Abstract
Nanomedicines have transformed promising therapeutic agents into clinically approved medicines with optimal safety and efficacy profiles. This is exemplified by the mRNA vaccines against COVID-19, which were made possible by lipid nanoparticle technology. Despite the success of nanomedicines to date, their design remains far from trivial, in part due to the complexity associated with their preclinical development. Herein, we propose a nanomedicine materials acceleration platform (NanoMAP) to streamline the preclinical development of these formulations. NanoMAP combines high-throughput experimentation with state-of-the-art advances in artificial intelligence (including active learning and few-shot learning) as well as a web-based application for data sharing. The deployment of NanoMAP requires interdisciplinary collaboration between leading figures in drug delivery and artificial intelligence to enable this data-driven design approach. The proposed approach will not only expedite the development of next-generation nanomedicines but also encourage participation of the pharmaceutical science community in a large data curation initiative.
Keywords: self-driving laboratories, materials acceleration platforms, nanomedicines, lipid-based nanoparticles, liposomes, polymer nanoparticles, drug delivery, machine learning, active learning, automation, high-throughput experimentation
Graphical abstract
Progress and potential
Recent advances in machine learning have led to the development of tools and techniques with the potential to make a transformative impact in the pharmaceutical sciences. In this perspective, we propose combining state-of-the-art machine-learning techniques with high-throughput experimentation to create a materials acceleration platform for nanomedicine development, NanoMAP. Development of such a platform requires interdisciplinary collaboration between the drug delivery and artificial intelligence communities. Currently, the lack of large robust datasets limits the use of these data-driven methods. To overcome this, NanoMAP includes a large data curation initiative made possible by a web-based application. We see the implementation of this platform as a means to improve bench-to-bedside translation of innovative medicines for patients who suffer from life-threatening diseases.
Blueprint for a materials acceleration platform for nanomedicine development.
Introduction
Nanomedicines have enabled significant advancement in pharmaceutical formulation due to their ability to confer improvements in drug safety and efficacy, thereby transforming promising active agents (such as small-molecule drugs and biologics) into viable drug products.1 , 2 The clinical success of nanomedicines has afforded global improvement in the treatments and lives of patients suffering from a range of diseases. For example, two of the clinically approved mRNA COVID-19 vaccines were formulated in lipid nanoparticles that provide stability to the mRNA cargo, allowing delivery to their sites of action in the body.3 , 4 Similarly, nanomedicines have been demonstrated to increase the efficacy and/or reduce the side effects (i.e., toxicity) of several chemotherapeutics.5 Prominent examples include DOXIL (liposomal doxorubicin) and VYXEOS (liposomal daunorubicin hydrochloride and cytarabine, encapsulated at a synergistic ratio). Indeed, numerous active agents require formulation in advanced drug delivery systems such as nanomedicines to be commercially viable. However, despite the great potential and clinical success of nanomedicines, their design and development remain challenging compared with the development of conventional pharmaceutical products such as tablets and capsules.6 As a result, we propose the use of a self-driving laboratory (SDL) or materials acceleration platform (MAP),7 , 8 , 9 , 10 , 11 , 12 named NanoMAP (nanomedicine MAP). NanoMAP will harness artificial intelligence (AI), machine learning (ML), and high-throughput experimentation to automate the design process and expedite the development of nanomedicines.
SDLs are loosely defined as an experimentation paradigm characterized by the union of automated laboratory equipment for both sample preparation and characterization (often with high-throughput capabilities) and ML-guided experiment planning. These two aspects are linked by software that facilitates the interexchange of information with limited or no human involvement. Specifically, the experiment planning algorithm sends instructions in the form of experimental parameters to be carried out by the autonomous laboratory, which in turn reports back the corresponding figures of merit or objective function values.
Researchers have been automating experiments using high-throughput strategies (such as factorial grid search or design of experiment) for decades,13 , 14 which has enabled reproducibility, parallelization, and miniaturization of experiments while freeing researchers from repetitive tasks.15 , 16 Although some demonstrations showed promising results,17 , 18 , 19 , 20 high-throughput experimentation strategies suffer from the curse of dimensionality, with higher-dimensional parameter search domains becoming exponentially costly to measure with uniform resolution. Early examples of SDLs, which focused on optimizing chemical reactions in continuous-flow systems,21 , 22 instead enlisted data-driven optimization techniques to navigate the parameter domain sequentially, thus leveraging feedback from previous measurements and enhancing the efficiency with which promising candidates are identified. Since then, SDLs have experienced cascading adoption in diverse research areas, including nanomaterials,23 , 24 , 25 catalysis,26 condensed matter physics,27 and photovoltaic materials,28 , 29 , 30 to name a few.
Rising to the challenge
NanoMAP addresses two of the key challenges that confront the field of nanomedicine development
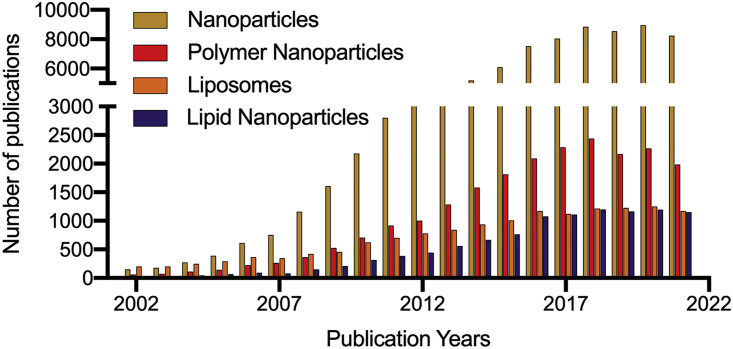
First, the application of AI and ML in the pharmaceutical sciences is currently challenged by the lack of high-quality data necessary for effective deployment of this approach.31 This may seem paradoxical given the tremendous investment into applications of nanotechnology in drug delivery in the last two decades. Indeed, there is a vast body of scientific literature on both nanotechnology-based formulation development and preclinical characterization of nanomedicines.32 A Web of Science search for either “nanoparticles,” “polymer nanoparticles,” “liposomes,” or “lipid nanoparticles” and the secondary keyword “drug delivery” returns more than 70,000, 20,000, 14,000, and 10,000 research articles in the last 20 years, respectively (Figure 1 ). However, the utility of this research for data-driven nanomedicine design is limited by poor publication practices and a lack of standardization in reporting key components or properties of drug formulations.33 , 34 For example, when one combs through the articles related to polymer nanoparticles and drug delivery to compile the properties of materials and active agents in an effort to build a database, the extent to which important meta-data is omitted from manuscripts becomes clear (Table 1 ). This lack of complete, high-quality data in scientific articles is recognized as a key challenge for researchers who rely on data mining to train ML models, with extracted datasets rarely exceeding 1,000 data instances and more often limited to <200 samples.31 In order to address this issue, NanoMAP will implement standardized high-throughput experimentation, open access databases, and ready-to-use experimental protocols.
Figure 1.
Summary of the number of publications on nanoparticles (yellow bars), polymer nanoparticles (red bars), liposomes (orange bars), and lipid nanoparticles (purple bars) and drug delivery in the past 20 years
Data sourced from the Web of Science using keyword combinations of “nanoparticles” and “drug delivery”; “polymer nanoparticles” and “drug delivery”; “liposomes” and “drug delivery”; or “lipid nanoparticles” and “drug delivery,” with the search results filtered by research articles only.
Table 1.
A snapshot of the information reported on drug-loaded polymer nanoparticle formulations
| Active agent | Polymer | Polymer PDI | Polymer molecular weight (kDa) | Particle size (nm) | Particle PDI | Zeta potential (mV) | Drug loading capacity (%) | Encapsulation efficiency (%) |
|---|---|---|---|---|---|---|---|---|
| Coumarin-6 | PLGA | – | 40–75 | 201 ± 2 | 0.04 | −37 | – | – |
| Docetaxel | PLGA (PEG) | – | 15 (3) | ∼50 | – | ∼ −29 | ∼4 | ∼28 |
| Paclitaxel | PLA | – | 106 | 589 ± 245 | 0.33 | – | – | 43 |
| Paclitaxel | PLGA | – | 22 | 112 ± 4 | 0.18 ± 0.005 | −0.6 ± 6 | 0.7 ± 0.04 | 70 ± 4 |
| Curcumin | alginate | – | – | 100 ± 20 | – | – | – | ∼11 |
| Tetanus toxoid | chitosan | – | >50 | 354 ± 27 | – | +37 ± 6 | – | 55 ± 3 |
| Silk peptide | chitosan | – | 80 | 206 ± 22 | – | +25 ± 3 | 8 | 82 |
| Gatifloxacin | chitosan | – | 65–90 | 205 ± 16 | – | +27 ± 4 | – | 78 ± 5 |
| Estradiol | PLGA | – | 15 | 98 ± 3 | 0.16 ± 0.008 | +79 ± 2 | – | 51 ± 6 |
| BSA | PLGA | – | 120 | 110 | – | – | 23 | 90 |
Data are extracted from a Web of Science literature search for publications on “polymer nanoparticles” and “drug delivery.” Specifically, the search results were sorted in order of the number of citations, and the top ten articles deemed relevant were inspected further are summarized in this table. PDI, polydispersity index.
Second, there are a plethora of active agent properties, material properties, and manufacturing parameters that must be considered in the development of nanomedicines. The currently employed design-build-test loop is largely reliant on an iterative, trial-and-error-based experimental approach. Owing to the combinatorial explosion of possible experiments, full resolution of the relationship between parameters and formulation performance is typically intractable. Moreover, it is well established that material-drug (or active agent) compatibility significantly influences the properties and performance of a formulation (i.e., loading capacity, stability, release kinetics, etc.). Yet, most current research efforts in this space focus on the use of the same small pool of materials that are clinically approved. This is largely due to the additional time and cost associated with potential regulatory hurdles for new materials and excipients.35 In order to justify the cost of bringing such a material to market, its inclusion would need to result in a formulation with a performance that is unachievable with any combination of currently approved materials or excipients. Additionally, as each active agent has its own unique physicochemical properties, it is understood that no one material can serve as the ideal candidate for the formulation of all active agents. Thus, restriction of the formulation design space clearly deters innovation. The proposed MAP harnesses advances in ML to enable data-driven development of innovative nanomedicines within the boundaries of the established, currently available and clinically approved materials. Moreover, NanoMAP may allow for the design of new materials that could afford formulations with leap-step advances in performance.
NanoMAP
Physical platform for high-throughput nanoparticle development
Nanoprecipitation for high-throughput nanomedicine screening
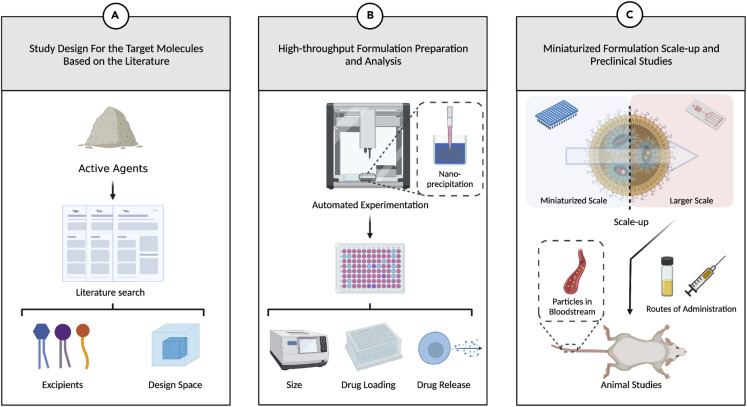
Among the various methods to prepare nanomedicines, nanoprecipitation is perhaps the most amenable to automation and high-throughput experimentation via liquid-handling robots. Briefly, this method involves the addition of molecularly dissolved excipients and active agents (within the organic phase) to an aqueous phase (often containing surfactant to stabilize the resulting nanoparticles). This process has been used to prepare lipid nanoparticles,36 polymer nanoparticles,37 and liposomes.38 Miniaturization of the nanoprecipitation process onto a 96-well plate format using a liquid-handling robot has been reported in an effort to screen the lipid composition for mRNA-loaded lipid nanoparticles.39 , 40 Moreover, it has also been demonstrated that nanomedicines screened in this manner can be scaled up on microfluidics platforms, thus eradicating potential downstream manufacturing concerns.39 , 40 It is straightforward to envision the development of similar workflows for small molecules, proteins, or peptides given the necessary equipment to characterize the resulting nanomedicines. For instance, drug loading capacity (DLC) and encapsulation efficiency (EE) can be automated with appropriate extraction methods and analysis via high-performance liquid chromatography (HPLC). High-throughput in vitro stability and release assays in biorelevant media are also amenable to automation using 96-well dialysis plates and similar sampling protocols to those mentioned above for DLC and EE. Dynamic light scattering plate readers enable the high-throughput measurement of particle size.39 , 40 Therefore, all of the necessary equipment and components to conduct such high-throughput experiments already exist. In the NanoMAP framework, we unite these components in order to systematically prepare and characterize nanomedicines in an automated fashion (Figure 2 ).
Figure 2.
A scheme summarizing the physical platform for high-throughput nanomedicine screening
(A) Candidate excipients and a formulation design space are derived from the existing literature.
(B) The design space can then be explored to identify nanomedicines with desirable performance (e.g., size, active agent loading, and release kinetics) using automated experimentation.
(C) This high-throughput approach will allow for lead candidate formulations that meet key design criteria (i.e., active agent loading levels, release kinetics) to be identified in a manner that saves time and resources. Potential lead candidate formulations can then be scaled up for further preclinical studies as required.
Figure created with BioRender.com.
ML strategies to enhance efficient formulation development
AL to efficiently generate informative datasets
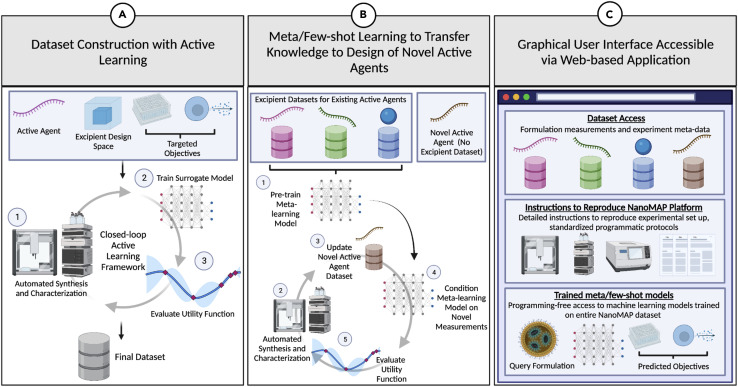
Active learning (AL)41 is a model-based sequential learning strategy in which an ML model can achieve greater predictive accuracy with fewer training data if it is allowed to choose the data on which it is trained. AL is a well-established framework for tasks in which unlabeled data are abundant (e.g., potential drug-lipid-surfactant formulations) but obtaining such labels incurs significant expense. Here, we intend to use an active learner to efficiently construct an informative dataset of nanomedicines (Figure 3 A). Our active learner will sequentially recommend formulations to be prepared and measured by the automated laboratory via a utility function, which prioritizes certain formulations based on their expected informativeness. Resulting formulation performance measurements will be passed to the model for retraining. This process will repeat until a predetermined experimental budget is exhausted. Indeed, AL strategies such as Bayesian optimization42 , 43 , 44 and variance-based sampling41 have recently been applied in the context of closed-loop design of nanomaterials, including inorganic nanoparticles,45 , 46 polymeric nanoparticles encapsulating nucleic acids,47 and polymer-protein hybrids,48 as well as being proposed to empower self-driving platforms for biologics formulation development.49
Figure 3.
Schematic of the machine-learning and software aspects of NanoMAP
(A) Given an active agent, excipient design space, and user-defined target objectives (e.g., active agent loading capacity and/or release kinetics), an active learner is employed to drive a closed-loop experiment, resulting in a dataset of parameter-objective pairs.
(B) After collection of formulation datasets for several active agents, meta/few-shot learning models are trained on existing data and used as the surrogate model in an active learning framework for a novel active agent.
(C) NanoMAP will consist of a graphical user interface accessible via a web-based application that provides facile access to formulation design (meta-)data and to detailed instructions on how to reproduce the physical NanoMAP platform and automated protocols, as well as programming-free access to pretrained machine-learning models.
Figure created with BioRender.com.
Nanomedicines commonly consist of a combination of polymer and/or lipid-based materials or excipients that encapsulate small molecules or biologic-based active agents. Representing these complex entities for AL therefore becomes a crucial challenge. Moreover, in the early days of NanoMAP, ML models will be forced to operate in the low-data regime (less than a few thousand training examples). In this regime, simple one-hot encodings of molecular parameters have been shown to produce reliable performance on reaction optimization tasks.50 Our group’s previous work also indicates that carefully selected physicochemical descriptors (e.g., via the Mordred package51) are a flexible representation that produces accurate and calibrated predictions of small-molecule properties when used in conjunction with probabilistic ML models in a low-data supervised learning setting52 and can also increase the optimization rate in an AL setting.53 The work of Tom et al.52 also suggests that graph-based representations54 , 55 of small molecules are not as effective as simple vector-valued representations in the low-data regime. However, as the corpus of information within NanoMAP increases, graph representations could be considered not only for small molecules but also for molecular ensembles.56
Few-shot learning for accurate prediction of nanomedicines for novel active agents
The availability of high-quality data in drug formulation and development is to some extent limited by a lack of standardization in experimental design and poor reporting practices. Although the present study will aid in the generation of a relatively large, standardized dataset of nanomedicines and will enable the rapid testing of conventional supervised ML models, the paramount practical advantage to such a dataset would be to enable accelerated nanomedicine development for novel active agents. However, extrapolation to out-of-distribution examples is a challenging task for conventional supervised ML. As such, we argue that nanomedicine design can be cast as a few-shot learning problem (Figure 3B).57 In this paradigm, the entire available dataset is organized by active agents into subsets called source tasks. Few-shot learners are then trained on all source tasks individually. Test time refers to commencement of a novel campaign in which an optimal formulation must be discovered for a novel active agent. Sequential experimentation can now commence but with the few-shot learner guiding experimentation with access to the novel campaign and the source task information. Such approaches have been shown to effectively transfer knowledge from source tasks onto novel campaigns, thus increasing predictive accuracy in the low-data regime and ultimately accelerating the discovery process.58 , 59 , 60 Despite the appeal of the aforementioned few-shot or meta-learning optimization procedures, they are limited to campaigns that share the same set of adjustable formulation parameters. Recently, developments in few-shot/meta-learning optimization based on large language models (LLMs), such as OptFormer,61 alleviate this restriction by learning from large-scale optimization campaign data using flexible text-based representations. Nevertheless, OptFormer was originally demonstrated on the task of ML hyperparameter optimization, for which results of millions of unique optimization campaigns are available. In the experimental sciences, such a corpus of curated data does not yet exist. In the distant future, we imagine that NanoMAP, along with self-driving platforms in general, could benefit from using optimization strategies based on LLMs.
Web-based application for data sharing
Graphical user interface for dissemination of research progress
Historically, in addition to the lack of freely available high-quality data to train models, one of the challenges with the integration of ML into the pharmaceutical sciences has been a lack of the necessary programming skills needed to implement these tools.12 , 31 To ensure the maximum impact and adoption of the data-driven tools described herein, the NanoMAP platform will include a user-friendly web-based application (Figure 3C). This application will allow access to the curated database and to detailed instructions on how to reproduce our low-cost physical platform, as well as access to the trained few-shot learning models via an interface that does not require programming expertise. Similar interfaces have been created to democratize data-driven molecular property prediction62 , 63 , 64 , 65 and chemical reaction optimization.66 , 67 Crucially, we will also provide an option to upload datasets and meta-data, which will promote community engagement and tackle the reproducibility crisis in the pharmaceutical sciences. The NanoMAP framework will also seek to abide by and promote adoption of the FAIR (findable, accessible, interoperable, and reusable) data principles for scientific data management.68
Roadmap to NanoMAP
NanoMAP will combine high-throughput experimentation with state-of-the-art advances in AI (including AL and few-shot learning) and a web-based application for data sharing to not only expedite the design of innovative next-generation nanomedicines but also to promote community participation in a larger data curation project. As a roadmap to the development of NanoMAP, we have identified three key milestones.
Milestone 1
An initial formulation design space of excipients and active agents will be established to develop the necessary automation workflows for NanoMAP. This initial design space will consist of commonly used excipients or materials and a panel of up to five active agents selected to have diverse physicochemical properties. The hardware required to conduct the proposed high-throughput experiments is commercially available. Most research labs skilled in nanomedicine development have the necessary analytical equipment and consumables to conduct this research. The key piece of equipment preventing automation is likely access to a liquid-handling robot. However, these have become significantly more affordable in recent years, with entry-level models available for as little as $5,000.
Milestone 2
Milestone 2 is the expansion of the initial design space with new active agents for the integration of active learners and few-shot learners into NanoMAP. At this point, key design criteria (i.e., figures of merit) for the nanomedicines will be established as target objectives for the active learners and few-shot learners. Various AL algorithms for optimization tasks have been developed and are freely available via GitHub, including several that have been developed by members of our research team.53 , 69 Conversely, few-shot learners are much less explored in the pharmaceutical sciences, and the full potential of such models has yet to be realized.
Milestone 3
Finally, there will be the construction of a web-based application to host a database of formulation design (meta-)data and automated protocols, as well as detailed instructions on how to reproduce the physical NanoMAP platform. Additionally, a graphical user interface will be accessible through the website that will host programming-free access to pretrained ML models. Various platforms exist to host such web-based applications. For example, the Aspuru-Guzik group has developed MOLAR, an open-source database management system for PostgreSQL that is tailored to the needs of materials scientists.70
Feasibility of approach
The necessary hardware to conduct the proposed high-throughput experimentation is commercially available. Moreover, advances in AI and ML research over the past two decades have afforded much of the software framework for the model-based optimizers necessary to close the design-build-test loop for NanoMAP. What remains is for the components to be assembled into a single platform. This is not a trivial task. The successful implementation of NanoMAP (and other such self-driving labs in the pharmaceutical sciences) requires interdisciplinary collaboration between leading figures in drug delivery and AI. Herein, we have assembled a team with the skills, experience, and knowhow necessary to build NanoMAP. However, measurable impact on the field requires widespread adoption of such an approach with open sharing of data, methods, and statistical models throughout the community. For this reason, we propose construction of a web-based application to enable and promote access and dissemination. Importantly, the platform will enable those who are not experts in formulation or computer science to deploy these data-driven tools.
Limitations
In this section, we identify and discuss several limitations to the proposed approach.
Software maintenance and supervision
NanoMAP proposes the construction of a web application intended to be interactive for the research community. Also, we propose to have state-of-the-art ML models available for prediction and trained periodically using the latest collective training set. These aspects will require at least part-time oversight by a programmer with software-engineering qualifications to allow the platform to operate as intended. For many academic research groups, financing one or more software engineers may be prohibitive.
Scale up of nanomedicine production
Our motivation in focusing on nanomedicines prepared via nanoprecipitation using liquid-handling robots stems from the amenability of this method to high-throughput experimentation. As well, this method is currently more economical on a per-experiment basis than other nanomedicine manufacturing techniques, such as microfluidics. For instance, an entry-level liquid-handling robot can be acquired for as little as $5,000, while an entry-level microfluidics system is much more expensive. Moreover, most microfluidics systems require expensive chips or disposable cartridges for operation, which further increases the cost associated with experiments. However, our proposed approach is not without its limitations. In general, nanoprecipitation affords lower active agent EEs and is a less scalable process than microfluidics. To balance this trade-off between experimental throughput and scalability, researchers have employed a hybrid approach wherein liquid-handling robots are used to conduct throughput nanoprecipitation experiments to identify lead formulation compositions. These lead candidates are then scaled up via microfluidics.39 , 40 , 71 Thus, it may be necessary for NanoMAP to adopt similar microfluidics-integrated approaches to scale lead candidate formulations.
Societal impact of NanoMAP
While the COVID-19 pandemic has challenged the operations of scientific laboratories around the globe, it is science and medicine that are leading us out of this pandemic. In fact, advanced pharmaceutical formulation is among the key areas that have played a critical role in the fight against COVID-19. NanoMAP will lead to a better understanding of the composition-performance relationships of nanomedicines, to the discovery of innovative nanomedicines, and to a decrease in the financial cost and time associated with bringing new nanomedicines to market. Moreover, once established, NanoMAP can be extended to generate biological data for nanomedicines by exploring applications that are amenable to automation, such as high-throughput cytotoxicity and cellular uptake assays, as well as evaluation in organ-on-a-chip technologies. This research will also result in the development of cutting-edge few-shot learning models that will help solve the “big data'' issue that has thus far limited the deployment of ML models in the pharmaceutical sciences. These models, as well as the curated datasets, will be shared openly online through open-access repositories such as GitHub for use by the global research community. To further increase the usability of the data-driven tools that will result from this research, we will also develop and deploy a free-to-use and user-friendly web-based application to allow programming-free access to pretrained models. While NanoMAP will focus on the development of nanomedicines, we see no reason why the foundational workflows and models cannot be improved or refined and made applicable to an extended range of sectors where formulation is critical to product development (e.g., diagnostic imaging agents, agriculture, cosmetics, paints, and coatings). Ultimately, we believe that the deployment of NanoMAP, and the development of similar self-driving labs, has the potential to make a transformative impact on the formulation development process and improve the bench-to-bedside translation of innovative medicines for patients who suffer from life-threatening diseases.
Acknowledgments
The authors are thankful to Dr. Matteo Aldeghi and Jonathan Zaslavsky for helpful discussions. R.J.H. gratefully acknowledges the Natural Sciences and Engineering Research Council of Canada (NSERC) for the provision of a Postgraduate Scholarship-Doctoral Program (PGSD3-534584-2019) as well as support from the Vector Institute. C.A. acknowledges an NSERC Discovery grant (RGPIN-2022-04910). A.A.-G. acknowledges support from the Canada 150 Research Chairs program and CIFAR, as well as the generous support of Anders G. Fröseth.
Author contributions
R.J.H., P.B., and Z.B. conceived of the idea and wrote and edited the manuscript. A.A.-G. and C.A. supervised the research and edited the manuscript.
Declaration of interests
A.A.-G. is the chief visionary officer and founding member of Kebotix, Inc.
References
- 1.Germain M., Caputo F., Metcalfe S., Tosi G., Spring K., Åslund A.K.O., Pottier A., Schiffelers R., Ceccaldi A., Schmid R. Delivering the power of nanomedicine to patients today. J. Control. Release. 2020;326:164–171. doi: 10.1016/j.jconrel.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J., Kantoff P.W., Wooster R., Farokhzad O.C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lázaro I., Mooney D.J. Obstacles and opportunities in a forward vision for cancer nanomedicine. Nat. Mater. 2021;20:1469–1479. doi: 10.1038/s41563-021-01047-7. [DOI] [PubMed] [Google Scholar]
- 7.Häse F., Roch L.M., Aspuru-Guzik A. Next-generation experimentation with self-driving laboratories. Trends Chem. 2019;1:282–291. doi: 10.1016/j.trechm.2019.02.007. [DOI] [Google Scholar]
- 8.Stein H.S., Gregoire J.M. Progress and prospects for accelerating materials science with automated and autonomous workflows. Chem. Sci. 2019;10:9640–9649. doi: 10.1039/C9SC03766G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores-Leonar M.M., Mejía-Mendoza L.M., Aguilar-Granda A., Sanchez-Lengeling B., Tribukait H., Amador-Bedolla C., Aspuru-Guzik A. Materials acceleration platforms: on the way to autonomous experimentation. Curr. Opin. Green Sustain. Chem. 2020;25:100370. doi: 10.1016/j.cogsc.2020.100370. [DOI] [Google Scholar]
- 10.Coley C.W., Eyke N.S., Jensen K.F. Autonomous discovery in the chemical sciences Part I: progress. Angew. Chem. Int. Ed. Engl. 2020;59:22858–22893. doi: 10.1002/anie.201909987. [DOI] [PubMed] [Google Scholar]
- 11.Coley C.W., Eyke N.S., Jensen K.F. Autonomous discovery in the chemical sciences Part II: outlook. Angew. Chem. Int. Ed. Engl. 2020;59:23414–23436. doi: 10.1002/anie.201909989. [DOI] [PubMed] [Google Scholar]
- 12.Stach E., DeCost B., Kusne A.G., Hattrick-Simpers J., Brown K.A., Reyes K.G., Schrier J., Billinge S., Buonassisi T., Foster I., et al. Autonomous experimentation systems for materials development: a community perspective. Matter. 2021;4:2702–2726. doi: 10.1016/j.matt.2021.06.036. [DOI] [Google Scholar]
- 13.Winicov H., Schainbaum J., Buckley J., Longino G., Hill J., Berkoff C.E. Chemical process optimization by computer — a self-directed chemical synthesis system. Anal. Chim. Acta X. 1978;103:469–476. doi: 10.1016/S0003-2670(01)83110-X. [DOI] [Google Scholar]
- 14.Lindsey J.S. A retrospective on the automation of laboratory synthetic chemistry. Chemometr. Intell. Lab. Syst. 1992;17:15–45. doi: 10.1016/0169-7439(92)90025-B. [DOI] [Google Scholar]
- 15.Fabry D.C., Sugiono E., Rueping M. Online monitoring and analysis for autonomous continuous flow self-optimizing reactor systems. React. Chem. Eng. 2016;1:129–133. doi: 10.1039/C5RE00038F. [DOI] [Google Scholar]
- 16.Fabry D.C., Sugiono E., Rueping M. Self-optimizing reactor systems: algorithms, on-line analytics, setups, and strategies for accelerating continuous flow process optimization. Isr. J. Chem. 2014;54:341–350. doi: 10.1002/ijch.201300080. [DOI] [Google Scholar]
- 17.Senkan S.M. High-throughput screening of solid-state catalyst libraries. Nature. 1998;394:350–353. doi: 10.1038/28575. [DOI] [Google Scholar]
- 18.Macarron R., Banks M.N., Bojanic D., Burns D.J., Cirovic D.A., Garyantes T., Green D.V.S., Hertzberg R.P., Janzen W.P., Paslay J.W., et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 19.Martin R.L., Simon C.M., Smit B., Haranczyk M. In silico design of porous polymer networks: high-throughput screening for methane storage materials. J. Am. Chem. Soc. 2014;136:5006–5022. doi: 10.1021/ja4123939. [DOI] [PubMed] [Google Scholar]
- 20.Cheng L., Assary R.S., Qu X., Jain A., Ong S.P., Rajput N.N., Persson K., Curtiss L.A. Accelerating electrolyte discovery for energy storage with high-throughput screening. J. Phys. Chem. Lett. 2015;6:283–291. doi: 10.1021/jz502319n. [DOI] [PubMed] [Google Scholar]
- 21.McMullen J.P., Stone M.T., Buchwald S.L., Jensen K.F. An integrated microreactor system for self-optimization of a heck reaction: from micro- to mesoscale flow systems. Angew. Chem. Int. Ed. Engl. 2010;49:7076–7080. doi: 10.1002/anie.201002590. [DOI] [PubMed] [Google Scholar]
- 22.Moore J.S., Jensen K.F. Automated multitrajectory method for reaction optimization in a microfluidic system using online IR analysis. Org. Process Res. Dev. 2012;16:1409–1415. doi: 10.1021/op300099x. [DOI] [Google Scholar]
- 23.Krishnadasan S., Brown R.J.C., deMello A.J., deMello J.C. Intelligent routes to the controlled synthesis of nanoparticles. Lab Chip. 2007;7:1434–1441. doi: 10.1039/B711412E. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaev P., Hooper D., Webber F., Rao R., Decker K., Krein M., Poleski J., Barto R., Maruyama B. Autonomy in materials research: a case study in carbon nanotube growth. NPJ Comput. Mater. 2016;2:16031. doi: 10.1038/npjcompumats.2016.31. [DOI] [Google Scholar]
- 25.Maruyama B., Decker K., Krein M., Poleski J., Barto R., Webber F., Nikolaev P. Autonomous experimentation applied to carbon nanotube synthesis. Microsc. Microanal. 2017;23:182. doi: 10.1017/S1431927617001593. [DOI] [Google Scholar]
- 26.Burger B., Maffettone P.M., Gusev V.V., Aitchison C.M., Bai Y., Wang X., Li X., Alston B.M., Li B., Clowes R., et al. A mobile robotic chemist. Nature. 2020;583:237–241. doi: 10.1038/s41586-020-2442-2. [DOI] [PubMed] [Google Scholar]
- 27.Wigley P.B., Everitt P.J., van den Hengel A., Bastian J.W., Sooriyabandara M.A., McDonald G.D., Hardman K.S., Quinlivan C.D., Manju P., Kuhn C.C.N., et al. Fast machine-learning online optimization of ultra-cold-atom experiments. Sci. Rep. 2016;6:25890. doi: 10.1038/srep25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLeod B.P., Parlane F.G.L., Morrissey T.D., Häse F., Roch L.M., Dettelbach K.E., Moreira R., Yunker L.P.E., Rooney M.B., Deeth J.R., et al. Self-driving laboratory for accelerated discovery of thin-film materials. Sci. Adv. 2020;6:eaaz8867. doi: 10.1126/sciadv.aaz8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langner S., Häse F., Perea J.D., Stubhan T., Hauch J., Roch L.M., Heumueller T., Aspuru-Guzik A., Brabec C.J. Beyond ternary OPV: high-throughput experimentation and self-driving laboratories optimize multicomponent systems. Adv. Mater. 2020;32:1907801. doi: 10.1002/adma.201907801. [DOI] [PubMed] [Google Scholar]
- 30.Hartono N.T.P., Ani Najeeb M., Li Z., Nega P.W., Fleming C.A., Sun X., Chan E.M., Abate A., Norquist A.J., Schrier J., Buonassisi T. Principled exploration of bipyridine and terpyridine additives to promote methylammonium lead iodide perovskite crystallization. Cryst. Growth Des. 2022;22:5424–5431. doi: 10.1021/acs.cgd.2c00522. [DOI] [Google Scholar]
- 31.Bannigan P., Aldeghi M., Bao Z., Häse F., Aspuru-Guzik A., Allen C. Machine learning directed drug formulation development. Adv. Drug Deliv. Rev. 2021;175:113806. doi: 10.1016/j.addr.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 32.Paull R., Wolfe J., Hébert P., Sinkula M. Investing in nanotechnology. Nat. Biotechnol. 2003;21:1144–1147. doi: 10.1038/nbt1003-1144. [DOI] [PubMed] [Google Scholar]
- 33.Lammers T., Storm G. Setting standards to promote progress in bio–nano science. Nat. Nanotechnol. 2019;14:626. doi: 10.1038/s41565-019-0497-8. [DOI] [PubMed] [Google Scholar]
- 34.Bhatia S.N., Chen X., Dobrovolskaia M.A., Lammers T. Cancer nanomedicine. Nat. Rev. Cancer. 2022;22:550–556. doi: 10.1038/s41568-022-00496-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingwell K. Excipient developers call for regulatory facelift. Nat. Rev. Drug Discov. 2020;19:823–824. doi: 10.1038/d41573-020-00201-1. [DOI] [PubMed] [Google Scholar]
- 36.Tao J., Chow S.F., Zheng Y. Application of flash nanoprecipitation to fabricate poorly water-soluble drug nanoparticles. Acta Pharm. Sin. B. 2019;9:4–18. doi: 10.1016/j.apsb.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan X., Bernard J., Ganachaud F. Nanoprecipitation as a simple and straightforward process to create complex polymeric colloidal morphologies. Adv. Colloid Interface Sci. 2021;294:102474. doi: 10.1016/j.cis.2021.102474. [DOI] [PubMed] [Google Scholar]
- 38.Shah S., Dhawan V., Holm R., Nagarsenker M.S., Perrie Y. Liposomes: advancements and innovation in the manufacturing process. Adv. Drug Deliv. Rev. 2020;154–155:102–122. doi: 10.1016/j.addr.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Fan Y., Yen C.-W., Lin H.-C., Hou W., Estevez A., Sarode A., Goyon A., Bian J., Lin J., Koenig S.G., et al. Automated high-throughput preparation and characterization of oligonucleotide-loaded lipid nanoparticles. Int. J. Pharm. 2021;599:120392. doi: 10.1016/j.ijpharm.2021.120392. [DOI] [PubMed] [Google Scholar]
- 40.Sarode A., Fan Y., Byrnes A.E., Hammel M., Hura G.L., Fu Y., Kou P., Hu C., Hinz F.I., Roberts J., et al. Predictive high-throughput screening of PEGylated lipids in oligonucleotide-loaded lipid nanoparticles for neuronal gene silencing. Nanoscale Adv. 2022;4:2107–2123. doi: 10.1039/D1NA00712B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Settles B. Vol. 6. 2012. Active learning; pp. 1–114. (Synthesis Lectures on Artificial Intelligence and Machine Learning). [DOI] [Google Scholar]
- 42.Môkus J. Proceedings of the IFIP Technical Conference. 1975. On Bayesian methods for seeking the extremum; pp. 400–404. [Google Scholar]
- 43.Mockus J., Tiesis V., Zilinskas A. The application of Bayesian methods for seeking the extremum. Glob. Optim. 1978;2:117–129. [Google Scholar]
- 44.Mockus J. Springer Netherlands; 1989. Bayesian Approach to Global Optimization: Theory and Applications. [Google Scholar]
- 45.Tao H., Wu T., Kheiri S., Aldeghi M., Aspuru-Guzik A., Kumacheva E. Self-driving platform for metal nanoparticle synthesis: combining microfluidics and machine learning. Adv. Funct. Mater. 2021;31:2106725. doi: 10.1002/adfm.202106725. [DOI] [Google Scholar]
- 46.Vaddi K., Chiang H.T., Pozzo L.D. Autonomous retrosynthesis of gold nanoparticles via spectral shape matching. Digit. Discov. 2022;1:502–510. doi: 10.1039/D2DD00025C. [DOI] [Google Scholar]
- 47.Dalal R., Leyden M., Oviedo F., Reineke T. Polymer design via SHAP and bayesian machine learning optimizes pDNA and CRISPR ribonucleoprotein delivery. Research square. 2022 doi: 10.21203/rs.3.rs-1785891/v1. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamasi M.J., Patel R.A., Borca C.H., Kosuri S., Mugnier H., Upadhya R., Murthy N.S., Webb M.A., Gormley A.J. Machine learning on a robotic platform for the design of polymer–protein hybrids. Adv. Mater. 2022;34:2201809. doi: 10.1002/adma.202201809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamasi M.J., Gormley A.J. Biologic formulation in a self-driving biomaterials lab. Cell Rep. Phys. Sci. 2022;3:101041. doi: 10.1016/j.xcrp.2022.101041. [DOI] [Google Scholar]
- 50.Pomberger A., Pedrina McCarthy A.A., Khan A., Sung S., Taylor C.J., Gaunt M.J., Colwell L., Walz D., Lapkin A.A. The effect of chemical representation on active machine learning towards closed-loop optimization. React. Chem. Eng. 2022;7:1368–1379. doi: 10.1039/D2RE00008C. [DOI] [Google Scholar]
- 51.Moriwaki H., Tian Y.-S., Kawashita N., Takagi T. Mordred: a molecular descriptor calculator. J. Cheminform. 2018;10:4. doi: 10.1186/s13321-018-0258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tom G., Hickman R.J., Zinzuwadia A., Mohajeri A., Sanchez-Lengeling B., Aspuru-Guzik A. Calibration and generalizability of probabilistic models on low-data chemical datasets with DIONYSUS. arXiv. 2022 doi: 10.48550/arXiv.2212.01574. Preprint at. [DOI] [Google Scholar]
- 53.Häse F., Aldeghi M., Hickman R.J., Roch L.M., Aspuru-Guzik A. Gryffin: an algorithm for Bayesian optimization of categorical variables informed by expert knowledge. Appl. Phys. Rev. 2021;8:031406. doi: 10.1063/5.0048164. [DOI] [Google Scholar]
- 54.Battaglia P.W., Hamrick J.B., Bapst V., Sanchez-Gonzalez A., Zambaldi V., Malinowski M., Tacchetti A., Raposo D., Santoro A., Faulkner R., et al. Relational inductive biases, deep learning, and graph networks. arXiv. 2018 doi: 10.48550/arXiv.1806.01261. Preprint at. [DOI] [Google Scholar]
- 55.Gilmer J., Schoenholz S.S., Riley P.F., Vinyals O., Dahl G.E. Neural message passing for quantum chemistry. arXiv. 2017 doi: 10.48550/arXiv.1704.01212. Preprint at. [DOI] [Google Scholar]
- 56.Aldeghi M., Coley C.W. A graph representation of molecular ensembles for polymer property prediction. Chem. Sci. 2022;13:10486–10498. doi: 10.1039/D2SC02839E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Yao Q., Kwok J.T., Ni L.M. Generalizing from a few examples: a survey on few-shot learning. ACM Comput. Surv. 2020;53:1–34. doi: 10.1145/3386252. [DOI] [Google Scholar]
- 58.Stanley M., Bronskill J., Maziarz K., Misztela H., Lanini J., Segler M., Schneider N., Brockschmidt M. FS-mol: a few-shot learning dataset of molecules. NeurIPS 2021 datasets and benchmarks track. 2021. https://openreview.net/forum?id=701FtuyLlAd
- 59.Altae-Tran H., Ramsundar B., Pappu A.S., Pande V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 2017;3:283–293. doi: 10.1021/acscentsci.6b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hickman R., Ruža J., Roch L., Tribukait H., García-Durán A. Equipping data-driven experiment planning for Self-driving Laboratories with semantic memory: case studies of transfer learning in chemical reaction optimization. ChemRxiv. 2022 doi: 10.26434/chemrxiv-2022-jt4sm. Preprint at. [DOI] [Google Scholar]
- 61.Chen Y., Song X., Lee C., Wang Z., Zhang Q., Dohan D., Kawakami K., Kochanski G., Doucet A., Ranzato M., et al. Towards learning universal hyperparameter optimizers with transformers. arXiv. 2022 doi: 10.48550/arXiv.2205.13320. Preprint at. [DOI] [Google Scholar]
- 62.Yang K., Swanson K., Jin W., Coley C., Eiden P., Gao H., Guzman-Perez A., Hopper T., Kelley B., Mathea M., et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 2019;59:3370–3388. doi: 10.1021/acs.jcim.9b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., French S., Carfrae L.A., Bloom-Ackermann Z., et al. A deep learning approach to antibiotic discovery. Cell. 2020;180:688–702.e13. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heid E., Green W.H. Machine learning of reaction properties via learned representations of the condensed graph of reaction. J. Chem. Inf. Model. 2022;62:2101–2110. doi: 10.1021/acs.jcim.1c00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MIT C.S.A.I.L. Chemprop. http://chemprop.csail.mit.edu/
- 66.Shields B.J., Stevens J., Li J., Parasram M., Damani F., Alvarado J.I.M., Janey J.M., Adams R.P., Doyle A.G. Bayesian reaction optimization as a tool for chemical synthesis. Nature. 2021;590:89–96. doi: 10.1038/s41586-021-03213-y. [DOI] [PubMed] [Google Scholar]
- 67.Torres J.A.G., Lau S.H., Anchuri P., Stevens J.M., Tabora J.E., Li J., Borovika A., Adams R.P., Doyle A.G. A multi-objective active learning platform and web app for reaction optimization. J. Am. Chem. Soc. 2022;144:19999–20007. doi: 10.1021/jacs.2c08592. [DOI] [PubMed] [Google Scholar]
- 68.Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aspuru-Guzik Group GitHub Organization Gryffin: bayesian optimization of continuous and categorical variables. https://github.com/aspuru-guzik-group/gryffin
- 70.Gaudin T., Benlolo I., Cui Z.Y., Hickmann R., Tamblyn I., Aspuru-Guzik A. 2022. Molar. [DOI] [Google Scholar]
- 71.Cui L., Pereira S., Sonzini S., van Pelt S., Romanelli S.M., Liang L., Ulkoski D., Krishnamurthy V.R., Brannigan E., Brankin C., et al. Development of a high-throughput platform for screening lipid nanoparticles for mRNA delivery. Nanoscale. 2022;14:1480–1491. doi: 10.1039/D1NR06858J. [DOI] [PubMed] [Google Scholar]