Abstract
Chronic exposure to arsenic (As) remains a global public health concern and our understanding of the biological mechanisms underlying the adverse effects of As exposure remains incomplete. Here, we used a high-resolution metabolomics approach to examine how As affects metabolic pathways in humans.
We selected 60 non-smoking adults from the Folic Acid and Creatine Trial (FACT). Inorganic (AsIII, AsV) and organic (monomethylarsonous acid [MMAs], dimethylarsinous Acid [DMAs]) As species were measured in blood and urine collected at baseline and at 12 weeks. Plasma metabolome profiles were measured using untargeted high-resolution mass spectrometry. Associations of blood and urinary As with 170 confirmed metabolites and >26,000 untargeted spectral features were modeled using a metabolome-wide association study (MWAS) approach. Models were adjusted for age, sex, visit, and BMI and corrected for false discovery rate (FDR).
In the MWAS screening of confirmed metabolites, 17 were associated with ≥1 blood As species (FDR<0.05), including fatty acids, neurotransmitter metabolites, and amino acids. These results were consistent across blood As species and between blood and urine As. Untargeted MWAS identified 423 spectral features associated with ≥1 blood As species. Unlike the confirmed metabolites, untargeted model results were not consistent across As species, with AsV and DMAs showing distinct association patterns. Mummichog pathway analysis revealed 12 enriched metabolic pathways that overlapped with the 17 identified metabolites, including one carbon metabolism, tricarboxylic acid cycle, fatty acid metabolism, and purine metabolism.
Exposure to As may affect numerous essential pathways that underlie the well-characterized associations of As with multiple chronic diseases.
Keywords: arsenic, metabolomics, one carbon metabolism, TCA cycle, folate, folic acid
Graphical Abstract
Introduction
Chronic exposure to inorganic arsenic (InAs) in drinking water is a global health concern that afflicts >140 million people in 70+ countries and contributes to multiple chronic diseases, including cancer1–3, cardiovascular disease4,5, and metabolic dysfunction6,7. In Bangladesh, exposure to As began in the 1970s, when nongovernmental organizations advocated a massive switch from drinking microbially contaminated surface water to groundwater in an effort to reduce infant mortality due to diarrheal disease. This led to excessive consumption of groundwater contaminated with naturally occurring As, with concentrations ranging from nondetectable to levels exceeding 1,000 μg/L8. This widespread exposure is further complicated by limited scalable, economical, and effective exposure prevention or mitigation strategies. The metabolism of InAs in the human body is well characterized. Arsenate (AsV) is reduced to the more toxic inorganic form, arsenite (AsIII), before being methylated and oxidized to monomethylarsonic acid (MMAs) by arsenic-3-methyltransferase, which is further methylated to dimethylarsinic acid (DMAs). Efficient methylation of InAs to DMAs is important because studies have shown that intermediates such as AsIII and trivalent MMAs are among the most toxic forms of As9–11.
Despite clear evidence that As contributes to many chronic diseases, the mechanisms underlying exposure-disease relationships remains poorly understood. Part of the reason for this knowledge gap is that mechanistic studies require model organisms, but rodents are remarkably efficient in methylation of As and are less susceptible to As toxicity than humans, with the exception of in utero exposure windows12. Thus, new approaches using human populations are needed to complement existing data to characterize As-affected biological pathways. Identification of these effects and pathways, especially among individuals without overt disease, will help us better identify individuals at risk and design effective mitigation strategies.
Metabolomic studies can be used to identify key metabolic changes and toxic mechanisms underlying environmental exposures and the associated health consequences. By profiling the metabolome, we can directly assess the functional effect of environmental exposures downstream from gene expressionand epigenetic changes and more proximal to health-relevant endpoints. Untargeted metabolomics, which aims to comprehensively measure metabolic changes in response to an exposure or disease outcome, provide a key strategy to test targeted hypotheses on biological changes underlying As exposure as well as generate new insight into systemic biological alterations underlying As toxicity.
Studies of As-impacted populations suggests that As exposure is associated with metabolic alterations in urine13–16 and blood14. For example, a study in Mexico found that urinary total As levels were associated with numerous changes in plasma and urinary metabolites among individuals with and without diabetes14. However, there were minimal overlapping metabolites differing between individuals with and without diabetes. This result suggests there may be specific metabolic responses that set certain exposed individuals on a path to diabetes or poor metabolic health. Overall, these studies highlight the need to comprehensively characterize metabolic patterns and pathways underlying As toxicity, particularly prior to disease onset.
In this study, we leveraged comprehensive metabolomic profiling to evaluate the impact of As exposure and characterize potential mechanisms underlying As toxicity using samples collected as part of the Folic Acid and Creatine Trial (FACT). The original FACT study showed that daily supplementation of folic acid (FA) decreased total As levels in blood17 and increased the efficiency of As metabolism18 after 6–12 weeks. In the current study, we used a subset of participants from the FACT study to investigate the relationships of As exposure with untargeted ultra-high-resolution mass spectrometry to identify key pathways underlying the relationship between As exposure and health outcomes.
Methods
Study Population and Study Design:
The Folic Acid and Creatine Trial (FACT) is a unique clinical trial of 610 Bangladeshi adults chronically exposed to As-contaminated drinking water. Starting in 2010, 610 participants were recruited from the Health Effects of Arsenic Longitudinal Study (HEALS) cohort based in Araihazar, Bangladesh, a parent cohort in which As exposure had been well characterized since its inception in 200019. FACT participants were randomly selected given the following criteria: inclusion criteria were adults between 20–75 years of age who had been drinking from their current well with water As > 50 μg/L for at least 3 years. We excluded women who were pregnant, individuals taking nutritional supplements, individuals with proteinuria, and individuals with known renal disease, diabetes, or gastrointestinal or other health problems. Once enrolled, FACT participants were given arsenic water filters and block randomized separately for men and women into one of five treatment groups: placebo, 400 or 800 μg FA, creatine, or creatine+FA. Compliance was monitored by observing pill ingestion, inquiring about compliance and through pill counts; the latter indicated a median compliance of 99.5%. For the current study, starting from all participants who did not smoke nor chewed betel nut, we randomly selected 60 FACT participants (30 each from the 800 μg FA and placebo groups), specifying 50/50% male/female in each group.
Informed consent was obtained by our Bangladeshi field staff physicians. Ethical approval was obtained from the Institutional Review Board of Columbia University Medical Center and the Bangladesh Medical Research Council.
Blood and Urine Arsenic Concentrations:
Details of blood collection and As measurement have been described elsewhere17. In brief, venous blood was collected in Ethylenediaminetetraacetic acid (EDTA) vacutainers from participants at baseline and week 12. Arsenic species (AsIII, AsV, MMAs, and DMAs) from both blood and urine were measured using high performance liquid chromatography coupled to dynamic reaction cell inductively coupled plasma mass spectrometer20,21. The method can detect AsIII and AsV, but samples can oxidize during processing, particularly urine samples, resulting in exposure misclassification of the absolute and relative levels of AsIII and AsV. Blood As levels, expressed in absolute concentrations, are less susceptible to oxidation and hydration status and better reflect As exposure to tissues. Thus, for the present analysis, blood As species were the primary exposures of interest while specific gravity-corrected urinary InAs levels were used in sensitivity analyses to validate the observed associations for blood As.
Blood levels of AsIII and AsV were summed to create a single variable reflecting total inorganic As concentrations (∑InAs). We also derived %InAs, %MMAs, and %DMAs in urine as biomarkers of As metabolism. These As metabolism biomarkers were used in sensitivity analyses to assess potential confounding of our associations by As metabolic efficiency.
High Resolution Metabolomic Profiling:
We measured plasma metabolomic profiles at baseline and week 12 using untargeted high-resolution metabolomics22. Plasma samples were thawed, treated with 130 μL of acetonitrile containing mixture of 14 stable isotope standards to remove proteins and centrifuged; the resulting extract was transferred to a low-volume vial that was placed in an autosampler maintained at 4°C23. Sample extracts were analyzed in triplicate by liquid chromatography-Fourier transform mass spectrometry using a Thermo Scientific Ultimate 3000 liquid chromatography system interfaced to a Fusion Tribrid Orbitrap high-resolution mass spectrometer.23,24 Analyte separation was accomplished using dual column chromatography with C18 and hydrophilic interaction liquid chromatography (HILIC). HILIC columns were run in positive electrospray ionization (ESI) mode while C18 was run in negative ESI mode. The resulting spectral data were processed to provide data tables containing mass m/z, retention time(s), intensity, coefficient of variation, and related descriptive characteristics, including minimal information standards for metabolomics data using apLCMS25–27 and xMSanalyzer28. The untargeted metabolomics analysis comprised 12992 HILIC+ and 15549 C18- features in total, of which 98 (HILIC+) and 74 (C18-) metabolite identities were confirmed by comparison to a database of authentic standards ran on the same platform and confirmed using MSMS. For features without confirmed identities, we used xMSannotator with the HMDB database29,30. In brief, xMSannotator allows feature clustering to combine the feature m/z (mass tolerance ±5 ppm), retention time (±5 seconds), ion intensity profiles, mass defect, and expected isotopic and adduct patterns to assign predicted metabolite annotations for detected features.
Metabolite Data Processing:
Raw feature tables were processed with established filter and normalization criteria. Daily batches were normalized using ComBat. Two individuals were identified as outliers via visualization of principal component analysis and were subsequently removed. Features that were not detected in at least 50% of samples (2000 features, 7%) were removed. For the remaining features, values below detection were replaced with the lowest observed value divided by square root of 2. All samples were then quantile normalized and log2 transformed. For confirmed metabolites, two C18 metabolites were dropped due to <50% detection rates, resulting in 98 HILIC+ and 72 C18- metabolites available for analysis. In total, 12413 HILIC+ and 14128 C18- m/z features without confirmed identities were available for analysis.
Statistical Analysis:
To estimate the association of blood As species (AsIII, AsV, ∑InAs, MMAs, and DMAs) with confirmed metabolites or unconfirmed features, we used generalized estimating equation (GEE) with identity link and Gaussian outcome likelihood to account for the repeated measures. In these models, blood As species were modeled as the predictor variables while log2-transformed metabolite abundance was modeled as the outcome variables. Because we log2-transformed the semi-quantitative metabolic data, the model coefficients were then converted to concentration ratios (CR) via exponentiation of regression coefficients, which can be interpreted as the relative change for that specific metabolite/feature for every μg/L increase in As species. Due to differences in the confidence of metabolite identification, we used this metabolome wide association study (MWAS) approach to analyze confirmed metabolites and unconfirmed features separately. All models were adjusted for age, sex, and BMI.
One primary finding of the parent FACT trial was that FA supplementation increased the metabolism of InAs to DMAs18. To test whether FA treatment impacted our MWAS results as a potential confounder, we conducted a sensitivity analysis comparing the estimates of the multivariable model with and without adjusting for treatment group, including an interaction term between treatment and time. Then, we tested whether FA supplementation may have altered the As related metabolic changes as a potential effect modifier. To do so, we used only week 12 metabolomic data and fitted multivariable linear regression models with blood As species as predictor variables, metabolite abundance as outcome, and an added interaction term between treatment group and As exposure at baseline. We tested for the statistical significance of the interaction term via likelihood ratio tests comparing models with and without this interaction term to calculate the p-values. All models in this analysis were adjusted for age, sex, and BMI. To minimize the possibility of spurious findings, we restricted this sub-analysis to only confirmed metabolites that were associated with at least one blood As species in the MWAS analysis.
While blood As species were our primary exposures of interest, we also compared model results using blood versus urinary InAs biomarkers as another sensitivity analysis. We did not compare methylated species in urine directly to blood concentrations due to well-known differences in their excretion patterns31. All urinary As biomarkers were corrected for specific gravity and the models were adjusted for age, sex, and BMI.
Finally, we specifically investigated phosphatidylcholine (PC) to phosphatidylethanolamine (PE) due to their abundance, previous known relationship with key As metabolism enzymes, link to chronic disease32,33, and dependence on methyl groups generated by one carbon metabolism (OCM). While our targeted panel does not contain PC and PE specifically, it includes lysoPCs and lysoPEs, derivatives of PC and PE, respectively. We summed all lysoPCs and lysoPEs and calculated the ratio of lysoPC to lysoPE as a proxy to PC/PE ratio. All models were adjusted for age, sex, and BMI.
Pathway Analysis
We performed pathway enrichment analysis using Mummichog34, which was developed specifically for untargeted high resolution metabolomics data. For each blood As species, we ran the Mummichog algorithm on the MetaboAnalyst platform35 specifying mixed mode (both positive and negative ion modes), 5 parts per million (ppm) mass tolerance, and included retention time, p-values, and Z-scores from the GEE models. Our p-value cutoff was 0.05 and only used pathways containing at least 3 entries. For visualization, putative metabolite Kyoto Encyclopedia of Genes and Genomes (KEGG) IDs for significant features generated through Mummichog pathway analysis were used to populate the KEGG human metabolic map using iPATH36.
Results
Descriptive Features and Arsenic Distribution
Age and BMI were similar between the placebo and treatment groups (Table 1). Baseline blood As distribution was also similar with respect to total metabolite concentrations (8.84 μg/L in placebo vs. 8.80 μg/L in FA supplementation) as well as relative concentrations of AsV, AsIII, MMAs, and DMAs (Table 1). Since we gave water filters to all participants to reduce As exposure, both groups showed somewhat lower blood As concentrations at week 12 (Supplemental Table 1). Arsenic metabolite concentrations showed strong correlation with one another (Spearman r ≥0.87), with the exception of AsV, which showed a moderate correlation with other metabolites (0.45 < Spearman r < 0.61) (Supplemental Figure 1). Consistent with the overall FACT study, the 800 μg/day FA treatment group selected for this study had lower concentrations of As species compared to the placebo group after 12 weeks (Supplemental Table 1), supporting that FA supplementation facilitates the elimination of As from blood17.
Table 1.
Baseline characteristics of the participants in the pilot study.
| Placebo (n=30) | Treatment (n=30) | p-value | |
|---|---|---|---|
| Mean (SD) | Mean (SD) | ||
| Age | 34.43 (6.60) | 36.83 (6.58) | 0.16 |
| BMI | 21.18 (2.97) | 20.00 (2.63) | 0.11 |
| Sex | |||
| Male | 15 (50.0) | 15 (50.0) | 1.00 |
| Female | 15 (50.0) | 15 (50.0) | |
| Baseline Blood Arsenic (ug/L) | |||
| ASV | 0.27 (0.25) | 0.36 (0.36) | 0.29 |
| ASIII | 1.99 (0.90) | 1.98 (0.79) | 0.95 |
| MMA | 3.95 (2.23) | 3.85 (1.80) | 0.86 |
| DMA | 2.63 (1.71) | 2.61 (1.79) | 0.97 |
| ∑inAs1 | 2.26 (1.06) | 2.34 (1.00) | 0.79 |
| ∑Total2 | 8.84 (4.87) | 8.80 (4.37) | 0.97 |
Sum of AsIII and AsV
Sum of all metabolites
Standard Confirmed Metabolites
MWAS with the 170 confirmed metabolites as outcomes identified 17 metabolites associated with at least one of AsV, AsIII, ∑InAs, MMAs, and DMAs at the FDR <0.05 when adjusting for age, sex, and BMI. Among all As species, AsIII, the trivalent inorganic As species, had the most statistically significant associations (n=12 at FDR < 0.05), followed by MMAs (n=7), AsV (n=4), and DMAs (n=2). Although the number of statistically significant associations differed by metabolite, when results were compared across different As species, model results overlapped and were largely consistent across all blood As species. As shown in Figure 1, 16 of the 17 identified metabolites were nominally associated with four of the five blood As species (p-value<0.10) and have the same direction of association. Full results of the MWAS models, including metabolite information, can be found in Supplemental Table 2.
Figure 1.
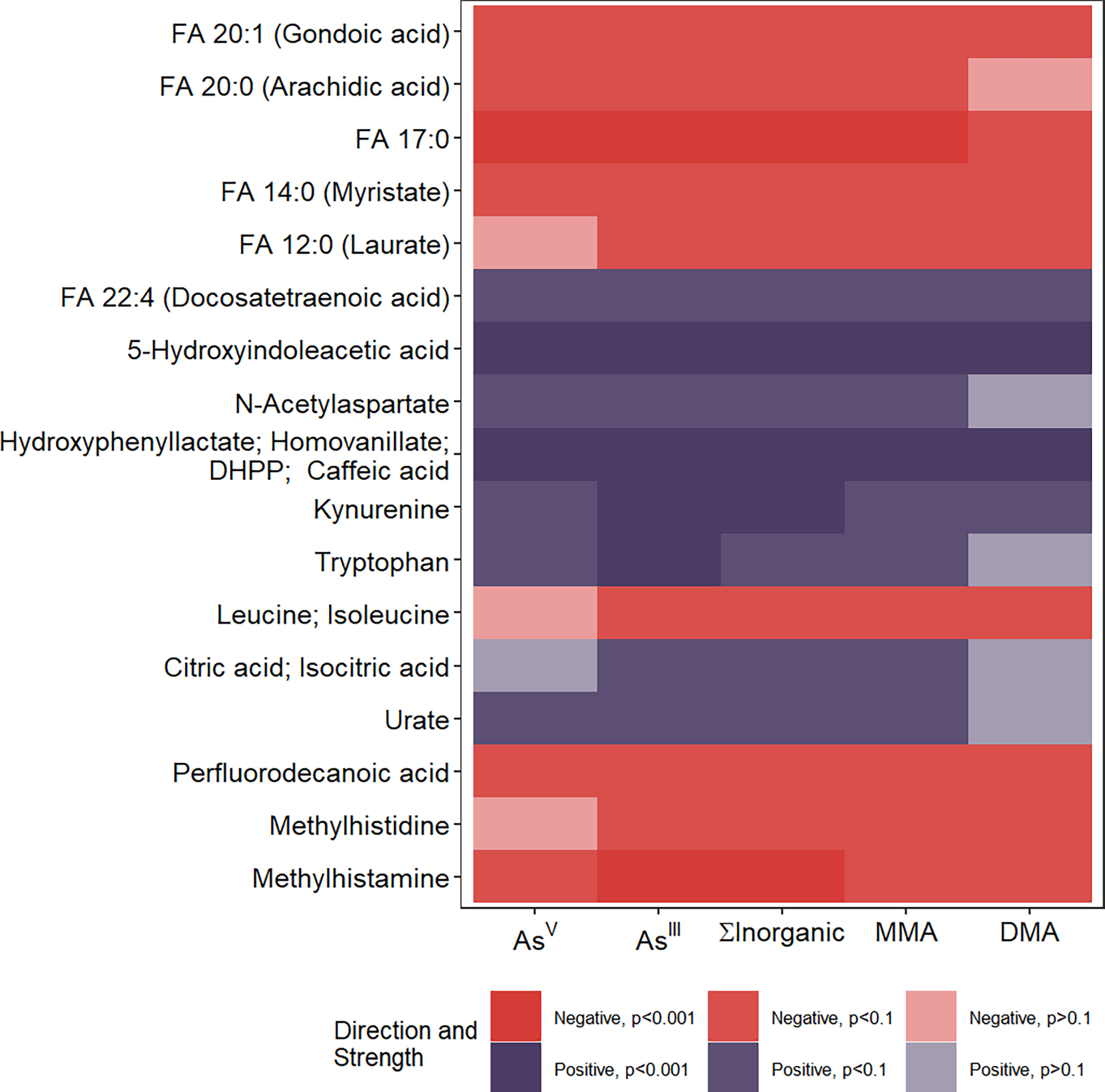
Heat map showing 17 metabolites with confirmed identities associated with at least one of the five blood Arsenic species at the False Discovery Rate (FDR) <0.05 level. Cells were colored based on the direction of the regression coefficient and nominal statistical significance. Overall, the plot shows that for all 17 metabolites, the models were consistent across As species in both statistical significance and direction of effect. FA: Fatty Acid; DHPP: 3, 4-Dihydroxyphenylpyruvic acid.
As a sensitivity analysis, adjustment for FA supplementation status in addition to age, sex, and BMI did not change our model estimates (Supplemental Figure 2). We found no strong evidence for effect modification by FA treatment in the 17 metabolites previously identified to be associated with blood As levels (Supplemental Table 3).
We also compared models using blood As to comparable models using urinary As biomarkers corrected for specific gravity and found that the results were similar between blood and urinary As species (Supplemental Figure 3).
Untargeted Metabolome Wide Association Study
We next applied an untargeted MWAS framework to evaluate systems-level metabolic alterations associated with As. MWAS of C18 and HILIC metabolite features as outcomes identified 423 unique features associated with at least one blood As species at FDR<0.05 after adjusting for age, sex, and BMI. Similar to the confirmed metabolites, inorganic As species had more statistically significant associations compared to methylated As species MMAs and DMAs (Supplemental Table 4). However, there were some differences across As species, as AsV had far more statistically significant associations than the rest of the four metabolites combined and the majority of these features uniquely associated with AsV were not associated with any other blood As species (Supplemental Table 4, Figure 2A). Results were more consistent across AsIII, ∑InAs, and MMAs, while the associations were weaker for blood DMAs (Figure 2B). Similar to the confirmed metabolites MWAS analysis, further adjustment for supplementation status did not change the model estimates (Supplemental Figure 2). Full MWAS results from the untargeted features can be found in Supplemental Table 5.
Figure 2.
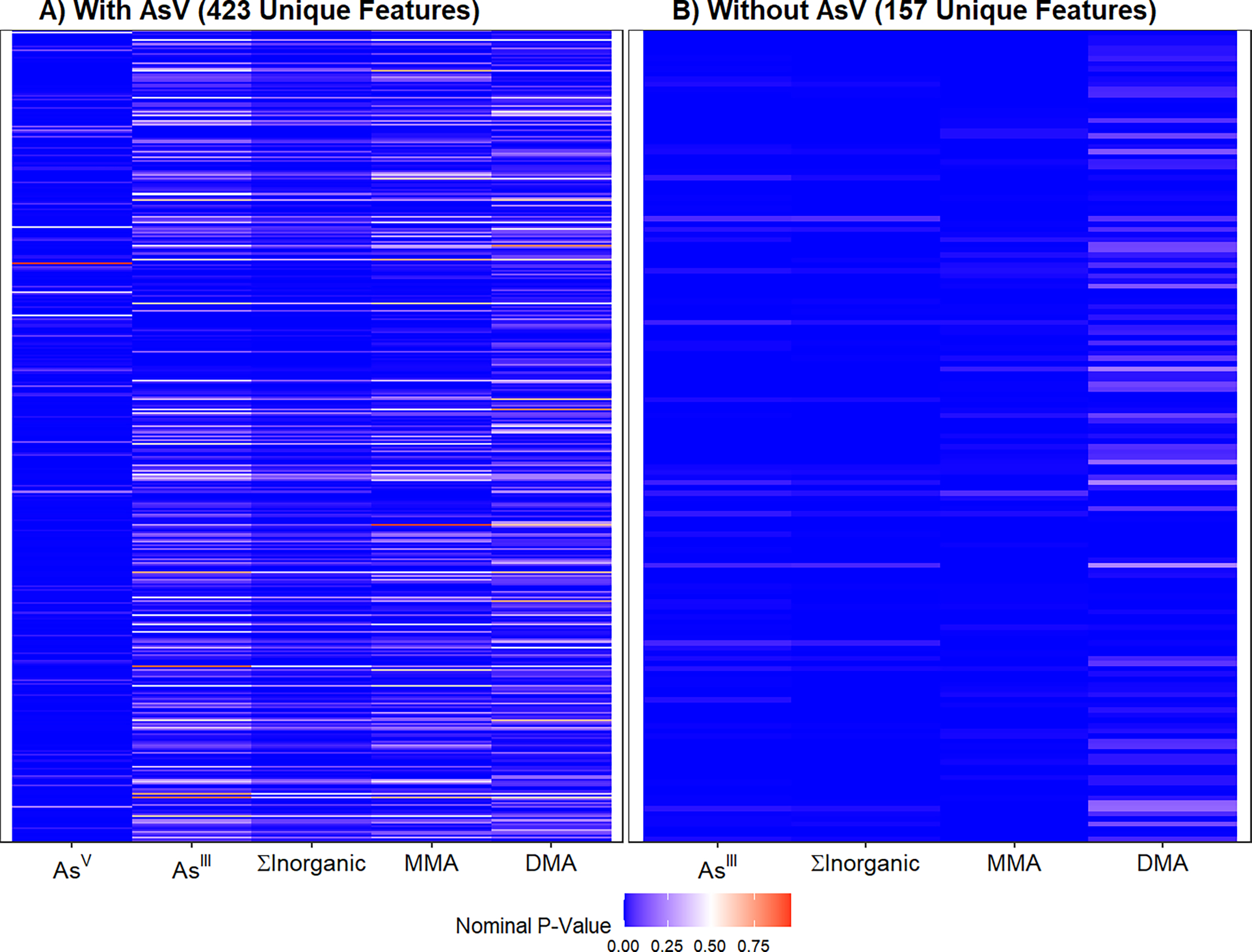
Heat map showing untargeted features associated with blood Arsenic species at the False Discovery Rate (FDR) <0.05 level A) with AsV and B) without AsV. Cells were colored based on nominal p-values. Panel A shows that many features were exclusively associated with AsV and no other As species. Panel B shows that AsIII, ∑inAs, and MMA were consistent and DMA is different from the other species.
Pathway enrichment analysis identified 12 metabolic pathways associated with at least one blood As species (Figure 3). Seven of the enriched pathways were related to lipid metabolism, and there was general agreement across all As species, which suggests that blood As was related to alterations in fatty acid metabolism. Metabolite annotations from Mummichog were evaluated using the KEGG human metabolic map (Figure 4). The results suggest multiple KEGG metabolites mapped to OCM pathway and lipid metabolism and were common across several As species. Additional mapped metabolites included xenobiotic metabolism associated with AsV and energy metabolism and tricarboxylic acid (TCA) cycle associated with DMAs.
Figure 3.
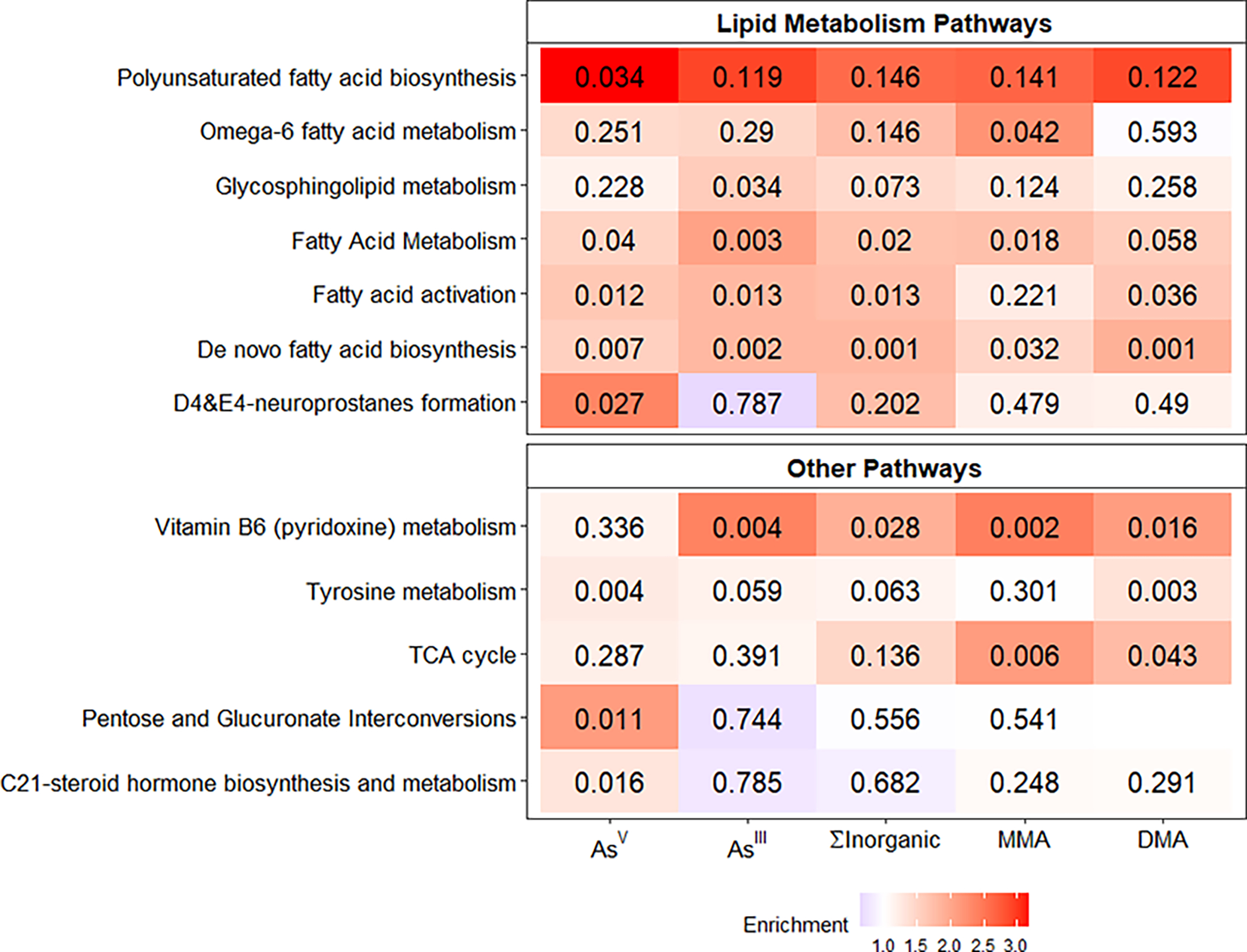
Heat map showing the 12 KEGG metabolic pathways that were enriched for at least one blood As species. The color represent the degree of fold enrichment while the number shows the Fisher Exact Test p-values from Mummichog pathway analysis. All pathways are also significant using a permutation based significant test.
Figure 4.
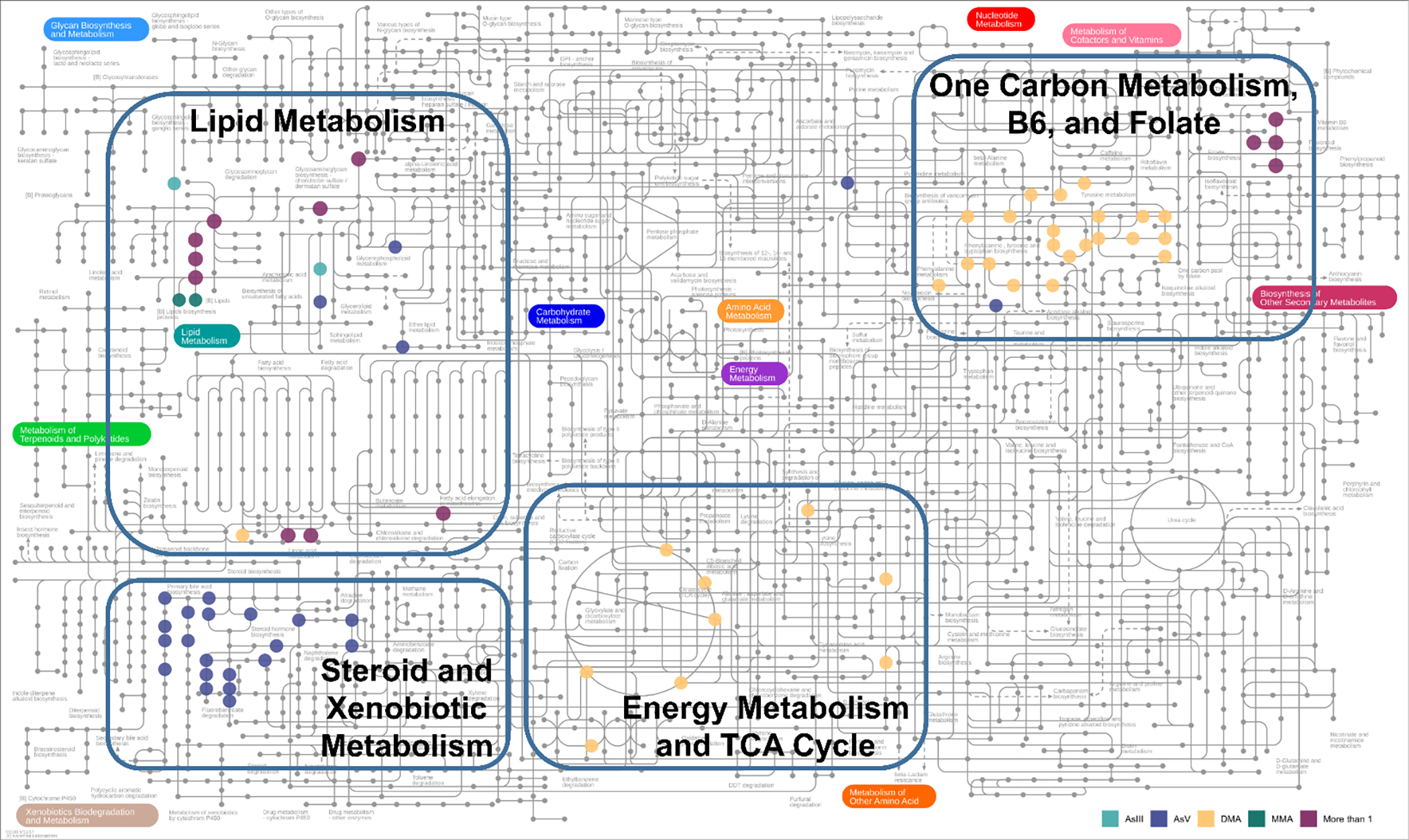
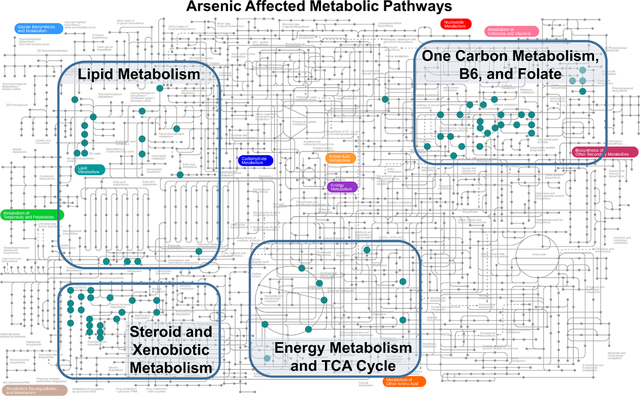
KEGG human metabolic map showing the clustering of related pathways associated with As exposure around four central themes. Each colored dot represents a metabolite associated with a specific As species. We identified four general metabolic paths, shown in rectangles and group labels. The lipid metabolism pathway was well represented by all As species. AsV was associated with xenobiotic metabolism. DMA was associated with energy metabolism and TCA cycle as well as metabolites related to one carbon metabolism such as vitamin B6 and folate.
To support our MWAS results, we extracted unique KEGG IDs identified from these enriched pathways and compared them across blood As species (Figure 5). AsV and DMAs were associated with numerous unique KEGG metabolites that were not shared with other blood As species while numerous KEGG metabolites were associated with ≥1 blood As species.
Figure 5.
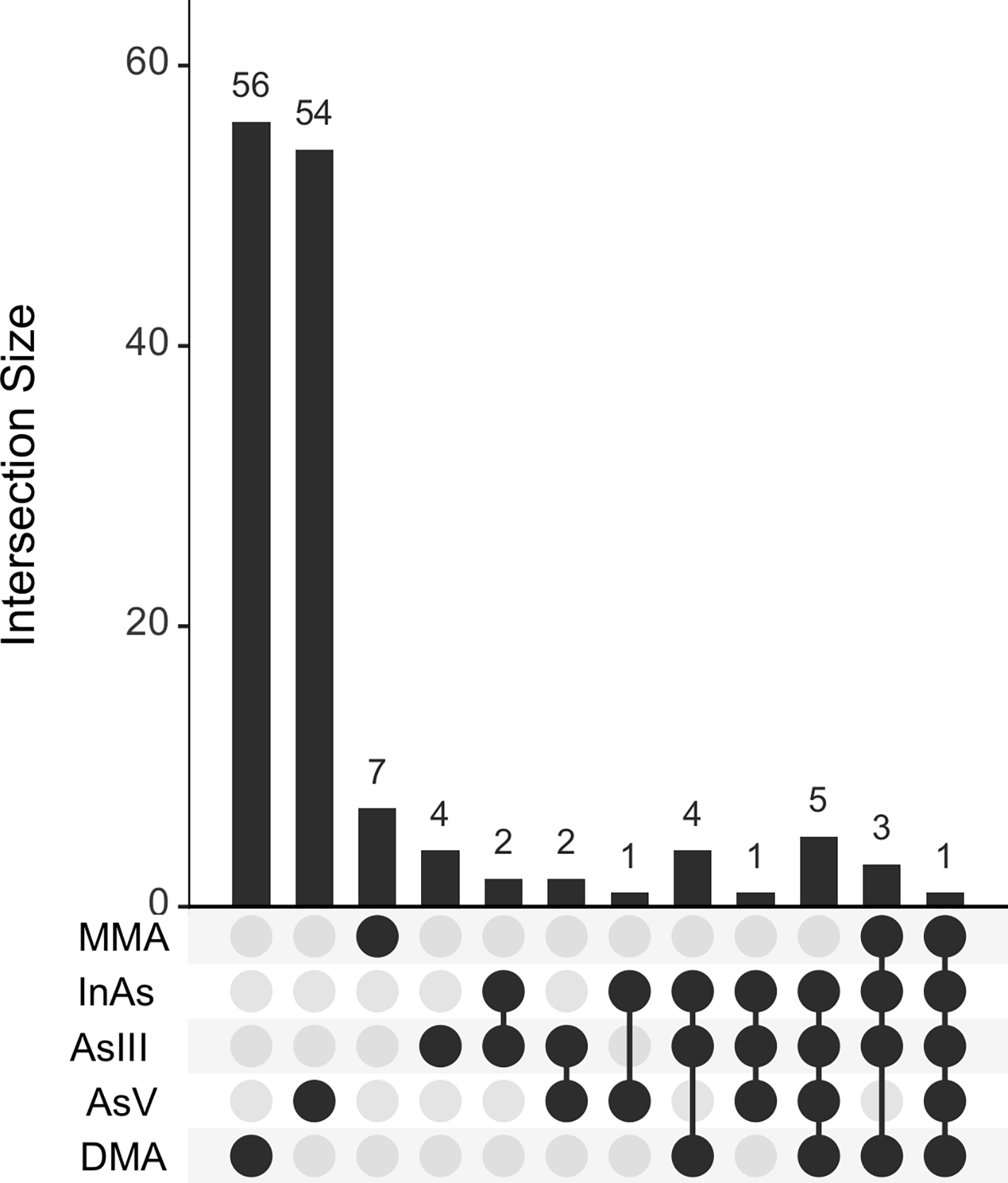
Upset plot showing the number of unique KEGG IDs associated with each As species and overlaps across species. The KEGG IDs were extracted from enriched pathways identified by Mummichog.
Lysophosphatidylcholine (lysoPC) and Lysophosphatidylethanolamine (lysoPE)
Given that OCM is a key regulator of As methylation, we examined known pathways that may connect OCM to lipid metabolism, which was observed to be associated with blood As levels in our analyses of both confirmed metabolites and untargeted metabolomic features. All blood As species were positively associated with lysoPC:lysoPE ratio in multivariable models adjusting for age, sex, BMI, and study visit (Table 2).
Table 2.
Associations of Blood Arsenic Metabolites with Lysophosphotidylcholine to Lysophosphatidylethanolamine Ratio
| Arsenic Species | ∑lysoPC:∑lysoPE Ratio | |||
|---|---|---|---|---|
| Regression β2 | 95% CI | P-value | IQR Estimate3 | |
| ASV | 6.18 | 1.78, 10.58 | 0.01 | 1.52 |
| ASIII | 1.15 | 0.14, 2.15 | 0.03 | 1.42 |
| ∑inAs1 | 1.14 | 0.30, 1.97 | 0.01 | 1.59 |
| MMA | 0.59 | 0.17, 1.02 | 0.01 | 1.53 |
| DMA | 0.59 | −0.001, 1.18 | 0.05 | 1.24 |
Sum of AsIII and AsV
Interpreted as the change in ratio per 1 ug/L increase in exposure
Effect estimate standardized to interquartile range
All models adjusted for age, sex, BMI, and visit
Arsenic Metabolism Biomarkers
To examine the impact of As metabolism on the metabolome, we conducted MWAS screening of 170 confirmed metabolites with urinary %InAs, %MMAs, and %DMAs. We identified 5 unique metabolites that were associated with at least one of the three urinary As metabolism biomarkers (Supplemental Figure 4). There were no overlaps with the results from blood As species.
Discussion
In our analysis of 60 individuals from Araihazar, Bangladesh, evaluated twice over a 12 week period, we found that all blood As species were associated with key metabolites broadly related to lipid metabolism (various fatty acids), neurotransmitters (N-acetylaspartate, 5-hydroxyindoleacetic acid [5-HIAA], homovanillate), amino acid metabolism (leucine/isoleucine, tryptophan, kynurenine), and purine metabolism (urate). Using untargeted analyses, we reaffirmed the association between As and lipid metabolism alterations common across all As species. In addition, we identified metabolic pathways specific to AsV (steroid and xenobiotic metabolism) and DMAs (TCA cycle). Mechanistically, As exposure has been linked to oxidative damage, mitochondrial dysfunction, inflammation37, and neurotransmitter dysregulation38–40. Our study was able to similar to observations from previous studies13–15,41–46 and identified novel pathways associated with As.
The observed enrichment of OCM metabolites and pathways in our analysis are consistent with existing knowledge of As metabolism and its relationship with OCM. OCM comprises a series of reactions in both the cytoplasm and mitochondria centered around folate and methionine cycles, involving numerous enzymes and co-factors, including several B vitamins47. OCM takes methyl groups from folate, making these methyl groups available for transmethylation reactions in the form of S-adenosylmethionine (SAM). The ability to metabolize InAs to methylated species requires SAM, and almost all methyltransferases are subject to inhibition by S-adenosyl-homocysteine (SAH), a product of all SAM-dependent methylation reactions. Our research group demonstrated in the original FACT study, and in an earlier RCT of folate-deficient participants, that FA supplementation increases As methylation, reducing blood MMAs burden by up to 50%17,48. Vitamin B6 (pyridoxine) is a key co-factor for multiple reactions in OCM, and its metabolic pathway was enriched in our untargeted metabolites analysis. Vitamin B6 was also in our panel of confirmed metabolites; while it did not cross the FDR threshold, it was positively associated with all As species except AsV (p-values = 0.003–0.07). Thus, our targeted and untargeted results both show the expected association between blood As and OCM related metabolites.
OCM is an essential process that is critical to numerous other biological processes, and disruption may have downstream consequences on metabolic pathways such as purine metabolism. In the methionine cycle, SAH is converted by S-adenosylhomocysteine hydrolase to homocysteine and adenosine. Related to the folate cycle, 10-formyl-tetrahydrofolate (10-formyl-THF) is required for the synthesis of inosine monophosphate (IMP), the precursor to both adenosine and guanine. The catabolism of adenosine and guanine, two purines, produces uric acid. In the present study, we observed that blood As is positively associated with plasma uric acid, which suggests that the interactions between As exposure and OCM may result in greater uric acid levels. This may also be related to the observed negative association between blood As and methylhistamine in our study as the catabolism of histidine to methylhistamine generates 10-formyl-THF. However, the potential mechanistic interplay involving As, OCM, histidine metabolism, and purine metabolism is ambiguous and merits deeper investigation in the future. Ultimately, As association with uric acid are supported by previous targeted studies that showed urinary arsenic levels were associated with higher serum uric acid levels and odds of hyperuricemia in adults from the US41 and Bangladesh42. Elevated uric acid is associated with gout49, diabetes50,51, cardiovascular disease52,53, kidney diseases54 and may be one of the biological pathways that underlies As-associated disease risk.
The broad impact of As exposure on pathways related to lipid metabolism and TCA cycle was evident throughout our results. All blood As species were associated with lower levels of leucine/isoleucine and fatty acids, higher citric acid level, and higher lysoPC:lysoPE ratio while pathway analysis of untargeted results reveled enrichment of lipid metabolism pathways and the TCA cycle pathway. Previous studies have reported separate evidence for associations of As exposure with different aspects of energy metabolism13,14,43–45; these results suggest broad alterations in metabolic effects that may arise from interactive crosstalk between As exposure, OCM, and energy metabolism. This is perhaps not surprising because As exposure can impact the TCA cycle and lipid metabolism in several ways. For example, As is a known inhibitor of pyruvate dehydrogenase55, a key enzyme that helps convert pyruvate to acetyl-CoA. Not only does this directly affect the TCA cycle, the regulation and availability of pyruvate dehydrogenase can directly impact glucose and fatty acid oxidation56.
It is also possible for As to influence lipid metabolism through OCM-related pathways. OCM is a regulator of lipid metabolism through its production of methyl-donor SAM. One potential pathway is through the SAM-dependent methylation of lysine to trimethyl-lysine, a precursor to carnitine, which in turn is a regulator of lipid metabolism because it transports long-chain fatty acids from cytosol into mitochondria for beta-oxidation57. However, our evidence does not support this particular connection because As was not associated with trimethyl-lysine or carnitine in our data. A more likely pathway is the SAM-dependent methylation of PE to PC. PE and PC are the two most abundant phospholipids and comprise a large portion of cellular lipid membranes. While we could not assess PC:PE ratio directly, we did observe an association of blood As levels with the ratio of their metabolic derivatives – lysoPCs and lysoPEs – as proxies. One previous study showed that AsIII exposure induced decreased PC and increased lysoPCs in rats46, but our observation in humans is novel. Not only are PE and PC important for cell membrane integrity, their balance has been associated with numerous chronic diseases32,33 that have also been linked to As exposure. Thus, it is possible that disruption to PE and PC balance partially underlies As-associated health effects.
OCM is also essential to maintaining redox state58 via production of NADPH and glutathione. The oxidation of 10-formyl-THF to CO2 generates an NADPH equivalent that is not consumed by the OCM cycle and this has been suggested to be important for mitochondrial redox homeostasis59,60. The redox balance crosstalk between OCM and TCA cycle is biologically important because excess NADPH/NADH, possibly from impaired cellular respiration, inhibits the TCA cycle while OCM remains uninhibited. In this state, the continued contribution of NADPH by OCM may result in redox imbalance and NADH toxicity in the mitochondria61,62. Indeed, As has been known to impair cellular respiration63, induce oxidative stress37, and lead to mitochondrial dysfunction64,65.
Arsenic is also known to be neurotoxic66 and has been previously linked to poor cognitive development67–69 and neurodegeneration37. In the present study, we see that As is associated with numerous neurotransmitter metabolites such 5-HIAA (metabolite of serotonin), tryptophan (precursor to serotonin and 5-HIAA), N-acetylaspartate (key metabolite and regulator of glutamate), and potentially homovanillate (metabolite of dopamine). Glutamate was also in our panel of confirmed metabolites and was nominally associated with AsV (p=0.06) and MMA (p=0.03). While our analytical method could not distinguish homovanillate from hydroxyphenyl lactate, 3,4-dihydroxyphenylpyruvic acid (DHPP), and caffeic acid due to their identical chemical formula (C9H10O4), our pathway analysis identified that As was associated with an overrepresentation of metabolites related to tyrosine metabolism, which is the precursor of dopamine, providing overall support for an association between As exposure and dopamine homeostasis. Tryptophan metabolism can be tied to OCM as the kynurenine pathway of tryptophan produces formate, a 1-carbon source and a key component of the folate cycle. However, the significance of this connection is unclear as formate is produced from numerous sources70. Overall, our results are consistent with previous human metabolic studies suggesting similar actions of As on pathways broadly related to serotonin13,14 and glutamate14. Importantly, our study was able to demonstrate these changes among healthy individuals and across multiple As species.
Despite the relatively small sample size, there was a clear observation that many metabolites and pathways were associated with multiple As species but some were uniquely associated with specific As species. The latter is to be expected given that 1) As species differ in their toxicity and their mechanism(s) of action, and 2) methyl-group availability is crucial for As metabolism, so the observed associations, e.g between blood DMAs levels and OCM related metabolites and pathways, may well be a product of reverse causation. However, As metabolism capacity is unlikely to be a major confounder in other observed associations of As with metabolites and pathways because there were no overlaps between metabolites associated with blood As species and biomarkers of As metabolism.
A key strength of our study was that we were able to utilize blood and urinary measures of As. We were able to leverage this to show that the plasma metabolome differences associated with As were generally consistent between blood and urinary As. Furthermore, we were able to utilize proportional As measures in urine (%InAs, %MMA, %DMA) as indicators of in As metabolic efficiency to show that our primary results with blood As concentrations were not a function of confounding by As metabolic efficiency. In addition, our study was able to leverage data on both targeted (i.e. confirmed metabolites) and untargeted metabolomic features. We were able to show consistency in the MWAS results from both approaches, which affords us greater confidence in our findings. We were also able to complement untargeted pathway analysis with specific known metabolites to gain additional insight toward the underlying biological actions.
While our study provides novel insights toward the underlying mechanisms behind As-related pathophysiology, there are some limitations. Our sample size was limited and given the high number of tests, it is possible that we failed to detect small or moderate associations. However, we did find several associations in both the confirmed metabolites and the untargeted mass spectrometry features. Importantly, our results were internally consistent not only between confirmed and untargeted analyses, but also across multiple types of analyses, As species, and biomatrices, showing the reliability and robustness of our results despite limited sample size. It is also unclear whether results from this Bangladeshi population is generalizable to other populations. It may be reasonable to expect that the results will be applicable to other As-afflicted regions because high As exposure is not exclusive to Bangladesh71, and we are examining biologic relationships between As exposure and metabolomic changes. However, we cannot know for certain without direct replication in other populations whether the strength of relationships may differ due to differences in genetics, exposure levels, relative As compositions, and other influences such as dietary folate intake. Another limitation is that while this is set within the context of a randomized trial, the present study is observational in nature and cannot strictly establish causality and temporality because the exposure predates our study and our analyses are cross-sectional in nature. Lastly, while this study includes metabolomic changes assessed at two time-points within a randomized clinical trial, we cannot asses the temporality of the observed associations. As exposure is chronic and ubiquitous in this population and predates the baseline of our study. OCM, lipid metabolism, and TCA cycle are all fundamental processes that are intricately linked to other metabolic pathways, and it is difficult to accurately detail the cascade of effects with only snapshots of the metabolome. Thus, more studies are needed to better understand the mechanisms of As action on these processes,
Conclusion
In this analysis of As biomarker concentrations and plasma metabolomic differences in adults from Bangladesh, we found that blood As concentrations were associated with multiple metabolic alterations. Specifically, we observed associations of As with neurotransmitter levels, expected alterations in OCM pathway, and changes in many pathways that directly connect to OCM such as purine metabolism, TCA cycle, and fatty acid metabolism. These results provide plausible pathways and explanations that enhances our understanding of the biological changes underlying As related health effects such as diabetes50,51, cardiovascular disease52,53, and neurotoxicity37,66. Future studies should not only seek to reaffirm these associations, but also identify the timescale and reversibility of these effects, as well as seek to identify unique effects in vulnerable windows such as childhood and pregnancy. As climate change affects global water resources while simultaneously increasing groundwater As levels72, our increasing global reliance on groundwater makes As exposure an urgent global health priority.
Supplementary Material
Targeted analyses identified 17 metabolites associated with blood arsenic levels
Untargeted screening identified 12 metabolic pathways associated with blood arsenic
Altered pathways were consistent with changes to one-carbon metabolomics
Most effects were common across all arsenic species
Some pathways and associations were unique to arsenate and dimethylarsinous acid
Acknowledgements and Funding:
This work was supported by grants from the National Institute of Environmental Health Sciences (R01ES030945, R01ES028805, R01ES032831, P30ES009089, P30ES019776, P42ES033719, and P42ES010349) and National Institute of Diabetes and Digestive and Kidney Diseases (R01DK123285). The funding source did not have any role in the interpretation of the study results, writing of the manuscript, or decision to submit for publication.
Footnotes
Competing Financial Interests: The authors declare they have no actual or potential competing financial interests.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This trial was registered at https://clinifscaltrials.gov as NCT01050556.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wei S, Zhang H & Tao S A review of arsenic exposure and lung cancer. Toxicol Res (Camb) 8, 319–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khanjani N, Jafarnejad A-B & Tavakkoli L Arsenic and breast cancer: a systematic review of epidemiologic studies. Rev Environ Health 32, 267–277 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Gamboa-Loira B, Cebrián ME, Franco-Marina F & López-Carrillo L Arsenic metabolism and cancer risk: A meta-analysis. Environ Res 156, 551–558 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Kononenko M & Frishman WH Association Between Arsenic Exposure and Cardiovascular Disease. Cardiol Rev 29, 217–221 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Moon KA et al. Association between Low to Moderate Arsenic Exposure and Incident Cardiovascular Disease. A Prospective Cohort Study. Ann Intern Med 159, 649–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navas-Acien A et al. Early-Life Arsenic Exposure, Nutritional Status, and Adult Diabetes Risk. Curr Diab Rep 19, 147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farkhondeh T, Samarghandian S & Azimi-Nezhad M The role of arsenic in obesity and diabetes. J Cell Physiol 234, 12516–12529 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Smith AH, Lingas EO & Rahman M Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull World Health Organ 78, 1093–1103 (2000). [PMC free article] [PubMed] [Google Scholar]
- 9.Moe B et al. Comparative cytotoxicity of fourteen trivalent and pentavalent arsenic species determined using real-time cell sensing. Journal of Environmental Sciences 49, 113–124 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE & Vasken Aposhian H Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol 163, 203–207 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Hughes MF, Beck BD, Chen Y, Lewis AS & Thomas DJ Arsenic Exposure and Toxicology: A Historical Perspective. Toxicological Sciences 123, 305–332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokar EJ, Qu W & Waalkes MP Arsenic, stem cells, and the developmental basis of adult cancer. Toxicol Sci 120 Suppl 1, S192–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozłowska L, Janasik B, Nowicka K & Wąsowicz W A urinary metabolomics study of a Polish subpopulation environmentally exposed to arsenic. J Trace Elem Med Biol 54, 44–54 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Martin E et al. Metabolomic characteristics of arsenic-associated diabetes in a prospective cohort in Chihuahua, Mexico. Toxicol Sci 144, 338–346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J et al. Urinary Metabolomics Revealed Arsenic Internal Dose-Related Metabolic Alterations: A Proof-of-Concept Study in a Chinese Male Cohort. Environ. Sci. Technol. 48, 12265–12274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H et al. Urinary metabolomics revealed arsenic exposure related to metabolic alterations in general Chinese pregnant women. Journal of Chromatography A 1479, 145–152 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Peters BA et al. Folic Acid and Creatine as Therapeutic Approaches to Lower Blood Arsenic: A Randomized Controlled Trial. Environ Health Perspect 123, 1294–1301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bozack AK et al. Folic acid supplementation enhances arsenic methylation: results from a folic acid and creatine supplementation randomized controlled trial in Bangladesh. Am J Clin Nutr 109, 380–391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahsan H et al. Health Effects of Arsenic Longitudinal Study (HEALS): description of a multidisciplinary epidemiologic investigation. J Expo Sci Environ Epidemiol 16, 191–205 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Vela NP, Heitkemper DT & Stewart KR Arsenic extraction and speciation in carrots using accelerated solvent extraction, liquid chromatography and plasma mass spectrometry. Analyst 126, 1011–1017 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Abuawad A et al. Association between body mass index and arsenic methylation in three studies of Bangladeshi adults and adolescents. Environ Int 149, 106401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu KH et al. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem. 92, 8836–8844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soltow QA et al. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 9, S132–S143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JM, Yu T, Strobel FH & Jones DP A practical approach to detect unique metabolic patterns for personalized medicine. Analyst 135, 2864–2870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu T & Jones DP Improving peak detection in high-resolution LC/MS metabolomics data using preexisting knowledge and machine learning approach. Bioinformatics 30, 2941–2948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu T, Park Y, Johnson JM & Jones DP apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 25, 1930–1936 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu T, Park Y, Li S & Jones DP Hybrid feature detection and information accumulation using high-resolution LC- MS metabolomics data. J Proteome Res 12, 1419–1427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uppal K et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 14, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uppal K, Walker DI & Jones DP xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem. 89, 1063–1067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uppal K et al. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 29, 1956–1975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchet JP, Lauwerys R & Roels H Comparison of the urinary excretion of arsenic metabolites after a single oral dose of sodium arsenite, monomethylarsonate, or dimethylarsinate in man. Int Arch Occup Environ Health 48, 71–79 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Law S-H et al. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int J Mol Sci 20, 1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Veen JN et al. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochimica et Biophysica Acta (BBA) - Biomembranes 1859, 1558–1572 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Li S et al. Predicting Network Activity from High Throughput Metabolomics. PLOS Computational Biology 9, e1003123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang Z et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Research 49, W388–W396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darzi Y, Letunic I, Bork P & Yamada T iPath3.0: interactive pathways explorer v3. Nucleic Acids Res 46, W510–W513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garza-Lombó C, Pappa A, Panayiotidis MI, Gonsebatt ME & Franco R Arsenic-induced neurotoxicity: A mechanistic appraisal. J Biol Inorg Chem 24, 1305–1316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mejía JJ, Díaz-Barriga F, Calderón J, Ríos C & Jiménez-Capdeville ME Effects of lead-arsenic combined exposure on central monoaminergic systems. Neurotoxicol Teratol 19, 489–497 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Nagaraja TN & Desiraju T Regional alterations in the levels of brain biogenic amines, glutamate, GABA, and GAD activity due to chronic consumption of inorganic arsenic in developing and adult rats. Bull Environ Contam Toxicol 50, 100–107 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Ramos-Chávez LA et al. Neurological effects of inorganic arsenic exposure: altered cysteine/glutamate transport, NMDA expression and spatial memory impairment. Front Cell Neurosci 9, 21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo C-C et al. Arsenic exposure, hyperuricemia, and gout in US adults. Environment International 76, 32–40 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Huda N et al. Elevated levels of plasma uric acid and its relation to hypertension in arsenic-endemic human individuals in Bangladesh. Toxicol Appl Pharmacol 281, 11–18 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Bi D et al. LC/MS/MS-Based Liver Metabolomics to Identify Chronic Liver Injury Biomarkers Following Exposure to Arsenic in Rats. Biol Trace Elem Res (2022) doi: 10.1007/s12011-021-03026-0. [DOI] [PubMed] [Google Scholar]
- 44.Qi Z, Wang Q, Wang H & Tan M Metallothionein Attenuated Arsenic-Induced Cytotoxicity: The Underlying Mechanism Reflected by Metabolomics and Lipidomics. J. Agric. Food Chem. 69, 5372–5380 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Wang C et al. Changes in metabolomics and lipidomics in brain tissue and their correlations with the gut microbiome after chronic food-derived arsenic exposure in mice. Ecotoxicol Environ Saf 228, 112935 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Rivas-Santiago C et al. Lipid Metabolism Alterations in a Rat Model of Chronic and Intergenerational Exposure to Arsenic. BioMed Research International 2019, e4978018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyon P, Strippoli V, Fang B & Cimmino L B Vitamins and One-Carbon Metabolism: Implications in Human Health and Disease. Nutrients 12, 2867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gamble MV et al. Folic acid supplementation lowers blood arsenic. Am J Clin Nutr 86, 1202–1209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalbeth N et al. Gout. Nat Rev Dis Primers 5, 69 (2019). [DOI] [PubMed] [Google Scholar]
- 50.King C et al. Uric Acid as a Cause of the Metabolic Syndrome. Contrib Nephrol 192, 88–102 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Lee SJ, Oh BK & Sung K-C Uric acid and cardiometabolic diseases. Clin Hypertens 26, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ndrepepa G Uric acid and cardiovascular disease. Clin Chim Acta 484, 150–163 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Kimura Y, Tsukui D & Kono H Uric Acid in Inflammation and the Pathogenesis of Atherosclerosis. Int J Mol Sci 22, 12394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung SW, Kim S-M, Kim YG, Lee S-H & Moon J-Y Uric acid and inflammation in kidney disease. Am J Physiol Renal Physiol 318, F1327–F1340 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Schiller CM, Fowler BA & Woods JS Effects of arsenic on pyruvate dehydrogenase activation. Environ Health Perspect 19, 205–207 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S, Hulver MW, McMillan RP, Cline MA & Gilbert ER The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutrition & Metabolism 11, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Longo N, Frigeni M & Pasquali M CARNITINE TRANSPORT AND FATTY ACID OXIDATION. Biochim Biophys Acta 1863, 2422–2435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ducker GS & Rabinowitz JD One-Carbon Metabolism in Health and Disease. Cell Metabolism 25, 27–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan J et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piskounova E et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L et al. Serine Catabolism Feeds NADH when Respiration Is Impaired. Cell Metab 31, 809–821.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maynard AG & Kanarek N NADH Ties One-Carbon Metabolism to Cellular Respiration. Cell Metabolism 31, 660–662 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Luz AL et al. From the Cover: Arsenite Uncouples Mitochondrial Respiration and Induces a Warburg-like Effect in Caenorhabditis elegans. Toxicol Sci 152, 349–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jomova K et al. Arsenic: toxicity, oxidative stress and human disease. J Appl Toxicol 31, 95–107 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Prakash C, Soni M & Kumar V Mitochondrial oxidative stress and dysfunction in arsenic neurotoxicity: A review. J Appl Toxicol 36, 179–188 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Mochizuki H Arsenic Neurotoxicity in Humans. Int J Mol Sci 20, E3418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tolins M, Ruchirawat M & Landrigan P The Developmental Neurotoxicity of Arsenic: Cognitive and Behavioral Consequences of Early Life Exposure. Annals of Global Health 80, 303–314 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Toxicological profile for arsenic. https://www.atsdr.cdc.gov/toxprofiles/tp2.pdf (2007). [PubMed]
- 69.Wasserman GA et al. A cross-sectional study of water arsenic exposure and intellectual function in adolescence in Araihazar, Bangladesh. Environment International 118, 304–313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brosnan ME, MacMillan L, Stevens JR & Brosnan JT Division of labour: how does folate metabolism partition between one-carbon metabolism and amino acid oxidation? Biochem J 472, 135–146 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Bagchi S Arsenic threat reaching global dimensions. CMAJ 177, 1344–1345 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lombard MA, Daniel J, Jeddy Z, Hay LE & Ayotte JD Assessing the Impact of Drought on Arsenic Exposure from Private Domestic Wells in the Conterminous United States. Environ. Sci. Technol. 55, 1822–1831 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


