Abstract
Trophic factor withdrawal induces cell death by mechanisms that are incompletely understood. Previously we reported that withdrawal of interleukin-7 (IL-7) or IL-3 produced a rapid intracellular alkalinization, disrupting mitochondrial metabolism and activating the death protein Bax. We now observe that this novel alkalinization pathway is mediated by the pH regulator NHE1, as shown by the requirement for sodium, blocking by pharmacological inhibitors or use of an NHE1-deficient cell line, and the altered phosphorylation of NHE1. Alkalinization also required the stress-activated p38 mitogen-activated protein kinase (MAPK). Inhibition of p38 MAPK activity with pharmacological inhibitors or expression of a dominant negative kinase prevented alkalinization. Activated p38 MAPK directly phosphorylated the C terminus of NHE1 within a 40-amino-acid region. Analysis by mass spectroscopy identified four phosphorylation sites on NHE1, Thr 717, Ser 722, Ser 725, and Ser 728. Thus, loss of trophic cytokine signaling induced the p38 MAPK pathway, which phosphorylated NHE1 at specific sites, inducing intracellular alkalinization.
The requirement for cytokines in hematopoiesis is partly attributable to a trophic activity, the protection of cells from programmed cell death (17, 44). Interleukin-7 (IL-7), a product of the thymic epithelium, protects lymphocyte progenitor cells from apoptotic death during T-cell development (19, 55). Survival of pro-T cells isolated from the thymus requires the presence of IL-7 (25), whereas disruption of the gene for IL-7 (51) or its receptor (35) dramatically reduces thymic cellularity (29). IL-7 also has trophic activities on cells of the developing brain (30). IL-3 has similar trophic activities on early hematopoietic precursors of the myeloid, lymphoid, and erythoid lineages. In IL3-dependent cell lines, withdrawal leads to apoptotic cell death (21).
The trophic action of cytokines has been partly attributed to the Bcl-2 family of proteins, which are important intracellular regulators of apoptosis (25, 37). Overexpression of the anti-apoptotic protein Bcl-2 in IL-7Rα−/− mice partially restored T-cell numbers (1), but complete restoration of a normal phenotype was not achieved (8, 13). Recent studies with IL-3-dependent cell lines have shown that the up-regulation of Bcl-2 and Bcl-XL and the down-regulation of proapoptotic proteins such as Bad are involved in survival (21); however, as with IL7, the trophic action of IL-3 involves more than the balance of Bcl-2 family members, since the overexpression of Bcl-2 extended life for only 1 day following IL-3 withdrawal (32).
The intracellular signaling pathways triggered by cytokine receptor engagement are yet to be fully defined. However, it is known that hematopoietic cytokine receptors can induce distinct members of the mitogen-activated protein kinase (MAPK) family, contributing to the processes of proliferation and survival (10, 11; E. Rajnavolgyi, N. Benbernou, K. Muegge, and S. K. Durum, unpublished data [for similar results with IL-7]). Three of the MAPK signaling units have been characterized in detail: the extracellular signal-regulated kinases (ERKs), the c-Jun amino-terminal kinases (JNK) or stress-activated protein kinases (SAPK), and the p38 MAPKs (p38) (20). Mitogens, inflammatory cytokines, and growth factors are known to activate various MAPK signaling pathways, whereas cellular stresses such as UV light, heat, or osmotic shock selectively induce the JNK/SAPK and p38 MAPK pathways. A common feature of all the MAPKs is that they are activated by the phosphorylation of both threonine and tyrosine residues by a dual-specificity serine-threonine MAPK (18). In turn, MAPKs frequently phosphorylate their substrates at serine or threonine residues adjacent to prolines (15, 28). Although the MAPKs have been implicated in the regulation of apoptotic death, for example following nerve growth factor withdrawal (26, 59), the spontaneous apoptosis of neutrophils (2), and loss of IL-7 signaling in a dependent cell line (Rajnavolgyi et al., unpublished) it is not known how these MAPKs contribute to the apoptotic process and their relevant substrates have not been identified.
The dysregulation of intracellular pH is part of the trigger for apoptotic cell death following cytokine withdrawal (23, 24) or ceramide treatment (3). Control of cytosolic pH in eukaryotic cells involves various proton pumps, proton channels, and ion transporters that drive H+ or H+ equivalents and HCO3− ions into and out of the cell. Among the better-characterized ion transporters or exchangers are the Na+/H+ exchanger (NHE), which has multiple isoforms with NHE1 being the most prevalent, the Na+-dependent and Na+-independent HCO3− transporters or anion exchangers (AE), the CL−/OH− exchanger (CHE), and a lactate-proton cotransporter (36). Through the use of specific inhibitors, the roles of these complex membrane proteins have been examined; however, much remains to be understood regarding the mechanisms of their action and regulation.
To dissect the complex physiological events that occur on trophic factor withdrawal, two cytokine-dependent cell lines, one requiring IL-7 and one requiring IL-3, were used. Our previous studies showed that following cytokine withdrawal, a novel intracellular alkalinization occurred and triggered a death pathway involving mitochondrial translocation of the proapoptotic protein Bax (23) and inhibition of mitochondrial ADP transport, ATP synthesis, and subsequent mitochondrial bioenergetics (24). In the present study we examine the mechanism of this cytokine withdrawal-induced alkalinization and show that it is caused by a specific membrane ion exchanger, which in turn is phosphorylated by a stress-activated MAPK.
MATERIALS AND METHODS
Cells and reagents.
The IL-3-dependent murine pro-B-cell line FL5.12A (a kind gift from James A. McCubrey, East Carolina University, Greenville, S.C.) was maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 2-β-mercaptoethanol (1,000×; Life Technologies), and 0.4 ng of recombinant mouse IL-3 (PeproTech Inc.) per ml. The IL-7-dependent cell line D1 was established from the CD4− CD8− subset sorted from p53−/− mouse thymocytes initially propagated in IL-7 and stem cell factor and maintained as previously described (23). Chinese hamster ovary cells (CHO) and the NHE−/−-deficient CHO cells (39) were maintained in Dulbecco Modified Eagle Medium DMEM (Life Technologies) with 10% fetal bovine serum.
FL5.12A cells were transiently transfected by electroporation (0.310 V, 960 μF capacitance) using a Bio-Rad GenePulser. Prior to pulsing, 5 × 107 cells in 0.4-cm cuvettes (Bio-Rad) were preincubated for 10 min with either 40 μg of pEGFP-C3 (Clontech) or 20 μg of pEGFP-C3 and 20 μg of pCMV-Flag-p38 (agf) (a kind gift from Roger Davis, University of Massachusetts Medical Center, Worcester, Mass.). To select for cells transfected with plasmid DNA, green fluorescent protein-containing (GFP+) cells were first sorted by fluorescence-activated cell sorting, and cell populations containing at least 98% GFP+ cells were cultured with or without IL-3 for 48 h (after which detection of GFP fluorescence declined). Viability and cell number were evaluated by trypan blue staining (Life Technologies). For detection of DNA fragmentation, cells were stained with a 2-μg/ml solution of propidium iodide in a detergent buffer (48) and analyzed by flow cytometry. CHO cells were transfected with 3 μg of pCMV-Flag-p38 (agf) using Fugene 6 (Roche) as specified by the manufacturer. Transfection efficiencies averaged 90%.
To measure intracellular pH, BCECF-AM (Molecular Probes) was used at 1 μM. Cells were treated as described in the figure legends with the following reagents: 1 mM ouabain (Sigma), 10 μg of cycloheximide (Sigma) per ml, 1 μg of antimycin A (Sigma) per ml, 200 μM 5-(N,N-dimethyl)amiloride (DMA) (Sigma), 200 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA) (Sigma), 1,000 and 200 μM 4,4′(diisothiocyanato)stilbene-2-2′-disulfonic acid (DIDS) (Sigma), 1,000 and 200 μM 4-acetamido-4′-isothiocyanato-stilbene-2-2′-disulfonic acid (SITS) (Sigma), 100 μg of anisomycin (Sigma) per ml, 20 μM PD169316, 20 μM SB202190, 20 μM SB203580, 20 μM SB202474, and 20 μM PD98059 (all from Calbiochem). The NaCl buffer contained 140 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1 mM KCl, 10 mM glucose, and 20 mM HEPES. For the buffer lacking Na+, 140 mM choline chloride replaced NaCl. Bicarbonate-free RPMI 1640 (Sigma) was used in the experiments requiring a solution lacking HCO3− ions.
Measurement of intracellular pH.
FL5.12A or D1 cells (106 cells/ml) deprived of IL-3 or IL-7, respectively, for various times were treated, after being washed in a HEPES buffer (25 mM HEPES, 140 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 5.0 mM glucose), with 1 μM BCECF-AM for 30 min at 37°C, and intracellular pH was determined by flow cytometry with excitation set at 488 nm and emissions filtered through 525-nm band pass (green fluorescence) and 610-nm long-pass (red fluorescence) fitters (16). Dead cells (less than 10%) were excluded by forward- and side-scatter gating. A pH calibration curve was generated by preloading cells with 1 μM BCECF-AM for 30 min followed by incubation for 30 min in a high-K+ HEPES buffer (25 mM HEPES, 145 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 5.5 mM glucose) at different pH values from 6.8 to 8.0 in the presence of the permeabilizing agent nigericin (10 μM) (23). After incubation, nigericin was absorbed with excess bovine serum albumin.
Detection of MAPKs by Western blot analyses.
Whole-cell lysates were made by lysing cells (107) in a modified RIPA buffer containing phosphatase inhibitors (150 mM NaCl, 80 mM NaF, 50 mM Tris-HCl, 5 mM EDTA, 1 mM EGTA, 25 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1.0% [wt/vol] NP-40, 0.5% [wt/vol] deoxycholate, 0.1% [wt/vol] sodium dodecyl sulfate [SDS]) (31). A protease inhibitor cocktail (Complete Mini; Roche) was added to the lysis buffer. Cell lysates were centrifuged at 20,000 × g and 4°C, and the recovered supernatant was used for protein analysis.
For detection of phosphorylated and nonphosphorylated forms of p38 MAPK, ATF-2, ERK1/2, SAPK/JNK, and p90RSK proteins by Western blot analysis, protein samples were heated to 90°C for 5 min in 2× Tris-glycine SDS loading buffer (Invitrogen) with 2.5% mercaptoethanol. Cell equivalent samples (20 μl containing approximately 40 μg of protein) were then separated by SDS-polyacrylamide gel electrophoresis on either 8 or 12% Tris-glycine gels (Invitrogen) run under denaturing conditions using Tris-glycine SDS running buffer (Invitrogen). Proteins were transferred to 0.2 μM polyvinylidene difluoride membranes (Invitrogen) using Tris-glycine transfer buffer (Invitrogen). The blots were probed with a rabbit polyclonal antiserum specific for phospho-p38 MAPK protein (D-8; Santa Cruz) and p38 MAPK protein (Santa Cruz), phospho-ATF-2 (Cell Signaling Technologies), phospho-ERK1/2 and ERK1/2 (Cell Signaling Technologies), phosphoJNK/SAPK (Cell Signaling Technologies), and phospho-p90RSK (Cell Signaling Technologies), as specified by the manufacturers, followed by anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP) (Santa Cruz), and then developed by enhanced chemiluminescence (ECL) (Pierce) and visualized on BioMax ML film (Kodak).
NHE1 phosphorylation and detection of complex with p38 MAPK.
To measure NHE1 phosphorylation, 2 × 107 cells were preincubated for 30 min at 37°C in a phosphate-free buffer (130 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgSO4, 5 mM glucose, 20 mM HEPES, 2 mM glutamine, and 1 g of bovine serum albumin per liter). The cells were then incubated, with or without cytokine, for 3 h (FL5.12A cells) or 5 h (D1 cells) with 1 mCi of [32P]orthophosphate per ml (stock, 10 mCi/ml) in the phosphate-free buffer at 37°C. Cell lysates were made as described above using the modified RIPA buffer containing phosphatase inhibitors (31). Protein lysates were immunoprecipitated overnight using an anti-NHE1 antibody against the C terminus (A7) (31) and protein A-Sepharose beads (Amersham), treated to reduce nonspecific binding. After extensive washing of beads, the recovered protein was analyzed by SDS-polyacrylamide gel electrophoresis on 8% Tris-glycine SDS gels (Invitrogen). Autoradiography was performed, and the results were visualized on BioMax MR film (Kodak).
To detect complex formation between NHE1 and p38 MAPK, 5 × 107 cells were first chemically cross-linked with 10 mM disuccinimidyl suberate (Pierce) for 15 min and then lysed in the modified RIPA buffer. Samples were immunoprecipitated overnight, after being precleared with rabbit immunoglobulin (Ig) and protein A-Sepharose, with the anti-NHE1 antibody (A7) as well as an antibody to Mapkap 2 (Santa Cruz) and rabbit Ig (Santa Cruz). The immunoprecipitates were washed in RIPA buffer, run on a 10% Tris-glycine SDS gel (Invitrogen), and blotted for p38 MAPK (Santa Cruz) as previously described.
NHE1 protein detection.
To detect NHE1 protein in cell lysates by Western blotting, whole-cell lysates, made from 2 × 107 cells using the modified RIPA buffer previously described, were precleared with mouse nonimmune sera (Santa Cruz) and protein A-Sepharose and immunoprecipiated with the anti-NHE (A7) antibody and protein A-Sepharose beads, and the recovered protein was analyzed on 8% Tris-glycine SDS gels (Invitrogen), transferred as described above, blotted using a commercially available anti-NHE1 mouse monoclonal antibody (diluted 1:1,000) (Chemicon) followed by the appropriate secondary mouse antibody conjugated to HRP (Santa Cruz), and then developed by ECL (Pierce) and visualized on BioMax ML film.
Kinase assays.
To determine whether in vivo-activated kinases could phosphorylate NHE1, FL5.12A cells, (2 × 107) were cultured with or without IL-3 for 1 or 3 h and lysates were made with an extraction buffer containing 50 mM sodium pyrophosphate, 5 mM sodium fluoride, 5 mM sodium chloride, 5 mM EDTA, 5 mM EGTA, 0.1 mM sodium orthovanadate, 0.1% Triton X-100, 10 mM HEPES (pH 7.2), and protease inhibitors (31). Equal amounts of protein were used for an in-gel kinase assay in which cell lysates were separated in a 10% gel containing 1 mg of either glutathione S-transferase (GST) or a carboxy-terminal wild-type (WT) NHE-GST fusion protein (described in Fig. 7C) per ml. Protocols for the in-gel kinase assay and the purification of the NHE-GST fusion proteins have been previously described (31). NHE-GST fusion proteins, WT and deletion mutants, were based on the rabbit NHE1 sequence, which is almost identical to the human NHE1 sequence.
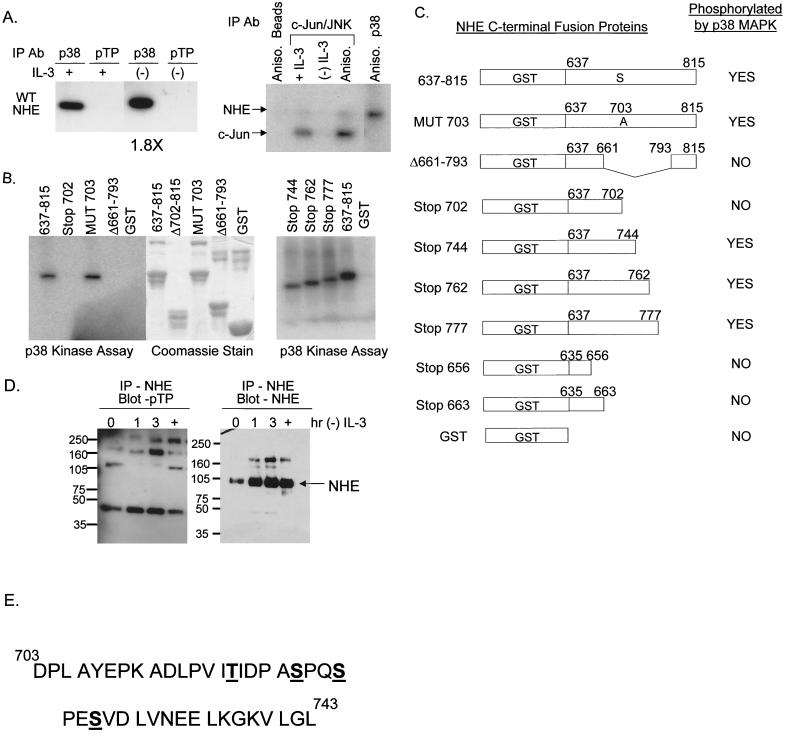
FIG. 7.
Kinase assays demonstrate that p38 MAPK directly phosphorylates NHE1 at a site(s) between amino acids 703 and 793. The numbering system used corresponds to the rabbit NHE1 sequence. (A) Protein lysates from FL5.12A cells cultured with or without IL-3 for 2 h were immunoprecipitated with either an antibody specific for phospho-p38 MAPK (p38) or an antibody specific for the motif, p-TP, and the ability of these kinases to phosphorylated the WT NHE-GST fusion protein was evaluated by an in-gel kinase assay as described in Materials and Methods. Only p38 phosphorylated the WT NHE-GST fusion protein, an activity which increased 1.8-fold during IL-3 withdrawal (left panel). Protein lysates from FL5.12A cells were also immunoprecipitated with c-Jun fusion beads to capture JNK, which was tested in an in vitro kinase assay for its ability to phosphorylate the WT NHEGST fusion protein. Unlike p38 MAPK, JNK did not phosphorylate NHE (right panel). (B) Protein lysates extracted from anisomycin-activated FL5.12A cells were immunoprecipitated with antibody to phospho-p38 MAPK and a kinase assay performed as described in Materials and Methods using the WT NHE-GST fusion protein and seven mutant NHE-GST fusion proteins as substrates. GST alone was included as a negative control. (C) The results of the kinase assay from panel B are summarized in table form. In addition, phospho-p38 MAPK did not phosphorylate NHE-GST fusion proteins containing amino acids 635 to 663. Possible phosphorylation sites by p38 MAPK on NHE are located in the region from amino acids 703 to 743, which excludes the proposed p90RSK phosphorylation site (Serine 703, numbering from human NHE1 sequence). (D) FL5.12A cells, cultured without IL-3 for 0, 1, 2, and 3 h or treated with anisomycin (+), were lysed, and the precleared protein extracts were immunoprecipitated with the A7 antibody specific for NHE1 and then run on an 8% Tris-glycine SDS gel and analyzed by Western blotting with the p-TP-specific antibody (left panel). The same blots were then reprobed with a commercially available anti-NHE1 antibody (right panel). NHE1 did not contain an activated p-TP motif. (E) Mass spectrometry analysis, as described in Materials and Methods, identified four sites at Thr 717, Ser 722, Ser 725, and Ser 728 as in vitro targets for p38 MAPK activity. Phosphorylation of one or more of these sites by p38 MAPK modulates alkalinization mediated by NHE during trophic factor withdrawal.
The p38 MAPK and SAPK/JNK kinase assays were performed with a commercially available kits (Cell Signaling Technologies) as specified by the manufacturer. Lysates extracted as described above were immunoprecipitated overnight with an antibody specific for phospho-p38 MAPK conjugated to beads or with c-Jun fusion beads for phosphorylated SAPK/JNK. Phosphorylated ATF-2 or c-Jun were detected by Western blotting with a specific antibody as described in the kit instructions (Cell Signaling Technologies). For detection of the kinase-phosphorylated NHE-GST fusion proteins, [γ-32P]ATP was incorporated in the kinase assay mix and samples were analyzed on 12% Tris-glycine SDS gels by autoradiography. Gel Code blue stain reagent (Pierce) was used to stain the total concentration of NHE-GST fusion proteins run in the 12% Tris-glycine SDS gels.
Phosphothreonine proline protein detection.
To detect proteins containing the phosphorylated threonine-proline motif in cell lysates by Western blotting, whole-cell lysates, made from 2 × 107 cells, were first precleared with nonimmune mouse sera and protein A-Sepharose and then immunoprecipiated with a mouse antibody targeting phosphothreonine proline (Cell Signaling Technologies); alternatively, to detect such proteins associated with NHE1, lysates were first immunoprecipitated with the anti-NHE1 (A7) antibody as described above. Using protein A-Sepharose beads to bind protein-antibody complexes, the recovered protein was analyzed on 8% Tris-glycine SDS gels, transferred, blotted as previously described above using first the anti-phosphothreonine antibody (diluted 1:1,000), then reblotted with a commercially available anti-NHE1 mouse monoclonal antibody (diluted 1:1,000) (Chemicon), as indicated in the figure legends, followed by the appropriate secondary mouse antibody conjugated to HRP (Santa Cruz), and then developed by ECL (Pierce) and visualized on BioMax ML film.
In-gel tryptic digestion or formic acid cleavage of gel-separated NHE-GST fusion proteins.
For mass spectrometry analysis, the WT NHE-GST fusion protein was phosphorylated in vitro by immunoprecipitated phospho-p38 MAPK as described above. Phosphorylated (or unphosphorylated) fusion protein was run on a 12% Tris–glycine–SDS gel and stained with Gel Code blue stain reagent, and the stained gel bands were excised, cleaned, dehydrated, dried, and digested with trypsin by standard procedures (57). Tryptic digestion was carried out at 37°C for 16 to 20 h. The extracted peptides were pooled and dried with a SpeedVac. A 6-μl volume of 50% acetonitrile in water was added to reconstitute the peptide solution for mass spectrometry analysis. Alternatively, approximately 200 μl of freshly diluted 2% formic acid was added to the dry protein gel band in a microcentrifuge tube. After complete rehydration of the gel, the mixture was incubated at 108°C in a Thermolyne Dri-Bath. The resulting supernatant was then transferred to a fresh microcentrifuge tube and dried for further analysis by mass spectrometry as described below.
Phosphatase treatment of peptides.
A 2-μl volume of the reconstituted trypsin digest or formic acid cleavage peptide products was dried under vacuum, mixed with 2 μl of 0.5-U μl of calf intestine phosphatase (New England Biolabs, Beverly, Mass.) in 50 mM NH4HCO3, and incubated at 37°C for 1.5 h. The reaction mixture was SpeedVac dried and reconstituted in 1.5 μl of 50% acetonitrile in water for mass spectrometry analysis.
Mass spectrometry.
A MALDI-TOF mass spectrometer with delayed extraction (Voyager-DE PRO; Perseptive Biosystems, Framingham, Mass.) was used. The matrix solution used was α-cyano-4-hydroxycinnamic acid in methanol (Agilent Technology). Aliquots of 0.5 μl of the peptide mixture and 0.5 μl of the matrix solution were mixed on the sample plate and air dried prior to analysis. All spectra were taken in reflectron mode. Masses were calibrated internally with peptides from trypsin autolysis or from NHE1.
RESULTS
Intracellular alkalinization occurs following trophic factor withdrawal (23, 24) or ceramide treatment (3). The mechanism of this alkalinization was examined using two cytokine-dependent cell lines, D1, a thymocyte cell line that is dependent on IL-7 for survival, and FL5.12A, a pro-B-cell line that is dependent on IL-3 for survival. Withdrawal of the required cytokine from either cell line results in cell death by 48 and 72 h, respectively.
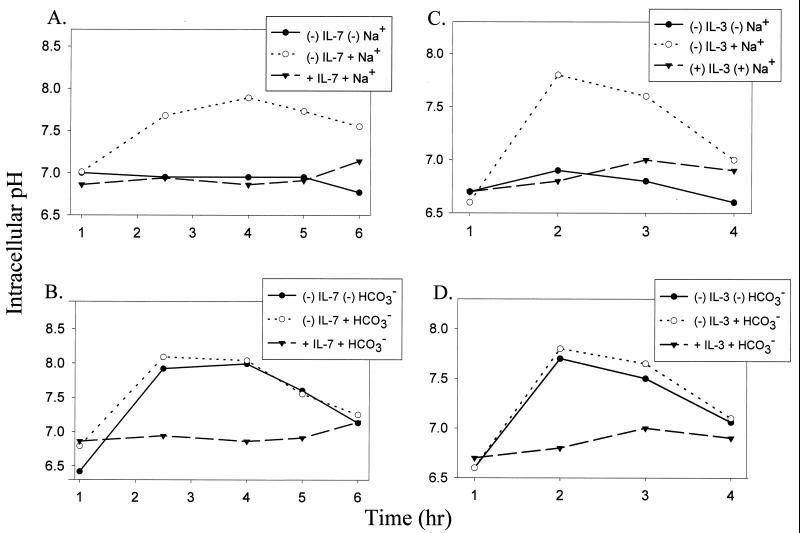
To distinguish among several membrane ion exchangers that could potentially cause the observed alkalinization, we first evaluated the requirement for sodium or bicarbonate. Intracellular pH was measured using the pH-sensitive fluorochrome BCECF, specifically reacts to pH within the cytosolic compartment (56). Figure 1 demonstrates the requirement for sodium in alkalinization following either IL-7 or IL-3 withdrawal (Fig. 1A and C), whereas removal of bicarbonate did not affect alkalinization (Fig. 1B and D). The requirement for sodium and independence of bicarbonate tentatively implicated the NHE.
FIG. 1.
Intracellular alkalinization following IL-7 or IL-3 withdrawal is dependent on sodium but not bicarbonate. (A) D1 cells were deprived of IL-7 for 0 to 6 h and incubated in either Na+-containing buffer or Na+-free buffer. D1 cells were maintained in Na+-containing buffer with IL-7 as a handling control. (B) D1 cells were deprived of IL-7 for 0 to 6 h and incubated in either HCO3−-containing medium or HCO3−-free medium. D1 cells were maintained in HCO3−-containing medium with IL-7 as a handling control. (C) FL5.12A cells, deprived of IL-3 for 0 to 4 h, were incubated in either Na+containing buffer or Na+-free buffer. FL5.12A cells were maintained in Na+-containing buffer with IL-3 as a handling control. (D) FL5.12A cells, deprived of IL-3 for 0 to 4 h, were incubated in either HCO3-containing medium or HCO3−-free medium. FL5.12A cells were maintained in HCO3−containing medium with IL-3 as a handling control. Intracellular pH was assayed using the pH-sensitive fluorescent probe BCECF, and pH values were derived from a pH calibration curve generated through the use of nigericin as described in Materials and Methods.
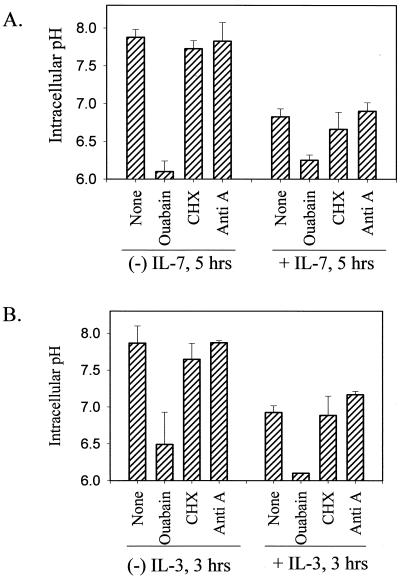
We next disrupted the sodium gradient, generated by the plasma membrane Na+/K+ ATPase, with ouabain. As shown in Fig. 2, ouabain prevented intracellular alkalinization during either IL-7 (Fig. 2A) or IL-3 (Fig. 2B) withdrawal. In contrast, inhibition of de novo protein synthesis with cycloheximide or inhibition of de novo ATP synthesis with antimycin A did not prevent the alkalinization process (Fig. 2). We further evaluated the sodium flux across the plasma membrane following cytokine withdrawal by using 22Na and observed that 22Na efflux (export versus import) increased during cytokine withdrawal and that this increase could be reversed by the addition of ouabain (data not shown). Hence, sodium and the maintenance of a sodium gradient across the plasma membrane are components in the process of alkalinization induced by cytokine withdrawal.
FIG. 2.
Disrupting the plasma membrane sodium gradient blocks IL-7- or IL-3-induced alkalinization, but inhibition of protein or ATP synthesis does not. (A) D1 cells were incubated with or without IL-7 for 5 h and not treated (None) or treated with either 1 mM ouabain or 10 μg of cycloheximide (CHX) per ml. To inhibit de novo ATP synthesis, cells were pretreated with 1 μg of antimycin A (Anti A) for 1 h. (B) FL5.12A cells were incubated with or without IL-3 for 3 h and treated or not treated as described above for D1 cells. The intracellular pH was assayed as described in Materials and Methods.
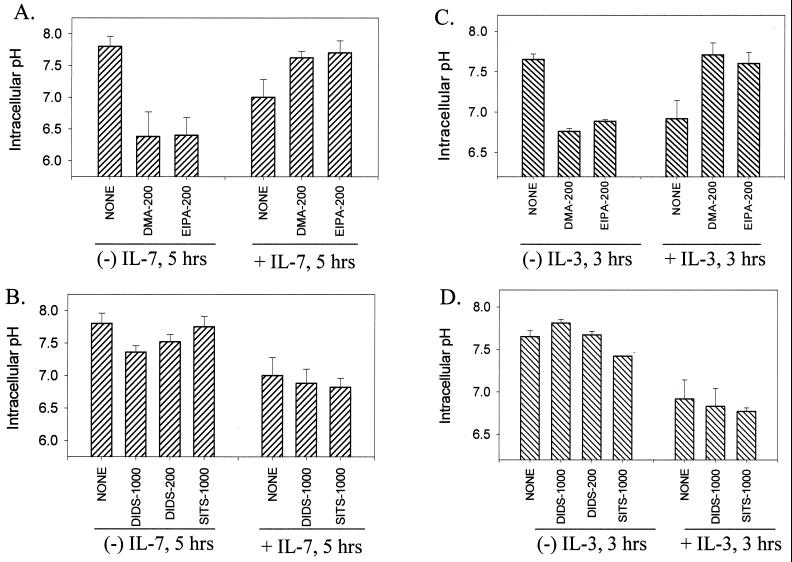
Regulators of intracellular pH include the NHEs as well as the sodium-dependent and -independent AEs. We tested several specific inhibitors of the ion exchangers for their effect on alkalinization induced by cytokine withdrawal. In Fig. 3 are shown the effect of either DMA or EIPA (NHE inhibitors) versus DIDS and SITS (AE inhibitors) in culture with the D1 or FL5.12A cells with or without cytokines. Only the NHE inhibitors, DMA and EIPA, prevented the pH rise induced by withdrawal of IL-7 (Fig. 3A) or IL-3 (Fig. 3C). Surprisingly, the same NHE inhibitors induced alkalinization when added to cells in the presence of either IL-7 (Fig. 3A) or IL-3 (Fig. 3C). This appears to result from overactivation of AE when NHE is blocked (data not shown), suggesting that the presence of a functional NHE is required for the regulation of HCO3−-dependent transporters, as has been shown in the CA1 neurons of NHE1 mutant mice (60). The use of the AE inhibitors DIDS (which blocks sodium-independent AE) and SITS (which blocks sodium-dependent AE), had no effect on alkalinization induced by either IL-7 withdrawal (Fig. 3B) or IL-3 withdrawal (Fig. 3D). Likewise, these inhibitors did not alter intracellular pH levels in the presence of either cytokine (Fig. 3B, 3D). Therefore, our results identify NHE as the mediator of alkalinization following cytokine withdrawal based on sodium dependence, requirement for a sodium gradient, and the effect of inhibitors of NHE.
FIG. 3.
Inhibitors of the NHE (but not the anion exchangers) prevent intracellular alkalinization following IL-7 or IL-3 withdrawal. (A) D1 cells were cultured in the presence or absence of IL-7 for 5 h and either not treated (None) or treated with 200 μM DMA or EIPA (inhibitors of the NHE). (B) D1 cells were cultured in the presence or absence of IL-7 for 5 h and either not treated (None) or treated with 200 or 1,000 μM DIDS or 1,000 μM SITS (inhibitors of the anion exchangers). (C) FL5.12A cells, incubated with or without IL-3 for 3 h, were either not treated (None) or treated with 200 μM DMA or EIPA (inhibitors of the NHE). (D) FL5.12A cells, incubated with or without IL-3, were either not treated (None) or treated with 200 or 1,000 μM DIDS or 1000 μM SITS (inhibitors of the anion exchangers). The intracellular pH was assayed as described in Materials and Methods.
Since various signaling pathways regulate NHE, including those involving the MAPKs, we next examined the pathway leading to activation of NHE. Using anisomycin, which activates stress-signaling pathways, we observed the induction of intracellular alkalinization in the presence of IL-3 (Fig. 4A) and an accelerated induction of alkalinization during withdrawal of IL-3 (Fig. 4A). Similar results were obtained with IL-7-dependent D1 cells (data not shown). This suggested that activation of stress pathways involving MAPKs could potentially mediate alkalinization following the withdrawal of trophic cytokines.
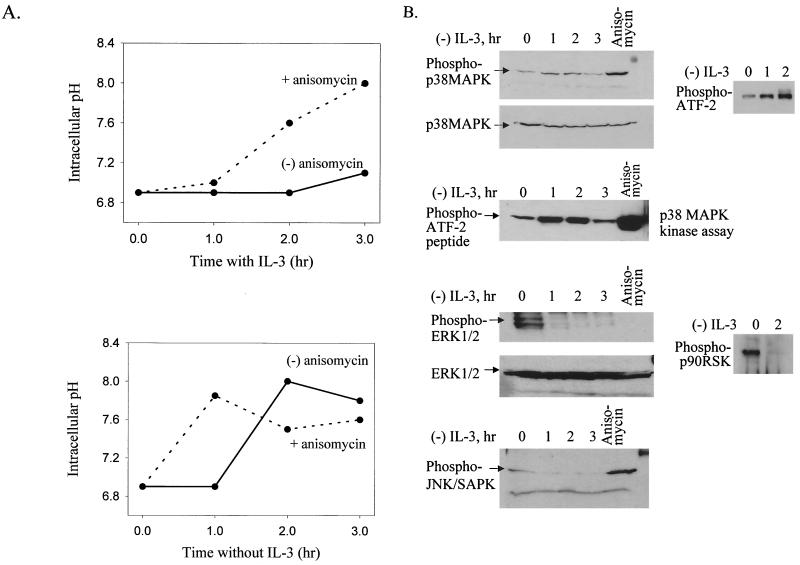
FIG. 4.
IL-3 withdrawal and anisomycin treatment both activate p38 MAPK and induce alkalinization. (A) Treatment with anisomycin (a p38 MAPK activator) induces alkalinization in the presence (top) or absence (bottom) of IL-3. FL5.12A cells were treated with 100 μg of anisomycin per ml. The intracellular pH was measured as described in Materials and Methods. (B) IL-3 withdrawal activates the phosphorylation of p38 MAPK and ATF-2 but not ERK1/2, p90RSK, or JNK. FL5.12A cells were incubated without IL-3 for 0 to 3 h, as well as stimulated with anisomycin, and levels of phosphorylated p38 and phosphorylated ATF-2 (top panel), phosphorylated ERK1/2 or phosphorylated p90RSK (second panel from bottom), and phosphorylated JNK (bottom panel) were evaluated from whole-cell lysates. Nonphosphorylated p38 and ERK were included as loading controls. In addition, p38 MAPK was immunoprecipitated from protein lysates, made from cells incubated without IL-3 for 0 to 3 h, and used in an in vitro kinase assay to phosphorylate an ATF-2 peptide (second panel from top). SDS-polyacrylamide gel electrophoresis and Western blot analysis were performed as described in Materials and Methods.
To determine which of the MAPK pathways could be involved in alkalinization, we evaluated the phosphorylation status of different MAPKs during IL-3 or IL-7 withdrawal. In Fig. 4B are shown the results of Western blot analyses for the phosphorylated (activated) and nonphosphorylated forms of p38 MAPK and its substrate ATF-2, ERK1/2 and its substrate p90RSK, and JNK. The levels of phospho-p38 MAPK increased following IL-3 withdrawal (Fig. 4B, top) at time points corresponding to the alkalinization period (Fig. 1C and D). Although a representative phospho-p38 blot is shown (Fig. 4B), these results were reproduced in numerous experiments. The levels of a downstream substrate of p38 MAPK, phospho-ATF-2, also increased in parallel to those of phospho-p38 (Fig. 4B, top), suggesting increased activation of the p38 MAPK pathway following IL-3 withdrawal. However, since ATF-2 is not a specific p38 MAPK substrate, we further confirmed that p38 MAPK activity is increased during IL-3 withdrawal by performing a p38 MAPK in vitro kinase assay with immunoprecipitated p38 MAPK and an ATF-2 peptide as substrate (Fig. 4B, second panel from the top). We also examined the phosphorylation of MKK3/6, upstream activators of p38 MAPK, and found elevated phosphorylated protein levels during the withdrawal of IL-3 (data not shown). The phosphorylation of p38 MAPK was transient, in that 3 to 4 h after IL-3 withdrawal the level of phospho-p38 MAPK declined (Fig. 4B, top) and was undetectable by 5 h (data not shown); this decline coincided with the decline in intracellular pH. As a positive control, anisomycin-treated cells showed increased phospho-p38 MAPK (Fig. 4B, top).
In contrast to the rise in phospho-p38 MAPK, the opposite pattern was observed in levels of phospho-ERK1/2, which rapidly declined following IL-3 withdrawal, as did the levels of its substrate, phospho-p90RSK (Fig. 4B, second panel from the bottom). Therefore, IL-3 stimulation (not withdrawal) activated phospho-ERK1/2 and its downstream substrate, p90RSK. The levels of phospho-JNK, like ERK1/2, were stimulated by IL-3 and decreased following IL-3 withdrawal; however, the effect of anisomycin was stimulatory, resembling that of phospho-p38 rather than ERK1/2 (Fig. 4B, bottom). JNK activity during trophic factor withdrawal was further assayed using an in vitro kinase assay with c-Jun as a substrate, and no significant kinase activity was detected (data not shown).
Similar to the IL-3-dependent FL5.12A cells, up-regulation of phospho-p38 MAPK occurred following IL-7 withdrawal in the D1 thymocyte cell line and was concurrent with alkalinization (4 to 6 h post-withdrawal) (data not shown). However, phospho-ERK1/2 was not detected in either the presence or absence of IL-7, as previously observed in this cell line (E. Rajnavolgyi et al., unpublished results). Therefore, in both IL-3-dependent and IL-7-dependent cells, cytokine withdrawal up-regulated phospho-p38 MAPK and induced alkalinization, suggesting a connection. Although the ERK pathway is potentially involved in IL3 (but not IL-7)-mediated NHE regulation, we conclude that the ERK and JNK pathways are not involved in the IL-3 or IL-7 withdrawal-induced alkalinization. Hence, the alkalinization induced by anisomycin treatment (Fig. 4A) was probably due to activation of p38 MAPK and not JNK, which is especially true during IL-3 withdrawal, when JNK is undetectable (Fig. 4B, bottom).
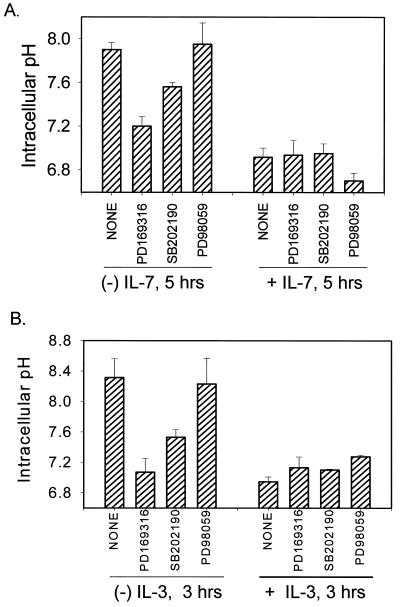
To examine the relationship between activation of p38 MAPK, the process of intracellular alkalinization and trophic factor withdrawal-induced death, we tested specific inhibitors of either p38 MAPK (PD169316 and SB202190) or MEK1 (PD98059), the upstream activator of ERK, to block alkalinization induced by cytokine withdrawal. The optimal dose response for the pharmacological inhibitors was previously established (data not shown). During IL-7 withdrawal, we observed maximal inhibition of alkalinization (peaking at 5 h after IL-7 withdrawal) with the p38 inhibitor, PD169316, and partial inhibition with SB202190, while the MEK1 inhibitor, PD98059, had no effect (Fig. 5A). Likewise, the p38 inhibitors PD169316 and SB202190 also blocked IL-3 withdrawal-induced alkalinization (peaking 3 hours post-IL-3-withdrawal) with greater efficacy (Fig. 5B). These inhibitors had little or no effect on cells maintained in the presence of the cytokines (Fig. 5A and B). We also tested the effects of dimethyl sulfoxide alone (carrier for the inhibitors) and found no artifactual variations in intracellular pH (data not shown). The different effects of the p38 MAPK inhibitors were correlated with their activity, as shown in Fig. 5C, in an in vitro p38 MAPK assay. PD169316 was the best inhibitor of p38 MAPK activity, with SB202190 being less effective and another inhibitor, SB203580, being the least effective (Fig. 5C). Hence, inhibition of p38 MAPK by pharmacological inhibitors blocked intracellular alkalinization induced by cytokine withdrawal, while inhibition of the ERK pathway did not block alkalinization (however, the ERK pathway did contribute to survival as part of IL-3 signaling [data not shown]).
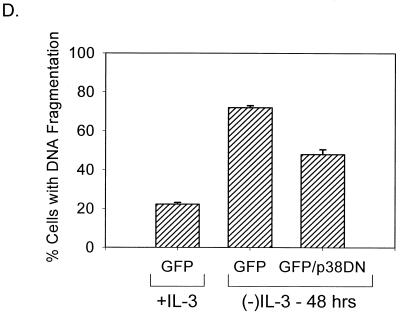
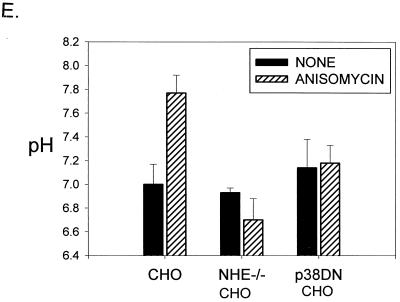
FIG. 5.
Inhibitors of p38 MAPK block alkalinization induced by IL-7 or IL-3 withdrawal and enhance survival. (A) D1 cells were cultured in the presence or absence of IL-7 for 5 h and either not treated (None) or treated with 20 μM PD169316, 20 μM SB202190 (inhibitors of p38 MAPK), or 20 μM PD98059 (inhibitor of MEK1). (B) FL5.12A cells were incubated with or without IL-3 for 3 h and either not treated (None) or treated with 20 μM PD169316, 20 μM SB202190 (inhibitors of p38 MAPK), or 20 μM PD98059 (inhibitor of MEK1). Intracellular pH was measured as described in Materials and Methods. (C) The efficacy of the different MAPK inhibitors (20 μM) was tested in a p38 MAPK in vitro kinase assay using p38 MAPK immunoprecipiatated from anisomycintreated FL5.12A cells. Of the three p38 MAPK inhibitors assayed, PD169316 was the most effective in blocking p38 MAPK activity (shown at two concentrations, 20 and 10 μM), SB202190 was the second best, and SB203580 had minimal effect; the ERK inhibitor (PD98059) or the nonspecific inhibitor (SB202474) had no effect at the 20 μM dose. (D) GFP+ cells or GFP+ cells cotransfected with a DN-p38 MAPK were cultured in the absence of IL-3 for 48 h, and the percentage of cells with DNA fragmentation was measured by propidium iodide staining and flow cytometry analysis. Expression of the DN-p38 MAPK decreased DNA fragmentation by 20% during IL-3 withdrawal. GFP+ cells cultured in the presence of IL-3 were included as a positive control for survival. (E) Loss of NHE or expression of a DN-p38 MAPK in CHO cells prevents alkalinization after anisomycin treatment. CHO cells were made deficient for NHE as previously described (39) and/or transfected with DN-p38 MAPK and evaluated 24 h later for pH changes after 1 h of anisomycin treatment by incorporation of BCECF as described in Materials and Methods.
To determine whether the p38 MAPK pathway was involved in inducing cell death following cytokine withdrawal, we first tested the pharmacological inhibitors of p38 MAPK, used in the preceding experiments for measuring pH, and found that these inhibitors, during longer treatments (i.e., 15-h cultures), were toxic to cells. We then expressed a dominant negative (DN) p38 MAPK in the FL5.12A cells and quantitated cell death, induced by IL-3 withdrawal, by measuring DNA fragmentation. Shown in Fig. 5D is the result of one such experiment in which cells were transfected with GFP or cotransfected with GFP and DN p38 MAPK and the GFP+ cells were cultured with or without IL-3 for 48 h. Expression of the DN p38 MAPK protected a significant proportion of cells (20%) from DNA fragmentation induced by IL-3 withdrawal. Complete protection was not achieved, perhaps due to the transient expression of the construct and the added stress of transfection and cell sorting. In addition, GFP expression alone had a modest effect on cell survival during cytokine withdrawal (data not shown). It also should be noted that assaying the effects of DN p38 MAPK expression is complicated by the fact that p38 MAPK regulates cell cycling; inhibition of p38 MAPK activity can increase cell division (12). In summary, despite the technical difficulties of transfection and expression of the DN p38 MAPK, we favor the interpretation that p38 MAPK contributes to cell death following trophic factor withdrawal.
We next examined whether expression of the DN p38 MAPK could inhibit alkalinization. Since the emission spectra of the fluorescent probe BCECF overlapped with that of GFP, we could not use the same approach as was employed in Fig. 5D to select transfected cells. Hence, we chose to use CHO cells, which can be readily transfected at efficiencies of 80 to 90%. Treatment with anisomycin was selected as the means of inducing alkalinization (previously evaluated in Fig. 4A). Multiple experiments are summarized in Fig. 5E, showing that CHO cells alkalinize after 1 h of anisomycin treatment. CHO cells made deficient for NHE (39) do not increase pH on anisomycin treatment, and CHO cells transfected with DN p38 MAPK also do not alkalinize on anisomycin treatment. These findings confirm the results obtained with pharmacological inhibitors for NHE (Fig. 3) and p38 MAPK (Fig. 5A).
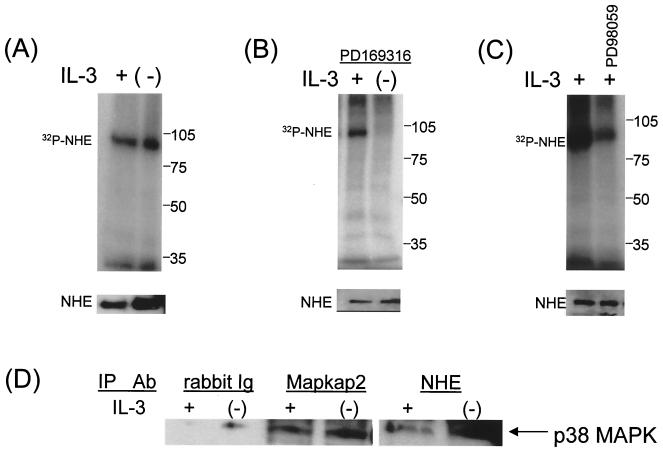
Since inhibition of either p38 MAPK or NHE prevented intracellular alkalinization following IL-7 or IL-3 withdrawal, we examined the phosphorylation of NHE for effects of cytokine withdrawal and MAPKs. Figure 6 displays the results, representative of multiple experiments, in which cells were radiolabeled with [32P]orthophosphate and lysed and NHE1 was immunoprecipitated with a specific antibody, confirming the presence of this isoform. As a control for the immunoprecipitation procedure, the lower panels show equivalent amounts of unlabeled NHE1 protein obtained using a different anti-NHE1 antibody for detection from that used for immunoprecipitation (Fig. 6). As shown, NHE1 was phosphorylated in either the presence or absence of either cytokine (data for IL-3-dependent cells are displayed in Fig. 6A). The total amount of NHE1 protein per cell was much greater in the IL-3-dependent FL5.12A cells than the IL-7-dependent D1 cells, as seen in a comparative Western blot for NHE1 (data not shown), so the subsequent phosphorylation studies are shown in the former cell type only. The p38 inhibitor PD169316 dramatically inhibited the phosphorylation of NHE1 following IL-3 withdrawal (Fig. 6B) but not in the presence of the cytokine. The MEK1 inhibitor PD98059 did not decrease NHE1 phosphorylation induced by cytokine withdrawal (data not shown), although it did inhibit the phosphorylation of NHE1 in the presence of IL-3 (Fig. 6C). This suggests that two different phosphorylation pathways act on NHE1: in the presence of IL-3, the ERK MAPK pathway results in normal NHE1 function, whereas in the absence of IL-3, the p38 MAPK pathway induces NHE1 to alkalinize the cytosol.
FIG. 6.
Phosphorylation of NHE1 occurs via different MAPKs in the presence and absence of IL-3. (A) FL5.12A cells were incubated for 3 h with [32P]orthophosphate in the presence (+) or absence (−) of IL-3. (B) 32P-labeled FL5.12A cells were incubated for 3 h with PD169316 with (+) or without (−) IL-3. (C) 32P-labeled FL5.12A cells were incubated with or without PD98059 in the presence (+) of IL-3. Cells were lysed, and NHE1 was immunoprecipitated and electrophoresed on SDS–8% polyacrylamide gels and autoradiography was performed as described in Materials and Methods. To confirm the specificity of the NHE antibody (A7) used for immunoprecipitation, parallel cell lysates (not labeled with [32P]orthophosphate) were immunoprecipitated with the anti-NHE antibody (A7), electrophoresed on SDS–8% polyacrylamide gels, analyzed by Western bloting using a different anti-NHE1 antibody, and shown in the lower panels. (D) In vivo coimmunoprecipitation of NHE and p38 MAPK. Protein lysates extracted from cells cultured with or without IL-3, and chemically cross-linked with DSS, were immunoprecipitated with either an antibody specific for NHE1, an antibody for Mapkap2 (positive control), or rabbit Ig (negative control). The lysates were blotted for p38 MAPK as described in Materials and Methods.
Having shown that p38 MAPK inhibitors blocked NHE1 phosphorylation following IL-3 withdrawal (Fig. 5A and B), we explored the possibility that p38 MAPK directly phosphorylated NHE1. To test whether p38 MAPK physically interacted with NHE1 in vivo, it was necessary to treat FL5.12A cells with a chemical crosslinker (disuccinimidyl suberate) to observe any associations. Cells treated with cross-linker were lysed, and NHE1 was immunoprecipitated and blotted for coassociation with p38 MAPK. As shown in Fig. 6D, NHE1 and p38 MAPK did coimmunoprecipitate, and the association increased following IL-3 withdrawal. Mapkap2 is shown as a positive control for a substrate known to associate with p38 MAPK, whereas normal rabbit Ig served as a negative control for immunoprecipitation.
Further evidence that p38MAPK could directly phosphorylate NHE1 was obtained by performing an in-gel kinase assay with an NHE-GST fusion protein as the substrate. The results demonstrated that lysates from FL512.A cells undergoing IL-3 withdrawal contained a kinase of approximately 40 kDa that was capable of phosphorylating the C terminal of NHE1 (data not shown). This kinase was then identified as p38 MAPK by immunoprecipitating phospho-p38 MAPK, which then phosphorylated the NHE-GST fusuin protein in an in-gel assay, as shown in Fig. 7A (left panel). Additionally, to determine if any other kinases in the MAPK pathway could phosphorylate NHE1 during IL-3 withdrawal, we tested an antibody specific for the motif, phosphorylated threonine-proline (p-TP), that is created on its substrates by MAPKs (28). Only the immunoprecipitated phospho-p38 MAPK phosphorylated the NHE-GST fusion protein, as shown in Fig. 7A (left panel), with a 1.8-fold increase in activity during IL-3 withdrawal. Therefore, in vitro, p38 MAPK, activated by IL-3 withdrawal, is capable of directly phosphorylating NHE1, and no other substrates of the MAPK pathway containing active p-TP sites (i.e., Mapkap2) are involved.
Anisomycin induced alkalinization, as shown in Fig. 4A. Since anisomycin activated both p38 MAPK and JNK (Fig. 4B), we next determined if JNK could phosphorylate the NHE-GST fusion protein in an in vitro kinase assay. Results shown in Fig. 7A (right panel) demonstrate that JNK, precipitated from cells treated or not treated with IL-3 and treated with anisomycin, does not phosphorylate NHE1 as does p38 MAPK. The levels of c-Jun phosphorylation are shown to indicate JNK activity. The slight phosphorylation of NHE1 by the JNK preparation is probably due to contamination by p38 MAPK, since elimination of p38 MAPK by preimmunoprecipitation decreased this activity.
To determine the phosphorylation site for p38 MAPK in the NHE1 C-terminal region, NHE-GST fusion proteins based on the rabbit NHE1 sequence were made containing various deletions. The results of these experiments are shown in Fig. 7B and summarized in Fig. 7C. Phospho-p38 MAPK phosphorylated the WT NHE-GST fusion protein containing amino acids 637 to 815. To test whether p38 MAPK phosphorylated the same site on NHE1 as shown for p90RSK (46), this residue, serine 703 (numbering from the human NHE1 sequence), was changed to alanine. This mutation did not block phosphorylation by phospho-p38 MAPK (Fig. 7B), implicating a different target site(s) for p38 MAPK on NHE1 than the proposed target of p90RSK. Deleting amino acids 702 to 815 or 661 to 793 eliminated phosphorylation of NHE1 (Fig.7B). This first group of NHE-GST deletion proteins defined the target site(s) on NHE1 for p38 MAPK phosphorylation to a region from amino acids 703 to 793 (Fig. 7C). In a second group of experiments, testing NHE-GST deletion proteins with stops at amino acids 744, 762, and 777, the location of the site for p38 MAPK phosphorylation of NHE was further narrowed to a region between amino acids 703 and 743 (Fig. 7B and C). Furthermore, NHE-GST fusion proteins spanning the regions from amino acids 635 to 663 (the proposed sites for calmodulin binding [54]) were not phosphorylated by p38 MAPK (Fig. 7C).
To characterize the p38 MAPK phosphorylation site further, we sought the p-TP motif that many of its substrates, like MapKap2, display. However, immunoprecipitation and Western blot analysis did not reveal the p-TP motif on NHE (Fig. 7D). The 40-amino-acid region defined by deletion analysis contained multiple threonine and serine residues. To determine which of these sites were the targets for p38 MAPK phosphorylation, we performed mass spectroscopy on the WT NHE-GST fusion protein phosphorylated in vitro by immunoprecipiated p38 MAPK. For comparison, the unphosphorylated NHE-GST fusion protein was also tested. Mass spectrometry analysis of NHE-GST peptides digested with either trypsin or formic acid revealed discrete phosphorylation sites on NHE1 between amino acids 711 and 739. Tryptic peptide fragments, corresponding to amino acids 711 to 739, contained one to three phosphates. Peptide fragments from formic acid cleavage, spanning amino acids 713 to 718, contained one phosphate, and those spanning amino acids 720 to 729 contained one to three phosphates. Treatment with calf intestinal phosphatase removed the phosphates from all these peptides, confirming the phosphorylated state. Therefore, we concluded, from both the deletion analysis and mass spectrometry, that there are four targets on NHE1 for in vitro phosphorylation by p38 MAPK-Thr 717, Ser 722, Ser 725, and Ser 728 (Fig. 7E). Two of the serines, Ser 722 and Ser 725, are located adjacent to prolines, representing common MAPK motifs. It remains to be determined how phosphorylation by p38 MAPK of these sites on NHE1 induces the intracellular alkalinization that is triggered by cytokine withdrawal.
DISCUSSION
Loss of cytokine receptor signaling induces apoptotic cell death in dependent cell lines. To understand this process, we have defined early events (i.e., pH rise) following cytokine withdrawal that lead to sequelae such as mitochondrial hyperpolarization, loss of bioenergetics, and the mitochondrial translocation of Bax, culminating in cell death (23, 24). Here we show that the rapid alkalinization of the cytosol was mediated by the membrane exchanger NHE1 through activation by the stress kinase p38 MAPK. Phosphorylation of p38 MAPK increased within the first hours of IL-3 or IL-7 withdrawal, while levels of other MAPKs, ERK or JNK, decreased. A DN-p38 MAPK could protect cells from DNA fragmentation induced by IL-3 withdrawal and alkalinization induced by anisomycin treatment. p38 MAPK directly phosphorylated NHE between amino acids 704 and 743, on one threonine (Thr 717) and three serines (Ser 722, Ser 725, and Ser 728), as defined by mass spectrometry analysis. Therefore, our data suggest a model in which cytokine withdrawal activates the p38 MAPK pathway leading directly to the phosphorylation and activation of NHE1, the membrane ion exchanger responsible for intracellular alkalinization, a critical trigger in the apoptotic process.
Although NHE is normally involved in maintaining physiological pH, there are other reports of NHE-mediated intracellular alkalinization. Stimulation of platelets with agonists like thrombin lead to a rapid rise in cytosolic pH, induced by increased transport activity of NHE, of approximately 0.4 pH unit (38). Thymocytes that underwent spontaneous apoptosis were also highly alkaline (pH > 7.6), and depletion of sodium could prevent the death process, as did inhibitors of the membrane ion exchangers (50). Our own studies show that alkalinization contributes to cell death in several ways. (i) Alkalinization activates Bax, inducing its mitochondrial translocation (23), also shown to occur during ceramide treatment of U937 cells (3). (ii) Alkalinization inhibits the import of ADP into mitochondria, which severely impairs the synthesis of ATP, and heightens the mitochondrial membrane potential (24).
The NHE, expressed by many cell types, is regulated by diverse stimuli. One means by which intracellular pH can be modulated is by cellular adherence mediated through integrins (42). However, pH changes induced by cytokine loss were independent of adherence (data not shown). Fluxes of ions through the NHE are driven by the sodium gradient across the plasma membrane and do not directly require metabolic energy, although ATP can influence it (33). A number of studies demonstrated that the NHE is regulated by the association of calmodulin, which causes autoinhibition of the transporter (4); deletion of the calmodulin binding domain in NHE renders the exchanger constitutively activated, perhaps by altering its set point for “pH sensing” (53, 54). In our studies, we did not identify any sites in the calmodulin binding domain of NHE as targets for phosphorylation by p38 MAPK. Hence, modification of calmodulin binding by p38 MAPK activity does not appear to account for alkalinization during cytokine withdrawal.
Phosphorylation of the NHE plays an essential role in its activation. NHE is constitutively phosphorylated in resting cells, and mitogenic stimulation leads to an increase in phosphorylation on serine residues in a time course that parallels the rise in intracellular pH (40). These results and others suggested that phosphorylation of the NHE directly triggered its activation (5, 41). Recently, MAPKs were examined as potential mediators of growth factor-induced activation of NHE1. Inhibition by either a dominant negative ERK or the ERK inhibitor PD98059 prevented growth factor-induced activation of the NHE (6, 31). However, it did not appear that ERKs were solely responsible for phosphorylating the NHE but, rather, that a downstream substrate of the ERK pathway, p90 RSK, could also in some instances be involved (31, 46). Therefore, ERKs, perhaps via p90RSK, are in the pathway from stimuli such as mitogens, serum, or growth factors leading to the activation of NHE. We found that IL-3 induced a similar pathway, in that IL-3 induced high levels of ERKs and p90RSK (Fig. 4B), and addition of the ERK inhibitor PD98059 decreased IL-3-stimulated NHE phosphorylation (Fig. 6C). However, the withdrawal-induced phosphorylation of NHE was not an ERK-mediated event but instead required the activation of a different kinase, p38 MAPK (Fig. 5 and 6B).
That intracellular alkalinization could activate the stress pathway was suggested in a study in which treatment of U937 cells with permeant weak bases increased the activity of JNK and p38 MAPK (43). The authors proposed that the activation of NHE and that of stress kinases were parallel pathways. However, our results clearly show that following cytokine withdrawal, activation of p38 MAPK is upstream of NHE phosphorylation (Fig. 6B) and alkalinization (Fig. 5A and E).
In another study, p38 MAPK was proposed to have an inhibitory effect on NHE (27) (in contrast to our findings, in which p38 MAPK has a stimulatory effect). In that report vascular smooth muscle cells were stimulated with angiotensin II, which activated both ERK and p38 MAPK, and ERK in turn activated NHE; it was suggested that p38 MAPK inhibited the function of ERK and therefore inhibited NHE. However, these results differ from the cellular responses to the two trophic cytokines studied here. The IL-7-dependent cell line used here lacks ERK (Rajnavolgyi et al., unpublished), and IL-7 was previously shown to not activate ERK in other cells (9, 14), whereas IL-3 withdrawal rapidly down-regulates ERK (and up-regulates p38 MAPK). Withdrawal of either IL-7 or IL-3 activated the p38 MAPK (independent of other MAPKs), and the pH rose. There are previous accounts of trophic-factor withdrawal causing a shift in the balance of p38 MAPK and ERK, with ERK activity decreasing as p38 MAPK activity increased (7, 59). Thus, our data show that following trophic factor withdrawal (and in the absence of ERK), p38 MAPK is an exceedingly potent inducer of NHE phosphorylation and activity.
The direct phosphorylation of NHE by MAPKs has been demonstrated by others in vitro. We initially detected in cell lysates a kinase of approximately 40 kDa that phosphorylated NHE in an in-gel kinase assay (data not shown). By immunoprecipitation, we confirmed that the kinase activity seen in the protein lysates from IL-3-deprived cells was p38 MAPK and not one of the other MAPK family member (i.e., ERK or JNK), nor was it a downstream substrate of p38 MAPK such as Mapkap2 (45) (data not shown and Fig. 7D). As seen in our results, p38 MAPK phosphorylated NHE at multiple sites in a region between amino acids 704 and 743 (Fig. 7B and C). In addition, p38 MAPK did not appear to phosphorylate the proposed p90 RSK target site, Serine 703 (human) (46). This suggests that whereas the RSK site on NHE activates NHE to accelerate the adjustment of acidic pH to physiological levels, the p38 site(s) activates NHE to create alkaline levels. Using mass spectrometry, we identified four in vitro phosphorylation sites for p38 MAPK on NHE at Thr 717, Ser 722, Ser 725, and Ser 728. We speculate that phosphorylation of the NHE in this region could create either a docking site for another protein, leading to deregulation and alkalinization, or repress an autoinhibitory regulatory domain.
One further feature of the cytokine withdrawal-induced alkalinization is its transience. Previously we reported that loss of IL-7 or IL-3 signaling led to a pH rise lasting 2 to 3 h (23, 24). We do not yet know the mechanism that returns the intracellular pH to neutrality. Since p38 MAPK is an upstream mediator of the alkalinization process, one of many possible mechanisms of normalization would be for alkaline pH to inhibit p38 MAPK activity. A recent study identified the docking sites in MAPKs and MAPK-interacting proteins and showed that such sites contain defined motifs characterized by the presence of charged amino acids (47). We and others have shown that charged amino acids in interacting peptides can be a basis for the pH- sensitivity of proteins (23, 34).
The upstream chain of events that activate the p38 MAPK pathway following cytokine withdrawal remain to be identified. The possibilities include G proteins, which have already been shown to regulate intracellular pH (52); RhoA, which activates the NHE (49); and other GTPases and signaling proteins that lie upstream of p38 MAPK activation (61). Another possibility is that cessation of Jak/STAT signaling following cytokine withdrawal may contribute to the loss of ERK and a compensatory activation of p38 MAPK observed during apoptotic death (58). It is also possible that induction of a dephosphorylation activity, as seen during cytomegalovirus infection, modulates p38 MAPK function (22). Whatever the initiating mechanism, the activation of p38 MAPK following IL-7 and IL-3 withdrawal, phosphorylation of specific sites, and subsequent activation of the NHE represents a novel pathway that links trophic factor deprivation to a death-promoting physiological consequence, intracellular alkalinization.
ACKNOWLEDGMENT
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-56000.
REFERENCES
- 1.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman I L. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 2.Aoshiba K, Yasui S, Hayashi M, Tamaoki J, Nagal A. Role of p38-mitogen-activated protein kinase in spontaneous apoptosis of human neutrophils. J Immunol. 1999;162:1692–1700. [PubMed] [Google Scholar]
- 3.Belaud-Rotreau M A, Leducq N, de Gannes F M P, Diolez P, Lacoste L, Lacombe F, Bernard P, Belloc F. Early transitory rise in intracellular pH leads to Bax conformation change during ceramideinduced apoptosis. Apoptosis. 2000;5:551–560. doi: 10.1023/a:1009693630664. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand B, Wakabayashi S, Ikeda T, Pouyssegur J, Shigekawa M. The Na+/H+ exchanger isoform 1 (NHE1) is a novel member of the calmodulin-binding proteins. Identification and characterization of calmodulin-binding sites. J Biol Chem. 1994;269:13703–13709. [PubMed] [Google Scholar]
- 5.Bianchini L, Woodside M, Sardet C, Pouyssegur J, Takai A, Grinstein S. Okadaic acid, a phosphatase inhibitor, induces activation and phosphorylation of the Na+/H+ antiport. J Biol Chem. 1991;266:15406–15413. [PubMed] [Google Scholar]
- 6.Bianchini L, L'Allemain G, Pouyssegur J. The p42/p44 mitogen-activated protein kinase cascade is determinant in mediating activation of the Na+/H+ exchanger (NHE1 isoform) in response to growth factors. J Biol Chem. 1997;272:271–279. doi: 10.1074/jbc.272.1.271. [DOI] [PubMed] [Google Scholar]
- 7.Birkenkamp K U, Dokter W H, Esselink M T, Jonk L J, Kruijer W, Vellenga E. A dual function for p38 MAP kinase in hematopoietic cells: involvement in apoptosis and cell activation. Leukemia. 1999;13:1037–1045. doi: 10.1038/sj.leu.2401447. [DOI] [PubMed] [Google Scholar]
- 8.Blom B, Spits H, Krimpenfort P. The role of the common gamma chain of the IL-2, IL-4, IL-7 and IL-15 receptors in development of lymphocytes. Constitutive expression of bcl-2 does not rescue the developmental defects in common gamma deficient mice. In: Smit Sibinga S, Das P, Loewenberg D, editors. Cytokines and growth factors in blood transfusion. Kluwer Academic Publishers, Dordrecht, The Netherlands. 1997. pp. 3–11. [Google Scholar]
- 9.Crawley J B, Willcocks J, Foxwell B M. Interleukin-7 induces T cell proliferation in the absence of Erk/MAP kinase activity. Eur J Immunol. 1996;26:2717–2723. doi: 10.1002/eji.1830261125. [DOI] [PubMed] [Google Scholar]
- 10.Crawley J B, Rawlinson L, Lali F V, Page T H, Saklatvala J, Foxwell B M. T cell proliferation in response to interleukins 2 and 7 requires p38MAP kinase activation. J Biol Chem. 1997;272:15023–15027. doi: 10.1074/jbc.272.23.15023. [DOI] [PubMed] [Google Scholar]
- 11.de Groot R P, Coffer P J, Koenderman L. Regulation of proliferation, differentiation and survival by the IL-3/IL-5/GM-CSF receptor family. Cell Signalling. 1998;10:619–628. doi: 10.1016/s0898-6568(98)00023-0. [DOI] [PubMed] [Google Scholar]
- 12.Diehl N L, Ensien H, Fortner K A, Merritt C, Stetson N, Charland C, Flavell R A, Davis R J, Rincon M. Activation of the p38 mitogen-activated protein kinase pathway arrests cell cycle progression and differentiation of immature thymocytes in vivo. J Exp Med. 2000;191:321–334. doi: 10.1084/jem.191.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Santo J P, Rodewald H R. In vivo roles of receptor tyrosine kinases and cytokine receptors in early thymocyte development. Curr Opin Immunol. 1998;10:196–207. doi: 10.1016/s0952-7915(98)80249-5. [DOI] [PubMed] [Google Scholar]
- 14.Dorsch M, Hock H, Diamantstein T. Gene transfer of the IL2 receptor beta chain into an IL-7-dependent pre-B cell line permits IL-2-driven proliferation: tyrosine phosphorylation of Shc is induced by IL-2 but not IL-7. Eur J Immunol. 1994;24:2049–2054. doi: 10.1002/eji.1830240917. [DOI] [PubMed] [Google Scholar]
- 15.English J, Pearson G, Wilsbacher J, Swantek J, Karandikar M, Xu S, Cobb M H. New insights into the control of MAP kinase pathways. Exp Cell Res. 1999;253:255–270. doi: 10.1006/excr.1999.4687. [DOI] [PubMed] [Google Scholar]
- 16.Franck P, Petitipain N, Cheriet M, Dardennes M, Maachi F, Schutz B, Poisson L, Nabet P. Measurement of intracellular pH in cultured cells by flow cytometry with BCECF-AM. J Biotechnol. 1996;46:187–195. doi: 10.1016/0168-1656(95)00189-1. [DOI] [PubMed] [Google Scholar]
- 17.Goldsmith M A, Mikami A, You Y, Liu K D, Thomas L, Pharr P, Longmore G D. Absence of cytokine receptor-dependent specificity in red blood cell differentiation in vivo. Proc Natl Acad Sci USA. 1998;95:7006–7011. doi: 10.1073/pnas.95.12.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herlaar E, Brown Z. p38 MAPK signaling cascades in inflammatory disease. Mol Med Today. 1999;5:439–447. doi: 10.1016/s1357-4310(99)01544-0. [DOI] [PubMed] [Google Scholar]
- 19.Hofmeister R, Khaled A R, Benbernou N, Rajnavolgyi E, Muegge K, Durum S K. Interleukin-7: physiological roles and mechanisms of action. Cytokine Growth Factor Rev. 1999;10:41–60. doi: 10.1016/s1359-6101(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 20.Ichijo H. From receptors to stress-activated MAP kinases. Oncogene. 1999;18:6087–6093. doi: 10.1038/sj.onc.1203129. [DOI] [PubMed] [Google Scholar]
- 21.Johnson D E. Regulation of survival pathways by IL-3 and induction of apoptosis following IL-3 withdrawal. Front Biosci. 1998;3:d313–d324. doi: 10.2741/a276. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R A. Activation of the mitogen-activated protein kinase p38 by human cytomegalovirus infection trhough two distinct pathways: a novel mechanism for activation of p38. J Virol. 2000;74:1158–1167. doi: 10.1128/jvi.74.3.1158-1167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaled A R, Kim K, Hofmeister R, Muegge K, Durum S K. Withdrawal of IL-7 induces Bax translocation from cytosol to mitochondria through a rise in intracellular pH. Proc Natl Acad Sci USA. 1999;96:14476–14481. doi: 10.1073/pnas.96.25.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaled A R, Reynolds D A, Young H A, Thompson C B, Muegge K, Durum S K. IL-3 withdrawal induces an early increase in mitochondrial membrane potential unrelated to the Bcl-2 family: roles of intracellular pH, ADP transport and FOF1-ATPase. J Biol Chem. 2001;276:6453–6462. doi: 10.1074/jbc.M006391200. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Lee C K, Sayers T J, Muegge K, Durum S K. The trophic action of IL-7 on pro-T cells: inhibition of apoptosis of pro-T1, -T2, and -T3 cells correlates with Bcl-2 and Bax levels and is independent of Fas and p53 pathways. J Immunol. 1998;160:5735–5741. [PubMed] [Google Scholar]
- 26.Kummer J L, Rao P K, Heidenreich K A. Apoptosis induced by withdrawal of trophic factors is mediated by p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:20490–20494. doi: 10.1074/jbc.272.33.20490. [DOI] [PubMed] [Google Scholar]
- 27.Kusuhara M, Takahashi E, Peterson T E, Abe J, Ishida M, Han J, Ulevitch R, Berk B C. p38 Kinase is a negative regulator of angiotensin II signal transduction in vascular smooth muscle cells: effects on Na+/H+ exchange and ERK1/2. Circ Res. 1998;83:824–831. doi: 10.1161/01.res.83.8.824. [DOI] [PubMed] [Google Scholar]
- 28.Lewis T S, Shapiro P S, Ahn N G. Signal transduction through MAP kinase cascades. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 29.Maraskovsky E, Teepe M, Morrissey P J, Braddy S, Miller R E, Lynch D H, Peschon J J. Impaired survival and proliferation in IL-7 receptor-deficient peripheral T cells. J Immunol. 1996;157:5315–5323. [PubMed] [Google Scholar]
- 30.Michaelson M D, Mehler M F, Xu H, Gross R E, Kessler J A. Interleukin-7 is trophic for embryonic neurons and is expressed in developing brain. Dev Biol. 1996;179:251–263. doi: 10.1006/dbio.1996.0255. [DOI] [PubMed] [Google Scholar]
- 31.Moor A N, Fliegel L. Protein kinase-mediated regulation of the Na(+)/H(+) exchanger in the rat myocardium by mitogen-activated protein kinase-dependent pathways. J Biol Chem. 1999;274:22985–22992. doi: 10.1074/jbc.274.33.22985. [DOI] [PubMed] [Google Scholar]
- 32.Nunez B, London L, Hockenbery D, Alexander M, McKearn J P, Korsmeyer S J. Deregulated Bcl-2 expression selectively prolongs survival of growth factor-deprived hemopoietic cell lines. J Immunol. 1990;144:3602–3610. [PubMed] [Google Scholar]
- 33.Orlowski J, Grinstein S. Na+H+ exchangers of mammalian cells. J Biol Chem. 1997;272:22373–22376. doi: 10.1074/jbc.272.36.22373. [DOI] [PubMed] [Google Scholar]
- 34.Papa S, Zanotti F, Cocco T, Perrucci C, Candita C, Minuto M. Identification of functional domains and critical residues in the adenosinetriphosphatase inhibitor protein of mitochondrial F0F1 ATP synthase. Eur J Biochem. 1996;240:461–467. doi: 10.1111/j.1432-1033.1996.0461h.x. [DOI] [PubMed] [Google Scholar]
- 35.Peschon J J, Morrissey P J, Grabstein K H, Ramsdell F J, Maraskovsky E, Gliniak B C, Park L S, Ziegler S F, Williams D E, Ware C B. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puceat M. pHi regulatory ion transporters: an update on structure, regulation and cell function. Cell Mol Life Sci. 1999;55:1216–1229. doi: 10.1007/s000180050368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinaudo M S, Su K, Falk L A, Haider S, Mufson R A. Human interleukin-3 receptor modulates bcl-2 mRNA and protein levels through protein kinase C in TF-1 cells. Blood. 1995;86:80–88. [PubMed] [Google Scholar]
- 38.Rosskopf D. Sodium-hydrogen exchange and platelet function. J Thromb Thrombolysis. 1999;8:15–24. doi: 10.1023/a:1008986329267. [DOI] [PubMed] [Google Scholar]
- 39.Rotin D, Norwood D, Grinstein S, Tannock I. Requirement of the Na+/H+ exchanger for tumor growth. Cancer Res. 1989;49:205–211. [PubMed] [Google Scholar]
- 40.Sardet C, Counillon L, Franchi A, Pouyssegur J. Growth factors induce phosphorylation of the Na+H+ antiporter, glycoprotein of 110 kD. Science. 1990;247:723–726. doi: 10.1126/science.2154036. [DOI] [PubMed] [Google Scholar]
- 41.Sardet C, Fafournoux P, Pouyssegur J. Alpha-thrombin, epidermal growth factor, and okadaic acid activate the Na+H+ exchanger, NHE-1, by phosphorylating a set of common sites. J Biol Chem. 1991;266:19166–19171. [PubMed] [Google Scholar]
- 42.Schwartz M A, Both G, Leohene C. Effect of cell spreading on cytoplasmic pH in normal and transformed fibroblasts. Proc Natl Acad Sci USA. 1989;86:4525–4529. doi: 10.1073/pnas.86.12.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrode L D, Rubie E A, Woodgett J R, Grinstein S. Cytosolic alkalinization increases stress-activated protein kinase/c-Jun NH2-terminal kinase (SAPK/JNK) activity and p38 mitogen-activated protein kinase activity by a calcium-independent mechanism. J Biol Chem. 1997;272:13653–13659. doi: 10.1074/jbc.272.21.13653. [DOI] [PubMed] [Google Scholar]
- 44.Socolofsky M, Lodish H F, Daley G Q. Control of hematopoietic differentiation: lack of specificity in signalling by cytokine receptors. Proc Natl Acad Sci USA. 1998;95:6573–6575. doi: 10.1073/pnas.95.12.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stokoe D, Campbell D G, Nakielny S, Hidaka H, Leevers S J, Marshall C, Cohen P. MAPKAP kinase-2: a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992;11:3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi E, Abe J, Gaills B, Aebersold R, Spring D J, Krebs E G, Berk B C. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J Biol Chem. 1999;274:20206–20214. doi: 10.1074/jbc.274.29.20206. [DOI] [PubMed] [Google Scholar]
- 47.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 48.Telford W G, King L E, Fraker P J. Evaluation of glucocorticoid-induced DNA fragmentation in mouse thymocytes by flow cytometry. Cell Prolif. 1991;24:447–449. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 49.Tominaga T, Barber D L. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol Biol Cell. 1998;9:2287–2303. doi: 10.1091/mbc.9.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsao N, Lei H Y. Activation of the Na(+)/H(+) antiporter, Na+/HCO3(−)/CO3(2−) cotransporter, or Cl(−)/HCO3(−) exchanger in spontaneous thymocyte apoptosis. J Immunol. 1996;157:1107–1116. [PubMed] [Google Scholar]
- 51.von-Freeden-Jeffry U, Vieira P, Lucian L A, McNeil T, Burdach S E, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voyno-Yasenetskaya T A. G proteins and Na+/H+ exchange. Biol Signals Recept. 1998;7:118–124. doi: 10.1159/000014537. [DOI] [PubMed] [Google Scholar]
- 53.Wakabayashi S, Bertrand B, Ikeda T, Pouyssegur J, Shigekawa M. Mutation of calmodulin-binding site renders the Na+/H+ exchanger (NHE1) highly H(+)-sensitive and Ca2+ regulationdefective. J Biol Chem. 1994;269:13710–13715. [PubMed] [Google Scholar]
- 54.Wakabayashi S, Ikeda T, Iwamoto T, Pouyssegur J, Shigekawa M. Calmodulin-binding autoinhibitory domain controls “pH-sensing” in the Na+/H+ exchanger NHE1 through sequence-specific interaction. Biochemistry. 1997;36:12854–12861. doi: 10.1021/bi9715472. [DOI] [PubMed] [Google Scholar]
- 55.Watson J D, Morrissey P J, Namen A E, Conlon P J, Widmer M B. Effect of IL-7 on the growth of fetal thymocytes in culture. J Immunol. 1989;143:1215–1222. [PubMed] [Google Scholar]
- 56.Weinllch M, Thelss C, Lin C T, Kinne R K. BCECF in single cultured cells: inhomogeneous distribution but homogeneous response. J Exp Biol. 1998;201:57–62. doi: 10.1242/jeb.201.1.57. [DOI] [PubMed] [Google Scholar]
- 57.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 58.Winston L A, Hunter T. Intracellular signaling: putting JAKs on the kinase MAP. Curr Biol. 1996;6:668–671. doi: 10.1016/s0960-9822(09)00445-x. [DOI] [PubMed] [Google Scholar]
- 59.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 60.Yao H, Ma E, Gu X Q, Haddad G G. Intracellular pH regulation of CA1 neurons in Na(+)/H(+) isoform 1 mutant mice. J Clin Investig. 1999;104:637–645. doi: 10.1172/JCI6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang S, Han J, Sells M A, Chernoff J, Knaus U G, Ulevitch R J, Bokooch G M. Rho family GTPases regulate p38 mitogenactivated protein kinase through the downstream mediator Pak1. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]