Abstract
Epigenetic modification of DNA via CpG methylation is essential for the proper regulation of gene expression during embryonic development. Methylation of CpG motifs results in gene repression, while CpG island-containing genes are maintained in an unmethylated state and are transcriptionally active. The molecular mechanisms involved in maintaining the hypomethylation of CpG islands remain unclear. The transcriptional activator CpG binding protein (CGBP) exhibits a unique binding specificity for DNA elements that contain unmethylated CpG motifs, which makes it a potential candidate for the regulation of CpG island-containing genes. In order to assess the global function of this protein, mice lacking CGBP were generated via homologous recombination. No viable mutant mice were identified, indicating that CGBP is required for murine development. Mutant embryos were also absent between 6.5 and 12.5 days postcoitum (dpc). Approximately, one-fourth of all implantation sites at 6.5 dpc appeared empty with no intact embryos present. However, histological examination of 6.5-dpc implantation sites revealed the presence of embryo remnants, indicating that CGBP mutant embryos die very early in development. In vitro blastocyst outgrowth assays revealed that CGBP-null blastocysts are viable and capable of hatching and forming both an inner cell mass and a trophectoderm. Therefore, CGBP plays a crucial role in embryo viability and peri-implantation development.
Cytosine methylation of the CpG dinucleotide is a major epigenetic modification of DNA which functions in a wide variety of cellular processes, including transcription repression, neoplasia, genomic imprinting, X-chromosome inactivation, and early mammalian development (48, 5, 43, 22). The presence of this dinucleotide is relatively rare, approximately 5 to 10% of its predicted frequency (48, 3, 4, 8). Approximately half of all genes in the mouse and human genomes contain small regions or islands of DNA that contain the expected frequency of the CpG dinucleotide (3). Promoters containing these CpG islands are relatively hypomethylated, and the associated gene is active (11).
The molecular mechanisms involved in hypomethylation and transcription activation have not been clearly defined. However, extensive data demonstrate the coupling of DNA methylation, chromatin condensation, and gene silencing (9, 16–18, 26, 36–38, 47, 55, 61). Methyl-CpG binding domain proteins 1 to 3 (MBD1 to -3) and methyl cytosine binding protein 2 (MeCP2) have been implicated in transcriptional repression (9, 16, 17, 36–38, 55, 61), while MBD4 is a mismatch repair protein (24). MeCP2 and MBD2 repress transcription through the recruitment of histone deacetylases (26, 37, 38). MBD3 is a member of the NuRD (Mi-2) histone deacetylase and nucleosome remodelling complex (55, 61), while MBD2 is capable of recruiting the NuRD complex to methylated DNA in vitro (61). In addition, mutations in MeCP2 and DNA methyltransferase 3B have been linked to Rett and immunodeficiency, centromere instability, and facial anomaly (ICF) syndromes, respectively (2, 10, 20, 21, 40, 59).
DNA methylation is critical for embryonic mammalian development (30, 40). A dramatic reduction in CpG dinucleotide methylation occurs during early development in pre-implantation embryos (35, 48). At the time of implantation, de novo methylation occurs at most CpG residues except for CpG islands, which remain unmethylated and transcriptionally active (27, 42). A small number of CpG island-associated genes on the inactive X chromosome and several parentally imprinted genes are methylated during development (14, 44, 51). In addition, DNA methylation represses expression of repetitive DNA elements during mouse development (56). Although the mechanisms involved in DNA methylation and gene repression have been extensively studied, little is known regarding the manner in which CpG island-containing genes are maintained in the unmethylated state.
Our laboratory recently identified a novel transcriptional activator, CpG binding protein (CGBP), which binds specifically to DNA elements containing unmethylated CpG motifs (54). CGBP is expressed in a wide variety of adult tissues and cell lines. In addition, analysis of expressed sequence tag databases revealed the presence of CGBP cDNA in embryonic tissues at various stages of development, indicating that CGBP is widely expressed in both adult and embryonic tissues. CGBP contains a cysteine-rich CXXC motif that comprises the DNA-binding domain (J.-H. Lee, K. S. Voo, and D. G. Skalnik, submitted for publication). This domain is highly conserved among several proteins, including DNA methyltransferase 1 (DNMT1) (6), human trithorax (HRX) (also known as MLL or ALL-1) (13, 19, 32, 41, 52, 60), MBD1 (11, 23), and MLL-2 (15). CGBP also contains two plant homeodomains which are characteristic of chromatin-associating proteins and/or regulators of gene expression (1).
A growing number of proteins that bind methylated CpG motifs and repress gene activity, including DNMT1, MBD1 to -3, and MeCP2, have been identified (9, 16–18, 23, 36–38, 55, 61). However, CGBP is the first identified protein that binds specifically to unmethylated CpG dinucleotides and activates gene expression. CGBP binding affinity increases with the number of CpG motifs (Lee et al., submitted), suggesting that CGBP may regulate CpG island-containing genes. Alternatively, binding of CGBP to unmethylated CpG motifs may prevent targeting of DNMT to CpG islands. Consequently, delineation of the functional role of CGBP may enable us to further understand the molecular mechanisms involved in the epigenetic control of gene expression during development.
This study describes the generation via homologous recombination of mice lacking CGBP. Results presented in this study demonstrate that CGBP is crucial for early embryogenesis. No viable CGBP-null animals were obtained, and no CGBP-null embryos were obtained between 6.5 and 12.5 days postcoitum (dpc). Approximately one-fourth of the implantation sites examined at 6.5 dpc appeared empty, with no intact embryos. Furthermore, histological examination of 6.5-dpc implantation sites revealed remnants of embryos in approximately one-fourth of the sites. Blastocyst outgrowth experiments revealed that CGBP-null blastocysts are viable and capable of hatching and forming both an inner cell mass and a trophectoderm. Therefore, CGBP is crucial for peri-implantation development in the mouse.
MATERIALS AND METHODS
Disruption of the CGBP locus in embryonic stem cells.
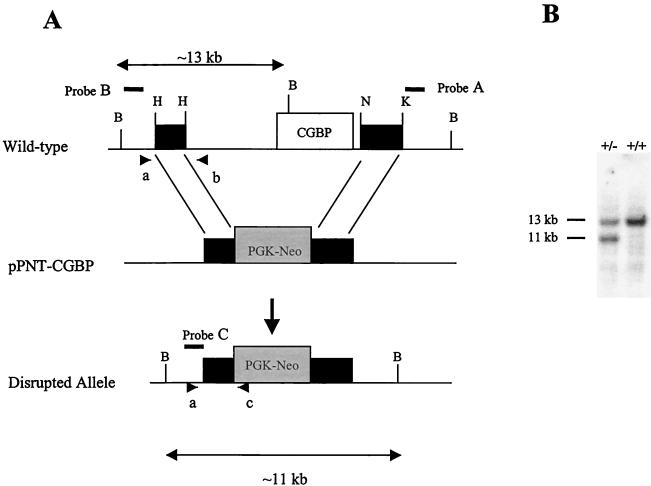
To generate a mutated CGBP allele, a 129SVJ mouse lambda genomic library (Stratagene, La Jolla, Calif.) was screened using a human expressed sequence tag cDNA (Genome Systems Inc., St. Louis, Mo.). A clone that contains the entire CGBP gene locus was isolated (data not shown, D. L. Carlone, S. R. L. Hart, P. D. Ladd, and D. G. Skalnik, unpublished data). The mutated allele was constructed by inserting a 2.3-kb HindIII fragment upstream and a 3.0-kb NcoI-KpnI fragment downstream of the neomycin gene in the pPNT vector (53) (Fig. 1). Homologous recombination results in a deletion of approximately 11 kb including the entire mouse CGBP gene as well as 5 kb of upstream sequence.
FIG. 1.
Targeted disruption of the murine CGBP gene. (A) Schematic of the disrupted CGBP allele generated by homologous recombination. The open box indicates the CGBP gene, while the black boxes denote the flanking genomic fragments. The gray box denotes PGK-neomycin (Neo). The arrows indicate the sizes of the wild-type (13-kb) and the disrupted (11-kb) BamHI fragment. Probes A to C are designated by solid lines. Oligonucleotide primers a to c used for PCR genotyping are designated by arrowheads. (B) Southern blot analysis was performed on genomic DNAs isolated from wild-type ES cells (+/+) and the ES cell clone (+/−) that contains the disrupted CGBP allele. DNA was digested with BamHI and hybridized with probe B (see above) corresponding to the 5′ flank of the CGBP locus.
Murine embryonic stem cells (CCE916) were transfected with the mutated construct by electroporation as previously described (45, 46). Approximately 500 clones were analyzed for the homologous recombination event by Southern blot analysis. Briefly, 10 μg of genomic DNA was digested with NcoI and separated on a 0.55% agarose gel in 0.5× Tris-borate-EDTA. Following transfer to a nylon membrane, blots were probed with a 500-bp KpnI-EcoRI fragment (Fig. 1A) (probe A) corresponding to the 3′ region of the locus outside the region of homologous recombination. A single positive clone was identified, and an additional Southern blot analysis was performed to confirm that the disrupted allele was in the correct chromosomal location. Genomic DNA isolated from wild-type (CCE916) or heterozygous ES cells was digested with BamHI, subjected to electrophoresis, and transferred to a nylon membrane. The blot was then probed with a 1.2-kb NotI-EcoRI fragment (Fig. 1A) (probe B) corresponding to the 5′ region of the locus (Fig. 1B).
Generation of CGBP mutant mice.
Cells heterozygous for the disrupted CGBP allele were injected into C57BL/6 blastocysts and implanted into pseudopregnant C57BL/6 females (Jackson Laboratories, Bar Harbor, Maine) by standard protocols. Four germ line-transmitting males were then backcrossed to C57BL/6 females to generate heterozygous animals. Heterozygous mating pairs were established, and the offspring were analyzed. Timed-pregnancy females were obtained by mating them with heterozygous animals and examining them for a plug the following day. Noon on the day of the plug was considered 0.5 day of gestation. Embryos were then collected at various time points.
Genotyping of animals and embryos.
Weaned pups and embryos were genotyped by either Southern blot or PCR analysis. Southern blot analysis was performed using 10 to 20 μg of genomic DNA digested with BamHI as described above. A competitive-PCR method was used to simultaneously detect the mutated and wild-type alleles. An oligonucleotide primer common to both alleles (primer a) was used in the same reaction with two primers specific for either the wild-type (primer b) or mutated (primer c) alleles (Fig. 1A). The following oligonucleotides were used: primer a, 5′-GGGCTCCCTTGTTCAAATAC-3′; primer b, 5′-GATCCTGACCATGCTGCTTG-3′; and primer c, 5′-GCTAAAGCGCATGCTCCAGACTG-3′ (31). The PCR products were analyzed on a 1.5% agarose gel in 0.5× Tris-borate-EDTA. For 6.5-dpc embryo genotyping, PCR products were transferred to a nylon membrane following electrophoresis and probed with an EcoRI-HindIII fragment (Fig. 1A) (probe C).
Histological analysis of embryos.
Implantation sites were surgically removed at 6.5 dpc, fixed in 10% formalin, and embedded in paraffin. Six-micrometer-thick sections were stained with hematoxylin and eosin.
Blastocyst outgrowths.
Six- to 8-week-old heterozygous females were superovulated (7.5 to 10 IU of pregnant mare serum gonadotropin [Sigma-Alrich Co., St. Louis, Mo.], followed 48 h later by 7.5 to 10 IU of human chorionic gonadotropin [Sigma-Alrich Co.]) and mated with heterozygous males. Blastocysts (3.5 dpc) were collected and cultured for 4 days on gelatin in ES culture medium containing leukemia inhibitory factor (LIF). The outgrowths were then photographed and genotyped by PCR. Briefly, outgrowths were washed with phosphate-buffered saline and lysed with 20 μl of PCR lysis buffer (10 mM Tris [pH 8.3] 50 mM KCl, 2.5 mM MgCl, 0.1 μg of gelatin per ml, 0.45% NP-40, 0.45% Tween 20, 200 μg of proteinase K per ml) at 55°C for 1 h and then at 95°C for 15 min. Two microliters was then used to genotype the outgrowths by PCR as described for the 6.5-dpc embryos.
RESULTS
Generation and characterization of CGBP-null mice.
To determine the cellular function of CGBP, mutant mice were generated following homologous recombination in ES cells. The mutated allele was constructed by deleting the entire CGBP gene as well as approximately 5 kb of the upstream sequence (Fig. 1A). A single ES clone exhibited the disrupted allele as observed by the presence of a smaller 11-kb BamHI fragment upon Southern blot analysis (Fig. 1B). Several restriction enzyme digestions and Southern blots were performed to confirm that the disrupted allele was in the correct chromosomal location (data not shown). Germ line transmission was obtained for four chimeric male mice. Each mouse was then backcrossed to a C57B/6 female, and the resulting heterozygous offspring were mated. Eleven heterozygous breeding pairs were established, and the resulting pups were genotyped by Southern blot analysis (Table 1). No CGBP-null mice were obtained of the 123 analyzed, indicating that animals lacking CGBP are not viable. In addition, there was no detectable difference in phenotype between wild-type and heterozygous animals. Both male and female heterozygous animals were fertile and transmitted the mutated allele equally (data not shown).
TABLE 1.
Genotype analysis of progeny from heterozygous matings
| Status or age (dpc) | No. of progeny with genotypea:
|
Total | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Weaned | 41 | 82 | 0 | 123 |
| 12.5 | 4 | 2 | 0 | 6 |
| 10.5 | 17 | 44 | 0 | 61 |
| 8.5 | 10 | 14 | 0 | 24 |
| 6.5 | 2 | 14 | 0 | 16 |
Genotypes were determined by Southern blot or PCR analysis.
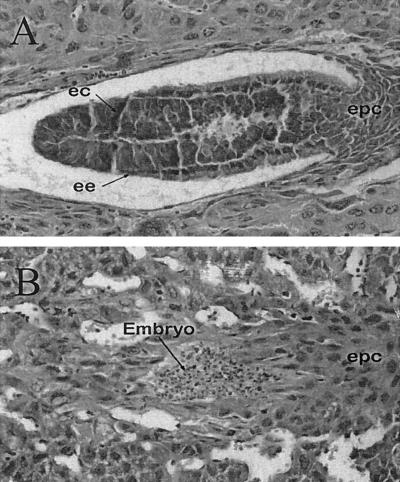
To assess at which stage of development the lethality occurred, 6.5- to 12.5-dpc embryos were collected and genotyped by either PCR or Southern blot analysis. CGBP-null embryos were not detected at any of the time points examined (Table 1). In addition, approximately 27% of the implantation sites (6 of 22) examined at 6.5 dpc appeared empty or reabsorbed (data not shown), indicating that CGBP-null embryos are incapable of developing to the gastrulation stage. Histological examination of 6.5-dpc implantation sites (Fig. 2) revealed the presence of abnormal embryos in approximately 28% of the sites examined (7 of 25). Both the embryonic ectoderm and the embryonic endoderm were evident in normal embryos (Fig. 2A). However, presumptive mutant embryos failed to exhibit these cell layers but rather appeared as a small mass of picnotic cells (Fig. 2B). In addition, the ectoplacental cones of normal embryos exhibited a cobblestone appearance, while in mutant embryos a region comparable to the ectoplacental cone was detectable but did not exhibit the cobblestone appearance (compare Fig. 2A and B). We conclude that failure to express CGBP results in early embryonic lethality.
FIG. 2.
Abnormal embryos are detected at 6.5 dpc. CGBP+/− animals were mated, and 6.5-dpc implantation sites were fixed, paraffin-embedded, and stained. (A) Normal 6.5-dpc embryo. The arrows indicate the presence of the embryonic ectoderm (ec) and endoderm (ee) as well as the ectoplacental cone (epc). (B) Abnormal (presumptively mutant) embryo. The lens objective was ×40.
Blastocyst outgrowth assays.
Failure of CGBP-null embryos to thrive may be due to the loss of cell viability or their inability to develop and/or properly implant. To begin to assess these possibilities, blastocysts were collected from heterozygous matings and their ability to form blastocyst outgrowths in vitro was examined. Blastocysts (3.5 dpc) were cultured for 4 days (in the presence of LIF), at which time they were photographed and genotyped. Mutant CGBP blastocysts were detected, indicating that CGBP-null cells are viable and that embryos lacking CGBP are capable of developing to the preimplantation stage. Of 38 blastocyst outgrowths from heterozygous matings, the PCR-determined genotypes of 10 were +/+, those of 24 were +/−, and those of 4 were −/−. In addition, mutant embryos hatched and formed both an inner cell mass and a trophectoderm in outgrowth assays (Fig. 3B to D). No discernible morphological difference was observed between the CGBP-null and heterozgyous outgrowths (compare Fig. 3A with B to D). Therefore, loss of CGBP results in peri-implantation embryonic lethality, indicating that this molecule is key for embryonic survival.
FIG. 3.
CGBP−/− blastocysts exhibit normal outgrowth characteristics. Blastocysts from heterozygous matings were collected at 3.5 dpc and cultured for 4 days. The presence of both the inner cell mass (ICM) and the trophectoderm (TE) was determined. Each outgrowth was genotyped by PCR. The lens objective was ×32.
DISCUSSION
Previously our laboratory demonstrated that CGBP binds specifically to unmethylated CpG motifs and transactivates reporter constructs containing this dinucleotide (54). However, little was known regarding the function of this molecule in the whole organism. This study has shown that CGBP is a critical player during early embryogenesis. No CGBP-null animals were detected, indicating that CGBP is critical for development. Upon further examination, mutant embryos were determined to be viable at 3.5 dpc (blastocyst), but analysis of embryos between days 6.5 and 12.5 dpc failed to detect CGBP-null embryos. However, histological examination of 6.5-dpc implantation sites revealed abnormal embryos, which identifies the time of embryonic lethality as being between 3.5 and 6.5 dpc. In addition, the detection of CGBP-null blastocysts indicates that CGBP is not essential for cell viability. Therefore, lethality of CGBP-null embryos is a peri-implantation defect, indicating that CGBP may be an important regulator during development.
The molecular mechanism(s) by which CGBP regulates normal embryogenesis remains unclear. We had previously postulated that CGBP regulates gene expression via binding to unmethylated CpG islands (54). However, the precise gene targets of CGBP action have yet to be delineated. Extensive evidence demonstrates the association of DNA methylation, histone deacetylation, chromatin remodelling, and gene silencing (9, 17, 18, 36, 37, 47, 55, 61). MeCP2 and MBD2 function as transcriptional repressors through binding to methylated DNA and associating with histone deacetylases (26, 37, 38, 55, 61). Although MBD3 has not been shown to bind methylated CpG motifs directly, it is a component of the NuRD (Mi-2) histone deacetylase and nucleosome remodelling complex (55, 61) and may be recruited to methylated DNA via its interaction with methyl-binding proteins. Whether or not CGBP regulates gene activity through association either with coactivators or directly with histone acetyltransferases remains to be determined. However, the early embryonic lethality of CGBP mutant embryos may indicate the loss of activation of key developmental genes.
The loss of embryonic viability of CGBP-null embryos coincides with the global switch in genome methylation. Following implantation, the murine genome normally becomes methylated and failure to adequately methylate genomic DNA results in embryonic lethality (30, 40). In addition, mutations in DNA methyltransferase 3B and MeCP2 results in the ICF and Rett syndromes, respectively (2, 21, 40, 59). Therefore, tight control of cytosine methylation is crucial for proper development and cell function. CpG islands, on the other hand, retain their hypomethylated state during development, with the exception of those associated with genes on the inactive X chromosome and some parentally imprinted genes (5, 22, 48). The molecular mechanisms involved in maintaining CpG islands as hypomethylated remains unclear. Demethylase activity has been identified (49), and the short form of MBD2 (23) was originally proposed to directly demethylate DNA through removal of the methyl group from 5-methylcytosine (7). However, this observation has not been substantiated (55). In addition, the transcription factor Sp1, which binds to GC-rich sequences, was proposed to protect genomic DNA from methylation. However, loss of Sp1 did not affect DNA methylation (33). The ability of CGBP to bind specifically to DNA containing unmethylated CpG dinucleotides and the loss of viability of CGBP-null embryos at the same time period as the switch in DNA methylation indicate that CGBP is a potential candidate for maintaining hypomethylation of CpG islands. Therefore, the loss of CGBP may result in hypermethylation and gene inactivation, thereby resulting in embryonic lethality. Interestingly, the failure to express CGBP results in an earlier phenotype compared to the loss of maintenance of de novo DNMTs, MeCP2, MBD2, or MBD3, which exhibit their phenotypes at mid-gestation or later (10, 20, 25, 30, 40, 50). Additional studies examining both global changes in DNA methylation and gene-specific changes in CGBP-null embryos are required before a role for CGBP in the maintenance of unmethylated DNA can be determined.
Finally, further study of CGBP may begin to elucidate the mechanisms involved in the regulation of CpG island-containing genes crucial for proper development and/or the establishment and maintenance of unmethylated CpG motifs. The early lethality of CGBP-null embryos makes deciphering these molecular events quite difficult. We are currently generating CGBP-null ES cell lines in order to delineate CGBP target genes as well as assess CGBP's contribution in the maintenance of unmethylated CpG islands.
ACKNOWLEDGMENTS
We acknowledge David Williams (Herman B Wells Center for Pediatric Research, Indiana University, Indianapolis, Ind.) and Celeste Simon (University of Pennsylvania, Philadelphia) for helpful discussions regarding construction of the targeting vector. We also thank Joseph Ruiz and John Critser (Herman B Wells Center for Pediatric Research) for their generous assistance in analysis of the mutant embryos and Gen-Sheng Feng (Burnham Insitute, La Jolla, Calif.) for providing the pPNT vector. We also acknowledge LeAnn Boldridge and the Center for Excellence in Molecular Hematology for histological sectioning; the Indiana University Cancer Center mouse core facility for ES cell transfections, blastocyst injections, and generation of chimeric animals; and Paula D. Ladd, Angela Nevins, and Mark Starr for their technical assistance.
This work was supported by the Riley Memorial Association, Public Health Service grant CA58947 from the National Cancer Institute (D.G.S), an NRSA from the NIH (D.L.C.), and an American Heart Association postdoctoral fellowship (D.L.C.).
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Amir R E, Van den Veyver I B, Wan M, Tran C Q, Francke U, Zoghbi H Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 3.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antequera F, Bird A. CpG islands. EXS. 1993b;64:169–185. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- 5.Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T H, Verdine G L. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya S K, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 8.Bird A P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 9.Bird A P, Wolffe A P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 11.Cross S H, Bird A P. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 12.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 13.Domer P H, Fakharzadeh S S, Chen C S, Jockel J, Johansen L, Silverman G A, Kersey J H, Korsmeyer S J. Acute mixed-lineage leukemia t(4;11) (q21;q23) generates an MLL-AF4 fusion product. Proc Natl Acad Sci USA. 1993;90:7884–7888. doi: 10.1073/pnas.90.16.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald K T, Diaz M O. MLL2: a new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- 16.Fujita N, Takebayashi S-I, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol Cell Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 20.Guy J, Hendrich B, Holmes M, Martin J E, Bird A. A mouse MeCP2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 21.Hansen R S, Wijmenga C, Luo P, Stanek A M, Canfield T K, Weemaes C M. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 23.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrich B, Hardeland U, Ng H H, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 25.Hendrich B, Guy J, Ramsahoye B, Wilson V A, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 27.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 28.Lei H, Oh S P, Okano M, Juttermann R, Goss K A, Jaenish R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 29.Leonhardt H, Page A W, Weier H-U, Bestor T H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 30.Li E, Bestor T H, Jaenish R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 31.Li K, Li Y, Shelton J M, Richardson J A, Spencer E, Chen Z J, Wang X, Williams R S. Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell. 2000;101:389–399. doi: 10.1016/s0092-8674(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 32.Ma Q, Alder H, Nelson K K, Chatterjee D, Gu Y, Nakamura T, Canaani E, Croce C M, Siracusa L D, Buchberg A M. Analysis of the murine ALL-1 gene reveals conserved domains with human ALL-1 and identifies a motif shared with DNA methyltransferase. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor SP1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 34.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 35.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 36.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 37.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenmann R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 38.Ng H H, Zhang Y, Heinrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 39.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 40.Okano M, Bell D W, Haber D A, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 41.Prasad R, Yano T, Sorio C, Nakamura T, Rallapalli R, Gu Y, Leshkowitz D, Croce C M, Canaani E. Domains with transcriptional regulatory activity within the ALL-1 and AF4 proteins involved in acute leukemia. Proc Natl Acad Sci USA. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 43.Reik W, Walter J. Imprinting mechanisms in mammals. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 44.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 45.Roberts A W, Kim C, Zhen L, Lowe J B, Kapur R, Petryniak B, Spaetti A, Pollock J D, Borneo J B, Bradford G B, Atkinson S J, Dinauer M C, Williams D A. Deficiency of the hematopoietic cell-specific rho family GTPase rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–196. doi: 10.1016/s1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- 46.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 47.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B-Z, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 48.Singal R, Ginder G D. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 49.Szyf M, Theberge J, Bozovic V. Ras induces a general DNA demethylation activity in mouse embryonal P19 cells. J Biol Chem. 1995;270:12690–12696. doi: 10.1074/jbc.270.21.12690. [DOI] [PubMed] [Google Scholar]
- 50.Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 51.Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 52.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocation in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 53.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 54.Voo K S, Carlone D L, Jacobsen B M, Flodin A, Skalnik D G. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 56.Walsh C P, Chaillet J R, Bestor T H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 57.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolffe A P, Jones P L, Wade P A. DNA demethylation. Proc Natl Acad Sci USA. 1999;96:5894–5896. doi: 10.1073/pnas.96.11.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu G L, Bestor T H, Bourc'his D, Hsieh C L, Tommerup N, Brugge M, Hulten M, Qu X, Russo J J, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 60.Zeleznik-Le N J, Harden A M, Rowley J. 11q23 translocation split the “AT-hook” cruciform DNA-binding region and the transcription repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Ng H-H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]