Abstract
In modern clinical decision-support algorithms, heterogeneity in image characteristics due to variations in imaging systems and protocols hinders the development of reproducible quantitative measures including for feature extraction pipelines. With the help of a reader study, we investigate the ability to provide consistent ground-truth targets by using patient-specific 3D-printed lung phantoms. PixelPrint was developed for 3D-printing lifelike computed tomography (CT) lung phantoms by directly translating clinical images into printer instructions that control density on a voxel-by-voxel basis. Data sets of three COVID-19 patients served as input for 3D-printing lung phantoms. Five radiologists rated patient and phantom images for imaging characteristics and diagnostic confidence in a blinded reader study. Effect sizes of evaluating phantom as opposed to patient images were assessed using linear mixed models. Finally, PixelPrint's production reproducibility was evaluated. Images of patients and phantoms had little variation in the estimated mean (0.03–0.29, using a 1–5 scale). When comparing phantom images to patient images, effect size analysis revealed that the difference was within one-third of the inter- and intrareader variabilities. High correspondence between the four phantoms created using the same patient images was demonstrated by PixelPrint’s production repeatability tests, with greater similarity scores between high-dose acquisitions of the phantoms than between clinical-dose acquisitions of a single phantom. We demonstrated PixelPrint's ability to produce lifelike CT lung phantoms reliably. These phantoms have the potential to provide ground-truth targets for validating the generalizability of inference-based decision-support algorithms between different health centers and imaging protocols and for optimizing examination protocols with realistic patient-based phantoms.
Classification: CT lung phantoms, reader study
Keywords: computed tomography, three-dimensional printing, phantoms, lung imaging, reader study
Significance Statement.
Diagnostic imaging is often the front-line method for diagnosis and treatment evaluation. Modern radiological workflows increasingly rely on analyzing medical images for quantifiable features. The variability associated with device/vendor, acquisition protocol, data processing, etc. is undesirable and can have a dramatic effect on quantitative measures, including the radiomics features. A clinically realistic evaluation strategy is necessary to understand the effects of such variabilities. Recently, we introduced a method to 3D-print lifelike patient-specific CT lung phantoms. The objective of this study is to compare the diagnostic performance in a reader study of our phantoms to ground-truth patient data. Our study demonstrates that there is no clinically significant difference, thereby providing the foundation for future standardization in diagnostic imaging.
Introduction
Quantitative imaging is receiving increased interest and acknowledgment from clinicians and healthcare providers as a supporting tool for data-driven, patient-specific clinical decision-making (1–3). However, variability in image acquisition and reconstruction techniques introduce heterogeneity in image characteristics and features that are independent of the underlying biology and pathophysiology (4). Modern medical imaging modalities, such as computed tomography (CT), magnetic resonance imaging, and positron emission tomography (PET), allow a wide variety of imaging parameters that are, in general, lacking standardization between different health centers and different scanner models. While these differences typically have little clinical impacts for routine radiological interpretation, they introduce biases when analyzed numerically to extract meaningful data (4). This hampers advancement of reproducible feature extraction pipelines, a critical pre-requisite for clinical translation (5). Despite ongoing efforts to account for factors originating from the recognized lack of imaging standardization, the problem of biases and variability persists. Experimental validation of image cohort normalization methods, such as ComBat (6–8), is currently limited due to an inability to repeat patient scans on multiple scanners or with multiple imaging protocols given logistical and risk-related considerations, e.g. the risks of ionizing radiation in CT and PET. There is therefore a growing need for realistic patient-based volumetric phantoms that can accurately mimic human anatomy and disease manifestations to provide consistent imaging ground-truth targets when comparing postprocessing image cohort normalization and feature extraction techniques.
Anthropomorphic phantoms are fundamental tools for developing, optimizing, and evaluating hardware and software advances in medical imaging research and clinical practice. Such phantoms are typically manufactured by machining, casting, or molding homogenous materials that mimic tissue properties relevant for the specific imaging modality, e.g. x-ray attenuation coefficients for CT (9). Realistic patient-based phantoms have additional advantages for clinical and development tasks, such as imaging protocol optimization, and provide ground-truth targets for denoizing or artifact correction artificial intelligence (AI) algorithms. Despite a wide range of commercially available phantoms, there is a lack of patient-based phantoms capable of reliably representing the quantitative imaging characteristics and textures found in clinical patient images. The academic and clinical radiology communities would greatly benefit from rapid, versatile, lifelike, as well as inexpensive phantom manufacturing processes, compared with commercial solutions currently available.
Throughout the last decade, 3D-printing of phantoms that represent the X-ray attenuations and textures of various tissues, anatomies, and disease has been widely explored. These studies focused on several developmental aspects, including 3D-printing of accurate attenuation profiles (10–13), manufacturing anatomically correct organ models (14–18), and generation of realistic tissue textures (19–21). Novel 3D-printing techniques, mainly using fused deposition modeling (FDM), have been proposed to generate variable material densities that mimic the imaging features observed in clinical CT images. These methods (22–24) include utilization of different infill printing patterns (25), variable voxel-dependent extrusion rates (12, 13), or interlacing two different materials with dual-extrusion printers (26).
Generation of 3D-printed anthropomorphic phantoms from clinical CT images typically involves (17, 24, 27–29): (i) automated or manual segmentation of specific tissues or organs, e.g. an entire lung or identified findings, (ii) conversion of the segmented volumes into triangulated surface geometry models, such as standard triangle/tessellation language (STL), and (iii) utilization of printer-specific slicing software to generate instructions (e.g. G-code) that determine relevant 3D-printing parameters, such as extrusion rate, printing speed, infill ratios, etc. While phantoms produced this way may approximate clinical imaging characteristics, they still have shortcomings. Most importantly, due to segmentation of regions followed by conversion to surface models, abrupt and unrealistic transitions between homogenous regions of different densities are created within the printed products, and spatial resolution and textural information are compromised.
In this work, we evaluate a promising alternative called PixelPrint that we recently developed to overcome the limitations described above. PixelPrint directly translates DICOM image data into printer instructions that continuously control the printed material density by varying the printer speed on a voxel-by-voxel basis, while maintaining a constant filament extrusion rate. Previously we reported on quantitative comparisons between a clinical CT slice and its corresponding 3D-printed phantom (30). Here we report on subjective (blinded) reader studies conducted to assess the correspondence of image quality and general imaging characteristics between images of three 3D-printed volumetric COVID-19 pneumonia lung phantoms with those of the original patient images used to produce these phantoms. The main purpose of these studies was to evaluate whether differences between patient and phantom images are of any clinical significance. We also report quantitative comparisons between four 3D-prints of the same patient for production reproducibility assessments.
Methods
Three patients were selected from the Hospital of the University of Pennsylvania PACS by a thoracic radiologist (L.R., 4 years of experience) under an IRB approved protocol. Patients were selected based on the assessed COVID-19 severity level (mild, moderate, and severe), patient habitus, and absence of significant metal artifacts. For each patient, clinical DICOM images reconstructed with a sharp kernel (Table 1) were converted into 3D-printer instructions using PixelPrint software. A complete technical background of the PixelPrint algorithm, pipeline, and quantitative evaluation is available in our previous publication (30). A primary advancement of PixelPrint presented in this study is the 3D-printing of phantoms based on volumetric patient data (Fig. 1).
Table 1.
Patient information together with the scan and reconstruction parameters that were used to generate the original diagnostic CT images and the images of the three corresponding 3D-printed phantoms.
| COVID-19 severity | Mild | Moderate | Severe |
|---|---|---|---|
| Patient sex | Male | Female | Male |
| Patient age | 71 years | 57 years | 68 years |
| CT manufacturer | Siemens | GE | Siemens |
| CT model | Sensation 64 | Revolution CT | Definition Edge |
| Tube voltage | 120 kVp | 100 kVp | 120 kVp |
| Collimation | 19.2 mm | 80 mm | 38.4 mm |
| Rotation time | 0.5 s | 0.5 s | 0.33 s |
| Spiral pitch factor | 1.5 | 0.992 | 1.45 |
| Dose modulation | XYZ | XYZ | XYZ |
| Exposure (at lungs) | 55–64 mAs | 75–90 mAs | 75–77 mAs |
| CTDIvol (at lungs) | 4.213–4.953 mGy | 5.081–6.125 | 5.112–5.219 mGy |
| Recon. kernel | B31f, B70f | CHEST, LUNG | Bf37f/3, Br51f/2 |
| Slice thickness | 1.0, 1.0 (mm) | 0.977, 0.625 (mm) | 1.0, 1.0 (mm) |
| Slice increment | 1.0, 1.0 (mm) | 1.0, 0.625 (mm) | 1.0, 1.0 (mm) |
| Recon. field of view | 425 mm | 500 mm | 365 mm |
| Matrix size | 512 × 512 | 512 × 512 | 512 × 512 |
| Pixel spacing (x/y) | 0.83/0.83 mm | 0.98/0.98 mm | 0.71/0.71 mm |
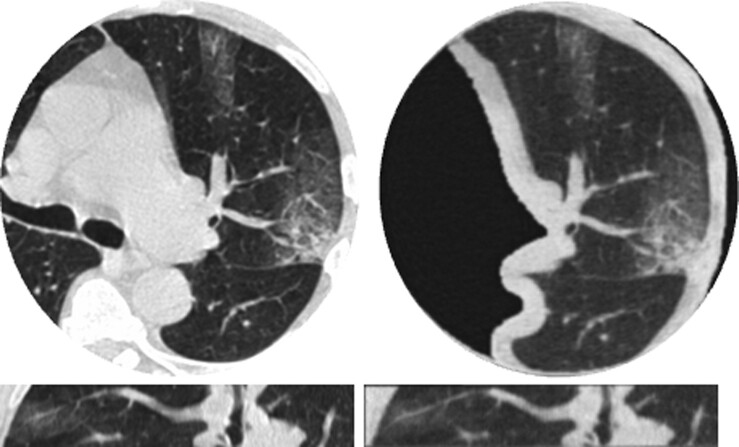
Fig. 1.
Comparisons between clinical CT lung images of a mild COVID-19 patient (left) and images of a corresponding 30 mm thick 3D-printed volumetric phantom (right), acquired with the same CT scanner and imaging parameters. Presented in two orthogonal views: axial (top), sagittal (bottom). Window level/width is −500/1,400 HU.
All phantoms presented in this work were printed using 1.75 mm diameter polylactic acid filament (MakeShaper; Keene Village Plastics, Cleveland, OH, USA) on a Lulzbot TAZ 6 fused-filament 3D-printer (Fargo Additive Manufacturing Equipment 3D, LLC Fargo, ND, USA) with a 0.25 mm brass nozzle. Phantoms were printed with a constant extrusion rate of 0.6 mm3/s and a layer height of 0.2 mm. Printing speeds varied from 3 to 30 mm/s, with acceleration and jerk (threshold velocity for applying acceleration) settings of 500 mm/s (2) and 8 mm/s, respectively, producing line widths from 0.1 to 1.0 mm.
Each phantom was scanned on the same scanner using the same acquisition and reconstruction settings as the input patient scan (Table 1). The phantoms were placed within the 20 cm bore of a 300 × 400 mm2 phantom (Gammex MECT, Sun Nuclear, Melbourne, FL, USA) to mimic attenuation profiles of a medium-sized patient. A preprocessing pipeline was developed for preparing images for a reader study using the following steps. First, lung segmentations obtained using a pretrained AI (31) from each of the original patient scans were dilated by eight pixels in every direction and manually positioned on the 3D-printed phantom image volumes. Next, an image registration algorithm (Simple-ITK (32)) was applied to accurately align phantom images with their corresponding patient images and a circular binary mask of 18 cm diameter was applied to both the segmented phantom and their corresponding patient images to hide their surroundings (patient anatomy or multi-energy CT phantom [MECT] phantom). Finally, images from both the phantoms and the corresponding patient images were randomized separately for each reader evaluation.
The reader study consisted of two parts. In the first part, radiologists were asked to review 120 randomized slices from patient and phantom scans, reconstructed with either a sharp or smooth kernel, and answer four questions regarding whether the presented slice had realistic imaging, contrast, noise, and resolution characteristics of a diagnostic quality CT lung scan (see Fig. S1 in the Supplemental Material). In the second part, radiologists were asked to review 90 randomized slices from patient and phantom scans, all reconstructed with a smooth kernel, and for each slice rate the severity of COVID-19 consolidations (none, mild, moderate, and severe) and whether there are sufficient details (e.g. resolution, contrast-to-noise ratios) for a confident COVID-19 diagnosis (see Fig. S2 in the Supplemental Material). The same five radiologists (1 with 8 years of experience, 3 with 4 years of experience, and 1 with 2 years of experience) were involved in assessing the images in these two parts. To simplify the analysis of the reader study, a higher rating indicates a better review score for all questions except for the COVID-19 severity question. A dedicated user interface was implemented to simplify the review process and to record the radiologists’ replies. Importantly, the participating radiologists were told that they were taking part in a “CT lung image evaluation study” and were completely unaware of the fact that the reviewed data sets included phantom images, which is why this study can be considered a “completely blinded” reader study.
Statistical analysis was performed by experts in statistics (K.D. and R.T.S.) to assess the mean difference in responses between patient and phantom images with the aid of linear mixed models. For this, each question was modeled (separately) using the following equation:
where i denotes the reader and j denotes the image, β0 and β1 denote the mean response across readers for patient scans and difference in mean response between patient and phantom across readers, respectively. The model allows estimation of the mean rating difference between phantom and patient images, while controlling for potential differences between readers in their responses through φi and . φi, which represents reader-level differences in mean response for a given question, and , which represents the remaining model errors, are assumed to be independent across scans and readers with equal variance and zero mean, as well as normally distributed.
Along with statistical significance, which was assessed through standard hypothesis testing, a measure of “clinical significance” is important to quantify the estimated difference between the two sets of images, i.e. phantom vs. patient, with respect to different measures of variance. This is because while differences may be “statistically” significant based on the resulting P-values, at the same time they may be clinically insignificant in terms of their magnitude relative to inter- and intraobserver variabilities. Moreover, if sample sizes are large, arbitrarily small differences will often be statistically significant (33). Thus, assessments of effect sizes are critical to fully assess the mean difference (34). In the two-sample context, Cohen's d is a commonly used measure of effect size (34). However, in the context of clustered data, where in this case, readers are the clusters, a different estimate for the pooled standard deviation (SD) is needed. An alternative for this context, proposed in Westfall et al., is , where denotes the variance in the error terms (within-reader variance) and denotes the between-reader variance (35). Another similar effect size measure is the ratio between the mean difference and within-reader variability, given by Both effect size calculations were assessed here as part of our analysis, together with R2 calculations to measure the proportion of response variation that is associated with the scanned object type (patient vs. phantom).
Finally, to assess the robustness and reproducibility of PixelPrint's phantom production process, three additional phantoms were 3D-printed based on the moderate COVID-19 patient images. The four theoretically equivalent phantoms were scanned on a dual-energy CT scanner (IQon; Philips Healthcare, Cleveland, OH, USA) using an axial protocol at 120 kVp and 0.75 s rotation time, both at clinical-dose exposure levels (6 mGy CTDIvol) and at high-dose exposure levels (18 mGy CTDIvol), and reconstructed with a smooth kernel and a 250 mm field of view at 1.0 mm slice thicknesses. Correspondence between the four phantoms was evaluated with the structural similarity index measure (36) (SSIM).
Results
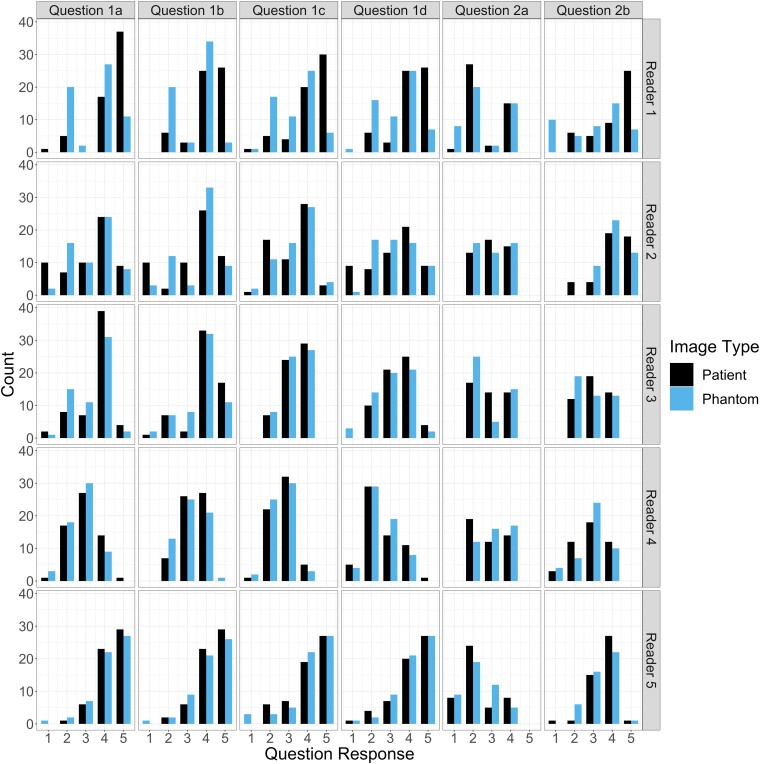
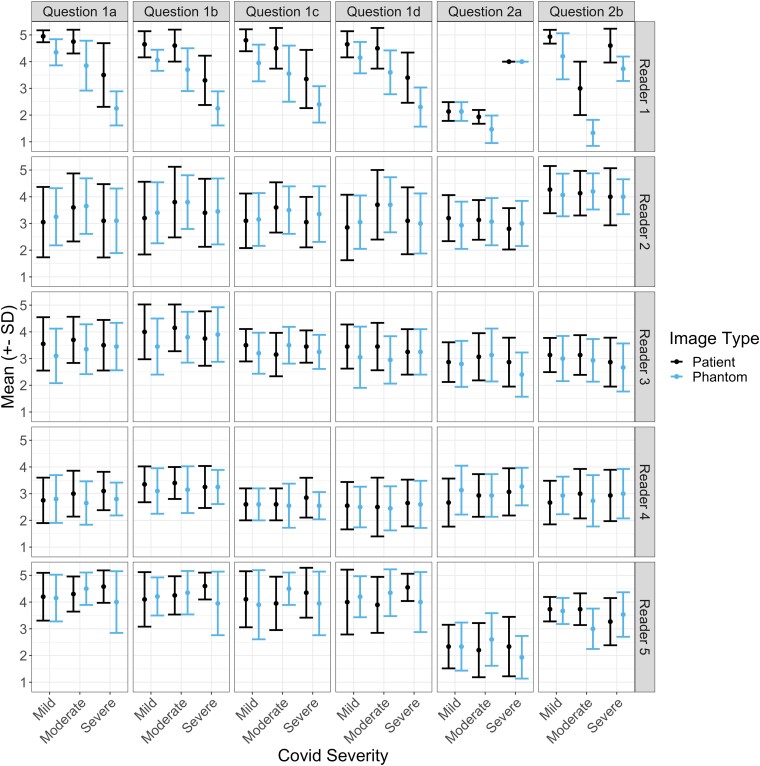
To visualize the data, frequency of reader ratings and mean response values are provided in Figs. 2 and 3. Fig. 2 provides the counts of each response score as values between “1” and “5,” where a higher rating indicates a better score, across all questions, and separated between readers. The figure reveals similar counts between the patient and phantom images, with a response of “4” being most common in both cases for both scan types. Fig. 3 presents calculated mean ± 1 SD response values for each reader and question, separated by the patient COVID-19 severity. Visually, the patient scans have a higher mean response across the different severity levels; however, these differences are small in all cases (<0.5), and are mainly driven by the responses of the first reader.
Fig. 2.
Counts of responses for phantom and patient images by reader (rows) and question (columns): (1a–d) imaging, contrast, noise, and resolution characteristics; (2a) COVID-19 severity; and (2b) diagnostic confidence. Except for the COVID-19 severity question, higher ratings indicate better review scores. Overall, the count frequencies portray a high correspondence between phantom and patient images.
Fig. 3.
Mean ± SD of responses for different COVID-19 severity levels on phantom and patient images by reader (rows) and question (columns): (1a–d) imaging, contrast, noise, and resolution characteristics; (2a) COVID-19 severity; and (2b) diagnostic confidence.
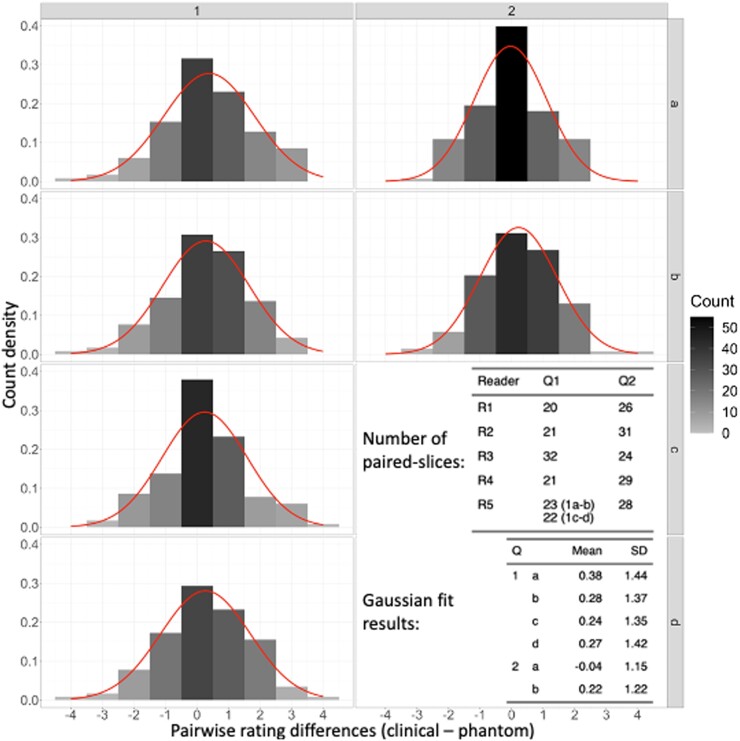
Fig. 4 presents differences in reader ratings between phantom slices that have corresponding (paired) patient slices, i.e. differences in rating between a phantom slice and its matching patient slice: same reader, COVID-19 severity, slice location, and convolution kernel (sharp/smooth), together with Gaussian fits to the data. In general, the data indicate rating differences that are centered between −0.04 and 0.38, implying that on average differences in reader ratings between phantom and patient images are much smaller than a single rating point.
Fig. 4.
Rating difference frequencies between corresponding (paired) patient and phantom images that were reviewed by the same radiologist, together with Gaussian fits to the distributions (overlaid curves). The analysis reveals average differences that are much smaller than a single rating point for all questions and nearly zero points for the COVID-19 severity question (2a).
Modeling results for the six questions that compose both parts of the reader study are provided in Table 2. Each row in the table reports the mean rating (β0), rating difference between patient and phantom images (β1), and R2 values that were obtained for each question separately. Within a question, for a given parameter the estimate, 95% CI, and P-value are provided. Since the rating scores are categorical, P-values for this parameter are not included. In all cases, while the estimated mean differences between patient and phantom were statistically significant (P < 0.005), these differences were very small in magnitude, ranging from 0.03 to 0.29. The magnitude of the difference was also evaluated using R2 measures, resulting in low values for all questions, with a maximum of 0.02 maximum, indicating that a low proportion of response variation is associated with replacing a patient image with a phantom image.
Table 2.
Modeling results for mean ratings and mean differences due to having a phantom in the images, rather than a patient, for each of the reader study questions. Results are accompanied by 95% confidence intervals (CIs), P-values, and R2 values.
| Question | Synopsis | Parameter | Estimate | 95% CI | P-value |
|---|---|---|---|---|---|
| 1a | Imaging characteristics | Patient mean | 3.71 | (3.21, 4.21) | |
| Phantom eff. | −0.29 | (−0.45, −0.13) | <0.005 | ||
| R2 | 0.02 | ||||
| 1b | Contrast characteristics | Patient mean | 3.85 | (3.52, 4.19) | |
| Phantom eff. | −0.27 | (−0.42, −0.11) | <0.005 | ||
| R2 | 0.02 | ||||
| 1c | Noise characteristics | Patient mean | 3.53 | (3.03, 4.03) | |
| Phantom eff. | −0.20 | (−0.35, −0.06) | 0.006 | ||
| R2 | 0.01 | ||||
| 1d | Resolution characteristics | Patient mean | 3.50 | (2.95, 4.05) | |
| Phantom eff. | −0.22 | (−0.39, −0.06) | 0.006 | ||
| R2 | 0.01 | ||||
| 2a | COVID-19 severity | Patient mean | 2.77 | (2.49, 3.05) | |
| Phantom eff. | −0.03 | (−0.20, 0.14) | 0.756 | ||
| R2 | 0.00 | ||||
| 2b | Diagnostic confidence | Patient mean | 3.56 | (3.10, 4.02) | |
| Phantom eff. | −0.29 | (−0.47, −0.12) | <0.005 | ||
| R2 | 0.02 |
Assessment of effect sizes with respect to both inter- and intrareader variabilities are presented in Table 3. The two calculated ratios that were used to estimate the clinical significance of the effect of having a phantom in the image, |d′| and |d|, are reported for each question separately. For each question, both resulting ratios have similar small magnitudes, with a maximal difference of 0.03, and none surpassing a maximal value of 0.31.
Table 3.
Assessment of effect sizes with respect to both inter- and intrareader variabilities reveals that the effect of having a phantom in the image, rather than a patient, is all smaller than one-third of inter/intrareader uncertainty, indicating the clinical insignificance of this effect.
| Q | Synopsis | Phantom effect (β1) | Inter-reader () | Intrareader () | ||
|---|---|---|---|---|---|---|
| 1a | Imaging characteristics | −0.29 | 0.99 | 0.31 | 0.29 | 0.26 |
| 1b | Contrast characteristics | −0.27 | 0.96 | 0.13 | 0.27 | 0.25 |
| 1c | Noise characteristics | −0.20 | 0.83 | 0.31 | 0.22 | 0.19 |
| 1d | Resolution characteristics | −0.22 | 1.01 | 0.37 | 0.22 | 0.19 |
| 2a | COVID-19 severity | −0.03 | 0.83 | 0.09 | 0.03 | 0.03 |
| 2b | Diagnostic confidence | −0.29 | 0.87 | 0.25 | 0.31 | 0.28 |
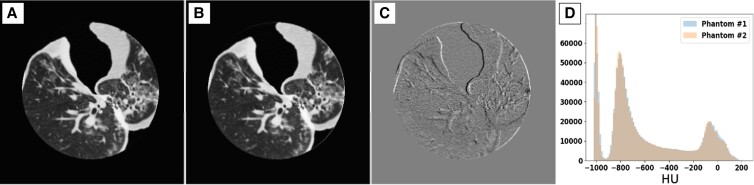
Results for the reproducibility of PixelPrint's production process are presented in Fig. 5 and Table 4. Fig. 5 presents images of two phantoms that were 3D-printed separately using the same patient input (the moderate severity patient), the difference image, and histograms of Hounsfield unit (HU) distribution within each image. As can be seen from the figure, differences in HU mainly arise from minor misalignments between the phantoms rather than offsets in attenuation of geometry (Fig. 5C). This can also be observed by the excellent overlap of histograms (Fig. 5D). Table 4 summarizes SSIM comparisons between the four 3D-printed phantoms. Normalized SSIM values, which were calculated by dividing SSIM values by the ratio of SSIM between the second high-dose acquisition of phantom #1 and the two other high-dose acquisitions of the same phantom, were between 0.928 and 0.979 with an average of 0.965. This value is higher than the normalized SSIM value of the low-dose acquisition for phantom #1 (same phantom that was used for normalization).
Fig. 5.
Comparison between two 3D-printed phantoms (A, B), both based on the moderate COVID-19 patient, scanned separately at a high (non-clinical) dose level show high structural similarities and imaging features, implying high reproducibility of the PixelPrint phantom production process. Window level/width is −400/1,000 HU. C) Difference image between the two sets of images reveals that most of the differences between the images are mainly due to slight misalignments between the two phantoms. Window level/width is 0/200 HU. D) Histograms of CT numbers within the entire phantom volume demonstrate excellent reproducibility.
Table 4.
Comparisons of structural similarity index measures (SSIM) between four 3D-printed phantoms that are al based on the same clinical images. Normalized SSIM values, calculated by dividing SSIM values by the ratio of SSIM between the second high-dose acquisition of phantom #1 and the two other high-dose acquisitions of the same phantom were between 0.928 and 0.979, with an average of 0.965. This value is higher than the normalized SSIM value of the low-dose acquisition for phantom #1 (same phantom that was used for normalization), demonstrating the high production reliability of PixelPrint.
| Phantom # | mAs | Acquisition # | SSIM | Norm. SSIM | Mean ± SD |
|---|---|---|---|---|---|
| 1 | 160 | 1 | 0.776 | 0.953 | 0.953 ± 0.000 |
| 2 | 0.776 | 0.953 | |||
| 3 | 0.777 | 0.953 | |||
| 480 | 1 | 0.811 | Used for normalization | ||
| 2 | 1.000 | → Reference acquisition | |||
| 3 | 0.819 | Used for normalization | |||
| 2 | 480 | 1 | 0.776 | 0.953 | 0.965 ± 0.022 |
| 2 | 0.756 | 0.928 | |||
| 3 | 0.757 | 0.929 | |||
| 3 | 480 | 1 | 0.798 | 0.979 | |
| 2 | 0.797 | 0.978 | |||
| 3 | 0.798 | 0.979 | |||
| 4 | 480 | 1 | 0.797 | 0.978 | |
| 2 | 0.798 | 0.979 | |||
| 3 | 0.797 | 0.978 | |||
Discussion
PixelPrint was developed to provide realistic phantoms that could serve as ground-truth targets for validating the generalizability of inference-based decision-support algorithms between different health centers and imaging protocols, e.g. by imaging the same phantom on multiple scanners, as well as for optimizing disease-targeting imaging protocols. We previously assessed the geometrical and attenuation accuracy of our 3D-printed phantoms for CT lung imaging (30). Here, we validated the adequacy of our phantoms for a specific clinical indication, i.e. diagnosis of COVID-19 consolidations, through a “completely blinded” reader study. As far as we know, this is the first time that 3D-printed CT lung phantoms were evaluated in a reader study. Statistical analysis of image quality ratings, e.g. imaging characteristics, diagnostic outcome, and diagnostic confidence, revealed that difference in replacing a patient image with a phantom image is, on average, smaller than one-third of a single rating point. Importantly, when examining the clinical significance of these differences by relating them to inter- and intrareader variability with effect sizes (Table 3), we conclude that the impact of reading a phantom image rather than a patient image is clinically insignificant. Additionally, tests of PixelPrint's production reproducibility resulted in very high correspondence between phantoms that were 3D-printed using the same patient input. This is based on the higher normalized SSIM values that were measured between high-dose acquisitions of four different phantoms (0.965 ± 0.022), compared with those measured between clinical-dose acquisitions of a single phantom (0.953 ± 0.000).
Several methods have been put forth over the past decade to create clinically applicable CT phantoms. 3D prints with different infill densities were described by Madamesila et al. for quality control purposes in CT (25). A technique to create a patient-based lung phantom was introduced by Kairn et al., where a tissue equivalent lung phantom is created by segmenting the lung's CT scans into three distinct sections (37). Their method, however, falls short of the resolution standards needed to portray the structures in the lung parenchyma. For the purpose of evaluating the quality of CT images, Hernandez-Giron et al. and Joemai and Geleijns developed printed lung phantoms; their prints have vascular systems, however, limited realistic lung textures (17, 38). High-density patient-based bone phantoms were produced by Tino et al. using a dual-head printer and segmentation to produce STL models (26). An alternate method of creating patient-based 3D phantoms was introduced by Jahnke et al. by layering radiopaque 2D prints (39, 40). Although this technique does not use FDM printing and may produce highly detailed head or abdomen phantoms, it is unable to support chest phantoms with density volumes inside the lung that are less than 200 HU. Okkalidis and Marinakis introduced an algorithm that can produce patient-specific skull and chest phantoms and translate DICOM data into printer instructions (12, 13). Their results indicated a trustworthy match in HU; however, specific lung features and textures, for instance, have not been preserved. In contrast to their method, PixelPrint uses unique print-speed control mechanism to change the infill ratio while maintaining a constant extrusion rate per unit of time. With this technique, subpixel linewidth responsiveness is achieved, and hence significantly greater resolution structures can be produced, allowing to preserve realistic lung textures such as small COVID-19 pathological alterations in HU. Notably, PixelPrint also gets rid of the need for morphological and contour detection processes.
With the help of our technique, clinical CT data may be transformed into actual ground truth, creating new prospects for clinical and academic study. Our phantom printing approach enables optimizing CT protocols for daily operations with a focus on specific clinical objectives. For instance, the ethical challenges of scanning patients twice and the limited clinical usefulness of technical phantoms make the clinical introduction of advanced nonlinear reconstruction algorithms (41) hard. With the help of our phantoms, a sizable parameter space may be assessed to choose the best scan and reconstruction parameters in terms of radiation exposure and diagnostic image quality. Accelerating clinical evaluations using patient-based phantoms could have a favorable impact on CT research and development. Early access to accurate clinical data can have a considerable positive impact on breakthroughs in AI and radiomics that are primarily data-driven (1–3). The impact of various CT methods and vendor-to-vendor variations on radiomic characteristics is a major unresolved issue. One would be able to properly assess this effect and identify a strong and rigorous operating environment for radiomic feature extraction with a representative sample of patient-based phantoms produced with PixelPrint. The same collection of phantoms might also be used as a tool to assess and validate harmonization plans (4).
Our study does have limitations. First, while the reader study included a large sample size of images (210 per reader), these images originated from only three clinical patient scans representing three levels of COVID-19 severity. Second, our study focused on a specific clinical indication, i.e. diagnosis of COVID-19 pneumonia. Further studies are required to validate the adequacy of PixelPrint for other lung imaging indications, e.g. lung nodule detection. Nevertheless, results from our study indicate that PixelPrint can potentially serve as an accurate tool for optimization of disease-targeting protocols and for experimental validation of novel inference algorithms, such as radiomics and predictive AI.
In conclusion, we have demonstrated PixelPrint's ability to produce realistic 3D-printed phantoms reliably. As the utilization of these phantoms will grow, they will become more beneficial to the entire community and enable standardization of tests and comparisons of evaluation of advanced medical inference algorithms. For this, we offer copies of the phantoms presented in this study, as well as phantoms based on specific CT images, for the larger medical, academic, and industrial CT community (visit https://www.pennmedicine.org/CTresearch/pixelprint).
Supplementary Material
Acknowledgment
This manuscript was posted on a preprint: https://doi.org/10.1101/2022.05.06.22274739
Contributor Information
Nadav Shapira, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Kevin Donovan, Penn Statistics in Imaging and Visualization Center, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania, 423 Guardian Drive, Philadelphia, PA 19104, USA.
Kai Mei, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Michael Geagan, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Leonid Roshkovan, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Grace J Gang, Department of Biomedical Engineering, Johns Hopkins University, 720 Rutland Avenue, Baltimore, MD 21205, USA.
Mohammed Abed, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA; Department of Radiology, College of Medicine, Ibn Sina University of Medical and Pharmaceutical Sciences, 79G3+3RR Qadisaya Expy, Baghdad, Iraq.
Nathaniel B Linna, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Coulter P Cranston, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Cathal N O'Leary, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Ali H Dhanaliwala, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Despina Kontos, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
Harold I Litt, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA.
J Webster Stayman, Department of Biomedical Engineering, Johns Hopkins University, 720 Rutland Avenue, Baltimore, MD 21205, USA.
Russell T Shinohara, Penn Statistics in Imaging and Visualization Center, Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania, 423 Guardian Drive, Philadelphia, PA 19104, USA; Center for Biomedical Image Computing and Analytics (CBICA), Perelman School of Medicine of the University of Pennsylvania, 3700 Hamilton Walk, Philadelphia, PA 19104, USA.
Peter B Noël, Department of Radiology, Perelman School of Medicine of the University of Pennsylvania, 3400 Civic Center, Philadelphia, PA 19104, USA; Department of Diagnostic and Interventional Radiology, School of Medicine and Klinikum rechts der Isar, Technical University of Munich, Arcisstraße 21, 80333 München, Germany.
Ethics Approval Statement
The data used in the paper was retrospective imaging data. All readers were authors of the paper and gave their informed consent.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
The authors acknowledge support through the National Institutes of Health (R01-CA-249538, R01-CA-264835-01, R01EB031592, and R01-EB-030494).
Author Contributions
N.S., K.M., M.G., L.R., G.J.G., D.K., H.I.L., J.W.S., P.B.N.: study design, phantom production, analysis, manuscript writing. K.D., R.T.S.: statistical analysis, manuscript writing. M.A., N.B.L., C.P.C., C.N.O., A.H.D.: image review.
Data Availability
The data sets used and/or analyzed during the current study available from the corresponding author on reasonable request (pending approval of the University of Pennsylvania Institutional Review Board).
References
- 1. Morin O, et al. 2018. A deep Look into the future of quantitative imaging in oncology: a statement of working principles and proposal for change. Int J Radiat Oncol Biol Phys. 102(4):1074–1082. [DOI] [PubMed] [Google Scholar]
- 2. Xu P, et al. 2021. Radiomics: the next frontier of cardiac computed tomography. Circ Cardiovasc Imaging. 14:256–264. [DOI] [PubMed] [Google Scholar]
- 3. Quantitative Imaging Biomarkers Alliance [accessed 2022 April 26]. https://www.rsna.org/research/quantitative-imaging-biomarkers-alliance
- 4. Rizzo S, et al. 2018. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reiazi R, et al. 2021. The impact of the variation of imaging parameters on the robustness of computed tomography radiomic features: a review. Comput Biol Med. 133:104400. [DOI] [PubMed] [Google Scholar]
- 6. Luna JM, et al. 2022. Radiomic phenotypes for improving early prediction of survival in stage III non-small cell lung cancer adenocarcinoma after chemoradiation. Cancers (Basel) 14(3):700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fortin J-P, et al. 2018. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167:104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127. [DOI] [PubMed] [Google Scholar]
- 9. Shapira N, et al. 2022. Pixelprint: three-dimensional printing of realistic patient-specific lung phantoms for CT imaging. In: Medical imaging 2022: physics of medical imaging. Vol. 12031. SPIE, 2022. 10.1117/12.2611805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ardila Pardo GL, et al. 2020. 3D Printing of anatomically realistic phantoms with detection tasks to assess the diagnostic performance of CT images. Eur Radiol. 30(8):4557–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pegues H, et al. 2019. Using inkjet 3D printing to create contrast-enhanced textured physical phantoms for CT. SPIE-Intl Soc Optical Eng. 10948:181. [Google Scholar]
- 12. Okkalidis N. 2018. A novel 3D printing method for accurate anatomy replication in patient-specific phantoms. Med Phys. 45(10):4600–4606. [DOI] [PubMed] [Google Scholar]
- 13. Okkalidis N, Marinakis G. 2020. Technical note: accurate replication of soft and bone tissues with 3D printing. Med Phys. 47(5):2206–2211. [DOI] [PubMed] [Google Scholar]
- 14. Dangelmaier J, et al. 2018. Experimental feasibility of spectral photon-counting computed tomography with two contrast agents for the detection of endoleaks following endovascular aortic repair. Eur Radiol. 28:3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kopp FK, et al. 2018. Evaluation of a preclinical photon-counting CT prototype for pulmonary imaging. Sci Rep. 8(1):17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muenzel D, et al. 2017. Spectral photon-counting CT: initial experience with dual-contrast agent K-edge colonography. Radiology 283:723–728. [DOI] [PubMed] [Google Scholar]
- 17. Hernandez-Giron I, den Harder JM, Streekstra GJ, Geleijns J, Veldkamp WJH. 2019. Development of a 3D printed anthropomorphic lung phantom for image quality assessment in CT. Phys Med. 57(November):47–57. [DOI] [PubMed] [Google Scholar]
- 18. Abdullah KA, McEntee MF, Reed W, Kench PL. 2018. Development of an organ-specific insert phantom generated using a 3D printer for investigations of cardiac computed tomography protocols. J Med Radiat Sci. 65(3):175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Medical Physics. https://aapm.onlinelibrary.wiley.com/journal/24734209.
- 20. Samuelson FW, Taylor-Phillips S, editors. 2020. Medical imaging 2020: image perception, observer performance, and technology assessment. Vol. 11316. SPIE-Intl Soc Optical Eng. p. 25. 10.1117/12.2550579. [DOI] [Google Scholar]
- 21. Solomon J, Ba A, Bochud F, Samei E. 2016. Comparison of low-contrast detectability between two CT reconstruction algorithms using voxel-based 3D printed textured phantoms. Med Phys. 43(12):6497–6506. [DOI] [PubMed] [Google Scholar]
- 22. Filippou V, Tsoumpas C. 2018. Recent advances on the development of phantoms using 3D printing for imaging with CT, MRI, PET, SPECT, and ultrasound. Med Phys. 45(9):e740–e760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tino R, Yeo A, Leary M, Brandt M, Kron T. 2019. A systematic review on 3D-printed imaging and dosimetry phantoms in radiation therapy. Technol Cancer Res Treat. 18:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leary M, et al. 2020. Additive manufacture of lung equivalent anthropomorphic phantoms: a method to control Hounsfield number utilizing partial volume effect. J Eng Sci Med Diagn Ther. 3(1):011001. [Google Scholar]
- 25. Madamesila J, McGeachy P, Villarreal Barajas JE, Khan R. 2016. Characterizing 3D printing in the fabrication of variable density phantoms for quality assurance of radiotherapy. Phys Med. 32(1):242–247. [DOI] [PubMed] [Google Scholar]
- 26. Tino R, Yeo A, Brandt M, Leary M, Kron T. 2021. The interlace deposition method of bone equivalent material extrusion 3D printing for imaging in radiotherapy. Mater Des. 199:109439. [Google Scholar]
- 27. Hamedani BA, et al. 2018. Three-dimensional printing CT-derived objects with controllable radiopacity. J Appl Clin Med Phys. 19(2):317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hazelaar C, et al. 2018. Using 3D printing techniques to create an anthropomorphic thorax phantom for medical imaging purposes. Med Phys. 45(1):92–100. [DOI] [PubMed] [Google Scholar]
- 29. Leary M, et al. 2015. Additive manufacture of custom radiation dosimetry phantoms: an automated method compatible with commercial polymer 3D printers. Mater Des. 86:487–499. [Google Scholar]
- 30. Mei K, et al. 2022. Three-dimensional printing of patient-specific lung phantoms for CT imaging: emulating lung tissue with accurate attenuation profiles and textures. Med Phys. 49(2):825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hofmanninger J, et al. 2020. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp. 4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beare R, Lowekamp B, Yaniv Z. 2018. Image segmentation, registration and characterization in R with SimpleITK. J Stat Softw. 86:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullivan GM, Feinn R. 2012. Using effect size—or why the P value is not enough. J Grad Med Educ. 4(3):279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen J. 2013. Statistical power analysis for the behavioral sciences. 2nd ed. New York. 10.4324/9780203771587. [DOI]
- 35. Westfall J, Kenny DA, Judd CM. 2014. Statistical power and optimal design in experiments in which samples of participants respond to samples of stimuli. J Exp Psychol Gen. 143(5):2020–2045. [DOI] [PubMed] [Google Scholar]
- 36. Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. 2004. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 13(4):600–612. [DOI] [PubMed] [Google Scholar]
- 37. Kairn T, et al. 2020. Quasi-simultaneous 3D printing of muscle-, lung- and bone-equivalent media: a proof-of-concept study. Phys Eng Sci Med. 43(2):701–710. [DOI] [PubMed] [Google Scholar]
- 38. Joemai RMS, Geleijns J. 2017. Assessment of structural similarity in CT using filtered backprojection and iterative reconstruction: a phantom study with 3D printed lung vessels. Br J Radiol. 90(1079):20160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jahnke P, et al. 2019. Paper-based 3D printing of anthropomorphic CT phantoms: feasibility of two construction techniques. Eur Radiol. 29(3):1384–1390. [DOI] [PubMed] [Google Scholar]
- 40. Jahnke P, et al. 2017. Radiopaque three-dimensional printing: a method to create realistic CT phantoms. Radiology. 282(2):569–575. [DOI] [PubMed] [Google Scholar]
- 41. Willemink MJ, Noël PB. 2019. The evolution of image reconstruction for CT—from filtered back projection to artificial intelligence. Eur Radiol. 29(5):2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and/or analyzed during the current study available from the corresponding author on reasonable request (pending approval of the University of Pennsylvania Institutional Review Board).