Abstract
Weissella bacteria are gram-positive, anaerobic, fermentative, and have probiotic potential. This study aimed to compare the genomes of W. cibaria YRK005 and W. confusa CCK931 isolated from young radish and kimchi, respectively. The genomic size of W. cibaria YRK005 and W. confusa CCK931 with GC content is 2.36 Mb (45%) and 2.28 Mb (44.67%), respectively. The genome study identified 92 and 83 CAZymes genes, respectively, for W. cibaria YRK005 and W. confusa CCK931, that are responsible for 26 and 27 glycoside hydrolases (GH) and 21 and 27 glycosyl transferases. Both species have one gene for carbohydrate esterases and three genes for carbohydrate-binding modules. The primary CAZymes found in both species that are involved in oligosaccharide utilization are GH1, GH2, GH30, GH13_30, GH13_31, GH42, GH43, and GH65. The study also details the production pathways for glycogen and folate. Both strains include a unique repertoire of genes, including hypothetical proteins, showing adaptability to diverse ecological niches and evolution over time.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-022-01232-7.
Keywords: Weissella cibaria, Weissella confusa, Comparative genomics, Carbohydrate-active enzymes, Oligosaccharide
Introduction
The Weissella genus comprises catalase-negative, gram-positive, non-spore-forming, obligate heterofermentative, coccoid to short rod-shaped bacterial species. The genus belonging to the family Leuconostocaceae was first described in 1993 and has 23 validated species (Panthee et al., 2019). These are classified as lactic acid bacteria (LAB) because they produce lactic acid during the fermentation of carbohydrates. Interestingly, several Weissella species and strains have been isolated from diverse habitats, including fermented foods and products and are known to play an essential role in the texture and flavor of food (Hernández-Oaxaca et al., 2021). Weissella species have also been reported in humans (Wang et al., 2020) and animals, such as rainbow trout (Mortezaei et al., 2020) and giant pandas (Xiong et al., 2019). Among all the species, W. confusa and W. cibaria are related and have been recently separated into two species (Bourdichon et al., 2021). Recently, Weissella has generated considerable interest from the research community because of its potential probiotic properties and ability to produce bacteriocins, although certain strains are also opportunistic pathogens (Kamboj et al., 2015).
Initially discovered in fermented Thai dishes, W. cibaria was later isolated from a variety of additional sources, including fermented milk, vegetables, fish, meat, sourdough, silage, and cheese, as well as animal feces and milk (Srionnual et al., 2007). The clinical sources of W. cibaria include human saliva, vagina, urine, blood, feces, and milk. In 2007, W. cibaria 110 was the first strain to produce bacteriocin against gram-positive bacteria (Srionnual et al., 2007). Numerous reports have described the production of various bacteriocins by different strains of this genus (Panthee et al., 2019). Moreover, W. cibaria species are used in starter cultures. They are known to produce a significant amount of dextran (homopolysaccharides), which can be used to enhance fresh bread softness and in the preparation of gluten-free baked foods. Interestingly, the anticancer activity of W. cibaria has also been reported (Kwak et al., 2014).
Formerly known as Lactobacillus confusus, W. confusa has been isolated from numerous fermented foods including sourdough, vegetables, cereals, cheese, and fermented milk (Fusco et al., 2015). Weissella confusa was isolated from human feces. An earlier study demonstrated that W. confusa is a good substitute for the commonly utilized Leuconostoc mesenteroides B512F strain to synthesize large volumes of linear dextran (Maina et al., 2008). Weissella confusa Ck15 has been used for exopolysaccharide (EPS) production in chickpea sourdough (Galli et al., 2020). Recently, W. confusa CGMCC 19308 demonstrated potential as an anti-aging and antibacterial agent (Wang et al., 2022). Dextran was also found in products manufactured from wheat and rye bran that had been fermented by two W. confusa strains isolated from fermented vegetables (Kajala et al., 2016).
Comparative genome sequence analysis of Weissella affiliates has increased over the last five years. Examples include W. cibaria 110, W. hellenica 0916-4-2 (Panthee et al., 2019), and W. ceti (Figueiredo et al., 2015). Despite its widespread importance, little attention has been paid to the genomic characterization of this genus. The first insect-derived and largest genome (2.53 Mb) of the W. confusa strain to be sequenced was W. confusa LM1.
Weissella cibaria YRK005, isolated from young radish, produces gluco-oligosaccharides with a molecular weight of 1.12 × 102 Da. and can be used as a potential prebiotic (Bang et al., 2019). Weissella confusa CCK931, isolated from kimchi, also produces gluco-oligosaccharides (Park and Park, 2021). To advance our understanding of genome diversity, whole-genome sequencing and genomic characterization of W. cibaria YRK005 and W. confusa CCK931 were conducted. Comparative genomic analysis was performed to reveal the evolutionary relationships between these species. The differences in the number and variety of carbohydrate-active enzymes (CAZymes) between W. cibaria YRK005 and W. confusa CCK931 were analyzed. The relationship between the number and variety of CAZymes and oligosaccharide utilization pathways was also examined. To the best of our knowledge, this is the first comparative genomic analysis of two different Weissella strains.
Materials and methods
Weissella confusa and Weissella cibaria isolation and DNA extraction
Two strains used in this study, including W. cibaria YRK005 and W. confusa CCK931, were isolated from young radish (Park and Park, 2021) and kimchi (Bang et al., 2019), respectively, in our laboratory using de Man, Rogosa, and Sharpe (MRS) medium and incubated at 37 °C for 24 h. Sequences of the 16S rRNA genes from both strains were analyzed and identified within the Weissella genus. The isolated strains were maintained in glycerol stocks and stored at -80 °C for future use.
Genome sequencing, assembly, and gene annotation
Purified DNA of W. cibaria YRK005 and W. confusa CCK931 was sent for sequencing to Sanigen Co., Ltd. (Anyang, Korea). The obtained whole-genome sequences were mapped at the default parameters using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/) and Clusters of Orthologous Groups (COG, www.ncbi.nlm.nih.gov/COG) databases for pathway analysis. Circular graphs of the genomes were created using Rapid Annotations using Subsystems Technology (RAST) software. Putative prophage insertion regions were detected using the PHAge Search Tool Enhanced Release (PHASTER) server (http://phast.wishartlab.com/index.html). Mauve analysis, which is used to discover genome-wide rearrangements and inversions in multiple genome alignments, was used to visualize various genomic sequence alignments. The occurrence of genomic islands (GIs) was estimated using IslandViewer 4 (a freely accessible web tool), which practices three prediction algorithms, namely IslandPath-DIMOB, score-based prediction of genomic islands (SIGI)-Hidden Markov model (HMM), and IslandPick, to compute codon usage, analyze dinucleotide bias inside a genome, and create GIs and non-GI datasets from phylogenetically linked organisms.
CAZyme annotation
The dbCAN2 database was used to annotate several CAZymes present in W. cibaria YRK005 and W. confusa CCK931 (Zhang et al., 2018). This database probes the genome using different algorithms, including DIAMOND, a HMM profile that uses HMMdb v7, and Hotpep to expand the prediction. Genes annotated by at least two of the three algorithms described above were included in this manuscript.
Results and discussion
Common features of genome analysis
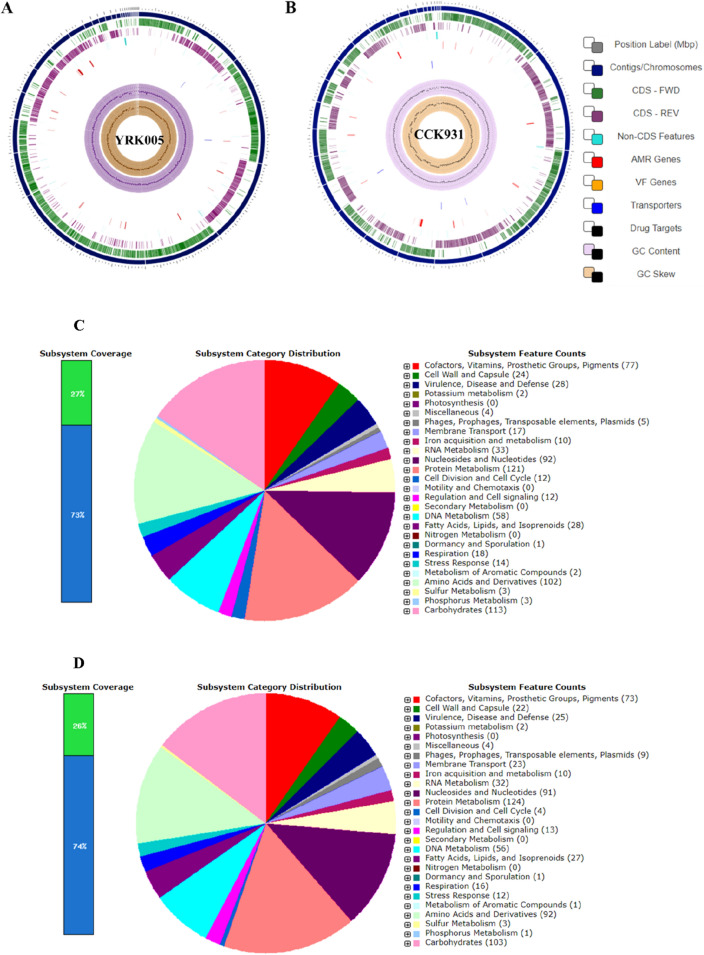
Weissella cibaria YRK005 had a 2.36 Mb circular chromosome with a 45% guanine-cytosine (GC) content and a base pair count of 1,061,398 (Fig. 1A). These results are consistent with those of previous studies (Lynch et al., 2015). The genome size of W. confusa CCK931 was 2.28 Mb with a GC content of 44.67% and 1,017,473 bp (Fig. 1B) (Table 1). The number of DNA scaffolds in W. cibaria YRK005 and W. confusa CCK931 were 31 and 27, respectively. A total of 2241 and 2245 open reading frames (ORFs) were predicted, with 2120 and 2105 protein-coding genes in W. cibaria YRK005 and W. confusa CCK931, respectively. The number of RNA and pseudogenes in W. cibaria YRK005 were 93 and 28, respectively. In W. confusa CCK931, these numbers were 95 and 45, respectively (Table 1). Previously, among three (AB3b, ff3PR, and MG1) strains of W. cibaria, the number of DNA contigs was 88, 60, and 44, with a genome size of 2.5 Mb, 2.4 Mb and 2.4 Mb, respectively (Lynch et al., 2015). Another study on two strains of W. confusa (WcL17 and WCP-3a) described genome sizes of 2.3 Mb and 2.2 Mb, respectively (Hernández-Oaxaca et al., 2021). Yuan et al. described the genome size and GC content of W. confusa LM1 as 2.5 Mb and 44.4%, respectively (Yuna et al., 2021).
Fig. 1.
Clusters of Orthologous Group (COG) analysis based circular map and pie-chart depicting the pathway distribution of the genes in the genome. Circular map view of W. cibaria YRK005 (A) and W. confusa CCK931 (B) genomes created using the CGView Comparison Tool. The concentric circles (from outside to inside) represent the following sequence categories: circle 1 in blue symbolizes contigs/chromosome; 2, the next circle symbolizes forward coding sequences; 3, reverse strand coding sequences; 4, non-CDS; 5, AMR; 6, VF; 7, transporters; 8: drug targets; 9, GC content; and 10, GC-skew. AMR antimicrobial resistance genes, VF virulence factors, COG Clusters of Orthologous Group, CDS coding sequence. Pie chart depicting the pathway distribution of the genes in the genome of W. cibaria YRK005 (C) and W. confusa CCK931 genomes (D), using the rapid annotation system technology (RAST) annotation 654 subsystem distribution. The pie chart displays the percentage distribution and subsystem category, as well as the subsystem count, in the right panel. The higher the value in the bar graph, the higher the proportion of genes in the subsystem, and the lower the value, the fewer the genes in the subsystem
Table 1.
General features of Weissella cibaria YRK005 and Weissella confusa CCK931 genomes
| Feature | W. cibaria YRK005 | W. confusa CCK931 |
|---|---|---|
| Genome size (bp) | 2,358,664 | 2,277,843 |
| DNA G + C (bp) | 1,061,398 | 1,017,473 |
| G + C content (%) | 45 | 44.67 |
| DNA scaffolds | 31 | 27 |
| N50 (bp) | 243,938 | 140,331 |
| Total genes | 2241 | 2245 |
| Protein coding genes | 2120 | 2105 |
| RNA genes | 93 | 95 |
| Pseudo genes | 28 | 45 |
A circular RAST annotation view for W. cibaria YRK005 and W. confusa CCK931 is depicted in Fig. 1C and D, respectively. For W. cibaria YRK005 and W. confusa CCK931, 200 and 198 subsystems, respectively, were observed and categorized into 27 subsystem features. Table S1 lists the Clusters of Orthologous Genes (COG) functions of different genes from two different species of Weissella. Almost 20 COG functions were identified, each with a different fraction of genes linked to it. The highest values of 183 (8.32%) and 182 (8.28%) were observed for COG class J and ascribed to W. cibaria YRK005 and W. confusa CCK931, respectively. This class describes translation, ribosomal structure, and biogenesis. Lynch et al. (2015) reported comparable results for the translation category in W. cibaria with high COG values. The second highest values were for COG class G describing carbohydrate transport and metabolism. The values were 148 (6.73%) for W. cibaria YRK005 and 156 (7.09%) for W. confusa CCK931. The genes involved in COG class E, amino acid transport and metabolism, had values of 123 (5.59%) and 138 (6.28%) for W. cibaria YRK005 and W. confusa CCK931, respectively. Cell wall/membrane/envelope biogenesis (COG class M) was higher for W. confusa CCK931 at 111 (5.05%) than for W. cibaria YRK005, at 96 (4.37%). The genes for replication, recombination and repair, transcription, and nucleotide transport and metabolism were 101 (4.59%) and 93 (4.23%) in COG class L; 103 (4.68%) and 105 (4.78%) in COG class K; and 96 (4.37%) and 91 (4.14%) in COG class F for W. cibaria YRK005 and W. confusa CCK931, respectively.
Of note, the other large fraction of genes without a specified function was 119 (5.41%) for W. cibaria YRK005 and 111 (5.05%) for W. confusa CCK931 (Table S1). This is similar to a recent study on W. cibaria, which reported that many protein fractions had unknown functions (Lynch et al., 2015). This also highlights the discrepancy in genomic information between this genus and other LAB, and the need for further genomic studies on this genus. COG analysis is a valuable method to characterize bacteria and archaea using functional and comparative genomics (Galperin et al., 2015). The existence or absence of related protein families is indicated by the COG functional categories, indicating that these genes are essential for survival, adaptation to harsh environmental conditions, and consumption of multifaceted carbohydrates for growth.
The genomes of W. cibaria YRK005 and W. confusa CCK931 have been deposited in GenBank under accession numbers NZ_JACGMM000000000 and NZ_JACGMJ000000000, respectively.
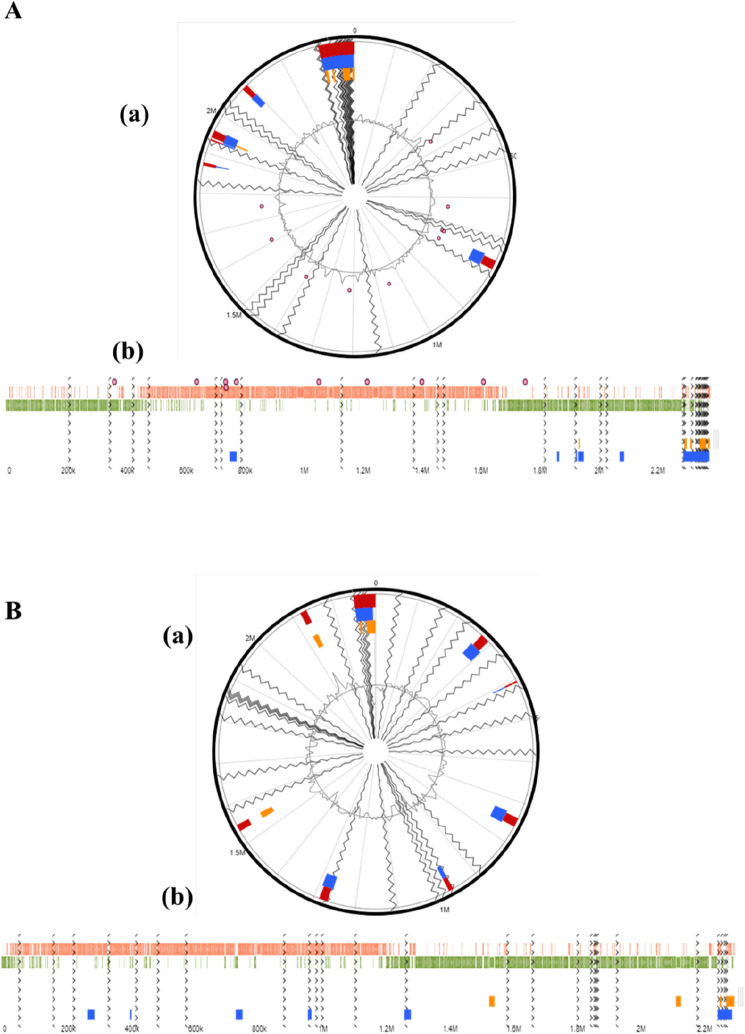
Genomic islands
GIs are distinct gene segments such as transposons, bacteriophages, and other mobile elements in the bacterial genome that are acquired via horizontal gene transfer and may provide a selective benefit to the host (da Silva Filho et al., 2018). The occurrence of genomic islands in the analyzed genomes was predicted using the closest complete (or closed) sequence of W. cibaria CH2 as a reference for the W. cibaria YRK005 genome, and W. confusa N17 for W. confusa CCK931. Genomic island analysis using the web tool, IslandViewer 4 revealed that the W. cibaria YRK005 genome contained five GIs (Fig. 2A), while the W. confusa CCK931 genome contained eight GIs (Fig. 2B).
Fig. 2.
Circular plots visualizing genome islands predicted in the (A) W. cibaria YRK005 and (B) W. confusa CCK931 genomes, aligned against the complete reference genome, with blocks colored according to the prediction method. IslandPath-DIMOB (blue), SIGI-HMM (orange), and the integrated results (dark red) by utilizing RAST annotation subsystems distribution. The GIs with (a) circular outlook and (b) horizontal outlook of (A) W. cibaria YRK005, (B) W. confusa CCK931 genomes analyzed by SIGI-HMM and IslandPath-DIMOB predictions, represented by different colors. GIs: genomic islands; SIGI-HMM, score-based prediction of genomic islands- Hidden Markov Model
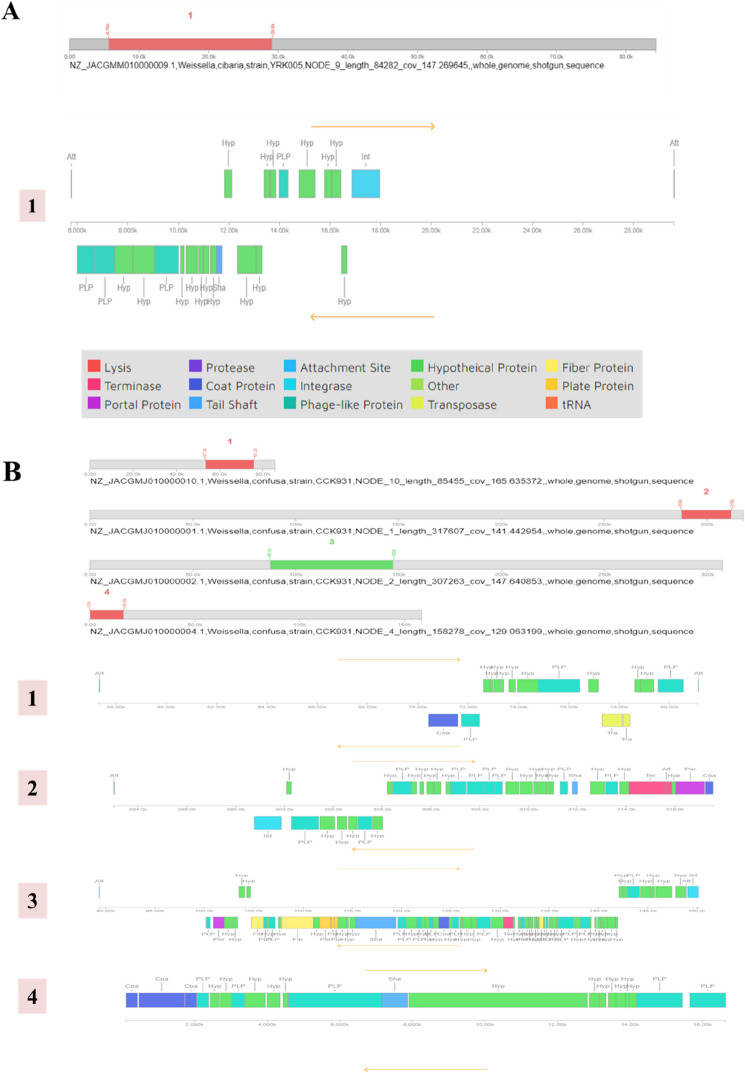
Prophage regions
Phages are an important part of the bacterial genome because they help bacteria adapt to a new environment and acquire bacterial resistance (Panthee et al., 2019). The W. cibaria YRK005 prophage region was divided into 16 hypothetical (Hyp) proteins, four shaft-like proteins, one integrase, and one tail shaft (Fig. 3A). Similar to a previous study [34] on W. cibaria (AB3b, ff3PR, and MG1 strains), no phage repressor protein was identified, indicating the need to study and characterize Hyp proteins that could act as repressors. As shown in Fig. 3B, four prophage-related regions were observed in W. confusa CCK931, whereas one prophage-related region was observed in W. cibaria YRK005. For W. confusa CCK931, the first prophage region consisted of nine Hyp proteins, three phage-like proteins (PLP), two transposases (TRA), and one coat protein (Coa), while the second region consisted of 20 hypo proteins, nine PLP, and one each portal protein (por), Coa, integrase (int), and terminase (ter). The third was a complex with maximum ORFs, including 44 hyp, 15 PLP, three fiber proteins, three plate proteins, and one protein each for Coa, int, por, ter, and shaft proteins. The fourth prophage segment consisted of 12 Hyp, three Coa, five PLP, and one shaft protein (Fig. 3B). Absence of a recognizable integrase protein in the third and fourth prophages suggests that these phages are lysogenically incapable (Lynch et al., 2015).
Fig. 3.
Prophage regions identified using PHASTER server in (A) W. cibaria YRK005 and (B) W. confusa CCK931. PHASTER, PHAge Search Tool Enhanced Release
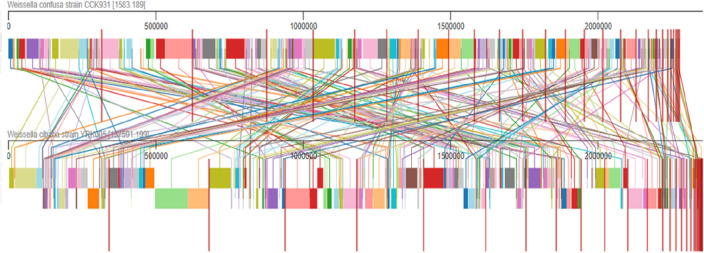
Mauve and Dot plot matrix analysis
As shown in Fig. 4, the Mauve-based analysis showed an overall collinear relationship across the genomes of W. confusa CCK931 and W. cibaria YRK005. Many different colored local collinear blocks (LCBs) were identified in the two genomes, with connecting lines joining the regions on the homologous chromosomes. The gaps shown in white between both genomes suggest the presence of species-specific regions. To analyze the genomic structures of W. confusa CCK931 strains, we compared all four complete genomes and subjected them to similarity analysis using the Mauve software. In a recent study, the W. confusa LM1 genome was used as a reference, compared to the four genomes, and showed 11 LCBs. A dot plot matrix analysis of the two species is presented in the supplementary data (Fig. S1). The pairwise alignment dot plot pattern revealed stronger low-level commonality among Weissella spp.
Fig. 4.
Mauve analysis of (A) W. cibaria YRK005 and (B) W. confusa CCK931. Colorful blocks of the first horizontal alignment connect with lines (lines of homologous) to the similar colored blocks in the genome present on the lower side. Breaks among the colored blocks represent genomic arrangements
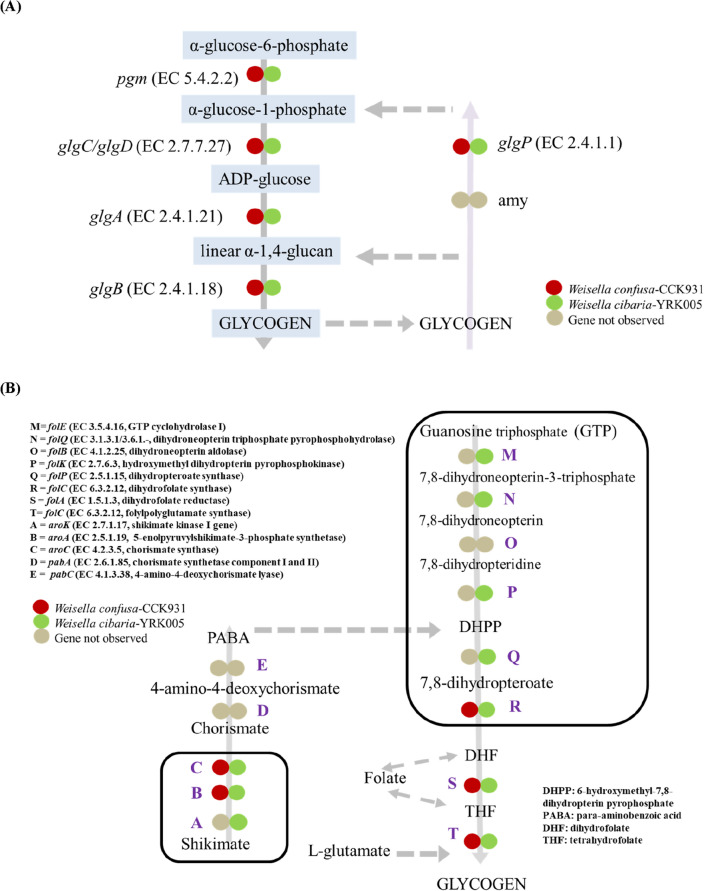
Glycogen pathway
Glycogen is one of the most prevalent water-soluble homogeneous polysaccharides found in bacteria, archaea, and eukaryotes, and plays a vital role in physiology. During unfavorable periods, glycogen is broken down to support long-term bacterial survival. In the present study, an intact classical glycogen biosynthesis pathway (glg operon) was found in both species of Weissella, which involves phosphoglucomutase (pgm), glucose-1-phosphate adenylyltransferase (glgC or glgD), ADP-glucose-specific glycogen synthase (glgA), and a branching enzyme (glgB) (Fig. 5A). Complete glycogen metabolic pathways are often found in species adapted to more diversified niches and variable lifestyles, according to a new study of 1,202 bacterial genomes (Wang and Wise, 2011). Furthermore, for glycogen catabolism, two enzymes are needed, namely glycogen phosphorylase (glgP) and a debranching enzyme (amy or glgX) (Alonso-Casajús et al., 2006). However, only glgP was observed in both the strains under investigation.
Fig. 5.
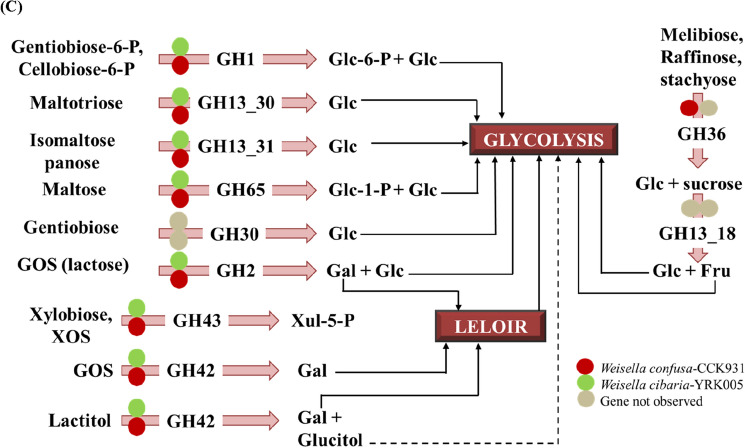
Outline of glycogen and folate biosynthesis pathways, and oligosaccharide utilization pathways in W. cibaria YRK005 and W. confusa CCK931. (A) Glycogen biosynthesis pathways. The presence of the enzyme in W. confusa is indicated in red, whereas green indicates that the enzyme is present in W. cibaria. Enzymes not observed are represented by colorless boxes. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to create the pathway. The light grey arrow shows the process of making glycogen, while the light purple arrow shows the process of breaking down glycogen. Dotted arrows represent multi steps in the pathway. Pgm- phosphoglucomutase; Amy-α, amylase. (B) Folate biosynthesis pathways in W. cibaria YRK005 and W. confusa CCK931. (C) Oligosaccharide utilization pathways and the information on relevant carbohydrate-active enzymes (CAZymes) predicted in W. cibaria YRK005 and W. confusa CCK931
Folate Biosynthesis in W. confusa CCK931 and W. cibaria YRK005
It is critical to understand the genes involved in folate biosynthesis to adjust the folate content of fermented products, such as LAB, which are used as starter cultures. Folate is a generic term for conjugated compounds formed by a pteridine ring linked to para-aminobenzoic acid (PABA) and one or more L-glutamates (Mahara et al., 2019). The presence or absence of enzymes involved in folate production was predicted in W. confusa CCK931 and W. cibaria YRK005. The enzymes involved in folate production are folE, folQ, folB, folK, folP, folC1, 2 and folA. In this study, W. confusa CCK931 appeared to lack the genes folE, folQ, folB, folK, and folP (Fig. 5B). Weissella cibaria YRK005 exhibits a complete folate biosynthetic pathway (Fig. 5B) and uses PABA, an intermediate in folate biosynthesis, via the shikimate pathway. A recent study reported the production of folate in both W. confusa and W. cibaria isolated from fermented Thai dishes (Deatraksa et al., 2018). The ability of LAB to synthesize folate is associated with three main genes, namely folB, folK, and folP (Mahara et al., 2019). These three genes were not identified in the present study except folK and folP in W. cibaria YRK005 However, the detection of these genes is insufficient to estimate their capacity for folate production (Greppi et al., 2017).
Types of CAZymes in W. cibaria YRK005 and W. confusa CCK931
The search for CAZymes indicated that the genomes of W. cibaria YRK005 and W. confusa CCK931 encode enzymes with 92 and 83 genes, respectively (Table S2). Only the genes predicted by at least two of the three algorithms were considered for further research. Further evaluation of specific CAZyme families revealed significant differences in CAZyme abundance and distribution between Weissella species. The glycoside hydrolase (GH) genes were 26 and 27 for W. cibaria YRK005 and W. confusa CCK931, respectively. A large percentage of glycosyl transferase (GT) (56%) and GH (31%) genes was found in a study on the CAZymes of W. confusa CCK931strains. There were 27 and 21 genes encoding GTs in W. cibaria YRK005 and W. confusa CCK931, respectively. Among the GH, an enzyme from the GH70 family was discovered in both species under investigation, both of which have been previously reported to produce EPS (Hernández-Oaxaca et al., 2021). Similarly, both species have different enzymes from the GH13 family that encode amylases, implying a potential role in starch degradation (Hernández-Oaxaca et al., 2021). Both species share 15 GH-related genes, including GH1, GH2, GH3, GH8, GH13, GH25, GH31, GH32, GH43, GH65, GH70, and GH73. GH23 was unique to W. cibaria YRK005, whereas GH36 and GH120 were only found in W. confusa CCK931 (Table S2). The GH1, GH3, GH8, and GH13 families comprise enzymes that break down starch or cellulose (Hernández-Oaxaca et al., 2021). GT2, GT0, GT4, GT5, GT28, GT35, and GT51 were shared by both species, whereas GT111 was unique to W. cibaria YRK005. However, GT4 (17%), GT2 (28%), GH23 (7%), and GH70 (6%) were unique to W. confusaCCK931 strains. Furthermore, both species contain three carbohydrate-binding module (CBM) genes and one carbohydrate esterases (CE) gene. However, we found no genes associated with the phosphorylase (PL) family in the genomes of either species (Table S2).
To further investigate oligosaccharide synthesis, we performed an analysis and discovered the presence of different CAZymes deciphered in the genomes of both species. CAZymes include a dynamic repertoire of enzymes in all living organisms and play an essential role in carbohydrate metabolism. An in-depth analysis of CAZymes, which have a role in oligosaccharide synthesis, including GT, glucansucrase, GH, glycosidases, and PL, was performed in both species of Weissella. In GT, a few subclass enzymes have been reported in both species including cell division protein FtsI (peptidoglycan synthetase), multimodular transpeptidase-transglycosylase, bactoprenol glucosyl transferase, glycosyltransferase, group 2 family protein, GT, lipopolysaccharide biosynthesis glycosyltransferase. However, two enzymes in the GT category, including glycosyltransferase LafB, responsible for the formation of Gal-Glc-diacylglycerol (DAG), and biofilm poly-glutamic acid (PGA) synthesis N-glycosyltransferase PgaC, were reported only in W. cibaria YRK005. Enzymes, including sucrose-6-phosphate hydrolase, family 13, and A/G-specific adenine glycosylase (mutY), have been found in glucansucrase, GH, and glycosidases, respectively. In the phosphorylase class, maltose phosphorylase, sugar kinase, transcription regulator, HPr kinase/phosphorylase, and glycogen phosphorylase have been discovered.
CAZymes and oligosaccharide utilization
Both W. cibaria YRK005 and W. confusa CCK931 strains were capable of using maltotriose, glucooligosaccharide (GOS), lactose (GOS), isomaltose, panose, melibiose, raffinose, stachyose, xylobiose, xylooligosaccharides (XOS), lactitol, maltose, gentibiose-6-phoshphate, and cellobiose-6-phosphate while they failed to utilize gentibiose and sucrose (Fig. 5C). Similar to a previous study based on in silico analysis (Lynch et al., 2015), Weissella species in this study also utilized galactose via the Leloir pathway. Both species possess GH1 (β-glucosidase), which can metabolize gentibiose-6-phoshphate and cellobiose-6- phosphate to glucose-6-phosphate and glucose, respectively.
GH43 (β-xylosidase) was observed in W. cibaria YRK005 and W. confusa CCK931, which is involved in the metabolism of xylooligosaccharides (XOS). This enzyme was characterized in a previous study on Weissella sp. strain 92. The ability of W. cibaria to utilize XOS in this investigation, as well as in a prior study (W. cibaria strain 92) (Månberger et al., 2020), highlights the relevance of the genus as a probiotic (da Silva Filho et al., 2018). The lactose-degrading enzyme (GOS) GH2 (β-galactosidase candidate) was discovered in both strains, which corroborates a recent study (Månberger et al., 2020). Both strains hydrolyze lactitol (a disaccharide and commercial sweetening ingredient) in the presence of GH42. Weissella cibaria YRK005 and W. confusa CCK931 contained the maltotriose-hydrolyzing enzyme GH13_30 (α-glucosidase), which cleaves the α-1,4-glycosidic bonds of oligosaccharides and starch from the non-reducing ends. The presence of this enzyme has recently been characterized at the biochemical and structural levels in W. cibaria BBK-1 (WcAG) (Wangpaiboon et al., 2021). Similarly, both species have the GH13_31 enzyme, which is responsible for the hydrolysis of isomaltose and panose. Weissella cibaria (strain 92) contains the GH 13 family enzymes (Månberger et al., 2020). Maltose phosphorylase (GH 65) has also been found in W. cibaria YRK005 and W. confusa CCK931. Maltose phosphorolysis by this dimeric enzyme leads to the conversion of maltose into glucose-1-phosphate and glucose. Natural substrates, commonly known as α-galactosides, including raffinose, melibiose, and stachyose, are hydrolyzed in the presence of α-galactosidase (Mn2+- and NAD+-dependent) (Morabbi Heravi et al., 2019). Mital et al. (1973) originally defined the α-galactosidase activity of Lactobacillus. Several lactic acid bacteria, including L. acidophilus and L. plantarum, harbor melA, which encodes α-galactosidase, a member of the GH36 family (Gänzle and Follador, 2012). In our study, α-galactosidase (GH 36) was found only in W. confusa CCK931 (Fig. 5C).
Unique enzyme repertoire compared to four Leuconosotc spp.
Additionally, we compared the W. cibaria YRK005 and W. confusa CCK931 strains with four Leuconostoc species isolated, characterized, and sequenced in our laboratory (Sharma et al., 2022). Surprisingly, we found that 220 genes were specific to W. cibaria YRK005 and W. confusa CCK931, as they did not have homologs in Leuconostoc spp. (Fig. S2 and S3) (Table S3), and more than half of them (120) were classified as either hypothetical or harboring a domain of unknown function. These genes may be responsible for the distinctive biological actions of the genus Weissella, and an in-depth characterization should offer insights into the strain’s miscellaneous physiognomies. Weissella cibaria has gained attention for its capability to manufacture large amounts of novel, undigestible oligosaccharides and EPS. These substances can be employed as probiotics or sweetening agents in food and feedstuff items. Weissella cibaria contained a gene for EPS biosynthesis, as reported for W. cibaria GA44 and W. cibaria W27 (with antibacterial applications). However, in the present study, a hypothetical protein was identified at this locus in W. confusa CCK931. EPS production with biological activity has recently been reported in W. confusa MD1 (Lakra et al., 2020). Neopullulanase was also detected in both strains. This enzyme can produce panose by hydrolyzing pullulan, indicating its ability to hydrolyze starch. This enzyme has been reported in Lactobacillus plantarum A6 (Humblot et al., 2014). Examples have been identified in W. cibaria YRK005 but reported as hypothetical proteins in W. confusa CCK931. These examples include permease of the drug/metabolite transporter (DMT) superfamily, dipeptide ATP-binding cassette (ABC) transporter, permease protein DppB, and immunodominant staphylococcal antigen A precursor. This suggests that further in-depth genomic studies should be conducted to reveal the full biotechnological potential of these strains.
The present research focuses on the comparative genomics of W. cibaria YRK005 and W. confusa CCK931 species, concentrating on the genome features, genomic islands, prophage regions, mauve analysis, and dot plot matrixes. Furthermore, this study highlights CAZymes types in both species, the glycogen pathway, oligosaccharide utilization routes, and the folate biosynthesis pathway. The study also highlights the unique genes present in the Weissella genus compared to the genus Leuconostoc. This indicates the miscellaneous physiognomies of the genus Weissella, which necessitate exploring the untapped potential applications of industrial significance.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIT) (No. 2019R1A2C1004950). Deepshikha Gupta, DG thanks DBT, Government of India for the RA fellowship.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Neha Sharma, Email: nehaworld92@gmail.com.
Deepshikha Gupta, Email: deepshikha.gupta1110@gmail.com.
Young-Seo Park, Email: ypark@gachon.ac.kr.
References
- Alonso-Casajús N, Dauvillée D, Viale AM, Munoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Eydallin G, Ball S, Pozueta-Romero J. Glycogen phosphorylase, the product of the glgP gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. Journal of Bacteriology. 2006;188:5266–5272. doi: 10.1128/JB.01566-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J, Jeong SY, Bang S, Park J, Kim S, Park YS. Production of gluco-oligosaccharides from Weissella confusa CCK931. In: Abstract: 2019 Fall Meeting and Symposium of Korean Society for Food Engineering. October 25, aT Center. Seoul, Korea. Korean Society for Food Engineering. p. 153 (2019)
- Bourdichon F, Patrone V, Fontana A, Milani G, Morelli L. Safety demonstration of a microbial species for use in the food chain: Weissella confusa. International Journal of Food Microbiology. 2021;339:109028. doi: 10.1016/j.ijfoodmicro.2020.109028. [DOI] [PubMed] [Google Scholar]
- da Silva Filho AC, Raittz RT, Guizelini D, De Pierri CR, Augusto DW, dos Santos-Weiss ICR, Marchaukoski JN. Comparative analysis of genomic island prediction tools. Frontiers in Genetics. 2018;9:619. doi: 10.3389/fgene.2018.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatraksa J, Sunthornthummas S, Rangsiruji A, Sarawaneeyaruk S, Suwannasai N, Pringsulaka O. Isolation of folate-producing Weissella spp. from Thai fermented fish (Plaa Som Fug) LWT. 2018;89:388–397. doi: 10.1016/j.lwt.2017.11.016. [DOI] [Google Scholar]
- Figueiredo HC, Soares SC, Pereira FL, Dorella FA, Carvalho AF, Teixeira JP, Azevedo VA, Leal CA. Comparative genome analysis of Weissella ceti, an emerging pathogen of farm-raised rainbow trout. BMC Genomics. 2015;16:1095. doi: 10.1186/s12864-015-2324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco V, Quero GM, Cho GS, Kabisch J, Meske D, Neve H, Bockelmann W, Franz CM. The genus Weissella: taxonomy, ecology and biotechnological potential. Frontiers in Microbiology. 2015;6:155. doi: 10.3389/fmicb.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli V, Venturi M, Coda R, Maina NH, Granchi L. Isolation and characterization of indigenous Weissella confusa for in situ bacterial exopolysaccharides (EPS) production in chickpea sourdough. Food Research International. 2020;138:109785. doi: 10.1016/j.foodres.2020.109785. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Research. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gänzle M, Follador R. Metabolism of oligosaccharides and starch in lactobacilli: a review. Frontiers in Microbiology. 2012;3:340. doi: 10.3389/fmicb.2012.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greppi A, Hemery Y, Berrazaga I, Almaksour Z, Humblot C. Ability of lactobacilli isolated from traditional cereal-based fermented food to produce folate in culture media under different growth conditions. LWT. 2017;86:277–284. doi: 10.1016/j.lwt.2017.08.007. [DOI] [Google Scholar]
- Hernández-Oaxaca D, López-Sánchez R, Lozano L, Wacher-Rodarte C, Segovia L, López Munguía A. Diversity of Weissella confusa in pozol and its carbohydrate metabolism. Frontiers in Microbiology. 2021;12:629449. doi: 10.3389/fmicb.2021.629449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humblot C, Turpin W, Chevalier F, Picq C, Rochette I, Guyot JP. Determination of expression and activity of genes involved in starch metabolism in Lactobacillus plantarum A6 during fermentation of a cereal-based gruel. International Journal of Food Microbiology. 2014;185:103–111. doi: 10.1016/j.ijfoodmicro.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Kajala I, Mäkelä J, Coda R, Shukla S, Shi Q, Maina NH, Juvonen R, Ekholm P, Goyal A, Tenkanen M. Rye bran as fermentation matrix boosts in situ dextran production by Weissella confusa compared to wheat bran. Applied Microbiology and Biotechnology. 2016;100:3499–3510. doi: 10.1007/s00253-015-7189-6. [DOI] [PubMed] [Google Scholar]
- Kamboj K, Vasquez A, Balada-Llasat JM. Identification and significance of Weissella species infections. Frontiers in Microbiology. 2015;6:1204. doi: 10.3389/fmicb.2015.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SH, Cho YM, Noh GM, Om AS. Cancer preventive potential of kimchi lactic acid bacteria (Weissella cibaria, Lactobacillus plantarum) Journal of Cancer Prevention. 2014;19:253–258. doi: 10.15430/JCP.2014.19.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakra AK, Domdi L, Tilwani YM, Arul V. Physicochemical and functional characterization of mannan exopolysaccharide from Weissella confusa MD1 with bioactivities. International Journal of Biological Macromolecules. 2020;143:797–805. doi: 10.1016/j.ijbiomac.2019.09.139. [DOI] [PubMed] [Google Scholar]
- Lynch KM, Lucid A, Arendt EK, Sleator RD, Lucey B, Coffey A. Genomics of Weissella cibaria with an examination of its metabolic traits. Microbiology. 2015;161:914–930. doi: 10.1099/mic.0.000053. [DOI] [PubMed] [Google Scholar]
- Mahara FA, Nuraida L, Lioe HN. Fermentation of milk using folate-producing lactic acid bacteria to increase natural folate content: a review. Journal of Applied Biotechnology Reports. 2019;6:129–136. doi: 10.29252/JABR.06.04.01. [DOI] [Google Scholar]
- Maina NH, Tenkanen M, Maaheimo H, Juvonen R, Virkki L. NMR spectroscopic analysis of exopolysaccharides produced by Leuconostoc citreum and Weissella confusa. Carbohydrate Research. 2008;343:1446–1455. doi: 10.1016/j.carres.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Mital B, Shallenberger R, Steinkraus K. α-Galactosidase activity of lactobacilli. Applied and Environmental Microbiology. 1973;26:783–788. doi: 10.1128/am.26.5.783-788.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morabbi Heravi K, Watzlawick H, Altenbuchner J. The melREDCA operon encodes a utilization system for the raffinose family of oligosaccharides in Bacillus subtilis. Journal of Bacteriology. 2019;201:e00109–e119. doi: 10.1128/JB.00109-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortezaei F, Royan M, Allaf Noveirian H, Babakhani A, Alaie Kordghashlaghi H, Balcázar J. In vitro assessment of potential probiotic characteristics of indigenous Lactococcus lactis and Weissella oryzae isolates from rainbow trout (Oncorhynchus mykiss Walbaum) Journal of Applied Microbiology. 2020;129:1004–1019. doi: 10.1111/jam.14652. [DOI] [PubMed] [Google Scholar]
- Panthee S, Paudel A, Blom J, Hamamoto H, Sekimizu K. Complete genome sequence of Weissella hellenica 0916-4-2 and its comparative genomic analysis. Frontiers in Microbiology. 2019;10:1619. doi: 10.3389/fmicb.2019.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Park YS. Gluco-oligosaccharides from Weissella cibaria as potential prebiotics. Food Engineering Progress. 2021;25:305–312. doi: 10.13050/foodengprog.2021.25.4.305. [DOI] [Google Scholar]
- Sharma A, Sharma N, Gupta D, Lee HJ, Park YS. Comparative genome analysis of four Leuconostoc strains with a focus on carbohydrate-active enzymes and oligosaccharide utilization pathways. Computational and Structural Biotechnology Journal. 2022;20:4771–4785. doi: 10.1016/j.csbj.2022.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srionnual S, Yanagida F, Lin LH, Hsiao KN, Chen YS. Weissellicin 110, a newly discovered bacteriocin from Weissella cibaria 110, isolated from plaa-som, a fermented fish product from Thailand. Applied and Environmental Microbiology. 2007;73:2247–2250. doi: 10.1128/AEM.02484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Liu W, Chu W. Isolation and preliminary screening of potentially probiotic Weissella confusa strains from healthy human feces by culturomics. Microbial Pathogenesis. 2020;147:104356. doi: 10.1016/j.micpath.2020.104356. [DOI] [PubMed] [Google Scholar]
- Wang L, Wise MJ. Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften. 2011;98:719. doi: 10.1007/s00114-011-0832-x. [DOI] [PubMed] [Google Scholar]
- Wang W, Li S, Heng X, Chu W. Weissella confusa CGMCC 19,308 Strain protects against oxidative stress, increases lifespan, and bacterial disease resistance in Caenorhabditis elegans. Probiotics and Antimicrobial Proteins. 2022;14:121–129. doi: 10.1007/s12602-021-09799-z. [DOI] [PubMed] [Google Scholar]
- Wangpaiboon K, Laohawuttichai P, Kim SY, Mori T, Nakapong S, Pichyangkura R, Pongsawasdi P, Hakoshima T, Krusong K. A GH13 α-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides. Acta Crystallographica Section D: Structural Biology. 2021;77:1064–1076. doi: 10.1107/S205979832100677X. [DOI] [PubMed] [Google Scholar]
- Xiong L, Ni X, Niu L, Zhou Y, Wang Q, Khalique A, Liu Q, Zeng Y, Shu G, Pan K. Isolation and preliminary screening of a Weissella confusa strain from giant panda (Ailuropoda melanoleuca) Probiotics and Antimicrobial Proteins. 2019;11:535–544. doi: 10.1007/s12602-018-9402-2. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wang Y, Zhao F, Kang L. Complete genome sequence of Weissella confusa LM1 and comparative genomic analysis. Frontiers in Microbiology. 2021;12:749218. doi: 10.3389/fmicb.2021.749218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.