Significance
Over 9,000 unique molecules have been isolated from marine sponges, including several bioactive molecules such as the calcium-channel blocker manoalide, antimalarial compounds kalihinol A and axisonitrile-3, and anticancer drug eribulin. Until now, all recent work on the origins of bioactive sponge-derived molecules has indicated that they are made by the symbiotic microorganisms that live within the sponge animal host. Herein, we describe type I terpene synthases characterized from the sponge holobiome and report that these enzymes originate from the sponge animal itself and not a microbial symbiont.
Keywords: natural products, terpene synthase, marine sponge, biosynthesis
Abstract
Sea sponges are the largest marine source of small-molecule natural products described to date. Sponge-derived molecules, such as the chemotherapeutic eribulin, the calcium-channel blocker manoalide, and antimalarial compound kalihinol A, are renowned for their impressive medicinal, chemical, and biological properties. Sponges contain microbiomes that control the production of many natural products isolated from these marine invertebrates. In fact, all genomic studies to date investigating the metabolic origins of sponge-derived small molecules concluded that microbes—not the sponge animal host—are the biosynthetic producers. However, early cell-sorting studies suggested the sponge animal host may play a role particularly in the production of terpenoid molecules. To investigate the genetic underpinnings of sponge terpenoid biosynthesis, we sequenced the metagenome and transcriptome of an isonitrile sesquiterpenoid-containing sponge of the order Bubarida. Using bioinformatic searches and biochemical validation, we identified a group of type I terpene synthases (TSs) from this sponge and multiple other species, the first of this enzyme class characterized from the sponge holobiome. The Bubarida TS-associated contigs consist of intron-containing genes homologous to sponge genes and feature GC percentage and coverage consistent with other eukaryotic sequences. We identified and characterized TS homologs from five different sponge species isolated from geographically distant locations, thereby suggesting a broad distribution amongst sponges. This work sheds light on the role of sponges in secondary metabolite production and speaks to the possibility that other sponge-specific molecules originate from the animal host.
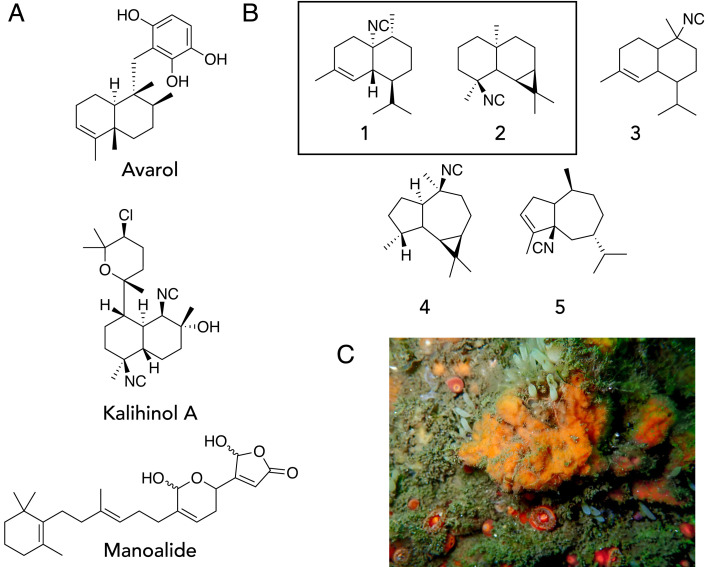
Marine sponges are the richest source of bioactive small molecules from the ocean, accounting for nearly 30% of all known marine natural products (1). Over 9,000 sponge-derived molecules have been characterized to date and represent nearly all biosynthetic molecular classes, such as polyketides, peptides, alkaloids, and terpenoids (1, 2). Not only do sponge-derived compounds serve important ecological roles as feeding deterrents, antioxidants, and antifouling agents, but they also feature impressive medicinal activity, such as the antimalarial activity of kalihinol A, the antitumor activity of avarol, and the anti-inflammatory, antimicrobial, and analgesic activities of manoalide. (Fig. 1A) (3–11). In recent times, the vast majority of investigations into the biosynthetic origins of sponge-derived molecules have focused on the sponge microbiome—the consortium of bacteria, fungi, archaea, and viruses that live in the sponge host—and not on the sponge animal host itself (SI Appendix, Table S1) (12–21). To date, all biosynthetic genes responsible for sponge-derived natural product production have been found in microbial symbionts, which has led to a paradigm that the sponge microbiome is the general source of these molecules.
Fig. 1.
Terpenoids from marine sponges. (A) Bioactive sponge-derived terpenoids: avarol from Dysidea avara, kalihinol A from Acanthella sp., and manoalide from Luffariella variabilis. (B) Reported isonitrile sesquiterpenes 1 to 5 from Bubarida sponges from San Diego. Boxed compounds are molecules validated from the uncharacterized Bubarida sample that was sequenced in this study and represent relative stereochemistry only (22). (C) in situ uncharacterized Bubarida sponge from San Diego.
Pregenomic era cell sorting studies determined that certain sponge metabolites are enriched in sponge animal cells, while others are concentrated in microbial symbiont cells. These experiments suggested that both the sponge animal and its microbiome control the production of sponge-derived metabolites (23–26); however, very few recent studies have followed up on the sponge animal’s role in specialized metabolite production. Notably, most of the sponge cell-associated metabolites were identified as terpenoids, making this class of sponge-derived molecules an appealing target for the study of sponge animal biosynthesis.
Terpenoids are the largest known class of secondary metabolites (12, 27), and sponges are an especially rich source, with approximately 4,100 of the 9,400 reported sponge natural products belonging to this compound class (2). Biosynthetically, terpenoids are derived from linear oligoprenyl diphosphate precursors, which are assembled from C5 isoprenoid building blocks by isopentyl diphosphate synthases (IDSs). These linear precursors are cyclized by terpene synthases (TSs) through cationic cascade reactions initiated by either diphosphate elimination (type I) or protonation (type II) (28). Across various lifeforms, type II TSs are generally more common and conserved than type I TSs, since type II TSs function in the production of key cellular molecules such as sterol triterpenoids, which are involved in membrane stabilization, hormonal signaling, and other essential processes (29). Accordingly, the majority of (>3,000) sponge terpenoids likely originate from type II terpene synthase chemistry, most of which are steroids, carotenoids, and meroterpenoids. We estimate 6% (~550) of all sponge natural products originate from type 1 TSs. Most type 1 TS-derived terpenoids from plants, fungi, and bacteria are usually oxidatively tailored by cytochrome p450s and short-chain dehydrogenases (28, 30, 31). In sponges, notably, roughly 2/3 of probable type I TS-type terpene products belong to the sponge-exclusive class known as nitrogenous terpenes, which feature nitrogen-containing functional groups like isonitriles, isothiocyanates, and formamides that originate from inorganic cyanide (32, 33).
Type I TSs are found in a wide variety of organisms but are scarce in the animal kingdom, where they have, so far, only been described in insects (34–37) and most recently octocorals (38, 39). While all type I TSs feature a conserved IDS-type α-helical fold and are thought to share an ancient evolutionary origin with IDSs (40), insect type I TSs appear to have more recently evolved independently from duplicated IDSs and, therefore, do not bear much sequence similarity to canonical type I TSs (34–37). On the other hand, coral TSs may be the result of ancient horizontal gene transfer from bacteria, given that they share a similar overall structure with key active site residues but still form their own distinct type I TS clade (34). While rare in animals, the presence of type I TSs in insects and corals does suggest that the sponge host animal could potentially house the genes necessary to produce terpenoids. Early radiolabeled feeding work investigated the biosynthesis of sponge nitrogenous terpenoids and, though key information about the origin of the nitrogen-containing functional groups was obtained, the origin of the terpenoid backbone could not be established (33, 41–48). As far as we know, no studies have successfully characterized a TS from a sponge or a sponge-associated microbe, leaving a gap in our understanding of the origin and enzymology underlying sponge terpene biosynthesis.
To address this fundamental gap, we investigated terpenoid biosynthesis in an undescribed San Diego sponge in the order Bubarida [formal species description in process (49), previously identified as Axinella sp. (50)], which is known to contain isonitrile sesquiterpenoids (Fig. 1 B and C) (50). By performing metagenomic and transcriptomic sequencing of the sponge and its microbiota, we identified several candidate TS genes resident on sponge genomic contigs. Heterologous expression of these putative TSs and homologs from other Agelas, Stylissa, and Phakellia sponges demonstrated the activity of seven type I sesquiterpene synthases, which are amongst the first biochemically characterized small molecule biosynthetic enzymes of sponge animal origin.
Results
Metagenome Sequencing and Assembly.
We first queried publicly available sponge metagenomes for canonical microbial and plant-type terpene synthases. We also queried all four sponge genomes available on National Center for Biotechnology Information (NCBI)—including the Amphimedon queenslandica reference genome (51)—as well as genomes from the widespread sponge symbiont genus Entotheonella (52). While we readily identified multiple type II TSs, we detected only one putative type I TS gene, from a Lamellodysidea herbacea sponge-derived “Candidatus Paraprochloron terpiosi SP5CPC1” Metagenome Associated Genome (accession number GCA_014323965.1) (53, 54). Given the broad distribution of characterized sponge terpenoids, we were surprised we did not observe more candidate type I TSs in sponge metagenomes, which we hypothesized was because of the small number of public sponge sequences and a lack of sequences with paired metabolomics data indicative of terpenoids. To address these challenges, we sequenced the metagenome and metatranscriptome of a sponge known to produce isonitrile sesquiterpenoids 1 to 5 (50), belonging to an undescribed species of the order Bubarida.
We collected the San Diego Bubarida sponge by SCUBA and performed gas chromatography mass spectrometry (GCMS) analysis of crude sponge extract, which indicated the presence of several previously reported isonitrile sesquiterpenoids 1 to 5 (SI Appendix, Figs. S1–S3) (50). We further independently established the structures of 1 and 2 using an isonitrile chemoselective chlorooxime probe (22). Next, we generated a hybrid metagenomic assembly from a combination of Illumina paired-end (PE250) and Oxford Nanopore reads, as well as a metatranscriptomic assembly from Illumina data (SI Appendix, Methods and Table S2). The hybrid assembly features 5.5k contigs larger than 5k bp, totaling 180M bp. The N50 value was 61 kb, and the maximum contig size was 960 kb (SI Appendix, Table S2 and Fig. S4).
Terpene Synthase Genome Mining and Identification.
To guide our bioinformatic analysis, we hypothesized that the TSs responsible for sesquiterpene production belonged to one of three previously described groups: 1. Canonical microbial type I TSs from a microbial symbiont; 2. IDS-like TSs akin to those found in insects; or 3. Coral-like type I TSs in the sponge animal. We first searched for canonical microbial type I TSs in our metagenome using BLAST and HMM searches with published TS HMMs, which were solely built from either plant or bacterial TSs (55, 56); however, these initial searches did not uncover any promising candidate TSs from the sponge microbiome. We next investigated the IDSs within the metagenome to search for evidence of neofunctionalization, like that seen in insect TSs. We identified three intron-containing IDS homologs in our metagenomic assembly that we heterologously expressed in Escherichia coli and found to perform typical IDS biochemistry. Two functioned as canonical farnesyl diphosphate (FPP) synthases, while the third functioned as a geranylgeranyl diphosphate (GGPP) synthase (SI Appendix, Figs. S5–S11). None had terpene cyclase-like activities.
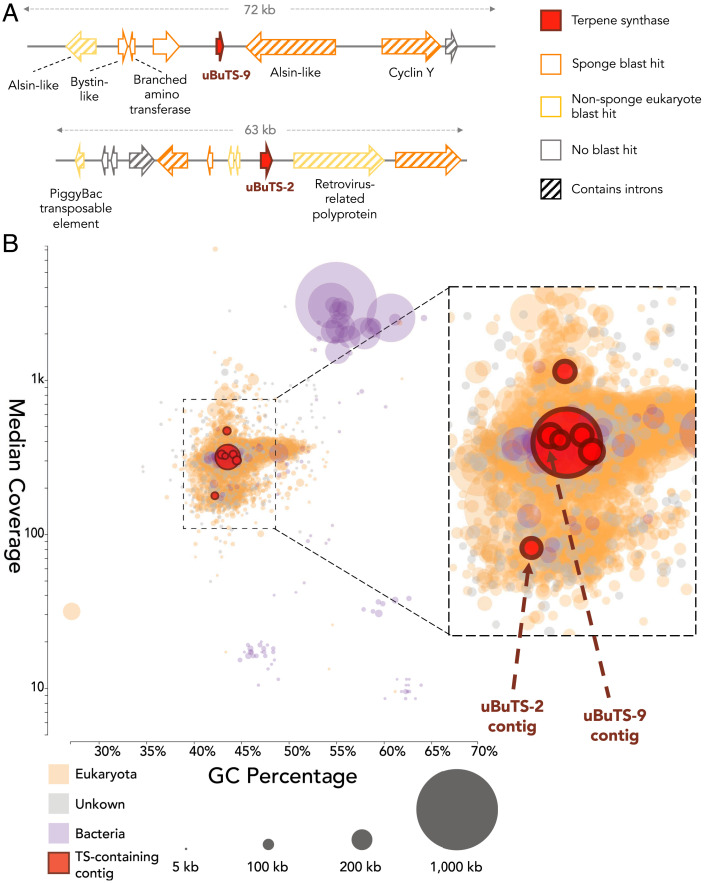
Next, we queried the sponge metatranscriptome with our custom type I TS HMM (TSHMM1), which we created from a diverse set of characterized fungal, bacterial, plant, and recently discovered coral TSs (SI Appendix, Table S3) (38). We hypothesized this more expansive TS HMM would cast a broader net than established TS HMMs to help uncover unusual TSs that were not revealed in earlier searches. Using TSHMM1, we identified 20 transcripts (representing eight unique ORFs) with relatively weak E-values (>10E-7) that contained key terpene synthase amino acid sequence motifs (i.e., DDXXD, NSD/DTE, and RY) and resembled other type I TSs based on homology modeling (SI Appendix, Tables S3 and S4 and Fig. S12). Notably, we detected transcripts derived from five unique putative type I TS genes in the polyadenylation-enriched transcriptome, one of which had a poly-A tail (uBuTS-10), suggestive of eukaryotic origin (SI Appendix, Fig. S13). We mapped four of the putative TSs from the transcriptome to our assembled metagenome, and we additionally identified six unique putative type I TSs only found in the metagenome (SI Appendix, Table S4). After identifying several single-exon candidate gene sequences in the metagenomic dataset, we examined the gene neighborhoods of these putative TSs on assembled metagenomic contigs. We determined that several of the surrounding genes contained introns and had homology to genes from A. queenslandica and other publicly available sponge animal genomes (Fig. 2A and SI Appendix, Table S5). The GC percentages and coverages of these TS-containing contigs were consistent with contigs from our assemblies predicted to belong to the sponge animal host (Fig. 2B). Taking all these features into consideration, we conclude that these genes are part of the sponge animal host genome.
Fig. 2.
Eukaryotic features of terpene synthase-containing contigs. (A) TS-containing contigs from the uncharacterized Bubarida sponge. Annotations based on closest blastx hit from the NCBI GenBank nonredundant database (SI Appendix, Table S5). (B) GC percentage vs. median coverage plot for all uncharacterized Bubarida metagenomic contigs greater than 5 kb in length. Each circle represents an individual contig. The size of the circle indicates the length of the contig, and for contigs not harboring a TS, the circle’s color indicates taxonomic assignment as inferred by homology. Total contig count, sum, contig length, and n50 are as follows: Eukaryota: 2,700, 140 Mb, 74 kb; Unknown: 433, 7.1 Mb, 21 kb; Bacteria: 122, 8.8 Mb, 180 kb. TS-containing contigs: 7, 650 kb, 74 kb. Maximum contig size is 960,860 kb.
We repeated our TSHMM1 search on several other public and in-house sponge metagenomic datasets. We identified two additional putative TSs from two in-house metagenomes of Caribbean Agelas clathrodes and Agelas tubulata, sponges which are not known to produce sesquiterpenoids. By searching all publicly available Sequence Read Archive sponge entries, we also identified a complete putative TS from a Norwegian Phakellia ventilabrum and two overlapping partial TS sequences from a Guamanian Stylissa massa (SI Appendix, Fig. S14) (57, 58). Neither of these sponges had paired metabolomics data available to verify the presence of terpenoids; however, both S. massa and several different sponge species from the Phakellia and Stylissa genera have previously been reported to contain isonitrile terpenoids (59–61). Interestingly, other Norwegian P. ventilabrum specimens from the same area as the sequenced TS-containing P. ventilabrum specimen have been shown to contain isonitrile sesquiterpenoids (62). P. ventilabrum has also recently been reported to form a subclade with the San Diego uncharacterized Bubarida sponge and Axinella cannabina, another nitrogenous sesquiterpene-producing sponge (49).
In Vitro Validation of Sponge Terpene Synthase Function.
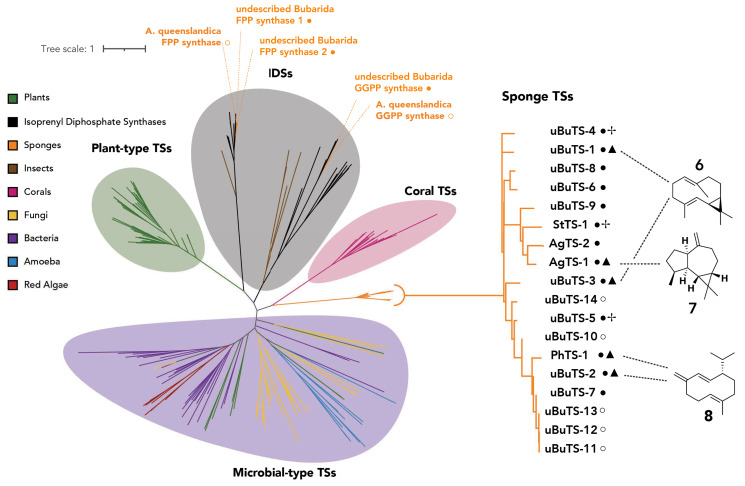
To investigate the activity of these candidate sponge TSs, we heterologously expressed 13 codon-optimized TS genes from five sponge species in E. coli BL21 (DE3). We purified the recombinant proteins and performed in vitro assays with geranyl diphosphate (GPP), FPP, and GGPP. Seven of the 13 TSs showed sesquiterpene synthase activity with selectivity for FPP, five showed low-level monoterpene synthase activity with GPP, and one showed diterpene synthase activity with GGPP (Fig. 3 and SI Appendix, Table S4 and Figs. S15–S35). Five of the active sesquiterpene synthases, which are named according to the taxonomy of their sponge of origin, (uBuTS-1, uBuTS-2, and uBuTS-3 from the undescribed Burarida, AgTS-1 from A. clathrodes, and PhTS-1 from P. ventilabrum) displayed high product specificity, while the other two (uBuTS-4 and uBuTS-5) produced a mixture of sesquiterpene products (SI Appendix, Figs. S15–S21). The products of sponge sesquiterpene synthases uBuTS-1, uBuTS-2, and AgTS-1 were isolated from scaled-up in vitro assays and purified, and their structures were characterized by NMR spectroscopy and polarimetry to be (+)-bicyclogermacrene (6), (–)-germacrene D (8), and (+)-alloaromadendrene (7), respectively (Fig. 3 and SI Appendix, Supplementary Note). The products of uBuTS-3 and PhTS-1 were determined via GCMS using purified 6 and 8 from uBuTS-1 and uBuTS-2, respectively, as standards. uBuTS-4 and uBuTS-5 produced several sesquiterpenes distinct from 6 to 8 which were observed by GCMS but not fully characterized (SI Appendix, Figs. S18 and S19). Four of the TSs with low-level monoterpene synthase activity produced only linear myrcene and ocimene type products (uBuTS-2, uBuTS-3, uBuTS-4, and AgTS-1) and one produced both cyclized and linear monoterpenes (PhTS-1) (SI Appendix, Figs. S24–S26, S32, and S34). StTS-1 from Stylissa produced a mixture of diterpenes when incubated with GGPP (SI Appendix, Fig. S35). The remaining five enzymes were not active with the tested substrates or had low levels of expression (SI Appendix, Figs. S22–S35).
Fig. 3.
Phylogenetic tree of selected type I TSs from several domains of life. Isoprenyl diphosphate synthases (IDSs) are included as an outgroup. The first two or three letters of each TS name represent the taxonomy of sponge from which it originated. Enzymes with ● have been biochemically tested, while those with ○ have not been tested. The major sesquiterpene products of enzymes marked with ▲ were fully characterized (SI Appendix, Supplementary Note), whereas enzymes with ✢ produce a mixture of terpenes identified only by GC-MS (SI Appendix, Figs. S18, S19, and S35). Entries marked with only ● did not produce terpenes under our experimental conditions.
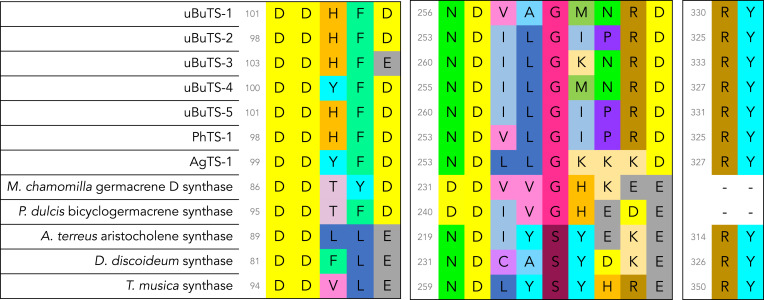
To better understand the sponge TSs in the context of other known type I TSs, we collected sequences of type I TSs with characterized activity from several domains of life, including sequences of several IDSs as an outgroup, and conducted a phylogenetic analysis. The sponge TSs distinctly clade apart from all known terpene synthases and form a monophyletic group (Fig. 3). Despite clading separately from other TSs, they still share many of the key residues that define canonical TSs (Fig. 4). The sponge TSs have the standard aspartate-rich DDXXD motif, which is characteristic of TSs and IDSs and critical for metal binding. Canonical TSs also contain an additional metal binding region exemplified by (N/D)DXX(S/T)XXX(D/E), commonly known as the NSD/DTE motif, which has been hypothesized to arise from the second aspartate-rich DDXXD motif found in IDSs (40, 63). Twelve of the 13 characterized sponge TSs share an unusual NDXXGXXXD motif which aligns with the traditional NSD/DTE domain. While rare in microbial TSs, this unusual Gly residue in the “middle” Ser/Thr position is seen in plant TSs (64). All of the active sponge TSs we characterized feature an RY dimer near the C terminus that’s also present in microbial and octocoral terpene synthases but is not present in known plant terpene synthases (27).
Fig. 4.
Multiple sequence alignment of key active site residues for sponge TSs and selected plant, microbial, and coral TSs.
The Role of Sponge TSs in Isonitrile Sesquiterpenoid Biosynthesis.
Given the predominant terpenoid natural products in the Bubarida sponge are isonitrile sesquiterpenoids, including 1 and 2 that resemble uBuTS products 9 and 7, respectively, we performed exploratory experiments to investigate the role of the newly discovered sponge TSs in the biosynthesis of isonitrile sesquiterpenoids. Garson and colleagues have proposed that inorganic cyanide is the source of the isonitrile and other nitrogenous functional groups unique to sponge terpenoids, by means of radiolabel feeding experiments (33, 41–48, 65). They proposed that a rearranged terpene carbocation intermediate generated by a TS during cyclization might react with the cyanide ion. To test this hypothesis, we performed a series of in vitro TS assays and controls with FPP and potassium cyanide. We tested the 13 sponge TS enzymes, including the five TS enzyme homologs that were not functional in the terpene cyclase assay. In all cases, we did not observe the production of nitrogenous sesquiterpenoids by GCMS in the enzymatic assays or noenzyme controls (SI Appendix, Figs. S36–S47). We also combined uBuTS1–9 and ran an in vitro assay with FPP and cyanide to test whether there was cross-reactivity between any of the nine uBuTS enzymes, but again we observed only the unmodified sesquiterpenes (SI Appendix, Fig. S48). These results suggest that a separate enzyme is required for the regio- and stereoselective installation of the isonitrile. We inspected the gene neighborhoods of the uBuTS genes for clustered genes that may be involved in cyanide generation or installation; however, no candidates were identified.
To further interrogate the role of cyanide in this system, we tested the sponge tissue for the presence of free cyanide using 4-(4,6-dimethoxy- 1,3,5-triazin-2-yl)-4-methylmorpholinium chloride, a selective reagent that allows for cyanide detection by GCMS (66). Freshly collected Bubarida sponge tissue did not contain a detectable amount of cyanide (SI Appendix, Fig. S49). Additional work to understand the biosynthesis of isonitrile sesquiterpenoids is ongoing.
Discussion
We have discovered a new class of type I TSs from marine sponges. The Bubarida TS genes are surrounded by intron-containing genes that closely resemble genes from publicly available sponge genomes and are located on contigs with nucleotide compositions (percent GC) and coverages characteristic of other contigs matching eukaryotic, rather than bacterial sequences. Additionally, the polyadenylation of a TS transcript in our RNA-seq data and the fact we identified type I TSs in multiple sponge species from geographically distant locations strongly support the origin of these TSs sequences in the sponge animal host and not a microbial symbiont. The discovery of sponge animal TSs contradicts the current paradigm that microbes are the only source of natural products isolated from sponges. Besides octocorals and insects, sponges are now the third class of animals recognized to biosynthesize specialized terpenes other than steroids and carotenoids.
Given that marine sponges have been studied as sources of bioactive natural products—including hundreds of terpenoids—for over 70 y, it is notable that TSs were identified in only a few sponges. We believe that this is due to a scarcity of sponge-specific genomic and transcriptomic data, as relatively few species of sponges have any sequencing data associated with them, and even fewer projects focus on the sponge animal as opposed to the associated microbiota. Additionally, nitrogenous terpene production is largely concentrated in a relatively small number of sponges belonging to the order Bubarida (67), for which sequencing data are especially sparse. Furthermore, the order Bubarida is a recently erected and highly debated order that arose because of genetic evidence conflicting with earlier morphology-based classifications of Axinella, Acanthella, Phakellia, Axinyssa, and other nitrogenous terpenoid-producing genera (49, 68). It has even been suggested that nitrogenous terpenes are a chemotaxonomic marker of Bubarida (67), and the monophyletic nature of the newly identified TS sequences points to one common ancestor sequence that was inherited with evolutionary radiation of sponge species. Therefore, our unexpected discovery of functional TSs encoded by sponges outside of Bubarida and not previously reported to produce terpenoids suggests either a need for further expansion of the Bubarida order or perhaps that this terpene producing capacity may be more widespread than currently appreciated. Unlike coral TSs, which are ubiquitous across all octocorals and are descended from an ancestral TS in the last common octocoral ancestor, sponge TSs are confined to a smaller group of sponge species. Our observation suggests that these TSs may have evolved more recently to serve a specific evolutionary need; however, more research is needed to access the greater distribution and evolutionary history of these genes. As such, further enzyme discovery efforts could greatly benefit from the increased pairing of public sequencing and metabolomics datasets (69).
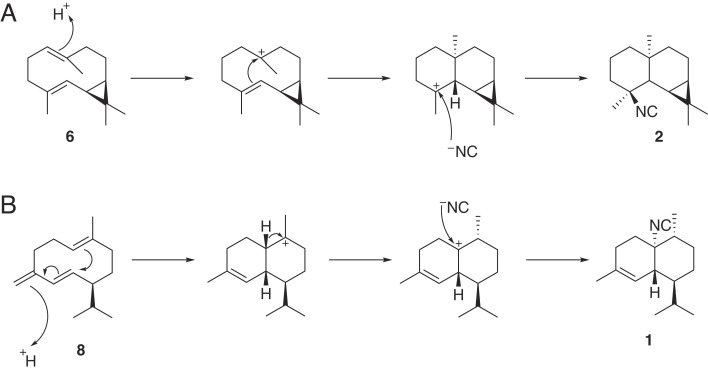
Many terpenoids found in sponges are uniquely functionalized with isonitrile and related nitrogen-containing functional groups like isothiocyanates, formamides, isocyanates, and amines, suggesting the presence of enzymes that stereoselectively install these functional groups onto terpene hydrocarbons. We hypothesize that the sesquiterpenes produced by the Bubarida TS enzymes are intermediates in the biosynthesis of the isonitrile natural products (22, 50) given their uncanny structural resemblance and the fact Axinella cannabina, another nitrogenous terpene-producing sponge closely related to the San Diego Bubarida, is reported to produce bicyclogermacrene 6 (70). The genes and enzymes responsible for these distinctive sponge functionalizations, however, remain unknown, though work from Garson, Scheuer, and others has suggested the origin of these functional groups is inorganic cyanide (44–47). Our observation that the sponge TSs do not install the isonitrile functionality upon the addition of cyanide to TS enzyme assays suggests that there is at least one additional enzyme involved in this biotransformation. We posit that the mechanism to generate 1 from 8 and 2 from 6 proceeds via protonation of an alkene, ring formation, and carbocation quenching by free cyanide to form the isonitrile (Fig. 5), a reaction that may be catalyzed by an enzyme akin to a type II TS, which are known to perform cyclizations on isoprenoid substrates (28). Currently, there is no known precedent for such an “isonitrile synthase” which utilizes cyanide as a substrate; however, the study of enzymes such as FlvF, a repurposed terpene cyclase involved in C-N bond formation, could provide insight into how an isonitrile synthase might evolve from isoprenoid biosynthesis machinery (71). While plant, bacteria, and fungi TS genes often have genes coding for tailoring enzymes like cytochrome P450s clustered near them on the genome (72), the Bubarida sponge TSs do not cluster with obvious candidate tailoring enzyme genes. There is still much to be learned about this fascinating pathway, of which the late D. John Faulkner once claimed that there is “no more interesting biosynthetic study among the marine natural products” (73).
Fig. 5.
Two proposed mechanistic routes to nitrogenous sesquiterpenoids from sponge TS products. (A) Proposed mechanism to yield undescribed Bubarida natural product 2 from uBuTS-1 product 6. (B) Proposed mechanism to yield undescribed Bubarida natural product 1 from uBuTS-2 product 8.
The discovery of sponge animal biosynthesis genes sheds new light on other classes of sponge natural products that have long been speculated to be sponge animal-derived, such as the pyrrole-imidazole alkaloids, bromotyrosine alkaloids, and other amino acid-derived sponge alkaloids. These sponge-specific alkaloids are also enriched in sponge cell cultures and sponge tissues (74, 75), and recent work has shown that bromotyrosine alkaloids are shared between sponges with vastly different microbiomes (76). Accordingly, the sponge terpene synthase work presented herein not only lends credence to the possibility of sponge animal biosynthesis in other natural product classes but also provides a framework for identifying novel biosynthetic genes from sponge animal hosts.
Methods and Materials
Methods for sponge collection, sequencing, enzyme discovery, and chemical analysis are included in the SI Appendix, Methods. The SI Appendix, Methods also includes methodologies for enzyme expression, purification, and activity assays. The structural characterization of enzyme products is detailed in the SI Appendix, Supplementary Note. Remaining data and methods are included in the SI Appendix. The accession numbers of sequences deposited and used in this study are listed in SI Appendix, Table S6.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Christian McDonald, Phil Zerofski, and Rich Walsh (Scripps Institution of Oceanography) for their support and assistance in sponge collection, Thomas Turner (UC Santa Barbara) for his assistance with sponge taxonomy, Kristen Jepsen (Institute for Genomic Medicine, UC San Diego) for guidance with sequencing, and Lihini Aluwihare (Scripps Institution of Oceanography) and Brendan Duggan (UC San Diego) for access to GCMS and NMR equipment, respectively. This research was supported by a UCSD Academic Senate Research Grant to B.S.M. and K.W., the NSF Graduate Research Fellowship Program to K.W., NIH grants R01-GM085770 to B.S.M., R01-ES030316 to B.S.M. and E.E.A., F32-GM129960 to T.d., the Marsden Fund, Royal Society Te Apārangi to T.d., a Leopoldina postdoctoral fellowship (LPDS 2019-04) to I.B., and a Swiss NSF postdoctoral fellowship (P2EZP3_195643) to R.J.B.S.
Author contributions
K.W., T.d.R., and B.S.M. designed research; K.W., T.d.R., I.B., T.S.S., and S.P. performed research; K.W., T.d.R., I.B., T.S.S., R.J.B.S., S.P., E.E.A., and B.S.M. analyzed data; and K.W. and B.S.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
DNA data have been deposited in NCBI Bioproject (PRJNA907134 and PRJNA824609) (77, 78).
Supporting Information
References
- 1.Mehbub M. F., Lei J., Franco C., Zhang W., Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs 12, 4539–4577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyu C., et al. , CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 49, D509–D515 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proksch P., Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon 32, 639–655 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Takamatsu S., et al. , Marine natural products as novel antioxidant prototypes. J. Nat. Prod. 66, 605–608 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fusetani N., Antifouling marine natural products. Nat. Prod. Rep. 28, 400–410 (2011). [DOI] [PubMed] [Google Scholar]
- 6.de Silva E. D., Scheuer P. J., Manoalide, an antibiotic sesterterpenoid from the marine sponge Luffariella variabilis (polejaeff). Tetrahedron Lett. 21, 1611–1614 (1980). [Google Scholar]
- 7.Ferrándiz M. L., et al. , Avarol and avarone, two new anti-inflammatory agents of marine origin. Eur. J. Pharmacol. 253, 75–82 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Amigó M., Payá M., Braza-Boïls A., De Rosa S., Terencio M. C., Avarol inhibits TNF-α generation and NF-κB activation in human cells and in animal models. Life Sci. 82, 256–264 (2008). [DOI] [PubMed] [Google Scholar]
- 9.de Freiras J. C., Blankemeier L. A., Jacobs R. S., In vitro inactivation of the neurotoxic action of β-bungarotoxin by the marine natural product, manoalide. Experientia 40, 864–865 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Chang C. W. J., Patra A., Roll D. M., Scheuer P. J., Kalihinol-A, a highly functionalized diisocyano diterpenoid antibiotic from a sponge. J. Am. Chem. Soc. 106, 4644–4646 (1984). [Google Scholar]
- 11.Miyaoka H., et al. , Antimalarial activity of kalihinol A and new relative diterpenoids from the Okinawan sponge, Acanthella sp. Tetrahedron 54, 13467–13474 (1998). [Google Scholar]
- 12.Blunt J. W., et al. , Marine natural products. Nat. Prod. Rep. 35, 8–53 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Hentschel U., Piel J., Degnan S. M., Taylor M. W., Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10, 641–654 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Fisch K. M., et al. , Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 5, 494–501 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Schmidt E. W., Obraztsova A. Y., Davidson S. K., Faulkner D. J., Haygood M. G., Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 136, 969–977 (2000). [Google Scholar]
- 16.Mori T., et al. , Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc. Natl. Acad. Sci. U.S.A. 115, 1718–1723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal V., et al. , Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat. Chem. Biol. 13, 537–543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storey M. A., et al. , Metagenomic exploration of the marine sponge Mycale hentscheli uncovers multiple polyketide-producing bacterial symbionts. MBio 11, 1–16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima Y., Egami Y., Kimura M., Wakimoto T., Abe I., Metagenomic analysis of the sponge Discodermia reveals the production of the cyanobacterial natural product kasumigamide by “entotheonella”. PLoS One 11, 1–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman M. F., Vagstad A. L., Piel J., Polytheonamide biosynthesis showcasing the metabolic potential of sponge-associated uncultivated “Entotheonella” bacteria. Curr. Opin. Chem. Biol. 31, 8–14 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Ueoka R., et al. , Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat. Chem. Biol. 11, 705–712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schäfer R., et al. , Identification of isonitrile-containing natural products in complex biological matrices through ligation with chlorooximes. Chem. Eur. J. 29, e202203277 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unson M. D., Faulkner D. J., Cyanobacterial symbiont biosynthesis of chlorinated metabolites from Dysidea herbacea (Porifera). Experientia 49, 349–353 (1993). [Google Scholar]
- 24.Flowers A. E., Garson M. J., Webb R. I., Dumdei E. J., Charan R. D., Cellular origin of chlorinated diketopiperazines in the dictyoceratid sponge Dysidea herbacea (Keller). Cell Tissue Res. 292, 597–607 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Uriz M. J., Turon X., Galera J., Tur J. M., New light on the cell location of avarol within the sponge Dysidea avara (Dendroceratida). Cell Tissue Res. 285, 519–527 (1996). [Google Scholar]
- 26.Garson M. J., et al. , Terpenes in sponge cell membranes: Cell separation and membrane fractionation studies with the tropical marine sponge Amphimedon sp. Lipids 27, 378–388 (1992). [Google Scholar]
- 27.Dickschat J. S., Bacterial terpene cyclases. Nat. Prod. Rep. 33, 87–110 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Christianson D. W., Structural and chemical biology of terpenoid cyclases. Chem. Rev. 117, 11570–11648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrader J., Bohlmann J., “Biotechnology of Isoprenoids” in Advances in Biochemical Engineering/Biotechnology, Scheper T., Ed. (Springer, 2015), pp. 3–475. [Google Scholar]
- 30.Rudolf J. D., Alsup T. A., Xu B., Li Z., Bacterial terpenome. Nat. Prod. Rep. 23, 9–10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quin M. B., Flynn C. M., Schmidt-Dannert C., Traversing the fungal terpenome. Nat. Prod. Rep. 31, 1449–1473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsermann J., Kauhl U., Opatz T., Marine isonitriles and their related compounds. Mar. Drugs 14, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garson M. J., Simpson J. S., Marine isocyanides and related natural products–Structure, biosynthesis and ecology. Nat. Prod. Rep. 21, 164–179 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Beran F., et al. , Novel family of terpene synthases evolved from trans-isoprenyl diphosphate synthases in a flea beetle. Proc. Natl. Acad. Sci. U.S.A. 113, 2922–2927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lancaster J., et al. , De novo formation of an aggregation pheromone precursor by an isoprenyl diphosphate synthase-related terpene synthase in the harlequin bug. Proc. Natl. Acad. Sci. U.S.A. 115, E8634–E8641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilg A. B., Bearfield J. C., Tittiger C., Welch W. H., Blomquist G. J., Isolation and functional expression of an animal geranyl diphosphate synthase and its role in bark beetle pheromone biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 9760–9765 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilg A. B., Tittiger C., Blomquist G. J., Unique animal prenyltransferase with monoterpene synthase activity. Naturwissenschaften 96, 731–735 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Burkhardt I., De Rond T., Chen P. Y., Moore B. S., Ancient plant-like terpene biosynthesis in corals. Nat. Chem. Biol. 18, 664–669 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scesa P. D., Lin Z., Schmidt E. W., Ancient defensive terpene biosynthetic gene clusters in the soft corals. Nat. Chem. Biol. 18, 659–663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei G., et al. , Evolution of isoprenyl diphosphate synthase-like terpene synthases in fungi. Sci. Rep. 10, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumdei E. J., Flowers E., Garson M., June M., The biosynthesis of sesquiterpene isocyanides and isothiocyanates in the marine sponge Acanthella cavernosa (Dendy); Evidence for dietary transfer to the dorid nudibranch Phyllidiella pustulosa. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 118, 1385–1392 (1997). [Google Scholar]
- 42.Garson M. J., Partali V., Liaaen-Jensen S., Stoilov I. L., Isoprenoid biosynthesis in a marine sponge of the Amphimedon genus: Incorporation studies with [l-14C]acetate [4-14C]cholesterol and [2-14C]mevalonate. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 91B, 293–300 (1988). [DOI] [PubMed] [Google Scholar]
- 43.Simpson J. S., Garson M. J., Biosynthetic pathways to isocyanides and isothiocyanates; precursor incorporation studies on terpene metabolites in the tropical marine sponges Amphimedon terpenensis and Axinyssa n.sp. Org. Biomol. Chem. 2, 939–948 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Garson M. J., Biosynthetic studies on marine natural products. Nat. Prod. Rep. 6, 143–170 (1989). [Google Scholar]
- 45.Simpson J. S., Garson M. J., Advanced precursors in marine biosynthetic study: The biosynthesis of diisocyanoadociane in Amphimedon terpenensis. Tetrahedron Lett. 40, 3909–3912 (1999). [Google Scholar]
- 46.Simpson J. S., Garson M. J., Thiocyanate biosynthesis in the tropical marine sponge Axinyssa n. sp. Tetrahedron Lett. 39, 5819–5822 (1998). [Google Scholar]
- 47.Chang C. W. J., Scheuer P. J., Biosynthesis of marine isocyanoterpenoids in sponges. Comp. Biochem. Physiol. B Biochem. 97, 227–233 (1990). [Google Scholar]
- 48.Karuso P., Scheuer P. J., Biosynthesis of isocyanoterpenes in sponges. J. Org. Chem. 54, 2092–2095 (1989). [Google Scholar]
- 49.Turner T. L., Lonhart S., The sponges of the carmel pinnacles marine protected area. bioRxiv [Preprint] (2022). 10.1101/2022.11.02.514922 (Accessed 9 November 2022). [DOI] [PubMed]
- 50.Thompson J. E., Walker R. P., Faulkner D. J., Screening and bioassays fro biologically-active substances from forty marine sponge species from San Diego, California, USA. Mar. Biol. 21, 11–21 (1985). [Google Scholar]
- 51.Srivastava M., et al. , The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson M. C., et al. , An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Podell S., et al. , A genomic view of trophic and metabolic diversity in clade-specific Lamellodysidea sponge microbiomes. Microbiome 8, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agger S. A., Lopez-Gallego F., Hoye T. R., Schmidt-Dannert C., Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J. Bacteriol. 190, 6084–6096 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada Y., et al. , Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 857–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mistry J., et al. , Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryu T., et al. , Hologenome analysis of two marine sponges with different microbiomes. BMC Genomics 17, 158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plese B., et al. , Mitochondrial evolution in the Demospongiae (Porifera): Phylogeny, divergence time, and genome biology. Mol. Phylogenet. Evol. 155, 107011 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Mitome H., Shirato N., Miyaoka H., Yamada Y., Van Soest R. W. M., Terpene isocyanides, isocyanates, and isothiocyanates from the Okinawan marine sponge Stylissa sp. J. Nat. Prod. 67, 833–837 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Chanthathamrongsiri N., Yuenyongsawad S., Wattanapiromsakul C., Plubrukarn A., Bifunctionalized amphilectane diterpenes from the sponge Stylissa cf. massa. J. Nat. Prod. 75, 789–792 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Wolf D., Schmitz F. J., New diterpene isonitriles from the sponge Phakellia pulcherrima. J. Nat. Prod. 61, 1524–1527 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Possner S. T., Isolierung und Strukturaufklärung neuer Naturstoffe aus marinen Invertebraten (University of Hamburg, Hamburg, Germany, 2005). [Google Scholar]
- 63.Aaron J. A., Christianson D. W., Trinuclear metal clusters in catalysis by terpenoid synthases. Pure Appl. Chem. 82, 1585–1597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou K., Peters R. J., Investigating the conservation pattern of a putative second terpene synthase divalent metal binding motif in plants. Phytochemistry 70, 366–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fookes C. J. R., Garson M. J., MacLeod J. K., Skelton B. W., White A. H., Biosynthesis of diisocyanoadociane, a novel diterpene from the marine sponge. J. Chem. Soc. Perkin Trans. 1, 1003–1011 (1988). [Google Scholar]
- 66.Yamaguchi A., Miyaguchi H., A screening method for cyanide in blood by dimethoxytriazinyl derivatization-GC/MS. J. Chromatogr. Sci. 59, 1–6 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Galitz A., Nakao Y., Schupp P. J., Wörheide G., A soft spot for chemistry–Current taxonomic and evolutionary implications of sponge secondary metabolite distribution. Mar. Drugs 19, 1–25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrow C., Cárdenas P., Proposal for a revised classification of the Demospongiae (Porifera). Front. Zool. 12, 1–27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schorn M. A., et al. , A community resource for paired genomic and metabolomic data mining. Nat. Chem. Biol. 17, 363–368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciminiello P., Fattorusso E., Magno S., Mayol L., Sesquiterpenoids based on the epi-maaliane skeleton from the marine sponge Axinella cannabina. J. Nat. Prod. 48, 64–68 (1985). [Google Scholar]
- 71.Yee D. A., et al. , Genome mining of alkaloidal terpenoids from a hybrid terpene and nonribosomal peptide biosynthetic pathway. J. Am. Chem. Soc. 142, 710–714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Medema M. H., de Rond T., Moore B. S., Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet. 22, 553–571 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faulkner D. J., Interesting aspects of marine natural products chemistry. Tetrahedron 33, 1421–1443 (1977). [Google Scholar]
- 74.Andrade P., Willoughby R., Pomponi S. A., Kerr R. G., Biosynthetic studies of the alkaloid, stevensine, in a cell culture of the marine sponge Teichaxinella morchella. Tetrahedron Lett. 40, 4775–4778 (1999). [Google Scholar]
- 75.Richelle-Maurera E., et al. , Localization and ecological significance of oroidin and sceptrin in the Caribbean sponge Agelas conifera. Biochem. Syst. Ecol. 31, 1073–1091 (2003). [Google Scholar]
- 76.Mohanty I., et al. , Presence of bromotyrosine alkaloids in marine sponges Is independent of metabolomic and microbiome architectures. mSystems 6, e01387-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson, et al. , Terpene biosynthesis in marine sponge animals. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/sra/PRJNA907134 Deposited 6 December 2022.
- 78.Podell S., Caribbean Agelas sponge metagenomes. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA824609 Deposited 7 April 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
DNA data have been deposited in NCBI Bioproject (PRJNA907134 and PRJNA824609) (77, 78).