Abstract
Background and aims
Asparagaceae subfamily Nolinoideae is an economically important plant group, but the deep relationships and evolutionary history of the lineage remain poorly understood. Based on a large data set including 37 newly sequenced samples and publicly available plastomes, this study aims to better resolve the inter-tribal relationships of Nolinoideae, and to rigorously examine the tribe-level monophyly of Convallarieae, Ophiopogoneae and Polygonateae.
Methods
Maximum likelihood (ML) and Bayesian inference (BI) methods were used to infer phylogenetic relationships of Nolinoideae at the genus level and above. The diversification history of Nolinoideae was explored using molecular dating.
Key results
Both ML and BI analyses identically recovered five clades within Nolinoideae, respectively corresponding to Dracaeneae + Rusceae, Polygonateae + Theropogon, Ophiopogoneae, Nolineae, and Convallarieae excluding Theropogon, and most deep nodes were well supported. As Theropogon was embedded in Polygonateae, the plastome phylogeny failed to resolve Convallarieae and Polygonateae as reciprocally monophyletic. Divergence time estimation showed that the origins of most Nolinoideae genera were dated to the Miocene and Pliocene. The youthfulness of Nolinoideae genera is well represented in the three herbaceous tribes (Convallarieae, Ophiopogoneae and Polygonateae) chiefly distributed in temperate areas of the Northern Hemisphere, as the median stem ages of all 14 genera currently belonging to them were estimated at <12.37 Ma.
Conclusions
This study recovered a robust backbone phylogeny, providing new insights for better understanding the evolution and classification of Nolinoideae. Compared with the deep relationships recovered by a previous study based on transcriptomic data, our data suggest that ancient hybridization or incomplete lineage sorting may have occurred in the early diversification of Nolinoideae. Our findings will provide important reference for further study of the evolutionary complexity of Nolinoideae using nuclear genomic data. The recent origin of these herbaceous genera currently belonging to Convallarieae, Ophiopogoneae and Polygonateae provides new evidence to support the hypothesis that the global expansion of temperate habitats caused by the climate cooling over the past 15 million years may have dramatically driven lineage diversification and speciation in the Northern Hemisphere temperate flora.
Keywords: Ancient hybridization, Convallarieae, incomplete lineage sorting, plastome, phylogeny, Polygonateae, Theropogon
INTRODUCTION
Advancements in molecular phylogenetics over the past few decades have driven extensive taxonomic revision in angiosperms, resulting in great changes in the boundaries of many families (Angiosperm Phylogeny Group, 1998, 2003, 2009, 2016). This is well represented in the monocotyledonous family Asparagaceae (Asparagales): inferences from molecular phylogenies (Chase et al., 1995, 2006, 2009; Fay et al., 2000; Rudall et al., 2000) have dramatically expanded Asparagaceae to accommodate numerous families that were previously circumscribed based on morphological characteristics (Chase et al., 2009). As a result, Asparagaceae sensuAngiosperm Phylogeny Group (2009, 2016), consisting of approximately 153 genera and 2900 species distributed across both the Old and New Worlds (Stevens, 2001), has become a morphologically diverse and species-rich family of Asparagales.
Asparagaceae is a plant group with significant economic importance. Numerous members of the family have medicinal properties (e.g. Asparagus, Dracaena and Polygonatum), have ornamental value (e.g. Convallaria, Hosta, Nolina, Ophiopogon and Ruscus), and are used as industrial raw materials due to their high fibre and starch contents (e.g. Agava and Yucca). The most recent classification of Asparagaceae (Chase et al., 2009) divided the family into seven subfamilies (Agavoideae, Aphyllanthoideae, Asparagoideae, Brodiaeoideae, Lomandroideae, Nolinoideae and Scilloideae), and previous phylogenetic studies consistently resolved each as a well-supported monophyletic lineage (Kim et al., 2010; Seberg et al., 2012; Steele et al., 2012; Chen et al., 2013). Among them, the components of Nolinoideae, which combines four families (Convallariaceae, Dracaenaceae, Nolinaceae and Ruscaceae) recognized by Dahlgren et al. (1985), exhibit high levels of heterogeneity in composition (Chase et al., 2009). As a result, the subfamily possesses considerable floral and vegetative diversity, leading to a lack of morphological synapomorphies that enables distinction from other Asparagaceae subfamilies (Chase et al., 2009; Meng et al., 2021).
Asparagaceae subfamily Nolinoideae, encompassing about 23 genera, is further divided into seven tribes, namely Convallarieae, Dracaeneae, Eriospermeae, Nolineae, Ophiopogoneae, Polygonateae and Rusceae (Stevens, 2001). Although Eriospermeae has consistently been resolved as sister to the rest of Nolinoideae with strong support, relationships within the remaining Nolinoideae remain poorly resolved (Rudall et al., 2000; Jang and Pfosser, 2002; Kim et al., 2010; Seberg et al., 2012; Steele et al., 2012; Chen et al., 2013; Meng et al., 2021). Additionally, uncertainty remains regarding whether Convallarieae, Ophiopogoneae and Polygonateae are monophyletic lineages, given that the intergeneric relationships of the three tribes, which are perennially rhizomatous herbs chiefly distributed in temperate areas of Eurasia and North America (Rudall et al., 2000), remain contentious (Rudall et al., 2000; Yamashita and Tamura, 2000; Kim et al., 2010; Seberg et al., 2012; Chen et al., 2013; Wang et al., 2014; Floden and Schilling, 2018; Meng et al., 2021).
Chen et al. (2013) proposed that Nolinoideae may represent a recently diversified clade. Due to insufficient phylogenetic information, reconstructing robust phylogenetic trees using single or a few sequence regions is difficult, particularly for plant lineages that have experienced rapid diversification (Rokas and Carroll, 2005; Whitfield and Lockhart, 2007; Philippe et al., 2011). Under these circumstances, employing alternative sequence data sets with more informative loci to reconstruct a robust phylogeny of Nolinoideae is necessary. Although Meng et al. (2021) used a transcriptomic data set to investigate deep relationships of Nolinoideae and which greatly improved our understanding of this phylogenetically problematic plant group, taxonomic sampling at the genus level within Convallarieae, Ophiopogoneae and Polygonateae was too low to satisfactorily address the issues of tribe-level monophyly or intergeneric relationships of these three herbaceous tribes.
With advancements in high-throughput DNA sequencing technologies, plastid genomes (plastomes), as well as genome-wide nuclear sequence data, have been increasingly used to infer phylogenetic relationships. In contrast to biparentally inherited nuclear genomes, phylogenetic analyses of uniparentally inherited plastomes usually recover only the maternal (or in some cases the paternal) evolutionary history rather than the complete relationships of the lineage. Nevertheless, plastid phylogenomic studies have provided valuable insights into the resolution of historically difficult problems in plant phylogenetics (e.g. Jansen et al., 2007; Moore et al., 2007, 2010; Parks et al., 2009; Huang et al., 2016; Carlsen et al., 2018; Ji et al., 2019a, 2021; Li et al., 2019; Yang et al., 2019). As plastomes have become widely used in phylogenetic studies, previously undetected conflicts between plastid and nuclear phylogenies (cytonuclear discordance) have been found in more plant lineages, providing crucial evidence for inferring complicated evolutionary events, such as incomplete lineage sorting (ILS) and hybridization (e.g. Folk et al., 2017; Morales-Briones et al., 2018; Ji et al., 2019a, 2019b; Stull et al., 2020; Wen et al., 2021; Li et al., 2022). Accordingly, plastomes are no less important for phylogenetic reconstruction than nuclear genome data sets and will continue to play an integral role in plant phylogenetics. Based on phylogenomic analyses of a large plastome data set including representatives from 18 out of the 23 genera currently accepted in Nolinoideae, the primary objectives of the present study are: (1) to better resolve the evolutionary relationships among the tribes Convallarieae, Dracaeneae, Nolineae, Ophiopogoneae, Polygonateae and Rusceae; and (2) to rigorously examine the tribe-level monophyly of Convallarieae, Ophiopogoneae and Polygonateae.
MATERIALS AND METHODS
Taxon sampling, shotgun sequencing, plastome assembly and annotation
A total of 88 plastomes from 80 species were sampled, including representatives of the six tribes (Convallarieae, Dracaeneae, Nolineae, Ophiopogoneae, Polygonateae and Rusceae) of Asparagaceae subfamily Nolinoideae. Among them, 37 plastomes were newly sequenced in this study (voucher information is presented in Supplementary Data Table S1), and the rest were obtained from the NCBI GenBank database (Table S2, last accessed 22 October 2022). Taxon sampling representing 18 out of the 23 genera of Asparagaceae subfamily Nolinoideae completely covering the currently accepted genera in Convallarieae, Ophiopogoneae and Polygonateae allows for critically exploring intergeneric relationships and testing for monophyly of the three herbaceous tribes.
Genomic DNA of newly collected samples was extracted from silica gel-dried leaf tissue using the CTAB method (Doyle and Doyle, 1987). Shotgun libraries with an average insert size of ~400 bp were constructed using a TruSeq DNA PCR-free prep kit (Illumina Inc., San Diego, CA, USA) following the manufacturer’s instruction. Prepared libraries were sequenced on an Illumina Novaseq 6000. For each sample, paired-end sequencing (2 × 150 bp) generated ~4 Gb of raw reads, and Trimmomatic v0.40 (Bolger et al., 2014) was used to remove adaptors and to filter low-quality reads with preset parameters. The GetOrganelle v1.7.5.0 pipeline (Jin et al., 2020) was used to recover plastomes from filtered Illumina sequencing reads with default parameters, using the complete plastome of Dracaena hokouensis (GenBank accession number: MN200197) as the reference. The assembled plastomes were annotated using the Plastid Genome Annotator (Qu et al., 2019) and further validated by performing a BLAST search against the NCBI protein data set with Geneious v10.2.3 (Kearse et al., 2012). The junctions of the large-single copy (LSC), small-single copy (SSC) and inverted-repeat (IR) regions for each plastome were visually examined and manually adjusted by comparison with the reference plastome using Geneious v10.2.3 (Kearse et al., 2012).
Phylogenomic analyses
In addition to representatives of Asparagaceae subfamily Nolinoideae, 35 publicly available plastomes (Supplementary Data Table S2) representing five Asparagales families (Amaryllidaceae, Asphodelaceae, Hypoxidaceae, Iridaceae and Orchidaceae) and the remaining Asparagaceae subfamilies (Agavoideae, Aphyllanthoideae, Asparagoideae, Brodiaeoideae, Lomandroideae and Scilloideae) were incorporated into the data set. Given the close relationship between Asparagales and Liliales (Givnish et al., 2018), six taxa from three Liliales families (Colchicaceae, Liliaceae and Melanthiaceae) were selected as the outgroup. Among the sampled plastomes, 68 commonly shared plastid protein-coding genes (PCGs) were extracted from the complete plastome data set using the software PhyloSuite v1.1.15 (Zhang et al., 2020). The PCGs were aligned and concatenated with MAFFT v7.402 (Katoh and Standley, 2013) using default parameters. The best partitioning schemes for the concatenated data set were determined using PartitionFinder v2.1.1 (Lanfear et al., 2017), using the ModelFinder (Kalyaanamoorthy et al., 2017) option to identify the optimal partitioning scheme and substitution models for among-site rate heterogeneity.
Based on the recommended partitioning schemes and substitution models, phylogenetic analyses were performed using both maximum likelihood (ML) and Bayesian inference (BI) methods. The ML phylogeny was reconstructed with RAxML-HPC BlackBox v8.1.24 (Stamatakis, 2006), estimating the support value for each node with 1000 bootstrap (BS) replicates. The BI phylogeny was inferred using MrBayes v3.2 (Ronquist et al., 2012). BI analysis comprised two simultaneous and independent Markov chain Monte Carlo (MCMC) runs of 10 million generations, sampling one tree every 1000 generations with the first 25 % of trees abandoned as burn-in. After reaching the stationary state when the average standard deviation of the split frequencies was <0.01, the two independent runs were combined to obtain the majority rule consensus trees and to calculate posterior probabilities (PP).
Estimation of divergence times
Based on the concatenated data set of 68 plastid PCGs, divergence times were estimated with BEAST v2.4.7 (Bouckaert et al., 2014). The molecular clock was calibrated with the incorporation of six secondary calibration priors provided by a previous study (Givnish et al., 2018): (1) 116.32 million years ago (Ma) for the crown age of Asparagales; (2) 68.72 Ma for the stem age of Iridaceae; (3) 59.38 Ma for the stem age of Asphodelaceae; (4) 52.09 Ma for the divergence between Amaryllidaceae and Asparagaceae; (5) 49.53 Ma for the crown age of Asparagaceae; and (6) 43.12 Ma for the divergence between Asparagaceae subfamilies Asparagoideae and Nolinoideae. We used the ML tree as a topological constraint in the BEAST analysis, with the uncorrelated log-normal relaxed clock approach with a Yule tree prior, and under the sequence substitution models recommended by PartitionFinder. The MCMC simulations were run for 500 million generations, sampling a tree every 5000 generations with the first 10 % of trees being discarded as burn-in. The convergence of the MCMC stimulations was inspected in TRACER v1.7.1 (Rambaut et al., 2018), and the maximum clade credibility tree with median ages and 95 % highest posterior density (HPD) intervals for all nodes was visualized in FIGTREE v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS
Illumina sequencing, plastome assembly and characteristics
A summary of Illumina sequencing and plastome assembly is presented in Supplementary Data Table S3: the reference-guided plastome assembly recovered the complete plastomes of all samples with sequence coverage ranging from 113.727× to 1256.114× (Table S3). These newly sequenced plastomes were deposited in NCBI GenBank with the accession numbers shown in Table S1. The newly sequenced plastomes varied from 153 883 to 162 227 bp in size, with the GC content ranging from 37.3 to 38.0 % (Table S4). Except for loss of the rps16 gene in the plastome of Ruscus aculeatus, each plastome identically possessed 114 unique genes, including 80 PCGs, 30 tRNA genes and four plastid rRNA genes (Table S5). Additionally, an insertion of ~3.3 kb in the IR regions was found in the plastomes of Convallaria majalis. This mutation was also observed in its congeneric species, C. keiskei, and was proposed to be caused by horizontal gene transfer between mitochondrial and plastid genomes (Raman et al., 2019).
Phylogenetic relationships
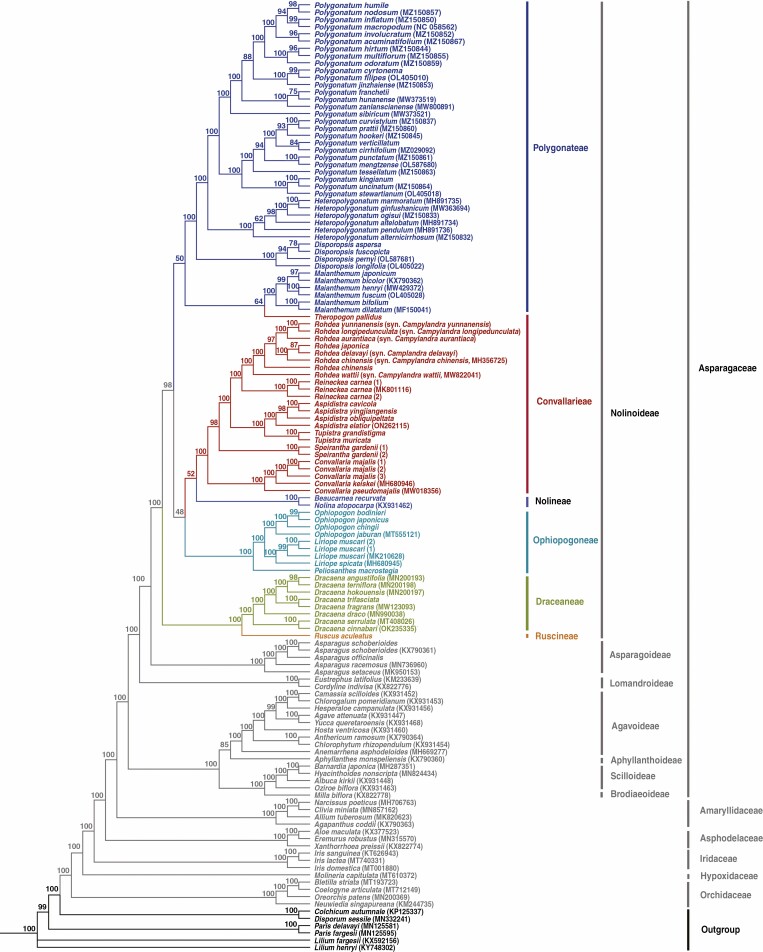
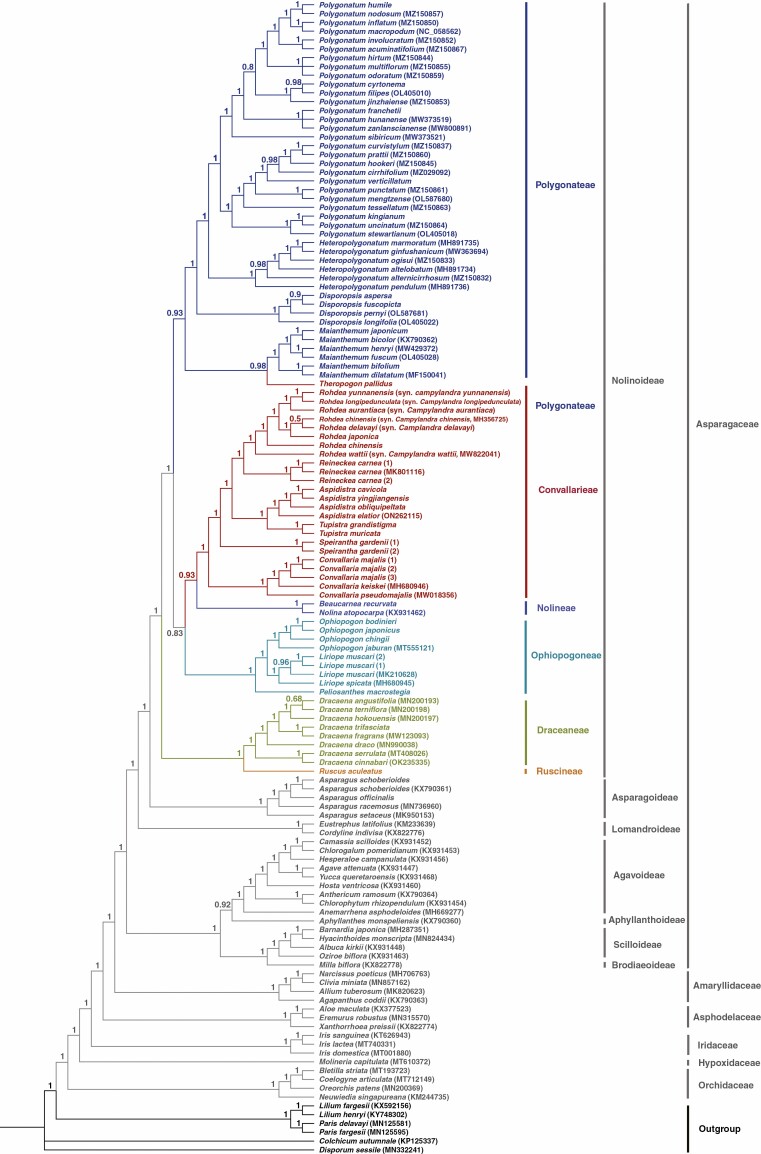
The concatenated matrix of 68 plastid PCGs was 65 859 bp in length, including 21 994 variable sites, of which 16 068 were parsimony-informative (Supplementary Data Table S6). Based on the concatenated matrix, the ML and BI phylogenies were almost identical in tree topologies, despite several nodes recovered with low support values in the ML phylogeny (Fig. 1) were well supported in the BI phylogeny (Fig. 2). All seven subfamilies of Asparagaceae outlined by Chase et al. (2009) were recovered as monophyletic and grouped in two well-supported major clades (BS = 100 %, PP = 1.00), within which the successive divergence of Brodiaeoideae + Scilloideae, Aphyllanthoideae and Agavoideae, as well as of Lomandroideae, Asparagoideae and Nolinoideae, were recovered. Except for the sister relationship between Aphyllanthoideae and Agavoideae (BS = 85 %, PP = 0.92), all nodes at the subfamily level were fully supported (BS = 100 %, PP = 1.00).
Fig. 1.
Phylogeny of Asparagaceae reconstructed by analyses of 68 plastid protein-coding genes (PCGs) using the maximum likelihood (ML) method. Numbers above branches indicate ML bootstrap (BS) percentages.
Fig. 2.
Phylogeny of Asparagaceae reconstructed by analyses of 68 plastid protein-coding genes (PCGs) using the Bayesian inference (BI) method. Numbers above branches indicate BI posterior probability (PP).
Within Asparagaceae subfamily Nolinoideae, our plastid phylogenomic analyses identically recovered five clades corresponding to (1) Dracaeneae + Rusceae (BS = 100 %, PP = 1.00), (2) Polygonateae + Theropogon (BS = 50 %, PP = 0.93), (3) Ophiopogoneae (BS = 100 %, PP = 1.00), (4) Nolineae (BS = 100 %, PP = 1.00) and (5) Convallarieae excluding Theropogon (BS = 100 %, PP = 1.00). Due to Theropogon being embedded in Polygonateae, both Convallarieae and Polygonateae were not resolved as reciprocally monophyletic by either the ML or BI phylogenies. In Convallarieae (excluding Theropogon), a highly resolved and well-supported intergeneric phylogeny was recovered: Convallaria was resolved as the earliest diverging clade, which was successively sister to Speirantha (BS = 98 %, PP = 1.00), Tupistra + Aspidistra (BS = 100 %, PP = 1.00), Reineckea (BS = 100 %, PP = 1.00) and Rohdea (BS = 100 %, PP = 1.00). In Ophiopogoneae, successive divergences of Peliosanthes, Liriope and Ophiopogon were recovered with robust branch support (BS = 100 %, PP = 1.00). The Polygonateae + Theropogon clade was resolved as two subclades. The first comprised Theropogon and Maianthemum (BS = 64 %, PP = 0.98); within the second subclade, Polygonatum was sister to Heteropolygonatum (BS = 100 %, PP = 1.00), and the two genera, in turn, were sister to Disporopsis (BS = 100 %, PP = 1.00).
Molecular dating
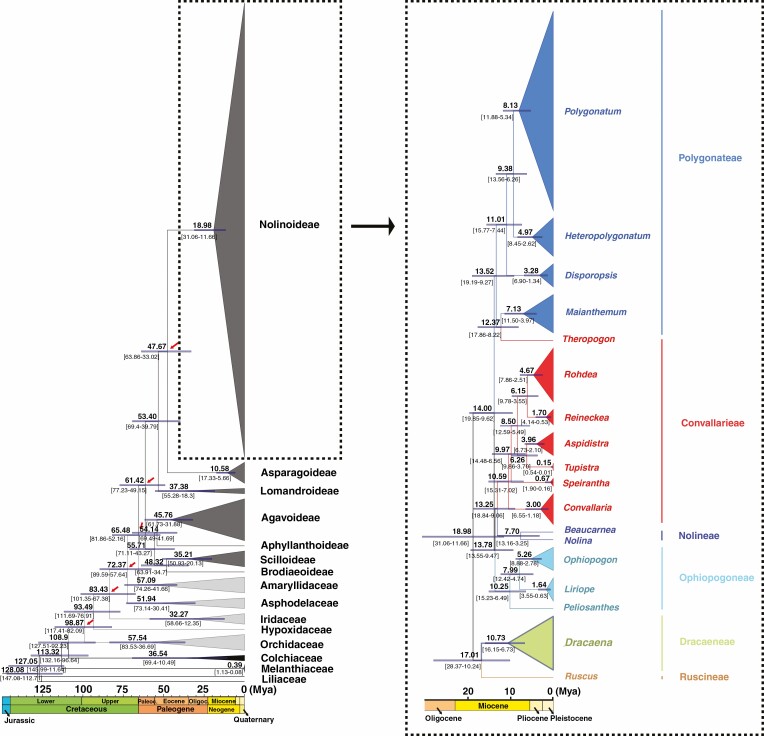
Among the five successively divergent clades recovered in Asparagaceae subfamily Nolinoideae, the early divergence of the Dracaeneae + Rusceae clade occurred ~18.89 Ma, and Polygonateae + Theropogon clade emerged at ~14.00 Ma followed by the divergence of Ophiopogoneae at ~13.78 Ma (Fig. 3). Subsequently, Nolineae diverged from Convallarieae (excluding Theropogon) at ~13.25 Ma (Fig. 3). The crown ages of the five successive diverging clades were dated to ~17.01 Ma (Cracaeneae + Rusceae), ~13.52 Ma (Polygonateae + Theropogon), ~10.25 Ma (Ophiopogoneae), ~10.59 Ma (Convallarieae excluding Theropogon) and ~7.70 Ma (Nolineae), respectively (Fig. 3).
Fig. 3.
Divergence time estimation based on 68 plastid protein-coding genes. Numbers above and below the branches represent mean divergence ages and 95 % confidence interval of each node. Red arrows show the calibration points for molecular dating. Divergence time and the timeline are indicated in million years ago.
DISCUSSION
Relationships among Asparagaceae subfamilies
Due to its great economic importance, the phylogeny of Asparagaceae has been extensively investigated (e.g. Chase et al., 1995, 2006; Fay et al., 2000; Rudall et al., 2000; Kim et al., 2010; Seberg et al., 2012; Steele et al., 2012; Chen et al., 2013), with previous studies identically resolving the seven subfamilies circumscribed by Chase et al. (2009) as well-supported monophyletic lineages (Pires et al., 2006; Seberg et al., 2012; Steele et al., 2012; Chen et al., 2013). The relationships among them, however, remain poorly resolved. This is mainly due to the ambiguous position of Aphyllanthoideae, which has been proposed to be sister to Agavoideae (Seberg et al., 2012; Steele et al., 2012), Brodiaeoideae (Chen et al., 2013) and Lomandroideae (Pires et al., 2006).
Our phylogenomic analyses not only recovered the seven subfamilies as monophyletic but also provided robust support for their relationships. At the subfamily level, the relationships recovered in this study are identical to that inferred from the combination of plastid, mitochondrial and nuclear ribosomal gene data sets (Steele et al. 2012), but with quite strong support for each node. Our results further confirm the sister relationship of Aphyllanthoideae and Agavoideae, and suggest that Agavoideae, Aphyllanthoideae, Brodiaeoideae and Scilloideae may have originated from a common maternal ancestor. This study recovered a robust backbone phylogeny of Asparagaceae at the subfamily level, providing new insights for elucidating the long-standing controversies over the deep phylogenetic relationships of this economically important plant group.
Phylogeny and evolution of Nolinoideae
Nolinoideae is a phylogenetically problematic subfamily within Asparagaceae, given that the tribe-level relationships (except for the early divergence of Eriospermeae) and the monophyly of Convallarieae, Ophiopogoneae and Polygonateae remain unresolved. In this study, except for Comospermum, Danae, Dasylirion, Eriospermum and Semele, representatives of 18 out of the 23 genera currently accepted in Nolinoideae were included in the phylogenetic analyses. Based on the comprehensive taxonomic sampling and the concatenated 68 plastid PCG data set that contains more variable sites and parsimony-informative variations than was available in previous studies, this study provides new insights for better understanding the relationships of phylogenetically problematic lineages at the genus level and above.
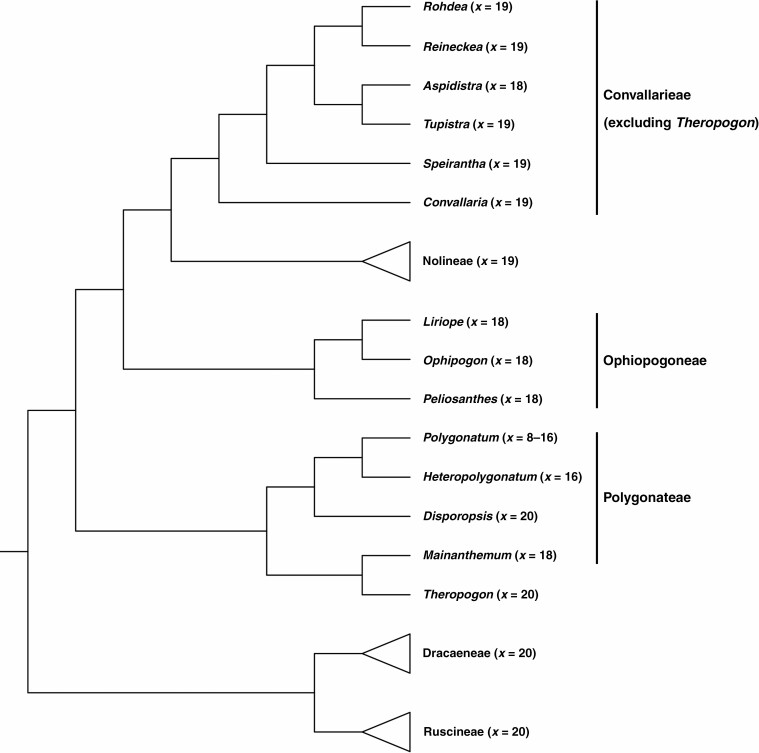
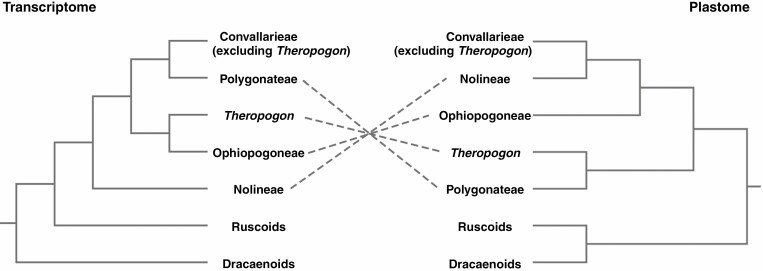
The close relationships between Ruscineae and Convallarieae (Rudall et al., 2000), as well as between Dracaeneae and Nolineae (Kim et al., 2010), were proposed by previous studies. However, our plastid phylogenomic analyses showed Ruscineae is more closely related to Dracaeneae than to Convallarieae (excluding Theropogon), and Nolineae is closely allied to Convallarieae (excluding Theropogon) rather than to Dracaeneae. Notably, the close relationships between Dracaeneae and Ruscineae (Chen et al., 2013; Meng et al., 2021), as well as between Convallarieae (excluding Theropogon) and Nolineae (Seberg et al., 2012), were also proposed by previous studies based on different sequence data sets. These affinities can be justified by cytological evidence (Fig. 4), given that Dracaeneae and Ruscineae have a basic chromosome number x = 20, in contrast to the basic chromosome number x = 19 of Convallarieae (excluding Theropogon) and Nolineae. Nevertheless, based on transcriptome data, high levels of gene tree conflict regarding the relationships of Nolineae, Ophiopogoneae, Polygonateae and Theropogon with other Nolinoideae were detected, and such discordance was hypothesized to have been caused by ancient hybridization or ILS (Meng et al., 2021). Since the close relationships between Convallarieae (excluding Theropogon) and Polygonateae, Ophiopogoneae and Theropogon, as well as Nolineae and the clade consisting of the three herbaceous tribes were strongly supported by phylogenomic analyses of transcriptome data (Meng et al., 2021), the relationships are quite different from those recovered by our data (Fig. 5). In addition to the nuclear gene tree conflict identified by Meng et al. (2021), there is also large discordance regarding the phylogenetic positions of Nolineae, Ophiopogoneae, Polygonateae and Theropogon between the transcriptome (Meng et al., 2021) and plastome (this study) tree topologies, providing good support to the hypothesis that the early evolution of these taxa may have undergone hybridization or ILS (Meng et al., 2021).
Fig. 4.
Basic chromosome number at the genus level mapped along the plastome phylogeny of Nolinoideae.
Fig. 5.
Comparison of deep relationships of Nolinoideae recovered from analyses of transcriptomic data (Meng et al., 2021) and plastomes (this study).
Regarding the three herbaceous tribes (Convallarieae, Ophiopogoneae and Polygonateae) within Nolinoideae, the non-monophyly of Convallarieae has been generally revealed in previous studies (Kim et al., 2010; Seberg et al., 2012; Chen et al., 2013; Meng et al., 2021). By contrast, only a few studies resolved Ophiopogoneae (Jang and Pfosser, 2002; Seberg et al., 2012) and Polygonateae (Seberg et al., 2012; Floden and Schilling, 2018; Wang et al., 2022) as non-monophyletic, and most studies support the monophyly of Ophiopogoneae (Rudall et al., 2000; Yamashita and Tamura, 2000; Kim et al., 2010; Wang et al., 2014; Floden and Schilling, 2018) and Polygonateae (Rudall et al., 2000; Yamashita and Tamura, 2000; Meng et al., 2008, 2014; Wang et al., 2022). Notably, previous studies based on genome-scale sequence data (e.g. Floden and Schilling, 2018; Meng et al., 2021; Wang et al., 2022) had limited generic sampling from the three herbaceous tribes, which may have resulted in phylogenetic errors or uncertainty in the tree topology (Rokas and Carroll, 2005; Philippe et al., 2011), and consequently led to ambiguity on the monophyletic nature of the three herbaceous tribes.
With a complete generic sampling of the three herbaceous tribes, our results showed that Ophiopogoneae is a well-supported monophyletic lineage. Consistent with morphological characteristics, the three genera (Liriope, Ophiopogon and Peliosanthes) traditionally assigned to Ophiopogoneae share the unusual morphologies that their capsules dehisce early to expose the immature seeds during development (Jessop, 1976) and their basic chromosome number is x = 19 (Rudall et al., 2000). Accordingly, recognizing Ophiopogoneae as a distinctive tribe is reasonable (Conran, 1989; Wang et al., 2014) rather than to place Peliosanthes in a separate tribe, Peliosantheae (Nakai, 1936; Dai and Liang, 1991; Liang and Dai, 1992). Although Mcharo et al. (2003) and Yamashita and Tamura (2004) proposed that Liriope is closely related to Peliosanthes but disparate from Ophiopogon, the present study proposes that Liriope has a sister relationship with Ophiopogon, and these two genera, in turn, are sister to Peliosanthes. The well-supported intergeneric relationships of Ophiopogoneae revealed by our data can be further restrengthened by previous studies (Rudall et al., 2000; Floden and Schilling, 2018), which generated identical results, as well as by morphological and palynological evidence (Chang and Hsu, 1974; Dai and Liang, 1991; Cutler, 1992; Liang and Dai, 1992; Rudall et al., 2000).
On the other hand, our plastid phylogenomic analyses recovered neither Convallarieae nor Polygonateae as monophyletic. As the tree topology indicated, Theropogon is phylogenetically disparate from the rest of Convallarieae but closely related to Maianthemum, and the two genera are sister to the clade including the remaining genera (Disporopsis, Heteropolygonatum and Polygonatum) of Polygonateae. Additionally, without the inclusion of Theropogon, the rest of Convallarieae formed a well-supported clade. The relationships are consistent with some morphological features. Specifically, Theropogon possesses ovarian nectaries, which resembles Polygonateae but differs from the absence of a flora nectary in the rest of Convallarieae (Vaikos et al., 1989). This supports the exclusion of Theropogon from Convallarieae and as a lineage closer to Polygonateae. Additionally, the stamens are free in Maianthemum and Theropogon but are adnate to tepals in the remaining genera of Convallarieae and Polygonateae (Rudall et al., 2000), which supports the close relationship between Maianthemum and Theropogon. Taken together, the reciprocally reinforcing evidence suggests that taxonomic work based on multidisciplinary data is needed to establish the monophyly of Convallarieae and Polygonateae.
With the exclusion of Theropogon, this study recovered a well-supported intergeneric phylogeny for the rest of Convallarieae, and resolved Convallaria and Speirantha as two early diverging lineages of the clade. The placement of these two genera can be justified based on some morphological features: they both possess underground rhizomes and long and slender creeping stems, unlike Aspidistra, Rohdea, Tupistra, and Reineckia, which have prostrate or ascending rhizomes above the ground and extremely shortened (or nearly absent) stems, respectively, providing support for the close relationship between Convallaria and Speirantha; as Convallaria is distinctive in having nodding flowers in contrast to the erect flowers of Aspidistra, Rohdea, Reineckia, Speirantha and Tupistra, this supports the transitional position of Speirantha between Convallaria and the subclade consisting of Rohdea, Reineckia, Speirantha and Tupistra. Additionally, this study also provides insightful evidence to resolve the disagreements over the generic circumscription of Campylandra (currently synonymized to Rohdea), Rohdea and Tupistra. Briefly, Campylandra and Tupistra were recognized as two distinct genera (Baker, 1875; Engler, 1888; Hutchinson, 1934; Liang and Tamura, 2000; Tamura et al., 2000), although some authors proposed that they are congeneric (e.g. Bentham, 1883; Hooker, 1892; Liang, 1978). Based on comprehensive morphological analyses, the morphological differences between Campylandra and Rohdea are unlikely to be robust enough for the recognition of the two as separate genera (Tanaka, 2003); accordingly, Yamashita and Tamura (2004) merged Campylandra with Rohdea. Our results show that Rohdea and Campylandra are not reciprocally monophyletic, while Tupistra is more closely related to Aspidistra than to Campylandra. This implies that Campylandra is not congeneric with Tupistra, and validates the taxonomic proposal that reduced Campylandra to a synonym of Rohdea (Yamashita and Tamura, 2004).
Previous studies have shown that the origins of some Asparagaceae genera, such as Agave sensu lato and allied genera (Good-Avila et al., 2006; Flores-Abreu et al., 2019; Jiménez-Barron et al., 2020), the Milla complex (Gándara et al., 2014), and Yucca (Smith et al., 2008), can be traced back to the Miocene or Pliocene. Similarly, our results suggest that the most extensive lineage divergence at the genus level, which resulted in the formation of genera in Asparagaceae subfamily Nolinoideae, took place in the Miocene. The youthfulness of genera is more evident in the three herbaceous tribes (Convallarieae, Ophiopogoneae and Polygonateae) chiefly distributed in the temperate areas of the Northern Hemisphere, as the median stem ages of all the 14 genera currently belonging to them were estimated to <12.37 Ma. The recent radiative divergence of these herbaceous genera is congruent with the speculation that the global expansion of temperate habitats caused by climate cooling over the past 15 million years has contributed greatly to lineage diversification and speciation in the Northern Hemisphere temperate flora (Folk et al., 2019; Sun et al., 2020).
CONCLUSIONS
The robust plastome phylogeny reconstructed in this study provides insightful perspectives for better understanding the deep relationships and classification of Nolinoideae, a phylogenetically problematic lineage. The significant incongruences between our plastome phylogeny and previous results from phylogenetic analyses of transcriptomic data (Meng et al., 2021) suggests that hybridization or ILS may have occurred in the early diversification of Nolinoideae. The findings provide new insights into the phylogeny and evolution of Nolinoideae. Nevertheless, the taxonomic sampling of Nolinoideae at the genus level in this study is incomplete, due to the absence of five genera, particular the enigmatic Comospermum. Additionally, both misspecification of the substitution model for plastid and nuclear data sets and erroneous assembly of polyploid transcriptome data probably result in phylogenetic errors, which may in turn lead to the incongruence between nuclear and plastid phylogenies of Nolinoideae. To critically explore the evolutionary complexity of Nolinoideae, a sampling strategy covering all genera currently recognized in Nolinoideae and the application of nuclear genomic data are needed.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1. Collection information of samples and GenBank accession numbers of newly sequenced plastomes in this study. Table S2: Publicly available plastomes obtained from GenBank. Table S3: Summary of Illumina sequence and plastome assembly. Table S4: Features of Nolinoideae plastomes. Table S5: List of genes identified in Nolinoideae plastomes. Table S6: Sequence characteristics of 68 protein-coding genes involved in the phylogenetic analyses.
ACKNOWLEDGEMENTS
The authors are grateful to two anonymous reviewers for their insightful comments that helped to improve the manuscript. Special thanks are given to Fu Gao, Haicheng An, Sirong Yi, Qiuping Tan, Qiliang Gan and Zhangming Wang for their help with collection of plant samples.
Contributor Information
Yunheng Ji, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Population, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Jacob B Landis, School of Integrative Plant Science, Section of Plant Biology and the L. H. Bailey Hortorium, Cornell University, Ithaca, NY 14850, USA; BTI Computational Biology Center, Boyce Thompson Institute, Ithaca, NY 14853, USA.
Jin Yang, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Shuying Wang, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Nian Zhou, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yan Luo, Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences & Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan 666303, China.
Haiyang Liu, State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
FUNDING
This study was financially supported by the NSFC-Joint Foundation of Yunnan Province (U1802287), and the National Natural Science Foundation of China (31872673).
LITERATURE CITED
- Angiosperm Phylogeny Group. 1998. An ordinal classification for the families of flowering plants. Annals of the Missouri Botanical Garden 85: 531–553. doi: 10.2307/2992015. [DOI] [Google Scholar]
- Angiosperm Phylogeny Group. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society 141: 399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x. [DOI] [Google Scholar]
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161: 105–121. doi: 10.1111/j.1095-8339.2009.00996.x. [DOI] [Google Scholar]
- Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- Baker JG. 1875. Revision of the genera and species of Asparagaceae. Botanical Journal of the Linnean Society 14: 508–632. doi: 10.1111/j.1095-8339.1875.tb00349.x. [DOI] [Google Scholar]
- Bentham G. 1883. Liliaceae. In: Bentham G, Hooker JD, eds. Genera Plantarum, Vol. 3. London: London L Reeve, 748–836. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic. A flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30: 2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen MM, Fér T, Schmickl R, Leong-Škorničková J, Newman M, Kress WJ. 2018. Resolving the rapid plant radiation of early diverging lineages in the tropical Zingiberales: pushing the limits of genomic data. Molecular Phylogenetics and Evolution 128: 55–68. doi: 10.1016/j.ympev.2018.07.020. [DOI] [PubMed] [Google Scholar]
- Chang HJ, Hsu CC. 1974. A cytotaxonomical study on some Formosan Liliaceae. Taiwania 19: 58–74. [Google Scholar]
- Chase MW, Duvall MR, Hills HG, et al. 1995. Molecular phylogenetics of Lilianae. In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, eds. Monocotyledons: systematics and evolution. London: Royal Botanic Gardens, Kew, 109–137. [Google Scholar]
- Chase MW, Fay MF, Devey DS, et al. 2006. Multigene analyses of monocot relationships: a summary. Aliso 22: 63–75. doi: 10.5642/aliso.20062201.06. [DOI] [Google Scholar]
- Chase MW, Reveal JL, Fay MF. 2009. A subfamilial classification for the expanded Asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Botanical Journal of the Linnean Society 161: 132–136. doi: 10.1111/j.1095-8339.2009.00999.x. [DOI] [Google Scholar]
- Chen SC, Kim DK, Chase MW, Kim JH. 2013. Networks in a largescale phylogenetic analysis: reconstructing evolutionary history of Asparagales (Lilianae) based on four plastid genes. PLoS One 8: e59472. doi: 10.1371/journal.pone.0059472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conran JG. 1989. Cladistic analysis of some net-veined Liliiflorae. Plant Systematics and Evolution 168: 123–141. doi: 10.1007/bf00936093. [DOI] [Google Scholar]
- Cutler DF. 1992. Vegetative anatomy of Ophiopogoneae (Convallariaceae). Botanical Journal of the Linnean Society 110: 385–419. doi: 10.1111/j.1095-8339.1992.tb00301.x. [DOI] [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. 1985. The families of the monocotyledons: structure, evolution and taxonomy. Berlin: Springer. [Google Scholar]
- Dai LK, Liang SY. 1991. Epidermal features of leaves and their taxonomic signification in subfamily Ophiopogonoideae (Liliaceae). Acta Phytotaxonomica Sinica 29: 335–346. [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin 19: 11–15. [Google Scholar]
- Engler A. 1888. Liliaceae. In: Engler A, Prantl K, eds. Die naturlichen Pflanzenfamilien, Vol. 2(5). Leipzig: Wilhelm Engelmann, 10–91. [Google Scholar]
- Fay MF, Rudall PJ, Sullivan S, et al. 2000. Phylogenetic studies of Asparagales based on four plastid DNA regions. In: Wilson KL, Morrison DA, eds. Monocots: systematics and evolution. Proceedings of the 2nd International Monocot Symposium. Melbourne: CSIRO Publishing, 360–371. [Google Scholar]
- Floden A, Schilling EE. 2018. Using phylogenomics to reconstruct phylogenetic relationships within tribe Polygonateae (Asparagaceae), with a special focus on Polygonatum. Molecular Phylogenetics and Evolution 129: 202–213. doi: 10.1016/j.ympev.2018.08.017. [DOI] [PubMed] [Google Scholar]
- Flores-Abreu IN, Trejo-Salazar RE, Sánchez-Reyes LL, et al. 2019. Tempo and mode in coevolution of Agave sensu lato (Agavoideae, Asparagaceae) and its bat pollinators, Glossophaginae (Phyllostomidae). Molecular Phylogenetics and Evolution 133: 176–188. doi: 10.1016/j.ympev.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Folk RA, Mandel JR, Freudenstein JV. 2017. Ancestral gene flow and parallel organellar genome capture result in extreme phylogenomic discord in a lineage of angiosperms. Journal of Bioinformatics and Systems Biology 66: 320–337. doi: 10.1093/sysbio/syw083. [DOI] [PubMed] [Google Scholar]
- Folk RA, Stubbs RL, Mort ME, et al. 2019. Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proceedings of the National Academy of Sciences of the United States of America 116: 10874–10882. doi: 10.1073/pnas.1817999116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gándara E, Specht CD, Sosa V. 2014. Origin and diversification of the Milla Clade (Brodiaeoideae, Asparagaceae): a Neotropical group of six geophytic genera. Molecular Phylogenetics and Evolution 75: 118–125. doi: 10.1016/j.ympev.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Zuluaga A, Spalink D, et al. 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. American Journal of Botany 105: 1888–1910. doi: 10.1002/ajb2.1178. [DOI] [PubMed] [Google Scholar]
- Good-Avila S, Souza V, Gaut B, Eguiarte LE. 2006. Timing and rate of speciation in Agave (Agavaceae). Proceedings of the National Academy of Sciences of the United States of America 103: 9124–9129. doi: 10.1073/pnas.0603312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker JD. 1892. Tupistra. In: Hooker JD, ed. Flora of British India, Vol. 6. London: London L Reeve, 324–325. [Google Scholar]
- Huang Y, Li X, Yang Z, et al. 2016. Analysis of complete chloroplast genome sequences improves phylogenetic resolution of Paris (Melanthiaceae). Frontiers in Plant Science 7: 1797. doi: 10.3389/fpls.2016.01797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson J. 1934. The families of flowering plants 2, Monocotledons. London: Macmillan. [Google Scholar]
- Jang CG, Pfosser M. 2002. Phylogenetics of Ruscaceae sensu lato based on plastid rbcL and trnL-F DNA sequences. Stapfia 80: 333–348. [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proceedings of the National Academy of Sciences of the United States of America 104: 19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop J. 1976. A revision of Peliosanthes (Liliaceae). Blumea 23: 141–159. [Google Scholar]
- Ji Y, Liu C, Yang Z, et al. 2019a. Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Molecular Ecology Resources 19: 1333–1345. doi: 10.1111/1755-0998.13050. [DOI] [PubMed] [Google Scholar]
- Ji Y, Yang L, Chase MW, et al. 2019b. Plastome phylogenomics, biogeography, and clade diversification of Paris (Melanthiaceae). BMC Plant Biology 19: 543. doi: 10.1186/s12870-019-2147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Liu C, Landis JB, Deng M, Chen J. 2021. Plastome phylogenomics of Cephalotaxus (Cephalotaxaceae) and allied genera. Annals of Botany 127: 697–708. doi: 10.1093/aob/mcaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Barron O, García-Sandoval R, Magallón S, et al. 2020. Phylogeny, diversification rate, and divergence time of Agave sensu lato (Asparagaceae), a group of recent origin in the process of diversification. Frontiers in Plant Science 11: 536135. doi: 10.3389/fpls.2020.536135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Yu W, Yang J, Song Y, Yi T, Li D. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biology 21: 241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772737–772780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim DK, Forest F, Michael FF, Chase MW. 2010. Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Annals of Botany 106: 775. doi: 10.1093/aob/mcq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HT, Yi TS, Gao LM, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461–470. doi: 10.1038/s41477-019-0421-0. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang Y, Ruhsam M, et al. 2022. Seeing through the hedge: Phylogenomics of Thuja (Cupressaceae) reveals prominent incomplete lineage sorting and ancient introgression for Tertiary relict flora. Cladistics 38: 187–203. doi: 10.1111/cla.12491. [DOI] [PubMed] [Google Scholar]
- Liang SY. 1978. Tupistra. In: Wang FT, Tang J, eds. Flora Reipublicae Popularis Sinicae, Vol. 15. Beijing: Science Press, 6–15. [Google Scholar]
- Liang SY, Dai LK. 1992. Pollen morphology and generic phylogenetic relationships in Ophiopogonoideae (Liliaceae). Acta Phytotaxonomica Sinica 30: 427–437. [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Liang SY, Tamura MN. 2000. Campplandra, Rohdea, Tupistra. In: Wu ZY, Raven PH, eds. Flora of China, Vol. 24. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press, 235–240. [Google Scholar]
- Mcharo M, Bush E, Bonte DL, Broussard C, Urbatsch L. 2003. Molecular and morphological investigation of ornamental liriopogons. Journal of the American Society for Horticultural Science 128: 575–577. [Google Scholar]
- Meng R, Luo LY, Zhang JY, Zhang DG, Nie ZL, Meng Y. 2021. The deep evolutionary relationships of the morphologically heterogeneous nolinoideae (Asparagaceae) revealed by transcriptome data. Frontiers in Plant Science 11: 584981. doi: 10.3389/fpls.2020.584981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Nie ZL, Deng T, Wen J, Yang YP. 2014. Phylogenetics and evolution of phyllotaxy in the Solomon’s seal genus Polygonatum (Asparagaceae: Polygonateae). Botanical Journal of the Linnean Society 176: 435–451. doi: 10.1111/boj.12218. [DOI] [Google Scholar]
- Meng Y, Wen J, Nie ZL, Sun H, Yang YP. 2008. Phylogeny and biogeographic diversification of Maianthemum (Ruscaceae: Polygonatae). Molecular Phylogenetics and Evolution 49: 424–434. doi: 10.1016/j.ympev.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences of the United States of America 104: 19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences of the United States of America 107: 4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Briones DF, Liston A, Tank DC. 2018. Phylogenomic analyses reveal a deep history of hybridization and polyploidy in the Neotropical genus Lachemilla (Rosaceae). The New Phytologist 218: 1668–1684. doi: 10.1111/nph.15099. [DOI] [PubMed] [Google Scholar]
- Nakai T. 1936. Subdivision of Convallariaceae Link. Journal of Japanese Botany 12: 145–150. [Google Scholar]
- Parks M, Cronn R, Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7: 84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, et al. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology 9: e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires JC, Maureira IJ, Givnish TJ, Sytsma KJ. 2006. Phylogeny, genome size, and chromosome evolution of Asparagales. Aliso. 22: 287–304. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15: 1–12. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman G, Park S, Lee EM, Park S. 2019. Evidence of mitochondrial DNA in the chloroplast genome of Convallaria keiskei and its subsequent evolution in the Asparagales. Scientific Reports 9: 5028. doi: 10.1038/s41598-019-41377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Alexei J, Drummond A, et al. 2018. Tracer, version 1.7.1. http://tree.bio.ed.ac.uk/software/tracer/. [Google Scholar]
- Rokas A, Carroll SB. 2005. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Molecular Biology and Evolution 22: 1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark PVD, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ, Conran JG, Chase MW. 2000. Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Botanical Journal of the Linnean Society 134: 73–92. doi: 10.1006/bojl.2000.0365. [DOI] [Google Scholar]
- Seberg O, Petersen G, Davis JI, et al. 2012. Phylogeny of the Asparagales based on three plastid and two mitochondrial genes. American Journal of Botany 9: 875–889. doi: 10.3732/ajb.1100468. [DOI] [PubMed] [Google Scholar]
- Smith CI, Pellmyr O, Althoff D, Balcázar-Lara M, Leebens-Mack J, Segraves K. 2008. Pattern and timing of diversification in Yucca (Agavaceae): specialized pollination does not escalate rates of diversification. Proceedings of the Royal Society of London. Series B, Biological Sciences 275: 249–258. doi: 10.1098/rspb.2007.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood–based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Steele PR, Hertweck KL, Mayfield D, McKain MR, Leebens-Mack J, Pires JC. 2012. Quality and quantity of data recovered from massively parallel sequencing: examples in Asparagales and Poaceae. American Journal of Botany 99: 330–348. doi: 10.3732/ajb.1100491. [DOI] [PubMed] [Google Scholar]
- Stevens PF. 2001. Angiosperm phylogeny website. Version 14. http://www.mobot.org/MOBOT/research/APweb/. July 2017. [Google Scholar]
- Stull GW, Soltis PS, Soltis DE, Gitzendanner MA, Smith SA. 2020. Nuclear phylogenomic analyses of asterids conflict with plastome trees and support novel relationships among major lineages. American Journal of Botany 107: 790–805. doi: 10.1002/ajb2.1468. [DOI] [PubMed] [Google Scholar]
- Sun M, Folk RA, Gitzendanner MA, et al. 2020. Recent accelerated diversification in rosids occurred outside the tropics. Nature Communications 11: 3333. doi: 10.1038/s41467-020-17116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura MN, Liang SY, Turland NJ. 2000. New combinations in Campylandra (Convallariaceae, Convallarieae). Novon 10: 158–160. doi: 10.2307/3393019. [DOI] [Google Scholar]
- Vaikos NP, Markandeya SK, Pai RM. 1989. Floral anatomy of the Liliaceae: Tribe Convallarieae. Proceedings of Indian Academy of Sciences (Plant Sciences) 22: 258–265. doi: 10.1007/BF03053520. [DOI] [Google Scholar]
- Whitfield JB, Lockhart PJ. 2007. Deciphering ancient rapid radiations. Trends in Ecology & Evolution 22: 258–265. doi: 10.1016/j.tree.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Yamashita J, Tamura MN. 2000. Molecular phylogeny of the Convallariaceae (Asparagales). In: Wilson KL, Morrison DA, eds. Monocots: systematics and evolution. Collingwood: CSIRO Publishing, 387–400. [Google Scholar]
- Yamashita J, Tamura MN. 2004. Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato), with some taxonomic treatments. Journal of Plant Research 117: 363–370. doi: 10.1007/s10265-004-0169-z. [DOI] [PubMed] [Google Scholar]
- Tanaka N. 2003. New combinations in Rohdea (Convallariaceae). Novon 13: 329–333. doi: 10.2307/3393269. [DOI] [Google Scholar]
- Wang GY, Meng Y, Huang JL, Yang YP. 2014. Molecular phylogeny of Ophiopogon (Asparagaceae) inferred from nuclear and plastid DNA sequences. Systematic Botany 39: 776–784. doi: 10.1600/036364414x682201. [DOI] [Google Scholar]
- Wang J, Qian J, Jiang Y, et al. 2022. Comparative analysis of chloroplast genome and new insights into phylogenetic relationships of Polygonatum and tribe Polygonateae. Frontiers in Plant Science 13: 882189. doi: 10.3389/fpls.2022.882189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Xie DF, Price M, et al. 2021. Backbone phylogeny and evolution of Apioideae (Apiaceae): new insights from phylogenomic analyses of plastome data. Molecular Phylogenetics and Evolution 161: 107183. doi: 10.1016/j.ympev.2021.107183. [DOI] [PubMed] [Google Scholar]
- Yang L, Yang Z, Liu C, et al. 2019. Chloroplast phylogenomic analysis provides insights into the evolution of the largest eukaryotic genome holder, Paris japonica (Melanthiaceae). BMC Plant Biology 19: 293. doi: 10.1186/s12870-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlic I, et al. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20: 348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.