Abstract
The yeast NuA4 complex is a histone H4 and H2A acetyltransferase involved in transcription regulation and essential for cell cycle progression. We identify here a novel subunit of the complex, Yng2p, a plant homeodomain (PHD)-finger protein homologous to human p33/ING1, which has tumor suppressor activity and is essential for p53 function. Mass spectrometry, immunoblotting, and immunoprecipitation experiments confirm the stable stoichiometric association of this protein with purified NuA4. Yeast cells harboring a deletion of the YNG2 gene show severe growth phenotype and have gene-specific transcription defects. NuA4 complex purified from the mutant strain is low in abundance and shows weak histone acetyltransferase activity. We demonstrate conservation of function by the requirement of Yng2p for p53 to function as a transcriptional activator in yeast. Accordingly, p53 interacts with NuA4 in vitro and in vivo, an interaction reminiscent of the p53-ING1 physical link in human cells. The growth defect of Δyng2 cells can be rescued by the N-terminal part of the protein, lacking the PHD-finger. While Yng2 PHD-finger is not required for p53 interaction, it is necessary for full expression of the p53-responsive gene and other NuA4 target genes. Transcriptional activation by p53 in vivo is associated with targeted NuA4-dependent histone H4 hyperacetylation, while histone H3 acetylation levels remain unchanged. These results emphasize the essential role of the NuA4 complex in the control of cell proliferation through gene-specific transcription regulation. They also suggest that regulation of mammalian cell proliferation by p53-dependent transcriptional activation functions through recruitment of an ING1-containing histone acetyltransferase complex.
In eukaryotes, different activities that alter nucleosomal structure or modify nucleosomal histones are involved in the modulation of transcription. These activities, mediated by multiprotein complexes, contribute to relieve or reinforce the chromatin inhibition of transcription process (49). Two types of chromatin-modifying activities were identified in various organisms. The first type includes the SWI2-related chromatin remodeling complexes, which use the energy of ATP hydrolysis to alter histone-DNA contacts, favoring nucleosome disruption, mobility, and transfer. The second type modifies the acetylation state of nucleosomal histone N-terminal tails. The balance between histone acetyltransferases (HATs) and histone deacetylases is a key factor in the determination of gene-specific transcription levels. Hyperacetylation of nucleosomal histones is correlated with increase of transcription, while hypoacetylation is linked to repression (49).
Histone hyperacetylation does not result in a dramatic change in nucleosomal structure but appears to increase DNase I sensitivity (26) and transcription factor accessibility (45), which correlate with facilitated RNA polymerase II transcription (33). Recently, an increasing number of HAT complexes were identified and linked to the transcription process (8). In the yeast Saccharomyces cerevisiae, various complexes with nucleosomal HAT activities were identified. These include Gcn5p-containing complexes, SAGA, and ADA (20). Recently, Sas3p, a MYST family HAT, was found as the catalytic subunit of the NuA3 complex (25). Nucleosomal HAT activity was also found in the mediator complexes (30).
NuA4 is another large multisubunit HAT complex, composed of 12 subunits (2). This complex is unique in yeast because it specifically acetylates nucleosomal histone H4 and, to lesser extent H2A (compared to the histone H3 preference of the other complexes). We have previously identified Esa1p, a HAT essential for cell growth (11, 40), as the catalytic subunit of the complex (2). Esa1p is required for cell cycle progression making NuA4 the only essential HAT complex known in budding yeast. Crystal structure analysis of the Esa1p HAT domain revealed a mechanism of catalysis and substrate binding related to other HATs and the presence of a MYST-family-specific zinc-finger domain (50). Esa1p is a member of a family that includes a large number of known or putative HATs and is closely related to human Tip60, which has the same substrate specificity, coactivates transcription, affects DNA repair and apoptosis, and is present in a complex related to NuA4 (6, 12, 23). Esa1p is also closely related to Drosophila MOF, which is the HAT subunit of the dosage-compensation (MSL) complex involved in hypertranscription and histone H4 acetylation of the X-chromosome in males (14, 41). NuA4 is able to stimulate in vitro transcription from chromatin substrate through specific recruitment by transcription activators, creating a large domain containing hyperacetylated histone H4 (44, 46).
Other known subunits of the NuA4 complex include the following: Tra1p, an essential ATM-family member highly related to an essential cofactor for c-Myc and E2F transcription and/or transforming potential (2); Act1p (cellular actin) and Act3p/Arp4p, an essential actin-related protein implicated in epigenetic control of transcription (15); Epl1p, an essential protein homologous to Enhancer of polycomb, E(Pc), a modifier of position effect of variegation in Drosophila (15); and Eaf3p, a two-chromodomain protein related to another subunit of the Drosophila MSL complex and to the human growth regulator MRG15 (14). Using esa1, act3/arp4, and eaf3 mutants, we demonstrated the role of the NuA4 complex and its HAT activity in gene-specific transcription regulation in vivo (14, 15). Importantly, a recent study showed that Esa1p is targeted in vivo to ribosomal protein promoters in an activator-dependent manner (35). Additionally, Esa1p seems important for global nontargeted or large domains of histone H4 acetylation in yeast chromatin which are not necessarily linked to transcription regulation (35, 47).
In this report, by using various biochemical approaches, we identified Yng2p as a stable stoichiometric subunit of the purified NuA4 complex. Yng2p is required for normal cell growth and contains a PHD-finger domain, which is commonly found in proteins involved in chromatin-mediated transcriptional regulation (1). Interestingly, Yng2p is closely related to p33/ING1, a human protein identified in a functional screen for tumor suppressors (17, 18). Furthermore, p33/ING1 cooperates with tumor suppressor p53 in cell growth control as it physically interacts with it and is necessary for p53-dependent transcriptional activation of the p21/WAF1/CIP1 gene (16). We demonstrate here the requirement of Yng2p for normal NuA4 activity since deletion of YNG2 reduces NuA4 abundance and specific HAT activity. The deletion also leads to decreased transcription of NuA4-dependent genes. We present data indicating that p53 function as a transcription activator in yeast depends on Yng2p, a link supported by direct in vitro and in vivo interaction detected between the NuA4 complex and p53. In agreement with a recruitment process, transcriptional activation by p53 provokes histone H4-specific hyperacetylation on the responsive gene in a NuA4-dependent manner. The primary functional role of Yng2p in the NuA4 complex argues for a conserved function in human cells, i.e., the presence of an ING1-containing NuA4 complex recruited by p53 for transcriptional activation of cell growth regulator genes.
MATERIALS AND METHODS
Yeast strains and plasmids.
The genotypes of all of the strains used in this study are presented in Table 1. Yeast culture, transformation, mating, sporulation, and dissection were done according to standard protocols. YNG2 was disrupted by transforming diploid strains with linear DNA fragment in which the selectable marker Kanr is flanked by YNG2 noncoding sequences (48). This fragment was obtained by PCR amplification with pFA6aKanMX4 as a template and with the primers 5′-TAACCCACCTACCGTTAGTTGAAATAGAAACAAAGAAGAAGGTTTAGCTTGCCTCGTCCCCGCCG-3′ and 5′-GGTATTTT TG T TCAGT TACG T T T TCT T T TCAGT T TGT T T T T T TCCATC TCGAC TCACTATAGGGAGACCGGCAG-3′. Disrupted clones and the spores of at least two tetrads were checked by PCR and Southern blotting.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BMA41 | MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 trp1Δ/trp1Δ ura3-1/ura3-1 leu2-3,112/leu2-3,112 | F. Lacroute |
| YBC76 | MATα/MATahis3Δ200/his3Δ200 ura3-52/ura3-52 trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 lys2-128δ/lys2-128δ | B. Cairns |
| BY4743 | MATα/MATahis3Δ1/his3Δ1 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 | Resgen |
| QY200 | MATα/MATahis3Δ1/his3Δ1 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 MET15/met15Δ0 LYS2/lys2Δ0 YNG2/yng2::Kanr | This study |
| QY201 | MATα/MATahis3Δ200/his3Δ200 ura3-52/ura3-52 trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 lys2-128δ/lys2-128δ YNG2/yng2::Kanr | This study |
| QY202 | MATahis3Δ200 trp1Δ63 ura3-52 leu2Δ1 lys2-128δ yng2::Kanr | This study |
| QY203 | MATα his3Δ200 trp1Δ63 ura3-52 leu2Δ1 lys2-128δ yng2::Kanr | This study |
| QY204 | MATα his3Δ200 trp1Δ63 ura3-52 leu2Δ1 lys2-128δ YNG2 | This study |
| QY205 | MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 trp1Δ/trp1Δ ura3-1/ura3-1 leu2-3,112/leu2-3,112 YNG2/yng2::Kanr | This study |
| QY206 | MATaade2-1 his3-11,15 trp1Δura3-1 leu2-3,1121 yng2::Kanr | This study |
| QY207 | MATα ade2-1 his3-11,15 trp1Δura3-1 leu2-3,1121 yng2::Kanr | This study |
| QY108a | MATahis3Δ1 ura3Δ0 leu2Δ0 met15Δ0 epl1::TRP1 pPHEU (HA-EPL1/URA3/ARS-CEN) | A. Boudreault |
To generate pAN100, the YNG2 open reading frame (ORF) was amplified from yeast genomic DNA with Resgen Gene Pairs, digested with EcoRI/SmaI, and ligated with pSK. The vector pAN102 is yeast low-copy (ARS, CEN) plasmid containing the YNG2 ORF, tagged at the N-terminal with two HA epitopes, under the control of the PGK promoter. It was constructed by cloning a PCR fragment, amplified from pAN100, into BamHI/SmaI of pAN101a. This last plasmid was obtained by inserting the HindIII fragment of TL38 (10) into pFL36 (5). The promoter of YNG2 was amplified by PCR and inserted into pAN101b, which was digested by SacI/NcoI. The new vector, pAN103, was digested and ligated with the NcoI/SmaI fragment from pAN102. The resulting vector, pAN104, expresses HA-Yng2p under the control of its own promoter. pAN105 and pAN106 are ARS-CEN vectors containing the YNG2 gene. They were generated by cloning YNG2 (−938/+849 bp with respect to initiation codon), amplified from genomic DNA, into SacI/SmaI of pFL38 and pFL36 (5). The vector pYD100, expressing HA-Yng2p deleted of the PHD-finger, was generated by inserting a PCR-amplified YNG2 sequence from pAN104 corresponding to the first 218 amino acids (aa) of the protein into the BamHI/SmaI sites of pAN103 by using the primers 5′-AAGGCTAGATCTATGGATCCAAGTTTAGTTTTAGAGCAAACG-3′ and 5′-ATTAGTCCCGGGGTCCTATTCCTCGTTTTCAGGGGAACCG-3′. The plasmid pYD101 expresses Yng2p C-terminal domain (from aa 154 to 282) containing the PHD-finger. It was obtained by subcloning a 389-bp BamHI/SmaI fragment of pAN104 into the respective restriction sites of pAN103. The two proteins, encoded by pYD100 and pYD101, contain a nuclear localization sequence and were expressed from the native YNG2 promoter. The pLS76 contains p53 under the control of ADH1 promoter. The reporter plasmid pSS1 has the HIS3 ORF under the control of a p21-ΔUAS/GAL1 promoter. These two plasmids were described earlier (13). In order to perform the FASAY with the different constructions of YNG2, we swapped the wild-type human p53 ORF under the control of the ADH1 promoter and followed by the CYC1 terminator into a URA3 marked vector. A 3.2-kb KpnI/SacI fragment derived from pLS76 was subcloned directly into the KpnI/SacI sites of pFL38 to produce pYD102.
The glutathione S-transferase (GST)–p53 fusion proteins were constructed as follows. The full-length mouse p53 coding sequence was amplified by PCR from pECM53 (kindly provided by A. Anderson) by using the following primers: 5′-AAGGCTGGATCCATGACTGCCATGGAGGAGTCACAGTCGG-3′ and 5′-ATTAGTCCCGGGTCAGTCTGAGTCAGGCCCCAC-3′. The activation domain (aa 1 to 292) was amplified with the primer pair 5′-AAGGCTGGATCCATGACTGCCATGGAGGAGTCACAGTCGG-3′ and 5′-ATTAGACCCGGGAGGTCAAAGGACTTCCTTTTTGCG-3′. These PCR products were then cloned into pGEX-4T-3 in BamHI/SmaI sites.
NuA4 purification and peptide sequencing.
Partial purification of the NuA4 complex by fractionation over Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen), MonoQ HR5/5, and Superose-6 HR10/30 columns (Pharmacia) and the HAT assay on oligonucleosomes or HeLa core histones and Western blotting were previously described (2, 20). The hemagglutinin (HA) antibody (Babco) was used at 1:3,000. Immunoprecipitation of the NuA4 complex was performed on the peak Superose-6 fraction (2). The NuA4 36- to 37-kDa sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) excised bands corresponding to Eaf4p were subjected to in gel reduction, carboxyamidomethylation, and tryptic digestion (Promega). Ion trap mass spectrometry and peptide sequencing were performed as described previously (14).
Coimmunoprecipitation and NuA4 pulldown assays.
Coimmunoprecipitations on purified fractions have been described elsewhere (2). For coimmunoprecipitation in whole-cell extracts, proteins were isolated from 150-ml cell cultures grown in yeast extract-peptone-dextrose (YPD) to an optical density (OD) of 2.5. Cells were washed in 20 mM HEPES [pH 7.5]–150 mM NaCl and resuspended in 1 ml of lysis buffer (40 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1% Tween 20, 2 μg of leupeptin and pepstatin A/ml, 5 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride). Two cell suspension aliquots of 500 μl were mixed with the same volume of glass beads and vortexed four times for 1 min. Cell lysates were clarified by two successive centrifugations (20 min at 7,000 rpm and then 30 min at 14,000 rpm) at 4°C. Ten milligrams of total proteins was used in 500-μl immunoprecipitation reactions. Then, 50 μl of protein A-Sepharose beads was used to preclear the lysate. Cross-linked anti-HA protein A-Sepharose beads (2) were then added, and the mixture was incubated overnight at 4°C. The beads were washed three times with 10 volumes of 100 mM NaCl lysis buffer. Input (0.3%) and immunoprecipitated proteins (22.5%) were analyzed by Western blot.
GST fusion proteins were expressed and purified on glutathione-Sepharose beads as described previously (44). GST pulldown assays with NuA4 and HAT reactions with nucleosomes were performed as described by Utley et al. (44) with equivalent amounts of input, supernatant, and beads.
Northern blot analysis.
Total yeast RNA was isolated by the hot-phenol method (38). Fifteen micrograms of RNA was separated by electrophoresis on a formaldehyde-agarose gel, blotted, and UV cross-linked to a nylon membrane (Amersham). Hybridization was performed in 0.5 M phosphate buffer (pH 6.8), 7% SDS, and 1% bovine serum albumin. The probes used were ORFs from HIS3, HIS4, PHO5, GAL1, and ACT1 obtained by PCR and radiolabeled by using the Multiprime Labeling System (Amersham).
Chromatin immunoprecipitations.
Chromatin was prepared as described previously (27) with few modifications. Cell lysates were sonicated three times by using Fisher Sonic dismembrator 150 set at 0.4 to 0.6 output for 10 s. Sonication yielded 1- to 3-kb chromatin fragments. Samples were then centrifuged for 1 h at 14,000 × g, and the supernatant was collected. Immunoprecipitation of that material was also performed as described previously (27), except that incubations with αAcH3, αhyperAcH4, or αAcH4 antibodies (Upstate Biotech) were done for 90 min at room temperature. PCRs were carried out in 25 μl by using 1/100 of the immunoprecipitated material and 1/10,000 of the input material as templates. A region of the pSS1 plasmid (24) spanning from the p53 binding site to HIS3 (+19 from start site) was amplified by using a specific primer pair. A total of 0.5 μCi of [32P]dATP (3,000 Ci mmol−1) was included in the PCRs, and amplified products were quantified by PhosphorImager (Molecular Dynamics). Two different chromatin preparations were assayed in duplicate experiments.
RESULTS
Yng2p, an ING1 family member, is a stable subunit of the yeast NuA4 acetyltransferase complex.
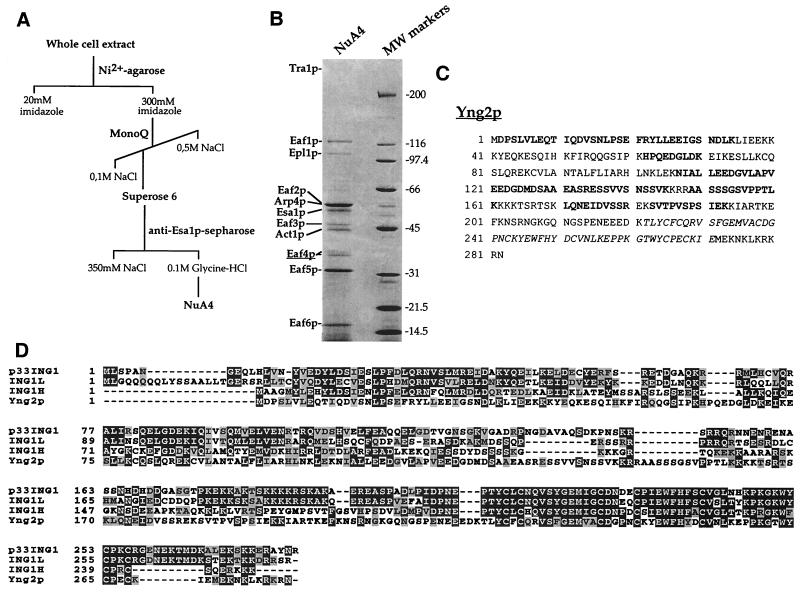
To determine the identity of additional NuA4 subunits, the complex was purified to homogeneity from yeast extract by using a combination of conventional and immunoaffinity chromatographic steps (Fig. 1A). The Coomassie blue-stained gel of the immunopurified complex is shown in Fig. 1B. Previously identified subunits are labeled with their respective names (Tra1p, Epl1p, Esa1p, Arp4p, and Act1p). The band corresponding to the ATM-related cofactor Tra1p is very faint because it did not solubilize efficiently in the sample buffer after trichloroacetic acid precipitation (data not shown). Unidentified polypeptides were named in order Eaf1p to Eaf6p for “Esa1p-associated factor.” Two bands, previously named p36 and p37 based on their size and referred to as Eaf4p (see below), were digested with trypsin and analyzed by microcapillary reversed-phase high-pressure liquid chromatography (HPLC) nanoelectrospray tandem mass spectrometry.
FIG. 1.
An ING1-related protein is present in the purified yeast NuA4 complex. (A) Chromatographic steps to obtain the purified NuA4 complex. (B) Coomassie stained gel of the affinity-purified NuA4 complex. The complex was eluted with glycine-HCl, precipitated, and loaded onto an SDS–10% PAGE gel. Specific NuA4 subunits are indicated by their respective names, and uncharacterized protein bands are labeled Eaf1 to Eaf6. Ion trap mass spectrometry of tryptic peptide obtained from the 36- to 37-kDa Eaf4p bands identified them as proteins encoded by the yeast ORF YHR090C, also recently called YNG2 (29). (C) Amino acid sequence of YNG2 gene product. The nine peptide sequences obtained by tandem mass spectrometry are shown in boldface; the PHD-finger region is shown in italics. (D) Multiple sequence alignment between yeast Yng2p and human ING1-family members. Accession numbers: p33/ING1, AAC00501; ING1L, inhibitor of growth 1-like, NP001555; ING1H, p33/ING1 homolog, NP057246; Yng2p, NP011958.
All nine peptides obtained correspond to sequences from the YHR090C gene product and cover 41% of the protein (see Fig. 1C, amino acids in boldface). Thus, the two protein bands are encoded by the same gene, which was named YNG2 in a recent report (29). The gene encodes a 282-aa protein with a predicted molecular mass of 32.1 kDa. Yng2p contains a PHD-finger domain in its C-terminal region (italicized in Fig. 1C). This Cys4-His-Cys3 domain is predicted to chelate two Zn2+ ions and is found in many different proteins throughout evolution, several of which are involved in chromatin structure and function and linked to transcription regulation (1). Yng2p PHD-finger is highly related to the one present in a group of human proteins, called the ING1 family (Fig. 1D). The founding member of the family is p33/ING1, a candidate tumor suppressor gene that is involved in the control of cell proliferation and apoptosis (17, 18, 22). p33/ING1 expression was found to be repressed in several cancer cells (17, 34, 42, 43), and tumor-specific missense mutations were identified in the nuclear localization motif and the PHD-finger domain (21). Furthermore, p33/ING1 was shown to cooperate with p53 tumor suppressor in cell growth control and apoptosis and physically associates with p53 in vivo (16, 39, 51). A multiple sequence alignment of Yng2p with p33/ING1 and -2 other highly related uncharacterized human proteins is shown in Fig. 1D. Clearly, these four proteins not only are highly homologous over the PHD region but also show significant homology over their entire sequence. Yng2p has higher homology over the N-terminal region with ING1H (accession number NP057246) versus to the two other human proteins; Yng2p shows 25% identity and 42% similarity to ING1H over the entire protein sequence.
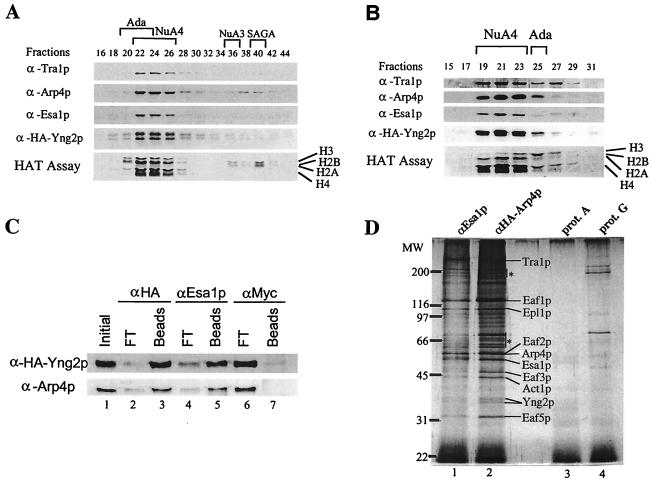
To demonstrate the stable association of Yng2p with NuA4, we confirmed its coelution by Western blotting with NuA4 HAT activity over MonoQ ion-exchange and Superose-6 gel filtration columns (Fig. 2A and B). Extracts were prepared from a strain expressing an HA-tagged version of Yng2p from a low-copy ARS/CEN plasmid. The MonoQ column separates the four previously identified native nucleosomal HATs (20). HA-Yng2p strictly coelutes with nucleosomal H4- and H2A-specific acetyltransferase activity and with the known NuA4 subunits, Esa1p, Tra1p, and Arp4p. Yng2p also specifically coelutes with NuA4 HAT activity and components from the subsequent gel filtration column, suggesting that Yng2p is associated with the 1.3-MDa complex. Similar to native Yng2p, HA-Yng2p migrates as two distinct protein bands (see native Yng2p/Eaf4p in Fig. 1B). The same doublet is obtained when Yng2p is produced in bacteria, arguing against differences in posttranslational modification. Furthermore, the two bands are not found when a version of Yng2p lacking the PHD-finger is produced in bacteria or yeast (data not shown; see Fig. 7). The ratio between the two bands seems to be variable between gels with the same sample, suggesting inefficient denaturation of the PHD-finger (data not shown).
FIG. 2.
Yng2p is a stable stoichiometric subunit of the NuA4 acetyltransferase complex. (A) Extract from BY4741 strain, transformed with HA-Yng2p-expressing plasmid (pAN102), was fractionated over nickel-agarose, followed by MonoQ column fractionation. MonoQ fractions were tested for HAT activity and by Western blotting with the indicated antisera. (B) MonoQ NuA4 peak fractions were pooled and loaded on a Superose-6 gel filtration column. HAT assays and Western blotting were performed as in panel A. (C) Coimmunoprecipitation of HA-Yng2p and Arp4p. Equal amounts of the Superose-6 fraction 21 described in panel B were incubated with anti-HA, anti-Esa1p, and anti-Myc. After washes, equivalent amounts of Initial, Beads (bound), and FT (flowthrough; unbound) were analyzed by Western blotting with anti-HA and anti-Arp4p. (D) Yng2p was also determined to be present in affinity-purified NuA4 complex by using HA-Arp4p as antigen. The Superose-6 peak fraction of NuA4 obtained from an HA-Arp4p-expressing strain (DY3558 [see reference 15 for details]) was immunoprecipitated with anti-Esa1p–protein A-Sepharose or with anti-HA–protein G-Sepharose. Silver staining of immunopurified complexes is shown. Specific NuA4 bands are indicated. Nonspecific bands present in the controls are indicated by asterisks.
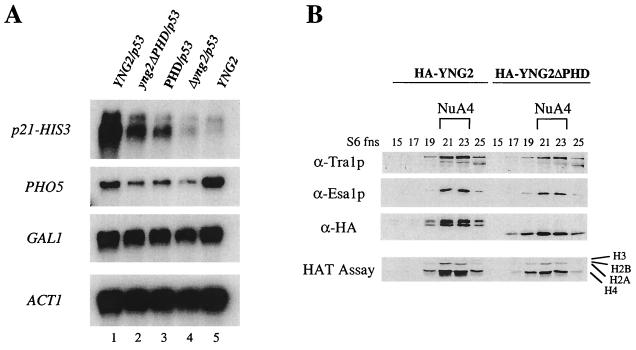
FIG. 7.
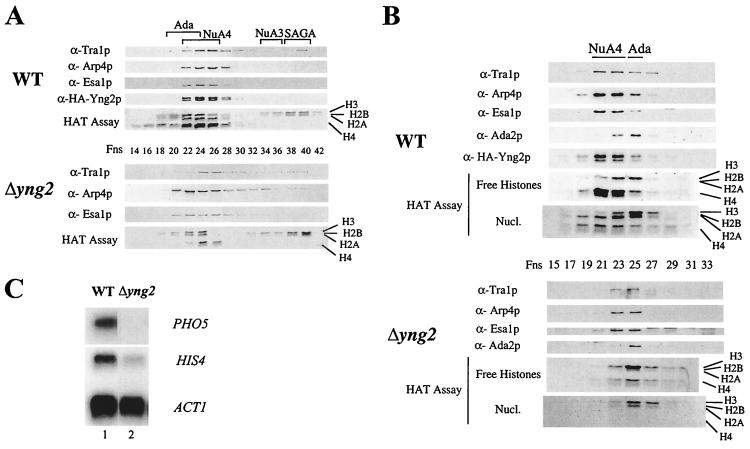
Yng2p PHD domain is important for gene-specific transcriptional regulation but is dispensable for NuA4 HAT activity. (A) p53-dependent transcriptional activation is affected in strains deleted of the PHD-finger or expressing only the PHD domain of Yng2p. Δyng2 strains (QY203) containing an episomal copy of wild-type YNG2 (pAN104), yng2ΔPHD (pYD100), or PHD (pYD101) or an empty vector (−) were also transformed with a p53-expressing plasmid (pYD102) and a reporter plasmid encoding His3p under the control of p21 promoter (pSS1). RNA samples were prepared as in Fig. 5, and Northern blots were hybridized with HIS3, PHO5, GAL1, and ACT1 probes. Expression of p21-HIS3 (activated by p53) and PHO5 (basal) is affected by the deletion of Yng2p PHD domain (compare lanes 1 and 2) or its N-terminal domain (compare lanes 1 and 3), while the expression of GAL1 and ACT1 remains unchanged. Equivalent levels of p53 protein between strains were confirmed by Western blot (data not shown). (B) Yng2p PHD-finger domain is not required for NuA4 complex integrity and HAT activity. Protein extracts from deleted strain QY203 expressing HA-Yng2p or HA-Yng2ΔPHD were fractionated over Ni-NTA, MonoQ, and Superose-6 columns. Fractions from the gel filtration were tested for the presence of NuA4 components by Western blot assayed for HAT activity on oligonucleosomes. Deletion of the Yng2p PHD domain does not affect the abundance, integrity, or activity of NuA4.
To further demonstrate a physical association of Yng2p with NuA4, immunoprecipitation from the Superose-6 fraction with Esa1p antibodies efficiently depleted HA-Yng2p from the supernatant and recovered it on the beads (Fig. 2C). Reciprocally, immunoprecipitation of Yng2p with HA antibodies efficiently brought down the Arp4p subunit of NuA4. Finally, immunopurified NuA4 complexes from an HA-Arp4p-expressing strain, by using HA-Arp4p or Esa1p as baits, contain the same specific protein bands, including the Yng2p doublet (Fig. 2D). Together, these data establish Yng2p as a stable stoichiometric subunit of the NuA4 HAT complex.
YNG2 is required for normal cell growth.
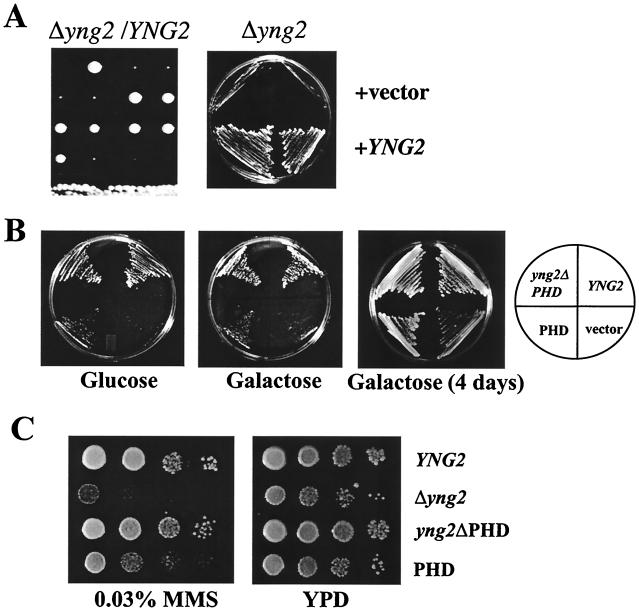
Given the lethality associated with the absence of NuA4 subunits Esa1p, Tra1p, Epl1p, Arp4p, and Act1p, we sought to determine whether YNG2 replacement would also be lethal. The gene was disrupted in diploid strains as described in Materials and Methods, and its requirement for cell growth was examined by tetrad analysis (Fig. 3A). Four representative tetrads are shown, each producing two wild-type and two very slow growing spores (left panel). The growth defect was suppressed by episomal expression of the wild-type gene (right panel). Since information in databases and an earlier report (32) indicated that the gene was essential for growth, we repeated the YNG2 disruption in other genetic backgrounds (W303, S288c, and BY; see Table 1) and obtained the extremely slow growth phenotype in each case (data not shown). A similar phenotype was also obtained recently in an independent study (29). Thus, the YNG2 gene is not essential for viability, but cells carrying its deletion show severe growth defects. Microscopy analysis of the mutant cells showed large multibudded cells, the majority of the buds lacking nuclei. On the other hand, fluorescence-activated cell-sorting analysis of exponentially growing cultures demonstrated that wild-type and mutant strains contained a similar distribution of G0/G1- versus G2/M-phase cells (data not shown). Importantly, the mutant strain can use glucose or galactose as carbon source, in agreement with NuA4 not being involved in GAL1 expression or induction (14; see also Fig. 5). We then investigated the ability of Yng2p subdomains to rescue wild-type growth (Fig. 3B). Low-copy episomal expression of Yng2p lacking its PHD-finger (aa 1 to 218, from natural YNG2 promoter) completely suppressed the growth defect of the mutant strain, indicating that the PHD-finger is not required for normal cell growth. Alternatively, episomal expression of the PHD-finger domain (aa 154 to 282) also suppressed the growth defect but at a much lower efficiency. The region used was larger than the PHD-finger itself (aa 222 to 271) to encompass the Yng2p putative nuclear localization motif and avoid an indirect effect due to lack of nuclear targeting. This could explain the discrepancy between our data and a recent report arguing that the PHD-finger could not suppress yng2::Kan growth defect (29). Nevertheless, the N-terminal region of Yng2p is sufficient to sustain normal growth. Mass spectrometric analysis of the 16-kDa Eaf6p band in immunopurified NuA4 notably turned up one peptide located in the N-terminal region of Yng2p (YLLEEIGSNDLK; aa 23 to 34). Since protein degradation is unlikely to occur after elution or disruption of the complex with glycine-HCl, this suggests that the N-terminal region of Yng2p is the domain responsible for its association with NuA4. This is also the domain required for normal cell growth (Fig. 3B), supporting a role for NuA4 in the control of cell proliferation.
FIG. 3.
Deletion of YNG2 results in slow-growth phenotype and MMS sensitivity that are rescued by the expression of Yng2p deleted of the PHD-finger. (A) The BMA41 diploid strain was disrupted for one copy of YNG2 (QY205) and subjected to sporulation and tetrad dissection. In each case, the spores deleted of YNG2 grew poorly. The deleted haploid strain (QY207: Δyng2) was transformed with low-copy plasmid containing YNG2 (pAN105) or empty vector (−). The wild-type YNG2 gene complements the growth defect phenotype of the disrupted strain. (B) Yng2p deleted for the PHD-finger rescues the growth defect phenotype on glucose and galactose. The Δyng2 strain (QY203) was transformed with plasmid containing wild-type YNG2 (pAN104), YNG2ΔPHD (pYD100), or PHD (pYD101) or an empty vector (−). These strains were plated on minimal medium with either glucose or galactose. After 3 days at 30°C, only the strain expressing Yng2ΔPHD shows normal wild-type growth on either carbon source. Further incubation on galactose shows that all of the strains are able to grow on this medium. (C) The YNG2-null strain is sensitive to MMS. Tenfold serial dilutions of the strains described above were spotted on YPD or YPD + 0.03% MMS plates and incubated at 30°C for 2 days. The strains deleted for YNG2 or expressing only the PHD domain of YNG2 grow poorly on the MMS plate.
FIG. 5.
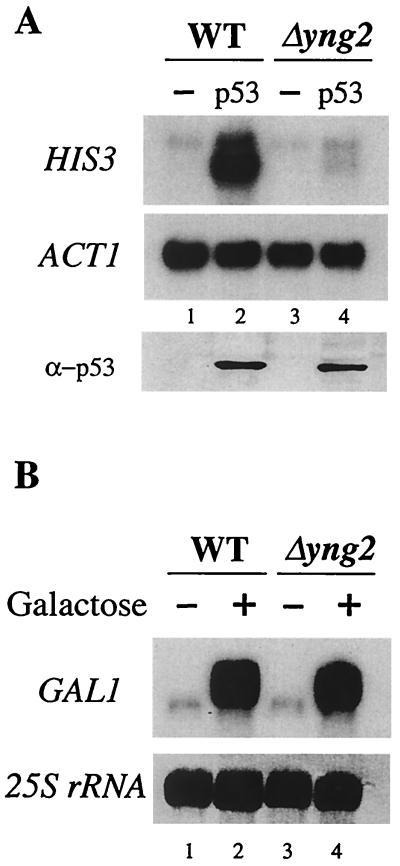
Yng2p is required for p53 transactivation function in S. cerevisiae but has no effect on Gal1 induction. (A) The strains QY204 (WT) and QY203 (Δyng2), each containing a reporter plasmid encoding His3p under the control of p21 promoter, were transformed with empty vector (−) or a plasmid constitutively expressing p53. Cultures of these strains were grown in minimal medium supplemented with the corresponding auxotrophy to an OD of 1 and then shifted to YPD (OD = 0.1) and grown to a final OD of 1. Total RNA was extracted and Northern blotting was performed using HIS3 and ACT1 probes. The p53-dependent p21-HIS3 activation is lost in the absence of Yng2p (compare lanes 2 and 4). Expression of p53 was monitored by Western blot on whole-cell extracts from each strain. (B) Transcriptional activation of the GAL1 gene does not require Yng2p. Cultures of strains QY204 (WT) and QY203 (Δyng2) were grown in YPD medium to an OD of 1 (lane 1 and 3) and then shifted to YP-galactose medium (lane 2 and 4) for 10 h. Aliquots were taken before and after galactose induction, and total RNA was extracted and analyzed by Northern blots by using a GAL1 probe. The membrane was stained with methylene blue, and 25S rRNA is shown to demonstrate equal loading between lanes. Induction of GAL1 was not affected by deletion of YNG2 (compare lanes 2 and 4).
Interestingly, cells harboring the yng2::Kan replacement showed sensitivity to the alkylating agent methyl methanesulfonate (MMS), suggesting that Yng2 is important for cells to survive DNA double-strand breaks (Fig. 3C). This further suggests conserved function of NuA4 throughout evolution since p53 and the Esa1-homolog Tip60 are important for mammalian DNA-repair processes (references 23 and 52 and references therein).
Yng2p is required for NuA4 HAT activity and function in vivo.
We next determined the role of Yng2p in NuA4 structure and HAT activity. Protein extracts from wild-type and Δyng2 strains were prepared, purified over nickel-agarose, MonoQ, and Superose-6 columns, and the fractions were analyzed for HAT activity and the presence of Esa1p, Tra1p, Arp4p, and HA-Yng2p (Fig. 4A and B). The wild-type strain is in fact disrupted at the YNG2 locus and carries a low copy ARS/CEN plasmid expressing HA-Yng2p from its natural promoter (as in Fig. 3A). The MonoQ fractions clearly show that the NuA4 complex lacking Yng2p is produced, but very weak nucleosomal H4 HAT activity is detected compared to the wild-type fractions (Fig. 4A). Histone H3-specific HAT activities from the ADA, NuA3, and SAGA complexes serve as good internal controls. Moreover, further fractionation over the Superose-6 column indicates that NuA4 elutes as a smaller complex when Yng2p is absent (fractions 23 to 25 versus 21 to 23 in wild type; Fig. 4B). Using the H3-specific HAT activity and the Western signal of Ada2p from the unaffected ADA complex (20) as an internal control, we observed that NuA4 lacking Yng2p has a smaller size, is less abundant (>3-fold) and has lower specific activity (>3-fold) than the wild-type complex (relative specific HAT activities were evaluated by using amounts standardized by Western blots with recombinant Esa1p and Arp4p [data not shown]). Altogether, these data support a primary role for Yng2p in NuA4 structure and activity. Indeed, there is a decrease of bulk H4 acetylation detected in both yng2 and esa1 mutant cells (11, 29). Importantly, Western analysis during the purification suggests that the majority of cellular Yng2p is associated with NuA4 in the cell (data not shown).
FIG. 4.
Deletion of YNG2 affects NuA4 activity and transcription of NuA4 target genes. (A) Protein extracts from deleted strain QY203 (Δyng2) or QY203, transformed with pAN104, and expressing HA-Yng2p (WT) were fractionated on a MonoQ column. Fractions were assayed for HAT activity and Western blotted for the indicated NuA4 components. (B) MonoQ NuA4 peak fractions of wild type (WT) and Δyng2, shown in panel A, were pooled and loaded on a Superose-6 gel filtration column. Eluted fractions were tested for the presence of NuA4 components and Ada2p by Western blot. HAT assays with nucleosomes and free histones as substrates were also performed. (C) The strains QY204 (WT) and QY203 (Δyng2) were grown in YPD to an OD of 1.0. Total RNA was extracted and Northern blots were done, using PHO5, HIS4, and ACT1 probes. The deletion of YNG2 significantly affected the transcript levels of PHO5 and HIS4 but not ACT1.
In separate reports we have shown that viable mutations in NuA4 subunit encoding genes, ESA1, ARP4, and EAF3, specifically affect PHO5 and HIS4 gene expression in vivo (14, 15). To see whether disruption of YNG2 creates such transcription defects, we performed Northern analysis with wild-type and mutant cells grown in rich medium and measured the mRNA levels of specific genes (Fig. 4C). The results indicate that PHO5 and HIS4 mRNA levels are decreased by 7- and 2.5-fold, respectively, in the yng2 mutant cells compared to the unaffected ACT1 signals. Thus, Yng2p plays a significant role in NuA4 activity and function in gene-specific transcription.
p53 function as a transactivator of the p21/WAF1 promoter requires Yng2p/NuA4.
Mammalian p53 can function as a transcription factor in yeast allowing the development of a S. cerevisiae-based functional assay to study the effects of germ line mutations found in p53 (24). Since p33/ING1 cooperates with p53 in mammalian cell growth control and apoptosis (16, 39, 51), we used the yeast-based system to analyze the requirement of Yng2p/NuA4 for p53-dependent transcriptional activation in vivo. The cells are transformed with two low-copy plasmids: one expressing mammalian p53 through the yeast ADH1 constitutive promoter and the other a reporter with p53-binding sites driving the HIS3 gene (13). In the genetic background used here only cells expressing the reporter gene will grow in the absence of histidine, reflecting p53-dependent activation of the HIS3 gene. Since p53-dependent transcriptional activation of the inhibitor of cyclin-dependent kinases p21/WAF1 depends on p33/ING1 (16), we used the reporter plasmid carrying the p53-binding sites found in the p21/WAF1 promoter (13).
Wild-type and Δyng2 cells were transformed with the plasmid mentioned above and p21-HIS3 expression was analyzed by Northern blot (Fig. 5A). In the wild-type strain, a strong signal is detected only when p53 is coexpressed (compare lanes 1 and 2). In contrast, the yng2 mutant cells show very limited transcription of the p21-HIS3 gene in presence of p53 (lanes 3 and 4). When we used the ACT1 signal as an internal control, the effect of the YNG2 deletion on p53-dependent transcriptional activation was >6-fold. This value is very close to the effect of p33/ING1 obtained on endogenous p21/WAF1 transcription in mammalian cells (16). These results are not due to indirect effect on p53 expression since ADH1 gene transcription is not affected by NuA4 and Western analysis shows equivalent p53 protein levels in both wild-type and mutant cells (Fig. 5A, lower panel). This striking role of Yng2p/NuA4 in transcriptional activation is activator specific since Gal4p-dependent GAL1 gene induction is not affected in mutant cells (Fig. 5B).
p53 physically interacts with the NuA4 complex in vitro and in vivo.
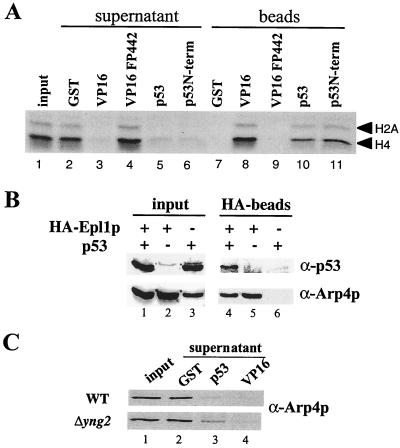
Since p33/ING1 is found physically associated with p53 in mammalian cells (16), we sought to determine whether the Yng2p-containing complex NuA4 could associate with p53 in vitro (Fig. 6A). GST pulldown assays were performed and clearly show that p53 efficiently binds NuA4 and brings down its H4 and H2A HAT activity on the beads (compare lanes 5 and 10 to the GST control in lanes 2 and 7). The very efficient binding is similar to samples containing the VP16 activation domain (compare lanes 3 and 8 to lanes 5 and 10), a functional interaction resulting in NuA4 recruitment to promoters for targeted chromatin acetylation and transcriptional activation in vitro (44, 46). The interaction is specific to the transactivator domain since a VP16 transcription mutant does not deplete NuA4 activity (see lanes 4 and 9). Furthermore, p53 binding to NuA4 occurs through its N-terminal region, known to be important for gene-specific transcriptional activation (aa 1 to 292; Fig. 6A, lanes 6 and 11). To confirm the biological relevance of the p53-NuA4 physical interaction, we performed coimmunoprecipitation studies in whole-cell extracts (Fig. 6B). We used isogenic strains carrying the p53 expression vector and expressing physiological levels of an HA-tagged version of Epl1p, an essential subunit of the NuA4 complex (15; A. Boudreault, D. Cronier, and J. Côté, unpublished data). HA-Epl1p immunoprecipitations with another subunit, Arp4p, as a marker indicating that p53 is indeed found specifically associated with NuA4 in the cell (see lanes 4 to 6). This argues that p53 activation domain is able to recruit NuA4 at the promoter region during transcriptional activation.
FIG. 6.
p53 interacts in vitro and in vivo with yeast NuA4 HAT complex. (A) Native NuA4 complex interacts with mammalian p53 protein. A fluorogram shows depletion (lanes 3, 5, and 6) and recovery (lanes 8, 10, and 11) of NuA4 HAT activity (Superose-6 fraction) on GST-p53, GST-p53N-terminal (aa 1 to 292), and GST-VP16 beads but no interaction with control GST or GST-VP16FP442 mutant beads (lanes 7 and 9). The lanes contain equivalent amount of fractions compared to the input. (B) p53 interacts with NuA4 in vivo. Total proteins from strains QY108a (expressing HA-Epl1p [lanes 1, 2, and 5]) and BY4741 (untagged isogenic to QY108a [lanes 3 and 6]) were transformed or not with p53-expressing plasmid (pLS76) as indicated and immunoprecipitated with HA antibody. Input proteins (lanes 1 to 3) and proteins bound to HA-beads (lanes 4 to 6) were analyzed by Western blot with anti-p53 and anti-Arp4p antibodies. The NuA4 subunit Arp4p coimmunoprecipitates with p53 only in the HA-Epl1-expressing extract (lane 4). (C) p53-NuA4 interaction is affected by the deletion of YNG2. An equal quantity of NuA4 complex purified from deleted strain (QY203, Δyng2) expressing HA-Yng2p (WT) or not (Δyng2) (Superose-6 peak fractions, Fig. 4B) was used in a pulldown experiment with GST, GST-p53, and GST-VP16. Supernatant proteins were analyzed by Western blot by using anti-Arp4p antibody. The Δyng2 complex is less depleted by GST-p53 than by GST-VP16. (Western analysis of the beads was not possible because of high nonspecific background signals.)
We then analyzed the role of Yng2p in NuA4 interaction with p53. GST pulldown assays were done with partially purified wild-type and Yng2-less complexes using VP16 and p53 as baits (Fig. 6C). GST-VP16 beads depleted as efficiently wild-type and mutant complexes (lane 4 versus lane 2), which most likely reflects the presence of Tra1p in both complexes (7). GST-p53 depletion of NuA4 was also seen in both cases, although a small but significant amount of Arp4p was still detected in the mutant complex supernatant compared to the wild type (compare lower and upper panels in lane 3 to lane 2). This weaker interaction could reflect a conformational change in the mutant complex. Attempts at detecting direct physical interaction between bacterially expressed Yng2p and p53 were unsuccessful (data not shown).
Yng2p PHD-finger domain is important for transcriptional activation but not for NuA4 HAT activity.
To investigate the role of Yng2p domains in transcription regulation and NuA4 function, we analyzed cells expressing truncated versions of the protein (as in Fig. 3). p53-dependent transcriptional activation was also crippled in cells expressing only the PHD-finger domain region (aa 154 to 282), albeit to a lower extent (threefold) than for Δyng2 cells. This is consistent with the growth phenotype and the idea that the protein lacks proper interface for its association with NuA4 (Fig. 7, compare lanes 1 and 3). Surprisingly, The yng2 mutant without the PHD-finger domain (aa 1 to 218) similarly cripples p53 transactivator function (lane 2), while growth is not affected (Fig. 3B). Both mutant forms of the protein also provoke lower expression of endogenous PHO5 gene while GAL1 is not affected. To further characterize the role of the Yng2p PHD-finger, we partially purified wild-type and mutant complexes (Fig. 7B). Strikingly, and in agreement with the growth phenotype, the NuA4 complex lacking the PHD-finger behaves similarly to the wild-type complex in amount, size, and specific activity. GST pulldown assays also indicate that the complex retains equivalent p53-binding affinity (data not shown). These data imply that Yng2p PHD-finger plays a role in transcriptional activation in an aspect that does not include the direct recruitment of NuA4 HAT activity.
Transcriptional activation by p53 is associated with targeted Yng2-dependent histone H4-specific hyperacetylation.
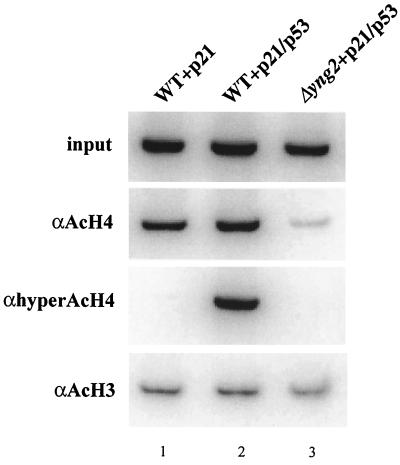
We found that Esa1p/NuA4 is not able to acetylate p53 in vitro, arguing against a role similar to GCN5/PCAF and CBP/p300 (data not shown) (49). Altogether, the present results demonstrate the requirement of Yng2p for p53 function in yeast, supporting a conserved role between the yeast protein and candidate tumor suppressor p33/ING1. Yng2p-dependent activation of transcription by p53 could occur through recruitment of NuA4 at the p21/WAF1 promoter and subsequent chromatin modification. To confirm this model, we performed chromatin immunoprecipitation experiments with specific antibodies recognizing acetylated or hyperacetylated forms of histone H4 and acetylated histone H3 (Fig. 8). Chromatin from three strains containing the p21-HIS3 low-copy-number plasmid was prepared: two wild-type strains either expressing or not p53 and a Δyng2 strain expressing p53. Primers amplifying the p21-HIS3 promoter region were used to show that the presence of p53 in the cell provokes a reproducible increase of acetylated histone H4 on the region (∼2-fold on average based on three repeats, compare lanes 1 and 2 in the AcH4 panel). The effect is a lot more dramatic when an antibody recognizing only the hyperacetylated isoforms of H4 is used (lanes 1 and 2, hyperAcH4 panel). Accordingly, we have shown in a previous report that NuA4 can create fully tetra-acetylated histone H4 isoform (2). As expected, deletion of YNG2 correlates with the disappearance of acetylated histone H4 on the p21-HIS3 chromatin (lane 3, AcH4 and hyperAcH4 panels). The decrease brings the level of AcH4 even below the level in the absence of p53 (compare lanes 1 and 3), in agreement with an effect of YNG2 deletion on bulk histone H4 acetylation (29). It is very interesting that the same p21-HIS3 chromatin does not show any variation in levels of histone H3 acetylation under the same conditions (lanes 1 to 3, AcH3 panel). These results illustrate that the NuA4 complex, such as the SAGA complex, can be recruited in vivo by a transactivator to create a chromatin region containing specific hyperacetylated histones important in the process of transcriptional activation.
FIG. 8.
Yng2-dependent specific targeting by p53 of histone H4 acetylation to the p21-HIS3 promoter. The Δyng2 strain (QY203) containing an episomal YNG2 gene (pAN104) or an empty vector and the reporter plasmid carrying the p21-HIS3 transcription unit (pSS1) was transformed or not with the p53-expressing plasmid (pYD102). Cultures were grown in minimal medium supplemented with the corresponding auxotrophies to an OD of 1 and then fixed with formaldehyde. The chromatin of each strain was extracted, sonicated, and immunoprecipitated with antiacetylated histone H4, antihyperacetylated histone H4, and antiacetylated histone H3 antibodies as indicated. Input (upper panel) and bound (lower three panels) fractions were assayed for the presence of p21-HIS3 promoter region by PCR amplification (in a ratio of 1:100 for input versus bound). Reactions were analyzed on a 1× TBE–6% polyacrylamide gel. An enhanced level of histone H4 acetylation at the p21-HIS3 promoter is observed in the strain expressing both Yng2p and p53 (compare lanes 1 and 2 for antiacetylated H4 and antihyperacetylated histone H4), while H3 acetylation levels remained constant for all strains. The absence of Yng2p caused a dramatic decrease of H4 acetylation (compare lanes 2 and 3) even below the level present in absence of p53 (compare lanes 1 and 3). These results were reproduced with a different chromatin preparation.
DISCUSSION
The NuA4 HAT complex has the unique specificity in yeast of modifying histone H4 and H2A N termini in chromatin, through its essential subunit Esa1p (2). Its activity can be recruited by DNA-bound activators to create a hyperacetylated chromatin region and stimulate in vitro transcription in an acetyl coenzyme A- and chromatin-dependent manner (44, 46). Indeed, mutations in three NuA4 subunits provoke gene-specific transcription defects in vivo (14, 15). Several lines of evidence suggest that NuA4 is involved in the control of cell proliferation through gene-specific transcription regulation. A number of NuA4 subunits are essential for cell growth, and conditional alleles of ESA1 arrest the cells at the G2/M border in the cell cycle when kept at a nonpermissive temperature (11, 15). Furthermore, mammalian homologs of NuA4 subunits have been implicated in the control of cell proliferation. Tra1p is highly related to human TRRAP, an ATM/phosphatidylinositol 3-kinase-related cofactor which associates with c-Myc and E2F transactivation domains and is essential for their oncogenic potential (31). Eaf3p is a close homolog of human MRG15, whose truncated form can induce senescent-like phenotype in immortal cell lines (4). We now have identified a novel subunit of NuA4, Yng2p, a PHD-finger protein highly related to the candidate tumor suppressor p33/ING1 that associates and cooperates with p53 in cell growth control. We demonstrate that Yng2p plays a primary role within NuA4 for transcription regulation and cell growth and is required for p53-dependent transcriptional activation in yeast and targeted histone H4 acetylation to p53-bound chromatin.
A recent independent study reported that disruption of the YNG2 gene creates a severe growth phenotype similar to the one reported here (29), except for the ability to use galactose as carbon source (see Fig. 3B). The report also showed, by using multicopy episomal overexpression, that Yng2p lacking the PHD-finger region was sufficient to suppress the growth defect and HA-Yng2p could coimmunoprecipitate HAT activity and two overexpressed subunits of NuA4, Tra1p and Esa1p. We now demonstrate with physiological protein levels and chromatography, mass spectrometry, immunoblotting, and immunoprecipitation experiments that Yng2p is a stable stoichiometric subunit of the purified NuA4 complex. Furthermore, characterization of the mutant complex lacking Yng2p illustrated its primary role in NuA4 structure and HAT activity. This concurs with a decrease of bulk H4 acetylation detected in esa1 and yng2 mutant cells and the loss of p53-dependent localized H4 hyperacetylation at the p21 promoter (11, 29, 35, 47) (Fig. 8). Although it was reported that overexpressed PHD-finger alone could not suppress growth defect of the mutant strain, we show that the physiological expression of Yng2p putative nuclear localization motif along with the PHD-finger can partially restore growth (Fig. 3B). Yng2p PHD-finger domain is clearly not involved in NuA4 HAT activity or recruitment by p53 but still has an important role in the level of transcriptional activation (Fig. 7). Interestingly, natural mutations of the AIRE gene, which lead to autoimmune disease affecting endocrine glands, produce a protein deleted for the PHD-finger domain, which provokes loss of the normal speckled nuclear localization (37). An extended PHD-finger present in human AF10 was also shown to mediate homo-oligomerization (28). Finally, tumor-specific mutations were found in Yng2p-related p33/ING1 PHD-finger and nuclear localization motif (21).
We demonstrated functional conservation between human p33/ING1 and yeast Yng2p for physical association and cooperativity with p53. Transcriptional activation by p53 of the p21/WAF1 promoter is dependent on Yng2p in yeast while it is affected by p33/ING1 in human cells. In vitro and in vivo association of p53 with the yeast NuA4 complex suggests that p33/ING1 or another human ING1 family member could be part of a human NuA4-related complex that is recruited by p53 for transcriptional activation of genes regulating cell proliferation, such as the cyclin-dependent kinase inhibitor p21/WAF1. Accordingly, overexpression of human p33/ING1 in yeast can suppress growth defects of yng2 mutant cells (29). Induction of p21/WAF1 gene transcription is known to be regulated, at least in part, by histone acetylation of the promoter-associated chromatin (36). On the other hand, activation of p21/WAF1 expression by histone deacetylase inhibitors is not dependent on promoter-bound p53 (3). E2F was also shown to bind p21/WAF1 promoter and to activate transcription (19), suggesting that it could still recruit a NuA4-like complex at this promoter through its TRRAP/Tra1p subunit (31). Most strikingly, a human HAT closely related to Esa1p, TIP60, was recently described as part of a multisubunit complex harboring other homologs of yeast NuA4 subunits, including TRRAP (23). It was also shown that ectopic expression of a dominant-negative mutant TIP60 lacking acetyltransferase activity results in defective DNA repair and inhibition of apoptosis (23). p53 transcriptional activity is essential for p53-dependent apoptosis after DNA damage (9). In agreement with a conserved role in DNA repair, Δyng2 mutant cells are sensitive to a DNA-damaging agent (Fig. 3C). This suggests a role for p33/ING1 or its human paralogs in the regulation of DNA repair and apoptosis through recruitment by p53 within a NuA4-like complex. Importantly, p33/ING1 is also involved in the regulation of p53-independent apoptosis (22). In conclusion, this study firmly links the NuA4 HAT complex to the control of cell proliferation and demonstrates for the first time in vivo the role of NuA4-dependent targeted histone H4 hyperacetylation in gene-specific transcriptional activation.
ACKNOWLEDGMENTS
We are grateful to C. Di Como and C. Prives for the ADH1:p53/p21:HIS3 plasmids; D. Stillman for the anti-Act3/Arp4 and the HA-Act3/Arp4-expressing yeast strain; A. Anderson for the anti-p53 and mouse p53 cDNA; A. Delahodde, S. Hermann, C. Jacq, and G. Hautbergue for sending us many reagents; K. Pierce for expert microcapillary HPLC-mass spectrometry; S. Berger for the anti-Ada2; J. Workman for the anti-Tra1; B. Cairns for the YBC76 diploid strain; and P. Philippsen for the plasmid pFaKanMX4. We also thank A. Boudreault for the HA-Epl1p expression vector and other members of our lab for their help and encouragement.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to J.C. A.N. and R.T.U. are CIHR postdoctoral fellows. Y.D. is a Natural Sciences and Engineering Research Council (NSERC) graduate student. J.C. is a CIHR scholar.
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Allard S, Utley R T, Savard J, Clarke A, Grant P, Brandl C J, Pillus L, Workman J L, Côté J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer S Y, Meng S, Shei A, Hodin R A. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc Natl Acad Sci USA. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram M J, Berube N G, Hang-Swanson X, Ran Q, Leung J K, Bryce S, Spurgers K, Bick R J, Baldini A, Ning Y, Clark L J, Parkinson E K, Barrett J C, Smith J R, Pereira-Smith O M. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonneaud N, Ozier-Kalogeropoulos O, Li G Y, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. colishuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 6.Brady M E, Ozanne D M, Gaughan L, Waite I, Cook S, Neal D E, Robson C N. Tip60 is a nuclear hormone receptor coactivator. J Biol Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 7.Brown C E, Howe L, Sousa K, Alley S C, Carrozza M J, Tan S, Workman J L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 8.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 9.Chao C, Saito S, Kang J, Anderson C W, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 11.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn F G, Scheidereit C, Leutz A. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene. 1999;18:3316–3323. doi: 10.1038/sj.onc.1202717. [DOI] [PubMed] [Google Scholar]
- 13.Di Como C J, Prives C. Human tumor-derived p53 proteins exhibit binding site selectivity and temperature sensitivity for transactivation in a yeast-based assay. Oncogene. 1998;16:2527–2539. doi: 10.1038/sj.onc.1202041. [DOI] [PubMed] [Google Scholar]
- 14.Eisen A, Utley R T, Nourani A, Allard S, Schmidt P, Lane W S, Lucchesi J C, Cote J. The yeast NuA4 and DrosophilaMSL complexes contain homologous subunits important for transcription regulation. J Biol Chem. 2001;276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- 15.Galarneau L, Nourani A, Boudreault A A, Zhang Y, Héliot L, Allard S, Savard J, Lane W S, Stillman D J, Côté J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 16.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 17.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 18.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gartel A L, Goufman E, Tevosian S G, Shih H, Yee A S, Tyner A L. Activation and repression of p21(WAF1/CIP1) transcription by RB binding proteins. Oncogene. 1998;17:3463–3469. doi: 10.1038/sj.onc.1202240. [DOI] [PubMed] [Google Scholar]
- 20.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 21.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 22.Helbing C C, Veillette C, Riabowol K, Johnston R N, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 23.Ikura T, Ogryzko V V, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 24.Ishioka C, Frebourg T, Yan Y X, Vidal M, Friend S H, Schmidt S, Iggo R. Screening patients for heterozygous p53 mutations using a functional assay in yeast. Nat Genet. 1993;5:124–129. doi: 10.1038/ng1093-124. [DOI] [PubMed] [Google Scholar]
- 25.John S, Howe L, Tafrov S T, Grant P A, Sternglanz R, Workman J L. The Something About Silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 26.Krajewski W A, Becker P B. Reconstitution of hyperacetylated, DNase I-sensitive chromatin, characterized by high conformational flexibility of nucleosomal DNA. Proc Natl Acad Sci USA. 1998;95:1540–1545. doi: 10.1073/pnas.95.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo M H, Allis C D. In vivo cross-linking and immunoprecipitation for studying dynamic protein-DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 28.Linder B, Newman R, Jones L K, Debernardi S, Young B D, Freemont P, Verrijzer C P, Saha V. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000;299:369–378. doi: 10.1006/jmbi.2000.3766. [DOI] [PubMed] [Google Scholar]
- 29.Loewith R, Meijer M, Lees-Miller S P, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorch Y, Beve J, Gustafsson C M, Myers L C, Kornberg R D. Mediator-nucleosome interaction. Mol Cell. 2000;6:197–201. doi: 10.1016/s1097-2765(00)00021-6. [DOI] [PubMed] [Google Scholar]
- 31.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 32.Niedenthal R, Riles L, Guldener U, Klein S, Johnston M, Hegemann J H. Systematic analysis of S. cerevisiaechromosome VIII genes. Yeast. 1999;15:1775–1796. doi: 10.1002/(SICI)1097-0061(199912)15:16<1775::AID-YEA496>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oki E, Maehara Y, Tokunaga E, Kakeji Y, Sugimachi K. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett. 1999;147:157–162. doi: 10.1016/s0304-3835(99)00288-8. [DOI] [PubMed] [Google Scholar]
- 35.Reid J L, Iyer V R, Brown P O, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 36.Richon V M, Sandhoff T W, Rifkind R A, Marks P A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinderle C, Christensen H M, Schweiger S, Lehrach H, Yaspo M L. AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered subcellular distribution of mutants lacking the PHD zinc fingers. Hum Mol Genet. 1999;8:277–290. doi: 10.1093/hmg/8.2.277. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinoura N, Muramatsu Y, Nishimura M, Yoshida Y, Saito A, Yokoyama T, Furukawa T, Horii A, Hashimoto M, Asai A, Kirino T, Hamada H. Adenovirus-mediated transfer of p33ING1 with p53 drastically augments apoptosis in gliomas. Cancer Res. 1999;59:5521–5528. [PubMed] [Google Scholar]
- 40.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith E R, Pannuti A, Gu W, Steurnagel A, Cook R G, Allis C D, Lucchesi J C. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokunaga E, Maehara Y, Oki E, Kitamura K, Kakeji Y, Ohno S, Sugimachi K. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett. 2000;152:15–22. doi: 10.1016/s0304-3835(99)00434-6. [DOI] [PubMed] [Google Scholar]
- 43.Toyama T, Iwase H, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, Riabowol K. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 44.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 45.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 46.Vignali M, Steger D J, Neely K E, Workman J L. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 2000;19:2629–2640. doi: 10.1093/emboj/19.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 48.Wach A, Brachat A, Rebischung C, Steiner S, Pokorni K, Heesen S, Philippsen P. PCR-based gene targeting in Saccharomyces cerevisiae. Methods Microbiol. 1998;26:67–81. [Google Scholar]
- 49.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 50.Yan Y, Barlev N A, Haley R H, Berger S L, Marmorstein R. Crystal structure of yeast esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 51.Zeremski M, Hill J E, Kwek S S, Grigorian I A, Gurova K V, Garkavtsev I V, Diatchenko L, Koonin E V, Gudkov A V. Structure and regulation of the mouse ing1 gene. Three alternative transcripts encode two phd finger proteins that have opposite effects on p53 function. J Biol Chem. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Ahn J, Wilson S H, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20:914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]