or GST-NICDΔTAD (Fig. 5B, upper panel). As expected, CSL was retained by GST-NICDΔTAD, but not by GST or GST-NICDΔRA (Fig. 5B, lower panel). Given the strong potentiation of LEF-1 by NICD, it was somewhat surprising that the NICD-LEF-1 interaction was so weak relative to that of NICD and CSL (see amounts retained versus input). Addition of ethidium bromide did not affect interactions between GST-NICDΔRA and LEF-1, indicating that nucleic acid is not mediating their association (data not shown). We therefore compared the interaction of LEF-1 with NICD to that of LEF-1 with ALY, a coactivator known to functionally interact with LEF-1 (7, 16). GST fusions of NICDΔRA or ALY were tested for their abilities to interact with LEF-1 from transfected whole-cell extracts (Fig. 5C). GST-NICDΔRA and GST-ALY retained similar amounts of LEF-1, suggesting comparable binding affinities. We could not compare these interactions with those involving β-catenin since we were unable to identify β-catenin-LEF-1 or NICD-LEF-1 complexes by immunoprecipitation (data not shown). Stable interactions between NICD and LEF-1 were also not seen using mobility shift assays of transfected-cell extracts or recombinant proteins (data not shown). While robust with respect to NICD's effect on LEF-1 activity in vivo, the interaction between NICD and LEF-1 is physically weak in vitro.
The region of LEF-1 that interacts with NICD was determined using radiolabeled in vitro-synthesized LEF-1 deletions (Fig. 5D). All deletions from the N terminus of LEF-1 retained the ability to interact with GST-NICDΔRA (Fig. 5E). While the HMG domain of LEF-1 was clearly sufficient to mediate an interaction with the Notch TAD, the observed interaction was weaker than that seen with the other deletion fragments. The diminished interaction between NICD and the HMG domain alone might suggest that sequences immediately N terminal to the DNA binding domain are involved in NICD-LEF interactions. This could explain the lower potentiation ability of NICD for TCF-1 (Fig. 2B), which differs from LEF-1 outside the HMG domain. Removal of the LEF-1 HMG domain eliminated the interaction with NICDΔRA. Thus, interactions are localized to the Notch TAD domain and the LEF-1 HMG domain. Since crude extracts were used to demonstrate NICD-LEF-1 interactions and these are physically weak, we cannot rule out the possibility that the interaction is indirect and mediated by an unknown bridging protein.
Stimulation of LEF-1 requires high levels of NICD.
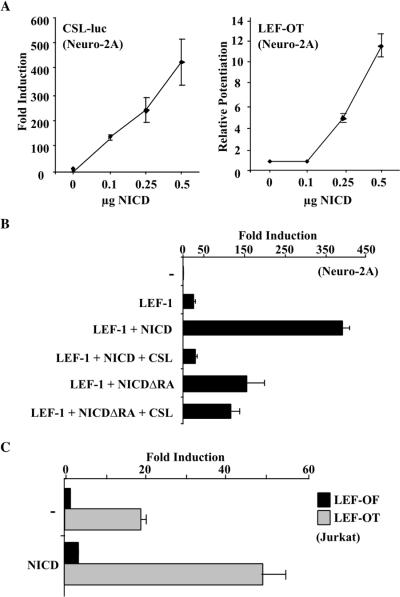
The apparent low in vitro affinity of NICD for LEF-1 prompted us to assess the relative in vivo responses of LEF-dependent and CSL-dependent promoters (Fig. 6A). The activity of NICD towards the CSL-dependent reporter was linear and apparent at low input concentrations (0.1 μg) of the NICD expression vector. By contrast, activity of the LEF-dependent reporter (LEF-OT) required higher amounts of NICD (0.25 μg) before the response was seen and became linear. We also carried out an experiment in which we transfected Jurkat cells with full-length Notch1 and mimicked ligand-mediated activation by treating cells with EDTA (32). Under these conditions, the CSL-dependent reporter was activated roughly sixfold, while the LEF-dependent reporter was unaffected (data not shown). One possible explanation for these results is that low cellular concentrations of NICD bind exclusively to CSL and binding to LEF occurs only when NICD concentrations functionally exceed those of CSL. Consistent with this, the ability of NICD to stimulate the LEF-responsive reporter was completely inhibited in the presence of overexpressed CSL (Fig. 6B).
FIG. 6.
High-level expression of NICD is required for LEF-1 activation. (A) Low levels of NICD stimulate a CSL reporter but do not potentiate LEF-1. Fold induction of CSL-luc, with increasing amounts of NICD expression plasmid, is shown relative to the reporter alone (left graph). Stimulation of LEF-1 in the presence of increasing amounts of NICD compared to the LEF-OT reporter and LEF-1 alone is shown (right graph). (B) Excess CSL inhibits NICD's ability to stimulate LEF-1 activity. Neuro-2A cells were transfected with the LEF-OT reporter and the indicated expression vectors. Results are given as fold induction relative to the reporter alone. (C) Activity of NICD in the presence of endogenous LEF-1. Jurkat cells were transfected with reporters containing wild-type (LEF-OT) or mutant (LEF-OF) LEF-1 binding sites, plus or minus an expression vector for NICD as indicated. Data are represented as fold induction relative to LEF-OF alone.
To examine the effect of NICD on endogenous LEF-1, we used Jurkat cells, a T-cell line that normally expresses LEF-1. In the absence of NICD the transfected LEF-OT reporter was approximately 20-fold more active than the promoter containing mutant LEF-1 sites (LEF-OF) (Fig. 6C). This is presumably due to endogenous LEF-1/TCF-1 activity. Upon transfection of an NICD expression plasmid, activity was increased to approximately 50-fold over that of LEF-OF. Similarly to what is depicted in Fig. 1A, the 7xLEF-luc reporter had no activity in Jurkat cells in the absence of β-catenin and was not induced with NICD (data not shown).
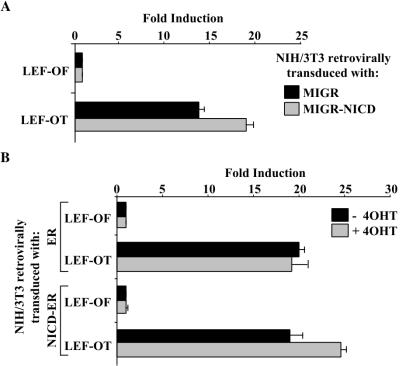
Retroviral expression of NICD has been used in a mouse model for Notch-induced leukemogenesis in humans (30, 31). The relatively high level of Notch expression obtained with retroviruses is necessary for the development of T-cell tumors and has been proposed to mimic the level obtained in human T-ALL carrying the t(7;9) translocation (17). We generated NIH 3T3 cells that harbor either a retrovirus that carries NICD linked to an IRES-GFP (MIGR-NICD) or a retrovirus that contains only the IRES-GFP (MIGR). When the two cell populations were transfected with LEF-1 and either LEF-OT or LEF-OF (to establish a baseline), we observed a 30% increase in activity in the cells stably expressing NICD (Fig. 7A). These data argue that there is a significant enhancement of LEF-1 activity in cells harboring NICD-expressing retroviruses. To better establish that the observed effect is direct, we generated cells that contain a retrovirus that expresses an NICD-ER fusion protein. In control experiments we showed that activity of a CSL-dependent reporter in those cells was stimulated roughly 10-fold by tamoxifen (data not shown). Consistent with the previous result using NICD-expressing retroviruses, LEF-OT activity was also increased approximately 30% by tamoxifen (Fig. 7B). Induction required the LEF binding sites in the promoter, did not occur in the absence of the fusion protein (Fig. 7B), and occurred within 12 h (data not shown). These data argue that the effect of tamoxifen is due to the activation of NICD and its binding to LEF-1.
FIG. 7.
Retrovirally expressed NICD stimulates LEF-1 activity. (A) NIH 3T3 cells transduced with either the MIGR (black bars) or MIGR-NICD (gray bars) retroviruses and then transfected with LEF-OF or LEF-OT reporters and a LEF-1 expression vector. Results for each transduced cell population are shown as fold induction relative to the LEF-OF plus LEF-1. (B) Tamoxifen-responsive NICD stimulates LEF-1 activity. NIH 3T3 cells were transduced with retroviruses expressing either the parental ER element (ER cells) or NICD fused to the estrogen receptor element (NICD-ER cells). LEF-1 activity was assayed in the absence (black bars) or presence (gray bars) of 25 nM tamoxifen, following the transfection of ER and NICD-ER cells with the LEF-OF or LEF-OT reporters and a LEF-1 expression vector. Results are shown as fold induction relative to LEF-OF and LEF-1 with no tamoxifen.
Our results describe a new activity for NICD and a new coactivator for LEF-1. To date, the only nuclear target for NICD has been the DNA binding protein CSL and the only coactivators for LEF-1 have been β-catenin and ALY. Our data therefore expand the repertoire of genes that may be influenced by Notch signaling and increase the known variety of ways in which genes can be activated through LEF-1. Numerous reports have described both negative and positive genetic interactions between the Notch and Wingless pathways in Drosophila melanogaster (reviewed in reference 26). It has been proposed, for example, that Wingless signaling can inhibit Notch through direct binding of Disheveled to the Notch C terminus (2). (We have been unable to demonstrate any effects of mouse Disheveled on NICD activity towards a CSL-dependent promoter [data not shown].) Wingless itself has also been reported to be a Notch ligand, thereby serving as a direct stimulator of Notch signaling (reviewed in references 26, 41, and 42). Our results with the various reporters imply that the promoters activated by NICD do not necessarily overlap with those activated by β-catenin; thus, our data do not explain how the two pathways may or may not interact. Although the Xtwn promoter can be activated by β-catenin (23, 28) and by NICD (Fig. 1), Xtwn gene activation during embryonic development occurs prior to the induction of Notch signaling. Thus, it remains to be determined if there are genes whose activity is influenced directly by both pathways. Interestingly, and perhaps directly relevant to our results, it has been shown that Notch can modulate the activity of the Drosophila UbxVMB enhancer through dTCF (24). Modulation of dTCF activity was shown to be independent of the components of the canonical Wingless pathway and of Su(H). Although this report showed that Notch can inhibit dTCF activity, deletion of the Notch RAM and Ankyrin domains resulted in stimulation of the UbxVMB enhancer and expression in regions where it was previously undetected. The latter set of results may reflect the type of activity we have described here.
Our experiments show that LEF-1 is likely to be activated only in those cells where NICD levels are high (i.e., with transfection experiments or transduced cell lines). Low levels of nuclear Notch are sufficient to activate CSL-dependent reporters (33) and may support the majority of Notch's signaling tasks during embryonic development. However, high levels of NICD are found in the nuclei of various cell types, including cortical neurons (34) and certain cancers (9, 44), and these have a higher likelihood of supporting the signaling pathway described here.
ACKNOWLEDGMENTS
We thank members of the Kadesch lab for their helpful comments and suggestions.
This work was supported by a grant from the National Institutes of Health to T.K. (RO1 GM58228); D.R. was supported by a National Institutes of Health National Cancer Institute training grant (T32 CA09140).
REFERENCES
- 1.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 2.Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 3.Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. doi: 10.1242/dev.126.17.3925. [DOI] [PubMed] [Google Scholar]
- 4.Beckman H, Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 5.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J R, Cumano A, Roux P, Black R A, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 7.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCR alpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson P, Waterman M L, Jones K A. The hLEF/TCF-1 alpha HMG protein contains a context-dependent transcriptional activation domain that induces the TCR alpha enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 9.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 10.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 11.Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 13.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–99. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu S C, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izon D J, Punt J A, Xu L, Karnell F G, Allman D, Myung P S, Boerth N J, Pui J C, Koretzky G A, Pear W S. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 2001;14:253–264. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 18.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 19.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopan R, Goate A. A common enzyme connects Notch signaling and Alzheimer's disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 21.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 22.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbe E, Letamendia A, Attisano L. Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-beta and wnt pathways. Proc Natl Acad Sci USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence N, Langdon T, Brennan K, Arias A M. Notch signaling targets the Wingless responsiveness of a Ubx visceral mesoderm enhancer in Drosophila. Curr Biol. 2001;11:375–385. doi: 10.1016/s0960-9822(01)00120-8. [DOI] [PubMed] [Google Scholar]
- 25.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah N G, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez Arias A. Interactions between Wingless and Notch during the assignment of cell fates in Drosophila. Int J Dev Biol. 1998;42:325–333. [PubMed] [Google Scholar]
- 27.Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Tian X, Pan D J, Ray W J, Kopan R. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 28.Nishita M, Hashimoto M K, Ogata S, Laurent M N, Ueno N, Shibuya H, Cho K W. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 29.Olave I, Reinberg D, Vales L D. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pui J C, Allman D, Xu L, DeRocco S, Karnell F G, Bakkour S, Lee J Y, Kadesch T, Hardy R R, Aster J C, Pear W S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 32.Rand M D, Grimm L M, Artavanis-Tsakonas S, Patriub V, Blacklow S C, Sklar J, Aster J C. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 34.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 35.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens S, Ordentlich P, Sen R, Kadesch T. HMG box-activating factors 1 and 2, two HMG box transcription factors that bind the human Ig heavy chain enhancer. J Immunol. 1996;157:3491–3498. [PubMed] [Google Scholar]
- 37.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 39.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 40.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 41.Wesley C S. Notch and wingless regulate expression of cuticle patterning genes. Mol Cell Biol. 1999;19:5743–5758. doi: 10.1128/mcb.19.8.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wesley C S, Saez L. Notch responds differently to delta and wingless in cultured drosophila cells. J Biol Chem. 2000;275:9099–9101. doi: 10.1074/jbc.275.13.9099. [DOI] [PubMed] [Google Scholar]
- 43.Xu L, Corcoran R B, Welsh J W, Pennica D, Levine A J. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 44.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou S, Fujimuro M, Hsieh J J, Chen L, Miyamoto A, Weinmaster G, Hayward S D. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]