Significance
Over 30 y ago, the phenylpropanoid dehydrodiconiferyl alcohol glucoside (DCG) was identified as a compound with cytokinin-substituting activity. The hormone-like activity of DCG has always intrigued the scientific community, and the initial studies are still frequently used to explain the growth defects of lignin-mutants. Despite general interest, no follow-up studies on DCG’s mode of action have been published, and the original activity claims were never scrutinized by an independent research group. We synthesized DCG and repeated the various activity assays as described in the original publications but could not replicate any of the reported results. Our conclusion is that DCG, unlike what has been accepted over the decades, has no cytokinin-substituting activity.
Keywords: phenylpropanoid, lignin, cytokinin, cell division, callus
Abstract
Dehydrodiconiferyl alcohol glucoside (DCG) is a phenylpropanoid-derived plant metabolite with reported cytokinin-substituting and cell-division-promoting activity. Despite its claimed activity, DCG did not trigger morphological changes in Arabidopsis seedlings nor did it alter transcriptional shifts in cell division and cytokinin-responsive genes. In reinvestigating the bioactivity of DCG in its original setting, the previously described stimulation of tobacco callus formation could not be confirmed. No evidence was found that DCG is actually taken up by plant cells, which could explain the absence of any observable activity in the performed experiments. The DCG content in plant tissue increased when feeding explants with the DCG aglycone dehydrodiconiferyl alcohol, which is readily taken up and converted to DCG by plant cells. Despite the increased DCG content, no activity for this metabolite could be demonstrated. Our results therefore demand a reevaluation of the often-quoted cytokinin-substituting and cell-division-promoting activity that has previously been attributed to this metabolite.
The phenylpropanoid pathway is a core metabolic pathway positioned at the interface between primary and secondary metabolisms in plants. Most of the carbon entering the pathway via the aromatic amino acid phenylalanine is channeled toward the building blocks for lignin, an aromatic polymer deposited mainly in the secondary-thickened cell wall. Any severe disruption of the pathway affects plant growth (1–8), with a severity linked to the degree of lignin reduction for a given gene. Besides the reduction in lignin content, the altered abundance of phenylpropanoid pathway-derived compounds could also contribute to the disturbed growth in phenylpropanoid pathway mutants. One of the claimed bioactive phenylpropanoids is dehydrodiconiferyl alcohol glucoside (DCG), a glycosylated dilignol derived from β-5-coupled coniferyl alcohol monomers (1–3, 7, 9, 10). Although this compound theoretically exists as four diastereomers due to the glucoside and the presence of two chiral carbon centers in the phenylcoumaran moiety, only the trans-configuration was found to be present in earlier studies (11), so there are only two physically distinct and NMR-distinct diastereomers. Different DCG isomers were originally isolated from tumor cells of Vinca rosea (12) and later found to accumulate in actively dividing tobacco cells (13). Interestingly, two of these could apparently substitute for cytokinin in callus growth assays (13, 14), making the authors conclude that cytokinin controls cell division through the accumulation of DCG.
One of the lignin mutants for which DCG depletion has been put forward as a possible cause for its growth defects is the Arabidopsis mutant cyp98A3 (9). The corresponding gene, CYP98A3, encodes a p-COUMAROYL SHIKIMATE/QUINATE 3′-HYDROXYLASE (C3′H) that catalyzes the conversion of 4-coumaroyl shikimate to caffeoyl shikimate (15), a crucial step in the phenylpropanoid pathway's shuttling of carbon flux toward DCG among other compounds (9). However, we were unable to complement the severe growth defect of this mutant with exogenous synthetic DCG. In addition, we found no indication for a cell-division or cytokinin response triggered by DCG in Arabidopsis. Driven by these unexpected results, we reinvestigated the previously described bioactivity of DCG in its original setting.
Results
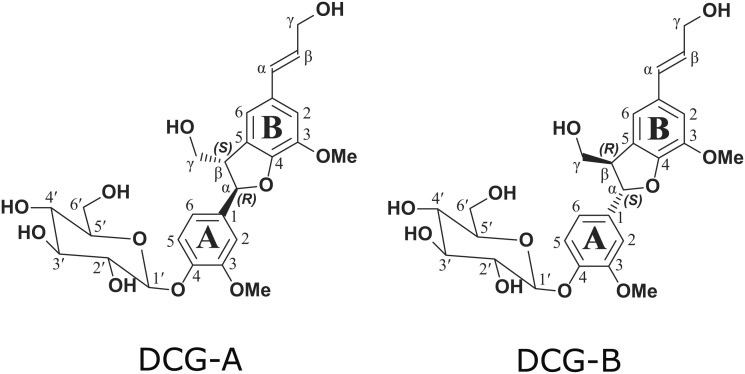
DCG diastereomers were synthesized as described in ref. 14 with minor modifications to increase the yield and purity of the end product (Materials and Methods). The identity of the synthesized compound was verified by Nuclear Magnetic Resonance spectrometry (NMR) providing evidence that our stock sample was authentic DCG (SI Appendix, Fig. S1A). In addition, we found there was clear splitting of A3, A5, and Aα 13C NMR peaks, and a tiny splitting in the glucoside anomeric carbon (1′) (SI Appendix, Fig. S1B), indicating the presence of DCG-A and DCG-B diastereomers (following the nomenclature introduced in ref. 13) (Fig. 1). As both diastereomers were previously described to have equal cytokinin-substituting activity (13), there was no need to further differentiate between the compounds. From here on, the synthesized racemic product will be referred to simply as “DCG”.
Fig. 1.
Structures of DCG-A and DCG-B. DCG-A and DCG-B representing the two DCG structures possessing the trans-configuration, respectively (AαR, AβS)-DCG and (AαS, AβR)-DCG, as previously defined (13). Because of the glucoside, these are two physically distinct compounds (diastereomers) that are not mirror images (i.e., not enantiomers) and are NMR-different (as seen by the splitting of carbon peaks A3, A5, and A1, as well as the anomeric carbon (1′) on the glucoside (SI Appendix, Fig. S1).
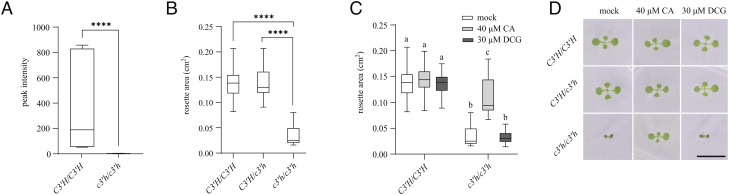
The synthesized DCG was subsequently used in an attempt to complement the growth defect of a lignin mutant. The transferred DNA (T-DNA) insertional knockout (KO) mutant c3′h was selected for this purpose, as this mutant has a dramatic growth phenotype for which the reduction in DCG content had been put forward as possible cause (9). As homozygous c3′h mutants are sterile, a segregating seed stock was used. After 10 d of growth on 0.5× MS medium, seedlings were genotyped by PCR and their DCG content was subsequently assessed by UPLC/MS. In line with the position of the affected step in the phenylpropanoid pathway, no DCG was detected in homozygous c3′h seedlings while being present in wild-type (WT) plants (Fig. 2A). For the feeding experiment, plants were grown for 10 d on 0.5× MS medium, after which the rosette area was quantified and plants were genotyped to determine zygosity of the T-DNA insert. Whereas no significant difference was found between the rosette area of WT and heterozygous plants, the rosette area of homozygous c3′h mutants was significantly smaller (reduction of 75% compared with WT and heterozygous plants) (Fig. 2B). This growth reduction could be partly restored by growing plants on medium supplemented with 40 µM coniferaldehyde, a downstream product of the C3′H depleted in the c3′h mutant (9). When coniferaldehyde was replaced by 30 µM DCG, the growth defect of homozygous c3′h mutants was not complemented (Fig. 2 C and D).
Fig. 2.
Effect of DCG on the growth of c3′h mutants. (A) Average DCG abundance in WT (C3′H/C3′H) and homozygous c3′h mutant seedlings 10 d after stratification (DAS) (n = 5). (B) Average projected rosette area of individual Arabidopsis plants (WT, heterozygous and homozygous c3′h mutants) grown on 0.5× MS medium 10 DAS (16 < n < 70 biological repeats). (C) Average projected rosette area of WT and c3′h grown on 0.5× MS medium supplemented with different compounds 10 DAS (16 < n < 37 biological repeats). (D) Pictures of representative plants used in C to measure the projected rosette area. For box and whisker plots, the box represents the interquartile range, the horizontal line represents the median, and the error bars represent min to max values. Asterisks and letters indicate significant differences as calculated by a Mann–Whitney test in A, Tukey’s multiple comparison test in B, and by Dunnett’s test in C. ****P < 0.0001. (Scale bar in D represents 1 cm.)
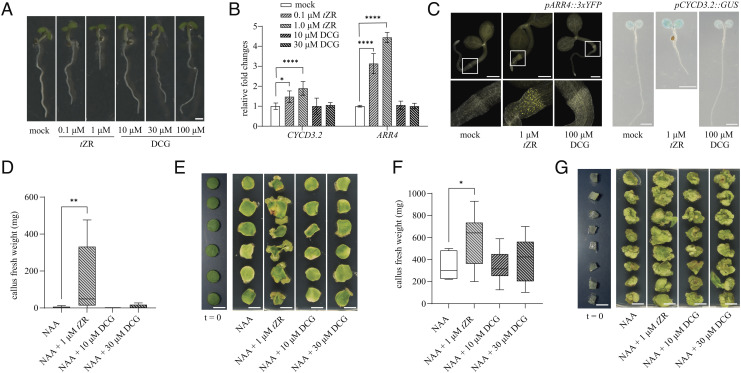
Besides the absence of complementation, it was equally surprising to observe no obvious phenotypic effects when the plants were treated with DCG (Fig. 3A and SI Appendix, Fig. S2), although this compound has been claimed to have cytokinin-substituting and cell-division-promoting activity. In contrast to DCG-treated seedlings, seedlings treated with the cytokinin trans-zeatin riboside (tZR), included as a positive control, showed a reduction in root length, a characteristic cytokinin-related phenotype (16). Also at the molecular level, RT-qPCR analysis showed no transcriptional response of cell division (CYCD3.2) (17) or cytokinin (ARR4) (18) marker genes in DCG-treated WT seedlings (Fig. 3B). To eliminate the possibility that the DCG response was overlooked because of its being minor and/or confined in space and time, cytokinin and cell division reporter lines were used. tZR consistently caused a clear response in both types of reporter lines (Fig. 3C). In contrast, no response was detected in either line upon treatment with a broad range of DCG concentrations (10 to 100 µM; SI Appendix, Fig. S2).
Fig. 3.
Response of plants and plant explants to DCG. (A) Arabidopsis seedlings 7 d after stratification (DAS) growing on 0.5× MS medium containing tZR (0.1 to 10 µm) or DCG (10 to 100 µM). (B) Relative expression of cytokinin (ARR4) and cell division (CYCD3.2) marker genes in Arabidopsis seedlings 7 DAS treated for 20 h in liquid 0.5× MS medium containing different compounds (n = 6 biological repeats). (C) pARR4::3xYFP and pCYCD3.2::GUS reporter line grown for 5 and 9 d, respectively, on 0.5× MS medium supplemented with either tZR or DCG. (D) Fresh weight of callus isolated from tobacco leaf, 21 d after explanting on 1× LS medium supplemented with either tZR or DCG in combination with NAA (10 < n < 14 biological repeats). (E) shows representative leaf explants used to quantify the fresh weight data presented in D. (F) Fresh weight of pith tissue, 21 d after explanting on 1× LS medium supplemented with either tZR or DCG in combination with NAA (9 < n < 15 biological repeats). (G) shows representative explants used to quantify the fresh weight data presented in F. For box plots and whisker plots in D and F, the box represents the interquartile range, the horizontal line represents the median, and the error bars represent min to max values. Error bars in B represent SEs. Asterisks indicate significant differences of the treated samples compared with the mock or NAA treatment as calculated by Dunnett’s test. *0.05 > P > 0.01; **0.01 > P > 0.001; ****P < 0.0001. (Scale bar in A represents 1 mm, in C, Left and Middle represents 0.5 mm, in C, Right represents 0.2 mm, and in E and G represents 1 cm.)
As the assumed activity of DCG was demonstrated previously in tobacco but never in Arabidopsis, the absence of any response in our assays could have pointed to a species-specific effect of DCG. To investigate this possibility, we repeated the original callus induction experiments with DCG on Nicotiana tabacum cv. Havana 425, the tobacco cultivar used in the original studies purportedly demonstrating the activity of DCG (13, 14). In line with the described experiments, both leaf and pith explants were used and the described protocols were closely followed. As expected, incubating freshly harvested leaf and pith explants on LS medium supplemented with both auxin and cytokinin (tZR) resulted in massive cell proliferation, and 3 wk after incubation large calli had formed on the explants. Withdrawing cytokinin from the growth medium resulted in significantly fewer calli, demonstrating the necessity of cytokinin to trigger the proliferation of callus in the presence of auxin. In contrast to what was described previously (13, 14), replacing cytokinin with DCG gave similar results as the medium depleted with cytokinin for leaf (Fig. 3 D and E and SI Appendix, Table S1) and pith (Fig. 3 F and G and SI Appendix, Table S2) explants, i.e., significantly fewer calli.
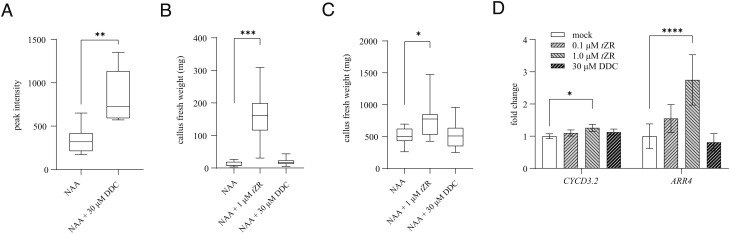
A study focusing on tobacco cell cultures tellingly suggested that DCG is not readily taken up by plant cells (19). Although a lack of uptake contradicts the apparently demonstrated activity of this metabolite on plant explants, it could explain the observed outcome of our experiments. To explore this hypothesis, the DCG levels of plants growing on DCG-containing medium were assessed. Tobacco explants were considered non-ideal for this purpose as endogenous DCG could mask DCG-uptake (20). In addition, the tissue is in direct contact with the DCG-containing medium, making it hard to exclude contamination. As an alternative, shoots of c3′h seedlings were used as these mutants do not produce DCG (Fig. 2A) and shoots are not in direct contact with the growth medium. In line with the experiments performed with tobacco cell cultures (19), no DCG was detected in shoots of seedlings growing on DCG-containing medium. DCG levels could be increased in plant tissues when feeding was performed with the DCG aglycone dehydrodiconiferyl alcohol (DDC) (Fig. 4A and SI Appendix, Fig. S3), a metabolite that is readily taken up and converted to DCG by plant cells (19). Despite the increase in DCG content, no differences were observed between DDC-treated leaf discs as compared with their corresponding controls (Fig. 4B and SI Appendix, Table S3). Similar results, although less clear, were obtained with pith explants (Fig. 4C and SI Appendix, Table S4). Also in Arabidopsis seedlings, no shift in expression of cytokinin and cell-division reporter genes could be demonstrated upon DDC treatment (Fig. 4D). Together, these data question the assumed cytokinin-substituting activity of DCG.
Fig. 4.
Response of plants and plant explants to DDC. (A) Average DCG abundance in tobacco leaf explants 21 d after explanting on 1× LS medium supplemented with either NAA or NAA and DDC (n = 6). (B) Fresh weight of callus isolated from tobacco leaf explants, 21 d after explanting on 1× LS medium supplemented with either tZR or DDC in combination with auxin (32 < n < 40). (C) Fresh weight of pith tissue, 21 d after explanting on 1× LS medium supplemented with either tZR or DCG in combination with NAA (8 < n < 19). (D) Relative expression of cell division (CYCD3.2) and cytokinin (ARR4) marker genes in Arabidopsis seedlings 7 d after stratification treated for 20 h in liquid 0.5× MS medium supplemented with either tZR or DDC (n = 6). The box represents the interquartile range, the horizontal line represents the median, and the error bars represent min to max values. Error bars in D represent SEs. Asterisks indicate significant differences of the treated samples compared with the mock or NAA treatment as calculated by Dunnett’s test. *0.05 > P > 0.01; **0.01 > P > 0.001; ***0.001 > P > 0.0001; ****P < 0.0001.
Discussion
The cytokinin-substituting activity of DCG is based on studies published over 30 y ago. Despite its apparently spectacular activity, DCG never took a foothold in the cytokinin field. The claimed activity was also never scrutinized by independent follow-up studies, and the underlying molecular mechanism remained unresolved, feeding the controversy concerning the bioactivity of this compound (10, 21). Nevertheless, a reduced DCG abundance in phenylpropanoid pathway mutants surged as a possible explanation for the growth defects characteristic of many lignin mutants (1–3, 7, 9, 10). The claimed contribution of DCG to the lignin mutant phenotypes is now challenged as we found no indications that DCG has bioactive, let alone cell-division-promoting or cytokinin-like, properties.
Reasons can be listed explaining the discrepancy between our results and those of other investigators, but none provides a satisfactory answer. First, biological and/or technical details that were unaccounted for in the previously published studies could have affected the results. Although plausible, this seems highly unlikely as the bioactivity of DCG has been demonstrated and retested in various assays (13, 14, 19), undermining the idea that DCG would only be active under a very strict experimental setting. Secondly, we could have used a different, inactive compound. This is equally unlikely as we unambiguously determined by NMR that our DCG was a mixture containing both DCG-A and -B, the two diastereomers with claimed activity (13, 14). A third possibility is that impurities present in our DCG stock interfered with the bioactivity of DCG. Although plausible, the presence of inhibitors is in our opinion irrelevant as we demonstrated that DCG was not taken up by plant cells, which is obviously a more logical explanation for why no activity could be demonstrated. The fact that DCG is not readily taken up (19) further questions the original studies that claimed cytokinin-substituting activity of this metabolite. It could still be argued that an enigmatic contaminant present in our DCG stock, but absent in the original stock, affected the uptake of DCG. However, this is equally unlikely as the lack of uptake was demonstrated previously (19) with the original DCG stock. A fourth possibility is that the claimed DCG activity was the consequence of a contaminant or degradation product in the DCG stock used in the original studies. The former seems improbable, as the authors confirmed their findings using synthetic DCG (14). The presence of a bioactive degradation product in previously used DCG stocks as the cause for the observed cell-division-promoting activity cannot, however, be excluded. Overall, the results in this study force us to revise any notions of the importance of DCG as a growth regulator, and to reconsider its bioactive properties altogether.
Materials and Methods
Chemicals.
All compounds were purchased from Sigma-Aldrich unless mentioned otherwise. All concentrated stock solutions were made in dimethylsulfoxide (commercial grade).
Chemical Synthesis.
Dehydrodiconiferyl alcohol glucoside (DCG).
DCG was prepared by coupling of diacetoxy-dehydrodiconiferyl alcohol and acetobromo-α-d-glucose (2,3,4,6-tetra-O-acetyl-α-d-glucopyranosyl bromide) followed by deacetylation.
Dehydrodiconiferyl alcohol (DDC).
To synthesize DDC, diethyl (8–5)-dehydrodiferulate was first prepared by radical coupling of ethyl ferulate using horseradish peroxidase and hydrogen peroxide in pH 4.0 acetate buffer (22). The dehydrodiferulate ester (1 g, 2.26 mmol) was completely dissolved in anhydrous toluene (200 mL), under nitrogen, and cooled in an ice bath for 30 min. Diisobutylaluminum hydride (1 M in toluene, 13.56 mmol) was slowly added via syringe and stirred for 1 h. The reaction was carefully quenched with ethanol. The reaction mixture was poured into EtOAc and washed with RO water (×1) and satd. NH4Cl (×3). The collected EtOAc layer was dried over anhydrous magnesium sulfate (MgSO4), the inorganics were filtered off, and the solution evaporated. The crude product was purified by flash column chromatography using hexane and EtOAc as eluant to obtain 414 mg (1.15 mmol, 51%) of the pure product as a colorless oil.
Diacetoxy-dehydrodiconiferyl alcohol.
DDC (850 mg, 2.37 mmol) was acetylated in acetic anhydride (15 mL) and pyridine (15 mL) overnight at room temperature. The reaction solution was directly dried on an evaporator to obtain a yellow oil in a quantitative yield of the peracetylated DDC. The triacetoxy-dehydrodiconiferyl alcohol (1.1 g, 2.27 mmol) was dissolved in pyrrolidine (20 mL) with shaking and poured into 250 mL EtOAc. The organic layer was washed with 1 M H2SO4 (×3), dried over anhydrous MgSO4, and the solvent was removed under vacuum. Purification by flash column chromatography provided 810 mg (1.82 mmol, 80%) of the final product as a colorless oil. 1H NMR (360 MHz, acetone-d6): δ 7.63 (br s, 1H, A4-OH), 7.05 (d, J = 1.9 Hz, 1H, A2), 7.033 (br s, 1H, B2), 7.017 (br s, 1H, B6), 6.89 (dd, J = 8.1, 1.9 Hz, 1H, A6), 6.83 (d, J = 8.1 Hz, 1H, A5), 6.63 (dt, J = 15.8, 1.1 Hz, 1H, Bα), 6.23 (dt, J = 15.8, 6.5 Hz, 1H, Bβ), 5.50 (d, J = 7.3 Hz, 1H, Aα), 4.66 (dd, J = 6.5, 1.1 Hz, 2H, Bγ), 4.42 (dd, J = 11.1, 5.5 Hz, 1H, Aγ1), 4.32 (dd, J = 11.1, 7.4 Hz, 1H, Aγ2), 3.86 (s, 3H, B-OMe), 3.82 (s, 3H, A-OMe), 3.77 (m, 1H, Aβ), 2.02 (s, 3H, Bγ-OAc), 1.99 (s, 3H, Aγ-OAc); 13C NMR: δ 170.9 (Aγ-OAc), 170.8 (Bγ-OAc), 149.3 (B4), 148.4 (A3), 147.5 (A4), 145.3 (B3), 134.8 (Bα), 133.3 (A1), 131.3 (B1), 129.2 (B5), 122.1 (Bβ), 119.9 (A6), 116.3 (B6), 115.7 (A5), 112.1 (B2), 110.5 (A2), 89.0 (Aα), 65.9 (Aγ), 65.5 (Bγ), 56.4 (B-OMe), 56.3 (A-OMe), 51.1 (Aβ), 20.8 (Bγ-OAc), 20.7 (Aγ-OAc).
Diacetoxy-dehydrodiconiferyl alcohol tetra-O-acetyl-β-d-glucopyranoside.
The coupling approach between diacetoxy-dehydrodiconiferyl alcohol and tetra-O-acetyl-β-d-glucopyranosyl bromide was based on a previous study (23). Diacetoxy-dehydrodiconiferyl alcohol (920 mg, 2.08 mmol) and 2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl bromide (2.56 g, 6.23 mmol) were dissolved in quinoline (10 mL), and silver oxide (Ag2O, 482 mg, 2.08 mmol) was added. The reaction mixture was stirred for 4 h at room temperature. The reaction mixture was then poured into EtOAc (200 mL) in a separatory funnel and washed with 1 M H2SO4 (×3) and sat. NH4Cl (×3) before being dried over anhydrous MgSO4. The inorganics were filtered off and the solvent evaporated under vacuum resulting in the crude product as an oil. Purification by flash-chromatography produced 1.37 g (1.77 mmol, 85%) of the final product. 1H NMR (360 MHz, acetone-d6): δ 7.19 (d, J = 8.3 Hz, 1H, A5), 7.11 (d, J = 2.0 Hz, 1H, A2), 7.05 (br s, 1H, B2), 7.03 (br s, 1H, B6), 6.96 (dd, J = 8.3, 2.0 Hz, 1H, A6), 6.64 (dt, J = 15.9 Hz, 1.2, 1H, Bα), 6.23 (dt, J = 15.9 Hz, 6.5, 1H, Bβ), 5.57 (d, J = 6.7 Hz, 1H, Aα), 5.35 (dd, J = 9.4, 9.3 Hz, 1H, 3′), 5.24 (dd, J = 7.9, 1.6 Hz, 1H, 1′), 5.18 (dd, J = 9.3, 7.9 Hz, 1H, 2′), 5.10 (dd, J = 9.7, 9.6 Hz, 1H, 4′), 4.66 (dt, J = 6.5, 1.2 Hz, 2H, Bγ), 4.44 (dd, J = 11.1, 5.5 Hz, 1H, Aγ1), 4.33 (dd, J = 11.1, 7.5 Hz, 1H, Aγ2), 4.28 (m, 1H, 6′b), 4.14 (dd, J = 12.3, 2.4 Hz, 1H, 6′a), 4.09 (m, 1H, 5′), 3.88 (s, 3H, B-OMe), 3.81 (s, 3H, A-OMe), 3.76 (m, 1H, Aβ), 2.03 to 1.95 (6s, 18H); 13C NMR: δ 170.9−169.6 (six acetate carbonyl peaks), 151.6 (A3), 149.3 (B4), 147.2 (A4), 145.4 (B3), 138.3 (A1), 134.7 (Bα), 131.6 (B1), 129.0 (B5), 122.3 (Bβ), 120.0 (A5), 119.1 (A6), 116.3 (B6), 112.2 (B2), 111.7 (A2), 100.9 (1′), 88.4 (Aα), 73.3 (3′), 72.6 (5′), 72.0 (2′), 69.4 (4′), 65.9 (Aγ), 65.5 (Bγ), 62.7 (6′), 56.6 (A-OMe), 56.4 (B-OMe), 51.3 (Aβ), 20.79−20.51 (six acetate methyl peaks).
Dehydrodiconiferyl alcohol 4-O-glucopyranoside.
[Glc-G(8–5)G, (E)-2-(hydroxymethyl)-6-(4-(3-(hydroxymethyl)-5-(3-hydroxyprop-1-en-1-yl)-7-methoxy-2,3-dihydrobenzofuran-2-yl)-2-methoxy-phenoxy)tetrahydro-2H-pyran-3,4,5-triol]
Diacetoxy-dehydrodiconiferyl alcohol tetra-O-acetyl-β-d-glucopyranoside (333 mg, 0.43 mmol) was dissolved in 100 mL THF, and the solution was kept in an ice bath for 30 min. The reaction flask was maintained under nitrogen before 1 M DIBAL-H (4.3 mmol) in toluene was slowly added, and stirred for 1 h. The reaction solution was quenched with ethanol, and the solvent directly removed on a rotary evaporator. The obtained yellow solid was treated with hot water (100 mL × 3), and the product was extracted from the solid by vacuum filtration. Following water removal by evaporation, a colorless oil was obtained. Purification by TLC (EtOAc: MeOH, 10:1, v/v) afforded the required dehydrodiconiferyl alcohol 4-O-glucopyranoside (139 mg, 0.26 mmol, 62%). 1H NMR (500 MHz, MeOH-d4): δ 7.14 (d, J = 8.4 Hz, 1H, A5), 7.02 (d, J = 1.9 Hz, 1H, A2), 6.95 (br s, 2H, B2 & 6), 6.92 (dd, J = 8.4, 1.9 Hz, 1H, A6), 6.53 (dt, J = 15.8, 1.1 Hz, 1H, Bα), 6.22 (dt, J = 15.8, 6.0 Hz, 1H, Bβ), 5.58 (d, J = 6.0 Hz, 1H, Aα), 4.89 (1H, 1′), 4.19 (dd, J = 5.8, 1.1 Hz, 2H, Bγ), 3.88 (s, 3H, B-OMe), 3.87 (m, 1H, 6′b), 3.83 (m, 1H, Aγ1), 3.82 (s, 3H, A-OMe), 3.76 (m, 1H, Aγ2), 3.67 (m, 1H, 6′a), 3.46 (m, 3H, Aβ & 2′ & 5′), 3.38 (m, 2H, 3′ & 4′); 13C NMR: δ 150.8 (A3), 149.1 (B4), 147.5 (A4), 145.4 (B3), 138.0 (A1), 132.7 (B5), 131.9 (Bα), 130.0 (B1), 127.7 (Bβ), 119.4 (A6), 118.0 (A5), 116.5 (B6), 112.0 (B2), 111.2 (A2), 102.6 (1′), 88.7 (Aα), 78.0 (3′), 77.7 (5′), 74.8 (2′), 71.2 (4′), 64.9 (Aγ), 63.8 (Bγ), 62.4 (6′), 56.8 (B-OMe), 56.7 (A-OMe), 55.2 (Aβ).
Solution-State NMR Experiments.
NMR experiments for the compounds were performed in acetone-d6 or MeOH-d4. NMR spectra were acquired on a Bruker Biospin (Billerica, MA) Avance 360 MHz instrument with a room-temperature probe or an Avance 500 MHz spectrometer equipped with a 5 mm Triple Resonance Inverse (TCI) 1H/13C/15N cryoprobe with inverse geometry (proton coils closest to the sample). The central acetone or MeOH solvent peaks were used as the internal reference (δC 29.8 and δH 2.04 ppm for acetone; δC 49.0 and δH 3.30 ppm for MeOH). Typical standard Bruker implementations of the traditional suite of one-dimensional (1D; 1H and 13C) and two-dimensional (2D) NMR experiments [multiplicity-edited 13C, Distortionless Enhancement by Polarization Transfer (DEPT-135), homonuclear COrrelation SpectroscopY (COSY), Heteronuclear Single-Quantum Coherence (HSQC), and Heteronuclear Multiple-Bond Correlation (HMBC)] were used to elucidate/validate the structures. A small exponential multiplication line-broadening value (0.1 Hz) and zero-filling to a processed spectrum size (SI) of 256 K datapoints were applied to see the clear splitting of A3, A5, and Aα peaks from the DDC moiety, as well as the glucoside 1′ peak.
Plant Growth Conditions.
Arabidopsis thaliana Col-0 seeds were surface-sterilized overnight with chlorine gas in a closed container. The chlorine gas was generated by mixing 150 mL NaOCl (12 to 14%) with 8 mL 37% HCl. Sterile seeds were sown on plates containing 0.5× Murashige and Skoog (MS) agar medium (1.5 g L−1 MS basal mixture powder (Duchefa), 10 g L−1 sucrose, 0.5 g L−1 MES monohydrate, 8 g L−1 Plant Tissue Culture Agar (Neogen); pH 5.7 with KOH). After sowing, seeds were stratified for 48 h at 4 °C, whereupon plates were transferred to a growth chamber (21 °C; 16 h light/8 h dark photoperiod). N. tabacum cv. Havana 425 seeds were sown in 13-cm pots containing universal potting soil (Saniflor). Plants were grown under long-day greenhouse conditions (21 °C; 16 h light/8 h dark photoperiod).
Projected Rosette Area Measurement.
A segregating c3′h population was grown on horizontal plates (12 × 12 cm) containing 0.5× MS medium as described above. After stratification for 10 d after stratification (DAS), pictures were taken using a Nikon D3200 camera mounted in a vertical setup and the projected rosette area of individual plants was measured using ImageJ software (24). The latter was done by converting the picture to binary values, after which the “magic stick” tool was used to select the pixels that represent the seedling’s rosette. Subsequently, one cotyledon of every plant was harvested for genotyping by PCR. DNA was extracted using the Edwards DNA extraction protocol (25). The primers used for genotyping are listed in SI Appendix, Table S5. Every PCR mixture had a total volume of 25 µL, consisting of 5 µL DNA template, 2.5 µL 5× Green GoTaq® Flexi Reaction Buffer (Promega), 1.25 µL MgCl2, 0.5 µL dNTP Mix (Promega), 1 µL of each primer (10 µM), 0.25 µL GoTaq® G2 Flexi DNA Polymerase (Promega), and 13.5 µL milli-Q water. Following PCR program was used; 5 min of initial denaturation (95 °C), followed by 37 consecutive cycles of 30-s denaturation (95 °C), 30-s annealing (55 °C), 1 min extending (72 °C). PCR products were visualized by agarose gel electrophoresis.
Measuring DCG Content.
After confirmation of the genotype as described above, plants homozygous for the presence and for the absence of the T-DNA insert were harvested for metabolic analysis. For wild-type and mutant samples 3 and 5 seedlings were pooled together, respectively, in 2-mL Eppendorf tubes. In case leaf explants were used, one leaf disc was transferred to a 2-mL Eppendorf tube. The tubes with plant biomass were flash-frozen in liquid nitrogen, and the plant material was subsequently ground with two 3-mm iron beads using a Retsch MM200 mixer mill (1 min, 30 Hz). For metabolite extraction 1 mL 100% methanol was added to the ground and frozen tissue, whereupon the samples were incubated for 20 min at room temperature in an Eppendorf Thermomixer (470 rpm). Next, samples were centrifuged for 3 min in a tabletop centrifuge (Eppendorf) at 14,000 rpm, whereupon 800 µL of the supernatant was transferred to a fresh 2-mL Eppendorf tube. Samples were subsequently dried in a SpeedVac 7810010 (Labconco), and the dried pellet was resuspended in 100 µL cyclohexane. MiliQ water (100 µL) was added and thoroughly mixed. Samples were centrifugated (15 min, 14,000 rpm), whereupon the liquid (lower) phase was used for metabolite analysis. Samples were subjected to Ultra Performance Liquid Chromatography–High Resolution Mass Spectrometry (UPLC–HRMS) at the VIB Metabolomics Core Ghent (VIB-MCG). Then, 10 µL was injected on a Waters Acquity Ultra-High-Performance Liquid Chromatography (UHPLC) device connected to a Vion Quadrupole Time-Of-Flight (Q-TOF) mass spectrometer (Waters). Chromatographic separation was carried out on an ACQUITY UPLC BEH C18 (150 × 2.1 mm, 1.7 μm) column (Waters), column temperature was maintained at 40 °C. A gradient of two buffers was used for separation: buffer A (100:0.1 water:formic acid, pH 3) and buffer B (100:0.1 acetonitrile:formic acid, pH 3), as follows: 99% A for 0 min decreased to 50% A in 30 min, decreased to 30% from 30 to 35 min, and decreased to 0% from 35 to 37 min. The flow rate was set to 0.35 mL min−1. Electrospray Ionization was applied, LockSpray ion source was operated in negative ionization mode under the following specific conditions: capillary voltage, 2.5 kV; reference capillary voltage, 3 kV; source temperature, 120 °C; desolvation gas temperature, 550 °C; desolvation gas flow, 800 L h−1; and cone gas flow, 50 L h−1. The collision energy for full MS scan was set at 6 eV for low-energy settings, for high-energy settings (MSe) it was ramped from 20 to 70 eV. For Data-Dependent Acquisition (DDA)–MS/MS energy settings were ramped from 10 to 35 eV for low mass, and from 35 to 70 eV for high mass. Mass range was set from 50 to 1,500 Da, scan time was set at 0.1s. Nitrogen (greater than 99.5%) was employed as desolvation and cone gas. Leucine-enkephalin (100 pg μL−1 solubilized in water:acetonitrile 1:1 [v/v], with 0.1% formic acid) was used for the lock mass calibration, with scanning every 2 min at a scan time of 0.1 s. Profile data were recorded through Unifi Workstation v2.0 (Waters). Data processing was done using Progenesis QI v2.4 (Waters). The area under a peak (peak area count) was used as a measure of the concentration of the compound it represents.
Identifying and Measuring DDC.
The method for DDC detection was as described for DCG detection.
RT-qPCR.
A. thaliana Col-0 plants were grown on 0.5× MS medium as described above. Subsequently, 7 DAS, seedlings were transferred to 12-well plates (VWR) containing liquid 0.5× MS medium supplemented with a compound of interest. DMSO was used for mock-treated controls. The liquid 0.5× MS medium was identical to the solid medium from which agar was omitted. Four hours after transfer, four plants of a particular treatment were pooled in a 2-mL Eppendorf tube flash-frozen in liquid nitrogen and subsequently ground with two 3-mm iron beads using a Retsch MM200 mixer mill (1 min, 30 Hz). For each treatment, eight biological replicates were used. RNA was extracted using Reliaprep™ RNA tissue miniprep kit (Promega) according to the manufacturer’s instructions, which included a DNase treatment. Extracted RNA was eluted from the spin column using 30 µL RNAse free water, whereupon the eluted total RNA was quantified using an ND-1000 Nanodrop spectrophotometer (Thermo Fisher Scientific). Then, 1 µg total RNA was converted to cDNA using the qScript® cDNA SuperMix (Quantabio) according to the manufacturer’s instructions; 4 µL qScript cDNA SuperMix (5×) was combined with a volume that contains 1 µg RNA, topped off with water so the solution’s total volume was 20 µL. Relative expression of the selected genes was determined with the Roche LightCycler 480 combined with the SYBR Green I master Kit (Roche Diagnostics) using the following PCR protocol: one activation cycle of 10 min (95 °C); 45 amplification cycles of 10 s (95 °C), 10 s (60 °C) and 10 s (72 °C). Each biological repeat sample was run in triplicate to identify potential technical variation that could have occurred during pipetting. Every sample had a total volume of 20 µL, consisting of 10 µL 2× SYBR Green mix, 8 µL primer mix (1 µM), and 2 µL cDNA. Fluorescence values were exported from the Lightcycler 480 program, after which Ct values, normalization factors, and primer efficiencies were calculated according to (26) using two reference genes: POLYUBIQUITIN10 (UBQ10; AT4G05320) and GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C-2 (GAPC2; AT1G13440). Primers used for RT-qPCR can be found in SI Appendix, Table S5.
Reporter Line Assays.
All reporter lines were grown on solid 0.5× MS medium containing the compound of interest as described above. After 5, 7, and 9 DAS, pARR4::3xYFP seedlings were fixated and cleared using a ClearSee solution as described in ref. 27. Cell walls of these samples were subsequently counterstained with 0.1% Calcofluor white M2R (Fluorescent brightener 28) (Polysciences, CAT#4359) solution for 1 h. After washing twice with ClearSee solution, Calcofluor White and YFP fluorescence signal was analyzed using 405 nm and 509 nm excitation and detection at 425 to 475 nm and 527 nm, respectively. pCYCD3.2::GUS seedlings were sampled at similar timepoints and cleared overnight in 80% ice-cold acetone. After washing the samples twice, seedlings were incubated for 20 min at 37 °C in X-gluc buffer (98% P-buffer, 1% 0.12 M X-glux, 2 mM K3[Fe(CN)6], 2 mM K4[Fe(CN)6]·3H2O). After overnight destaining in 70% ethanol, GUS staining was inspected using the Keyence VHX-7000 digital microscope.
Explant Assays.
The callus assays were performed according to ref. 13 following the technical details as described in the original paper as close as possible. Briefly, 12 to 15 cm long leaves were cut from 35 to 45 cm tall soil-grown N. tabacum cv. Havana 425 plants. Youngest leaves and oldest leaves were not sampled to ensure uniform sample collection. After washing the leaves with soap-containing milliQ (MQ) water, they were surface sterilized by three cycles of consecutive 7% bleach (NaOCl), water, and 70% ethanol (1 min each). The sterilization was followed by three consecutive rinses with sterile MQ water. To create leaf explants, 1-cm diameter circular explants were taken from the sterilized leaves, avoiding major veins. Fresh explants were immediately transferred to round Petri dishes (diameter 3 cm) containing solid LS medium (4.4 g L−1 LS basal mixture powder (Duchefa), 30 g L−1 sucrose, 10 g L−1 Plant Tissue Culture Agar (Neogen); pH 5.7 with KOH) supplemented with naphthalene acetic acid (NAA) and trans-zeatin riboside (tZR) or the DCG as indicated. The plates were sealed with two layers of parafilm (Bemis) and subsequently transferred to a tissue culture room where they were incubated under a 16-h light/8-h dark regime at 24 °C. After a 3-wk incubation period, explants were photographed using a Nikon D3200 camera whereupon the callus tissue was removed from the leaf discs, and the callus biomass was quantified using a micro balance (Mettler Toledo). For the pith assays, pith tissue was obtained from mature internodes of 35 and 45 cm tall soil-grown N. tabacum cv. Havana 425 plants. Internode tissue was sterilized as described above for leaves. Pith tissue was dissected from sterilized internodes, and subsequently cut into sections of approximately 10 mg. After measuring the weight of the explants using a micro balance (Mettler Toledo), they were transferred to solid LS medium, on which they were incubated as described earlier for the leaf explants. Three weeks after explanting, explants were imaged using a Nikon D3200 camera whereupon their biomass was quantified using a microbalance (Mettler Toledo). The start and end values allowed to calculate the net increases in biomass during the 3-wk incubation period.
Statistical Analysis.
Statistical analysis was performed using GraphPad Prism 9.2 software. In case of two groups of independent data, we first checked the equality of variance using an F-test, and subsequently performed a two-sided Student t test. When given more than two groups of independent data, determined by one factor, we checked the normality and equality of variance. If the data fulfilled both these criteria, an ordinary one-way ANOVA test, followed by a post hoc test (Tukey or Dunnett multiple comparison) was performed. If not, nonparametric tests were used to test the null hypothesis whereupon multiple comparison tests were performed. For normal data determined by more than one factor, a two-way ANOVA test combined with a Dunnett’s multiple comparisons test was performed.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the Research Foundation Flanders under project number G008116N and by the interuniversity Bijzonder Onderzoeksfonds iBOF “Next-Bioref” (BOFIBO2021000902-iBOF). H.K. and J.R. were funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409). We would like to thank the following people for their willingness to providing us seeds; Chiaraluce Moretti (Universita degli Studi di Perugia) for N. tabacum cv. Havana 425 seeds, Daniele Werck-Reichhart (CNRS, Université de Strasbourg) for c3′h Arabidopsis seeds, Bert De Rybel (VIB-UGent Center for Plant Systems Biology) for the pARR4::3xYFP reporter line, and Lieven De Veylder (VIB-UGent Center for Plant Systems Biology) for the CYCD3.2::GUS reporter line. Tijl Vanholme is acknowledged for technical assistance.
Author contributions
K.W., J.R., W.B., and B.V. designed research; K.W., H.K., B.V., and G.G. performed research; H.K. and J.R. contributed new reagents/analytic tools; K.W., H.K., R.V., J.R., and B.V. analyzed data; J.R. contributed writing; W.B. complemented writing; and K.W. and B.V. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Wout Boerjan, Email: Wout.Boerjan@psb.vib-ugent.be.
Bartel Vanholme, Email: Bartel.Vanholme@ugent.be.
Data, Materials, and Software Availability
All data are included in the article and SI Appendix.
Supporting Information
References
- 1.Ha C. M., Rao X., Saxena G., Dixon R. A., Growth–defense trade-offs and yield loss in plants with engineered cell walls. New Phytol. 231, 60–74 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Muro-Villanueva F., Mao X., Chapple C., Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 56, 202–208 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Li X., Chapple C., Understanding lignification: Challenges beyond monolignol biosynthesis. Plant Physiol. 154, 449–452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schilmiller A. L., et al. , Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60, 771–782 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Goujon T., et al. , Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability. Planta 217, 218–228 (2003). [DOI] [PubMed] [Google Scholar]
- 6.El Houari I., et al. , Seedling developmental defects upon blocking CINNAMATE-4-HYDROXYLASE are caused by perturbations in auxin transport. New Phytol. 230, 2275–2291 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Bonawitz N. D., Chapple C., The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Voelker S. L., Lachenbruch B., Meinzer F. C., Strauss S. H., Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents. New Phytol. 189, 1096–1109 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Abdulrazzak N., et al. , A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiology 140, 30–48 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Houari I., Boerjan W., Vanholme B., Behind the scenes: The impact of bioactive phenylpropanoids on the growth phenotypes of Arabidopsis lignin mutants. Front. Plant Sci. 12, 734070 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai N., et al. , Absolute-configuration of dehydrodiconiferyl alcohol. Biosci. Biotechnol. Biochem. 58, 1679–1684 (1994). [Google Scholar]
- 12.Lynn D. G., Chen R. H., Manning K. S., Wood H. N., The structural characterization of endogenous factors from Vinca rosea crown gall tumors that promote cell division of tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 84, 615–619 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binns A. N., Chen R. H., Wood H. N., Lynn D. G., Cell division promoting activity of naturally occurring dehydrodiconiferyl glucosides: Do cell wall components control cell division? Proc. Natl. Acad. Sci. U.S.A. 84, 980–984 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teutonico R. A., Dudley M. W., Orr J. D., Lynn D. G., Binns A. N., Activity and accumulation of cell division-promoting phenolics in tobacco tissue cultures. Plant Physiol. 97, 288–297 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoch G., et al. , CYP98A3 from Arabidopsis thaliana is a 3’-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 276, 36566–36574 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., et al. , Adenine phosphoribosyl transferase 1 is a key enzyme catalyzing cytokinin conversion from nucleobases to nucleotides in Arabidopsis. Mol. Plant 6, 1661–1672 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Zhang H., Dong Y. X., Hao Y. J., Zhang X. S., DNA METHYLTRANSFERASE 1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis. New Phytol. 217, 219–232 (2018). [DOI] [PubMed] [Google Scholar]
- 18.D’Agostino I. B., Deruere J., Kieber J. J., Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant physiol. 124, 1706–1717 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamagnone L., et al. , Inhibition of phenolic acid metabolism results in precocious cell death and altered cell morphology in leaves of transgenic tobacco plants. Plant Cell 10, 1801–1816 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr J. D., Lynn D. G., Biosynthesis of dehydrodiconiferyl alcohol glucosides–Implications for the control of tobacco cell-growth. Plant Physiol. 98, 343–352 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis N. G., Davin L. B., “Lignans: Biosynthesis and function” in Comprehensive Natural Products Chemistry, Barton D. H. R., Nakanishi K., Eds. (Elsevier, 1999), vol. 1, pp. 639–712. [Google Scholar]
- 22.Ralph J., Garcia-Conesa M. T., Williamson G., Simple preparation of 8-5-coupled diferulate. J. Agric. Food Chem. 46, 2531–2532 (1998). [Google Scholar]
- 23.Terashima N., Ralph S. A., Landucci L. L., New facile syntheses of monolignol glucosides; p-glucocoumaryl alcohol, coniferin and syringin. Holzforschung 50, 151–155 (1996). [Google Scholar]
- 24.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards K., Johnstone C., Thompson C., A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. M., Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Kurihara D., Mizuta Y., Sato Y., Higashiyama T., ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data are included in the article and SI Appendix.